Abstract
Sulfurifustis variabilis and Sulfuricaulis limicola are autotrophic sulfur-oxidizing bacteria belonging to the family Acidiferrobacteraceae in the order Acidiferrobacterales. The type strains of these species, strain skN76T and strain HA5T, were isolated from lakes in Japan. Here we describe the complete genome sequences of Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T. The genome of Sulfurifustis variabilis skN76T consists of one circular chromosome with size of 4.0 Mbp including 3864 protein-coding sequences. The genome of Sulfuricaulis limicola HA5T is 2.9 Mbp chromosome with 2763 protein-coding sequences. In both genomes, 46 transfer RNA-coding genes and one ribosomal RNA operon were identified. In the genomes, redundancies of the genes involved in sulfur oxidation and inorganic carbon fixation pathways were observed. This is the first report to show the complete genome sequences of bacteria belonging to the order Acidiferrobacterales in the class Gammaproteobacteria.
Keywords: Bacteria, Gram-negative, Sulfur-oxidizing bacteria, Acidiferrobacterales, Acidiferrobacteraceae
Introduction
Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T are gammaproteobacterial sulfur-oxidizing bacteria isolated from sediments of Lake Mizugaki and Lake Harutori, respectively [1, 2]. They both belong to the family Acidiferrobacteraceae in the order Acidiferrobacterales. In this order, only three species have been isolated in pure culture. They are all chemolithoautotrophs and can grow by oxidation of inorganic sulfur compounds. Sulfurifustis variabilis and Sulfuricaulis limicola are neutrophilic, whereas the other species, Acidiferrobacter thiooxydans, is acidophilic [3]. Taxonomy of Acidiferrobacter thiooxydans has been revised several times, and the family Acidiferrobacteraceae and order Acidiferrobacterales were recently established to accommodate the species [1, 3–5]. The members of the family Acidiferrobacteraceae have been frequently detected in various environments as gene sequences [2, 3, 6].
Here we show the complete genome sequences of Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T as the first genomes of the order Acidiferrobacterales.
Organism information
Classification and features
The cells of Sulfurifustis variabilis skN76T are rod-shaped or filamentous form with varying length, and 0.3–0.5 μm in width (Fig. 1a, Table 1). The cells of Sulfuricaulis limicola HA5T are rod-shaped, 1.2–6.0 μm in length and 0.3–0.5 μm in width (Fig. 1b, Table 1). They are both Gram-stain-negative. Sulfurifustis variabilis and Sulfuricaulis limicola belong to the family Acidiferrobacteraceae within the class Gammaproteobacteria (Fig. 2). They both utilized thiosulfate, tetrathionate and elemental sulfur as electron donors for chemolithoautotrophic growth under aerobic conditions [1, 2].
Fig. 1.
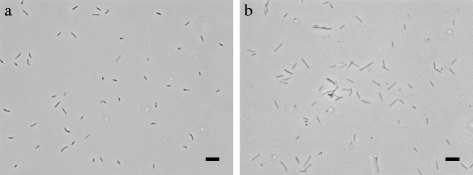
Phase-contrast micrographs of Sulfurifustis variabilis skN76T (a) and Sulfuricaulis limicola HA5T (b), grown with thiosulfate at 45 and 28 °C, respectively. Bars, 5 μm
Table 1.
Classification and general features of Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T according to MIGS recommendations
| MIGS ID | Property | Sulfurifustis variabilis skN76T | Sulfuricaulis limicola HA5T | ||
|---|---|---|---|---|---|
| Term | Evidence code a | Term | Evidence code a | ||
| Classification | Domain Bacteria | TAS [23] | Domain Bacteria | TAS [23] | |
| Phylum Proteobacteria | TAS [24] | Phylum Proteobacteria | TAS [24] | ||
| Class Gammaproteobacteria | TAS [25] | Class Gammaproteobacteria | TAS [25] | ||
| Order Acidiferrobacterales | TAS [1] | Order Acidiferrobacterales | TAS [1] | ||
| Family Acidiferrobacteraceae | TAS [1] | Family Acidiferrobacteraceae | TAS [1] | ||
| Genus Sulfurifustis | TAS [1] | Genus Sulfuricaulis | TAS [2] | ||
| Species Sulfurifustis variabilis | TAS [1] | Species Sulfuricaulis limicola | TAS [2] | ||
| Type strain skN76 | Type strain HA5 | ||||
| Gram stain | negative | TAS [1] | negative | TAS [2] | |
| Cell shape | rod or filaments | TAS [1] | rod | TAS [2] | |
| Motility | motile | TAS [1] | not reported | ||
| Sporulation | not reported | not reported | |||
| Temperature range | 28–46 °C | TAS [1] | 8–37 °C | TAS [2] | |
| Optimum temperature | 42–45 °C | TAS [1] | 28–32 °C | TAS [2] | |
| pH range; Optimum | 6.3–8.9; 6.8–8.2 | TAS [1] | 6.1–9.2; unknown | TAS [2] | |
| Carbon source | bicarbonate | TAS [1] | bicarbonate | TAS [2] | |
| MIGS-6 | Habitat | Sediment of a lake | TAS [1] | Sediment of a lake | TAS [2] |
| MIGS-6.3 | Salinity | <2.6 % NaCl (w/v) | TAS [1] | <1.2 % NaCl (w/v) | TAS [2] |
| MIGS-22 | Oxygen requirement | aerobic | TAS [1] | aerobic | TAS [2] |
| MIGS-15 | Biotic relationship | free-living | TAS [1] | free-living | TAS [2] |
| MIGS-14 | Pathogenicity | non-pathogen | NAS | non-pathogen | NAS |
| MIGS-4 | Geographic location | Lake Mizugaki, Japan | TAS [1] | Lake Harutori, Japan | TAS [2] |
| MIGS-5 | Sample collection | November 30, 2010 | NAS | April 26, 2012 | NAS |
| MIGS-4.1 | Latitude | 35°51.5′ N | TAS [26] | 42°58.4′ N | NAS |
| MIGS-4.2 | Longitude | 138°30.0′ E | TAS [26] | 144°23.9′ E | NAS |
| MIGS-4.4 | Altitude | not reported | not reported | ||
a Evidence codes–IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project
Fig. 2.
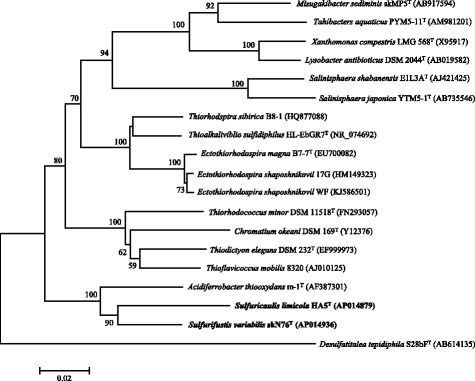
Phylogenetic tree showing the relationships of Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T with other members of the class Gammaproteobacteria based on 16S rRNA gene sequences aligned by using CLUSTAL W. Desulfatitalea tepidiphila S28bFT was used as an outgroup. This tree was reconstructed using 1412 sites with the neighbor-joining method by using MEGA6 [27]. Percentage values of 1000 bootstrap resamplings are shown at nodes; values below 50 % were not shown
Genome sequencing information
Genome project history
Sulfurifustisvariabillis skN76T and Sulfuricaulis limicola HA5T were selected for sequencing as representatives of sulfur-oxidizing bacteria belonging to the order Acidiferrobacterales, to reveal characteristics of their genomes. A summary of the project information is shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Sulfurifustis variabilis skN76T | Sulfuricaulis limicola HA5T |
|---|---|---|---|
| Term | Term | ||
| MIGS 31 | Finishing quality | Completed | Completed |
| MIGS-28 | Libraries used | 15–20 kb SMRTbellTM library | 10–20 kb SMRTbellTM library |
| MIGS 29 | Sequencing platforms | PacBio RS II | PacBio RS II |
| MIGS 31.2 | Fold coverage | 210 × | 142 × |
| MIGS 30 | Assemblers | RS_HGAP Assembly.2 | RS_HGAP Assembly.3 |
| MIGS 32 | Gene calling method | Microbial Genome Annotation Pipeline | Microbial Genome Annotation Pipeline |
| Locus Tag | SVA | SCL | |
| Genbank ID | AP014936 | AP014879 | |
| GenBank Date of Release | July 29, 2016 | July 29, 2016 | |
| BIOPROJECT | PRJDB4108 | PRJDB3927 | |
| MIGS 13 | Source Material Identifier | DSM 100313 | DSM 100373 |
| Project relevance | Environmental | Environmental |
Growth conditions and genomic DNA preparation
Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T were grown with 20 mM thiosulfate as an energy source in a bicarbonate-buffered medium previously described [1], at 45 and 28 °C, respectively. Genomic DNA samples were prepared by using Wizard® genomic DNA purification kit (Promega, Madison, WI, USA) from approximately 0.2 ml (skN76) or 0.1 ml (HA5) of cell pellets. Amounts of the obtained DNA assessed by spectrophotometry were ca. 270 μg (skN76) and 90 μg (HA5) respectively, and the UV absorption ratio of 260/280 nm was greater than 1.8 in both samples.
Genome sequencing and assembly
The genomic DNA was sheared into approximately 20 kb using g-TUBE (Covaris, Inc., Woburn, MA, USA). The SMRTbellTM templates were prepared from the fragments using SMRTbellTM Template Prep Kit 1.0 (Pacific Biosciences, Menlo Park, CA, USA). The size-selected libraries for sequencing were prepared by using BluePippin (Sage Science, Baverly, MA, USA). The libraries were sequenced on a PacBio RS II instrument (Pacific Biosciences) with P6-C4 chemistry (for Sulfurifustisvariabillis skN76T) or P5-C3 chemistry (for Sulfuricaulis limicola HA5T). De novo assembly was performed by using RS_HGAP Assembly.3 (for Sulfurifustisvariabillis skN76T) or RS_HGAP Assembly.2 (for Sulfuricaulis limicola HA5T), implemented within the SMRT Analysis v2.3 (Pacific Biosciences) software environment. By assembling 79,017 subreads (837,333,548 bp) of Sulfurifustisvariabillis skN76T, two contigs with the lengths of ca. 4.0 Mbp and ca. 5.4 kbp were obtained. The shorter one was identical to a partial sequence of the larger one, and a circular chromosome was manually constructed from the larger contig by finding self-overlapping regions using the in silico Molecular Cloning (R) Genomic Edition (In Silico Biology, Inc., Yokohama, Japan) application. As for Sulfuricaulis limicola HA5T, a single contig (ca. 2.9 Mbp) was obtained by assembling 61,565 subreads (409,124,339 bp), and circular chromosome was manually constructed in the same manner.
Genome annotation
The genomes were annotated automatically using the Microbial Genome Annotation Pipeline [7]. Further manual annotation of the predicted protein-coding sequences was performed on the basis of BLASTP searches against the NCBI nonredundant database. CDSs were annotated as hypothetical protein-coding genes when they met any of the following four criteria in the top hit of the BLASTP analysis: (1) E-value >1e-8, (2) length coverage <60 % against query sequence (3) sequence identity <30 % or (4) function of the hit was unidentified. The WebMGA server was used to assign the genes to Clusters of Ortholog Groups and Protein family domains [8–11]. The Phobius server was used to predict signal peptides and transmembrane helices [12]. Clustered Regularly Interspaced Short Palindromic Repeat loci were detected using CRISPRfinder [13].
Genome properties
The basic statistics of the genomes are shown in Table 3. Both genomes contained 46 tRNA genes and one rRNA operon. The genome size of Sulfurifustisvariabillis skN76T was approximately 1.4 times larger than that of Sulfuricaulis limicola HA5T. CRISPR loci were found only in the genome of Sulfurifustisvariabillis skN76T (Table 3). The distribution of genes into COGs functional categories is presented in Table 4.
Table 3.
Genome statistics of Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T
| Attribute | Sulfurifustis variabilis skN76T | Sulfuricaulis limicola HA5T | ||
|---|---|---|---|---|
| Value | % of Total | Value | % of Total | |
| Genome size (bp) | 3,958,814 | 100.00 | 2,864,672 | 100.00 |
| DNA coding (bp) | 3,565,567 | 90.06 | 2,567,493 | 89.63 |
| DNA G + C (bp) | 2,670,566 | 67.46 | 1,759,557 | 61.42 |
| DNA scaffolds | 1 | 100.00 | 1 | 100.00 |
| Total genes | 3913 | 100.00 | 2812 | 100.00 |
| Protein coding genes | 3864 | 98.75 | 2763 | 98.26 |
| RNA genes | 49 | 1.25 | 49 | 1.74 |
| Pseudo genes | unknown | unknown | ||
| Genes in internal clusters | unknown | unknown | ||
| Genes with function prediction | 2930 | 75.83 | 2036 | 73.69 |
| Genes assigned to COGs | 2921 | 75.60 | 2165 | 78.36 |
| Genes with Pfam domains | 2970 | 76.86 | 2208 | 79.91 |
| Genes with signal peptides | 893 | 23.11 | 562 | 20.34 |
| Genes with transmembrane helices | 845 | 21.87 | 622 | 22.51 |
| CRISPR repeats | 6 | 0 | ||
Table 4.
Number of genes associated with general COG functional categories
| Code | Sulfurifustis variabilis skN76T | Sulfuricaulis limicola HA5T | Description | ||
|---|---|---|---|---|---|
| Value | %age | Value | %age | ||
| J | 164 | 4.24 | 159 | 5.75 | Translation, ribosomal structure and biogenesis |
| A | 5 | 0.13 | 2 | 0.07 | RNA processing and modification |
| K | 191 | 4.94 | 130 | 4.71 | Transcription |
| L | 154 | 3.99 | 117 | 4.23 | Replication, recombination and repair |
| B | 1 | 0.03 | 1 | 0.04 | Chromatin structure and dynamics |
| D | 36 | 0.93 | 31 | 1.12 | Cell cycle control, Cell division, chromosome partitioning |
| V | 43 | 1.11 | 29 | 1.05 | Defense mechanisms |
| T | 283 | 7.32 | 218 | 7.89 | Signal transduction mechanisms |
| M | 265 | 6.86 | 210 | 7.60 | Cell wall/membrane biogenesis |
| N | 66 | 1.71 | 64 | 2.32 | Cell motility |
| U | 123 | 3.18 | 98 | 3.55 | Intracellular trafficking and secretion |
| O | 185 | 4.79 | 142 | 5.14 | Posttranslational modification, protein turnover, chaperones |
| C | 265 | 6.86 | 192 | 6.95 | Energy production and conversion |
| G | 148 | 3.83 | 101 | 3.66 | Carbohydrate transport and metabolism |
| E | 201 | 5.20 | 150 | 5.43 | Amino acid transport and metabolism |
| F | 63 | 1.63 | 59 | 2.14 | Nucleotide transport and metabolism |
| H | 167 | 4.32 | 129 | 4.67 | Coenzyme transport and metabolism |
| I | 90 | 2.33 | 65 | 2.35 | Lipid transport and metabolism |
| P | 189 | 4.89 | 127 | 4.60 | Inorganic ion transport and metabolism |
| Q | 56 | 1.45 | 35 | 1.27 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 394 | 10.20 | 247 | 8.94 | General function prediction only |
| S | 346 | 8.95 | 230 | 8.32 | Function unknown |
| - | 943 | 24.40 | 598 | 21.64 | Not in COGs |
Insights from the genome sequences
In both the genomes of Sulfurifustisvariabillis skN76T and Sulfuricaulis limicola HA5T, genes involved in the sulfur oxidation pathway were identified. The genomes of both strains contain genes of the DSR system related to the oxidation of elemental sulfur to sulfite [14, 15]. They contain a dsr gene cluster of identical composition, dsrABEFHCMKLJOPNR (SVA_1954-1967, SCL_1274-1261). There are some dsr genes outside of the gene cluster, dsrAB (SVA_0258-0259, SCL_0256-0257), dsrS (SVA_2921, SCL_0781) and dsrC (SVA_0281, SVA_0284, SVA_0358, SVA_0917, SVA_0969, SVA_1205, SVA_1793, SVA_1949, SVA_2832, SVA_3655; SCL_0275, SCL_0524, SCL_0785, SCL_1279, SCL_1423, SCL_2646).
As genes encoding proteins involved in oxidation of sulfite to sulfate in the cytoplasm, both genomes contain two copies of the aprAB genes encoding an adenosine-5’-phosphosulphate reductase (SVA_2607-2608, SVA_3565-3564; SCL_0600-0601, SCL_2474-2473), along with the sat gene encoding a sulfate adenylyltransferase (SVA_3563, SCL_2472) and the aprM gene (SVA_2609, SCL_0602). In addition, the genome of Sulfuricaulis limicola HA5T contains the hdrAACB genes encoding a Hdr (SCL_2523-2520), but that of Sulfurifustisvaliabilis skN76T does not. The AprM and Hdr complex are thought to have similar function that interacts with the adenosine-5’-phosphosulphate reductase [16–18]. The genomes also contain the soeABC genes (SVA_2734, SVA_2736-2737; SCL_0523-0521), encoding a membrane-bound polysulfide reductase-like iron-sulfur molybdoprotein, which is suspected to be involved in sulfite oxidation in the cytoplasm [19]. Further, the genome of Sulfurifustisvaliabilis skN76T contains the sorAB genes (SVA_1391-1390) related to the direct oxidation of sulfite to sulfate in the periplasm [20].
For thiosulfate oxidation, both genomes contain the soxXYZAB gene cluster (SVA_2999-3003, SCL_2229-2233). Although sulfide oxidation by these bacteria has not been demonstrated, genes related to sulfide oxidation were identified; the fccAB (soxEF) genes encoding a flavocytochrome c/sulfide dehydrogenase (SVA_0067-0066, SVA_3594-3595; SCL_0078-0077) and the sqr gene encoding a sulfide:quinone oxidoreductase (SVA_1781, SVA_2675, SVA3205).
Sulfurifustisvariabillis skN76T and Sulfuricaulis limicola HA5T are autotrophic bacteria. They both have two copies of the rbcL and rbcS genes, encoding large and small subunits of ribulose bisphosphate carboxylase/oxygenase (SVA_3460-3459, SVA_3471-3470; SCL_2417-2416, SCL_2425-2424), which is the key enzyme in the Calvin-Benson-Bassham cycle to catalyze inorganic carbon fixation. The two copies of RuBisCO in each genome are phylogenetically distinct, and belong to lineages referred to as green-like form IA and red-like form IC (Fig. 3) [21]. In the form IC RuBisCO coded by rbcL gene (SVA_3460, SCL_2417), Sulfurifustisvariabillis skN76T and Sulfuricaulis limicola HA5T have six-amino-acid inserts at the same position where a similar insert was reported from Nitrosospira sp. 40KI [22]. There are two other RuBisCO sequences which have six-amino-acid inserts at the same position, and these sequences with inserts formed a monophyletic cluster in the tree of RuBisCO (Fig. 3). In general, RuBisCO of form IA and IC have different properties which are thought to be advantageous to fix inorganic carbon under different concentrations of carbon dioxide and/or oxygen [21]. Possession of the genes for these two distinct RuBisCO forms may be beneficial to cope with changing environmental conditions, or to thrive in various types of ecosystems.
Fig. 3.
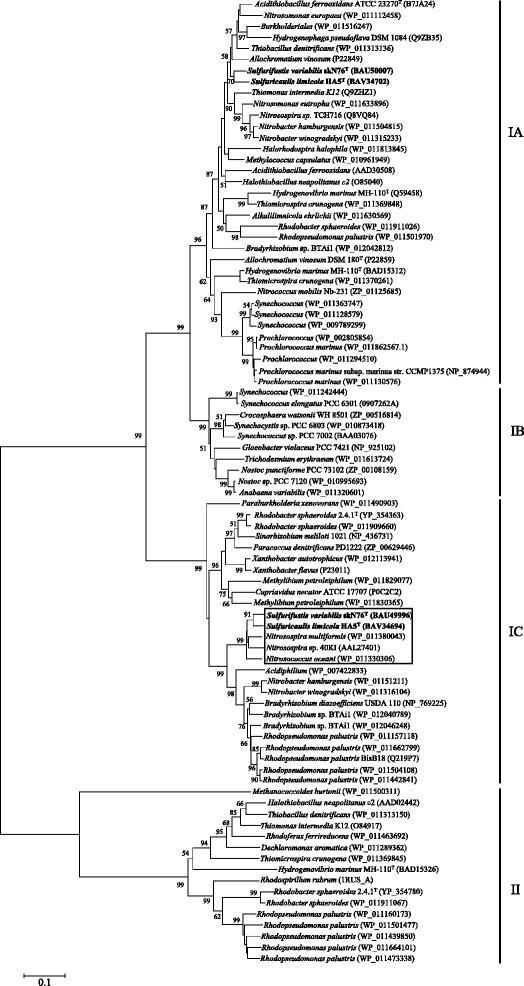
Neighbor-joining tree showing the phylogenetic positions of RuBisCO amino acid sequences coded in the genomes of Sulfurifustis variabilis skN76T and Sulfuricaulis limicola HA5T. The sequences aligned by using CLUSTAL W. This tree was reconstructed using 421 sites with MEGA6 [27]. Percentage values of 1000 bootstrap resamplings are shown at nodes; values below 50 % were not shown. The sequences shown in box have six-amino-acid inserts at the same position
Conclusion
This is the first report on complete genome sequences of bacteria belonging to the order Acidiferrobacterales. The genome analysis of Sulfurifustisvariabillis skN76T and Sulfuricaulis limicola HA5T revealed that they have similar sets of genes involved in sulfur oxidation pathways. In the both genomes, redundancies of the genes for sulfur oxidation and inorganic carbon fixation were observed, as represented by multiple copies of dsrAB, aprAB and rbcLS. Such redundancies may provide physiological flexibility to the chemolithotrophic sulfur oxidizers which are fully depending on these functions to obtain energy and carbon source for growth.
Acknowledgements
This study was supported by JSPS KAKENHI Grant Number 15 K07209 to H. Kojima. We thank R. Tokizawa and A. Shinohara for their technical assistance.
Authors’ contribution
MF and HK designed the study. HK characterized the strains and prepared genomic DNA. KU, TW and AM performed the bioinformatics analysis. KU and HK wrote the draft of manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- MiGAP
Microbial Genome Annotation Pipeline
- Hdr
Heterodisulfide reductase
- DSR
Dissimilatory sulfite reductase
References
- 1.Kojima H, Shinohara A, Fukui M. Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int J Syst Evol Microbiol. 2015;65:3709–3713. doi: 10.1099/ijsem.0.000479. [DOI] [PubMed] [Google Scholar]
- 2.Kojima H, Watanabe T, Fukui M. Sulfuricaulis limicola gen. nov., sp. nov., a sulfur oxidizer isolated from a lake. Int J Syst Evol Microbiol. 2016;66:266–270. doi: 10.1099/ijsem.0.000709. [DOI] [PubMed] [Google Scholar]
- 3.Hallberg KB, Hedrich S, Johnson DB. Acidiferrobacter thiooxydans, gen. nov. sp. nov.; an acidophilic, thermo-tolerant, facultatively anaerobic iron- and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles. 2011;15:271–279. doi: 10.1007/s00792-011-0359-2. [DOI] [PubMed] [Google Scholar]
- 4.Kelly DP, Wood AP. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int J Syst Evol Microbiol. 2000;50:511–516. doi: 10.1099/00207713-50-2-511. [DOI] [PubMed] [Google Scholar]
- 5.Williams KP, Kelly DP. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63:2901–2906. doi: 10.1099/ijs.0.049270-0. [DOI] [PubMed] [Google Scholar]
- 6.Dyksma S, Bischof K, Fuchs BM, Hoffmann K, Meier D, Meyerdierks A, et al. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 2016;10(8):1–15. doi: 10.1038/ismej.2015.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugawara H, Ohyama A, Mori H, Kurokawa K. Microbial Genome Annotation Pipeline (MiGAP) for diverse users. The 20th International Conference on Genome Informatics (GIW2009) Poster and Software Demonstrations (Yokohama). 2009;S001-1-2.
- 8.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kall L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pott AS, Dahl C. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology. 1998;144:1881–1894. doi: 10.1099/00221287-144-7-1881. [DOI] [PubMed] [Google Scholar]
- 15.Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, et al. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J Bacteriol. 2005;187:1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parey K, Demmer U, Warkentin E, Wynen A, Ermler U, Dahl C. Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum. PLoS One. 2013;8:e7407. doi: 10.1371/annotation/fab66ad6-bdfa-4f76-9c39-08f28a92494d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pires RH, Lourenço AI, Morais F, Teixeira M, Xavier AV, Saraiva LM, et al. A novel membrane-bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim Biophys Acta. 2003;1605:67–82. doi: 10.1016/S0005-2728(03)00065-3. [DOI] [PubMed] [Google Scholar]
- 18.Meyer B, Kuever J. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology. 2007;153:3478–3498. doi: 10.1099/mic.0.2007/008250-0. [DOI] [PubMed] [Google Scholar]
- 19.Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology. 2013;159:2626–2638. doi: 10.1099/mic.0.071019-0. [DOI] [PubMed] [Google Scholar]
- 20.Kappler U, Bennett B, Rethmeier J, Schwarz G, Deutzmann R, McEwan AG, et al. Sulfite:cytochrome c oxidoreductase from Thiobacillus novellus — purification, characterization and molecular biology of a heterodimeric member of the sulfite oxidase family. J Biol Chem. 2000;275:13202–13212. doi: 10.1074/jbc.275.18.13202. [DOI] [PubMed] [Google Scholar]
- 21.Badger MR, Bek EJ. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot. 2008;59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 22.Utåker JB, Andersen K, Aakra Å, Moen B, Nes IF. Phylogeny and functional expression of ribulose 1,5-Bisphosphate carboxylase/oxygenase from the autotrophic ammonia-oxidizing bacterium Nitrosospira sp. Isolate 40KI. J Bacteriol. 2002;184:468–478. doi: 10.1128/JB.184.2.468-478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology, Volume 2, Part B. 2nd ed. New York: Springer; 2005. p. 1.
- 25.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology, Volume 2, Part B. 2nd ed. New York: Springer; 2005. p. 1.
- 26.Kojima H, Iwata T, Fukui M. DNA-based analysis of planktonic methanotrophs in a stratified lake. Freshw Biol. 2009;54:1501–1509. doi: 10.1111/j.1365-2427.2009.02199.x. [DOI] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]


