Significance
Cancer progression typically involves inactivation of the p53 tumor suppressor. In testicular teratocarcinoma, there exist unusually high protein levels of wild-type p53, but strikingly, without normal activation of genes up-regulated by p53, such as cell cycle arrest and DNA repair pathways. This suggests that posttranslational mechanisms, such as protein modification, may repress activity of the high levels of wild-type p53. Our findings demonstrate an important role in teratocarcinoma of previously characterized repressive p53 methylation, which down-regulates normal p53 activation functions. The results suggest the potential for novel approaches to reactivate wild-type p53 in teratocarcinoma via therapeutic inhibition of the methyltransferase enzymes that modify p53, which could be similarly appropriate for treatment of other cancers expressing high levels of wild-type p53.
Keywords: p53, methylation, teratocarcinoma, cancer, pluripotency
Abstract
TP53 (which encodes the p53 protein) is the most frequently mutated gene among all human cancers, whereas tumors that retain the wild-type TP53 gene often use alternative mechanisms to repress the p53 tumor-suppressive function. Testicular teratocarcinoma cells rarely contain mutations in TP53, yet the transcriptional activity of wild-type p53 is compromised, despite its high expression level. Here we report that in the teratocarcinoma cell line NTera2, p53 is subject to lysine methylation at its carboxyl terminus, which has been shown to repress p53’s transcriptional activity. We show that reduction of the cognate methyltransferases reactivates p53 and promotes differentiation of the NTera2 cells. Furthermore, reconstitution of methylation-deficient p53 mutants into p53-depleted NTera2 cells results in elevated expression of p53 downstream targets and precocious loss of pluripotent gene expression compared with re-expression of wild-type p53. Our results provide evidence that lysine methylation of endogenous wild-type p53 represses its activity in cancer cells and suggest new therapeutic possibilities of targeting testicular teratocarcinoma.
The potent tumor suppressor p53 protein is encoded by the TP53 gene, which is genetically mutated in nearly half of human cancers (1). The remaining cases of cancer that retain wild-type TP53 gene often use various alternative mechanisms to interfere with wild-type p53 tumor-suppressive function. For example, amplification of the MDM2 gene encoding a negative regulator of p53 is found in multiple tumor types without p53 mutation to keep a low expression level of wild-type p53 protein (2, 3). Recently, posttranslational modifications on p53 have emerged as an additional mechanism to modulate p53 transcriptional activity. These modifications can either be activating or repressing to p53 transcriptional activity (4, 5). Among them, methylation of carboxyl-terminal lysines, in particular, monomethylation at K370 (K370me1, catalyzed by the methyltransferase SMYD2) and monomethylation at K382 (K382me1, catalyzed by the methyltransferase PR-Set7, encoded by KMT5A) have been associated with repression of p53 function (6, 7). A physiological role of repressive p53 lysine methylation was revealed through quantitative time-lapse microscopy of single cells, indicating that lysine methylation attenuates p53 transcriptional activity upon intrinsic transient damage during normal cell cycles and prevents p53 from inducing cell-cycle arrest (8). This mechanism can likely be coopted by cancer cells to suppress wild-type p53 activity. However, to our knowledge, there have been few studies demonstrating the functional importance of lysine methylation in repressing p53 activity in the context of cancer.
Teratocarcinomas consist of embryonal carcinoma stem cells and various layers of differentiated cells. It is one type of testicular germ cell tumor, and therefore indicates that true pluripotent cells have become cancerous. Teratocarcinoma has been historically linked to p53 because the cancerous cells express high levels of p53. In fact, when p53 was initially misinterpreted as an oncogenic tumor antigen, teratocarcinoma served as an illustration of cancer with elevated “oncogene” expression (9–12). It is now well recognized that p53 is a master tumor suppressor, yet it continues to be mysterious that p53 is rarely mutated in teratocarcinoma (13). Wild-type p53 appears to be transcriptionally compromised in teratocarcinoma despite high expression levels (13); hence, apparently, there is an absence of selection pressure for p53 mutations. However, upon stress signals, p53 can become rapidly activated (13, 14), suggesting an otherwise suppressed basal activity that can be switched on or overcome by stress signals. Although several explanations have been proposed, indicating a repressive domain of p53 that acts in trans (15), or micro RNAs that function to interfere with p53 downstream pathways (16), the mechanism of p53 repression in teratocarcinoma remains largely elusive.
Here we propose that carboxyl-terminal lysine methylation on p53 contributes to the repression of endogenous wild-type p53 activity in teratocarcinoma cells. Our results provide a mechanism of wild-type p53 repression in teratocarcinoma. Other types of cancer with wild-type p53 may use similar mechanisms to repress p53 tumor-suppressive activity. Hence, our findings may suggest potential new therapeutic opportunities for reactivating wild-type p53 in teratocarcinoma, as well as other cancers.
Results
Elevated SMYD2 and PR-Set7 Levels in NTera2 Cells.
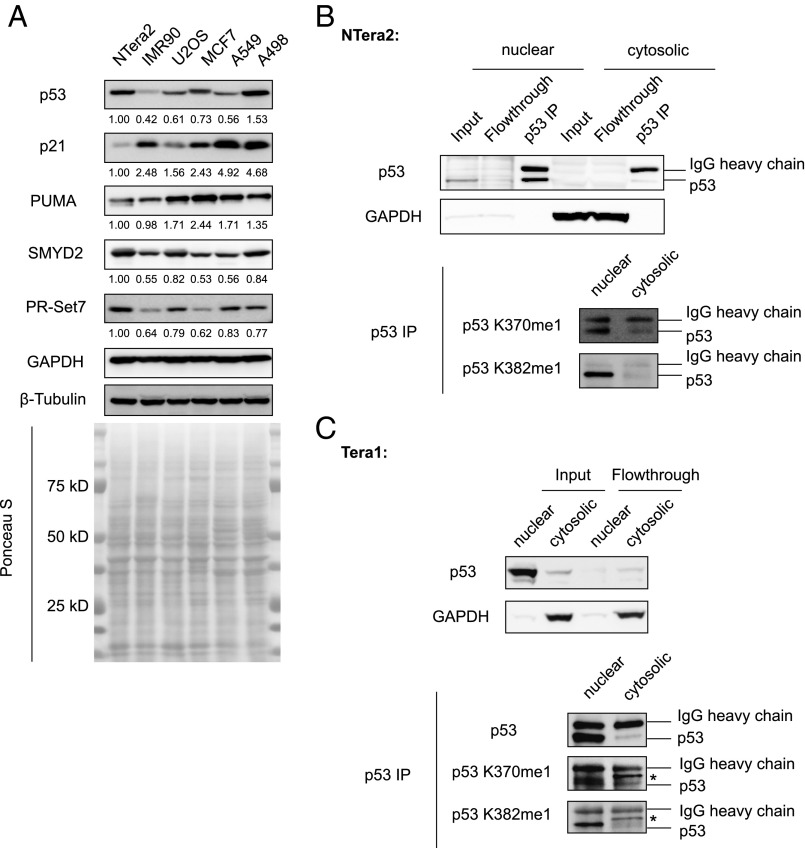
We first performed Western blot analyses in the teratocarcinoma cell line NTera2 and compared protein levels in parallel with multiple cell lines bearing wild-type p53. As previously noted, the teratocarcinoma cell line NTera2 has higher protein levels of p53 than that in most other wild-type p53 cell lines we examined, including a primary lung fibroblast line IMR90 and cancer cell lines U2OS, MCF7, A549, and A498 from various tissues of origin (with the exception of A498 cells having comparable amount of p53 expression level) (Fig. 1A). However, the expression levels of canonical p53 targets, including CDKN1A (also known as p21) and PUMA, are the lowest in NTera2 compared with these other cell lines (Fig. 1A). These observations are consistent with previous reports indicating high levels of transcriptionally compromised wild-type p53 in teratocarcinoma cells (13, 14).
Fig. 1.
p53 is monomethylated at K370 and K382 in teratocarcinoma cells. (A) Western blot analysis and quantification of protein levels in NTera2 and other cell lines with wild-type p53. For each blot, protein amount was set to 1 in NTera2 cells, and relative protein levels were denoted in other cell lines for comparison. (B) Nuclear-cytosolic fractionation and examination of p53 methylation in NTera2 cells by p53 immunoprecipitation, followed by p53 K370me1 and p53 K382me1 Western blot analysis. (C) Nuclear-cytosolic fractionation and examination of p53 methylation in Tera1 cells by p53 immunoprecipitation followed by p53 K370me1 and p53 K382me1 Western blot analysis. *Nonspecific band.
We then examined protein levels of the p53 methyltransferases SMYD2 and PR-Set7. Intriguingly, among all these cell lines we examined, NTera2 cells appeared to have the highest expression level of PR-Set7 (Fig. 1A). Similarly, the amount of SMYD2 protein in NTera2 cells is higher than that in most other cell lines, except for a comparable amount in A498 cells (Fig. 1A). Elevated p53 methyltransferases protein levels in NTera2 cells suggest a potential role of corresponding p53 methylation in regulating p53 activity.
p53 Is Monomethylated at K370 and K382 in Teratocarcinoma Cells.
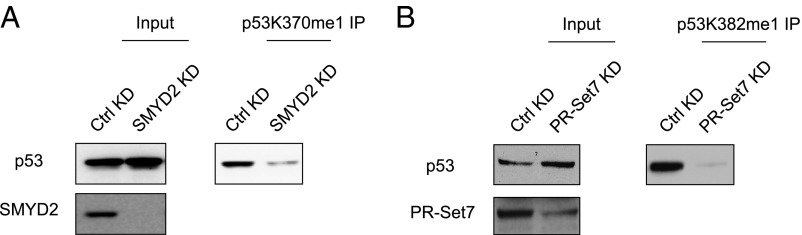
The elevated protein levels of SMYD2 and PR-Set7 led us to examine whether p53 is correspondingly monomethylated at K370 and K382 in NTera2 cells. We separated the nuclear fraction from the cytosolic fraction of NTera2 cells and found that p53 is localized predominantly in the nucleus (Fig. 1B), consistent with previous reports and indicating that the compromised p53 transcriptional activity is not simply a result of deregulation of p53 cellular compartmentalization (13). We then immunoprecipitated total p53 and detected both K370me1 and K382me1 signals in the nuclear fraction (Fig. 1B). A consistent pattern was observed in another teratocarcinoma cell line, Tera1: that p53 mainly resides in the nucleus, and that p53 monomethylation was detected at both K370 and K382 (Fig. 1C). We decreased the protein level of SMYD2 in NTera2 cells using short-hairpin RNA (shRNA)-mediated gene knockdown and observed a concomitant reduction of p53 K370me1 signal (Fig. 2A). Similarly, reduction of PR-Set7 resulted in decreased p53 K382me1 level (Fig. 2B). We conclude that p53 is methylated in NTera2 cells and that p53 methylation is dependent on the methyltransferase protein levels.
Fig. 2.
p53 methylation at K370 and K382 is dependent on the methyltransferases SMYD2 and PR-Set7. (A) p53 K370me1 immunoprecipitation followed by p53 Western blot analysis in NTera2 cells with control (Ctrl) or SMYD2 knockdown mediated by shRNA. (B) p53 K382me1 immunoprecipitation followed by p53 Western blot analysis in NTera2 cells with control (Ctrl) or PR-Set7 knockdown mediated by shRNA.
SMYD2 or PR-Set7 Knockdown Activates p53 Transcriptional Activity and Promotes a Differentiation Feature of NTera2 Cells.
p53 function is rigorously regulated in pluripotent cells to keep a balance between self-renewal and differentiation (17, 18). Increased p53 activity generally leads to enhanced differentiation phenotype, mainly through induction of the cell cycle arrest pathways (19, 20). As a result, reducing the activity of p53 improves the efficiency of generating induced pluripotent stem cells (21–25). In the context of cancer, the absence of p53 activity has also been linked to stem cell transcriptional signatures (26). Consistently, it has been previously inferred that repressed p53 activity is required for the maintenance of teratocarcinoma pluripotency and that activated p53 correlates with the loss of stemness (13, 14).
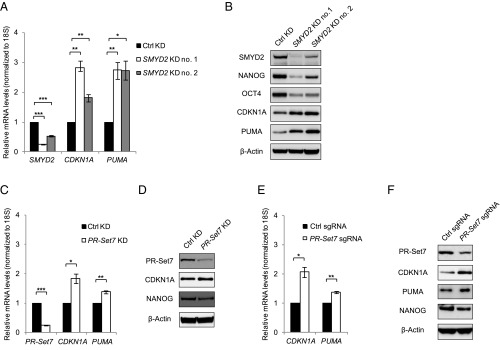
To investigate the functional importance of lysine methylation to p53-mediated transcriptional activity, we tested whether decreasing the level of p53 methyltransferases affects the expression of p53 downstream targets. Reduction of SMYD2 protein levels using two independent shRNA constructs resulted in increased expression of p53 target genes CDKN1A and PUMA, measured by both mRNA levels (Fig. 3A) and protein levels (Fig. 3B), and indicative of enhanced p53 transcription activity. Further, reduction of SMYD2 levels also led to decreased expression of pluripotent genes NANOG and OCT4 (also known as POU5F1) (Fig. 3B), suggesting a precocious loss of cell pluripotency that is consistent with the augmented p53 activity.
Fig. 3.
SMYD2 or PR-Set7 knockdown activates p53 transcriptional activity and promotes a differentiation feature of NTera2 cells. (A) Reverse transcription followed by qPCR (RT-qPCR) analysis of mRNA levels in NTera2 cells with control (Ctrl) or SMYD2 knockdown mediated by shRNA. No. 1 and no. 2 indicate two different shRNA constructs. (B) Western blot analysis of protein levels in NTera2 cells with control or SMYD2 knockdown mediated by shRNA. (C) RT-qPCR analysis of mRNA levels in NTera2 cells with control or PR-Set7 knockdown mediated by shRNA. (D) Western blot analysis of protein levels in NTera2 cells with control or PR-Set7 knockdown mediated by shRNA. (E) RT-qPCR analysis of mRNA levels in NTera2 cells with control sgRNA or sgRNA targeting PR-Set7 for gene “knockdown” at a cell population level. (F) Western blot analysis of protein levels in NTera2 cells with control sgRNA or sgRNA targeting PR-Set7 for gene “knockdown” at a cell population level. (Error bars represent mean ± SEM; n = 3; two-tailed Student’s t test: *P < 0.05; **P < 0.02; ***P < 0.01).
Similarly, we assessed the effect of PR-Set7 knockdown using shRNA and observed increased CDKN1A and PUMA expression (Fig. 3 C and D), as well as decreased NANOG level (Fig. 3D). To further demonstrate the role of PR-Set7 and to rule out off-target effects that a single shRNA construct may have, we used the CRISPR/Cas9 system (27) to achieve genome editing and gene “knockdown” at the cell population level (gene knockout in a subpopulation of the entire pool of cells). Along with the reduction of PR-Set7 at the average level, CDKN1A was also elevated and NANOG level was slightly decreased compared with a control small guide RNA (sgRNA) targeting a silent genomic region (Fig. 3 E and F). These results were consistent with the shRNA-mediated gene knockdown experiment, and therefore further support a role of PR-Set7 in p53 suppression, which in turn contributes to sustaining the pluripotent state of NTera2 cells.
Methylation-Deficient p53 Mutants Phenocopy the Effect of Methyltransferase Knockdown.
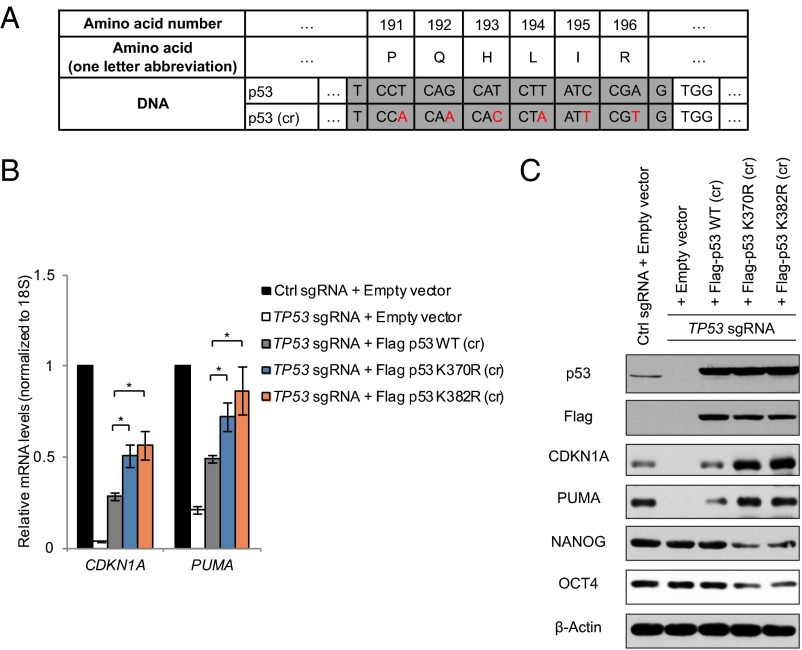
To demonstrate a direct role of lysine methylation specifically on p53 itself in repression of transcriptional activation of p53 target genes, we generated substituted forms of p53 with lysine 370 or lysine 382 mutated to arginine (p53 K370R or p53 K382R, respectively), and therefore unable to be methylated by SMYD2 or PR-Set7. We reconstituted expression of Flag-tagged wild-type or the mutant forms of p53 in an otherwise p53-depleted background by CRISPR/Cas9 mediated genome editing. The ectopic wild-type or mutant p53 were engineered with silent mutations in the region corresponding to sgRNA sequence, and hence were not targeted by the Cas9 enzyme cutting (Fig. 4A). CRISPR/Cas9-mediated TP53 gene knockout resulted in a complete loss of p53 protein, as well as great reduction in expression of p53 target genes CDKN1A and PUMA, compared with that with a control sgRNA (Fig. 4 B and C). Transient expression of wild-type p53 into p53-inactivated cells, as expected, partially rescued the expression of CDKN1A and PUMA (Fig. 4 B and C). Strikingly, compared with wild-type p53, the expression of p53 K370R or p53 K382R resulted in further increased expression of CDKN1A and PUMA at both mRNA and protein levels (Fig. 4 B and C). Consistently, p53 K370R or p53 K382R mutant also led to a reduction of NANOG and OCT4 levels (Fig. 4C), indicative of precocious differentiation, and is in line with previous results when SMYD2 or PR-Set7 methyltransferase levels were decreased (Fig. 3 B, D, and F). Taken together, these experiments further demonstrate that p53 monomethylation on K370 and K382 repress p53 transcriptional activity and contribute to the pluripotent state of NTera2 cells.
Fig. 4.
Methylation-deficient p53 mutants phenocopy the effect of methyltransferase knockdown. (A) Design of p53 with silent mutations in sequence targeted by p53 sgRNA therefore is resistant to CRIPSR/Cas9 enzyme cutting. cr, CRISPR-resistant. (B and C) NTera2 cells with control sgRNA or sgRNA targeting TP53 gene were transfected with vector control or vector expression CRISPR-resistant versions of p53 as indicated in A. RT-qPCR analysis (B) or Western blot analysis (C) was performed to examine mRNA levels or protein levels, respectively. (Error bars represent mean ± SEM; n = 3; two-tailed Student’s t test: *P < 0.05; **P < 0.02; ***P < 0.01).
Discussion
Here we show a key role of repression of p53 in cancer cells via posttranslational modification of the p53 protein. We discovered that, in the teratocarcinoma cell line NTera2, p53 is transcriptionally compromised at least in part through monomethylation at K370 and K382 via the modification enzymes SMYD2 and PR-Set7, respectively (Figs. 2 and 3). High levels of p53 normally lead to reduced proliferation because of p53-mediated activation of transcription of genes whose role is to slow cell cycle progression. However, although p53 is expressed at a high level in NTera2 cells, because of these modifications of p53 at K370me1 and K382me1, p53 has lowered activity; hence, NTera2 cells rapidly proliferate with stem-cell-like properties. We show that either reduction of these enzymes or direct substitution mutation of either K370 or K382 within p53 reactivates p53 and leads to escape from a stem-cell-like cancer state into a more differentiated state (Figs. 3 and 4). The fact that either reduced SMYD2 or PR-Set7, which methylate specific lysines in p53, or substitution of their cognate lysine methylation sites to arginine (which cannot be methylated by these enzymes) lead to similar outcome of induced p53-activated genes strongly implicates p53 as the key target of the enzymes in NTera2 cells.
Current management of testicular cancer is largely mediated by combined chemotherapeutics (28). Effective treatments and high cure rate can be achieved partly as a result of endogenous wild-type p53 activation (29), suggesting that chemotherapy induction of p53 can overcome the repressive mechanism under basal conditions. Our findings suggest an alternative possibility that p53 can be reactivated in certain cancers by inhibiting p53 modifying enzymes, including SMYD2 and PR-Set7. There is much effort currently underway to develop inhibitors of epigenetic enzymes as anticancer therapeutics. One group of targets is epigenetic enzymes that are highly up-regulated in cancers. In this regard, SMYD2 and PR-Set7 both are of therapeutic interest (30, 31), in part because of their roles in p53 modifications (6, 7, 32). Our results add further importance to SMYD2 and PR-Set7 inhibitors as potential anticancer drug targets.
These results reveal a posttranslational modification mechanism that mediates p53 repression in teratocarcinoma and potentially other tumors with wild-type p53. Lysine methylation, among other potential posttranslational modifications, could represent a general paradigm whereby these modifications play important roles in regulating p53 activity in cancer; in particular, cancer types that have low p53 mutation frequencies, such as leukemia, cervical cancer, kidney cancer, melanoma, and prostate cancer (1). Other posttranslational modifications, especially those that repress p53 activity, are yet to be discovered and may provide additional opportunities to reactivate wild-type p53 in cancer.
Materials and Methods
Cell Culture.
The NTera2 cells were obtained from American Type Culture Collection and were cultured in a 37 °C incubator at 20% (vol/vol) oxygen. NTera2 were maintained in complete DMEM culture medium [DMEM with 10% (vol/vol) FBS, 100 units/mL penicillin, and 100 mg/mL streptomycin] supplied with sodium pyruvate. MCF7, A549, and A498 cells were maintained in complete DMEM culture medium in a 37 °C incubator at 20% (vol/vol) oxygen. IMR90 cells were maintained in complete DMEM culture medium in a 37 °C incubator at 3% (vol/vol) oxygen. U2OS cells were maintained in complete McCoy’s culture medium [McCoy’s with 10% (vol/vol) FBS, 100 units/mL penicillin, and 100 mg/mL streptomycin] in a 37 °C incubator at 20% (vol/vol) oxygen.
Western Blot and Antibodies.
Cells were lysed in modified RIPA buffer containing 150 mM NaCl, 1% Nonidet P-40, 50 mM Tris⋅Cl at pH 8.0, and 1% SDS, supplemented with protease inhibitors (Life Technologies, no. 78446) before use. Protein concentration was determined by BCA protein assay (Life Technologies, no. 23227), following which equal amount of proteins were loaded and separated in polyacrylamide gels. Proteins were then transferred to nitrocellulose membrane and assessed by corresponding antibodies. Quantifications were performed using the MultiGauge software (Fujifilm). p53 K370me1 antibody was generated and characterized as previously reported (6). p53 K382me1 antibody was generated and tested similarly. Other antibodies used in this study were as follows: SMYD2 (Abcam, ab108712), PR-Set7 (abcam, ab3798), NANOG (abcam 21624), OCT4 (abcam ab19857), CDKN1A/p21 (Abcam, ab7960), PUMA (Cell Signaling Technology, number 4976), p53 monoclonal antibody DO-1 (Calbiochem EMD), Flag (Sigma, M2, F1804).
Immunoprecipitation.
Cells were lysed in buffer containing 20 mM Tris at pH 8.0, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% Nonidet P-40, with protease and phosphatase inhibitors, and 12.5 U⋅mL−1 benzonase (Novagen, 70746), at 4 °C for 1 h. The lysate was then added onto Protein G beads that had antibody conjugated. After overnight incubation at 4 °C, the immunoprecipitated product was washed, eluted, and subjected to Western blot analysis.
Nuclear-Cytosolic Fractionation.
In brief, cells were first lysed in buffer containing 10 mM Tris at pH 7.9, 1.5 mM MgCl2, 10 mM KCl, and freshly prepared 0.5 mM DTT, 50 mM β-glycerol-phosphate, as well as protease inhibitors, to isolate cytoplasmic extract. The nuclear extract was then further lysed in buffer containing 20 mM Tris at pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 25% (vol/vol) glycerol, 200 mM EDTA, and freshly prepared 0.5 mM DTT, 50 mM β-glycerol-phosphate, as well as protease inhibitors. Both cytoplasmic and nuclear extracts were then subjected to ultracentrifugation for 1 h at 60,000 × g. Extracts were collected for downstream analyses.
RNA Isolation and RT-qPCR.
RNA was isolated from cells using RNeasy kit (Qiagen, no. 74106), with a DNase I (Qiagen, no. 79254) digestion step to minimize genomic DNA contamination. RNA was then reverse transcribed to cDNA (Life Technologies, number 4387406), and then quantitative PCR (qPCR) was performed for quantification using standard procedures on a 7900HT Fast-Real-Time PCR platform (ABI). Primer sequences are available in Table S1.
Table S1.
Primer sequences for RT-qPCR
| Sequence (5′ to 3′) | Name |
| AGCGATGGAACTTCGACTTTG | CDKN1A F |
| CGAAGTCACCCTCCAGTGGT | CDKN1A R |
| CTCAGGAAAGGCTGTTGTGCT | PUMA F |
| AGTGTCACCCCTGCAGCTG | PUMA R |
| GTAACCCGTTGAACCCCATT | 18S rRNA F |
| CCATCCAATCGGTAGTAGCG | 18S rRNA R |
| TCATAGCTGTTGCCCCAATG | SMYD2 F |
| TCTGCCAGGGTCCCTTTGTA | SMYD2 R |
| GATTCCACCAATGCAGCCA | PR-Set7 F |
| CTTGATGGGCTTTTTCAGGG | PR-Set7 R |
Gene Knockdown and CRISPR/Cas9 Genome Editing.
All sequences for shRNA-mediated gene knockdown and for CRISPR/Cas9-mediated genome editing are available in Table S2. The control short-hairpin is scrambled nontargeting sequence. The control sgRNA targets the nonexpressed PRM2 gene to control for Cas9-induced DNA damage.
Table S2.
Targeted sequences by shRNA and sgRNA
| Sequence (5′ to 3′) | Name |
| GCTCTGTGTTTGAGGACAGTA | SMYD2 no.1 shRNA |
| CGATATTTCCTGATGTTGCAT | SMYD2 no.2 shRNA |
| CGCAACAGAATCGCAAACTTA | PR-Set7 shRNA |
| ACTGAGTTCTCTTCCTGAA | PR-Set7 sgRNA |
| TCCTCAGCATCTTATCCGAG | TP53 sgRNA |
Acknowledgments
We thank J. Huang for help on optimizing the use of p53 methylation antibodies and D. Shultz and the high-throughput screening core at the University of Pennsylvania for assistance in shRNA-mediated gene knockdown experiments. This study is supported by NIH Grant R01 CA078831 (to S.L.B.) and a Postdoctoral Fellowship from the American Cancer Society (to M.A.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610387113/-/DCSupplemental.
References
- 1.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 3.Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993;53(12):2736–2739. [PubMed] [Google Scholar]
- 4.Berger SL. Keeping p53 in check: A high-stakes balancing act. Cell. 2010;142(1):17–19. doi: 10.1016/j.cell.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai C, Gu W. p53 post-translational modification: Deregulated in tumorigenesis. Trends Mol Med. 2010;16(11):528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 7.Shi X, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27(4):636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142(1):89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bártková J, et al. p53 protein alterations in human testicular cancer including pre-invasive intratubular germ-cell neoplasia. Int J Caner. 1991;49(2):196–202. doi: 10.1002/ijc.2910490209. [DOI] [PubMed] [Google Scholar]
- 10.Chandrasekaran K, Mora PT, Nagarajan L, Anderson WB. The amount of a specific cellular protein (p53) is a correlate of differentiation in embryonal carcinoma cells. J Cell Physiol. 1982;113(1):134–140. doi: 10.1002/jcp.1041130122. [DOI] [PubMed] [Google Scholar]
- 11.Oren M, Reich NC, Levine AJ. Regulation of the cellular p53 tumor antigen in teratocarcinoma cells and their differentiated progeny. Mol Cell Biol. 1982;2(4):443–449. doi: 10.1128/mcb.2.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich NC, Oren M, Levine AJ. Two distinct mechanisms regulate the levels of a cellular tumor antigen, p53. Mol Cell Biol. 1983;3(12):2143–2150. doi: 10.1128/mcb.3.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutzker SG, Levine AJ. A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nat Med. 1996;2(7):804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- 14.Curtin JC, et al. Retinoic acid activates p53 in human embryonal carcinoma through retinoid receptor-dependent stimulation of p53 transactivation function. Oncogene. 2001;20(20):2559–2569. doi: 10.1038/sj.onc.1204370. [DOI] [PubMed] [Google Scholar]
- 15.Curtin JC, Spinella MJ. p53 in human embryonal carcinoma: Identification of a transferable, transcriptional repression domain in the N-terminal region of p53. Oncogene. 2005;24(9):1481–1490. doi: 10.1038/sj.onc.1208130. [DOI] [PubMed] [Google Scholar]
- 16.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124(6):1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 17.Rivlin N, Koifman G, Rotter V. p53 orchestrates between normal differentiation and cancer. Semin Cancer Biol. 2015;32:10–17. doi: 10.1016/j.semcancer.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Zhao T, Xu Y. p53 and stem cells: New developments and new concerns. Trends Cell Biol. 2010;20(3):170–175. doi: 10.1016/j.tcb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Jain AK, et al. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10(2):e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi L, Lu C, Hu W, Sun Y, Levine AJ. Multiple roles of p53-related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res. 2012;72(21):5635–5645. doi: 10.1158/0008-5472.CAN-12-1451. [DOI] [PubMed] [Google Scholar]
- 21.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci USA. 2010;107(52):22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fung C, Vaughn DJ. Complications associated with chemotherapy in testicular cancer management. Nat Rev Urol. 2011;8(4):213–222. doi: 10.1038/nrurol.2011.26. [DOI] [PubMed] [Google Scholar]
- 29.Voutsadakis IA. The chemosensitivity of testicular germ cell tumors. Cell Oncol (Dordr) 2014;37(2):79–94. doi: 10.1007/s13402-014-0168-6. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen H, et al. LLY-507, a cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J Biol Chem. 2015;290(22):13641–13653. doi: 10.1074/jbc.M114.626861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma A, et al. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase SETD8. J Med Chem. 2014;57(15):6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, et al. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. J Biol Chem. 2011;286(44):38725–38737. doi: 10.1074/jbc.M111.262410. [DOI] [PMC free article] [PubMed] [Google Scholar]