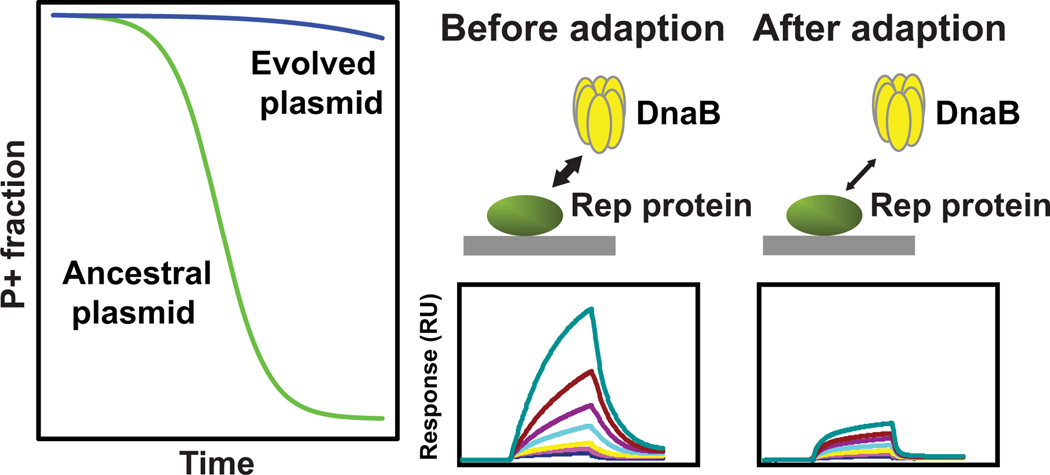
SUMMARY
Antibiotic selection drives adaptation of antibiotic resistance plasmids to new bacterial hosts, but the molecular mechanisms are still poorly understood. We previously showed that a broad-host-range plasmid was poorly maintained in Shewanella oneidensis, but rapidly adapted through mutations in the replication initiation gene trfA1. Here we examined if these mutations reduced the fitness cost of TrfA1, and whether this was due to changes in interaction with the host’s DNA helicase DnaB. The strains expressing evolved TrfA1 variants showed a higher growth rate than those expressing ancestral TrfA1. The evolved TrfA1 variants showed a lower affinity to the helicase than ancestral TrfA1 and were no longer able to activate the helicase at the oriV without host DnaA. Moreover, persistence of the ancestral plasmid was increased upon overexpression of DnaB. Finally, the evolved TrfA1 variants generated higher plasmid copy numbers than ancestral TrfA1. The findings suggest that ancestral plasmid instability can at least partly be explained by titration of DnaB by TrfA1. Thus under antibiotic selection resistance plasmids can adapt to a novel bacterial host through partial loss of function mutations that simultaneously increase plasmid copy number and decrease unfavorably high affinity to one of the hosts’ essential proteins.
Keywords: Experimental evolution, DNA replication, fitness cost, DNA helicase, plasmid, Rep protein
Graphical Abstract
INTRODUCTION
Horizontal gene transfer can facilitate rapid adaptation of bacterial populations to changes in their environment such as the presence of antibiotics, and plasmids are important players in this evolutionary process (Thomas and Nielsen, 2005; Frost et al., 2005). These mobile genetic elements often encode all the functions required for their replication, stable maintenance and horizontal transfer, in addition to host-beneficial traits such as antibiotic and metal resistance, virulence or catabolism of recalcitrant compounds (Reuter et al., 2014; Conlan et al., 2014; Nojiri et al., 2004; Heuer and Smalla, 2012). However, some plasmids do not carry any identifiable genes that confer a benefit to their host, or they are not under any known positive selection for prolonged periods of time. In theory, even such plasmids can successfully persist as genetic parasites when the following three parameters are balanced: the fitness cost imposed by the plasmid, the segregational loss rate, and the rate of conjugative transfer to plasmid-free cell (Stewart and Levin, 1977; Ponciano et al., 2007; Levin and Stewart, 1980; San Millan et al., 2014). Since plasmids are reservoirs of antibiotic resistance genes, including those that encode resistance to last-resort antibiotics such as colistin (Liu et al., 2015), it is important to understand how temporal selection for plasmid-carrying cells drives plasmid evolution and influences plasmid persistence in the long term.
In the past few decades, experimental evolution studies applied to plasmid-carrying hosts were performed to better understand short-term evolutionary mechanisms of plasmid-host association (Bouma and Lenski, 1988; Modi et al., 1992; De Gelder et al., 2008; Dahlberg and Chao, 2003; San Millan et al., 2014; Harrison et al., 2015). When plasmid-carrying bacteria are grown continuously, mutations that increase their fitness can occur in the host chromosome, plasmid, or both (Bouma and Lenski, 1988; Modi et al., 1991; Sota et al., 2010; De Gelder et al., 2008; Dahlberg and Chao, 2003; San Millan et al., 2014; Harrison et al., 2015; Loftie-Eaton et al., 2016). However, the detailed molecular mechanisms behind plasmid-host adaption were not clarified in most of these studies. This was due to either lack of available chromosomal sequences of the evolved hosts, or because the studies simply did not go beyond determining DNA sequence or transcriptome changes. Recently a few particular chromosomal mutations were shown to reduce the cost of a non-transmissible or self-transmissible plasmid through changes in gene expression (San Millan et al., 2014; Harrison et al., 2015), but the details of the underlying molecular mechanisms remain to be determined. In our recent study a mini-replicon evolved to reduce its loss rate in diverse hosts by acquiring functions that aid in vertical inheritance, such as a multimer-resolution and toxin-antitoxin systems (Loftie-Eaton et al., 2016). In the coevolved host epistatic interactions with chromosomal mutations were required for improved persistence, but the mechanism behind this epistasis has not yet been revealed.
Some knowledge about the molecular mechanism of plasmid-host adaptation has been obtained from in vitro mutagenesis studies. The best example is the adaptation between a Pseudomonas plasmid and E. coli through mutations in either the plasmid replication initiation protein (RepA) gene (Fernández-Tresguerres et al., 1995) or the host DNA replication initiation protein DnaA (Maestro et al., 2003). Variants obtained in each protein by mutagenesis showed enhanced binding affinity. This typical gain of function mutation most likely reduced the segregational loss rate by increasing plasmid copy number.
When conjugative plasmids have been evolved, impaired transferability as a trade-off for decreased fitness cost was observed in some studies (Turner et al., 1998; Dahlberg and Chao, 2003) but not all (Harrison et al., 2015; De Gelder et al., 2008). A transferability-fitness trade-off was also demonstrated in head-to-head competition assays involving fin+ and fin− R1 plasmid derivatives (Haft et al., 2009). Although the number of studies is still limited and the molecular details weak, there seems to be a variety of evolutionary pathways towards improved plasmid-host interactions among different plasmid-host systems, and reduction in plasmid cost is often the prime mechanism for improving plasmid persistence.
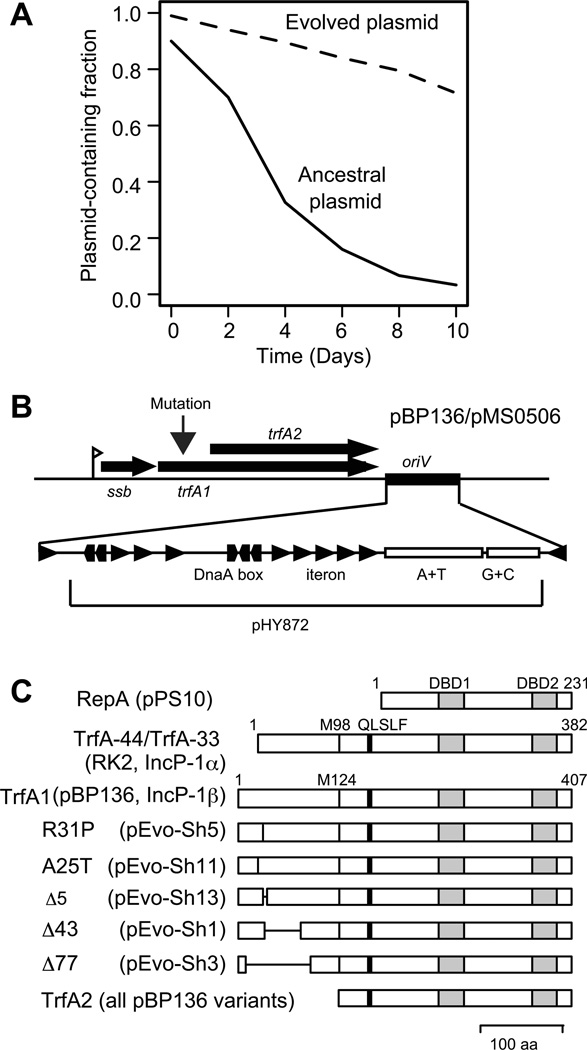
To understand the adaptation mechanisms of broad host range plasmids and their novel hosts, we previously evolved a non-self-transferrable derivative of IncP-1β plasmid pBP136, namely pMS0506, in a Shewanella oneidensis MR-1 host in which the plasmid was poorly maintained (Sota et al., 2010). The mutations that improved plasmid persistence occurred in the 5’-end of the replication initiation protein gene trfA1, which encodes one of the two replication initiation proteins conserved in most IncP-1 plasmids (Fig. 1). The evolved plasmids carrying in-frame mutations within trfA1 showed a ca. 1.5-fold increase in plasmid copy number compared to the ancestral plasmid, whereas the plasmid copy number of frame-shift mutants remained unchanged. In the evolving populations, multiple trfA alleles carrying a frame-shift mutation in the 5’-end arose in the early stages of evolution, but trfA alleles with an in-frame mutation followed and subsequently swept through the populations (Hughes et al., 2012). Therefore, there are at least two ways by which the plasmid was able to adapt to this host: either through a mutation in the TrfA1 N-terminus that resulted in simple loss of TrfA1 function, or through in-frame mutations in trfA1 whose effects remained elusive.
Fig 1. Improvement of plasmid persistence after plasmid adaptation to S. oneidensis.
(A) Representative plasmid persistence curves in the absence of selection for the plasmid. Curves were redrawn based on data from our previous study (Sota et al., 2010). (B) Genes and sites around the trfA locus in wild-type pBP136 and its derived mini-replicon pMS0506, used in this study. Triangles indicate iterons and pentagons indicate DnaA-boxes. (C) Locations of TrfA1 mutations and conserved motif in Rep proteins. DBD1 and DBD2 both indicate iteron-binding motifs, M98 and M124 are start methionines for TrfA-33 and TrfA2, respectively. QLSLF is a motif associated with binding to sliding clamp (Kongsuwan et al., 2006).
During initiation of replication, the initiator proteins of iteron-containing plasmids like those of the IncP-1 group need to interact with host proteins such as DnaA, DnaB, sliding clamp, and protein chaperones (DnaKJ/GrpE, ClpB, ClpX, ClpA) (Lu et al., 1998; Konieczny et al., 1997; Konieczny and Helinski, 1997a; Wickner et al., 1994; Konieczny and Liberek, 2002; Wickner et al., 1992; Zhong et al., 2003; Kongsuwan et al., 2006; Wawrzycka et al., 2015). The mutations that occurred in the trfA1 gene during evolution of pMS0506 in S. oneidensis were concentrated in the region coding for the predicted second helix within the TrfA1 N-terminus region (Hughes et al., 2012). This region corresponds to the N-terminal part of TrfA-44 of RK2 (TrfA1 ortholog), which was shown to interact with the DnaB helicase of Pseudomonas aeruginosa (Jiang et al., 2003; Zhong et al., 2003). Therefore we hypothesize (1) that the initial instability of mini-IncP-1 plasmid pMS0506 can be explained by a negative fitness effect of the TrfA1 protein itself due to the interaction of its N-terminus with DnaB, and (2) that the mechanism of plasmid adaptation to S. oneidensis centers on a decrease in the DnaB-binding function of the TrfA1 N-terminus. Here we address these hypotheses using in vivo and in vitro experiments.
RESULTS
Negative fitness effect of replication protein TrfA1
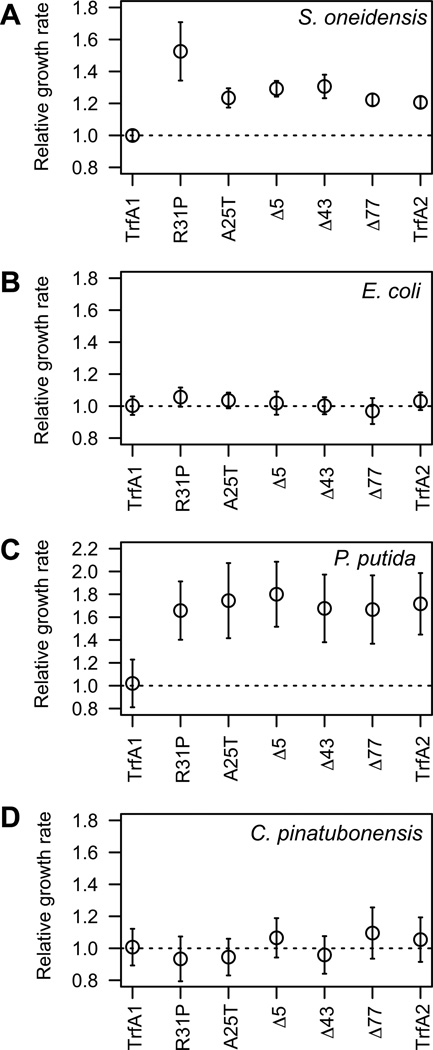
It was previously shown that the ancestral host S. oneidensis MR-1 had a higher exponential growth rate when carrying evolved pMS0506 variants with trfA1 mutations than when containing the ancestral plasmid (Sota et al., 2010). This is likely due to the evolved plasmids imposing a lower fitness cost than ancestral pMS0506. Here, we first tested our hypothesis that the observed mutations in trfA1 affect the fitness cost of the TrfA1 protein itself. To do so we cloned the ancestral trfA1 gene, the evolved in-frame deletion variants (Δ5, Δ43, Δ77), the variants with point mutations (R31P, A25T) or trfA2 (Fig. 1c) behind the constitutive lac promoter in vector pBBR1-MCS2. The growth rates of MR-1 carrying the constructs with evolved genes and trfA2 were compared to that of MR-1 carrying the ancestral TrfA1. The S. oneidensis clone producing TrfA1 showed a lower growth rate than all the clones producing TrfA1 variants or TrfA2 (Fig. 2A). These findings indicate that all five in-frame mutations in the TrfA1 N-terminus ameliorated the fitness cost of the TrfA1 protein itself.
Fig 2. Effect of TrfA1 mutations on maximum growth rates.
Relative maximum growth rates of bacterial cells carrying a cloned trfA gene are shown. A: S. oneidensis; B. E. coli; C, P. putida; D, C. pinatubonensis. Open circles represent means and error bars standard deviations. The M124L mutation was introduced into trfA1 and all its variants. For S. oneidensis and P. putida, the strain carrying wild-type trfA1 had a significantly lower growth rate than the same strain with trfA1 variants (P < 0.001, in multiple comparisons in Dunnett’s test), but this was not the case for the other two hosts.
To determine whether the observed fitness advantage of the in-frame mutations in S. oneidensis was general across species we conducted additional growth rate analyses for the Gammaproteobacteria Pseudomonas putida, E. coli, and the Betaproteobacterium Cupriavidus pinatubonensis containing the pBBR1-MCS2 vector with the ancestral or evolved trfA genes. The evolved TrfA proteins and TrfA2 imposed a lower cost than TrfA1 on P. putida (Fig. 2C), but there were no measurable differences in E. coli (Fig. 2B) and C. pinatubonensis (Fig. 2D). Thus, the negative fitness effect of the TrfA1 N-terminus, and amelioration of its cost through mutations in this region was not unique to strain MR-1. However, the extent of the difference in cost depends on the host.
To exclude that the positive fitness effects of the TrfA1 mutations in pMS0506 were due to the changes in the TrfA1 and TrfA2 protein expression levels, we determined the TrfA levels produced from pMS0506 variants in MR-1 by Western blotting (Fig. S1). In all instances the expression levels were similar or even higher for the TrfA1 variants and TrfA2 than for the ancestral TrfA1. Thus, improved host fitness was not due to the reduction of protein expression levels.
It was recently shown that a plasmid replication protein can induce an SOS response (San Millan et al., 2015). If TrfA1-mediated cellular stress would lead to DNA damage and thereby induce SOS response, cell elongation should be observed (Dwyer et al., 2012; Huisman et al., 1984). To test this we measured the length of cells in exponential phase using phase contrast microscopy (Fig. S2). The mean length of cells producing TrfA1 tended to be longer than that of cells producing evolved TrfA1 variant Δ43 or TrfA2. This was true for strains carrying the actual IncP-1 replicons and strains containing the cloned trfA genes. Furthermore, the coefficient of variation (CV) was generally larger in ancestral TrfA1-producing cells, partly due to the presence of unusually long cells, which were never observed in cells containing evolved TrfA1 or TrfA2 (Fig. S2). This suggests that the nature of the N-terminus of the ancestral TrfA1 may cause DNA damage in a small fraction of the MR-1 population through mechanisms other than plasmid replication.
Effect of TrfA1 N-terminus mutations on the helicase activation activity
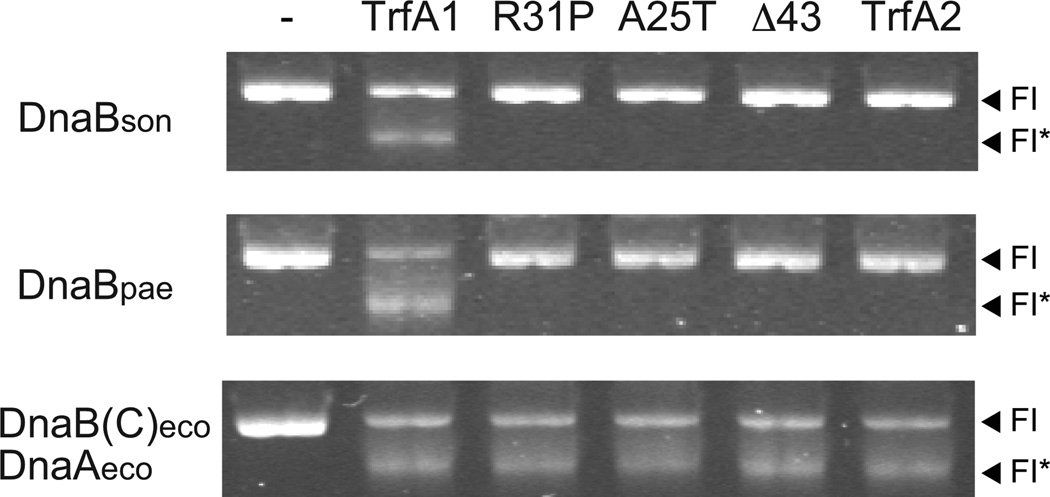
It was previously demonstrated in vitro for some hosts that the TrfA-44 protein of RK2 (an ortholog of TrfA1) can activate the DnaB helicase at the plasmid’s origin of vegetative replication (oriV) without the aid of host DnaA (Jiang et al., 2003), and that the second helix in the N-terminus of TrfA-44RK2 is essential for this process (Zhong et al., 2003). Since all mutations in the TrfA1 protein of pMS0506 were in this N-terminus we hypothesized that the plasmid adapted to S. oneidensis by decreasing the DnaB-binding function of its TrfA1 N-terminus. We first asked whether TrfA1 of pMS0506 also possesses DnaB-loading and activation activity using an FI* assay (also called helicase unwinding assay). When ancestral TrfA1 protein and DnaB of Shewanella oneidensis (DnaBson) were provided, supercoiled substrate DNA containing oriV was unwound (Fig. 3). This indicates that TrfA1 was able to load and activate DnaBson at the oriV without the aid of DnaA (Fig. 3). This TrfA1 activity was similar to that observed on DnaB of Pseudomonas aeruginosa (DnaBpae), whereas for activation of DnaBeco from E. coli, host DnaAeco was needed (Fig. 3). Strikingly, the evolved TrfA1 variants were unable to activate DnaBson and DnaBpae in vitro in the absence of DnaA, thus making replication of the evolved plasmids DnaA dependent.
Fig 3. DnaB-loading and activation activity of TrfA1 and its variants.
FI indicates the position of supercoiled substrate, pMS0506, containing oriV. FI* indicates the position of unwound substrate, and is indicative of DnaB-loading and activation activity. The 1st lane (left) indicates a control reaction without TrfA protein. To make constitutively monomeric TrfA, Q279D/S292L mutations were introduced into all TrfA proteins. A M124L mutation was also introduced into all TrfA1 variants to avoid production of TrfA2.
Effect of TrfA1 N-terminus mutations on DnaB binding
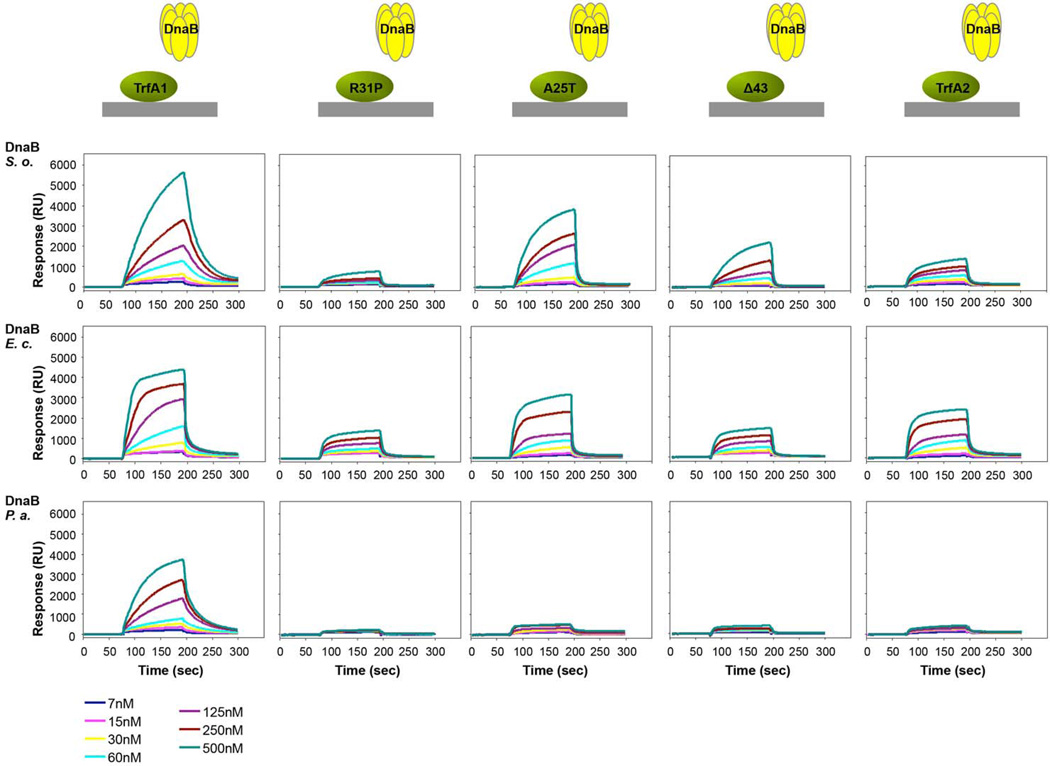
As our results suggested that the mutations cause a change in the binding affinity between TrfA1 and DnaB, we further tested this by conducting surface plasmon resonance (SPR) analysis. TrfA1 or its variants were immobilized to the sensor chip and DnaBson was provided at different concentrations (Fig. 4). TrfA1 bound DnaBson more efficiently than the TrfA1 variants or TrfA2. The reduced binding affinity between the TrfA1 variants and DnaBson was also confirmed by ELISA tests (Fig. S4). All these results together indicate that mutations in the replication initiation protein impaired interaction between TrfA1 N-terminus and the host-encoded DnaB. Our results are thus consistent with the findings from Zhong et al. (Zhong et al., 2003) that the second helix of TrfA-44 RK2 was essential for the in vitro activation of DnaB at the oriV in the absence of DnaA, and our hypothesis that decreasing this function would improve plasmid persistence in S. oneidensis was supported.
Fig 4. Analysis of DnaB helicases interactions with TrfA proteins.
Interactions of DnaB helicase from S. oneidensis, E. coli or P. putida with monomeric forms of TrfA1, R31P, A25T, Δ43 and TrfA2 proteins were analyzed with Surface Plasmon Resonance. TrfA proteins were immobilized on the surface of CM5 sensorchip. Increasing amounts of DnaB proteins (7, 15, 30, 60, 125, 250, and 500 nM) were flown over the sensor chip. TrfA proteins contain the M124L/Q279D/S292L mutations.
We previously found that the evolved pMS0506 variants cannot establish replication in P. aeruginosa (Sota et al., 2010). In the case of RK2 replication, the DnaBpae-TrfA-44 N-terminus interaction was shown to be required for plasmid replication in P. aeruginosa (Jiang et al., 2003). These previous observations suggest that TrfA1 variants cannot sufficiently interact with DnaBpae. Here, the ancestral TrfA1 was indeed able to bind DnaBpae, but TrfA1 variants barely did (Fig. 4). This result is consistent with the analysis of DnaB-loading activity and evolved plasmid phenotypes, in that such weak binding explains the inability of the evolved plasmids to load DnaB and thus replicate in P. aeruginosa.
It should be noted that the TrfA1 variants and TrfA2 were still able to bind DnaBson from S. oneidensis, and E. coli to some extent (Fig. 4), and were still able to replicate in these hosts (Sota et al., 2010). These results suggest that the TrfA2 domain itself interact with DnaBeco and DnaBson, but little with DnaBpae.
DnaB overproduction improves plasmid persistence
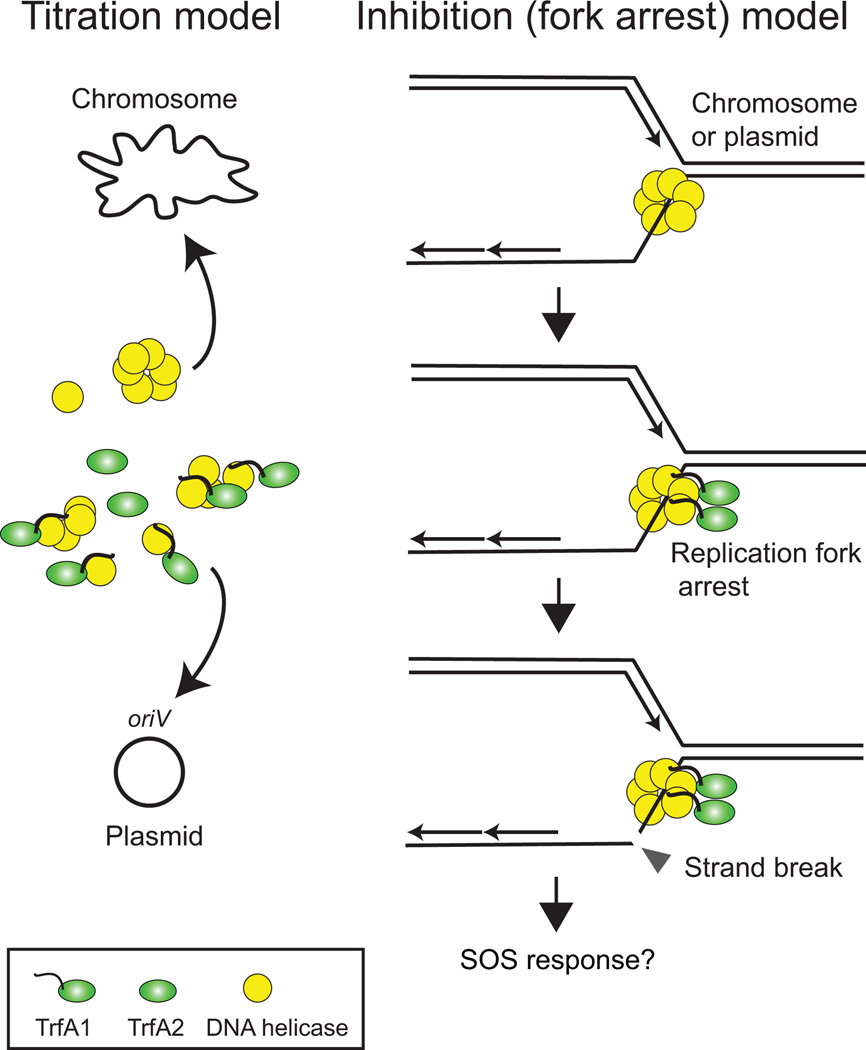
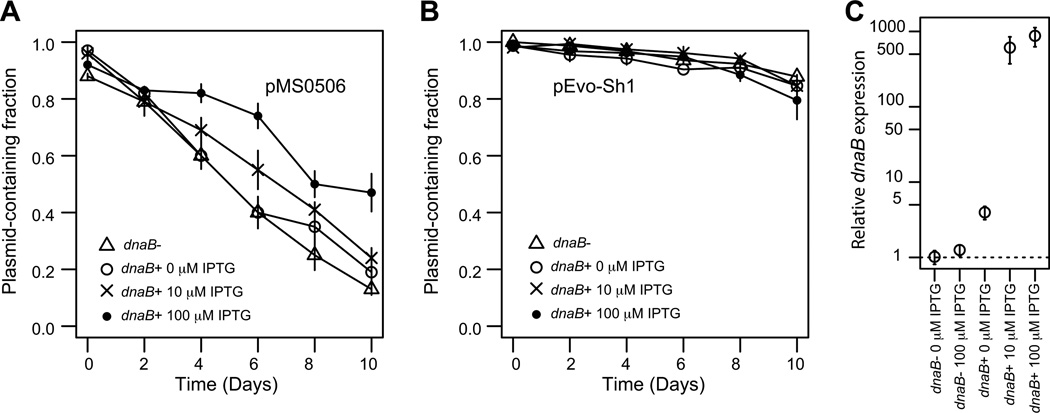
Based on the results above we postulate that the improved persistence of the evolved pMS0506 variants was due to amelioration of the plasmid fitness cost caused by relaxing the binding of TrfA1 to DnaBson. The fitness cost of the ancestral plasmid could either arise from reducing the concentration of free DnaB in the cell (titration model), or from inhibiting essential cellular functions, such as chromosomal DNA replication by formation of a TrfA1-DnaB complex (inhibition model) (Fig. 5). If the titration model is true, overproduction of DnaB should reduce the negative fitness effect of the ancestral TrfA1, and thus improve persistence of the ancestral plasmid in S. oneidensis MR-1. If the latter model is more likely, overproduction of DnaB may increase the amount of TrfA-DnaB complex and thereby increase plasmid cost and negatively affect plasmid persistence. These models are not necessarily mutually exclusive. We determined the effect of DnaB overproduction on the persistence of ancestral plasmid pMS0506 and evolved plasmid pEvo-Sh1 by comparing two S. oneidensis MR-1 strains wherein we chromosomally inserted a mini-Tn7 transposon either with or without an extra copy of dnaB inducible by IPTG (dnaB+ vs dnaB−). Different concentrations of IPTG were used (0 µM, 10 µM, 100 µM; average curves are shown in Fig. 6; individual curves in Fig. S4 and Fig. S5). When IPTG was added at 10 µM and 100 µM, the ancestral plasmid showed significantly different persistence dynamics in the dnaB+ strain than in the dnaB− strain (ΔBIC was −24.8 for 10 µM, and −119.9 for 100 µM; statistical analysis results are summarized in Table S3). In a set of follow-up experiments we even observed a significant difference between the dnaB+ strain and the dnaB− strain at 0 µM IPTG (ΔBIC: −240.8; Fig. S5). For the three paired assays the quasi extinction time of the ancestral plasmid was higher in the dnaB+ strain than when no additional DnaB was expressed, (Table S4), indicating that DnaB overproduction improves plasmid persistence.
Fig.5. Two models for the possible cause of plasmid interference cost associated with the replication initiation protein.
Left: Titration model. TrfA1, which shows high affinity to DnaB DNA helicase, reduces the amount of free DnaB available for chromosomal replication and repair. Right: Inhibition model. The TrfA1-DnaB complex occurring at the replication fork arrests replication or repair, leading to exposure of single strand DNA followed by strand break. This may occur on both the plasmid and chromosome.
Fig 6. Effect of DnaB overproduction on plasmid persistence in S. oneidensis.
The data points indicate means and standard deviations from triplicate assays. (A) Persistence of pMS0506. In this data set, the dnaB+ strain with 10 µM IPTG, and 100 µM IPTG showed improved persistence compared to the dnaB− strain (See ΔBIC test in Table S3, and quasi-extinction time in Table S4). In a second set of experiments for 0 µM IPTG condition, a significant difference was observed between dnaB+ and dnaB− strains (Fig. S5). (B) Persistence of pEvo-Sh1. (C) Relative dnaB mRNA levels determined by qPCR; the mean value for the dnaB− strain, 0 µM IPTG, was represented as 1 in this plot. Error bars indicate SD. The host used were S. oneidensis HY1014 (chr::mini-Tn7-tacp-dnaB; dnaB+), and S. oneidensis HY0759 (chr::mini-Tn7; dnaB−). For the dnaB− strain, only IPTC 100 µM conditions were shown for simplicity.
To assess which underlying parameters differentiated the persistence dynamics of the evolved and ancestral plasmid, we estimated the fitness cost, loss rate, and initial plasmid-free cell fraction by model fitting. Although not significant due to too much variation, the maximum likelihood estimates (MLE) of the fitness cost of pMS0506 were lower when dnaB was induced. This is consistent with improved persistence and supports our titration model, (Table S4). Moreover, in the dnaB+ strain the inferred fitness cost of pMS0506 (0.043) was not different from that of pEvo-Sh1 (0.043) at 100 µM IPTG (Table. S4). However, pEvo-Sh1 was still more stable than pMS0506 even at the highest DnaB induction levels (Fig. 6A, 6B and Fig S6). It should be noted that in the presence of 100 uM IPTG the dnaB mRNA levels were >800-fold higher in the dnaB+ strain according to qPCR (Fig. 6C). We were unable to draw conclusions on comparisons of the plasmid loss rates because the spread on the parameter estimates was too large (Table S4). Finally, the MLE of the initial plasmid-free cell fraction was significantly higher for pMS0506 (0.073) than pEvo-Sh1 (0.012) at 100 µM IPTG, even though the plasmid-containing cells were grown in the presence of antibiotics to prevent plasmid loss before time point 0 (Fig. S6). Together these results suggest that the titration of DnaB by TrfA1 and the resulting interference cost partially but not entirely explains the poor persistence of pMS0506 in host MR-1.
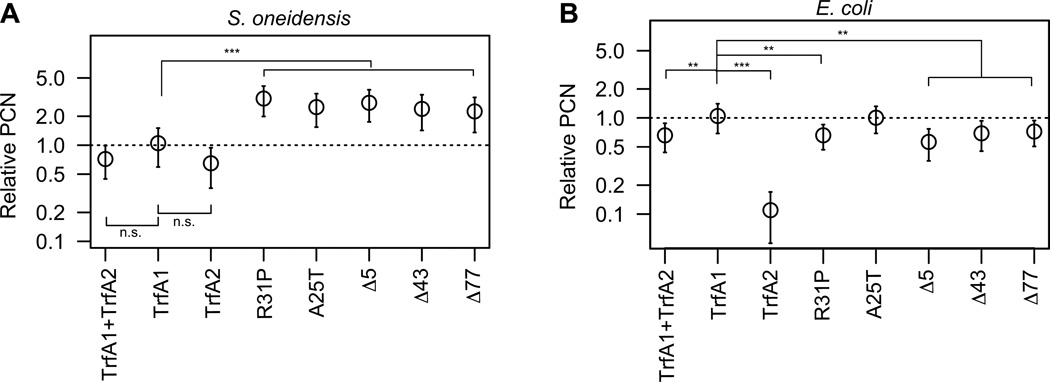
Effect of TrfA1 mutations on plasmid copy number is host specific
In S. oneidensis MR-1, a higher plasmid copy number has been observed for the evolved pMS0506 variants with in-frame trfA1 mutations compared to ancestral pMS0506, yet there was no difference for the evolved plasmids with frame-shift mutations in trfA1 (thus expressing only TrfA2) (Sota et al., 2010). Such an increase in plasmid copy number can be caused either by TrfA-host factor interactions or by TrfA-plasmid factor interactions (interaction between TrfA molecules or between TrfA and oriV). If the first mechanism were true, the increase in copy number could be specific to host S. oneidensis MR-1. To test this, we analyzed the effect of the in-frame trfA1 mutations on plasmid copy number in both S. oneidensis and E. coli. A copy number reporter plasmid carrying oriV was supplied to the strains, which carried either an ancestral or evolved trfA1 gene in the chromosome (see ‘plasmid and strain construction method’ section in the supporting information for details). The copy number of the reporter plasmid relative to the chromosome was then determined by qPCR. In S. oneidensis MR-1 the copy number was significantly higher for the evolved TrfA1 variants than for either of the three other strains, expressing either TrfA1, TrfA2, or both (the latter three being not significantly different from each other) (Fig. 7A). In contrast, in E. coli the plasmid copy number was highest for the ancestral TrfA1, and the evolved TrfA1 variants generated a similar or slightly lower copy number (Fig. 7B). This suggests that the increase in plasmid copy number due to trfA1 in-frame mutations was specific to host MR-1 and that the plasmid copy number shift is thus likely due to the TrfA1-host interactions. These results suggest that the ability of TrfA1 to generate a higher plasmid copy number than TrfA2 was suppressed in S. oneidensis MR-1, possibly due to tight binding to DnaB, and that the in-frame mutations counteracted this. In summary, antibiotic selection for improved plasmid persistence favored a decrease in the ability of TrfA1 N-terminus to bind DnaB, thereby decreasing the interference cost imposed by TrfA1 and increasing plasmid copy number for the trfA variants with in-frame mutations.
Fig 7. Effect of TrfA1 mutations on plasmid copy number.
(A) Relative plasmid copy number (PCN) in S. oneidensis. The copy number of pHY872 (Fig.1) for the TrfA1 condition is represented as 1. The data are shown on a logarithmic scale. (B) Relative PCN in E. coli. TrfA proteins were expressed from the chromosome (see materials and methods for details) ‘TrfA1+TrfA2’ indicates expression of two proteins from the wild-type trfA1 gene. The M124L mutation was introduced into TrfA1 and TrfA1 variants. In (B), the data for TrfA1+TrfA2, TrfA1, and TrfA2 were previously published (Yano et al., 2012). The trfA-tagged strains used were HY0321, HY0323, HY0325, HY0329, HY0333, HY0337, HY0341, HY0345 for panel (A), and HY0414, HY0390, HY0391, HY0392, HY0395, HY0408, HY0409, HY0475 for panel (B). Comparisons were made using the post-hoc Tukey test for the data before normalization to relative PCN (**, P <0.01; ***, P <0.001; n.s., not significant). Data are obtained from at least four independently grown cultures for each clone.
DISCUSSION
Experimental evolution of plasmid-host interactions
When a plasmid can replicate in a bacterial host, its persistence in the population is determined by various plasmid-encoded functions such as copy number control, partitioning, multimer-resolution, post-segregational-killing, and conjugation. These are expected to improve plasmid persistence by reducing segregational loss rate or increase the frequency of plasmid uptake after plasmid loss. Moreover, any of the plasmid-encoded genes may affect the fitness cost of plasmid carriage, which in turn influences plasmid persistence in the presence of plasmid-free cells. Selection for faster growing plasmid-carrying cells has been shown to drive the evolution of plasmid-encoded functions by either gain of function mutations (Maestro et al., 2002; Maestro et al., 2003; De Gelder et al 2008), acquisition of functions such as toxin-antitoxin and plasmid multimer-resolution systems (Loftie-Eaton et al., 2016), or loss of function mutations (Dahlberg and Chao, 2003). Recently putative loss of function mutations in chromosomal genes was also shown to reduce the fitness cost of plasmids (San Millan et al., 2014; Harrison et al., 2015). In long-term experimental evolution studies with E. coli (without plasmids), fitness gain was accomplished not only by simple gain or loss of function of a gene, but also by fine-tuning of protein expression levels (Philippe et al., 2009; Woods et al., 2006; Herring et al., 2006). However, the molecular mechanisms that explain the beneficial mutations observed during plasmid-host coevolution are still poorly understood. In this study, we examined how protein interactions changed during long-term host-plasmid interactions. We found that mutations in the plasmid replication initiation protein TrfA1 improved plasmid persistence by reducing the protein’s ability to bind with the host DNA helicase, DnaB. We propose several mechanistic models for how this decreased binding could affect the persistence of the plasmid in the absence of antibiotics.
In the course of adaptation of pMS0506 to the host S. oneidensis, both frame-shift mutations and in-frame mutations occurred in the 5’-end of trfA1 within a plasmid population (Hughes et al., 2012). Frame-shift mutations were the majority in the initial stage of evolution, but in-frame mutations eventually swept through the populations. This implies that the characteristics of in-frame and frame-shift mutants were distinct. While in-frame mutants showed slightly higher plasmid copy number than frame-shift mutants, we were not able to detect significant differences in fitness effects between them on the basis of growth rate analysis (Sota et al., 2010). In this study, we found that the in-frame mutations improved the fitness of the host that carried only the trfA1 locus cloned into a plasmid vector (Fig. 2), suggesting that the cost of TrfA1 itself can explain the poor persistence of pMS0506 (Fig. 6), and that a decrease in that cost was the primary target of selection. This is consistent with a recent experimental evolution study that focused on the interaction of a non-transferrable plasmid with its Pseudomonas aeruginosa host (San Millan et al., 2015). The fitness cost of plasmid carriage was due to enhanced expression of the replication initiation protein, which was in turn caused by the presence of a chromosomally encoded UvrD family DNA helicase, as the only mutation in one of the evolved clones was a nonsense mutation in this helicase gene (San Millan et al., 2015). In our model system the plasmid mutations in trfA1 alone could explain the improved plasmid cost and persistence (Sota et al., 2010).
Origin of fitness cost of replication protein
We demonstrated that the reduced cost of evolved TrfA1 variants was concomitant with reduced binding affinity to the helicase DnaBson (Fig. 4). This resulted in loss of DnaA-independent DnaB-activation activity in vitro, and most likely explains previously observed loss of replication activity of the evolved pMS0506 variants in P. aeruginosa (Sota et al., 2010). This suggests that the tight binding of the ancestral replication initiator TrfA1 to DnaBson is the cause of the high plasmid cost imposed by TrfA1 on S. oneidensis, leading to poor plasmid persistence. We propose two models that can explain this cost of tight binding. One is a simple titration model in which TrfA1 titrated free DnaB available for chromosome replication. The other is the replication fork arrest, or inhibition model, where the plasmid replication protein blocks progression of DNA helicase on chromosome and plasmids (Fig. 5). This may result in generating single strand DNA that is prone to DNA cleavage and induces SOS response. Interestingly, San Millan et al. (San Millan et al., 2015) observed induction of an SOS response in the presence of a chromosome encoded UvrD helicase and a plasmid replication protein. During SOS response, the induced protein SulA inhibits cell division, resulting in elongated cells (Huisman et al., 1984). We found that the fraction of elongated cells was larger in the S. oneidensis population that expressed the ancestral TrfA1 than in populations with TrfA2 or evolved TrfA1 variants (Fig. S2). While the means of cell lengths were not always significantly different, the spread in cell length was significantly higher in TrfA1 expressing cells. Thus, SOS response due to strand break after replication fork arrest may be induced in a subset of TrfA1-producing cell populations.
Overexpression of DnaB improved persistence of pMS0506. However, the persistence was not improved to the level of the evolved plasmid tested (Fig. 6), implying that not only the amount of free DnaB but also the TrfA1-DnaB complex negatively affected plasmid persistence. Even when DnaB was overproduced in vivo at the highest level, only the initial plasmid-free cell fraction was significantly different between ancestral and evolved plasmid, and not the plasmid cost (Table S4). According to our simulation, the difference in the estimated cost alone cannot account for the 6% of difference in initial plasmid free fraction between pMS0506 and pEvo-Sh1 at time point 0. Thus we speculate that a TrfA1-DnaB complex had a negative effect on plasmid segregation, and not just on fitness cost. Occurrence of partially replicated plasmid molecules caused by replication fork arrest would lead to aberrant plasmid segregation in the presence of a partitioning system, and lead to lower than normal plasmid copy number. Consistent with this the copy number in S. oneidensis MR-1 of pMS0506 and its minimal replicon was lower than their respective evolved variants (Fig. 7, see the discussion below). Therefore, although the effect on fitness is unclear, the speculated events in the fork arrest model may occur in the host carrying the ancestral plasmid.
The in-frame mutations in trfA1 are partial loss of function mutations
The change in plasmid characteristics other than its reduced fitness cost (Fig. 2) was a slightly elevated plasmid copy number in S. oneidensis MR-1 (Fig. 6). We previously found that in E. coli, P. putida, C. pinatubonensis, and Sphingobium japonicum pMS0506, which encodes both TrfA1 and TrfA2, can generate higher plasmid copy numbers than its trfA1 frame-shift mutants that no longer produce TrfA (Yano et al., 2012). However this was not the case in S. oneidensis (Sota et al., 2010). Moreover, the positive effect of the trfA in-frame mutations on plasmid copy number was observed only in host S. oneidensis MR-1 (Fig. 6). It is thus reasonable to think that the typical characteristic of TrfA1 to elevate plasmid copy number is suppressed in S. oneidensis MR-1, possibly due to the tight binding between the TrfA1-N terminus and DnaB. It therefore follows that the in-frame mutations in trfA1 are essentially partial-loss of function mutations that were selected to simultaneously ameliorate the cost of TrfA1 and slightly increase the plasmid copy number. The TrbC point mutation we previously observed in adaption of IncP-1 plasmid pB10 to Stenotrophomonas maltophilia had a similar dual function, as it simultaneously reduced the cost and increased the transferability of the plasmid (De Gelder et al., 2008). Is currently difficult to explain why the DnaB-TrfA1 interactions have a negative effect on host fitness and plasmid copy number in S. oneidensis MR-1 and not in other hosts tested. No conclusions can be drawn so far based on the DnaB sequence alone, because of the high number of amino-acid substitutions (see alignment in Fig. S7).
Significance of TrfA1
The question remains why broad host range plasmids possess such a costly TrfA1 protein. According to our growth assay (Fig. 2), not encoding TrfA1 may be beneficial for plasmids to persist in P. putida. This was supported by experimental data that used the self-transmissible plasmid pBP136Km, from which pMS0506 was derived (Yano et al., 2012) (Fig. S3). IncP-1 plasmids are unique among iteron-containing plasmids in that most of them encode two forms of replication initiation protein, namely TrfA1 (TrfA-44 in RK2) and TrfA2 (TrfA-33 in RK2). The TrfA2 region is equivalent to Rep proteins from other iteron-containing plasmids such as R6K, P1, F, and pPS10, while TrfA1 has an extended N-terminus in addition to the TrfA2 domain (Zhong et al., 2003). It has been shown that the TrfA1 N-terminus has two distinct intrinsic functions: DnaA-independent DnaB activation (Zhong et al., 2003), and increasing plasmid copy number (Yano et al., 2012). The former function allows replication of RK2 in a Pseudomonas aeruginosa strain where DnaA-TrfA-33 interaction is not functional (Caspi et al., 2001; Jiang et al., 2003). This trend seems to be the same in IncP-1β plasmid pBP136 (Fig. 3). The plasmid copy number function on the other hand helps plasmid establishment in recipient cells, thereby contributing to plasmid transmission in bacterial communities where inter-species transfer is common (Klumper et al., 2014). Both TrfA1 functions are important to support promiscuity of IncP-1 plasmids, but the instability of pMS0506 and plasmid copy number suppression in S. oneidensis suggests that the former function can interfere with the latter. DnaA-independent DnaB activation was lost during specialization to S. oneidensis and the plasmid replication became dependent on host DnaA (Fig. 3). Thus, the role of the DnaA-independent DnaB-activation function of TrfA1/TrfA-44 N-terminus in the host range expansion needs to be reevaluated, because P. aeruginosa is by far the only bacterial species where IncP-1 plasmids can benefit from that function. Furthermore our results may explain why most iteron-containing plasmids use DnaA for helicase recruitment.
Concluding remarks
Several experimental evolution studies have shown that plasmids can adapt to their hosts (Sota et al., 2010; De Gelder et al., 2008), hosts adapt to their plasmids (San Millan et al., 2015; Harrison et al., 2015), and plasmids and hosts can coevolve (Loftie-Eaton et al., 2016). Beneficial mutations that occurred in S. oneidensis (pMS0506) during experimental evolution were diverse (Hughes et al., 2012), yet here we showed that the genotypes that swept through the populations were likely selected based on more than one functional improvement, i.e. decreasing interference cost and increasing plasmid copy number (Fig. 2, and Fig. 7). The selection was host-specific, thus pointing to a mechanism for plasmid-host specialization. There have been three plasmid-host experimental evolution studies now wherein the host’s DNA helicases and host-plasmid interactions were highlighted as a cause of high plasmid cost (San Millan et al., 2015; Sota et al., 2010; Loftie-Eaton et al., 2016). Future studies on the molecular mechanisms behind these interactions are needed to provide a clear picture of the factors that can limit plasmid persistence. Understanding these factors and possibly determining common mechanisms across bacterial species and plasmids is critical in our attempts to limit the rapid spread of antibiotic resistance among bacterial pathogens.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
The bacterial strains used were as follows: E. coli EC100 (Epicentre, Madison, WI, USA); BL 21(DE3) (EMD Millipore, Billerica, Massachusetts, USA); BW25113 (Baba et al., 2006); Shewanella oneidensis MR-1 (ATCC700550); Pseudomonas putida KT2440 (Bagdasarian et al., 1981); Cupriavidus pinatubonensis JMP228 (Amy et al., 1985). Strains EC100 and BL21 (DE3) were used as DNA cloning host and protein expression host for affinity protein purification, respectively. Strains BW25113, MR-1, KT2440, and JMP228 were used to assess fitness effects of TrfA proteins or plasmid persistence. We constructed MR-1 derivatives carrying trfA or an additional copy of dnaB in the chromosome using the method described in the Supporting Information. A complete list of strains and plasmids used in this study is shown in Table S1.
E. coli, S. oneidensis, and P. putida strains were grown in Luria-Bertani (LB) medium. C. pinatubonensis JMP228 was grown in 1/10-TSB medium (one-tenth dilution of Bacto tryptic soy broth, [BD Bioscience, Sparks, MD, USA]). All strains were incubated at 30°C except for DNA-cloning experiments.
Plasmid pMS0506 is a non-transferable deletion derivative of pBP136, a plasmid originally discovered in Bordetella pertussis (Kamachi et al., 2006). The evolved pMS0506 variants, pEvo-Sh1, pEvo-Sh3, pEvo-Sh5, pEVo-Sh11, pEvo-Sh13, pEvo-Sh14, pEvo-Sh15 were obtained after growing strain S. oneidensis MR-1 carrying pMS0506 for 1000 generations in the presence of antibiotic selection for the plasmid (Sota et al., 2010). All evolved plasmids encode the shorter replication initiation protein TrfA2, and pEvo-Sh1, pEvo-Sh3, pEvo-Sh13, pEvo-Sh11 and pEvo-Sh5 additionally encode TrfA1 variant Δ43, Δ77, Δ5, A25T, and R31P, respectively (Fig.1). To address the fitness effects of the expressed wild-type trfA gene and its variants, each gene was cloned into pBBR1-MCS2 (Kovach et al., 1995). The pBBR1-MCS2 derivatives were designated, pHY987 (wild-type TrfA1), pHY988 (TrfA2), pHY1010 (R31P), pHY1011 (A25T), pHY1012 (Δ43), pHY1014 (Δ5), pHY1015 (Δ77).
Growth rate analysis
The strains carrying pMS0506 derivatives or pBBR1-MCS2 derivatives were initially streaked on LBA with kanamycin. Single colonies (n=8) were inoculated into 5 ml LB containing Km, and then incubated for 24 hours. The cultures were diluted 100-fold in fresh medium, and 100 ul was transferred to each well of a 96-well microtiter plate. The plate was incubated for 24 hours with agitation at 30°C in BIO-TEK Power Wave HT (BioTek instruments Inc., Winooski, VT, USA). OD600nm was recorded every 10 min. The maximum growth rate of each strain was determined using smoothing function of the Grofit package of R (Kahm et al., 2010). Maximum growth rate (µ) was used for multiple comparisons (ANOVA followed by Dunnett’s test) among samples.
Plasmid persistence
The strains carrying pMS0506 or pEvo-Sh1 were first grown in LB with antibiotics for 24 hours, and the culture was subsequently diluted 1024-fold in fresh medium without antibiotics and incubated for 24 hours. This serial batch culture transfer was repeated 10 times. Under this protocol, each 24-h period resulted in 10 generations. Each day the cultures were diluted and spread on non-selective plates. The next day, 52 randomly selected colonies were replicated on selective and non-selective LB agar to determine the number of colonies retaining a plasmid. Where necessary the data were fit to a mathematical segregation and selection model to estimate the initial plasmid-free cell fraction (0), fitness cost (σ) and segregational loss rate (λ) of the plasmids as described by (Ponciano et al., 2007) using a custom software package written for R (Loftie-Eaton et al., 2016). The β0, σ and λ parameters were then used to estimate the time in days till only one 1% (T1%) of the bacteria retained the plasmid. In addition the statistical significance of the difference between two persistence dynamics was evaluated using Bayesian Information Criterion (BIC) as described by us recently (Loftie-Eaton et al., 2016). When comparing two persistence dynamics, negative ΔBIC values (smaller than −2) suggest significant differences, with more negative values pointing to larger differences.
Phase contrast microscopy
Cells from exponential phase cultures (OD600nm = 0.4–0.6 in 1 cm gap cuvette) were suspended in PBS (pH 7.0), then prepared as wet mounts on glass slides. The cells were observed using a Nikon/Andor Spinning Disk confocal microscope with 100× magnification. Images were captured and analyzed using Andor Zyla sCMOS camera and NIS-elements acquisition software, respectively. Cell debris and chained cells were manually removed from the analysis.
Proteins
N-terminally histidine-tagged TrfA proteins were expressed from pET11a derivatives, pHY915 (TrfA1), pHY916 (R31P), pHY917 (A25T), pHY919 (Δ43), and pHY921 (TrfA2). In these proteins, the M124L mutation was introduced to eliminate co-expression of TrfA2 (for details, see supplemental materials). It was previously shown that two point mutations (G254D/S267L) make TrfA-33RK2 monomeric and constitutively active in iteron-binding (Blasina et al., 1996). The equivalent mutations (Q279D/S292L) were also introduced into TrfA1 and its variants. Thus, purified TrfA1 and its variant commonly contained the mutations M124L/Q279D/S292L, and TrfA2 contained Q279D/S292L. C-terminally histidine-tagged S. oneidensis DnaB (DnaBson) was expressed from pET22b derivative pHY1032. These proteins and E. coli DnaA, DnaB, DnaC, and P. aeruginosa DnaB (referred to as DnaBeco, DnaCeco, DnaBpae respectively) were overexpressed in E. coli BL21(DE3) and purified using affinity chromatography as described previously (Blasina et al., 1996; Caspi et al., 2000; Caspi et al., 2001).
Western blotting
To quantitate TrfA1 production levels in S. oneidensis MR-1, we conducted Western blotting for whole cell lysates made from strain MR-1 carrying pMS0506 or its derivatives, using purified TrfA1, TrfA2, Polyclonal anti-TrfA2 antibodies from rabbit, IRDye 800CW goat anti-rabbit IgG (Rockland, Gilbertsville, PA). Densitometry analysis was performed with an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE), as described in detail in our previous work (Yano et al., 2012).
Quantification of dnaB transcripts by RT-qPCR
To estimate dnaB transcription levels, we performed RT-qPCR on total RNA purified from three replicate stationary cultures of strain HY759 and HY1041, using the PureLink RNA Mini Kit and DNA-free Kit (Life Technologies, Carlsbad, CA, USA). Reaction mixtures were made using the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (ThermoScientific, Waltham, MA, USA), and qPCR was conducted using a one-step RT-qPCR protocol on the ABI 7900HT thermal cycler (Life Technologies). The mRNA copy number was estimated for dnaB and gap (Glyceraldehyde-3-phosphate dehydrogenase gene) based on standard curves. The dnaB to gap ratio was determined for each total RNA, and compared between experimental conditions.
Helicase unwinding assay
The helicase unwinding assay [Form I* (FI*) formation assay] was performed as described by (Konieczny and Helinski, 1997b). Reactions contained supercoiled plasmid pMS0506 DNA (1.4 nM), appropriate His-6-TrfA proteins (TrfA1, TrfA1 R31P, TrfA1 A25T, TrfA1 Δ43 or TrfA2) (180 nM), HU (110 nM), SSB (115 nM), DNA gyrase (13 nM) and DnaB helicases (200 nM) from E. coli, S. oneidensis or P. aeruginosa, as noted. Additionally, reactions with E. coli DnaB protein also contained DnaA (7.5 nM) and DnaC (1.1 µM) proteins. The samples were electrophoresed at 25 V for 22 h, and the gel was stained with ethidium bromide.
Surface plasmon resonance analysis
Standard SPR analyses using a BIAcore 2000 were performed essentially as described in the manufacturer’s manual. Binding of TrfA proteins by DnaB helicases from E. coli, S. oneidensis or P. aeruginosa was studied using a CM5 Sensor Chip. TrfA proteins (TrfA1, TrfA1 R31P, TrfA1 A25T, TrfA1 Δ43 and TrfA2) were immobilized on a Sensor Chip surface. Increasing amounts of DnaB proteins (7, 15, 30, 60, 125, 250, and 500 nM) were flown over the Sensor Chip surface in running buffer HBS-EP (150 mM NaCl, 10 mM HEPES pH 7.4, 3 mM EDTA, 0.005% Surfactant P20). In all experiments the buffer flow was set to 15 µl/min with all injections at a volume of 30 µl. The results are presented as sensorgrams obtained after subtraction of the background response signal from control experiments with buffer injections.
Plasmid copy number analysis
E. coli strains and S. oneidensis strains were tagged with mini-Tn7 carrying the tac promoter, trfA, and lacIq (see Supporting Information). Then, the constructed strains were transformed with pHY872 that carries oriVpBP136, but not trfA (Fig 1B). The strains harboring pHY872 were grown up to stationary phase. Total DNA was extracted and linearized by EcoRI digestion, ethanol precipitated, and diluted to 5 to 10 ng/ul. This DNA was used as template for qPCR, which was performed using an ABI 7900HT thermal cycler, Fast SYBR Green Master Mix (Life technologies), primer sets, tetAF and tetAR for pHY872, Ecoli_atpF and Ecoli_atpR for E. coliatpB in the oriC region, atpF and atpR for MR-1 atpB (Table S2). Vectors pHY873 or pHY924 digests were used as copy number control. The tetA to atpB ratio was used to represent plasmid copy number.
Supplementary Material
Acknowledgments
This work was funded by NIH R01 grant AI084918 from the National Institute of Allergy and Infectious Diseases (NIAID), with additional support from COBRE NIH grants P20RR16448 and P20GM103397 and the Idaho INBRE Program, NIH grants P20RR016454 and P20GM103408, through the IBEST Genomics and Computational Resources Cores, and from Japan Science and Technology (JST) - Exploratory Research for Advanced Technology (ERATO) Nomura Microbial Community Control Project. We thank the National BioResource Project at National Institute of Genetics Japan for E. coli strain BW25113, and Dr. Jose Ponciano at the University of Florida for advice on applying and interpreting his plasmid population dynamics model.
Footnotes
None of the authors have a conflict of interest.
Author contributions: EMT, IK, HY designed the research; HY, KW, WLE, JJ, LMR, GED conducted experiments; HY, WLE analyzed data; HY, EMT wrote the manuscript.
REFERENCES
- Amy PS, Schulke JW, Frazier LM, Seidler RJ. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1985;49:1237–1245. doi: 10.1128/aem.49.5.1237-1245.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in. Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Blasina A, Kittell BL, Toukdarian AE, Helinski DR. Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc Natl Acad Sci U S A. 1996;93:3559–3564. doi: 10.1073/pnas.93.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- Caspi R, Helinski DR, Pacek M, Konieczny I. Interactions of DnaA proteins from distantly related bacteria with the replication origin of the broad host range plasmid RK2. J Biol Chem. 2000;275:18454–18461. doi: 10.1074/jbc.M000552200. [DOI] [PubMed] [Google Scholar]
- Caspi R, Pacek M, Consiglieri G, Helinski DR, Toukdarian A, Konieczny I. A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J. 2001;20:3262–3271. doi: 10.1093/emboj/20.12.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med. 2014;6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg C, Chao L. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics. 2003;165:1641–1649. doi: 10.1093/genetics/165.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder L, Williams JJ, Ponciano JM, Sota M, Top EM. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics. 2008;178:2179–2190. doi: 10.1534/genetics.107.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012;46:561–572. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Tresguerres ME, Martín M, García de Viedma D, Giraldo R, Díaz-Orejas R. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J Bacteriol. 1995;177:4377–4384. doi: 10.1128/jb.177.15.4377-4384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Haft RJ, Mittler JE, Traxler B. Competition favours reduced cost of plasmids to host bacteria. ISME J. 2009;3:761–769. doi: 10.1038/ismej.2009.22. [DOI] [PubMed] [Google Scholar]
- Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr Biol. 2015;25:2034–2039. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BO. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- Heuer H, Smalla K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev. 2012;36:1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- Hughes JM, Lohman BK, Deckert GE, Nichols EP, Settles M, Abdo Z, Top EM. The role of clonal interference in the evolutionary dynamics of plasmid-host adaptation. MBio. 2012;3:e00077-12. doi: 10.1128/mBio.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O, D’Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Pacek M, Helinski DR, Konieczny I, Toukdarian A. A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin. Proc Natl Acad Sci U S A. 2003;100:8692–8697. doi: 10.1073/pnas.1532393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahm M, Hasenbrink G, Lichtenberg-Fraté H, Ludwig J, Kschischo M. grofit: fitting biological growth curves with R. Journal of Statistical Software. 2010;33:1–21. [Google Scholar]
- Kamachi K, Sota M, Tamai Y, Nagata N, Konda T, Inoue T, Top EM, Arakawa Y. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1beta plasmids without accessory mobile elements. Microbiology. 2006;152:3477–3484. doi: 10.1099/mic.0.29056-0. [DOI] [PubMed] [Google Scholar]
- Klumper U, Riber L, Dechesne A, Sannazzarro A, Hansen LH, Sorensen SJ, Smets BF. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2014 doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsuwan K, Josh P, Picault MJ, Wijffels G, Dalrymple B. The plasmid RK2 replication initiator protein (TrfA) binds to the sliding clamp β subunit of DNA polymerase III: implication for the toxicity of a peptide derived from the amino-terminal portion of 33-kilodalton TrfA. J Bacteriol. 2006;188:5501–5509. doi: 10.1128/JB.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny I, Doran KS, Helinski DR, Blasina A. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J Biol Chem. 1997;272:20173–20178. doi: 10.1074/jbc.272.32.20173. [DOI] [PubMed] [Google Scholar]
- Konieczny I, Helinski DR. The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc Natl Acad Sci U S A. 1997a;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny I, Helinski DR. Helicase delivery and activation by DnaA and TrfA proteins during the initiation of replication of the broad host range plasmid RK2. J Biol Chem. 1997b;272:33312–33318. doi: 10.1074/jbc.272.52.33312. [DOI] [PubMed] [Google Scholar]
- Konieczny I, Liberek K. Cooperative action of Escherichia coli ClpB protein and DnaK chaperone in the activation of a replication initiation protein. J Biol Chem. 2002;277:18483–18488. doi: 10.1074/jbc.M107580200. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Levin BR, Stewart FM. The population biology of bacterial plasmids: a priori conditions for the existence of mobilizable nonconjugative factors. Genetics. 1980;94:425–443. doi: 10.1093/genetics/94.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2015 doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, Hunter SS, Settles ML, Forney LJ, Ponciano JM, Top EM. Evolutionary Paths That Expand Plasmid Host-Range: Implications for Spread of Antibiotic Resistance. Mol Biol Evol. 2016;33:885–897. doi: 10.1093/molbev/msv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YB, Datta HJ, Bastia D. Mechanistic studies of initiator-initiator interaction and replication initiation. EMBO J. 1998;17:5192–5200. doi: 10.1093/emboj/17.17.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro B, Sanz JM, Diaz-Orejas R, Fernandez-Tresguerres E. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J Bacteriol. 2003;185:1367–1375. doi: 10.1128/JB.185.4.1367-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro B, Sanz JM, Faelen M, Couturier M, Diaz-Orejas R, Fernandez-Tresguerres E. Modulation of pPS10 host range by DnaA. Mol Microbiol. 2002;46:223–234. doi: 10.1046/j.1365-2958.2002.03155.x. [DOI] [PubMed] [Google Scholar]
- Modi RI, Castilla LH, Puskas-Rozsa S, Helling RB, Adams J. Genetic changes accompanying increased fitness in evolving populations of Escherichia coli. Genetics. 1992;130:241–249. doi: 10.1093/genetics/130.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi RI, Wilke CM, Rosenzweig RF, Adams J. Plasmid macro-evolution: selection of deletions during adaptation in a nutrient-limited environment. Genetica. 1991;84:195–202. doi: 10.1007/BF00127247. [DOI] [PubMed] [Google Scholar]
- Nojiri H, Shintani M, Omori T. Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl Microbiol Biotechnol. 2004;64:154–174. doi: 10.1007/s00253-003-1509-y. [DOI] [PubMed] [Google Scholar]
- Philippe N, Pelosi L, Lenski RE, Schneider D. Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J Bacteriol. 2009;191:909–921. doi: 10.1128/JB.01419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponciano JM, De Gelder L, Top EM, Joyce P. The population biology of bacterial plasmids: a hidden Markov model approach. Genetics. 2007;176:957–968. doi: 10.1534/genetics.106.061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S, Connor TR, Barquist L, Walker D, Feltwell T, Harris SR, Fookes M, Hall ME, Petty NK, Fuchs TM, Corander J, Dufour M, Ringwood T, Savin C, Bouchier C, Martin L, Miettinen M, Shubin M, Riehm JM, Laukkanen-Ninios R, Sihvonen LM, Siitonen A, Skurnik M, Falcao JP, Fukushima H, Scholz HC, Prentice MB, Wren BW, Parkhill J, Carniel E, Achtman M, McNally A, Thomson NR. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc Natl Acad Sci U S A. 2014;111:6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan A, Pena-Miller R, Toll-Riera M, Halbert ZV, McLean AR, Cooper BS, MacLean RC. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun. 2014;5:5208. doi: 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan A, Toll-Riera M, Qi Q, MacLean RC. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat Commun. 2015;6:6845. doi: 10.1038/ncomms7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota M, Yano H, Hughes JM, Daughdrill GW, Abdo Z, Forney LJ, Top EM. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 2010;4:1568–1580. doi: 10.1038/ismej.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FM, Levin BR. The population biology of bacterial plasmids: a PRIORI conditions for the existence of conjugationally transmitted factors. Genetics. 1977;87:209–228. doi: 10.1093/genetics/87.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- Turner PE, Cooper VS, Lenski RE. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution. 1998;52:315–329. doi: 10.1111/j.1558-5646.1998.tb01634.x. [DOI] [PubMed] [Google Scholar]
- Wawrzycka A, Gross M, Wasaznik A, Konieczny I. Plasmid replication initiator interactions with origin 13-mers and polymerase subunits contribute to strand-specific replisome assembly. Proc Natl Acad Sci U S A. 2015;112:E4188–E4196. doi: 10.1073/pnas.1504926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Skowyra D, Hoskins J, McKenney K. DnaJ, DnaK, and GrpE heat shock proteins are required in oriP1 DNA replication solely at the RepA monomerization step. Proc Natl Acad Sci U S A. 1992;89:10345–10349. doi: 10.1073/pnas.89.21.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Deckert GE, Rogers LM, Top EM. Roles of long and short replication initiation proteins in the fate of IncP-1 plasmids. J Bacteriol. 2012;194:1533–1543. doi: 10.1128/JB.06395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Helinski D, Toukdarian A. A specific region in the N terminus of a replication initiation protein of plasmid RK2 is required for recruitment of Pseudomonas aeruginosa DnaB helicase to the plasmid origin. J Biol Chem. 2003;278:45305–45310. doi: 10.1074/jbc.M306058200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.