Abstract
The tumor microenvironment is a complex network of cells that support tumor progression and malignancy. It has been demonstrated that tumor cells can educate the immune system to promote a tumor-friendly environment. Among all these immune cells, tumor-associated macrophages (TAMs) are well represented and their presence in mouse models has been shown to promote tumor progression and metastasis. These effects are through the stimulation of angiogenesis, enhancement of tumor cell invasion and intravasation, immunosuppression, and at the metastatic site tumor cell extravasation and growth. However, the precise mechanisms are not fully understood. Furthermore there is limited information on TAMs derived from human cancers. For this reason it is important to be able to extract TAMs from tumors in order to compare their phenotypes, functions, and transcriptomes with normal resident tissue macrophages. Isolation of these cells is challenging due to the lack of markers and standardized protocols. Here we show an optimized protocol for the efficient isolation and extraction of resident macrophages and TAMs from human and mouse tissues by using multicolor flow cytometry. These protocols allow for the extraction of thousands of macrophages in less than 5 h from tissues as small as half a gram. The isolated macrophages can then be used for both “omics” and in vitro studies.
Keywords: Tumor-associated macrophages, Macrophages, TAM, Breast cancer, Flow cytometry, Tissue, Mouse, Human
12.1 Introduction
Whole tissues are highly complex environments comprised of extracellular matrix and many cells types whose diversity depends on the organ but always includes epithelial cells and cells of mesenchymal origin such as blood vessels and fibro-blasts as well as a diversity of immune cells. In solid cancers the epithelial cells are transformed and subsequently progress to malignancy that can lead to metastasis. The tumor cells recruit around them a specialized microenvironment that is often characterized by an intense infiltration of cells of the immune system such as T and B cells, neutrophils, and macrophages. These tumors resemble organs but with an overall lack of organization and thus have been referred to as “outlaw organs.” In both tissues and tumors stromal cells can carry out a myriad of functions and different cell types within tissues can have opposing roles.
Most cancer studies have assessed whole tumor tissues but recent work has shown that isolating stromal cells can be more effective for identifying prognostic markers [1]. Hence it is important to study these cell types individually to better understand how they are involved in tumor progression. Indeed, in most cases, high macrophage density correlates with poor prognosis [2–10]. Tumor-associated macrophages (TAMs) have been shown to regulate many critical processes associated with malignancy including promotion of tumor cell migration [11–13], invasion and metastasis [14, 15], induction of the angiogenesis [16, 17], and seeding of metastatic cells, and support of their growth, at distant sites [18–20].
There are a number of methods to assess macrophage and TAM function and phenotype; these include (1) gene expression analysis [21–23]; (2) functional studies; (3) assessment of cytokine and chemokine production [24, 25]; and (4) analysis of protein expression and cell surface markers [26]. In order to carry out these analyses macrophages/TAMs must first be isolated from the tissue.
The most common methods to isolate macrophages include (1) magnetic bead-conjugated antibody cell isolation [27], (2) density gradient separation [28], (3) laser capture microdissection [29], and (4) digestion foll owed by fluorescence-activated cell sorting (FACS) [21]. Magnetic cell isolation is a relatively fast technique but is limited in regard to how many markers can be used to separate macrophages and TAMs from the other stromal cell types. Many immune cells share the same markers and it is rare to find only one marker that labels a particular immune cell type. While the density gradient method is cheap, it relies on differences in cell size and density to separate the cells leading to problems with purity. Laser capture microdissection, although allowing for isolation of cells according to spatial position and morphology, can only utilize 2–3 markers, requires the fixation of cells, is very slow, and yields low numbers of cells. These methods are a particular problem for macrophages as they are extremely heterogeneous and express a range of cell surface markers as well as being easily damaged. Therefore, FACS is particularly useful for isolating resident macrophages and TAMs as it allows for the use of multiple cell surface markers simultaneously; this not only leads to a higher purity of the isolated macrophages/TAMs but also allows for phenotypic analysis. Commonly used markers for mouse macrophage isolation are CD11b, F4/80, and Ly6G/C antigens [30, 31] while CD14, CD11b, HLA-DR, CD163, and CX3CR1 antigens are used to identify human resident macrophages and TAMs [32–34].
The first step to isolating macrophages and TAMs from tissues is to digest the tissue to get a single-cell suspension. A number of different digestion methods exist that involve different incubation periods, agitation speeds, and enzymes [35–37], but no protocol so far has been optimized for primary macrophage isolation. Moreover some digestion methods can result in loss of cell surface markers [38–40]; therefore we have developed a protocol where the cell surface markers remain intact. In this chapter we review the FACS methods developed in our laboratory over the last few years for isolation and analysis of macrophages and TAMs from mouse and human normal mammary tissue and breast tumors.
12.2 Protocol for Isolation of Resident Macrophages and TAMs from Mouse and Human Tumors
12.2.1 Before You Start
Mouse care and husbandry—all mouse work must be performed after approval and according to in-house institute protocols and appropriate national guidelines.
Human clinical samples—clinical samples must be collected following local ethics regulations and only after appropriate approval from the institution.
This protocol has been used extensively for the isolation of TAMs from mouse mammary tumors (PyMT model, [41]) and human breast tumors, but it can be used for isolation of TAMs from other solid tumors; this protocol has been optimized for 150–750 mg tumors and therefore volume adjustments need to be made for analysis of smaller or larger tumors (see troubleshooting).
It is recommended to use tumor-bearing mice that are within normal parameters in all other aspects including weight, injury, necrotic tissue, and infection.
For cellular assays and in vivo experiments sterile conditions must be maintained. The protocols must be carried out in a biohazard level 2 hood and all buffers must be filtered using 0.22 μm filters.
For multicolor staining, fluorescence compensation is required; we recommend using compensation beads (see troubleshooting).
This protocol is optimized for the identification and the isolation of resident tissue macrophages and TAMs; therefore we included only a set of basic extracellular markers; additional TAM markers, cytokines, and transcription factors or extracellular markers may be incorporated to the basic protocol in order to identify TAM subpopulations or particular functions.
Analysis of rare immune cell populations requires the collection of at least 5–10,000 events of the smallest population in order to ensure statistical power for analysis.
In addition to the experimental samples, negative unstained controls, isotype controls, and FMOs (fluorescence minus one) must be prepared (see troubleshooting).
This protocol requires 4–5 h of time.
12.2.2 Materials
– 60 and 100 mm petri dishes (Thermoscientific)
– Razor blades
– Forceps and surgical scissors for animal dissection (Fine Surgical Tools)
– DMEM (Gibco)
– 15 and 50 ml tubes (Falcon)
– 2 ml DNase and RNase-free collection tubes (Simport)
– 5 ml Polypropylene tubes (with and without filter cap) (Falcon)
– 10× Red blood cell (RBC) lysis buffer (eBioscience)
– Enzymes: Liberase TL and Liberase DL (Roche)
– DNase I (Sigma)
– Phosphate-buffered saline (PBS) (Gibco)
– Bovine serum albumin (BSA) (Sigma)
– 100 μm Cell strainers (Corning)
– Ethanol (EtOH)
– Isoflurane
– Blocking antibody (Ab) CD16/CD32 (eBioscience)
– Antibodies (Abs) for flow cytometry (see Tables 12.1 and 12.2)
– DAPI (eBioscience)
– Human AB serum (Lonza)
– Penicillin/streptomycin (Gibco)
Table 12.1.
Antibody master mix for FACS analysis and sorting, mouse tissue
| Antigen and fluorochrome | Clone |
|---|---|
| CD45 APC | 30-F11 |
| CD11b eFluor 605 | M1/70 |
| F4/80 PerCp-Cy5.5 | BM8 |
| Ly6C PE-Cy7 | HK1.4 |
| Ly6G APC-Cy7 | 1A8 |
| DAPI |
Table 12.2.
Antibody master mix for FACS analysis and sorting, human tissue
| Antigen and fluorochrome | Clone |
|---|---|
| CD45 PE-Texas Red | HI30 |
| CD11b BV605 | ICRF44 |
| CD14 BV510 | M5E2 |
| CD163 APC | GHI/61 |
| CX3CR1 FITC | 2A9-1 |
| HLA-DR BV650 | L243 |
| CD3 BV711 | OKT3 |
| CD19 BV711 | HIB19 |
| CD56 BV711 | HCD56 |
| DAPI |
12.2.3 Lab Eq uipment
– Vortex
– Lab centrifuge (TX-1000 rotor, 290 mm radius)
– Lab bench centrifuge (FA 45-30-11 rotor)
– Lab scale
– Flow cytometer
– Flow sorter
– Pipetman
– Pipettes
12.2.4 Solutions
– Staining buffer: PBS 2 % w/v BSA.
– Sorting buffer: PBS 0.1 % w/v BSA.
– Collection medium: DMEM 10 % v/v FBS, 1 % penicillin/streptomycin.
– 1× RBC lysis buffer: Dilute 10× RBC buffer in distilled water.
– Enzymes: Resuspend the lyophilized powder following the manufacturer's instructions. Make aliquots and avoid freeze and thawing.
12.2.5 Isolation of Resident Macrophages and TAMs from Mouse Primary Tissue
Tissue type: mouse mammary carcinomas, normal mammary gland
Average tissue weight: 0.5–1 g
Macrophages isolated with this protocol: resident tissue macrophages and TAMs
12.2.5.1 Tissue Extraction and Digestion
Anesthetize mice using CO 2 or isoflurane, followed by cervi cal dislocation.
In the case of mammary tumors gently remove the abdominal skin without tearing the peritoneal cavity (for other solid tumors remove the tumor according to an appropriate protocol).
Locate the local draining lymph node and remove it using forceps.
Gently separate the tumor from the skin using forceps and surgical scissors.
Transfer the tumor to a 60 mm petri dish and record the tumor weight.
Add 1 ml of medium without serum (if more than one tumor is processed keep on ice until further use; it is not recommended to keep the tumors at room temperature for any period of time).
Using a razor blade cut the tumor into small pieces (the efficiency of the digestion is dependent on how well the tumor is minced; large pieces will not digest properly and may need extra digestion time) (Fig. 12.1a).
Transfer the tumor lysates into a 15 ml tube using a P1000 pipette (cut the top of the tip in order to be able to take all the tissue) and adjust the volume to 4 ml of serum-free PBS.
Add 40 μl of Liberase DL stock solution 28U/ml (Roche), 80 μl of Liberase TL stock solution 14 U/ml (Roche), and 40 μl of DNase I (15 mg/ml stock solution Sigma 100×). Mix using a vortex and incubate for 45 min at 37 °C under continuous rotation.
Check the sample visually; the cell suspension should look smooth (if the cell suspension still contains tissue fragments digest for an additional 15 min). If after 15 min there are still tissue fragments remaining this is connective tissue that will not digest and there is no need for further digestion.
To end the enzymatic digestion add 10 ml PBS 1 % w/vBSA (staining buffer).
Filter the cells using a 100 μm cell strainer in a 50 ml tube and adjust the volume to 20 ml with staining buffer.
Centrifuge for 7 min at 500 rcf and 4 °C. Discard the supernatant.
Proceed to red blood cell lysis (Sect. 12.10.2.5.2).
Fig. 12.1.
Isolation of TAMs from mouse mammary tumors. (a) Representative images of mouse mammary tumor excision and processing; tumor-bearing mouse is euthanized and mammary tumors are exposed, and draining lymph nodes removed with forceps (left panel); tumors are excised from the skin using forceps and surgical scissors and transferred in a 60 mm petri dish (central panel); 1 ml of serum-free medium is added and tumor is cut into small pieces using a razor blade (right panel). (b) Representative gating strategy for the identification of TAMs in mouse tumors. TAMs are defined as DAPI−, CD45+, CD11b+, F4/80+, Ly6C−, and Ly6G− . First cells that are viable (DAPI negative) and are CD45 positive (to exclude nonimmune cells) are selected. All cells that are CD11b+ and F4/80+ are then selected (both markers are expressed by macrophages). In this particular analysis we gated also some CD11b+ F4/80− in order to show the Ly6G+ granulocyte population. Finally cells that are Ly6G− and Ly6C− (markers that are not expressed by mature macrophages, but mainly on monocytes and granulocytes) are selected
12.2.5.2 Red Blood Cell Lysis
Add 3 ml of 1× RBC lysis buffer to the tumor pellet fro m Step 1.14, mix well, and incubate on ice for 10 min.
Add 30 ml of staining buffer.
Centrifuge for 7 min at 500 rcf and 4 °C. Discard the supernatant and resuspend the pellet in 1 ml of staining buffer.
Transfer to a 5 ml polypropylene tube.
Proceed to blocking (Sect. 10.2.5.3).
12.2.5.3 Blocking
Add 2 μl (1:500) of blocking Ab CD16/CD32 (clone 93), mix well, and inc ubate on ice for a minimum of 30 min (mix every 10 min).
Add 4 ml of staining buffer and centrifuge for 5 min at 500 rcf and 4 °C. Discard the supernatant, resuspend the pellet in 1 ml of staining buffer, and proceed to FACS staining for analysis (Sect. 10.2.5.4) or FACS staining for sorting (Sect. 10.2.5.5).
12.2.5.4 FACS Staining and Analysis
Count the cells (trypan blue exclusion method) and adjust the number to 1–2 × 106 cells/ml.
Transfer 1 ml into a 5 ml polystyrene tube.
Centrifuge for 5 min at 500 rcf and 4 °C. Discard the supernatant and resuspend the pellet in 100 μl of staining buffer.
Add the antibody master mix (see Table 12.1).
Incubate on ice for 1 h protected from the light.
Add 4 ml of staining buffer.
Filter using filter top 5 ml polystyrene tubes (40 μm) to prevent clogging the machine.
Centrifuge for 5 min at 500 rcf and 4 °c.
Resuspend the pellet in 2 ml of staining buffer.
Analyze using flow cytometer using the gating strategy (shown in Fig. 12.1b).
12.2.5.5 FACS Staining and Cell Sorting
After blocking add 9 ml of staining buff er and centrifuge for 7 min at 500 rcf and 4 °C. Discard the supernatant and resuspend the pellet in 100 μl of staining buffer.
Add the antibody master mix (see Table 12.1).
Incubate for 1 h on ice protected from light (mix every 10 min).
Wash once with a large volume of staining buffer, around 30–40 ml.
Centrifuge for 7 min at 500 rcf and 4 °C.
Discard the supernatant and resuspend the pellet in 2–5 ml (depending on the initial tumor size) of sorting buffer.
Sort TAMs as DAPI−, CD45+, CD11b+, F4/80+, Ly6C−, Ly6G−, CD3−, CD19−, and NK1.1−.
Sort cells into collection tubes that contain 1–2 ml of collection medium.
Sorted cells can be used for DNA, RNA, and protein extraction, in vitro experiments such as migration analysis, angiogenesis, T cell activation, or suppression, etc.
12.2.6 Isolation of TAMs and Resident Macrophages from Human Breast Tumor and Normal Adjacent Tissue
Tissue type: needle biopsies, surgical surplus
Tissue weight: 0.5–1 g
Macrophages isolated with this protocol: resident tissue macrophages and TAMs
Before starting: collect tissue in collection medium and keep it/store it at 4 °C.
12.2.6.1 Tissue Processing and Digestion
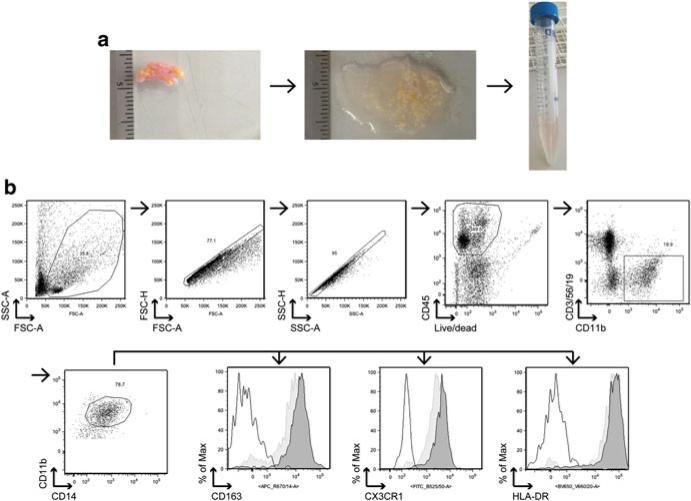
Transfer the tumor to a 60 mm petri dish, remove all medium with a pipette, and measure the tumor weight (Fig. 12.2a).
Add 1 ml of serum-free PBS (if more than one tumor is processed keep on ice until further use; it is not recommended to keep the tumors at room temperature for any time).
Using a razor blade cut the tumor into small pieces (the efficiency of the digestion is dependent on how well the tumor is minced, large pieces will not digest properly and may need more digestion time).
Transfer the tumor lysates to a 15 ml tube using a P1000 pipette (cut the top of the tip in order to be able to take all the tissue) and adjust the volume to 3 ml of serum-free PBS.
Add 30 μl of Liberase DL stock solution 28 U/ml (Roche), 120 μl of Liberase TL stock solution 14 U/ml (Roche), and 30 μl of DNase I (15 mg/ml stock solution Sigma 100×).
Mix using a vortex and incubate for 45 min at 37 °C applying continuous rotation or, if using a thermomixer, at 700/800 rpm.
Check digestion visually; the cell suspension should look smooth (if cell suspension is too lumpy digest for an additional 15 min). Do not incubate for more than 1 h to avoid overdigestion.
To end the enzymatic digestion add 10 ml of staining buffer.
Filter the cells using 100 μm cell strainer in a 50 ml tube and adjust the volume to 40 ml with staining buffer.
Centrifuge for 5 min at 500 rcf and 4 °C. Discard the supernatant.
Resuspend the pellet in 1 ml of staining buffer and transfer the residual volume to a 5 ml polypropylene tube.
Proceed to red blood cell lysis (Sect. 12.10.2.6.4)
Fig. 12.2.
Isolation of TAMs from human mammary tumors. (a) Representative images of human mammary tumor processing; after sample collection, medium is completely removed and the tumor is transferred in a 60 mm petri dish (left panel); 1 ml of serum-free PBS is added and tumor is cut into small pieces using a razor blade (right panel); after the digestion the mixture should look cloudy and >80 % of tissue fragments should be dissolved (right panel). (b) Representative gating strategy for the identification of resident and tumor-associated macrophages from human tissue. Tissue macrophages are defined as DAPI−, CD45+, CD11b+, HLA-DR+ CD14+, CD3−, CD19−, and CD56− . In addition, TAMs are CD163high and CX3R1High as shown in the histograms (black-lined empty histogram = isotype control, light grey-colored histogram = resident macrophages from normal mammary tissue, dark grey-colored histogram = TAMs from breast tumor). After adding a gate based on physical parameters (size and granularity) and after doublet removal, cells that are viable and CD45 positive are selected. A gate on CD11b+ cells and negative for CD3 (T cells), CD56 (NK cells), and CD19 (B cells) identifies the myeloid cell populations in the tumor (granulocytes and macrophages mainly). Cells that are CD11b+ CD14+ CD163+ CX3CR1+ HLA-DR+ are defined as macrophages
12.2.6.2 Isolation of Resident Macrophages from Normal Tissue (Mammoplasty Reductions)
Tissue type: mammoplasty reductions
Tissue weight: 15–20 g
Macrophages isolated: resident tissue macrophages
Before you start:
Collect the tissue into collection medium and keep it/store it at 4 °C.
Sterilize the razor blades and forceps with ETOH, then rinse, and cool in a 100 cm dish of PBS.
12.2.6.3 Tissue Processing and Digestion
Transfer the breast tissue to a 100 cm dish and wash several times with serum-free PBS.
Transfer the tissue several times to fresh 100 mm dishes, washing with serum-free PBS each time. The goal is to remove all traces of blood. Be generous with the PBS: use the entire 500 ml bottle if needed.
Once the washes are complete, place the tissue into a dry 100 mm dish (some PBS carryover is preferred).
Using the forceps in one hand (to hold the tissue in place) and your other hand to hold the razor blade or scalpel, gently divide the tissue into three parts and place each part into a fresh 100 cm dish in order to be able to cut easily.
Using a razor blade cut the tissue into small pieces (the efficiency of the digestion is dependent on how well the tissue is minced; large pieces will not digest properly and may need more digestion time). This procedure can take a considerable amount of time. Keep the dishes on ice at all times.
Combine the tissues fragment and then separate into two 50 ml tubes. Make up the volume to 50 ml with serum-free PBS and centrifuge for 5 min at 500 rcf and 4 °C.
Due to the nature of the specimen, there will be a pellet as well as an upper layer of yellow fat. Use forceps to transfer the yellow fat layer to a new 50 ml Falcon tube.
Discard the supernatant and resuspend the pellet in 25 ml of serum-free DMEM together with the fat.
Add 10 μl of Liberase DL stock solution 28 U/ml, 20 μl of Liberase TL stock solution 14 U/ml, and 10 μl of DNase I (15 mg/ml stock solution 100×) per ml of solution.
Wrap the top of the tube and the cap with parafilm, mix using a vortex, and incubate for 10–18 h at 37 °C, applying continuous rotation.
Check the digestion status. After the incubation, the mixture should look like a greasy, brown broth. If >80 % of the tissue is digested add 25 ml of staining buffer. If not then continue digestion for an additional hour and check the digestion status again.
Filter the entire volume through a 100 μm cell strainer into a new 50 ml tube.
Centrifuge for 7 min at 500 rcf and 4 °C (if there is a lot of fat floating, discard it and take ONLY the pellet).
Discard the supernatant and proceed to red blood cell lysis (Sect. 12.10.2.6.4).
12.2.6.4 Red Blood Cell Lysis
Resuspend the pellet in 3 ml of 1X RBC lysis buffer, mix well, and incubate on ice for 10 min.
Add 30 ml of staining buffer.
Centrifuge for 5 min at 500 rcf and 4°c. Discard the supernatant and resuspend the pellet in 900 μl of staining buffer.
Proceed to blocking (Sect. 10.2.6.5).
12.2.6.5 Blocking
Add 10 % (100 μl in PBS; total volume of 1 ml) of human AB serum, mix well, and incubate on ice for a minimum 30 min (mix every 10 min).
Add 5 ml of staining buffer, filter using filter top 5 ml polystyrene tubes (40 μm) to prevent clogging the machine.
Centrifuge for 5 min at 500 rcf and 4°c. Discard the su pernatant and resuspend the pellet in 1 ml of staining buffer.
Proceed to FACS staining for analysis (Sect. 10.2.6.6) or FACS staining for sorting (Sect. 10.2.6.7).
12.2.6.6 FACS Staining and Analysis
Count the cells, adjust t he number to 1–2 × 106 cells/ml, and transfer 1 ml to a 5 ml polystyrene tube.
Centrifuge for 5 min at 500 rcf and 4°c.
Discard the supernatant and resuspend the pellet in 100 μl of staining buffer.
Add the antibody master mix (see Table 12.2).
Incubate for 1 h on ice.
Add 4 ml of staining buffer.
Centrifuge for 5 min at 500 rcf and 4°c.
Resuspend the pellet in 350 μl staining buffer.
Analyze using a flow cytometer using the gating strategy shown in Fig. 12.2b.
12.2.6.7 FACS Staining and Cell Sorting
Count the cells in order to adjust antibodies’ quantity accordingly.
Add 4 ml staining buffer.
Centrifuge for 5 min at 500 rcf and 4°c. Discard the supernatant and resuspend the pellet in 100 μl of staining buffer.
Add the antibody master mix (see Table 12.2).
Incubate for 1 h on ice protected from light (mix every 10 min).
Add 5 ml of staining buffer.
Centrifuge for 5 min at 500 rcf and 4°c. Discard the supernatant and resuspend in 2–5 ml (depending on the initial tumor size) sorting buffer.
Sort TAMs as DAPI−, CD45+, CD11b+, CD14+, CD163+, CX3CR1+, HLA− DR+, CD3−, CD19−, and CD56−.
Sorted cells can be used for DNA, RNA, and protein extraction, in vitro experiments such as migration analysis, angiogenesis, T cell activation or suppression, in vivo assays, etc.
12.2.7 Expected Cellular Yield
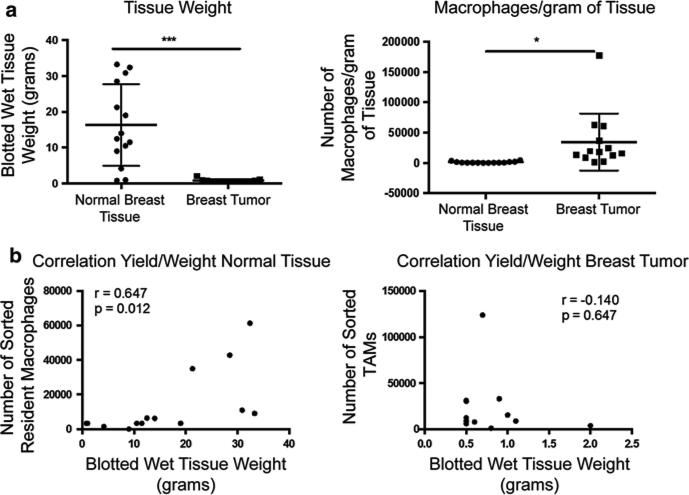
As shown in Fig. 12.3a, left panel, the weights of the mammoplasty reductions were generally higher than those of the tumor samples. However, we were able to sort significantly higher number of TAMs per gram of tumor tissue compared to resident macrophages (Fig. 12.3a, right panel). Our data confirmed that the density of TAMs is significantly higher than normal resident tissue macrophages. As expected there was a significant correlation between dry tissue weight and the number of cells extracted in normal tissue. However there was no such correlation in tumor samples, due to the extreme variability of tumor tissue composition (Fig. 12.3b, left and right panels, respectively).
Fig. 12.3.
TAM and resident macrophage yield from tumor and normal tissue. (a) Left panel represents the individual blotted wet tissue weights of normal mammary tissue and tumor samples collected (N = 15); right panel shows the number of macrophages extracted per gram of tissue in normal mammary tissue and tumor samples (N = 15). * = p < 0.01, *** = p < 0.0001, Student's T test. (b) Correlation between blotted wet tissue weight and cellular yield in normal tissue (left panel) and tumor tissue (right panel). Pearson correlation coefficient was calculated using Graphpad prism software. Each dot represents one sample. (N = 15, r = correlation coefficient, p = statistical value)
12.2.8 Troubleshooting/Useful Tips
As with any scientific protocol there are caveats and important considerations to take into account, in order to avoid artifacts and the generation of unreliable results.
12.2.8.1 Tissue Digestion
Most TAM extracellular markers are stable and sustain enzymatic digestion, but it is fundamental to optimize the digestion protocol before starting.
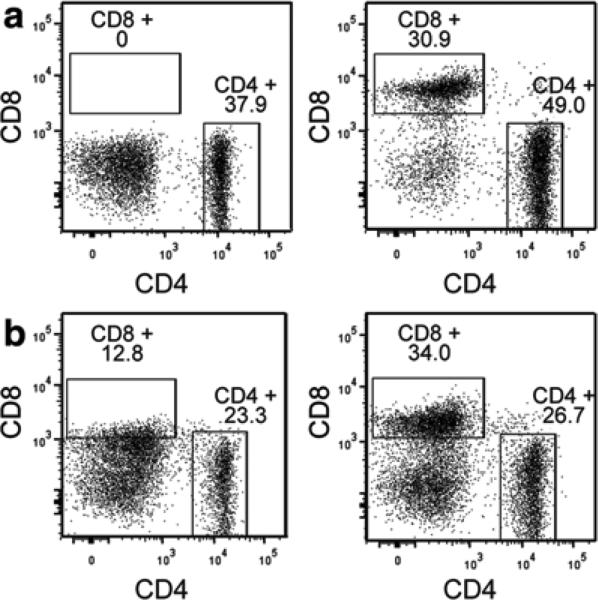
In this protocol we use Liberase enzyme blends (HPLC-purified blend); other compounds using collagenase mixtures are available but they may contain additional proteases that can cleave extracellular markers. In addition, long digestion times and impurities of the enzyme mix can degrade extracellular markers of other immune cell populations such as T and B cells (see Fig. 12.4).
Fig. 12.4.
Cleavage of extracellular markers as a result of overdigestion. (a) Loss of CD8 staining in single-cell suspensions—incubation of splenocytes with Liberase mix for 45 min (left panel) in comparison to splenocytes that were incubated with medium only (right panel). Data is presented after selection of CD45+, DAPI−, F4/80−, CD3+ cells. (b). Loss of CD8 staining in tumors—tumors were digested with a high concentration (twice higher than recommended, left panel) or with the recommended concentration (right panel) of Liberase mix for 45 min. Data is presented after selection of CD45+, DAPI−, F4/80−, CD3+ cells
Excess enzymes can also degrade/cleave certain surface markers, leading to artifacts. An example of this is reported in Fig. 12.4; CD8 is rapidly lost on the surface of splenocytes and T cells from mouse mammary tumors if high concentrations of enzymes are used [40].
Take-home message: Check your markers and do not overdigest the samples; carefully select the digestion enzymes that suit your experiment. If you are worried about marker degradation reduce the enzyme concentration and digestion time accordingly.
12.2.8.2 FACS Staining
Multicolor flow cytometry requires a number of internal controls and optimization steps that are necessary for the performance of a valid experiment.
For general information about flow cytometry staining:
https://www.bdbiosciences.com/us/support/training/s/itf_launch
Compensation
It is important that each individual fluorochrome is well compensated against all the other fluorochromes. Make sure that you have a validated compensation matrix before starting the experiment.
Internal controls
We suggest to use isotype antibody controls and FMO controls in order to verify the specificity of the signal for each cell surface marker. It is extremely important that all antibodies are titrated before their use in the multicolor panel.
Make sure that at least 5–10,000 events of the smallest population are acquired, in order to ensure statistical power for the analysis.
Choice of fluorochromes
This is an important choice, even if sometimes limited by the antibodies available on the market. Our protocol shows examples of fluorochromes for a typical seven-color experiment. We suggest using the brightest and most common fluorochromes for the markers that are expected to have a low density, and to use the remaining fluorochromes for highly expressed markers. Particular attention must be given to tandem dyes, including the Brilliant Violet ones, because they are more prone to degradation or nonspecific binding if left in a mix for a prolonged time prior to analysis. Moreover the Brilliant Violet dyes, if used in combination, will require a specific staining buffer to be added to the antibody mix.
Blocking
Myeloid cells, macrophages especially, express high levels of Fc receptors. Make sure to use an effective blocking reagent (serum or CD16/CD32 antibodies) before antibody staining.
Staining
For multicolor staining we suggest to prepare an antibody master mix (if possible) rather than adding single-antibody aliquots to all samples; this procedure will avoid potential mistakes and will reduce experimental bias.
Sample filtration
Primary digested tissues are particularly prone to clogging the machines. Before analysis we strongly recommend filtration of all the samples using a 40 μm filter cap polypropylene tube.
Take-home message
Make sure to carefully titrate your antibodies, check your compensation, and choose your fluorochromes wisely based on cytofluorimeter laser settings.
12.2.8.3 Cell Sorting
Sorting TAMs from primary tissue can be very difficult, as the amount of tissue available is the limiting step.
Sample concentration
Resuspend the sample in sorting buffer and do not exceed the optimal sorting concentration (1 × 106 cells/ml) in order to avoid clogging of the instrument.
Nozzle
It is important that a 100 μm nozzle is used to sort primary macrophages; smaller nozzles (85, 70 μm) can reduce the cellular yield and cell viability.
Sorting temperature
If possible, perform cell sorting at 4 °C.
Flow rate
Monitor the flow rate and sheer pressure constantly; if the flow rate is too high you will lose cells and cell viability will be reduced.
Sort protocol
Based on your experiment you may be interested in maximizing cellular yield or purity; to do so you will need to choose the optimal in-built sorting protocol of the machine. If the final goal is to perform “omics” studies, the “purity-001” or “two-way purity” protocols are preferable (BD Facs Aria II and III).
Sorting purity check
Make sure to perform a sorting purity check during setup of the sorting protocol settings; after sorting the population take an aliquot of cells and acquire it to verify the purity level of your sort.
Collection tubes
You can use 15, 5, and 1.5 ml collection tubes. Make sure to put an adequate volume of serum-containing medium. Importantly, the sides of the collection tubes need to be coated with medium to avoid drying out the sorted cells; to do this vortex the collection tubes just prior to sorting. For small-cell populations we suggest to use V-bottom 2 ml collection tubes filled with at least 750 μl of medium. At the end of sorting the vial should be spun using a lab bench centrifuge for 10 min at 450 Gav and 4 °C. Make sure to draw a line on the border of the vial before centrifugation in order to identify the pellet later (sometimes it is almost invisible). Remove the supernatant without touching the pellet, and resuspend the pellet in complete medium for tissue culture or lysis buffer for RNA or DNA extraction.
Take-home message
Check cell concentration, use 100 μm nozzle, sort at 4 °C, check purity of sorted cells, and adjust sort protocol (yield vs. purity).
12.2.9 Macrophage Plasticity and Tissue Extraction
One of the main hallmarks of macrophages is plasticity, i.e., the ability to respond to stimuli and to rapidly adapt to changes in the tissue microenvironment [42]. Pathological conditions such as cancer, inflammation, and infection can profoundly modify macrophage phenotype and function [18, 43], but any external stimulus (mechanical, physical, and chemical) introduced during their extraction from tissue could rapidly introduce additional changes not related to the microenvironment they were extracted from. For these reasons particular attention should be paid when macrophage extraction from tissue is performed, in order to minimize any extraction-induced bias.
Isolation method
Gene expression profiling of different leukocyte subsets recently demonstrated that FACS sorting is the recommended method for isolation of leukocyte subsets for gene expression studies since this method results in the purest subset populations and does not appear to induce a stress response [44]. This is the best available isolation method at the moment for human tissue macrophage extraction. However mouse tissue macrophages can also be extracted using specific genetic labeling (fluorescent macrophages) and mechanical tissue disruption [21].
Temperature
Another important aspect to consider is the temperature; prolonged incubations of samples at 37 °C and room temperature can introduce modification in macrophage phenotype as cellular transcriptional machinery is on and RNase, DNase, and proteases are active. It is recommended to keep the samples at 4 °C where possible.
Fresh isolation
It is recommended to extract tissue macrophages from fresh tissue in order to increase cellular yield and viability; we do not recommend cultivating macrophages in vitro if the purpose of the study is to characterize their pathological associated phenotype or transcriptome as this step will likely modify their activation status.
Matched controls
It is important that the phenotype and the transcriptome of macrophages extracted from human tissues are compared with matched controls, ideally from the same tissue type; in vitro monocyte-derived macrophages, frequently used as normal negative controls, are an heterogeneous population of macrophages with different phenotypes and functions [45] and do not represent optimal controls for these kind of studies.
12.3 Conclusion
In conclusion this chapter explains the best protocol currently available for the isolation of resident and tissue-associated macrophages from human and mouse samples. The main limitations are related to the amount of starting material and the availability of matched normal controls, especially for human samples.
Currently the lack of specific TAM-associated markers renders the identification of this population a real challenge, especially for human samples where it is not possible to genetically label this population. For this reason there is the need to identify new markers unique to TAMs that we hope will be discovered in a near future.
Acknowledgments
We would like to thank Dr Lisa Coussens, OHSU, for valuable discussions. This work was supported by Wellcome Trust (101067/Z/13/Z) and NIH R01CA172451 to JWP and Department of Defense (DOD W81XWH-111-0702) to Dr Lisa Coussens and JWP.
Contributor Information
Luca Cassetta, MRC Centre for Reproductive Health, Queen's Medical Research Institute, The University of Edinburgh, Edinburgh EH16 4TJ, UK, Luca.Cassetta@ed.ac.uk.
Roy Noy, Department of Developmental and Molecular Biology,, New York, NY 10461, USA, roy.noy@einstein.yu.edu.
Agnieszka Swierczak, MRC Centre for Reproductive Health, Queen's Medical Research Institute, The University of Edinburgh, Edinburgh EH16 4TJ, UK, A.Swierczak@ed.ac.uk.
Gaël Sugano, MRC Centre for Reproductive Health, Queen's Medical Research Institute, The University of Edinburgh, Edinburgh EH16 4TJ, UK, gsugano@staffmail.ed.ac.uk.
Harriet Smith, Department of Obstetrics and Gynecology and Women's Health, New York, NY 10461, USA, hasmith@montefiore.org.
Lisa Wiechmann, Department of Surgery, Albert Einstein College of Medicine, New York, NY 10461, USA, lisawiechmann@gmail.com.
Jeffrey W. Pollard, MRC Centre for Reproductive Health, Queen's Medical Research Institute, The University of Edinburgh, Edinburgh EH16 4TJ, UK Department of Developmental and Molecular Biology, New York, NY 10461, USA.
References
- 1.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 2.Yuan ZY, Luo RZ, Peng RJ, Wang SS, Xue C. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014;7:1475–80. doi: 10.2147/OTT.S61838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Wang F, Wang L, Huang L, Wang J, Zhang B, et al. CD163+ tumor-associated macrophage is a prognostic biomarker and is associated with therapeutic effect on malignant pleural effusion of lung cancer patients. Oncotarget. 2015;6:10592–603. doi: 10.18632/oncotarget.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Liu H, Dong X, Wu S, Zeng H, Liu Z, et al. High CD204+ tumor-infiltrating macrophage density predicts a poor prognosis in patients with urothelial cell carcinoma of the bladder. Oncotarget. 2015;6(24):20204–14. doi: 10.18632/oncotarget.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Li P, Wang L, Xia Z, Huang H, Lu Y, et al. High numbers of CD68+ tumor- associated macrophages correlate with poor prognosis in extranodal NK/T-cell lymphoma, nasal type. Ann Hematol. 2015;94(9):1535–44. doi: 10.1007/s00277-015-2401-4. [DOI] [PubMed] [Google Scholar]
- 6.Nabeshima A, Matsumoto Y, Fukushi J, Iura K, Matsunobu T, Endo M, et al. Tumour-associated macrophages correlate with poor prognosis in myxoid liposarcoma and promote cell motility and invasion via the HB-EGF-EGFR-PI3K/Akt pathways. Br J Cancer. 2015;112:547–55. doi: 10.1038/bjc.2014.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oishi K, Sakaguchi T, Baba S, Suzuki S, Konno H. Macrophage density and macrophage colony-stimulating factor expression predict the postoperative prognosis in patients with intrahepatic cholangiocarcinoma. Surg Today. 2015;45:715–22. doi: 10.1007/s00595-014-0989-y. [DOI] [PubMed] [Google Scholar]
- 8.Richardsen E, Uglehus RD, Johnsen SH, Busund LT. Macrophage-colony stimulating factor (CSF1) predicts breast cancer progression and mortality. Anticancer Res. 2015;35:865–74. [PubMed] [Google Scholar]
- 9.Smith HO, Anderson PS, Kuo DY, Goldberg GL, DeVictoria CL, Boocock CA, et al. The role of colony-stimulating factor 1 and its receptor in the etiopathogenesis of endometrial adeno-carcinoma. Clin Cancer Res. 1995;1:313–25. [PubMed] [Google Scholar]
- 10.Zhang QW, Liu L, Gong CY, Shi HS, Zeng YH, Wang XZ, et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One. 2012;7:e50946. doi: 10.1371/journal.pone.0050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–6. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–9. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 13.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeo EJ, Cassetta L, Qian BZ, Lewkowich I, Li JF, Stefater JA, et al. Myeloid WNT7b mediates the angiogenic switch and metastasis in breast cancer. Cancer Res. 2014;74:2962–73. doi: 10.1158/0008-5472.CAN-13-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Palma M, Naldini L. Angiopoietin-2 TIEs up macrophages in tumor angiogenesis. Clin Cancer Res. 2011;17:5226–32. doi: 10.1158/1078-0432.CCR-10-0171. [DOI] [PubMed] [Google Scholar]
- 18.Porta C, Riboldi E, Totaro MG, Strauss L, Sica A, Mantovani A. Macrophages in cancer and infectious diseases: the ‘good’ and the ‘bad’. Immunotherapy. 2011;3:1185–202. doi: 10.2217/imt.11.116. [DOI] [PubMed] [Google Scholar]
- 19.Biswas SK, Allavena P, Mantovani A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin Immunopathol. 2013;35:585–600. doi: 10.1007/s00281-013-0367-7. [DOI] [PubMed] [Google Scholar]
- 20.De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–86. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Ojalvo LS, King W, Cox D, Pollard JW. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am J Pathol. 2009;174:1048–64. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chittezhath M, Dhillon MK, Lim JY, Laoui D, Shalova IN, Teo YL, et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity. 2014;41:815–29. doi: 10.1016/j.immuni.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Pucci F, Venneri MA, Biziato D, Nonis A, Moi D, Sica A, et al. A distinguishing gene signature shared by tumor-infiltrating Tie2-expressing monocytes, blood “resident” monocytes, and embryonic macrophages suggests common functions and developmental relationships. Blood. 2009;114:901–14. doi: 10.1182/blood-2009-01-200931. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest. 2006;116:2132–41. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sica A. Role of tumour-associated macrophages in cancer-related inflammation. Exp Oncol. 2010;32:153–8. [PubMed] [Google Scholar]
- 26.Qian B, Deng Y, Im JH, Muschel RJ, Zou Y, Li J, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4:e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol. 2005;174:4880–91. doi: 10.4049/jimmunol.174.8.4880. [DOI] [PubMed] [Google Scholar]
- 28.Lin EY, Li JF, Bricard G, Wang W, Deng Y, Sellers R, et al. Vascular endothelial growth factor restores delayed tumor progression in tumors depleted of macrophages. Mol Oncol. 2007;1:288–302. doi: 10.1016/j.molonc.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trogan E, Fisher EA. Laser capture microdissection for analysis of macrophage gene expression from atherosclerotic lesions. Methods Mol Biol. 2005;293:221–31. doi: 10.1385/1-59259-853-6:221. [DOI] [PubMed] [Google Scholar]
- 30.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–5. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose S, Misharin A, Perlman H. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A. 2012;81:343–50. doi: 10.1002/cyto.a.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A. 2012;109:2796–801. doi: 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau SK, Chu PG, Weiss LM. CD163: a specific marker of macrophages in paraffin-embedded tissue samples. Am J Clin Pathol. 2004;122:794–801. doi: 10.1309/QHD6-YFN8-1KQX-UUH6. [DOI] [PubMed] [Google Scholar]
- 34.Zheng J, Yang M, Shao J, Miao Y, Han J, Du J. Chemokine receptor CX3CR1 contributes to macrophage survival in tumor metastasis. Mol Cancer. 2013;12:141. doi: 10.1186/1476-4598-12-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–52. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mroue R, Bissell MJ. Three-dimensional cultures of mouse mammary epithelial cells. Methods Mol Biol. 2013;945:221–50. doi: 10.1007/978-1-62703-125-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalley MJ, Kendrick H, Sheridan JM, Regan JL, Prater MD, Lindeman GJ, et al. Isolation of mouse mammary epithelial subpopulations: a comparison of leading methods. J Mammary Gland Biol Neoplasia. 2012;17:91–7. doi: 10.1007/s10911-012-9257-1. [DOI] [PubMed] [Google Scholar]
- 38.Hines WC, Su Y, Kuhn I, Polyak K, Bissell MJ. Sorting out the FACS: a devil in the details. Cell Rep. 2014;6:779–81. doi: 10.1016/j.celrep.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Hines WC, Yaswen P, Bissell MJ. Modelling breast cancer requires identification and correction of a critical cell lineage-dependent transduction bias. Nat Commun. 2015;6:6927. doi: 10.1038/ncomms7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur J Microbiol Immunol (Bp) 2012;2:112–20. doi: 10.1556/EuJMI.2.2012.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassol E, Cassetta L, Alfano M, Poli G. Macrophage polarization and HIV-1 infection. J Leukoc Biol. 2010;87:599–608. doi: 10.1189/jlb.1009673. [DOI] [PubMed] [Google Scholar]
- 44.Beliakova-Bethell N, Massanella M, White C, Lada SM, Du P, Vaida F, et al. The effect of cell subset isolation method on gene expression in leukocytes. Cytometry A. 2014;85:94–104. doi: 10.1002/cyto.a.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eligini S, Crisci M, Bono E, Songia P, Tremoli E, Colombo GI, et al. Human monocyte-derived macrophages spontaneously differentiated in vitro show distinct phenotypes. J Cell Physiol. 2013;228:1464–72. doi: 10.1002/jcp.24301. [DOI] [PubMed] [Google Scholar]