Significance
Clathrin forms coats on vesicles that control receptor endocytosis, thereby influencing cell signaling. The contribution of the clathrin light chain subunits to this process in mice was addressed by deleting the gene encoding the predominant light chain form (CLCa) present in B lymphocytes. CLCa-null mice have B cells with altered antibody production and aberrant development due to defects in endocytosis of signaling receptors that influence these processes. Only a subset of signaling receptors were found to depend on clathrin light chains for their endocytosis, demonstrating that light chain subunits contribute to selection of particular clathrin-coated vesicle cargo. This study clarifies a role for the light chains in vivo, showing that they contribute properties to the clathrin coat that regulate cargo uptake.
Keywords: clathrin light chain, endocytosis, antibody isotype switch, TGFβ, G protein-coupled receptors
Abstract
Clathrin, a cytosolic protein composed of heavy and light chain subunits, assembles into a vesicle coat, controlling receptor-mediated endocytosis. To establish clathrin light chain (CLC) function in vivo, we engineered mice lacking CLCa, the major CLC isoform in B lymphocytes, generating animals with CLC-deficient B cells. In CLCa-null mice, the germinal centers have fewer B cells, and they are enriched for IgA-producing cells. This enhanced switch to IgA production in the absence of CLCa was attributable to increased transforming growth factor β receptor 2 (TGFβR2) signaling resulting from defective endocytosis. Internalization of C-X-C chemokine receptor 4 (CXCR4), but not CXCR5, was affected in CLCa-null B cells, and CLC depletion from cell lines affected endocytosis of the δ-opioid receptor, but not the β2-adrenergic receptor, defining a role for CLCs in the uptake of a subset of signaling receptors. This instance of clathrin subunit deletion in vertebrates demonstrates that CLCs contribute to clathrin’s role in vivo by influencing cargo selectivity, a function previously assigned exclusively to adaptor molecules.
Clathrin-coated vesicles (CCVs) influence cell signaling by selecting membrane cargo during endocytosis (1). Clathrin consists of three clathrin heavy chain (CHC17) subunits, each associated with a clathrin light chain (CLC), configured into a three-legged triskelion. Clathrin triskelia self-assemble into a latticed coat that traps membrane-associated adaptor molecules, which recruit cargo into the CCV through recognition of defined protein motifs. Cargo specificity is therefore determined by adaptor binding, but recent studies in vitro have suggested that the CLC subunits can also influence cargo selection. Vertebrates have two CLC isoforms, CLCa and CLCb, encoded by separate genes, CLTA and CLTB (1). Depletion of both isoforms from tissue culture cells by siRNA treatment showed that CLCs are not required for clathrin-mediated uptake of classic CCV cargoes, such as transferrin receptor (TfR), epidermal growth factor receptor, or low-density lipoprotein receptor (2–4). CLCs were, however, implicated in uptake of three G protein-coupled receptors (GPCRs) (5). CLCs are also required for uptake of large particles, including some viruses and bacteria (6, 7). These roles for CLCs in cargo selection and their general contribution to clathrin function have been a matter of speculation for some time, with indications from in vitro experiments that CLCs regulate clathrin assembly (8) and the tensile strength of the clathrin lattice (1, 9). To assess such roles for CLCs in clathrin function in vivo, we made mutant mice that lack the CLTA gene, and we report their phenotype here.
In mammals, CLCa and CLCb are ∼60% identical in protein sequence (1). Shared sequences of 22 and 10 residues, respectively, mediate binding to the actin-organizing huntingtin-interacting proteins (mammalian Hip1 and Hip1R, yeast Sla2p) (10–12) or the leucine-rich repeat kinase 2 (LRRK2) (13). Mammalian cell culture experiments and genetic studies in yeast and flies have shown that, through these interactions, CLCs participate in several pathways that could significantly affect clathrin function in vertebrates. These pathways include clathrin-mediated endocytosis from membranes under tension (14, 15), formation of clathrin–actin interfaces during cell adhesion (16), clathrin-mediated recycling in cell migration (2), and endosome function during Drosophila eye development (13).
In vertebrates, both CLTA and CLTB gene products undergo alternative mRNA splicing to generate four possible forms of CLCa and two of CLCb (1). Neurons express the highest molecular weight forms of both CLCs. The lowest molecular weight forms predominate in nonneuronal tissue (17, 18), and vertebrate tissues maintain characteristic levels of CLCa and CLCb (17, 19). Here, using an updated quantification approach, we established that there are two CLC expression patterns, with most tissues expressing equal levels of CLCa and CLCb whereas CLCa expression is dominant in lymphoid tissue. Consequently, B cells in our CLTA knockout (KO) mice were effectively CLC-deficient, allowing us to address the functional consequence of CLC loss in vertebrates through analysis of immunological phenotypes. We observed that B cells from CLTA knockout mice had defects in internalization of transforming growth factor β receptor 2 (TGFβR2) and C-X-C chemokine receptor 4 (CXCR4). Although not all signaling receptors were affected by CLCa loss, defects in receptor internalization accounted for phenotypes observed in the knockout animals. Thus, CLCs play a significant role in cargo selection by CCVs in vivo by influencing uptake of specific signaling receptors.
Results
Tissues Maintain Characteristic CLCb Levels upon CLCa Loss, Resulting in CLC-Deficient Lymphocytes.
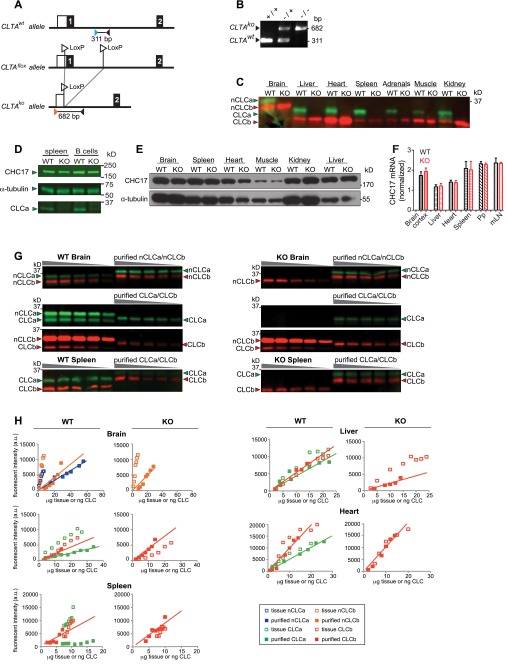
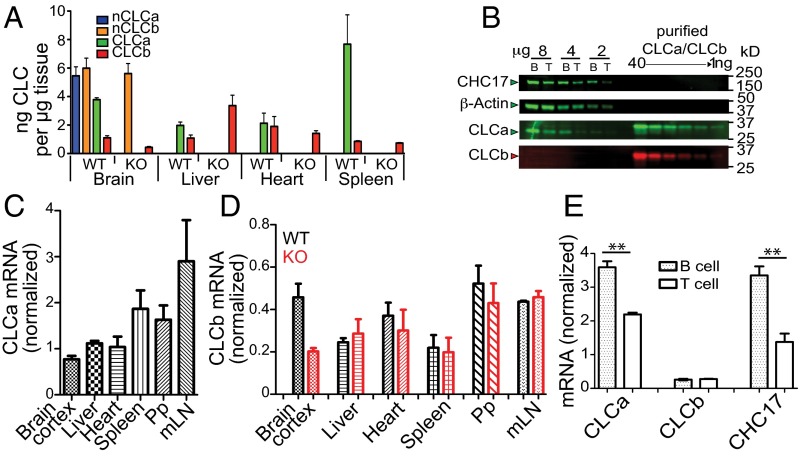
To investigate the physiological function of CLCs in vivo, we generated a CLCa-null heterozygote (CLTAko/+) by crossing mice with exon 1 of CLTA flanked by LoxP sites to mice expressing Cre recombinase under transcriptional control of the ACTB gene promoter (Fig. S1 A and B). Half of the homozygous CLTAko/ko mice produced by heterozygote mating died within a week of birth. Surviving CLTAko/ko homozygotes (KO mice) had no detectable CLCa protein in all tissues analyzed (Fig. S1 C and D), and loss of CLCa did not affect protein levels of the CHC17 subunit (Fig. S1 D and E). For WT and KO mice, we quantified the amount of each CLC isoform in brain, heart, liver, and spleen tissue by immunoblotting using isotype-specific antibodies (Fig. 1A and Fig. S1 G and H). As previously observed for bovine brain tissue (17), WT murine brain tissue expressed neuronal splice variants of both CLCa and CLCb in excess of the other spliced forms of CLCs. Brain, heart, and liver had equal amounts of CLCa and CLCb whereas spleen expressed predominantly CLCa. Analysis of purified B and T lymphocytes detected CLCa in these cells and no CLCb protein (Fig. 1B), suggesting that the small amount of CLCb detected in spleen derived from nonlymphoid cells. B cells had more clathrin than T cells, as revealed by CHC17 and CLCa levels (Fig. 1B). We did not see compensatory increase of CLCb protein levels in the KO mice, except in liver tissue (Fig. 1A). Tissue levels of mRNA encoding each isoform and CHC17 were consistent with protein levels detected (Fig. 1 C–E and Fig. S1F). Overall, these results indicate that there are two types of tissue, one with clathrin coats formed from triskelia with CLCa and CLCb, and the other in which CLCa is the predominant light chain of CCVs. Furthermore, expression of each CLC isotype seems to be independently regulated, such that B and T lymphocytes from the KO animals are effectively CLC-deficient.
Fig. S1.
Genetic deletion of CLCa in mice and quantification of CLC isoforms in murine tissues. (A) CLTAflox genetic construct for targeted deletion of CLTA by ACTB-Cre–mediated deletion of exon 1 flanked by LoxP sites (CLTAko). Exons are numbered and are indicated by black boxes. A white box indicates the 5′ untranslated region. Primers are designated by orange, blue, and black triangles. Black lines below each allele denote PCR products generated from the CLTA+ (311 bp) and CLTAko (682 bp) alleles. (B) PCR analysis of genomic DNA from WT (+/+, CLTA+/+), heterozygous (−/+, CLTAko/+), and homozygous knockout (KO) (−/−, CLTAko/ko) mice. (C) Indicated tissues from homozygous CLCa WT and KO (CLTAko/ko) littermates were homogenized and analyzed by immunoblotting for CLCa and CLCb using isoform-specific antibodies with CLCa in green and CLCb in red. nCLCa and nCLCb are the neuron-specific splice variants. Migration position of molecular mass of marker [kilodaltons (kDa)] is indicated on the right. (D) Total splenocytes and B cells isolated from spleens of WT or CLCa-null (KO) mice were lysed and analyzed by immunoblotting for proteins indicated by arrowheads. Migration positions of molecular mass of markers [kilodaltons (kDa)] is indicated on the right. (E) Indicated tissues from homozygous WT and KO littermates were homogenized and immunoblotted for CHC17 and α-tubulin. For each tissue analyzed, equal protein amounts of homogenate from WT or KO mice were compared. (F) Level of CHC17 mRNA measured by quantitative PCR analysis of the indicated tissues (mLN, mesenteric lymph node; Pp, Peyer’s patches) from WT and KO mice. Expression level was normalized to mRNA encoding hypoxanthine guanine phosphoribosyl transferase 1 (mean ± SEM of n = 3, except n = 2 for brain cortex). (G) Representative immunoblots for quantification of CLC isoforms in homogenates of brain tissue and spleen cells from WT and KO mice, detected using isoform-specific antibodies with CLCa in green and CLCb in red. nCLCa and nCLCb are the neuron-specific splice variants. Tissue samples were serially diluted and compared with serial dilutions of purified recombinantly produced CLCs to calibrate the blotting signal. The His-tag on the recombinant CLCs slows their migration compared with the untagged equivalents in the homogenates. Migration position of molecular mass markers [kilodaltons (kDa)] is indicated on the left. (H) Representative plots of fluorescence signals generated from isoform-specific immunoblots of the indicated tissues (no lines) or purified proteins (lines drawn for slope calculation). The concentrations reported in Fig. 1A were established from n = 3 of these analyses for each tissue.
Fig. 1.
CLC isoform expression in tissues from WT and CLCa-null mice. (A) Concentrations of CLC isoforms in indicated tissues from WT and CLCa-null (KO) mice (mean ± SEM, n = 3) determined by quantitative immunoblotting (Fig. S1 G and H). nCLCa and nCLCb are neuronal splice variants. (B) B and T lymphocytes isolated from WT murine spleens were lysed and analyzed by immunoblotting for expression of proteins indicated on the left. CLCa (green) and CLCb (red) signals were compared with dilutions of purified human CLCs (total protein per lane indicated above). Migration position of molecular mass markers [kilodaltons (kDa)] is indicated. (C) CLCa mRNA levels in indicated tissues from WT mice, normalized to mRNA levels for hypoxanthine guanine phosphoribosyl transferase 1 (HPRT1) (mean ± SEM, n = 3). mLN, mesenteric lymph node; Pp, Peyer’s patches. (D) CLCb mRNA levels in indicated tissues from WT and KO mice, normalized to mRNA levels for HPRT1 (mean ± SEM of n = 3, except n = 2 for brain cortex). (E) CLCa, CLCb, and CHC17 mRNA levels in B and T cells isolated from WT spleens, normalized to HPRT1 (mean ± SEM of n = 3, **P < 0.01; P values, unpaired t test).
Genetic Loss of CLCa Increases the Proportion of Germinal Center B Cells Expressing IgA but Reduces Numbers of Germinal Center B Cells.
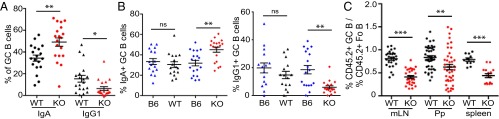
We focused on B-cell function in the KO animals to define the role of CLCs in vivo. A sign of B-cell function is the formation of germinal centers (GCs) in follicles within the spleen and other lymphoid organs, such as Peyer’s patches (Pp) and lymph nodes (LNs) (20). In GCs, B cells proliferate and differentiate to produce higher affinity antibodies and switch isotypes (21, 22). We found that KO mice had an increased frequency of IgA-expressing GC B cells in Pp compared with WT controls, as well as a variable but, on average, twofold reduction in IgG1-expressing B cells (Fig. 2A; gating shown in Fig. S2 A and B). To determine whether the observed isotype switch is an intrinsic B-cell defect or is caused by changes in the stroma, we studied mixed bone marrow (BM) chimeras produced by reconstitution of sublethally irradiated C57BL/6 mice (CD45.1+). For each transplant, KO or WT (CD45.2+) BM cells were combined 1:1 with WT C57BL/6 (CD45.1+CD45.2+) BM cells (B6), producing KO/B6 or WT/B6 mixed chimeras. The cotransplanted B6 (CD45.1+CD45.2+) cells served as a control for transplant variation, to which properties of the donor KO or WT (CD45.2+) cells were compared. As for full KO mice, the KO donor (CD45.2+) cells generated a greater proportion of IgA-expressing GC B cells but fewer IgG1-expressing GC B cells than cotransplanted B6 (CD45.1+CD45.2+) cells in KO/B6 mixed chimeras (Fig. 2B; gating shown in Fig. S2 C and D). In WT/B6 mixed chimeras, donor WT (CD45.2+) cells had similar proportions of IgA- and IgG1-expressing GC B cells to cotransplanted B6 (CD45.1+CD45.2+) cells (Fig. 2B). The contribution of KO cells to the GC B-cell population was about 50% of their contribution to the naive follicular compartment in three lymphoid organs examined (Fig. 2C; gating in Fig. S2C) whereas, in WT/B6 chimeras, the percentage of WT (CD45.2+) donor B cells in the GCs was similar to the follicular compartments (Fig. 2C). This reduced presence of KO B cells in the GCs suggested a defect in GC B-cell development or maintenance, upon CLCa loss. These data from BM chimeras further indicate that the perturbations in isotype switching by KO GC B cells are lymphocyte-intrinsic defects and that CLCa is required for normal isotype class switching to IgA and IgG1.
Fig. 2.
CLCa-null mice have an elevated frequency of IgA-expressing B cells and a reduced proportion of germinal center (GC) B cells. (A) Frequency of IgA- and IgG1-expressing GC B cells in Peyer's patches (Pp) of WT and CLCa-null (KO) littermates (mean ± SEM, n = 19, *P < 0.05, **P < 0.01; P values, unpaired t test). (B) Frequency of IgA- and IgG1-expressing GC B cells derived from C57BL/6 (B6) donor or KO and WT littermate donors in Pp of mixed chimeric mice transplanted with 50% WT B6 plus 50% KO or WT littermate bone marrow (mean ± SEM, n > 15 from more than four pairs of matched KO and WT donors; **P < 0.01, ns, not significant; P values, one-way ANOVA). (C) Percentage of donor (WT or KO CD45.2+) cells in the GC B compartment compared with their percentage in the corresponding follicular (Fo B) compartment in mixed bone marrow chimeras [mean ± SEM; n > 27 for mesenteric lymph node (mLN), n > 42 for Pp (33), n > 11 for spleen, **P < 0.01, ***P < 0.001; P values, unpaired t test].
Fig. S2.
Flow cytometry gating strategy for analysis of follicular and germinal center B cells. (A) Lymphocytes isolated from Peyer’s patches of WT and CLCa-null (KO) littermates were immunolabeled for the markers indicated. B220-positive cells were first gated on CD95high and IgDlow expression and then gated on GL7high expression to identify germinal center B cells (GC B). Follicular B cells (Fo B) were identified by CD95low and IgDhigh expression. (B) Representative flow cytometric analysis of the frequency of IgA- and IgG1-expressing GC B cells (gated as in A) in Peyer's patches of WT and KO littermates, detected by staining for each antibody isotype. (C) Mixed bone marrow chimeras (WT/B6 and KO/B6) were generated by injection of cells from WT or KO mice (CD45.2+) mixed in a 1:1 ratio with bone marrow cells from C57BL/6 (B6) mice (CD45.1+ CD45.2+) into irradiated recipient B6 mice (CD45.1+). GC and Fo B-cell populations from Peyer’s patches of chimeric animals were identified based on the gating scheme shown in A, and a representative analysis of the percentage of cells derived from each donor, assessed by labeling for CD45.1 and CD45.2 surface markers, is shown. (D) Representative flow cytometric analysis of the frequency of IgA- and IgG1-producing GC B cells in Peyer's patches shown for each donor population in the WT/B6 (Top pair) or KO/B6 (Bottom pair) mixed chimeras. For B–D, numbers indicate the percentage of cells in the respective quadrant or gate.
CLCa Regulates Internalization and Signaling of TGFβR2.
Selective in vivo deletion of TGFβR2 from murine B cells was shown to reduce IgA-expressing cells and increase IgG1-expressing B cells (23), suggesting that our KO phenotype might result from increased TGFβR2 signaling. Consistent with this hypothesis, B cells from Pp and spleen of KO mice had higher TGFβR2 surface levels than WT mice (Fig. 3A and Fig. S3A) whereas surface levels of the B-cell marker B220 were unchanged in KO mice (Fig. S3 A and B). Relative to cotransplanted B6 (CD45.1+CD45.2+) cells, elevated surface TGFβR2 was also observed for transplanted CD45.2+ KO B cells in the KO/B6 mixed chimeras, but not for B cells from the WT (CD45.2+) donor in WT/B6 chimeras (Fig. 3B and Fig. S3C). Again, a difference in surface level was not seen for B220 on KO-derived B cells (Fig. S3 C and D). These BM chimera phenotypes demonstrated that dysregulation of TGFβR2 surface expression is a cell-intrinsic defect and confirm that this defect does not affect B220.
Fig. 3.
CLCa regulates TGFβR2 internalization and signaling. (A) Mean fluorescence intensity (MFI) of TGFβR2 surface labeling of follicular (Fo) and germinal center (GC) B cells from Peyer’s patches (Pp) and total B cells from spleen of WT and CLCa-null (KO) littermates (n > 11 for Pp and n = 5 for spleen, *P < 0.05, **P < 0.01; P values, unpaired t test). (B) Ratio of the MFI of WT or KO donor cells (CD45.2+) to the MFI of B6 donor cells (CD45.1+CD45.2+) for surface labeling of TGFβR2 for Fo B and GC B from WT/B6 and KO/B6 chimeras (mean ± SEM of n = 10 from at least three pairs of different donors for mixed chimera mice,**P < 0.01, ***P < 0.001; P values, unpaired t test). (C) TGFβR1 and TGFβR2 mRNA levels in spleen B cells of WT and KO littermates, normalized to mRNA levels for hypoxanthine guanine phosphoribosyl transferase 1 (mean ± SEM of n = 3). (D) Lysates (equal protein loading) of B cells from WT and KO littermates were analyzed by immunoblotting for TGFβR2 and other proteins indicated by arrowheads on the left. Migration position of molecular mass markers [kilodaltons (kDa)] is indicated. (E) HEK293T cells were transiently transfected with TGFβR2-IRES-GFP and siRNA targeting CHC17 (blue), CLCa and CLCb (red, CLCab), or scrambled siRNA (control). Percent TGFβR2 internalization at 37 °C over time was quantified after labeling with primary antibody at 4 °C and using a secondary antibody to detect residual surface receptor relative to cells maintained at 4 °C (mean ± SEM of n = 5 independent experiments, ***P < 0.001; P values, two-way ANOVA followed by Bonferroni post test). (F) Internalization of endogenous transferrin receptor (TfR) at 37 °C over time analyzed by flow cytometry for cells treated as in E (mean ± SEM of n = 5 independent experiments, ***P < 0.001; P values, two-way ANOVA followed by Bonferroni post test). (G) Levels of pSmad2/3 and Smad2/3 in lysates of spleen or B cells from WT and KO littermates quantified by immunoblotting, normalized to CHC17 levels relative to total protein loaded (representative blots in Fig. S3 E and F) (n = 5 for total splenocytes and n = 2 for purified spleen B cells, *P < 0.05; P values, one-way ANOVA).
Fig. S3.
Expression of TGFβR2 and B220 detected by flow cytometry and analysis of TGFβR2 signaling. (A) Representative flow cytometry analysis of surface expression of TGFβR2 and B220 on germinal center (GC) and follicular (Fo) B cells from Peyer's patches (left) and total B cells from spleen (right) of WT and CLCa-null (KO) littermates (gating as in Fig. S2A). The x axes represent fluorescence intensities, and y axes are relative cell counts (normalized to the maximum cell count in each experiment). (B) Quantification of B220 surface fluorescent labeling from Peyer’s patches (left) and spleen (right) of WT and KO littermates. Data are presented as geometric mean fluorescent intensity (MFI) (n > 11 for Peyer’s patches and n = 7 for spleen. (C) Representative flow cytometry analysis of surface levels of TGFβR2 and B220 on GC B and Fo B cells in Peyer's patches from WT/B6 and KO/B6 chimera mice (produced as in Fig. S2C), separated by donor genotype (blue, B6; black, WT; red, KO; tinted peak, background staining). The x axes depict fluorescence intensities, and y axes depict cell count normalized to the maximum count. (D) The ratio of the MFI of WT or KO donor cells (CD45.2+) to the MFI of B6 donor cells (CD45.1+CD45.2+) for staining of B220 is shown for GC and Fo B cells (mean ± SEM of n = 10 from at least three pairs of different donors for WT/B6 and KO/B6 chimeras. (E and F) Total splenocytes and purified spleen B cells from WT and KO littermates were lysed and analyzed by immunoblotting for CHC17, CLCa, phosphorylated Smad2/3 (pSmad2/3) in E and total Smad2/3 in F. Representative blots are shown. Migration position of molecular mass markers is indicated [kilodaltons (kDa)].
In the KO B cells, increased TGFβR2 at the B-cell surface was not due to increased production of mRNA or protein (Fig. 3 C and D), suggesting that clathrin-mediated endocytosis of TGFβR2 (24, 25) is CLC-dependent. This pathway could not be studied using B cells from the KO mice because the TGFβR2 antibody signal was insufficient to assess receptor internalization. Therefore, we tested TGFβR2 uptake (Fig. 3E) in HEK293T cell lines transiently transfected with TGFβR2-IRES-GFP and cotransfected with siRNA targeting both CLCa and CLCb to recapitulate the CLC-deficient state of the KO B cells. TGFβR2 internalization was significantly reduced in CLC-depleted HEK293T cells compared with cells treated with nontargeting (control) siRNA, and we confirmed that TGFβR2 endocytosis was impaired in cells treated with siRNA targeting CHC17 to deplete all clathrin (24). In the same siRNA-treated TGFβR2-transfected cells, uptake of the endogenous transferrin receptor (TfR) was sensitive to CHC17 depletion but not CLC-dependent, as reported (3) (Fig. 3F). Together, our FACS and internalization studies indicate that TGFβR2, like several GPCRs (5), is CLC-dependent cargo for CCVs whereas B220 is like the CCV cargo whose surface expression is CLC-independent.
We assessed TGFβR2 signaling through Smad2/3 in the KO B cells because this signaling regulates class switch to IgA in B cells (23, 26–28). Smad signaling is initiated by phosphorylation of R-Smad2 and R-Smad3 when TGFβR1 binds a ligand from the TGFβ superfamily and then interacts with TGFβR2 (24). Consistent with activation of this pathway, we observed elevated levels of phospho-Smad2/3 in splenic B cells and total splenocytes isolated from KO mice relative to cells isolated from WT mice (Fig. 3G and Fig. S3 E and F). Thus, in the presence of endogenous concentrations of TGFβ, the higher levels of surface TGFβR2 lead to enhanced Smad signaling, establishing a molecular mechanism for the observed increase in B cells expressing IgA in the KO mice.
CLCa Influences Ligand-Induced Uptake of CXCR4, but Not CXCR5.
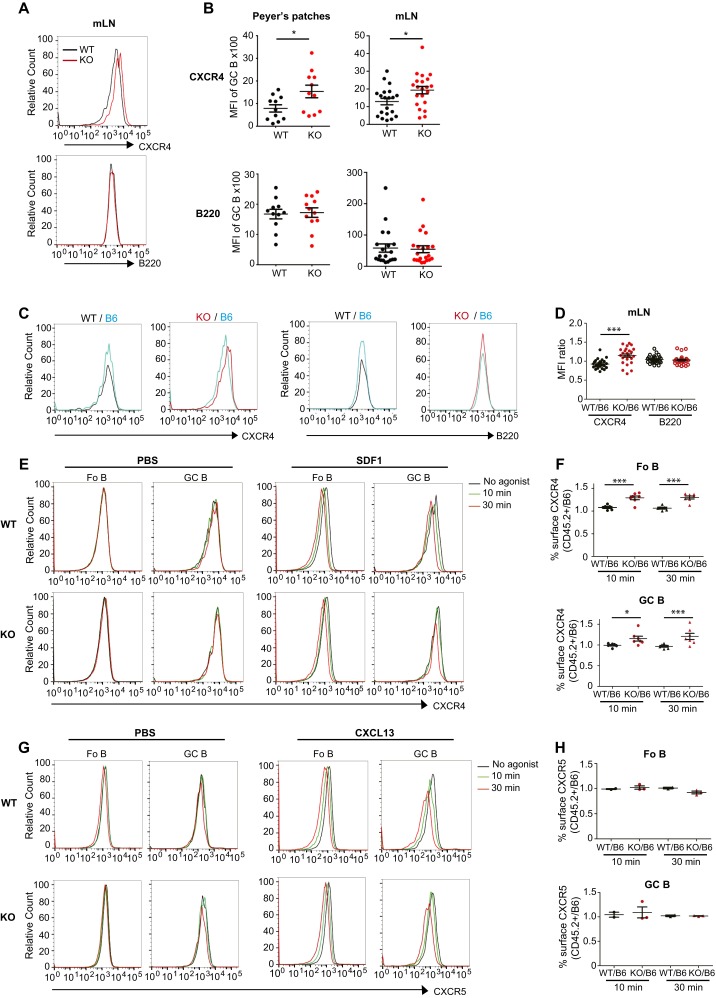
Because we observed increased surface TGFβR2 on KO B cells, we assessed surface levels of other receptors critical for B-cell function in GCs (29). GC B cells from Pp and mesenteric LNs (mLNs) of KO mice had higher surface levels of CXCR4, compared with equivalent B cells from WT controls, whereas B220 levels were not affected in the KO (Fig. S4 A and B), as previously noted (Fig. S3 A and B). Analysis of B cells from lymphoid compartments of BM chimeras also showed that donor KO (CD45.2+) cells had elevated CXCR4 levels whereas donor WT (CD45.2+) cells had similar levels (Fig. 4A and Fig. S4 C and D) compared with cotransplanted B6 (CD45.1+CD45.2+) cells. In contrast, neither KO nor WT (CD45.2+) donors displayed increased surface CXCR5 or B220 (Fig. 4 A and B). Increased surface CXCR4 was explained by reduced internalization in response to stromal cell-derived factor 1 (SDF1 or CXCL12) observed for both GC and follicular B cells from the mLNs of KO mice (Fig. 4C and Fig. S4E) compared with equivalent WT cells. Upon SDF1 exposure, KO-derived B cells from BM chimeras had consistently higher CXCR4 levels than cotransplanted B6 cells whereas WT-derived GC and follicular cells had levels comparable with cotransplanted B6 cells (Fig. S4F). In contrast, B cells from KO mice exposed to CXCR5 ligand (CXCL13) showed no difference in the internalization response compared with B cells from WT mice (Fig. 4D and Fig. S4G), and KO-derived B cells showed CXCR5 levels comparable with B6-derived donor cells from BM chimeras after CXCL13 treatment, indicating no difference in CXCR5 uptake behavior by KO cells (Fig. S4H).
Fig. S4.
Assessment of CXCR4 and CXCR5 surface levels and internalization by flow cytometry. (A) Representative flow cytometry analysis of surface levels of CXCR4 or B220 on germinal center (GC) B cells from mesenteric lymph nodes (mLNs) of WT and CLCa-null (KO) littermates. (B) Mean fluorescence intensities (MFIs) of CXCR4 and B220 surface labeling of GC B cells from Peyer’s patches and mLN from multiple animals as in A (n > 11 for Peyer’s patches, n > 20 for mLN; *P < 0.05; P values, unpaired t test, KO compared with WT mice). (C) Representative flow cytometry analysis of surface levels of CXCR4 or B220 on GC B cells from Peyer’s patches in WT/B6 and KO/B6 chimeras, separated by donor genotype (blue, B6; black, WT; red, KO). (D) Ratios of the mean fluorescence intensity (MFI) of CXCR4 on WT or CLCa-null (KO) donor cells (CD45.2+) to CXCR4 on C57BL/6 (B6) (CD45.1+CD45.2+) donor cells for germinal center (GC) B cells from mLNs of WT/B6 and KO/B6 chimeras (mean ± SEM; n > 24, from at least three pairs of different donors for mixed chimera mice, ***P < 0.001; P values, unpaired, t test). (E) B cells isolated from mLNs of WT and KO littermates were treated with PBS for 30 min at 37 °C (no agonist) or 100 ng/mL SDF1 stromal cell-derived factor 1 (SDF1) for 10 or 30 min at 37 °C and then immediately chilled, and CXCR4 surface levels on GC and follicular (Fo) B cells were assessed by flow cytometry. One representative experiment is shown. The x axes represent fluorescence intensities, and y axes represent cell count normalized to the maximum count. (F) B cells isolated from WT/B6 or KO/B6 chimeras were treated with 100 ng/mL stromal cell-derived factor 1 (SDF1) for 10 or 30 min at 37 °C and then immediately chilled, and CXCR4 surface levels on GC and Fo B cells were assessed by flow cytometry. Ratio of MFIs of surface CXCR4 on WT or KO donor cells (CD45.2+) to surface CXCR4 on B6 donor cells (CD45.1+CD45.2+) from each chimera is shown for the indicated time points (mean ± SEM of n = 6 WT and n = 7 KO mice from four independent experiments, *P < 0.05, ***P < 0.001; P values, two-way ANOVA with Bonferroni post tests). (G) B cells isolated from the mLNs of WT and KO littermates were treated with PBS for 30 min at 37 °C (no agonist) or 1 μg/mL CXCL13 for 10 or 30 min at 37 °C, after which they were immediately chilled, and CXCR5 surface levels on GC and Fo B cells were assessed by flow cytometry. One representative experiment is shown. The x axes indicate fluorescence intensities, and y axes represent relative cell counts normalized to the maximum count. (H) B cells isolated from WT/B6 or KO/B6 chimeras were treated with CXCL13 for 10 or 30 min at 37 °C, after which they were immediately chilled, and CXCR5 surface levels on GC and Fo B cells were assessed by flow cytometry. Ratio of MFIs of surface CXCR5 on WT or KO donor cells (CD45.2+) to surface CXCR5 on B6 donor cells (CD45.1+CD45.2+) from each chimera is shown for the indicated time points (mean ± SEM of n = 2 and n= 3 KO mice from two independent experiments).
Fig. 4.
CLCa-null B cells have increased CXCR4 surface levels and impaired ligand-induced CXCR4 internalization. (A) Ratios of the mean fluorescence intensity (MFI) of CXCR4 and B220 on WT or CLCa-null (KO) donor cells (CD45.2+) to MFIs on C57BL/6 (B6) (CD45.1+CD45.2+) donor cells for germinal center (GC) B cells from lymphoid tissues indicated from WT/B6 and KO/B6 chimeras (mean ± SEM; n = 42 for Peyer’s patches, n > 11 for spleen, from at least three pairs of different donors for mixed chimera mice, **P < 0.01, ***P < 0.001; P values, unpaired t test). (B) MFI ratios for CXCR5 and B220 on WT or KO donor follicular (Fo B) or GC B cells (CD45.2+) relative to B6 donor B-cell populations (CD45.1+CD45.2+) from mesenteric lymph nodes (mLNs) (mean ± SEM; n > 11). (C) Percent CXCR4 internalized at 37 °C over time after addition of SDF1 (relative to surface CXCR4 on control cells similarly treated with PBS) for Fo B and GC B cells from mLNs of WT or KO mice (mean ± SEM of n = 5 WT and n = 6 KO from two independent experiments, *P < 0.05; P values, two-way ANOVA with Bonferroni post tests). (D) Percent CXCR5 internalized at 37 °C over time after addition of CXCL13 (relative to surface CXCR5 on control cells similarly treated with PBS) for Fo B and GC B cells from mLNs of WT or KO mice (mean ± SEM of n = 5 WT and n = 5 KO mice, from two independent experiments).
CLC Depletion from Cell Lines Establishes Selectivity for Different CCV Cargo.
Finding that CLCa regulates the B-cell surface levels of TGFβR2 and CXCR4, but not CXCR5 and B220, inspired us to investigate CLC dependence for uptake of other cargoes. Given that three GPCRs were previously shown to depend on CLCs for uptake (5), we focused on two additional GPCRs, β2-adrenergic receptor and δ-opioid receptor, known to depend on clathrin for uptake (30, 31), but for which the role of CLCs had not been examined. Using siRNA, HEK293 cells that stably express either the N-terminally FLAG-tagged β2-adrenergic receptor (F-β2AR) or the N-terminally FLAG-tagged δ-opioid receptor (F-DOR) were made CLC-deficient by CLCa and CLCb knockdown, leaving residual CHC17, or made completely clathrin-deficient by CHC17 knockdown (Fig. S5 A and B). Ligand-induced internalization of the tagged receptors was then assessed. CLC depletion, as well as CHC17 depletion, impaired ligand-induced endocytosis of F-DOR (Fig. 5A and Fig. S5C). As expected, uptake of endogenous TfR, although sensitive to CHC17 depletion, was not affected by CLC depletion (Fig. 5B and Fig. S5D). Surprisingly, ligand-induced endocytosis of F-β2AR was not affected by CLC depletion, similar to the behavior of TfR in the same cells, although internalization of both receptors was affected by CHC17 depletion (Fig. 5 C and D and Fig. S5 E and F). Together, our data reveal that CLCs are required for uptake of a subset of signaling receptors, such as TGFβR2, CXCR4, and DOR, but are dispensable for internalization of other receptors, such as β2AR and CXCR5.
Fig. S5.
siRNA treatment of HEK293 stable transfectants expressing FLAG-tagged G protein-coupled receptors and representative flow cytometry data for receptor internalization assays. (A and B) HEK293 cells that stably expressed (A) FLAG-tagged β2-adrenergic receptor (FLAG-β2AR) or (B) FLAG-tagged δ-opioid receptor (FLAG-DOR) were treated with the siRNA targeting CHC17 or CLCa and CLCb (CLCab) or scrambled siRNA (Control) for 72 h, and then cell lysates were analyzed by immunoblotting for levels of the indicated proteins on the left. Migration position of molecular mass markers is indicated at the right [kilodaltons (kDa)]. (C and E) Representative flow cytometry analysis of receptor levels detected with anti-FLAG antibody on HEK293 cells stably expressing FLAG-tagged δ-opioid receptor (FLAG-DOR) or FLAG-tagged β2-adrenergic receptor (FLAG-β2AR) at indicated times after treatment at 37 °C with agonists (DPDPE for FLAG-DOR cells or isoproterenol for FLAG-β2AR cells). Black, time 0; green, 5 min; red, 20 min. Before exposure to agonist, cells were pretreated for 72 h with control siRNA or siRNA targeting CHC17 or CLCab, as indicated above each plot. The x axes indicate fluorescence intensities, and y axes represent relative cell counts normalized to the maximum count. (D and F) Representative flow cytometry analysis of endogenous transferrin receptor (TfR) surface levels on the siRNA-treated stable transfectants analyzed, respectively, in C and E (not agonist-treated), labeled with primary antibody to TfR at 4 °C at time 0, detected with secondary antibody at the indicated times after incubation at 37 °C. The x axes indicate fluorescence intensities, and y axes represent relative cell counts normalized to the maximum count.
Fig. 5.
CLC depletion impairs δ-opioid receptor internalization but does not affect β2-adrenergic receptor uptake. (A) Percent FLAG-tagged δ-opioid receptor (FLAG-DOR) or (B) transferrin receptor (TfR) internalized at 37 °C over time by stable transfectants expressing FLAG-DOR, treated with indicated siRNA targeting CHC17 (blue), CLCa and CLCb (red, CLCab), or scrambled siRNA (black, control) (mean ± SEM of n = 4 independent experiments, **P < 0.01, ***P < 0.001; P values, two-way ANOVA followed by Bonferroni post test). (C) Percent FLAG-tagged β2-adrenergic receptor (FLAG-β2AR) or (D) TfR internalized at 37 °C over time by stable transfectants expressing FLAG-β2AR, treated with siRNA indicated as in A and B (mean ± SEM of n = 3 independent experiments, **P < 0.01, ****P < 0.0001; P values, two-way ANOVA followed by Bonferroni post test). FLAG-tagged receptor uptake was measured on cells treated with agonist (DPDPE for FLAG-DOR and isopreteronol for FLAG-β2AR) relative to cells treated with PBS, and TfR uptake was relative to untreated cells labeled with primary antibody maintained at 4 °C.
Discussion
We produced a CLCa knockout mouse to probe the physiological function of CLCs. This analysis of clathrin subunit function in vertebrates by gene deletion defines a key role for CLCs in selection of cargo for CCVs in vivo. B cells from CLCa-null mice are effectively CLC-deficient because CLCa expression is dominant in lymphoid cells and their phenotypes revealed that cargoes requiring CLCs for endocytosis are important for B-cell function. Reduced internalization of TGFβR2 by CLCa-null B cells increased Smad signaling at the plasma membrane, generating a higher percentage of IgA-producing B cells in KO animals compared with WT. Though IgA switching is a known consequence of TGFβ signaling (23, 26–28), the role of TGFβR2 endocytosis in regulating signaling has been debated (24, 32). Our results support a role for CLC-dependent clathrin-mediated endocytosis in attenuating TGFβR2 signaling because increased surface levels had the functional effect of promoting isotype switching. Our results further suggest that increased switching to IgA in the Peyer’s patch occurs at the expense of switching to IgG1. We speculate that the elevated TGFβR signaling caused by CLCa loss dominates over signaling from factors that promote IgG1 switching (e.g., IL4), which may have been most evident in the Peyer’s patch where sufficient amounts of TGFβ are available to stimulate elevated surface TGFβRs.
The reduced frequency of germinal center (GC) B cells in CLCa-null mice could be explained by increased surface expression and reduced internalization of CXCR4, which plays a role in cell distribution within the GC (29). Additionally, CLCa deficiency might affect internalization of sphingosine-1-phosphate receptor 2 (S1PR2), a GPCR that negatively regulates GC B-cell numbers (33), but lack of reagents to track S1PR2 surface expression prevented us from testing this hypothesis. Thymocytes from the KO mice also had elevated levels of CXCR4, but no obvious changes in T-cell populations in spleen, lymph nodes, or bone marrow were observed (not shown). These results are consistent with the possibility that T cells compensate more readily for surface receptor changes, having more redundant regulatory pathways than B cells as a result of their more ancient origins (34).
Loss of CLCa from B cells did not affect CXCR5 levels or internalization. Thus, CLCs are dispensable for uptake of some GPCRs, as further observed in transfected cells, displaying CLC-dependent internalization of DOR, but not β2AR. Our findings plus earlier observations that CLCs are required for internalization of some GPCRs (5), but not several classical CCV cargoes (3, 4), suggest that CLCs are needed for endocytosis depending on the type and/or configuration of cargo present in a clathrin-coated pit. Variations in cargo crowding and local lipid organization affect membrane-bending properties, which in turn require different coat properties for deformation into a vesicle (35). CLCs contribute tensile strength to the clathrin lattice through controlling the rigidity of the hub region of the triskelion (8, 9) and, in cells, by recruiting the actin-regulating Hip proteins (2, 6, 8, 15). Thus, CLCs may contribute to uptake of cargoes that pose particular membrane-bending challenges. Analogously, Sec13p in COPII-coated vesicles contributes structural rigidity that is required to bend membranes with asymmetric cargo, such as GPI-anchored receptors (36). Two GPCR cargoes (μ-opioid receptor and CXCR4) that require CLCs for endocytosis are modified by ubiquitin, which attracts specific endocytic adaptors to promote uptake by clathrin-coated pits (5, 31, 37). Such aggregation, mediated by ubiquitination or other pathways, is likely to change membrane-bending properties that could then require extra tensile strength provided by CLCs in the clathrin coat. Mutagenesis studies suggest that CLC phosphorylation by GRK2 may contribute to regulation of CLC’s role in GPCR cargo selection (5). Whether CLC phosphorylation is also involved in uptake of the CLC-dependent receptors identified here is not yet established. However, CLC-dependent TGFβR internalization was observed in the absence of ligand engagement without associated signaling.
Past studies indicated that vertebrate tissues have characteristic patterns of CLCa and CLCb expression (17, 19). These studies used methods (SDS/PAGE analysis) that could not distinguish forms of CLCa and CLCb that comigrate. Using isotype-specific antibodies, calibrated with recombinantly expressed protein, we show here that mice have tissues that express either equal amounts of CLCa and CLCb (nonlymphoid) or predominantly CLCa (lymphoid). A remaining mystery is why, within all vertebrates, CLCa and CLCb genetic clades are clearly distinguishable and each highly conserved (1). Recently, expression of an alternatively spliced variant of CLCa during postnatal murine heart development was reported (18), suggesting that CLCa has specialized tissue functions, which may explain the 50% postnatal mortality rate of the CLCa-null mice. It is notable that, of the cell types analyzed here, lymphocytes are distinguished from other differentiated cell types by their lack of CLCb, which must change the nature of their clathrin. Major distinctions between lymphocytes and the other tissues examined are the lack of adherens junction or tight junction formation by lymphocytes and their persistent motility, as well as extensive cortical actin. Yeast with similar properties function with only one CLC, which is most homologous to CLCa and is a key player in endocytosis through its association with the actin-binding Hip homolog Sla2p, which suggests that CLCa could confer a specialized actin-interacting activity that is critical for clathrin function in lymphocytes. Indeed, both lymphocyte endocytosis and immune synapse formation depend on cooperation between clathrin and actin (38, 39). An alternative possible explanation for the conservation of CLC isoforms is that CLCa and CLCb influence cargo selection differently, which could also have tissue-specific and physiological consequences.
Materials and Methods
Production of CLCa-Null Mice.
Our BAC-targeting construct, produced by homologous recombination in Escherichia coli, contained PGKneo flanked by FRT sites and a loxP site in intron 1 of the CLTA gene, with a second loxP site cloned into the 5′ UTR. ES cells were targeted by transfection and G418 selection to produce a floxed CLTA gene (CLTAflox) and then transfected with Flpe-recombinase to excise the PGKneo cassette. Targeted ES cells were injected into C57BL/6 mouse blastocysts and transferred into foster mothers. Chimeric offspring were mated with C57BL/6 females, and germ-line transmission of the CLTAflox allele was established. CLTAflox/+ heterozygotes were mated to ACTB-Cre deleter mice, excising exon 1 to generate the CLTA-null allele (CLTAko). Heterozygous CLTAko/+ mice were backcrossed onto the C57BL/6 background and bred to produce CLTAko/ko homozygous mice (Fig. S1B). The University of California, San Francisco (UCSF) Institutional Animal Care and Use Committee approved all animal experiments.
Mixed Bone Marrow Chimeras.
Mixed bone marrow (BM) chimeric mice were generated by i.v. transfer of a 50:50 mixture of BM cells from two different lines of mice into CD45.1+ congenic C57BL/6 mice (3 × 106 to 5 × 106 cells transferred per mouse). Fifty percent of the transferred BM was derived from CLTAko/ko (CD45.2+) (KO) mice or CLTA+/+ CD45.2+congenic WT C57BL/6 (WT) mice and 50% was derived from CD45.1+CD45.2+ congenic WT C57BL/6 (B6), to generate KO/B6 or WT/B6 chimeric bone marrow mice. Before transfer, recipient mice were sublethally irradiated (2 × 450 rad, 3 h apart). Mice were analyzed at least 8 wk after reconstitution.
Splenic B-Cell and T-Cell Purification.
Splenic B and T cells were purified by negative selection using magnetic beads. For B-cell purification, spleen cells were exposed to anti-CD43–coated beads (MACS; Miltenyi Biotech). For T-cell purification, spleen cells were exposed to a mixture of biotin-conjugated antibodies (anti-Ter119, anti-B220, anti-CD19, anti-CD11b, anti-Gr1, and anti-CD11c), followed by binding to Streptavidin MicroBeads (MACS; Miltenyl Biotech). Cell purity was confirmed by FACS.
Flow Cytometry.
Spleen, mesenteric lymph node, and Peyer's patch cells were isolated and labeled for FACS analysis as described previously (29). Antibodies used for GC B-cell identification, Ig isotype detection, mixed bone chimera analysis, and labeling of TGFβR2, CXCR4, and CXCR5 are specified in SI Materials and Methods.
Internalization Assays.
Internalization assays measured residual receptors remaining on the cell surface after incubation at 37 °C. Percent internalization was established relative to surface labeling of cells maintained at 4 °C for the duration of the experiment or relative to cells with no ligand treatment at 37 °C, as specified. The background signal of secondary labeling was subtracted separately from experimental and control signals. See SI Materials and Methods for a detailed protocol.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism (GraphPad Software, Inc.). Parametric data were analyzed using two-tailed Student t tests and one-way or two-way ANOVA, followed by Bonferroni post hoc tests for multiple comparisons as appropriate (95% confidence interval).
Additional Methods.
Details of siRNAs, plasmids, and methods for tissue culture, transfection, quantitative immunoblotting, and mRNA quantification are in SI Materials and Methods.
SI Materials and Methods
Mice.
C57BL/6 (B6) mice were purchased from The Jackson Laboratory. CD45.1+ congenic B6 mice were obtained from the National Cancer Institute (01B96; B6-LY5.2/Cr). CD45.1+CD45.2+ mice were generated by crossing B6 and Boy/J (002014; B6.SJL-Ptprc a Pepc b/BoyJ; The Jackson Laboratory) mice. Mice were bred and maintained at the Laboratory Animal Research Center at the University of California, San Francisco (UCSF). The UCSF Institutional Animal Care and Use Committee approved all animal experiments.
siRNAs and Plasmids.
siRNA duplexes were synthesized by Qiagen. Targeting sequences for CHC17, CLCa, and CLCb were as published (2, 3). Human TGFβR2 (subcloned from pCMV5B-TGFbeta receptor II wt (N-term HA), Addgene plasmid no. 24801) was cloned in front of IRES-GFP in the vector PCL6IEGWO, and named as TGFβR2-IRES-GFP (40).
Tissue Culture and Transfections.
HEK293 cells were cultured in DMEM, supplemented with 10% (vol/vol) FBS (HyClone) and antibiotics. For siRNA treatment, cells were plated at ∼70% confluency and the following day were transfected with siRNA using the jetPRIME reagent (Polyplus-transfection), according to the manufacturer’s protocol. Cells were analyzed 72 h after transfection. For TGFβR2 internalization assays, HEK293T cells were cotransfected with siRNA and TGFβR2-IRES-GFP using the jetPRIME reagent (Polyplus-transfection).
Immunoblotting.
Transfected HEK293 cells were solubilized in lysis buffer [50 mM Tris (pH 7.2), 150 mM NaCl, 20 mM EDTA, 1% Triton X-100, and EDTA-free protease inhibitors (Roche)], and lysate was produced by centrifugation to remove nuclei. The protein content of lysates was determined using the Pierce BCA Protein Assay kit (23225; Thermo Scientific). Proteins were resolved on precast SDS/PAGE gels [4–12% (wt/vol) acrylamide; Invitrogen], transferred onto nitrocellulose membranes (Bio-Rad), and detected by labeling with primary and then secondary antibodies. Blots were scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences) or by using the ECL system (Amersham Biosciences) according to the manufacturers’ protocols.
Amounts of the CLC isoforms in tissues were determined by quantitative immunoblotting using a previously described protocol (19). Murine tissues were first homogenized in lysis buffer (150 mM NaCl, 1 mM EGTA, 10 mM Hepes, 0.5 mM MgCl2, 0.02% NaN3, 0.05% PMSF, pH 7.2) containing EDTA-free protease inhibitors (Roche) on ice and centrifuged at 1,000 × g for 30 min at 4 °C. Resulting supernatants were boiled for 10 min and centrifuged at 10,000 × g for 10 min at 4 °C. This protocol leaves boiling-resistant CLCs in solution and causes CHC and almost all other proteins to precipitate. To analyze lymphocyte subpopulations, purified splenic B and T cells were incubated in lysis buffer and then centrifuged to produce supernatant, which was not boiled. Protein concentrations of boiled or unboiled tissue or cell lysate supernatants (homogenates) were determined. Homogenates were then reduced in 4× sample buffer and alkylated by addition of 20 mM iodoacetamide for 1 h at room temperature. Serial dilutions of known concentrations of homogenates and of purified recombinantly expressed His-tagged human CLC proteins were run next to one another on 12% (wt/vol) polyacrylamide-SDS gels and then transferred onto nitrocellulose membranes (Bio-Rad). Membranes were first incubated for 1 h at room temperature with Odyssey blocking buffer (LI-COR Biosciences) diluted 1:1 in PBS and then incubated in primary antibodies overnight, followed by incubation with secondary antibodies for 1–2 h after three to four washes. Signal intensities of immunoblot bands were quantified using ImageJ. The amount of CLC in each tissue or cell homogenate was then determined using a standard curve produced from signals of adjacent purified protein samples quantified in the same way. Analysis was restricted to signals from dilutions in the linear range with an R2 value of at least 0.8, except for spleen homogenate where values of 0.73 were used due to the limited availability of samples for titrations. For quantification of pSmad2/3 and Smad2/3, the signal intensities were normalized to the intensity of the CHC17 signals per total protein of lysate.
The following primary antibodies, at the noted concentrations and dilutions, were used: mouse monoclonal antibody (mAb) anti-CHC17 (2 μg/mL, TD.1) (41); mAb anti-CLCa (5 μg/mL, X16) (41) or rabbit anti-CLCa (1:1,000; Proteintech); rabbit anti-CLCb (1:1,000; Proteintech); mAb anti-CLCb (2 μg/mL, LCB.1) (41); mouse anti–β-actin (1:5,000; Sigma); mouse anti–α-tubulin (1:10,000; Sigma); biotin-conjugated goat anti-mouse TGFβR2 (1:1,000; R&D); biotin-conjugated mouse anti–α-tubulin (1:3,000; eBiosciences); rabbit anti-pSmad2/3 (1:1,000; Cell Signaling Tech); and rabbit anti-Smad2 (1:1,000; Life Technologies). Primary antibodies were detected with the following secondary antibodies or streptavidin at the dilutions indicated: donkey anti-rabbit IgG IRDye700DX (1:10,000; Rockland), donkey anti-mouse IgG IRDye800 (1:10,000; Rockland), and Streptavidin IRDye 800 (1:10,000, LiCor). For detection of anti-pSmad2/3 and anti-Smad2, stabilized goat anti-rabbit IgG HRP-conjugated antibody (1:4,000; Pierce) was used.
Flow Cytometry.
DAPI or Fixable Viability Dye eFluor 780 (eBioscience) was used to exclude the dead cells. For GC B-cell identification, cells were labeled with APC-Cy7- or PerCPCy5.5-conjugated anti-B220 (clone RA3-6B2; BioLegend), PerCPCy5.5- or Pacific Blue-conjugated anti-IgD (clone IA6-2; BioLegend), PE-Cy7–conjugated anti-CD95 (clone Jo2; BD Biosciences), and FITC or Alexa Fluor 647-conjugated anti-GL7 antigen (T- and B-cell activation antigen or Ly-77, clone GL7; BioLegend). For Ig isotype detection, the following antibodies were used: anti-IgG1 (Bio-IgG1; BioLegend) with Streptavidin-Brilliant Violet 605 (BioLegend) or FITC-IgG1 (BioLegend) and anti-IgA (PE-IgA; Southern Biotech). To analyze mixed bone marrow chimeras, Pacific Blue-, PE-, FITC-, BV605-conjugated anti-CD45.1 (clone A20; BioLegend) or anti-CD45.2 (clone 104; BioLegend) antibodies were used. For labeling TGFβR2 on murine B cells, PE-conjugated (FAB532P; R&D) or biotin-conjugated (BAF532; R&D) anti-mouse TGFβR2 antibodies were used. Streptavidin-Brilliant Violet 605 (BioLegend) was used to detect the biotin-conjugated primary antibody. For CXCR4 and CXCR5 labeling, biotin-labeled rat anti-mouse CXCR4 (BD Biosciences) and anti-mouse CXCR5 (BD Biosciences) antibodies were used. Streptavidin-Brilliant Violet 605 (BioLegend) was used to detect the biotin-conjugated primary antibody. Data were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo (Tree Star).
Internalization Assays.
To study internalization of DOR and β2AR, stable transfectants of HEK293 cells expressing FLAG-tagged versions of these receptors (30, 31) were first washed with serum-free medium and then incubated with serum-free medium for 15 min at 37 °C. Agonist ligand (10 μM DPDPE for DOR or isoproterenol for β2AR) diluted in serum-free medium was then added to the cells at specified time points, after which cells were placed at 37 °C and allowed to internalize the receptor for the indicated times. Control cells were left untreated and remained at 37 °C. Then, agonist-treated and control cells were immediately placed on ice and washed once with cold PBS, after which they were labeled with anti-FLAG M1 antibody (Sigma) directly conjugated to Alexa Fluor-647 (using a kit from Life Technologies) for 30 min at 4 °C. Cells were transferred to tubes by gentle pipetting, washed once with 4 °C PBS, and then suspended in 0.2% BSA in PBS at 4 °C for FACS analysis.
For analysis of transferrin receptor internalization, stably transfected HEK293 cells above were serum starved and then labeled with mouse anti-TfR antibody (1:300; BD) diluted in 0.2% BSA in PBS for 30 min at 4 °C. Cells were then placed on ice, washed twice with 4 °C PBS, and then maintained in 0.2% BSA in PBS at 4 °C for the remainder of the experiment. Cells were removed from ice and placed at 37 °C for specified time points to allow labeled receptor to internalize, after which cells were placed back on ice and then incubated with Alexa Fluor-555 goat anti-mouse Ig antibody (1:500; Life Technologies) for 30 min at 4 °C. Cells were transferred to tubes by gentle pipetting, washed once with 4 °C PBS, and then suspended in 0.2% BSA in PBS for FACS analysis.
For CXCR4 or CXCR5 internalization by mesenteric lymph node (mLN) B cells, the mLNs were harvested into prewarmed internalization buffer (RPMI supplemented with 0.5% fatty acid-free BSA, 10 mM Hepes, glutamine, and antibiotics). Lymphocytes were then isolated by mechanical disaggregation through an 80-μm nylon sieve at room temperature. After two washes with internalization buffer, cells were resuspended in internalization buffer at 107/mL. Cells were incubated in a 37 °C water bath for 30 min. Then, 90-μL cell suspensions were added to tubes containing 10 μL of internalization buffer only, SDF1 (1 μg/mL), or CXCL13 (10 μg/mL) to reach the working concentration of SDF1 (100 ng/mL) or CXCL13 (1 μg/mL). Cells were incubated for indicated times, and then 100 μL of 4 °C internalization buffer was added to the tubes, which were immediately placed on ice. After internalization, cells were pelleted, washed, and labeled with biotinylated anti-CXCR4 (Bio-CXCR4, 1:100; BD Biosciences) or biotinylated anti-CXCR5 (Bio-CXCR5, 1:50; BD Biosciences). After staining with Streptavidin-Brilliant Violet 605 (1:200; BioLegend), the cells were analyzed by FACS. DAPI labeling was used to exclude dead cells by gating.
For TGFβR2 internalization, HEK293T cells cotransfected with plasmid encoding TGFβR2-IRES-GFP and siRNA were labeled with goat anti-human TGFβR2 antibody (1:250; R&D Systems) diluted in DMEM supplemented with 10 mM Hepes for 30 min on ice and then washed twice with 4 °C DMEM plus Hepes and placed at 37 °C for specified times to allow labeled receptor to internalize, after which cells were placed on ice and then incubated with Alexa Fluor-568–conjugated secondary donkey anti-goat IgG antibody (1:500; Life Technologies) for 30 min on ice in the dark and washed twice with 4 °C DMEM plus Hepes. Cells were transferred to tubes by gentle pipetting, washed once with 4 °C PBS, and then suspended in 0.2% BSA in PBS plus DAPI for FACS analysis. GFP-positive cells were gated to identify cells for TGFβR2 internalization analysis.
mRNA Quantification.
Total RNA was isolated with the RNeasy Micro Kit or RNeasy Mini Kit (Qiagen). cDNA was synthesized by using MMLV reverse transcriptase and random primers (Life Technologies). Real-time PCR was carried out using a StepOnePlus real-time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems) and the appropriate primer pairs. Relative mRNA abundance of target genes was determined by subtracting the threshold cycle for the internal reference mRNA encoding hypoxanthine guanine phosphoribosyl transferase 1 (HPRT1) from that of the target. The primer pairs used for real-time PCR were as follows: for CLTA, forward primer 5′-ATGCTGTTGACGGAGTGATGA-3′ and reverse primer 5′-CCACTTACGGATACTTTCAGGCT-3′; for CLTB, forward primer 5′-GAAAGCGAGATTGCTGGCATC-3′ and reverse primer 5′-CGTTAGCCTCCTGAAACACATC-3′; for CLTC (gene encoding CHC17), forward primer 5′- AGATTCTGCCCATTCGCTTTC-3′ and reverse primer 5′-TCAGTGCAATCACTTTGCTGG-3′; and for HPRT1, forward primer 5′-AGGTTGCAAGCTTGCTGGT-3′ and reverse primer 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′ (Integrated DNA Technologies).
Acknowledgments
We thank Nigel Killeen (formerly UCSF) for help with mouse production and Mark von Zastrow (UCSF) for cell lines. This work was supported by NIH Grants GM038093 (to F.M.B.), AI45073 (to J.G.C.), AI17892 (to P.P.), T32 GM07175 (to S.R.M.), and NCI F31 CA171594 (to S.R.M.), an Arthritis Foundation postdoctoral fellowship (to T.M.E.), and Wellcome Trust Investigator Award 107858/Z/15/Z (to F.M.B.). J.G.C. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611189113/-/DCSupplemental.
References
- 1.Brodsky FM. Diversity of clathrin function: New tricks for an old protein. Annu Rev Cell Dev Biol. 2012;28:309–336. doi: 10.1146/annurev-cellbio-101011-155716. [DOI] [PubMed] [Google Scholar]
- 2.Majeed SR, et al. Clathrin light chains are required for the gyrating-clathrin recycling pathway and thereby promote cell migration. Nat Commun. 2014;5:3891. doi: 10.1038/ncomms4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279(16):16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 4.Poupon V, et al. Clathrin light chains function in mannose phosphate receptor trafficking via regulation of actin assembly. Proc Natl Acad Sci USA. 2008;105(1):168–173. doi: 10.1073/pnas.0707269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira F, et al. Endocytosis of G protein-coupled receptors is regulated by clathrin light chain phosphorylation. Curr Biol. 2012;22(15):1361–1370. doi: 10.1016/j.cub.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 6.Bonazzi M, et al. Clathrin phosphorylation is required for actin recruitment at sites of bacterial adhesion and internalization. J Cell Biol. 2011;195(3):525–536. doi: 10.1083/jcb.201105152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cureton DK, Massol RH, Whelan SP, Kirchhausen T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010;6(9):e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilbur JD, et al. Conformation switching of clathrin light chain regulates clathrin lattice assembly. Dev Cell. 2010;18(5):841–848. doi: 10.1016/j.devcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dannhauser PN, et al. Effect of clathrin light chains on the stiffness of clathrin lattices and membrane budding. Traffic. 2015;16(5):519–533. doi: 10.1111/tra.12263. [DOI] [PubMed] [Google Scholar]
- 10.Chen CY, Brodsky FM. Huntingtin-interacting protein 1 (Hip1) and Hip1-related protein (Hip1R) bind the conserved sequence of clathrin light chains and thereby influence clathrin assembly in vitro and actin distribution in vivo. J Biol Chem. 2005;280(7):6109–6117. doi: 10.1074/jbc.M408454200. [DOI] [PubMed] [Google Scholar]
- 11.Legendre-Guillemin V, et al. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280(7):6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- 12.Newpher TM, Idrissi FZ, Geli MI, Lemmon SK. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol Biol Cell. 2006;17(10):4343–4352. doi: 10.1091/mbc.E06-07-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreij AM, et al. LRRK2 localizes to endosomes and interacts with clathrin-light chains to limit Rac1 activation. EMBO Rep. 2015;16(1):79–86. doi: 10.15252/embr.201438714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11(8):1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13(9):1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonazzi M, et al. A common clathrin-mediated machinery co-ordinates cell-cell adhesion and bacterial internalization. Traffic. 2012;13(12):1653–1666. doi: 10.1111/tra.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodsky FM, Parham P. Polymorphism in clathrin light chains from different tissues. J Mol Biol. 1983;167(1):197–204. doi: 10.1016/s0022-2836(83)80041-2. [DOI] [PubMed] [Google Scholar]
- 18.Giudice J, et al. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat Commun. 2014;5:3603. doi: 10.1038/ncomms4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acton SL, Brodsky FM. Predominance of clathrin light chain LCb correlates with the presence of a regulated secretory pathway. J Cell Biol. 1990;111(4):1419–1426. doi: 10.1083/jcb.111.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cyster JG. B cell follicles and antigen encounters of the third kind. Nat Immunol. 2010;11(11):989–996. doi: 10.1038/ni.1946. [DOI] [PubMed] [Google Scholar]
- 21.Green JA, Cyster JG. S1PR2 links germinal center confinement and growth regulation. Immunol Rev. 2012;247(1):36–51. doi: 10.1111/j.1600-065X.2012.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Germinal center B cells govern their own fate via antibody feedback. J Exp Med. 2013;210(3):457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cazac BB, Roes J. TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13(4):443–451. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 24.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5(5):410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell H, Choudhury A, Pagano RE, Leof EB. Ligand-dependent and -independent transforming growth factor-beta receptor recycling regulated by clathrin-mediated endocytosis and Rab11. Mol Biol Cell. 2004;15(9):4166–4178. doi: 10.1091/mbc.E04-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein J, et al. B cell-specific deficiency for Smad2 in vivo leads to defects in TGF-beta-directed IgA switching and changes in B cell fate. J Immunol. 2006;176(4):2389–2396. doi: 10.4049/jimmunol.176.4.2389. [DOI] [PubMed] [Google Scholar]
- 27.van Ginkel FW, et al. Partial IgA-deficiency with increased Th2-type cytokines in TGF-beta 1 knockout mice. J Immunol. 1999;163(4):1951–1957. [PubMed] [Google Scholar]
- 28.Gros MJ, Naquet P, Guinamard RR. Cell intrinsic TGF-beta 1 regulation of B cells. J Immunol. 2008;180(12):8153–8158. doi: 10.4049/jimmunol.180.12.8153. [DOI] [PubMed] [Google Scholar]
- 29.Bannard O, et al. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 2013;39(5):912–924. doi: 10.1016/j.immuni.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temkin P, et al. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13(6):715–721. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry AG, White IJ, Marsh M, von Zastrow M, Hislop JN. The role of ubiquitination in lysosomal trafficking of δ-opioid receptors. Traffic. 2011;12(2):170–184. doi: 10.1111/j.1600-0854.2010.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CL, et al. Inhibitors of clathrin-dependent endocytosis enhance TGFbeta signaling and responses. J Cell Sci. 2009;122(Pt 11):1863–1871. doi: 10.1242/jcs.038729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muppidi JR, et al. Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma. Nature. 2014;516(7530):254–258. doi: 10.1038/nature13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 35.Stachowiak JC, Brodsky FM, Miller EA. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat Cell Biol. 2013;15(9):1019–1027. doi: 10.1038/ncb2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Copic A, Latham CF, Horlbeck MA, D’Arcangelo JG, Miller EA. ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p. Science. 2012;335(6074):1359–1362. doi: 10.1126/science.1215909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 38.Calabia-Linares C, et al. Endosomal clathrin drives actin accumulation at the immunological synapse. J Cell Sci. 2011;124(Pt 5):820–830. doi: 10.1242/jcs.078832. [DOI] [PubMed] [Google Scholar]
- 39.Stoddart A, et al. Lipid rafts unite signaling cascades with clathrin to regulate BCR internalization. Immunity. 2002;17(4):451–462. doi: 10.1016/s1074-7613(02)00416-8. [DOI] [PubMed] [Google Scholar]
- 40.Bosch J, et al. Comparing the gene expression profile of stromal cells from human cord blood and bone marrow: Lack of the typical “bone” signature in cord blood cells. Stem Cells Int. 2013;2013:631984. doi: 10.1155/2013/631984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Näthke IS, et al. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68(5):899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]