Abstract
New strategies are needed to diagnose and target human melanoma. To this end, genomic analyses was performed to assess somatic mutations and gene expression signatures using a large cohort of human skin cutaneous melanoma (SKCM) patients from The Cancer Genome Atlas (TCGA) project to identify critical differences between primary and metastatic tumors. Interestingly, pyrimidine metabolism is one of the major pathways to be significantly enriched and deregulated at the transcriptional level in melanoma progression. In addition, dihydropyrimidine dehydrogenase (DPYD) and other important pyrimidine-related genes: DPYS, AK9, CAD, CANT1, ENTPD1, NME6, NT5C1A, POLE, POLQ, POLR3B, PRIM2, REV3L, and UPP2 are significantly enriched in somatic mutations relative to the background mutation rate. Structural analysis of the DPYD protein dimer reveals a potential hotspot of recurring somatic mutations in the ligand binding sites as well as the interfaces of protein domains that mediated electron transfer. Somatic mutations of DPYD are associated with upregulation of pyrimidine degradation, nucleotide synthesis, and nucleic acid processing while salvage and nucleotide conversion is downregulated in TCGA SKCM.
Keywords: TCGA, melanoma, DPYD, pyrimidine, systems biology
Introduction
Cancer cells take advantage of distinct metabolic pathways promoting cellular proliferation or oncogenic progression. Emerging evidence highlights central metabolic pathways including glucose- and glutamine-dependent biomass production to support tumor growth (1). However, complex metabolic requirements of dividing, migrating, or nutrient and oxygen limited cancer cells suggest that tumor cells have much more complex metabolic requirements than previously appreciated (2). The Cancer Genome Atlas (TCGA) puts an even-handed view on tissue-specific genomic determinants, revolutionizing our perspective on malignancies by next-generation sequencing (3). Here we describe cross-talk between signatures of somatic mutations and gene expression of skin cutaneous melanoma (SKCM) based on RNASeq data from 471 TCGA melanoma samples. By connecting pattern of somatic mutations with responses of gene expression at a pathway level, new features of melanoma metabolism and progression are elucidated.
Pyrimidine synthesis is a key metabolic bottleneck important for DNA replication in tumor cells and, therefore, represents a valuable diagnostic and therapeutic target. Early success in cancer metabolism took advantage of this characteristic by making cancer cells vulnerable to inhibition of this pathway. Heidelberger and colleagues designed fluorinated uracil-based pyrimidine analogues, which disrupted tumor DNA biosynthesis and which are to this day used to treat colorectal and breast cancer (4, 5).
To analyze TCGA SKCM dataset we have employed a bottom-up strategy involving pathway enrichment analysis of RNASeq data and structural analysis of somatic mutations. The approach identifies DPYD (dihydropyrimidine dehydrogenase, Gene ID: 1806) as a pivotal factor of pyrimidine metabolism and offers a comprehensive view on how a hypermutated metabolic gene deregulates pyrimidine and nucleic acid synthesis and promotes malignant progression of melanoma.
Methods
Patient cohort
The TCGA SKCM cohort includes RNASeq data for 471 samples allowing us to extract statistical significant pattern of differential expression between solid primary tumors (TP; 103 patients) and metastatic tumors (TM; 367 patients), while there is only one dataset for blood derived normal tissue (NB; 1 patient) (Supplementary table 1). In addition, we utilized files from whole-exome datasets of 339 patients (61 TP; 278 TM) (Supplementary table 2) (6). Clinical data including a history of drug treatment was available for 447 patients (Supplementary table 3). The study was carried out as part of IRB approved study dbGap ID 5094 “Somatic mutations in melanoma” and conducted in accordance with the Helsinki Declaration of 1975. The results shown are based upon next generation sequencing data generated by the TCGA Research Network http://cancergenome.nih.gov. Restricted access clinical, RNASeq, and whole-exome sequences were obtained from the TCGA genome data access center and the data portal.
Identification of somatic mutations
Identification of somatic mutations took advantage of components of the modular multi-step filter as described (6). TCGA data portal was used for cohort selection and CGHub for access of raw data. Whole-exome sequencing data for 339 patients with primary tumor or metastatic tumor were matched with blood-derived normal reference. For the MuTect 1.1.4 analysis (7) GrCh37 (Broad Institute variant of HG19), dbSNP build 132.vcf, and COSMIC_54.vcf library were referenced. Somatic incidences file was queried in bash prompt to retain all the statically significant KEEP mutations. The coverage.wig files served as input to model and account for Intron vs Exon functional mutation burden in InVEx 1.0.1 (8). In addition, MutSig 2.0 assessed the clustering of mutations in hotspots as well as conservation of the sites (9). It is noted that the SKCM cohort contains an interesting case, patient TCGA-FW-A3R5, who has more than 20,000 mutations and an APOBEC signature (10). This patient shows multiple missense mutations in DPYD with nucleotide transitions according to canonical UVB signature, C>T and G>A. Including or excluding this patient had no implications on the outcome of this study.
Structural model and molecular dynamics simulation
The structural model of human DPYD was based on Sus scrofa X-ray structure (PDB entry 1gth) using swiss-model. Mutations were plotted on the modeled human structure and ligand proximity was evaluated by a 5A cut-off. The solvent accessible surface of each residue of DPYD was determined based on a molecular dynamics simulation over a 5 ns trajectory using GROMACS 5.0.2 (11).
Gene expression analysis and statistical analysis
Level 3 RNASeq Log2 transformed expression levels for 18,086 genes were collected for each sample. Differential expression was determined by DESeq in the R package and Students T-test was used to determine significant differences in expression between TP and TM samples and onto metabolic pathways (12). The probability of the test statistics (p-values) were adjusted for multiple hypotheses testing (13). When referred to genomic information, gene symbols are italicized and upper case, while protein names are upper case but not italicized. All used gene symbols are listed with gene description in the glossary in the supplementary tables.
Results
Pathway enrichment of differential RNASeq gene expression data identifies shift in metabolism
Differential expression analysis by DESeq showed 4383 and 4811 to be significantly down- and upregulated, respectively. KEGG Pathway enrichment analysis highlights three distinct sets of pathways—metabolism, cancer signaling, and epidermal developmental markers—to be central to the changes occurring in the metastatic transition. Metabolic pathways include global metabolism (KEGG ID:01100), oxidative phosphorylation (ID:00190), pyrimidine metabolism (ID:00240), purine metabolism (ID:00230), glycosphingolipid biosynthesis (ID:00601), metabolism of cytochrome P450 (ID:00980), tyrosine metabolism (ID:00350), as well as glutathione metabolism (ID:00480) to be significantly enriched pathways with deregulated gene expression with p-values lower than 0.001. Interestingly, metabolic pathways show comparably high enrichment as pathways known to be closely associated with an invasive, metastatic phenotype. Next to pyrimidine metabolism, focal adhesion, actin cytoskeleton regulation, and tight junctions are highly enriched in the metastatic melanoma cohort with p-values below 1.0E-04. Pyrimidine metabolism stands out as highly enriched pathway (enrichment ratio down 3.60, ratio up 2.19, adjusted p-value down 3.49E-10 and adjusted p-value up 4.00E-04). There are 34 and 23, in total 57 genes in pyrimidine metabolism, which are significantly down- and upregulated, respectively (Supplementary table 4). Pyrimidine enzymes undergoing differential expression between skin cutaneous primary and metastatic tumors include all steps in nucleoside triphosphate synthesis, DNA and RNA polymerases, as well as pyrimidine degradation. The top down-regulated enzyme of pyrimidine metabolism is CDA (cytidine deaminase, Gene ID: 978, log2 change between primary and metastatic tumor: 1.66, p-value 5.06E-15), the highest up-regulated enzyme is DPYD (log2 difference between primary and metastatic tumor: +1.43, p-value 2.85E-13). Genes differentially expressed in pathways in cancer include important signaling molecules in MAPK signaling like BRAF, NRAS, KRAS, MAPK8, MAP2K1, in WNT signaling, CTNNB1, FZD1/3/4/8, in STAT signaling, PIAS1, STAT1, STAT5B, in AKT signaling AKT3, MTOR, and others like RB1, NFKB1. Another remarkable theme of enriched genes during metastatic progression is dedifferentiation of melanogenesis, keratinocytes, and Wingless (WNT) signaling (Table 1). Such gene sets are important for cell differentiation of normal melanocytes, epidermal development, and pigmentation.
Table 1.
KEGG pathway enrichment analysis of RNASeq data of 470 TCGA SKCM patients with adjusted p-values below 1.00E-03. Pathway enrichment on pyrimidine metabolism is highlighted in italic.
| Pathway name | KEGG ID | Number of deregulated genes in TCGA SKCM RNASeq | Expected number | Ratio of enrichment | p-value from hypergeometric test | adjusted p-value by multiple test adjustment | Number of reference genes in pathway | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| down | up | down | up | down | up | down | up | down | up | |||
| Metabolic pathways | ID:01100 | 359 | 216 | 107.77 | 119.87 | 3.33 | 1.80 | 3.16E-99 | 5.34E-18 | 6.79E-97 | 9.19E-17 | 1130 |
| Pathways in cancer | ID:05200 | 76 | 108 | 31.09 | 34.58 | 2.44 | 3.12 | 1.64E-13 | 3.70E-28 | 5.88E-12 | 1.94E-26 | 326 |
| Pyrimidine metabolism | ID:00240 | 34 | 23 | 9.44 | 10.50 | 3.60 | 2.19 | 1.30E-11 | 2.00E-04 | 3.49E-10 | 4.00E-04 | 99 |
| Purine metabolism | ID:00230 | 42 | 44 | 15.45 | 17.19 | 2.72 | 2.56 | 1.37E-09 | 3.14E-09 | 3.27E-08 | 1.35E-08 | 162 |
| Melanogenesis | ID:04916 | 31 | 27 | 9.63 | 10.71 | 3.22 | 2.52 | 2.41E-09 | 4.49E-06 | 5.18E-08 | 1.18E-05 | 101 |
| Endocytosis | ID:04144 | 47 | 60 | 19.17 | 21.32 | 2.45 | 2.81 | 5.73E-09 | 5.40E-14 | 1.03E-07 | 4.73E-13 | 201 |
| Phagosome | ID:04145 | 38 | 50 | 14.59 | 16.23 | 2.60 | 3.08 | 2.86E-08 | 1.40E-13 | 4.10E-07 | 1.09E-12 | 153 |
| Peroxisome | ID:04146 | 24 | 21 | 7.53 | 8.38 | 3.19 | 2.51 | 1.84E-07 | 5.44E-05 | 2.20E-06 | 1.00E-04 | 79 |
| Spliceosome | ID:03040 | 32 | 34 | 12.11 | 13.47 | 2.64 | 2.52 | 2.41E-07 | 2.68E-07 | 2.73E-06 | 8.28E-07 | 127 |
| Focal adhesion | ID:04510 | 43 | 68 | 19.07 | 21.22 | 2.25 | 3.21 | 3.12E-07 | 5.81E-19 | 3.35E-06 | 1.11E-17 | 200 |
| Wnt signaling pathway | ID:04310 | 35 | 46 | 14.31 | 15.91 | 2.45 | 2.89 | 5.01E-07 | 1.59E-11 | 4.49E-06 | 1.04E-10 | 150 |
| MAPK signaling pathway | ID:04010 | 52 | 86 | 25.56 | 28.43 | 2.03 | 3.02 | 6.02E-07 | 1.11E-21 | 5.18E-06 | 2.91E-20 | 268 |
| Neurotrophin signaling | ID:04722 | 31 | 36 | 12.11 | 13.47 | 2.56 | 2.67 | 7.79E-07 | 2.45E-08 | 6.44E-06 | 8.72E-08 | 127 |
| Actin cytoskeleton regulation | ID:04810 | 43 | 68 | 20.31 | 22.60 | 2.12 | 3.01 | 1.86E-06 | 2.51E-17 | 1.45E-05 | 3.51E-16 | 213 |
| Protein processing in ER | ID:04141 | 36 | 58 | 15.74 | 17.50 | 2.29 | 3.31 | 1.91E-06 | 3.70E-17 | 1.45E-05 | 4.86E-16 | 165 |
| Tight junction | ID:04530 | 30 | 32 | 12.59 | 14.00 | 2.38 | 2.29 | 5.59E-06 | 6.03E-06 | 3.43E-05 | 1.54E-05 | 132 |
| Ubiquitin mediated proteolysis | ID:04120 | 30 | 43 | 12.87 | 14.32 | 2.33 | 3.00 | 9.00E-06 | 1.80E-11 | 5.09E-05 | 1.15E-10 | 135 |
| Calcium signaling pathway | ID:04020 | 36 | 52 | 16.88 | 18.78 | 2.13 | 2.77 | 1.03E-05 | 4.91E-12 | 5.68E-05 | 3.33E-11 | 177 |
| GnRH signaling pathway | ID:04912 | 24 | 27 | 9.63 | 10.71 | 2.49 | 2.52 | 2.13E-05 | 4.49E-06 | 1.00E-04 | 1.18E-05 | 101 |
| Small cell lung cancer | ID:05222 | 21 | 26 | 8.11 | 9.02 | 2.59 | 2.88 | 3.67E-05 | 4.06E-07 | 2.00E-04 | 1.24E-06 | 85 |
| Oocyte meiosis | ID:04114 | 25 | 29 | 10.68 | 11.88 | 2.34 | 2.44 | 4.48E-05 | 4.05E-06 | 2.00E-04 | 1.10E-05 | 112 |
| ECM-receptor interaction | ID:04512 | 21 | 24 | 8.11 | 9.02 | 2.59 | 2.66 | 3.67E-05 | 5.36E-06 | 2.00E-04 | 1.39E-05 | 85 |
| N-Glycan biosynthesis | ID:00510 | 15 | 16 | 4.67 | 5.20 | 3.21 | 3.08 | 3.24E-05 | 2.71E-05 | 2.00E-04 | 6.39E-05 | 49 |
| Renal cell carcinoma | ID:05211 | 18 | 33 | 6.68 | 7.43 | 2.70 | 4.44 | 7.37E-05 | 1.18E-14 | 3.00E-04 | 1.13E-13 | 70 |
| Acute myeloid leukemia | ID:05221 | 16 | 23 | 5.44 | 6.05 | 2.94 | 3.80 | 5.84E-05 | 5.29E-09 | 3.00E-04 | 2.10E-08 | 57 |
| VEGF signaling pathway | ID:04370 | 19 | 25 | 7.25 | 8.06 | 2.62 | 3.10 | 7.13E-05 | 1.41E-07 | 3.00E-04 | 4.42E-07 | 76 |
| Fc gamma phagocytosis | ID:04666 | 21 | 38 | 8.96 | 9.97 | 2.34 | 3.81 | 2.00E-04 | 5.66E-14 | 7.00E-04 | 4.75E-13 | 94 |
| RNA transport | ID:03013 | 29 | 49 | 14.40 | 16.02 | 2.01 | 3.06 | 2.00E-04 | 3.31E-13 | 7.00E-04 | 2.48E-12 | 151 |
| Adipocytokine signaling | ID:04920 | 17 | 22 | 6.49 | 7.21 | 2.62 | 3.05 | 2.00E-04 | 1.07E-06 | 7.00E-04 | 3.16E-06 | 68 |
| Phosphatidylinositol signaling | ID:04070 | 18 | 31 | 7.44 | 8.27 | 2.42 | 3.75 | 3.00E-04 | 1.94E-11 | 9.00E-04 | 1.20E-10 | 78 |
| Gap junction | ID:04540 | 20 | 30 | 8.58 | 9.55 | 2.33 | 3.14 | 3.00E-04 | 5.82E-09 | 9.00E-04 | 2.26E-08 | 90 |
| Melanoma | ID:05218 | 17 | 20 | 6.77 | 7.53 | 2.51 | 2.66 | 3.00E-04 | 3.29E-05 | 9.00E-04 | 7.51E-05 | 71 |
| Natural killer cell cytotoxicity | ID:04650 | - | 61 | - | 14.43 | - | 4.23 | - | 2.13E-24 | - | 7.46E-23 | 136 |
| Cell adhesion molecules | ID:04514 | - | 54 | - | 14.11 | - | 3.83 | - | 2.46E-19 | - | 5.17E-18 | 133 |
| Jak-STAT signaling pathway | ID:04630 | - | 59 | - | 16.44 | - | 3.59 | - | 2.37E-19 | - | 5.17E-18 | 155 |
| Toll-like receptor signaling | ID:04620 | - | 41 | - | 10.82 | - | 3.79 | - | 7.37E-15 | - | 7.37E-14 | 102 |
| TGF-beta signaling pathway | ID:04350 | - | 31 | - | 8.91 | - | 3.48 | - | 1.81E-10 | - | 9.27E-10 | 84 |
| NOD-like receptor signaling | ID:04621 | - | 24 | - | 6.15 | - | 3.90 | - | 1.34E-09 | - | 5.99E-09 | 58 |
| Allograft rejection | ID:05330 | - | 18 | - | 3.93 | - | 4.59 | - | 6.72E-09 | - | 2.52E-08 | 37 |
| Antigen processing presentation | ID:04612 | - | 26 | - | 8.06 | - | 3.22 | - | 3.22E-08 | - | 1.13E-07 | 76 |
| Basal transcription factors | ID:03022 | - | 15 | - | 3.93 | - | 3.82 | - | 2.26E-06 | - | 6.33E-06 | 37 |
| Oxidative phosphorylation | ID:00190 | 76 | - | 12.59 | - | 6.04 | - | 5.25E-43 | - | 5.64E-41 | - | 132 |
| Proteasome | ID:03050 | 19 | - | 4.20 | - | 4.53 | - | 5.22E-09 | - | 1.02E-07 | - | 44 |
| RNA polymerase | ID:03020 | 14 | - | 2.77 | - | 5.06 | - | 9.76E-08 | - | 1.31E-06 | - | 29 |
| Glycosphingolipid biosynthesis | ID:00601 | 13 | - | 2.48 | - | 5.24 | - | 1.67E-07 | - | 2.11E-06 | - | 26 |
| Metabolism of xenobiotics P450 | ID:00980 | 22 | - | 6.77 | - | 3.25 | - | 4.02E-07 | - | 4.12E-06 | - | 71 |
| Tyrosine metabolism | ID:00350 | 16 | - | 3.91 | - | 4.09 | - | 4.56E-07 | - | 4.40E-06 | - | 41 |
| Glutathione metabolism | ID:00480 | 17 | - | 4.77 | - | 3.57 | - | 1.95E-06 | - | 1.45E-05 | - | 50 |
| Arginine and proline metabolism | ID:00330 | 17 | - | 5.15 | - | 3.30 | - | 6.44E-06 | - | 3.85E-05 | - | 54 |
| Glycosylphosphatidylinositol | ID:00563 | 11 | - | 2.38 | - | 4.61 | - | 7.33E-06 | - | 4.26E-05 | - | 25 |
| Drug metabolism P450 | ID:00982 | 20 | - | 6.96 | - | 2.87 | - | 1.09E-05 | - | 5.86E-05 | - | 73 |
| Fructose mannose metabolism | ID:00051 | 12 | - | 3.43 | - | 3.50 | - | 7.82E-05 | - | 3.00E-04 | - | 36 |
| Sulfur relay system | ID:04122 | 6 | - | 0.95 | - | 6.29 | - | 1.00E-04 | - | 4.00E-04 | - | 10 |
| Arachidonic acid metabolism | ID:00590 | 16 | - | 5.63 | - | 2.84 | - | 9.20E-05 | - | 4.00E-04 | - | 59 |
| Sphingolipid metabolism | ID:00600 | 12 | - | 3.81 | - | 3.15 | - | 2.00E-04 | - | 7.00E-04 | - | 40 |
| Glycosaminoglycan degradation | ID:00531 | 8 | - | 1.81 | - | 4.41 | - | 2.00E-04 | - | 7.00E-04 | - | 19 |
| Hedgehog signaling pathway | ID:04340 | 15 | - | 5.34 | - | 2.81 | - | 2.00E-04 | - | 7.00E-04 | - | 56 |
| Drug metabolism | ID:00983 | 14 | - | 4.96 | - | 2.82 | - | 3.00E-04 | - | 9.00E-04 | - | 52 |
Hypermutation of dihydropyrimidine dehydrogenase in pyrimidine metabolism
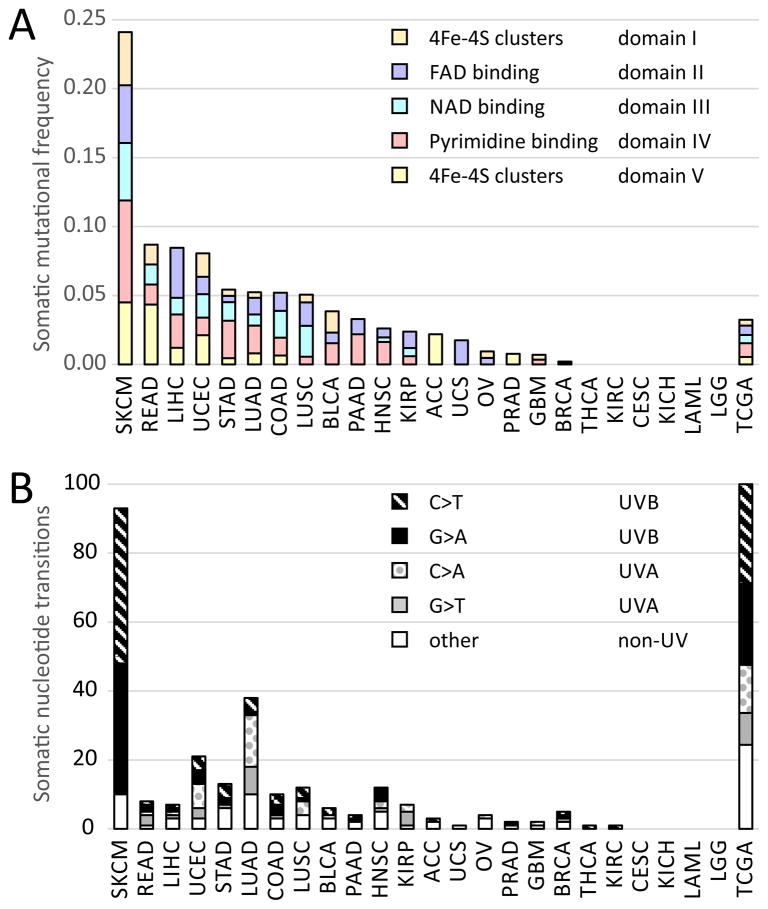
In order to identify potential melanoma driver genes in pyrimidine metabolism, we assessed recurrence and statistical enrichment of somatic mutations for all pyrimidine genes in melanoma. DPYD stands out for its highest mutation rate above 20% for all melanoma patients and significant enrichment of somatic mutations above background mutation rate with a q-value of 4.40E-06 (Table 2). Twelve other pyrimidine genes show high somatic mutation rates, p-values of recurrence and conservation, or q-values of enrichment above background mutation rate including DPYS (dihydropyrimidinase, Gene ID: 1807), AK9 (adenylate kinase domain containing 1, Gene ID: 221264), CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase, Gene ID: 790), CANT1 (calcium activated nucleotidase 1, Gene ID: 124583), ENTPD1 (ectonucleoside triphosphate diphosphohydrolase 1, Gene ID: 953), NME6 (NME/NM23 nucleoside diphosphate kinase 6, Gene ID: 10201), NT5C1A (5′-nucleotidase, cytosolic IA, Gene ID: 84618), POLE (polymerase (DNA directed), epsilon, Gene ID: 5426), POLQ (polymerase (DNA directed), theta, Gene ID: 10721), POLR3B (polymerase (RNA) III (DNA directed) polypeptide B, Gene ID: 55703), PRIM2 (primase, DNA, polypeptide 2 (58kDa), Gene ID: 5558), REV3L (REV3-like, polymerase (DNA directed), zeta, Gene ID: 5980), and UPP2 (uridine phosphorylase 2, Gene ID: 151531). The majority of these genes show statistically significant somatic mutations in other cancers of the TCGA Pan-cancer cohort (Table 2). For example, PRIM2 has recurrent somatic mutations at residue E221, G334, P391, and is also significantly mutated in HNSC (p=0.00558; q=3.34E-06) or LUSC (p=0.000339; q=2.27E-04). Somatic DPYD mutations coincide with deleterious mutations of gatekeeper and caretaker genes TP53, BRCA1, FAT3, FAT4, PTPRD, and SPEN with p-values below 1.0E-06 and q-values below 1.0E-04 connecting to DNA maintenance and stability. In comparison to other TCGA tissues, DPYD is the top somatically mutated gene in pyrimidine metabolism, affecting 67 patients of 278 whole-exome sequenced metastatic melanoma (Figure 1A, Supplementary table 5). There are in total 74 non-synonymous mutations in DPYD detected, including incidents where two or three residues of the same polypeptide chain are affected. Examples of multiple mutations coinciding in DPYD are S204F and D949N in patient TCGA-EE-A2MI, or V396I, G851R, E937K in patient TCGA-FW-A3R5. The nucleotide signature of somatic transitions of DPYD tracks with the validated mutational signature of melanoma identified across human cancers (6, 10), and is governed by UVB-associated C>T/G>A transitions (Supplementary table 6) (Figure 1B).
Table 2.
Analysis of somatic mutations in pyrimidine metabolism of 339 whole-exome sequenced TCGA SKCM patients (KEGG pathway ID:00240). P-values above 0.05 or q-values above 0.10 are in italic. The column TCGA Pan-cancer lists significant somatic events in pyrimidine metabolism in other cancer tissues.
| Symbol | Gene ID | Mutation rate | p-value MutSig | q-value MutSig | q-value InVEx | TCGA Pan-cancer Tissue (p- and q-value MutSig) |
|---|---|---|---|---|---|---|
| AK9 | 221264 | 5.00% | 4.30E-01 | 2.66E-01 | 8.80E-04 | LAML (p=NA;q=1.97E-02) |
| CAD | 790 | 5.00% | 6.77E-03 | 4.06E-02 | 2.71E-02 | LGG (p=2.87E-02;q=8.81E-02) |
| CANT1 | 124583 | 3.00% | 4.45E-02 | 2.86E-02 | 2.71E-02 | PRAD (p=4.13E-02;q=4.62E-03) |
| DPYD | 1806 | 20.50% | 1.00E+00 | 7.13E-01 | 4.40E-06 | HNSC (p=1.40E-02;q=7.36E-02) |
| DPYS | 1807 | 7.00% | 4.87E-01 | 5.32E-01 | 8.25E-03 | PAAD (p=3.02E-02;q=3.14E-07) |
| ENTPD1 | 953 | 4.00% | 1.53E-01 | 1.61E-01 | 1.58E-02 | - |
| NME6 | 10201 | 1.00% | 1.22E-01 | 1.76E-01 | 1.58E-02 | COAD (p=1.77E-01;q=3.68E-02) |
| NT5C1A | 84618 | 1.00% | 2.60E-02 | 9.72E-02 | 6.44E-01 | STAD (p=2.68E-02;q=9.90E-02) |
| POLE | 5426 | 6.00% | 2.69E-01 | 6.22E-01 | 1.51E-03 | ACC (p=5.86E-03;q=2.02E-02) KIRC (p=2.85E-02;q=9.37E-02) UCEC (p=1.29E-02;q=6.90E-02) |
| POLQ | 10721 | 8.00% | 3.55E-01 | 7.01E-01 | 8.14E-04 | LUSC (p=3.40E-02;q=1.43E-01) |
| POLR3B | 55703 | 5.00% | 7.59E-01 | 9.47E-01 | 1.32E-03 | UCEC (p=2.24E-03;q=3.96E-03) |
| PRIM2 | 5558 | 6.00% | 3.52E-02 | 1.00E-05 | 7.63E-04 | HNSC (p=5.58E-03;q=3.34E-06) LUSC (p=3.40E-04;q=2.27E-04) |
| REV3L | 5980 | 6.00% | 1.47E-01 | 4.29E-01 | 2.20E-01 | BRCA (p=6.48E-03;q=2.62E-02) |
| UPP2 | 151531 | 2.00% | 1.22E-02 | 2.99E-02 | 7.63E-04 | LUAD (p=3.90E-01;q=3.40E-02) |
Figure 1. Skin cutaneous melanoma (SKCM) stands out in frequency and nucleotide signature of somatic mutations of dihydropyrimidine dehydrogenase (DPYD) across Pan-cancer patients of The Cancer Genome Atlas (TCGA).
A) Somatic mutational frequency of DPYD mutations in melanoma and across TCGA PAN-cancer patients. B) Nucleotide signature of somatic transitions of DPYD mutations analyzed by UV-type for melanoma (UVB associated with C>T and G>A, and UVA-type with G>T and C>A) shown as absolute count of mutational incidences. Fraction of nucleotide signature of somatic transitions of DPYD mutations is given at the right across all TCGA Pan-cancer tissues in TCGA.
Structural hotspots of somatic hypermutation of DPYD in ligand binding sites as well as interfaces of protein domains
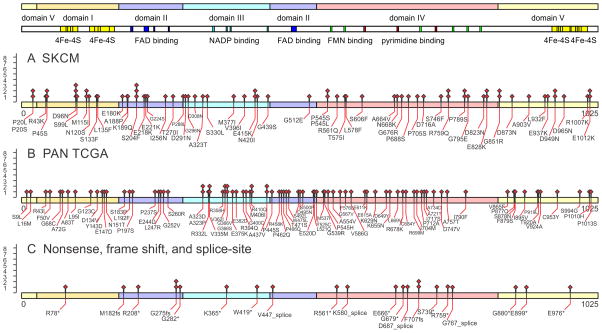
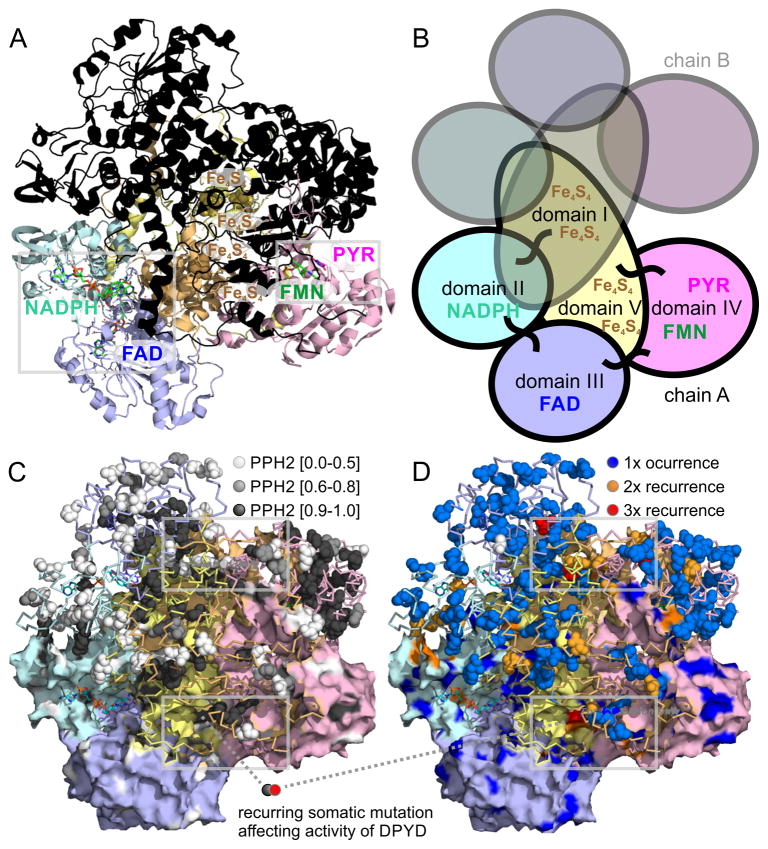
In order to decipher functional implications of somatic mutations of DPYD, we analyzed the domain distribution, polymorphism phenotyping v2 (PPH2) scores, solvent accessible surface, proximity to ligands, and mutational recurrence of all identified somatic mutations (Supplementary table 7). The cytosolic dihydropyrimidine dehydrogenase (EC 1.3.1.2; OMIM 612779 and 274270) is the initial and rate-limiting enzyme in the catabolism of pyrimidines. It reduces the pyrimidine bases thymine and uracil in a NADPH-dependent manner. The highly conserved homodimeric 1025-residue protein contains four 4Fe-4S-clusters, one FAD, and one FMN in the active site cavity of each subunit (Figure 2, Figure 3A–B). A special electron transfer pathway involves the 4Fe-4S-clusters of both subunits, so that DPYD comprises two independent electron transfer chains and is active just as a dimer (14, 15). The somatic mutations affect 153 residues of 1025 in the TCGA Pan-cancer dataset (TCGA: 181 mutation affecting 153 unique residues; SKCM: 74 mutations affecting 60 unique residues). Recurrent somatic mutations are detected in all five functional domains of DPYD (Figure 2). While the pyrimidine binding domain has the highest mutational count with 55 mutations in total, correction for domain length shows that domains II–IV, which bind metabolite substrates and cofactors, show enriched mutation frequency (37 mutations over 197 residues in FAD binding domain II domain; 33 mutations over 155 residues in NADPH binding domain III domain; 55 mutations over 323 residues in the FMN and pyrimidine binding domain IV). The functional impact of somatic missense mutations was quantified using PPH2 scores and plotted onto the protein structure (Figure 3C). Molecular dynamics simulations in combination with computation of solvent accessible surface revealed an accumulation of damaging, missense somatic mutations in the core of the protein (PPH2 scores > 0.95; making up 52% of somatic mutations; solvent accessible surface values of affected residues below 0.50). In contrast, possibly benign somatic changes are surface-bound (PPH2 scores < 0.50; making up 37% of somatic mutations; elevated average value above 0.65 for the solvent accessible surface, Figure 3C). The location of somatic mutations predicted to be damaging based on high functional PPH2 scores and lack of surface accessibility coincides with detected somatic recurrence in SKCM as well as in the TCGA Pan-cancer cohort (Figure 3A,C–D).
Figure 2. Somatic mutational landscape of DPYD mutations in melanoma and across TCGA PAN-cancer patients.
Somatic mutations are indicated on the protein sequence of DPYD, NCBI Gene ID 1806, for A) skin cutaneous melanoma (SKCM) and B) 24 TCGA tissues with missense DPYD mutations. C) Non-sense, frame-shift and splice-site mutations are indicated separately. Functional domains are annotated according uniprot entry Q12882 and 11179210: Domain I N-terminal 4Fe-4S clusters (27–172, yellow); domain II FAD binding domain (173–286, 442–524, blue); domain III NADPH binding domain (287–441, cyan); domain IV FMN and pyrimidine binding domain (525–848, red; FMN binding in green); domain V C-terminal 4Fe-4S clusters (1–26, 848–1025, yellow).
Figure 3. Structural analysis of DPYD dimer reveals hotspots of somatic mutations in ligand binding sites as well as interfaces of protein domains.
A) The 4Fe-4S cluster domain I of DPYD (uniprot entry Q12882 and PDB entry 1gth) is shown in orange, the FAD binding domain II in blue, the NADPH binding domain III in cyan, the pyrimidine and FMN binding domain IV in red, and the 4Fe-4S cluster domain V is shown in yellow. The lower monomer is color-coded according to domains and contains small-molecule ligands as sticks, the upper monomer is overlaid in black. B) The five domains of each DPYD monomer are color coded according to protein domains showing that the electron transfer chain crosses the dimer interface twice. The 4Fe-4S cluster domains I and V are intertwined and form extended inter-domain contacts. C) Functional impact of somatic missense mutations plotted as PPH2 scores onto surface (lower monomer; showing surface-bound mutations) and ribbon of DPYD structure (emphasizing accumulation of deleterious mutations in protein core in black). D) Recurrent missense somatic mutations in ligand binding sites as well as interfaces of protein domains of DPYD. Grey frames indicate regions of coincidence of high functional PPH2 scores, lack of surface accessibility coincides, and somatic recurrence shown enlarged in Figure 4.
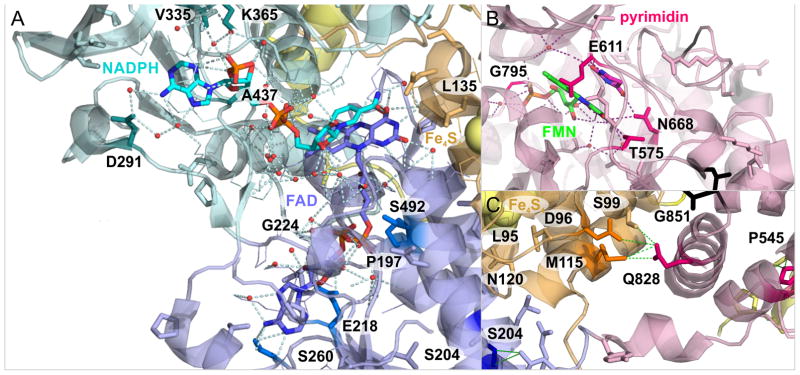
Recurring somatic missense mutations D291N, V335M, and A437 frame the nucleotide binding site (Figure 4A). In addition, the NADPH binding domain houses mutations A323D/P/T, V362I, G366I/S/V, as well as V365 nonsense mutation. Somatic mutations involved in the hydrogen bond network and within less than 3.5A to the FAD ligand are P197S, E218K, G224S/V, S260R, and S492L. The mutation L135F is located at the catalytic route between FAD and N-terminal 4Fe-4S cluster (Figure 4A). There are 20 non-recurring mutations that populate residues involved in the electron transfer between the four 4Fe-4S cluster. Between C-terminal 4Fe-4S cluster and electron entry site of FMN, there is the somatic mutation E611K (Figure 4B). Mutations D949N, D965N, A554V, and E615A line up between C-terminal 4Fe-4S domain V and pyrimidine binding domain IV. The pyrimidine substrate binding pocket is framed by recurrent somatic mutations T575I, E611K, N668K, and G795E (Figure 4B). A hotspot of recurring somatic mutations D96N (3x), S204F (3x), M115I (2x), G851R (2x), E828K (2x), and P545H/L/S is at the interface between domain I, II, and IV (Figure 3C,D). S204F is a structural residue of FAD binding domain II, linking the three domains forming the large cleft of DPYD. S204F is part of an alpha-helix which directly bridges to residues L95I and N120S, affected by somatic mutations. Similarly, Q828 is an anchor at domain IV and spans a hydrogen bond network to domain I via D96N, S99L, and M115I, affected by somatic mutations.
Figure 4. Recurring somatic missense mutations frame ligand binding sites of DPYD modulating its enzymatic activity.
A) Recurring somatic mutations in ligand binding sites of NADPH, FAD, and 4Fe-4S clusters at the interface of domain I–III. B) Recurring somatic mutations in pyrimidine and FMN binding site affect hydrogen bond network of enzymatic effector domain IV. C) Accumulation of recurring somatic mutations at domain interface shows hinge-residue Q828 in domain IV and its connectivity to domain I via hydrogen bonds. Location of expanded regions on global map of protein structure of the DPYD dimer is indicated as grey frames in Figure 3.
Cross-talk between DPYD mutations and gene expression of the pyrimidine pathway
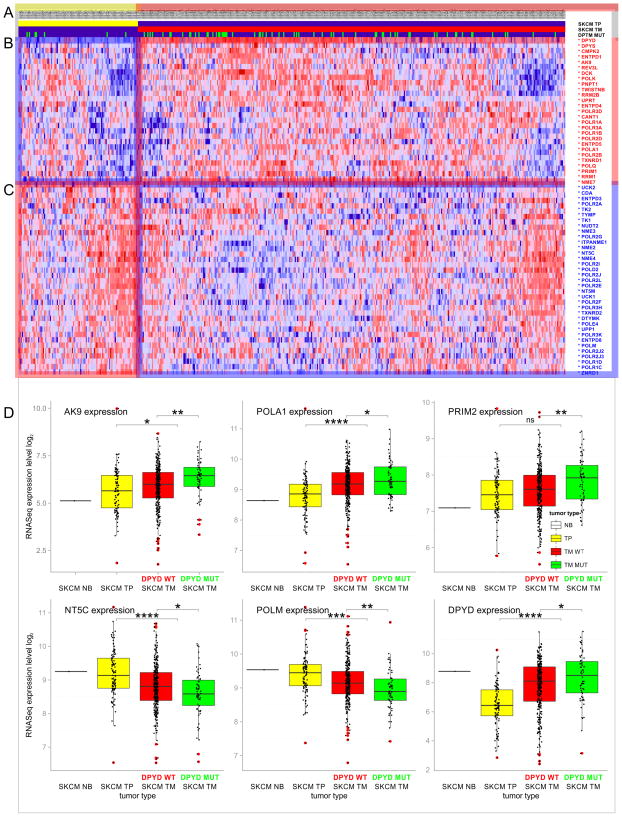
Next, we addressed whether the mutational and transcriptional signature of pyrimidine metabolism in melanoma follows a distinct pattern. More than half of the pyrimidine enzymes are differentially expressed between primary and metastatic tumor, showing distinct clusters in key steps of pyrimidine nucleoside triphosphate synthesis, DNA and RNA synthesis, as well as pyrimidine degradation (Figure 5A). At the mutational level there is also a progressive enrichment of somatic mutations in the SKCM cohort comparing primary and metastatic tumors. DPYD is mutated in 11.5% of primary tumors, while in metastatic tumors somatic mutations are detected in 22.5% of all whole exome-sequenced samples (Figure 5B). Comparison of gene expression and mutational data of DPYD and other key pyrimidine enzymes in SKCM shows that enrichment of somatic mutations in metastatic tumors coincides with elevated expression levels (Figure 5). The gene expression signature is significantly enhanced by somatic DPYD mutation. The expression level of pyrimidine enzymes changes in SKCM metastatic tumor samples with DPYD wild-type status in comparison to metastatic tumor samples with DPYD mutation with p-value below 0.05. In addition, the direction of expression change in melanoma progression (up or down from tumor to metastasis) is the same as the difference between DPYD mutation and wild-type (up or down from wild-type to mutation, respectively). The observed deregulated gene expression of pyrimidine enzymes—including DPYD itself—correlates with metastatic progression and is enhanced by somatic DPYD mutations (Figure 5B, C). Almost all differentially expressed pyrimidine enzymes (with the exception of POLR3D), which show up- or downregulation between primary and metastatic tumors, show progressive increase or decrease with DPYD mutation, respectively (Supplementary table 8). Somatic mutations of DPYD enhance the metastatic progression signature of melanoma (Figure 5D).
Figure 5. Gene expression signature of pyrimidine metabolism in the progression of metastatic melanoma.
A) Tumor status of TCGA skin cutaneous melanoma (SKCM) patients. Solid primary tumors (TP, yellow) are marked in the first row; metastatic tumors (TM, red, second row) are marked in the second row. Mutational status of DPYD is indicated in the third row (DPYD MUT, green). B) Significant upregulation and C) downregulation of genes in pyrimidine metabolism between skin cutaneous primary and metastatic tumors. Genes with p-values below 0.05 are marked with an asterisk next to the gene symbol. D) Impact of DPYD mutations on gene expression of pyrimidine enzymes is shown in green. Tumor progression of skin cutaneous melanoma (SKCM) cohort is shown for normal tissue (NB, black), solid primary tumor (TP, yellow), metastatic tumor with DPYD WT status (TM, DPYD WT, red), and metastatic tumor with DPYD mutations (TM, DPYD MUT, green). Box plots depict data distributions through quartiles. Asterisks above plots indicate results of statistical significance tests (Students T-test: * p-value ≤ 0.05; * p-value ≤ 0.01; * p-value ≤ 0.001; * p-value ≤ 0.0001; ns p-value ≥ 0.05).
Bifurcation of pyrimidine metabolism in metastatic melanoma
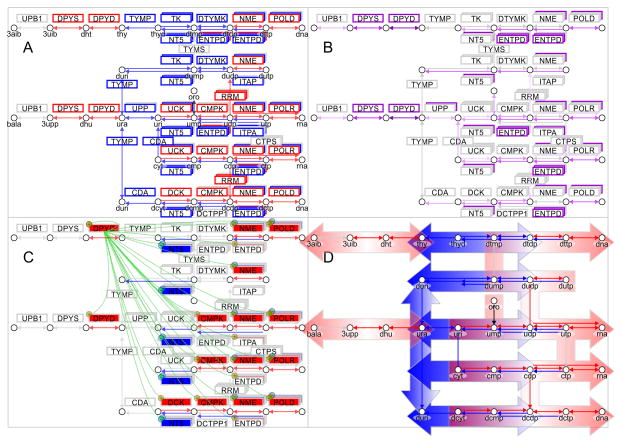
Somatic mutations and differential gene expression have severe implications for the metabolic network of pyrimidine metabolism. Mapping of gene expression data onto a pathway map of pyrimidine metabolism (modeled after KEGG pathway ID:00240) revealed a two-fold separation. Pyrimidine degradation initiated by enzymes DPYD and DPYS is significantly up-regulated (Figure 6A). Enzymes TYMP (thymidine phosphorylase, Gene ID: 1890), UPP1 (uridine phosphorylase 1, Gene ID: 7378), CDA, TK1 (thymidine kinase 1, soluble, Gene ID: 7083), TK2 (thymidine kinase 2, mitochondrial, Gene ID: 7084), UCK1 (uridine-cytidine kinase 1, Gene ID: 83549), and DTYMK (deoxythymidylate kinase (thymidylate kinase), Gene ID: 1841) salvaging pyrimidines are significantly down-regulated. Enzymes DCK (deoxycytidine kinase, Gene ID: 1633), CANT1, AK9, and NME7 (NME/NM23 family member 7, Gene ID: 29922) providing pyrimidine nucleoside triphosphate and nucleic acid building enzymes POLA1 (polymerase (DNA directed), alpha 1, Gene ID: 5422), POLK (polymerase (DNA directed) kappa, Gene ID: 51426), POLQ, POLR1A (polymerase (RNA) I polypeptide A, Gene ID: 25885), POLR1B (polymerase (RNA) I polypeptide B, Gene ID: 84172), POLR2B (polymerase (RNA) II (DNA directed) polypeptide B, Gene ID: 5431), POLR2D (polymerase (RNA) II (DNA directed) polypeptide D, Gene ID: 5433), POLR3A (polymerase (RNA) III (DNA directed) polypeptide A, Gene ID: 11128), POLR3D, REV3L, PRIM1 (primase, DNA, polypeptide 1, Gene ID: 5557), PNPT1, and TWISTNB (TWIST neighbor, Gene ID: 221830) are upregulated. This is enforced by a significant downregulation of pyrimidine nucleoside triphosphate degrading enzymes ENTPD3 (ectonucleoside triphosphate diphosphohydrolase 3, Gene ID: 956), ENTPD8 (ectonucleoside triphosphate diphosphohydrolase 8, Gene ID: 377841), ITPA (inosine triphosphatase (nucleoside triphosphate pyrophosphatase), Gene ID: 3704), NT5C (5′, 3′-nucleotidase, cytosolic, Gene ID: 30833), and NT5M (5′,3′-nucleotidase, mitochondrial, Gene ID: 56953). In addition, enzymes RRM1 (ribonucleotide reductase M1, Gene ID: 6240), RRM2B (ribonucleotide reductase M2 B (TP53 inducible), Gene ID: 50484) required for anabolic conversion of uracil nucleosides to thymidine diphosphate nucleosides are upregulated, while enzymes TYMP or CDA mediating production of uracil, uridine, and deoxyuridine are downregulated. At a pathway level, somatic mutations in SKCM patients are most frequently observed in enzymes DPYD, DPYS, ENTPD1, CANT1, and UPP2 of pyrimidine degradation, as well as degradation of pyrimidine nucleoside triphosphates (q-value below 0.03) (Figure 6B, Supplementary Table 5). Somatic mutations of DPYD significantly enhance the signature of pyrimidine nucleoside triphosphates and nucleic acid generating enzymes CMPK1, AK9, NME7, POLA1, POLD3, POLK, POLR3F, POLR3G, PRIM2, and TWISTNB (p-value < 0.05) (Figure 6C, Supplementary Table 8). In addition, DPYD mutated samples show significantly increased DPYD transcript levels (p-value < 0.05). Taken together, the combined mutational and gene expression analysis shows a shift towards pyrimidine nucleoside triphosphates and nucleic acid synthesis, and disconnection from pyrimidine salvage and degradation (Figure 6D, Supplementary Table 4).
Figure 6. Somatic mutations of DPYD in TCGA SKCM enhance metastatic signature of melanoma and promote deregulation of the pyrimidine pathway toward malignant cancer progression.
A) Gene expression signature is plotted onto pathway map of pyrimidine metabolism in metastatic melanoma (KEGG pathway ID:00240). On the left side enzymes DPYD, DPYS and UPB1 are responsible for pyrimidine degradation, in the center TYMP connects uracil derivatives, and at the right pyrimidine kinases DTYMK and CMPK, nucleoside diphosphate kinases NME, and polymerases POLD and POLR provide synthesis of DNA and RNA nucleic acids. B) Frequency of somatic mutations in pyrimidine enzymes is color coded from 0% in grey to 25% in purple in metastatic melanoma (Supplementary Table 5). C) Impact of DPYD mutations on enzymes of pyrimidine metabolism is indicated by regulatory symbols and shading of enzyme boxes (red, plus) for enhancement and suppression (blue, minus) (Supplementary Table 8). Somatic frequency of mutations, enhanced gene expression signature of patients with DPYD mutations are provided in supplementary tables. D) Pathway map shows separation and directionality of upregulated pyrimidine degradation (red, left) and nucleic acid synthesis (red, right) by down-regulated uracil and thymidine salvage (blue, center). Concerted dysregulation of metabolic enzymes at a pathway level contributes to bifurcation of pyrimidine metabolism. The systems biology maps depict metabolites as circles, reactions with their respective directionality as arrows, and enzymes as boxes. Enzymes of anabolic direction are shown above reactions and enzymes of catabolic direction below reactions. Staggered boxes indicate metabolic redundancy that multiple genes encode enzymes for the reaction.
Discussion
Enzymes in pyrimidine metabolism undergo a significant deregulation at the gene expression level in the transition from skin cutaneous primary tumors toward metastatic tumors (Table 1, Figure 5). This transition is accompanied by an enrichment of somatic mutations of DPYD (Table 2, Figure 1–4).
The mutational analysis identified more than 130 unique and novel recurrent somatic mutations in DPYD, including recurrent missense, nonsense and splice site mutations (Supplementary table 7). In addition, we were able to confirm frequently recurring deleterious mutations S204F and G275 frame shift (Figure 4A,C) (8, 16). The mutational burden of DPYD after correction for background rate is equally high as established melanoma drivers and shows significant enrichment with a q-value of 4.40E-06 (Table 2) (8). An emerge theme in cancer genomics—facilitated by the advent of deep sequencing data of large patient cohorts—is that the mutational landscape of proto-oncogenes and tumor suppressors is more diverse than anticipated (6). Structural analysis of cancer driver BRAF showed unprecedented events in the RAS binding domain interface and the ATP binding pocket aside from established p.V600E/K/R/D substitutions. Detailed topological analysis of the DPYD dimer reveals structural hotspots in ligand binding sites and interfaces of protein domains of DPYD. Events with three-time recurrence are detected in each of the functional domains with p.D96N (Fe-S cluster I–II), p.S204F (FAD binding domain), p.A323T/P/D (NADPH binding domain), and p.P545S/L/H (Pyrimidine binding domain). There are distinct areas of interest with high density of somatic recurrence of mutations in DPYD (Figure 4). Two NADPH binding loops between V335 and G366 positions the nucleotide and initiate the electron transfer. Somatic mutations V335M, A437M, D291N, V362I, and G366I/S/V closely frame the nucleotide binding site and are expected to have reduced NADPH binding, similarly to the reduced affinity of reported variant G366A (Figure 4A) (17). Recurring somatic mutations T575I, E611K, N668K, and G795E in pyrimidine and FMN binding site affect hydrogen bond network of enzymatic effector domain IV (Figure 4B).
The interface between FAD binding domain II, N-terminal 4Fe-4S cluster domain I, and pyrimidine binding domain IV stands out for high frequency recurrences of somatic mutation (Figure 3C,D). E828 is engaged in a tight hydrogen bonding network to D96, S99, and M115, which are also affected by somatic mutations (Figure 4C). E828K has demonstrated higher DPYD activity (18), stressing the importance of this hydrogen bonding network. Somatic mutations in highly recurring sites D96N, S204F, P545L/S/H is associated with high PPH2 values of 1.0 indicating possibly damaging outcome of DPYD function. In contrast the somatic mutation A323T has been shown to be benign with enzymatic activity close to wild type (18). Overall, the functional analysis of somatic mutations shows strong agreement with a comparative in vitro analyses of DPYD variants that somatic mutations have reduced DPYD activity (Supplementary table 7) (17, 18).
Pyrimidine enzymes undergo differential upregulation between skin cutaneous primary and metastatic tumors in key steps of nucleoside triphosphate, DNA and RNA synthesis, as well as pyrimidine degradation. Somatic mutations can generate metabolic bottlenecks and reroute metabolic paths. Visualization of somatic incident at a pathway level, helps identifying such bottlenecks (Figure 6). The intricate network pyrimidine metabolism has built-in redundancy, where enzymatic steps can be encoded by different genes or enzymes can recognize and process multiple substrates. Furthermore, the pathway contains steps for conversion between uridine and thymidine nucleotides as well as for salvage of pyrimidine bases. However, if the transition between skin cutaneous primary and metastatic tumors relies on distinct isoenzymes, new therapeutic targets might open. Distinct over-expression of nucleoside diphosphate kinases AK9 or NME7 in metastatic cancer puts emphasis on pyrimidine nucleotide synthesis while pyrimidine deamination is down-regulated (Figure 5B and Figure 6C). Based on the metabolic maps another potential melanoma drug target is RRM1. Established efficacy of nucleoside analogues in acute leukaemias might facilitate new treatment regimens in skin cancer (19). Given a strong reliance on biosynthetic building blocks, the upregulation of pyrimidine degradation lowers the pool of nucleotide bases available for salvage. The ribonucleotide reductases RRM1 responds to DPYD alteration, is significantly upregulated in metastatic melanoma, and bridges bottlenecks between deoxyribo- and ribonucleotides (Supplementary table 8, Figure 6C–D).
The systems biology analysis of melanoma data in TCGA revealed a strong separation of pyrimidine degradation and nucleotide synthesis, which is important for effective nucleic acid synthesis (Figure 6D). The mechanism of DPYD controlling pyrimidine metabolism is unknown (17). A likely possibility is the existence of metabolic feedback loops of other enzymes shifting metabolism into a different gear within the progression of cancer (20). Elevated DPYD expression results in low metabolite pools of the pyrimidine nucleobases thymine and uracil, which could allosterically bind metabolic enzymes or signaling molecules. Moreover dihydropyrimidins and deoxypyrimidins are allosteric inhibitors of thymidine kinase (21, 22), which enhance the importance of TYMS for de novo pyrimidine synthesis.
Nucleotide synthesis is closely linked to production as well as stability of nucleic acids. Not surprisingly, purine metabolism scored equally high in the enrichment study, since both pathways share important enzymes in nucleoside salvage and nucleic acid processing (Table 1). However, mutational signature, correlation between somatic alterations and gene expression, and metabolic bottlenecks were unique to pyrimidine metabolism motivating further studies of DPYD. Remarkably, mutated DPYD was found to be overexpressed in metastatic cells promoting synthesis of DNA and RNA (Figure 5). In addition, somatic DPYD alterations co-occurred with mutations of tumor suppressors and DNA caretakers in melanoma patients. Deregulated pyrimidine catabolism may not only be connected to nucleotide anabolism but also negatively affect DNA maintenance and stability. Further experiments will be needed to decipher the cellular mechanisms responsible for the development and the progression of melanoma.
In addition to executing the epithelial-mesenchymal transition program, metastatic cells acquire traits associated with high-grade malignancy, including resistance to apoptosis and chemotherapy. Patients with a complete or partial DPYD deficiency have been reported as suffering from lethal toxicity after the administration of 5-FU (23). Based on the pathway analysis of SKCM samples we established a gene expression signature of pyrimidine enzymes, which grants drug sensitivity while limiting toxicity. 5-FU has to be processed by TYMP, TK, CMPK, and NME enzymes to produce the active drug-metabolite FdUMP, which is a tight-binding inhibitor of TYMS. As TYMS represents the sole intracellular source of de novo TMP, the inhibition of TS exploits a metabolic bottlenecks in the biosynthesis of DNA (Figure 6). In addition, UPP, UCK, CMPK, NME enzymes facilitate production of 5-FUTP causing nucleic acid damage and apoptosis. Low levels of NT5 support accumulation of 5-FUMP and FdUMP and cell toxicity. Despite DPYD degrades 5-FU to DHFU, pyrimidine degradation is necessary and causes systemic failure if absent or partially dysfunctional (24). None of the metastatic melanoma patients show compatible signatures of gene expression (Figure 5A, 5B, 6A). Based on the mutation rate of DPYD of more than 20% in melanoma in combination with downregulation of TK and UPP, we predict high risks of 5-FU toxicity in melanoma. For these reasons fluorinated uracil-based pyrimidine analogues cannot considered to be a safe treatment regime for melanoma patients. While knockdown experiments will be necessary to identify more efficient therapeutic regimen in the pyrimidine pathway, the systems biology analysis provides a diagnostic insight at the pathway level. Importantly, the increased genotyping coverage achieved by a comprehensive description of the mutational landscape of DPYD improves predictive value for 5-FU toxicity.
Conclusion
The structure-based analysis of detected somatic events highlights vulnerabilities in DPYD. Recurring missense mutations accumulate in ligand binding sites as well as at domain interface between Fe4S4 clusters, FAD and pyrimidine binding. The transcriptional data shows that mutated DPYD selectively activates components of pyrimidine metabolism. The crosstalk between somatic mutations and gene expression promotes proliferative aggressiveness. Taken together, the transition from primary to metastatic tumors reconfigures the pyrimidine metabolism and emphasizes nucleic acid synthesis required for rapid cellular proliferation.
Supplementary Material
Implications.
At a systems biology level, somatic mutations of DPYD cause a switch in pyrimidine metabolism and promote gene expression of pyrimidine enzymes toward malignant progression.
Acknowledgments
Financial support
F.V.F. is grateful for the support of grant CA154887 from the National Institutes of Health, National Cancer Institute.
Footnotes
Conflicts of interest
The authors declare that there is no competing financial interest as part of the submission process of this manuscript.
Author contribution statement
F.V.F. designed the study and wrote the text. L.E., R.G., F.V.F. performed the data analysis of the SKCM TCGA dataset, and reviewed the final manuscript.
Competing financial interest
The authors declare that there is no competing financial interest as part of the submission process of this manuscript. We are thankful to all members of the TCGA Research Network for biospecimen collection, data acquisition, and benchmark analyses. F.V.F. is grateful for the support of grant CA154887 from the National Institutes of Health, National Cancer Institute.
References
- 1.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–9. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipp FV, Ratnikov B, De Ingeniis J, Smith JW, Osterman AL, Scott DA. Glutamine-fueled mitochondrial metabolism is decoupled from glycolysis in melanoma. Pigment Cell Melanoma Res. 2012;25:732–9. doi: 10.1111/pcmr.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 4.Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, et al. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–6. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 5.Wilson PM, Danenberg PV, Johnston PG, Lenz HJ, Ladner RD. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol. 2014;11:282–98. doi: 10.1038/nrclinonc.2014.51. [DOI] [PubMed] [Google Scholar]
- 6.Guan J, Gupta R, Filipp FV. Cancer systems biology of TCGA SKCM: efficient detection of genomic drivers in melanoma. Sci Rep. 2015;5:7857. doi: 10.1038/srep07857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–54. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 14.Podschun B, Cook PF, Schnackerz KD. Kinetic mechanism of dihydropyrimidine dehydrogenase from pig liver. J Biol Chem. 1990;265:12966–72. [PubMed] [Google Scholar]
- 15.Dobritzsch D, Ricagno S, Schneider G, Schnackerz KD, Lindqvist Y. Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer. J Biol Chem. 2002;277:13155–66. doi: 10.1074/jbc.M111877200. [DOI] [PubMed] [Google Scholar]
- 16.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amstutz U, Froehlich TK, Largiader CR. Dihydropyrimidine dehydrogenase gene as a major predictor of severe 5-fluorouracil toxicity. Pharmacogenomics. 2011;12:1321–36. doi: 10.2217/pgs.11.72. [DOI] [PubMed] [Google Scholar]
- 18.Offer SM, Fossum CC, Wegner NJ, Stuflesser AJ, Butterfield GL, Diasio RB. Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity. Cancer Res. 2014;74:2545–54. doi: 10.1158/0008-5472.CAN-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonate PL, Arthaud L, Cantrell WR, Jr, Stephenson K, Secrist JA, 3rd, Weitman S. Discovery and development of clofarabine: a nucleoside analogue for treating cancer. Nat Rev Drug Discov. 2006;5:855–63. doi: 10.1038/nrd2055. [DOI] [PubMed] [Google Scholar]
- 20.Filipp FV. Cancer metabolism meets systems biology: Pyruvate kinase isoform PKM2 is a metabolic master regulator. J Carcinog. 2013;12:14. doi: 10.4103/1477-3163.115423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Roy V, Wang L, You L, Agrofoglio LA, Deville-Bonne D, et al. 3′-(1,2,3-Triazol-1-yl)-3′-deoxythymidine analogs as substrates for human and Ureaplasma parvum thymidine kinase for structure-activity investigations. Bioorg Med Chem. 2010;18:3261–9. doi: 10.1016/j.bmc.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaul YD, Freinkman E, Comb WC, Cantor JR, Tam WL, Thiru P, et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Kuilenburg AB, Haasjes J, Richel DJ, Zoetekouw L, Van Lenthe H, De Abreu RA, et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: identification of new mutations in the DPD gene. Clin Cancer Res. 2000;6:4705–12. [PubMed] [Google Scholar]
- 24.van Kuilenburg AB. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur J Cancer. 2004;40:939–50. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.