Significance
Telomerase synthesizes the telomeric DNA at the 3′ ends of chromosomes and maintains genome integrity. Telomerase RNA (TR) provides the template for telomere-repeat synthesis within a template/pseudoknot (t/PK) domain that is essential for activity. We investigated the structure and dynamics of the t/PK from medaka fish, which contain the smallest vertebrate TR, using NMR and modeling. Despite differences in length, sequence, and predicted secondary structure with human TR, the remarkable similarities between subdomains, including one newly identified in medaka, reveal a conserved architecture for vertebrate t/PK. Combining our model of the full-length pseudoknot and information from the 9-Å structure of Tetrahymena telomerase, we propose models for the interaction of medaka and human t/PK with telomerase reverse transcriptase, providing insight into function.
Keywords: TERT, NMR, RNP, medaka, RNA structure
Abstract
Telomerase is an RNA–protein complex that includes a unique reverse transcriptase that catalyzes the addition of single-stranded telomere DNA repeats onto the 3′ ends of linear chromosomes using an integral telomerase RNA (TR) template. Vertebrate TR contains the template/pseudoknot (t/PK) and CR4/5 domains required for telomerase activity in vitro. All vertebrate pseudoknots include two subdomains: P2ab (helices P2a and P2b with a 5/6-nt internal loop) and the minimal pseudoknot (P2b–P3 and associated loops). A helical extension of P2a, P2a.1, is specific to mammalian TR. Using NMR, we investigated the structures of the full-length TR pseudoknot and isolated subdomains in Oryzias latipes (Japanese medaka fish), which has the smallest vertebrate TR identified to date. We determined the solution NMR structure and studied the dynamics of medaka P2ab, and identified all base pairs and tertiary interactions in the minimal pseudoknot. Despite differences in length and sequence, the structure of medaka P2ab is more similar to human P2ab than predicted, and the medaka minimal pseudoknot has the same tertiary interactions as the human pseudoknot. Significantly, although P2a.1 is not predicted to form in teleost fish, we find that it forms in the full-length pseudoknot via an unexpected hairpin. Model structures of the subdomains are combined to generate a model of t/PK. These results provide evidence that the architecture for the vertebrate t/PK is conserved from teleost fish to human. The organization of the t/PK on telomerase reverse transcriptase for medaka and human is modeled based on the cryoEM structure of Tetrahymena telomerase, providing insight into function.
Telomeres are the physical ends of linear chromosomes, composed of telomeric DNA and associated proteins. After each round of cell replication, telomeres shorten because of incomplete 3′-end replication of telomere DNA repeats and nucleolytic processing. Telomerase, a specialized reverse transcriptase, is a large ribonucleoprotein that plays essential roles in protecting the integrity of linear chromosomes in most eukaryotic species (1). Most somatic cells do not contain detectable telomerase activity, and consequently, after 40–60 cell divisions, their telomeres shorten below a critical length [∼12.8 repeats (2)] that leads to replicative senescence or to apoptosis (3). Most cancer cell lines, on the other hand, have significant telomerase activity, which continuously replenishes their telomeres and allows them to divide indefinitely; this dichotomy has made telomerase inhibition an attractive target for the development of anticancer drugs (4, 5). Mutations in human telomerase proteins and RNA cause a number of inherited diseases, such as autosomal dominant dyskeratosis congenita, idiopathic pulmonary fibrosis, myelodysplasia, and aplastic anemia (6–10).
The telomerase holoenzyme consists of a unique reverse transcriptase protein (telomerase reverse transcriptase; TERT), which contains the active site for nucleotide addition, an essential telomerase RNA (TR), which contains the template for telomere DNA synthesis, and other proteins required for its proper functioning in vivo (11). TERT is a conserved multidomain protein that usually contains four major evolutionarily conserved functional domains: the TERT essential N-terminal (TEN) domain that interacts with the primer–template duplex (12–14), the telomerase RNA-binding domain (TRBD) required for binding multiple sites of TR with high affinity (15), the reverse transcriptase (RT) domain including the enzyme active site (fingers and palm) where the TR template is located, and the C-terminal extension (CTE) (thumb), which may promote telomerase processivity (16, 17). The crystal structure of Tribolium castaneum (flour beetle) TERT, which intrinsically lacks the TEN domain, revealed that TRBD, RT, and CTE form a ring. The TRBD domain has high-affinity binding sites for two domains of TR, and the RT and CTE domains bind the template/telomeric DNA hybrid and catalyze the addition of DNA repeats onto the 3′ end (18, 19).
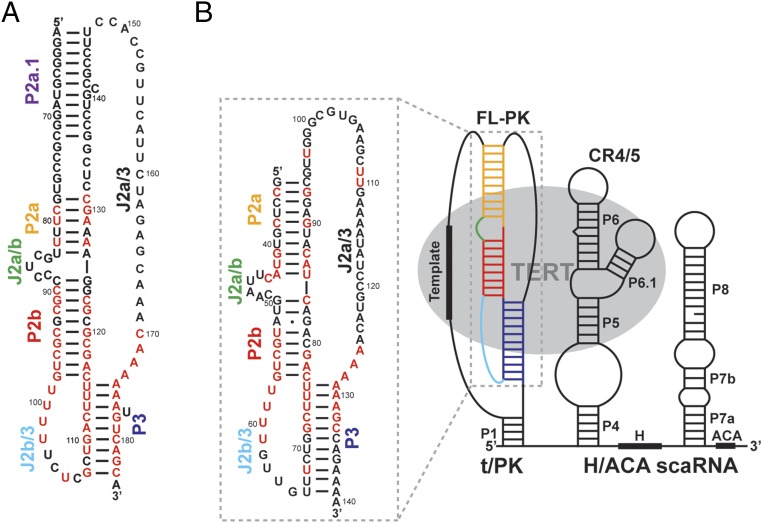
In contrast to the conserved TERT, TRs are extensively divergent in sequence and length (20–22). However, TRs almost universally contain a template/pseudoknot (t/PK), the core domain, and a separate TERT-activating domain, which together bind TERT and are required for catalysis (23). The t/PK contains three conserved regions: the template, a template boundary element (TBE), and a pseudoknot (Fig. 1) (24, 25). In the pseudoknot of mammalian vertebrates (e.g., human), there are three subdomains, the minimal P2b–P3 pseudoknot (P2b–P3 and associated loops J2b/3 and part of J2a/3), P2ab (helices P2a and P2b with a 5/6-nt bulge, J2a/b), and the P2a extension, P2a.1 (Fig. 1A). The minimal P2b–P3 pseudoknot contains most of the highly conserved nucleotides in the full-length pseudoknot (FL-PK) (Fig. 1); most of the nucleotide identities in the other subdomains are not essential for telomerase activity (26, 27). The solution NMR structures of a minimal pseudoknot from human TR (hTR), comprising the P2b–P3 stems and loops linked in a single strand, revealed extensive interactions between the stems and loops to form successive base triplets, and these tertiary interactions were shown to be important for activity (26, 28). Biochemical and structural studies have shown that the triple helix is a conserved and essential element of human and yeast pseudoknots (28–30). The solution NMR structure of human P2ab (hP2ab) revealed that the bulge induces an ∼90° bend between P2a and P2b. A combined residual dipolar coupling (RDC) and computational analysis (RDC–MC-Sym) showed that the internal loop between P2a.1 and P2a forms an irregular helix of mismatched base pairs and that P2a.1 and P2a are coaxially stacked (31). However, P2a.1 is not predicted to be present in nonmammalian vertebrate TRs (20). In vitro telomerase activity assays on P2a.1 have shown that it is important for human telomerase activity and processivity (12, 26). The minimal pseudoknot, P2ab, and P2a.1 domains were computationally combined to obtain a model of the human full-length pseudoknot (hFL-PK) domain, which showed that the P2ab bend defines the overall topology of t/PK (31). Modeling of Tetrahymena TR and TERT in a recent 9-Å–resolution cryoelectron microscopy (cryoEM) map revealed that its smaller t/PK domain encircles the TERT ring approximately perpendicular to its plane, with the template and pseudoknot on opposite sides (32).
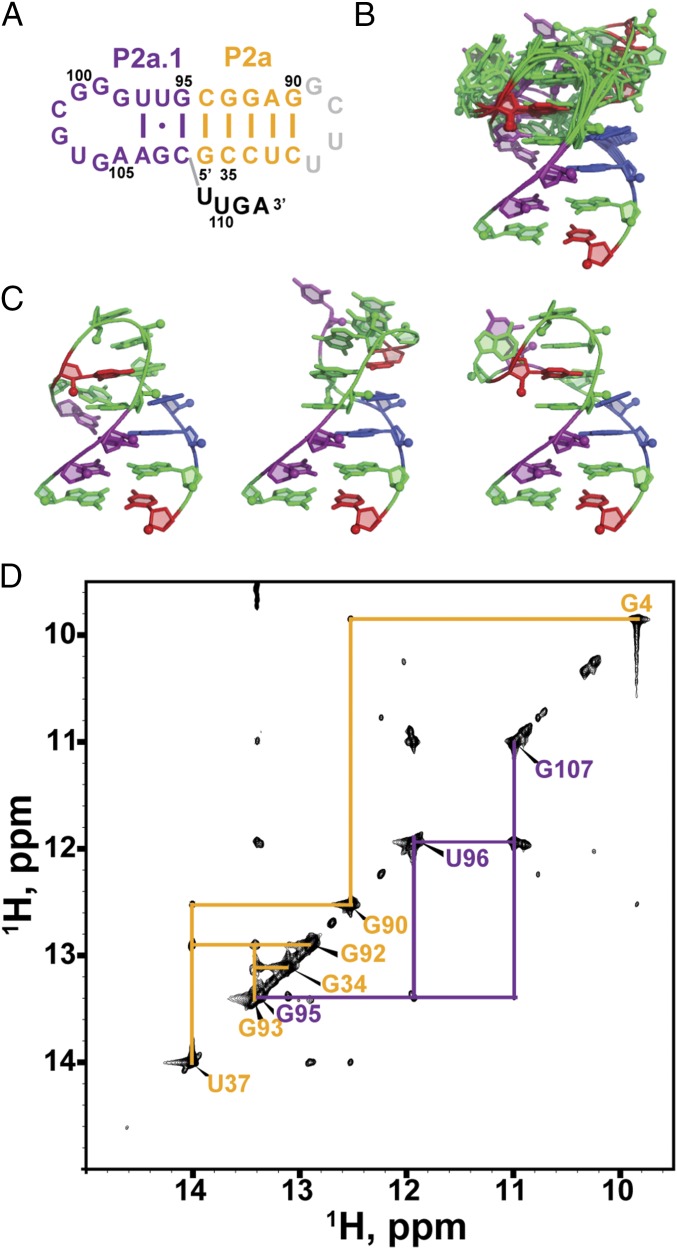
Fig. 1.
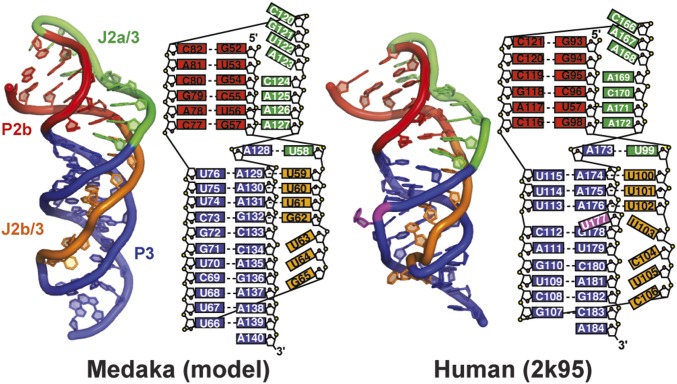
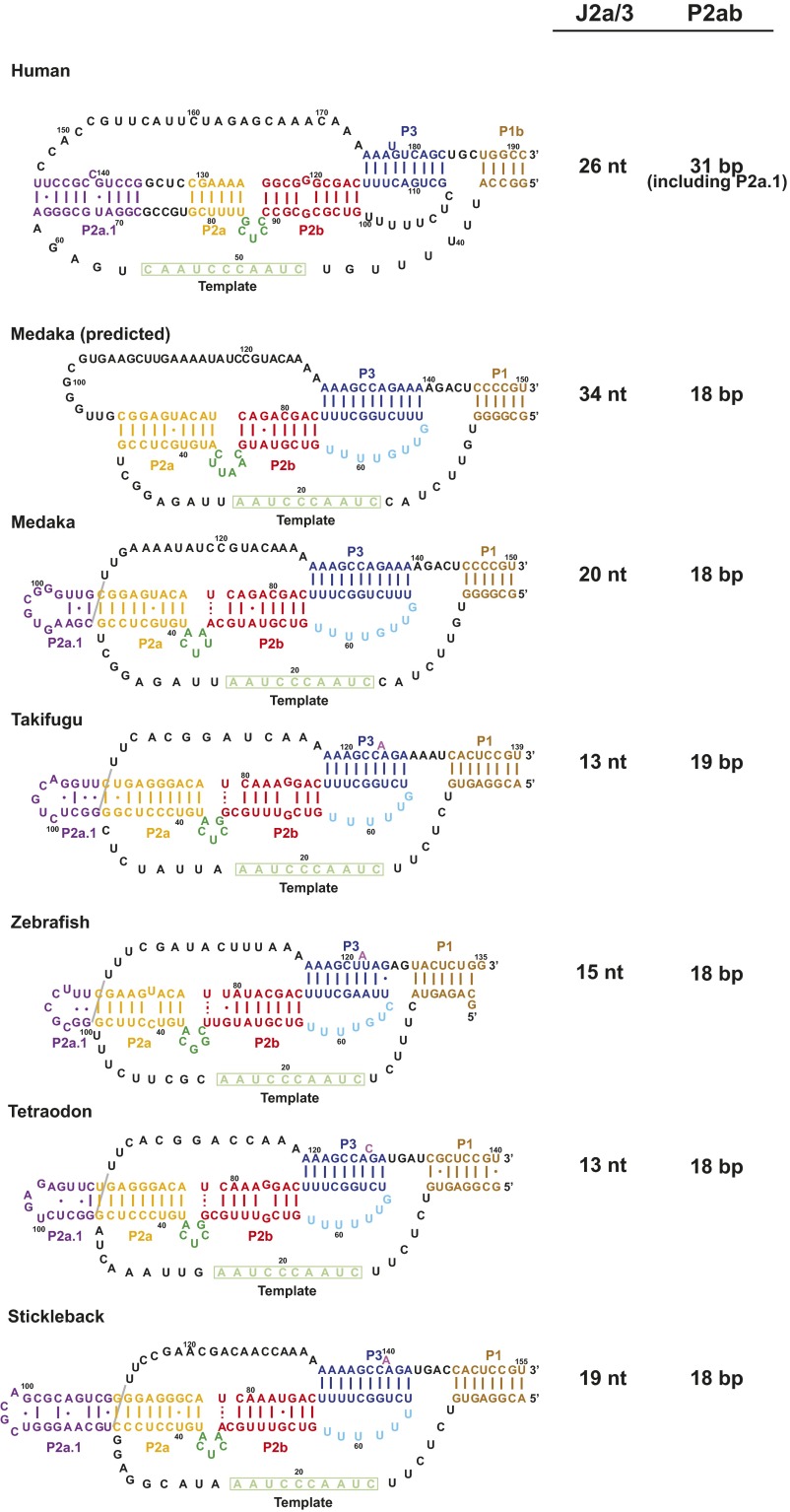
Medaka and human TR. (A) The secondary structure of the hFL-TR pseudoknot. The red letters indicate nucleotides with >80% sequence conservation among vertebrates. (B) Predicted secondary structure of mdTR containing the t/PK, CR4/5, and H/ACA domains. The FL-PK sequence and base pairs predicted by phylogenetic comparative analysis are shown in the box on the left. The 100%-conserved nucleotides identified in the five teleost fish TR are highlighted in red. TERT (gray ellipse) interacts with the t/PK and CR4/5 domains. The stems and loops are labeled and colored individually: P2a.1, purple; P2a, orange; P2b, red; P3, blue; J2a/b, green; J2a/3, black; and J2b/3, cyan.
Among the vertebrate species with identified TR sequences, the nonmammalian vertebrate Oryzias latipes (Japanese medaka fish) has the smallest TR (Fig. 1B) (33). A solution NMR structure of the medaka TR (mdTR) activating domain (CR4/5) and the crystal structure of the CR4/5–TRBD complex have been reported (34, 35). These structures show that phylogenetically conserved bases interact with the TRBD, suggesting that the mode of interaction is conserved among vertebrates (34). Based on sequence comparisons, the medaka t/PK domain is predicted to have the minimal pseudoknot and P2ab, but not P2a.1, and its J2a/3 loop is much longer than in hTR (Fig. 1B). We have investigated the structure of the medaka FL-PK (mdFL-PK) and subdomains using NMR and compared them with hTR. We identified the base pairs and tertiary interactions in medaka minimal pseudoknot (mdPK), determined the NMR solution structure of P2ab, and discovered that in the mdFL-PK the 5′ end of the J2a/3 loop folds to form a hairpin that we propose is a cryptic P2a.1. All the base pairs identified in the isolated subdomains are also identified in the mdFL-PK, and a model structure is presented. Comparison of medaka and human t/PK domains shows that, despite differences in sequence and length, the tertiary structures are highly similar, providing evidence for conserved vertebrate TR t/PK architecture. Based on the recent cryoEM structure of Tetrahymena holoenzyme (32), we modeled the medaka and human t/PK on TERT.
Results and Discussion
The Minimal MdPK Has No Bulge and Has an Additional Triple.
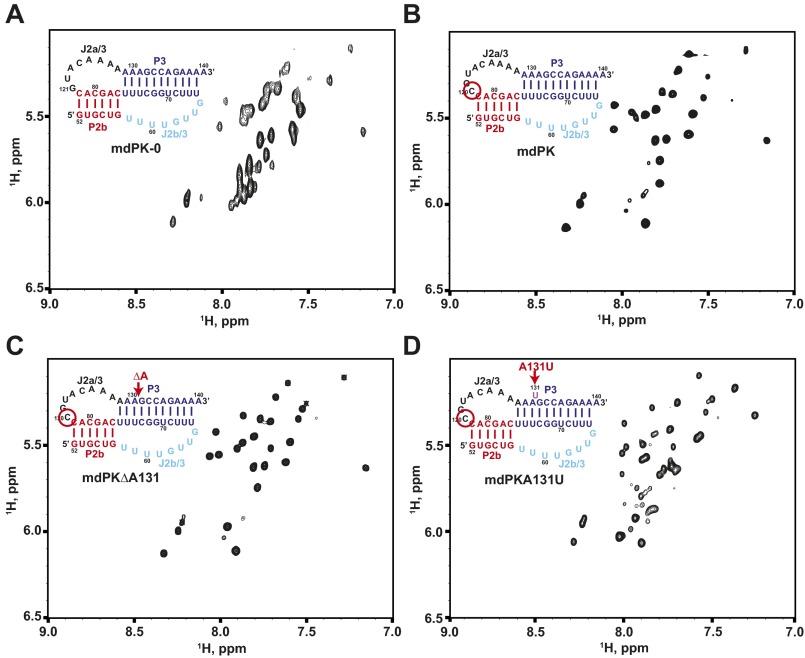
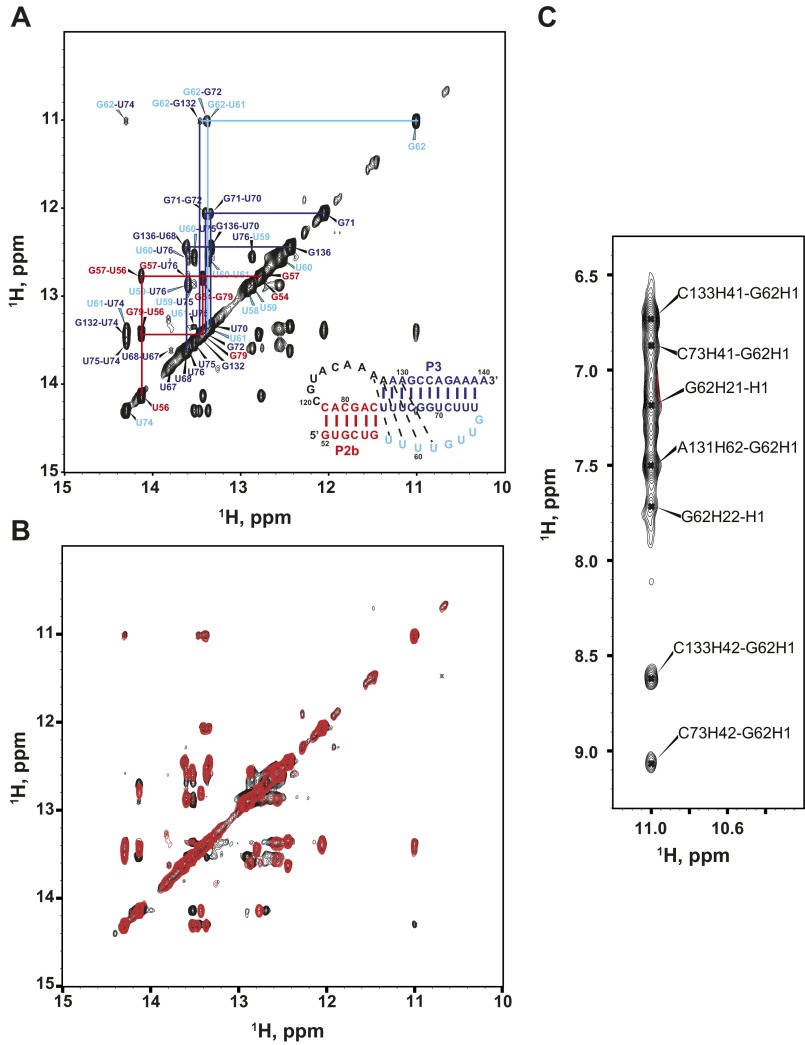
Previous phylogenetic analysis of teleost fish TRs predicted that the minimal P2b–P3 pseudoknot has the same tertiary interactions as the human minimal P2b–P3 pseudoknot (hPK) but without the single-nucleotide bulge in the P3 helix adjacent to the base triplets (Fig. 1B) (33). Based on the hPK NMR solution structure (28, 36), a minimal mdPK construct (mdPK-0; nucleotides 52–82 and 121–140) with the same number of base pairs in P2b (6 bp) and same length J2a/3 (8 nt) was synthesized by in vitro transcription (Fig. S1A). Surprisingly, the total correlation spectroscopy (TOCSY) spectrum of mdPK-0 shows evidence of exchange line-broadening, indicative of more than one conformation under the conditions in which hPK forms one dominant conformation (Fig. S1A). However, if the J2a/3 loop is extended by one C nucleotide (mdPK; nucleotides 52–82 and 120–140) (Fig. 2A), the RNA is in a single conformation, as determined by analysis of TOCSY spectra (Fig. S1B). Analysis of imino cross-peaks in the 2D NOESY spectrum in H2O indicates that this stable conformation is a pseudoknot with two stacked stems, a junction Hoogsteen U–A base pair, and loop J2b/3 interactions with P3 to form three U–A–U triplets (Fig. 2B), as discussed in detail below. An RNA construct with a C nucleotide at the start of the J2a/3 loop and a deletion of one of the seven consecutive A nucleotides (mdPKΔA131) also forms a stable pseudoknot that exhibits TOCSY and imino proton spectra very similar to those of mdPK (Figs. S1C and S2). Based on analysis of these spectra and also on comparisons with hPK and hPKΔU, in which the J2a/3 loop also starts with a C nucleotide, we conclude that the G nucleotide at the start of the J2a/3 loop in mdPK-0 destabilizes the folding of the pseudoknot, likely by stacking on P2b.
Fig. S1.
TOCSY spectra of the minimal mdPK constructs. (A) mdPK-0. (B) mdPK. (C) mdPKΔA131. (D) mdPKA131U. Spectra were acquired at 800 MHz, 10 °C. Sequence and secondary structures of each construct are shown in the upper left corners. Secondary structure elements are colored as in Fig. 1.
Fig. 2.
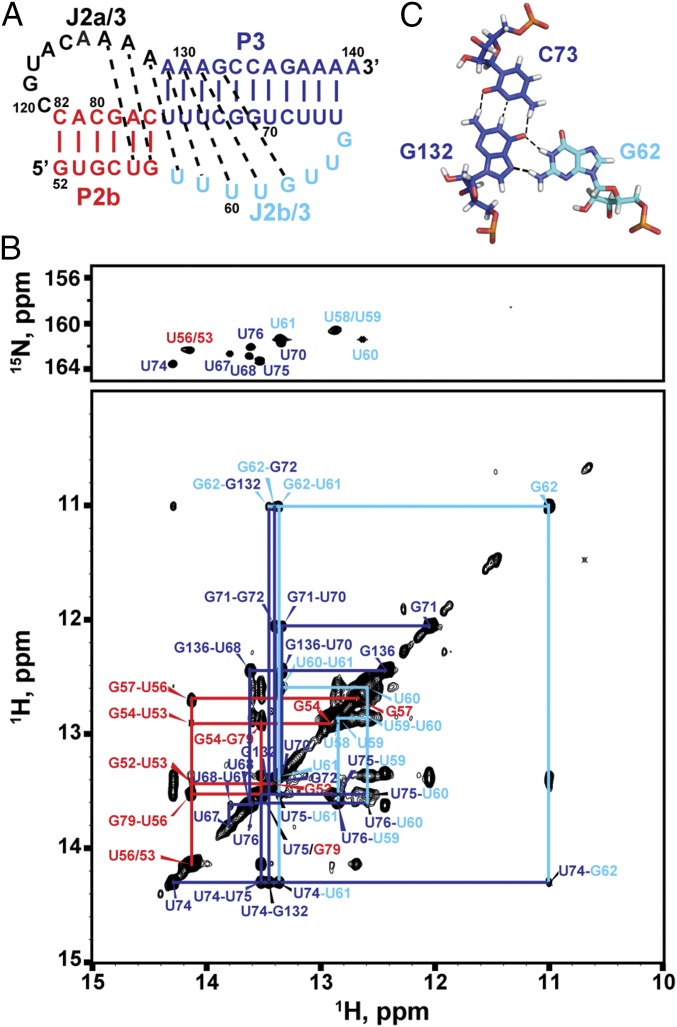
MdTR minimal (P2b–P3) pseudoknot. (A) Sequence and secondary structure of the minimal pseudoknot construct mdPK used in the solution NMR study. (B) Imino region of HNN-COSY (Upper) aligned with imino NOESY spectrum of mdPK (Lower). NOE cross-peaks of base pairs are connected and colored as in Fig. 1. (C) Stick representation of the G62–C73–G132 base triple. Secondary structure elements are colored as in Fig. 1.
Fig. S2.
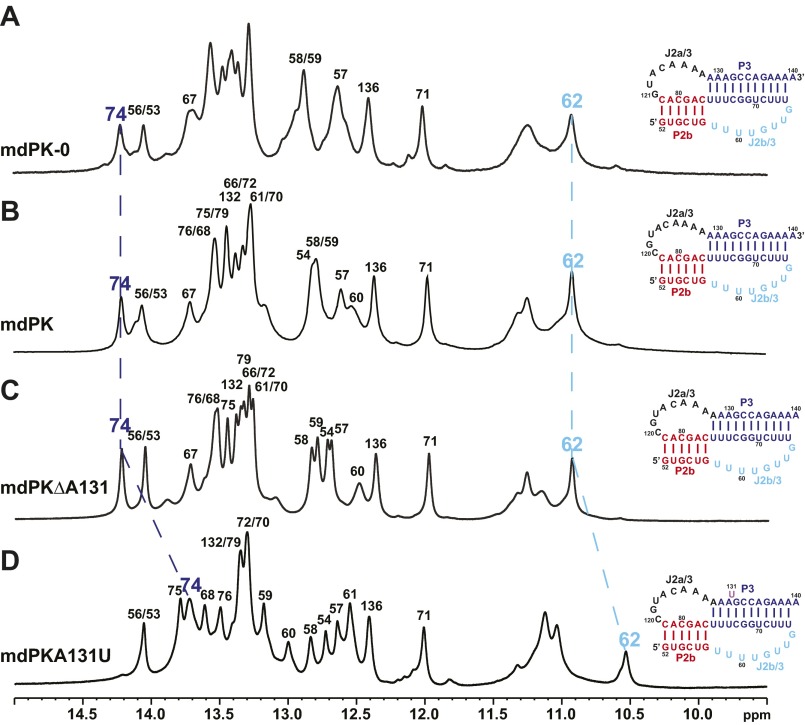
1D imino proton of the minimal mdPK constructs at 800 MHz and 10 °C. (A) mdPK-0. (B) mdPK. (C) mdPKΔA131. (D) mdPKA131U. The blue and cyan dashed lines connect imino resonances (62, 74) with different chemical shifts in different constructs. Sequence and secondary structures of each construct are shown at right. Secondary structure elements are colored as in Fig. 1.
The hTR pseudoknot has a bulge U nucleotide adjacent to the three U–A–U triplets; this nucleotide has been proposed to contribute to catalysis (30). In hPK, deletion of the bulge U177 to form hPKΔU stabilizes the pseudoknot (28, 36). Sequence alignment between medaka and human TRs suggests that medaka P2b–P3 could form a structure identical to hTR with a bulge A131 (instead of the U nucleotide) and with the CAAA nucleotides at the 5′ end of the J2a/3 loop at the same location relative to P2b found in hPK (Fig. 1B); however, comparison of the NOESY spectra of mdPK and mdPKΔA131 (Fig. S3) with hPKΔU confirms that medaka has no bulge A nucleotide, because the imino proton resonances for the tertiary interactions have nearly identical chemical shifts (Fig. S2). Furthermore, the results show that the alignment of the C nucleotide in the CAAA stretch of J2a/3 relative to P2b is not important for stability. In the structure of hPK, only the last two A nucleotides of the J2a/3 loop have hydrogen bond interactions with the minor groove of P2b. Last, we investigated an RNA construct in which A131 was replaced with a U nucleotide (mdPKA131U) (Figs. S1D and S2). This nucleotide change should induce a bulge in the position found in human telomerase. This construct also formed a pseudoknot, but the spectra showed evidence for conformational heterogeneity, as expected because of destabilization by a bulge base. We conclude that mdPK (as well as mdPKΔA131) folds into the biologically relevant structure in terms of tertiary interactions, and therefore we investigated its tertiary interactions in detail, as discussed below. These interactions are confirmed by telomerase activity assays using TRs with nucleotide substitutions, presented in the next section.
Fig. S3.
Comparison of spectra of mdPKΔA131 and mdPK. (A) The imino region of the H2O NOESY spectrum of mdPKΔA131. (B) Superimposition of the imino region of the H2O NOESY spectra of mdPK (black) and mdPKΔA131 (red). (C) Region of the H2O NOESY spectrum of mdPK showing cross-peaks of G62 imino.
Fig. 2B shows the imino region of the HNN correlation spectroscopy (HNN-COSY) spectrum of 13C,15N-A,U–labeled mdPK aligned with the imino proton region of the 2D H2O NOESY. The imino protons were assigned by experiments using sequential NOE connectivities in the NOESY and HNN-COSY spectra (37), which directly detect hydrogen bonds within Watson–Crick and Hoogsteen base pairs. Analysis of these spectra shows that the uridine-rich J2b/3 loop interacts with the major groove of P3. Specifically, the first four U nucleotides are all involved in Hoogsteen base pairs with J2a/3 (A128) and P3 stem adenines (A129–A131). U58 at the 5′ end of J2b/3 and A128 at the 3′ end of J2a/3 form a Hoogsteen U58–A128 base pair that inserts at the junction between the two stems (Fig. 2A). U59, U60, and U61 pair with the A129–U76, A130–U75, and A131–U74 Watson–Crick base pairs, respectively, to form U59–A129–U76, U60–A130–U75, and U61–A131–U74 triplets. These interactions are identical to those observed in hPK. There is an additional unexpected G–G–C triple following the three U–A–U triplets. This triple was identified on the basis of an upfield shifted imino resonance assigned to G62 (Fig. S2) and NOE resonances between G62 and its nearby residues (C73, A131, G132, and C133) (Fig. 2B and Fig. S3C). The imino of G62 is hydrogen bonded with O6 of G132, forming a G62–G132–C73 base triple adjacent to the U61–A131–U74 triple (Fig. 2C).
Because the NMR data indicated that almost all the tertiary interactions are conserved between mdPK and hPK, a model structure of mdPK was generated based on hPK without the bulge (hPKΔU177; PDB ID code 2K96), using the mdPK sequence and replacing the hPKΔU177 U–G–C triple with the mdPK G–G–C triple. A comparison of the model mdPK structure and the NMR structure of hPK is shown in Fig. 3. The central core that forms the triple helix is the same in both RNAs: There are two minor groove triplets (A127–G57–C77 and A126–U56–A78 in medaka) between P2b and J2a/3, three major groove U–A–U triplets between P3 and J2b/3, and a loop–loop Hoogsteen base pair at the junction between the stems formed by the last nucleotide (A128) in the J2a/3 loop and the first nucleotide (U58) in the J2b/3 loop (equivalent to the hPK A173–U99 Hoogsteen base pair). Medaka P3 is slightly longer (11 bp) than human P3 (9 bp plus a bulge U nucleotide), but the J2b/3 loops are the same length, although the sequence is different outside the conserved triple-helix core: Medaka has an extra triple (G–G–C), whereas human has the bulge U nucleotide which prevents the formation of the U–G–C triple observed when the bulge U is deleted (Fig. 3) (36). Despite the longer P3 stem, the medaka J2b/3 loop fits smoothly into the major groove. In comparison, human J2b/3 is more kinked, perhaps because of the shorter P3 stem and the slight bend in the helix caused by the bulge U (36).
Fig. 3.
Comparison of minimal mdPK and hPK (PDB ID code 2K95) structures. Secondary structure elements are P2b (red), P3 (blue), J2a/3 (green), and J2b/3 (gold).
Effect of Pseudoknot Mutations on Telomerase Activity.
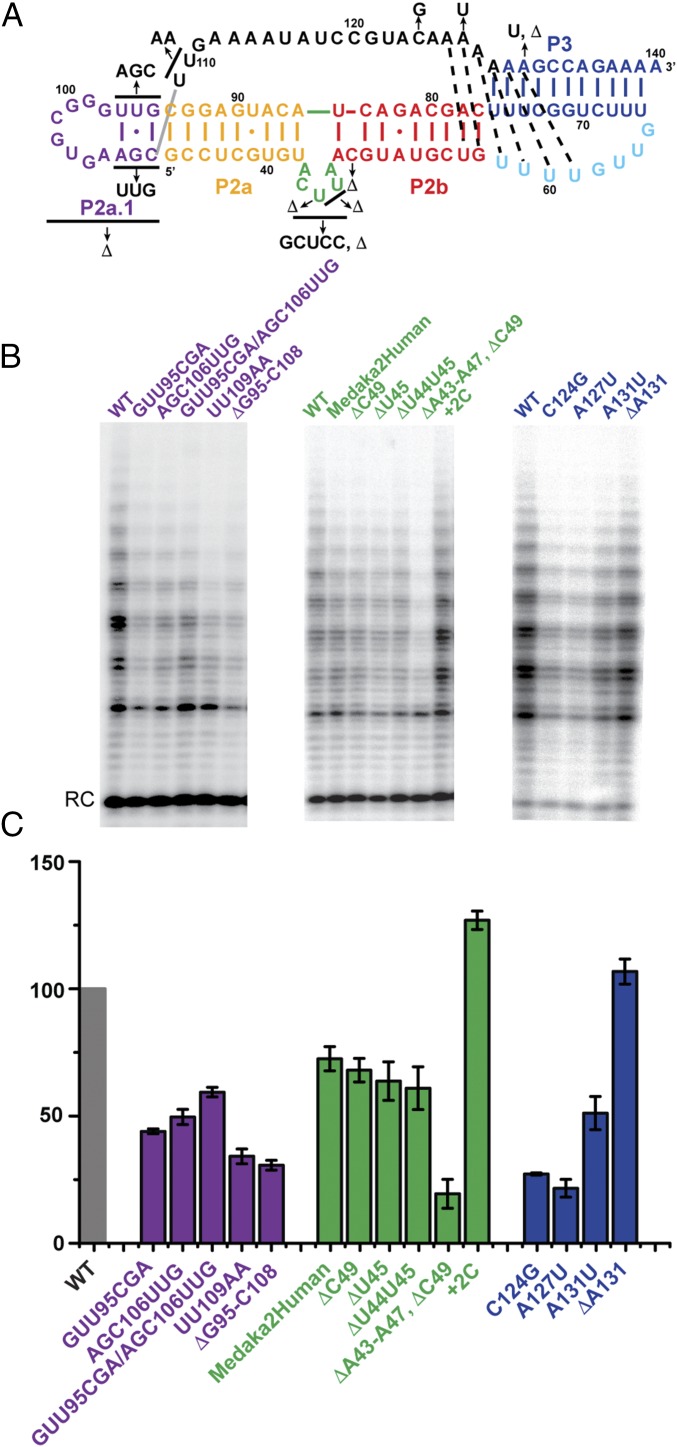
To confirm the presence and functional importance of the pseudoknot tertiary interactions observed by NMR for mdPK, telomerase activity was measured by direct assays. Medaka TERT was transcribed and translated in rabbit reticulocyte lysate (RRL) in the presence of full-length mdTR containing single-nucleotide substitutions in the pseudoknot (Fig. 4A) (35). Deletion of A131 has no effect on the telomerase activity (Fig. 4 B and C), as is consistent with the NMR data showing that the deletion of A131 has no effect on the folding of the minimal pseudoknot (Fig. S3A). In contrast, replacing A131 by U (A131U) decreases telomerase activity by half. Analysis of TOCSY spectra showed that mdPKA131U introduces a bulge U adjacent to the triple helix that destabilizes the pseudoknot (Fig. S1D). Deletion of the bulge in the hPK decreases activity by 50% in vitro (28, 38). These data support a model in which folding of the pseudoknot is important for assembly (32), and the relative stability of the folded and partially folded (hairpin vs. pseudoknot) forms might be important (39). The model mdPK structure shows that J2a/3 nucleotides A126 and A127 interact with the minor groove of P2b, as seen for the equivalent conserved residues in hTR. Replacing medaka A127 by a U nucleotide decreases activity by 80%, as is consistent with the A127–G57–C77 base triple having a role in stabilizing the tertiary structure of the pseudoknot, as previously observed for hTR (39). Somewhat surprisingly, the results of the medaka telomerase activity assay showed that the C124G substitution decreases activity to 27% of that of WT. Other teleost fish have a U or C nucleotide at this position, and the ΔA131 deletion that has no effect on activity shifts an A nucleotide to this position. There is also an A nucleotide at this position in hTR. Perhaps the G nucleotide destabilizes the interactions of the loop with the minor groove of P2b or in some way interferes with folding of the pseudoknot.
Fig. 4.
Telomerase activity assays of mdTR mutants. (A) Secondary structure of FL-PK with nucleotide substitutions indicated by arrows. (B) Effect of mdP2a1a, mdP2ab, and mdPK mutations (from left to right) on telomerase activity. Medaka TERT synthesized in RRL was assembled with full-length WT (first lane for each subgroup) or mutant mdTR. RC, recovery control. (C) Plot of the activity of each mutant relative to that of WT mdTR. The error bars indicate the difference or SD calculated from two or three independent assay reactions, respectively. Secondary structure elements and corresponding labels and bars in the plot are colored as in Fig. 1.
Solution Structure of Medaka P2ab Indicates That It Has a 5-nt Bulge.
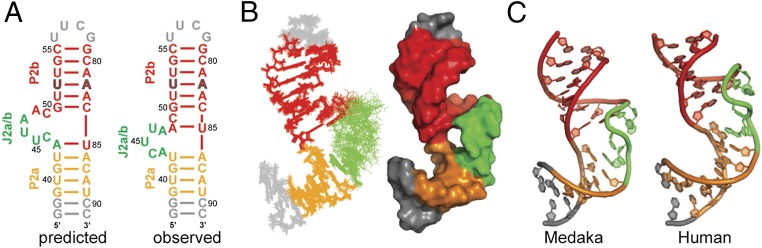
The J2a/b sequence is not highly conserved among vertebrate TRs, but it is usually a 5-nt bulge in mammalian TRs. The consensus sequence for the bulge is R87Y78Y87Y96Y87, where R is purine, Y is pyrimidine, and the subscripts are the per cent in 23 mammalian TRs (40). In hTR, the 5-nt J2a/b bulge induces a 90° bend between P2a and P2b, thus affecting the overall topology of the FL-PK (31). However, the mdTR J2a/b bulge is predicted to be a 6-nt loop that starts with a C nucleotide (Fig. 1B). To investigate whether in medaka the J2a/b bulge forms a similar structure and induces a bend similar to that in hP2ab, we determined the solution NMR structure of the medaka P2ab subdomain (mdP2ab). For the structural study, we designed a construct (nucleotides 39–55 and 79–89) that includes P2a, P2b, and the predicted 6-nt J2a/b bulge. The P2a was extended with two additional G–C base pairs to facilitate in vitro transcription, the A52–G82 mismatch in P2b was replaced with a U–A base pair to facilitate resonance assignment, and the P2b was capped by a UUCG tetraloop (Fig. 5A). A U–A base pair is found at this position in most other teleost fish TR.
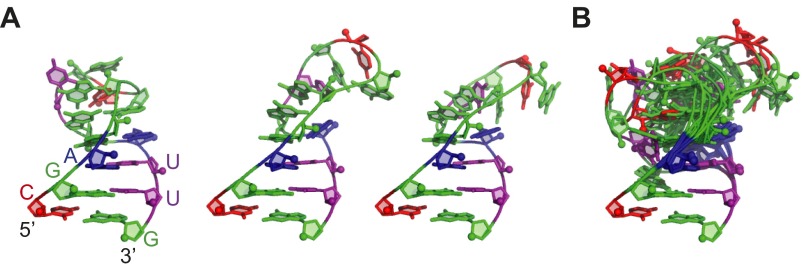
Fig. 5.
Solution structure of the mdP2ab (P2a–J2a/b–P2b). (A) Sequence and secondary structure of predicted (Left) and observed (Right) mdP2ab. (B) Superposition of the 20 lowest-energy structures over all heavy atoms and space-filling rendering of the lowest-energy structure of P2ab. (C) Comparison of the lowest-energy structure of mdP2ab (Left) and hP2ab (PDB ID code 2L3E) (Right). In all panels, P2a is gold, J2a/b is green, P2b is red, and the UUCG tetraloop is gray.
The solution structure of the mdP2ab was solved using 802 NOE-derived distance restraints (an average of 22 NOEs per nucleotide), 459 dihedral angle restraints, and 95 RDCs (Materials and Methods and Table S1). mdP2ab forms a well-defined structure, except for the loop, with an rmsd to the mean of 1.24 Å for all heavy atoms of the 20 lowest-energy structures. P2a and P2b are A-form helices with an rmsd of 0.16 and 0.37 Å, respectively. The structure shows that A48 and U85 form a base pair at the end of P2b and the C49 inserts between A48–U85 and G50–C84, resulting in a 5-nt bulge (ACUUA) that starts with a purine, instead of the predicted 6-nt bulge (CUUAAC) (Fig. 5B). Generally, purines tend to stack into a helix, whereas bulge pyrimidines tend to flip out (41). The nucleotide at the 5′ end of the bulge is well defined, with A43 stacking above P2a U42. The positions of the other four loop residues are less well defined, but all are flipped out of the loop. As shown below, these residues are dynamic on the pico- to nanosecond timescale. The medaka J2a/b bulge introduces a 79 ± 3° axial bend between P2a and P2b (Fig. 5B), which is similar to the bend angle of 89 ± 3° in hP2ab. Comparison of the tertiary structures of mdP2ab and hP2ab reveals that, despite the lack of the sequence identity, the overall structure and interhelical angle are similar (Fig. 5C), suggesting that the bend angle and direction in P2ab is conserved among vertebrates.
Table S1.
Restraints and structure statistics for the 20 lowest-energy structures of mdP2ab (36 nt)
| Restraints/statistics | number/value |
| Restraints | |
| Total NOE | 802 |
| Intraresidue | 339 |
| Interresidue | 463 |
| Sequential, |i − j| = 1 | 337 |
| Nonsequential, |i − j| > 1 | 126 |
| Hydrogen bond | 65 |
| Total RDCs, 1DCH and 1DNH | 95 |
| Total dihedral angle | 459 |
| Structure statistics | |
| Violations | |
| Distance restraints, (Å) | 0.019 ± 0.001 |
| Dihedral angle restraints (°) | 0.11 ± 0.02 |
| Dipolar coupling (Hz) | 0.29 ± 0.01 |
| Deviations from idealized geometry | |
| Bond length (Å) | 0.0049 ± 0.0001 |
| Bond angle (°) | 0.74 ± 0.01 |
| Impropers (°) | 0.51 ± 0.01 |
| Average pairwise rmsd from mean (Å) | |
| All heavy atoms (residues in P2a and P2b) | 0.43 |
| All heavy atoms | 1.24 |
| Number of NOE violations >0.5 (Å) | 0 |
| Number of NOE violations >0.2 (Å) | 0 |
| Number of dihedral violations >5 (°) | 0 |
| Number of RDC violations >2 (Hz) | 0 |
| Average MolProbity clashscore | 13.3 |
P2ab Dynamics Are Similar for Medaka and Human.
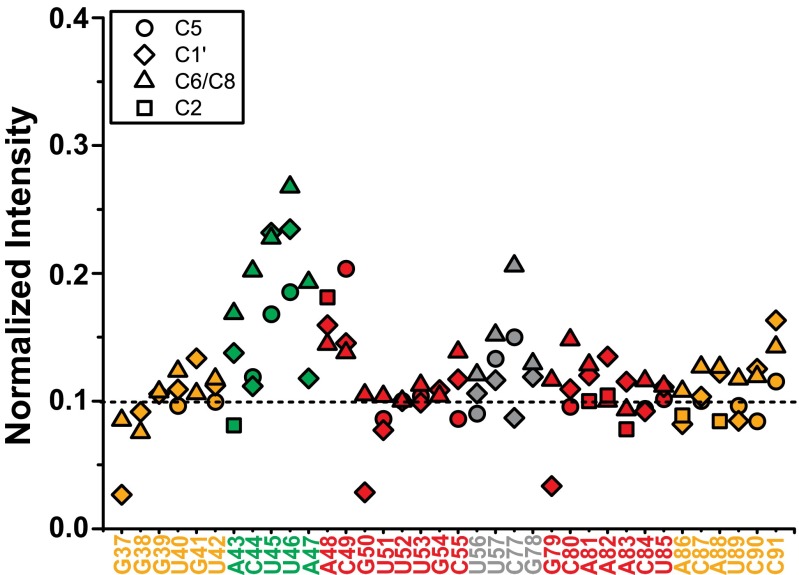
Previous studies on hP2ab showed that the J2a/b bulge is highly dynamic on the pico- to nanosecond timescale (31). To investigate whether the bulge in mdP2ab exhibits similar dynamics, the nonexchangeable resonance intensities from non–constant-time 1H-13C heteronuclear single-quantum coherence spectroscopy (HSQC) spectra of the mdP2ab were acquired. These data show that J2a/b nucleotides have significantly higher peak intensities than the residues from the helix region, indicating a highly flexible interface between P2a and P2b (Fig. S4). Among the J2a/b residues, U46 exhibits the most unrestricted motion, and A43 is the least dynamic, as is consistent with its stacking on top of P2a. We also obtained RDCs to characterize interhelical motion that is faster than the millisecond time scale by comparing the degree of order (ϑ) for individual stems (Tables S2 and S3). The ratio between the degree of order for each stem, defined as the interhelical generalized degree of order (ϑint= ϑP2a/ϑP2b), provides a measure of interhelical motions, with ϑint = 1 corresponding to complete rigidity and ϑint = 0 to maximum interhelical motion. The determined interhelical motion of mdP2ab (ϑint = 0.70) is the same as that of hP2ab (ϑint = 0.69), indicating that medaka and human P2ab bulges are similarly flexible.
Fig. S4.
Normalized resonance intensities of mdP2ab from non–constant-time 1H-13C HSQC experiments. The resonance intensities of each type of C–H spin were normalized against the lowest intensity from the helix to the reference value of 0.1. The resonances and nucleotide labels are colored by subdomain as in Fig. 5.
Table S2.
RDCs measured for mdP2ab
| Residue | Atom | RDC, Hz | Residue | Atom | RDC, Hz |
| G37 | C8H8 | −30.0 | G54 | N1H1 | −9.5 |
| G37 | C1′H1′ | −6.3 | C55 | C5H5 | 19.0 |
| G38 | C8H8 | −12.0 | C55 | C6H6 | 11.9 |
| G38 | C1′H1′ | −13.6 | C55 | C1′H1′ | 14.6 |
| G38 | N1H1 | −2.5 | U56 | C5H5 | 14.6 |
| G39 | C8H8 | 6.6 | U56 | C6H6 | 12.7 |
| G39 | C1′H1′ | −12.0 | U56 | C1′H1′ | 5.6 |
| G39 | N1H1 | −7.2 | U57 | C5H5 | −9.0 |
| U40 | C5H5 | −0.2 | U57 | C6H6 | 18.5 |
| U40 | C6H6 | 11.3 | U57 | C1′H1′ | −3.8 |
| U40 | C1′H1′ | 2.9 | C77 | C5H5 | −2.4 |
| U40 | N3H3 | −4.9 | C77 | C6H6 | −1.1 |
| G41 | C8H8 | 3.0 | C77 | C1′H1′ | −18.9 |
| G41 | C1′H1′ | 12.9 | G78 | C8H8 | 2.9 |
| G41 | N1H1 | 7.0 | G78 | C1′H1′ | −4.8 |
| U42 | C5H5 | 12.4 | G78 | N1H1 | −3.5 |
| U42 | C6H6 | −18.4 | G79 | C8H8 | 15.7 |
| U42 | C1′H1′ | 18.2 | G79 | C1′H1′ | −27.0 |
| A43 | C2H2 | −16.2 | G79 | N1H1 | −6.6 |
| A43 | C8H8 | −5.0 | C80 | C5H5 | 18.7 |
| A43 | C1′H1′ | 13.5 | C80 | C6H6 | 10.1 |
| C44 | C5H5 | −12.9 | C80 | C1′H1′ | −9.1 |
| C44 | C6H6 | 8.7 | A81 | C2H2 | 12.9 |
| C44 | C1′H1′ | 5.5 | A81 | C8H8 | 15.5 |
| U45 | C5H5 | −3.4 | A81 | C1′H1′ | 1.2 |
| U45 | C6H6 | 4.7 | A82 | C2H2 | 13.9 |
| U45 | C1′H1 ′ | −0.1 | A82 | C8H8 | 23.9 |
| U46 | C5H5 | −3.2 | A82 | C1′H1′ | 7.3 |
| U46 | C6H6 | 1.7 | A83 | C2H2 | 23.0 |
| U46 | C1′H1′ | −4.1 | A83 | C8H8 | 28.7 |
| A47 | C2H2 | 11.9 | A83 | C1′H1′ | 12.9 |
| A47 | C8H8 | 7.6 | C84 | C5H5 | 27.9 |
| A47 | C1′H1′ | −2.6 | C84 | C6H6 | 12.5 |
| A48 | C2H2 | 3.5 | C84 | C1′H1′ | 14.2 |
| A48 | C8H8 | 3.9 | U85 | C5H5 | 21.9 |
| A48 | C1′H1′ | −2.0 | U85 | C6H6 | −3.6 |
| C49 | C5H5 | 0.2 | U85 | C1′H1′ | 11.9 |
| C49 | C6H6 | −10.8 | A86 | C2H2 | −2.2 |
| C49 | C1′H1′ | −14.9 | A86 | C8H8 | −17.2 |
| G50 | C8H8 | 5.9 | A86 | C1′H1′ | 10.6 |
| G50 | C1′H1′ | −41.7 | C87 | C5H5 | −6.2 |
| G50 | N1H1 | −11.9 | C87 | C6H6 | −10.0 |
| U51 | C5H5 | −5.6 | C87 | C1′H1′ | 9.8 |
| U51 | C6H6 | 29.7 | A88 | C2H2 | −16.9 |
| U51 | C1′H1′ | −30.7 | A88 | C8H8 | −2.7 |
| U51 | N3H3 | −15.7 | A88 | C1′H1′ | 12.0 |
| U52 | C5H5 | 13.1 | U89 | C5H5 | −1.4 |
| U52 | C6H6 | 27.2 | U89 | C6H6 | 12.5 |
| U52 | C1′H1′ | −15.4 | U89 | C1′H1′ | 15.5 |
| U52 | N3H3 | −15.8 | U89 | N3H3 | −9.1 |
| U53 | C5H5 | 26.1 | C90 | C5H5 | 8.7 |
| U53 | C6H6 | 20.9 | C90 | C6H6 | 2.6 |
| U53 | C1′H1′ | 2.1 | C90 | C1′H1′ | 13.7 |
| U53 | N3H3 | −10.4 | C91 | C5H5 | 11.9 |
| G54 | C8H8 | 13.9 | C91 | C6H6 | −18.0 |
| G54 | C1′H1′ | 9.8 | C91 | C1′H1′ | 14.2 |
Table S3.
Order tensor analysis of RDCs measured in helical regions of mdP2ab using idealized A-form helices as input helix coordinates
| Helix | n | Rmsd, Hz | Q, % | R | CN | η | ϑ, ×10−4 | ϑint |
| P2a | 40 | 3.7 | 30.5 | 0.99 | 2.6 | 0.56 ± 0.09 | 6.1 ± 0.3 | 0.7 ± 0.3 |
| P2b | 39 | 2.6 | 14.2 | 0.99 | 2.5 | 0.36 ± 0.04 | 9.2 ± 0.2 |
CN, condition number; n, number of RDCs; Q, quality factor; R, correlation coefficient; η, asymmetry parameter; ϑ, generalized degree of order; ϑint, internal generalized degree of order.
Effect of mdP2ab Mutations on Telomerase Activity.
We made a series of deletions and substitutions in J2a/b in mdTR to investigate the effect of its sequence and the importance of the bend angle and dynamics on telomerase activity by direct telomerase activity assays. Replacing the 5-nt bulge with the sequence of human J2a/b (GCUCC) decreased telomerase activity only moderately, to ∼76% of WT (Fig. 4), as is consistent with the findings in human J2a/b that swapping the bulge sequence 5′→3′ or replacement by the sequence from the mouse TR counterpart had little effect on human telomerase activity (26, 42). Deletion of C49 had a comparable effect (∼71% of WT) on telomerase activity. We also shortened the length of the J2a/b bulge by deleting nucleotides from the center of the bulge. Deletion of residue U45, the central nucleotide in the J2a/b bulge, and of C44 and U45 reduced the telomerase activity moderately, to ∼70% of WT. However, deletion of the entire loop decreased activity to ∼15% of WT; these results are comparable to observations for J2a/b deletions in hTR (31). Insertion of two additional C nucleotides at the 3′ end of the bulge (+2C), which is expected to increase the interhelical freedom between P2a and P2b, actually increased activity to 130%. A similar increase in activity was observed for this substitution in hP2ab (31). Overall, these results are consistent with P2ab being required to induce a directional bend in the P2 stem of the vertebrate t/PK. This bend is likely important for the proper association of TR with TERT, but the flexibility of the bend also could be important for conformational changes in the TERT–TR complex during nucleotide addition or translocation.
FL-PK Has the Same Triple Helix as mdPK.
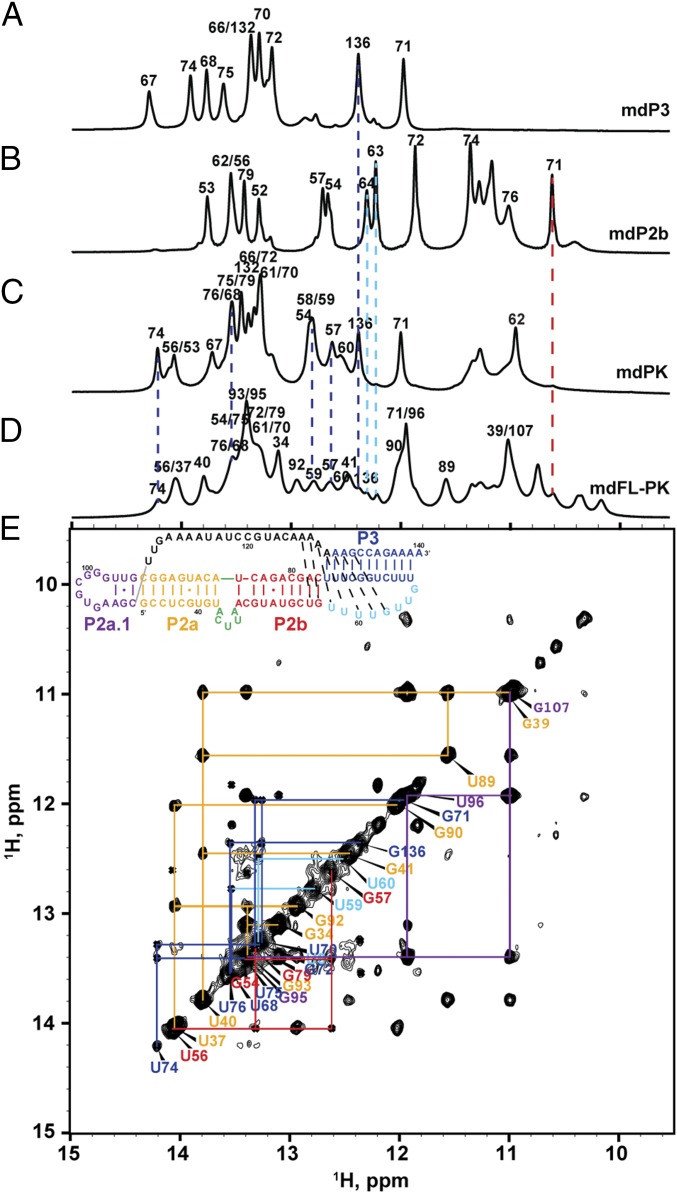
To confirm that the structures of the mdPK and mdP2ab subdomains are relevant in the context of the mdFL-PK, we synthesized an RNA containing nucleotides 34–140 comprising the FL-PK (Fig. 4A) and investigated the base pairing by NMR (Fig. 6). Imino proton resonances with cross-peak patterns and chemical shifts corresponding to those observed for mdPK and mdP2ab were identified, indicating that the structures of these subdomains were present in the context of mdFL-PK (Fig. 6E and Fig. S5).
Fig. 6.
The FL-PK. (A–D) 1D imino proton spectra of mdP3 (A), mdP2b (B), mdPK (C), and mdFL-PK (D). (E) Imino proton region of the NOESY spectrum of mdFL-PK. (Inset) Sequence and secondary structure. Secondary structure elements are colored as in Fig. 1.
Fig. S5.
Superimposition of the imino region of H2O NOESY spectra of mdFL-PK (black), mdPK (blue), and mdP2a1a (purple). The assignments of mdFL-PK are labeled along the diagonal using the color scheme in Fig. 1.
Previous studies showed that, depending on sample conditions, hPK is in equilibrium with a small amount of a partially folded structure in which P2b–J2b/3 forms a hairpin and the remaining P3–J2a/3 nucleotides are single stranded. We synthesized the two potential hairpins, P2b–J2b/3 (mdP2b; nucleotides 52–82) (Fig. S6A) and P3–J2a/3 (mdP3; nucleotides 67–82 and 120–138) (Fig. S6B), and investigated their secondary structure by NMR. In hP2b, the U nucleotides in the J2b/3 loop next to P3 base pair to form an extended helix with three U–U base pairs and one U–C base pair (39). Analysis of the imino proton region of the NOESY spectra shows that mdP2b similarly forms a series of U–U and G–U base pairs that extend the P2b, although their exact arrangement is not determined here (Fig. 6B and Fig. S6A). In contrast, only the predicted P3 stem residues are base paired in mdP3 (Fig. 6A and Fig. S6B). Comparison of the imino proton spectra of mdPK and mdP2b reveals that a small amount of the P2b hairpin is present in equilibrium with the folded minimal PK (Fig. 6 B and C). Cross-peaks equivalent to those from mdP2b were also present in the spectra of mdFL-PK (Fig. 6 B and D). Based on the relative intensities, it appears that about 20% of mdFL-PK does not fully fold into the pseudoknot but instead forms the P2b hairpin. Of note, the proportion of P2b hairpin conformation is higher in mdFL-PK than in mdPK, where resonance peaks corresponding to the P2b hairpin are barely above noise (Fig. 6 C and D). A pseudoknot–hairpin equilibrium was previously observed for the hPK by NMR, and single-molecule FRET studies indicated that in the human t/PK a significant portion of the molecules adopt an alternative conformation to the pseudoknot (28, 39, 43). These results are consistent with a model in which stable folding of FL-PK requires its protein partner TERT and may take place during catalytic core assembly.
Fig. S6.
Comparison of the imino regions of H2O NOESY spectra of mdP2b, mdP3, and mdFL-PK. (A) mdP2b. (B) mdP3. (C) Superimposition of mdFL-PK (black) and mdP2b (red) spectra. The assignments of mdFL-PK are labeled along the diagonal using the color scheme in Fig. 1. Resonances from mdP2b are labeled in red to demonstrate the presence of a small amount of the P2b hairpin in mdFL-PK.
The 5′ Nucleotides of the J2a/3 Loop Form a Hairpin in the Presence of P2a.
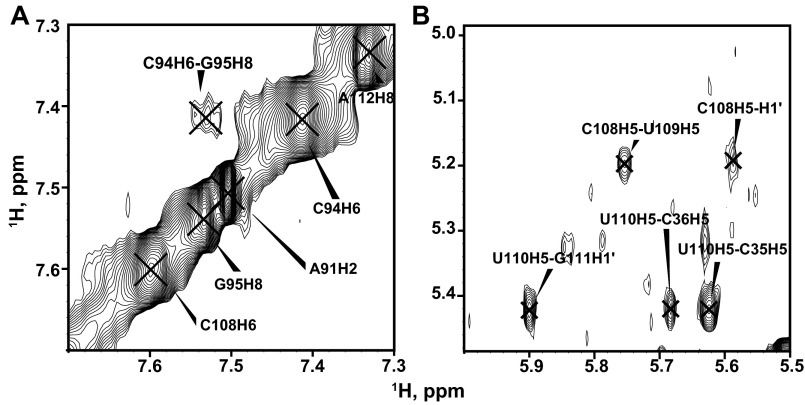
In mdFL-PK, there are several imino resonances that cannot be assigned to the mdPK and P2ab subdomains or to the minor conformation that forms the P2b hairpin instead of the pseudoknot (Fig. 6E). Among these iminos, NOE cross-peaks connecting sequential A–U, G–U, and G–C base pairs were observed. We hypothesized that these might arise from base pairing at the 5′ end of the J2a/3 loop (G95–C108, U96–G107, and U97–A106 base pairs), effectively forming a P2a.1 hairpin. We investigated this possibility, as described below. P2a.1 has been identified as a mammalian-specific helical extension to P2a (20). We enzymatically synthesized an RNA comprising the sequence of nucleotides 95–112 from the 5′ end of the J2a/3 loop and the adjacent P2a stem (nucleotides 34–38 and 90–94) capped with a UUCG tetraloop, here named “mdP2a1a” (Fig. 7A). The P2a stem was included to provide insight into the junction between P2a and the putative P2a.1 hairpin, and the four extra 3′ nucleotides (UUGA) after the hairpin were included because U109 and U110 are conserved in teleost fish. Imino resonances corresponding to the P2a stem and the UUCG tetraloop were sequentially assigned from cross-peaks in the NOESY spectrum, and another set of cross-peaks was assigned to iminos from three consecutive base pairs in the putative P2a.1 hairpin (Fig. 7D). These imino cross-peaks align well with those from mdFL-PK, confirming the formation of P2a in this construct and the putative P2a.1 hairpin assignment in the mdFL-PK spectrum (Fig. S5). The P2a.1 sequence (nucleotides 95–108) forms a duplex rather than a hairpin even at low salt; the isolated hairpin is not expected to be very stable, based on mFold (ΔG = 1.1). Formation of a single conformation for P2a1a also required the additional 3′ UUGA nucleotides (109–112) (Fig. S7).
Fig. 7.
MdTR P2a1a. (A) Proposed secondary structure of mdP2a1a, composed of a P2a.1 hairpin and part of the P2a stem capped with a UUCG tetraloop. (B) The ensemble of the 20 lowest-energy structures. (C) The three lowest-energy models for the P2a.1 hairpin. (D) The imino proton region of the NOESY spectrum of P2a1a.
Fig. S7.
Regions of the NOESY spectrum of mdP2a1a showing aromatic, H5, and H1′ cross-peaks. (A) The cross-peak between C94H6 and G95H8 indicates that P2a.1 stacks on P2a. (B) The cross-peaks of C35H5–U110H5 and C36H5–U110H5 indicate that 3′ UUGA (nucleotides 109–112) interacts with P2a.
In mdP2a1a, NOE cross-peaks observed between C94H6–G95H8 and C94H1′–G95H8, indicating that G95 stacks on C94, together with the presence of iminos for base pairs at the ends of P2a and P2a.1, suggest that P2a and P2a.1 stack on each other (Fig. S7). There are NOEs between P2a.1 (C108H6, C108H5) and 3′ single-strand U109H6 and U109H5, between P2a (C36H5) and 3′ single-strand U110H6, and between U110H5–G111H8 and U110H6–G111H8, together indicating that the 3′ UUGA residues (the 5′ end of the J2a/3 loop) are in the major groove of P2a. The 3′ nucleotides apparently stabilize the stacking of P2a.1 on P2a via their tertiary interactions with the P2a major groove.
Only three sequential base pairs were observed in the P2a.1 hairpin, resulting in an 8-nt loop at the end of the P2a.1. No other iminos or evidence for stable base pairs in the loop were observed in the NMR spectra. Our hypothesis is that the conformation of the loop is highly dynamic, excluding the possibility of determining its structure by NMR. To investigate this hypothesis further, we calculated 2,500 model structures of the P2a.1 loop using an optimized stepwise Monte Carlo (SWM) approach within Rosetta developed in the Das laboratory (SI Materials and Methods) (44). The predicted models of the P2a.1 hairpin (nucleotides 95–108) show that the hairpin loop (nucleotides 98–105) adopts multiple conformations of approximately the same estimated energy (Fig. 7B). These structures generally show partial stacking of bases in the loop and one or two non-Watson–Crick base pairs. For example, the lowest-energy structure has additional G98–A105 and G99–G104 base pairs, G102 stacks on G104, and the U103 base is flipped out of the loop (Fig. 7C). The modeling predicts that the isolated hairpin is of low stability (Rosetta energy −10.8), as is consistent with the NMR data on the P2a.1 sequence alone.
A Potential P2a.1 Hairpin in Other Teleost Fish.
We examined the sequences of TR in other teleost fish to see if they also could form a P2a.1 hairpin. We found that the 5′ end of the J2a/3 loop in all teleost fish could potentially form a P2a.1 hairpin (Fig. S8). Furthermore, the conserved UU nucleotides (109–110 in medaka) are always at the P2a–P2a.1 junction in the predicted secondary structures (Fig. S8) (33), consistent with a role in stabilizing the P2a.1 hairpin fold. In the human t/PK model, the J2a/3 loop binds along the P2a, P2b, and P2a.1 stems. The single-strand length of the human J2a/3 loop (26 nt) is comparable to the number of base pairs (31 bp) within these three stems (Fig. S8). However, the predicted single-strand length of the medaka J2a/3 loop is 34 nt, significantly longer than needed to span the length of P2 (18 bp). Formation of medaka P2a.1 shortens the length of the J2a/3 loop from 34 to 20 nt, making the distance that medaka J2a/3 loop must span similar to the length of P2. Similar ratios are observed for other teleost fish TRs, with the predicted number of nucleotides in the J2a/3 loop similar to the number of P2 base pairs.
Fig. S8.
Proposed secondary structure of teleost fish TR t/PK with the P2a.1 hairpin. At right is comparison of the estimated length of the J2a/3 loop (in nucleotides) vs. P2ab (in base pairs). Non-Watson--Crick pairs, but not bulge nucleotides, are included in the base-pair count.
Effect of Mutations in the J2a/3 Loop on Telomerase Activity.
We investigated the effect of nucleotide substitutions in the putative P2a.1 and flanking nucleotides on telomerase activity. We made mutations that disrupt the base pairs in the putative P2a.1 stem and made compensatory mutations to restore base pairing. Substitution of nucleotides 95–97, GUU, by CGA or nucleotides 106–108, AGC, by UUG, expected to abolish the formation of the P2a.1 stem, decreases telomerase activity by 57% and 48%, respectively (Fig. 4). Combining these in a compensatory mutant to re-pair the stem (CGA:UUG) restores telomerase activity only slightly, to 61% of WT. We used the SWM method to calculate the structure of a P2a.1 hairpin in which the stem sequence was swapped (CGA:UUG) and found that it is predicted to be significantly less stable than the WT P2a.1 hairpin (Rosetta energy of −8.0 vs. −10.8 for WT) (Fig. S9). Therefore the results using the compensatory mutant that repairs the putative P2a.1 stem do not distinguish between single-strand or P2a.1 hairpin formation in the TERT–TR complex. Phylogenetic analysis indicates that U97 is conserved among teleost fish TRs (33). Consistent with human telomerase activity results, substitution of the GUU sequence (nucleotides 95–97) in the stem upper strand has a larger effect on telomerase activity than the substitution of the AGC sequence (nucleotides 106–108) in the lower strand (26). Deletion of residues G95–C108, corresponding to the entire P2a.1 hairpin, decreases activity to ∼33% of WT, indicating that these residues, whether single-stranded or hairpin, are important for activity. In human telomerase, deletion of all J2a/3 residues except the conserved AAA sequence at the 3′ end has little effect on activity (27). Collectively, these results suggest that both the structure and sequence of the 5′ half of the J2a/3 loop are important for medaka telomerase activity and suggest that a P2a.1 hairpin may be formed in the TERT–TR complex.
Fig. S9.
MdTR P2a.1 mutant hairpin (GUU95–97CGA, AGC106–108UUG) with 5' and 3' stem sequence swapped. (A) The three lowest-energy models from the ∼2,500 models. (B) The ensemble of the 20 lowest-energy structures. Notice the lack of model convergence for the loops, suggesting that there is not a single stable lowest-energy structure.
The sequence of base pairs in P2a.1 is not conserved (26, 42). Mutagenesis studies have shown that some nucleotide substitutions in human P2a.1 and a C72G mutation associated with aplastic anemia result in a decrease in telomerase activity (26, 42). The results presented here provide evidence that the 5′ end of the medaka J2a/3 loop forms a P2a.1 hairpin that stacks on the P2a. This conformation is similar to hTR, in which P2a.1 is a linear extension of P2a and there is no significant motion between P2a.1 and P2a (31). Taken together, these results support the hypothesis that P2a.1 is not just a mammalian-specific domain but is conserved among vertebrate TRs.
Model Structure of the mdFL-PK.
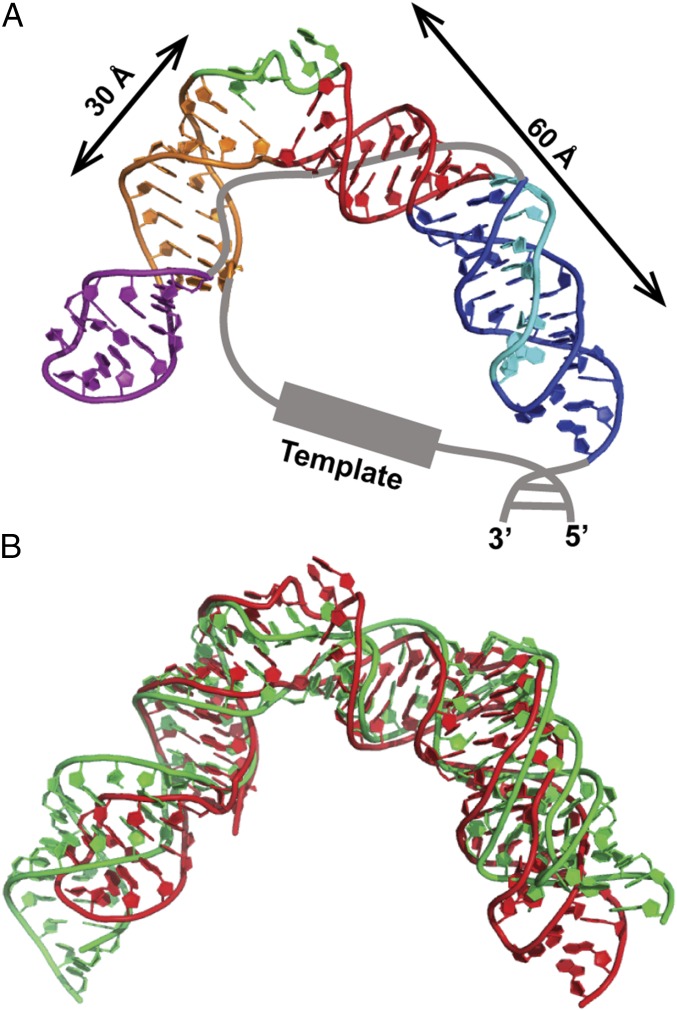
A model structure of the mdFL-PK (excluding the single-strand region of the J2a/3 loop) was made by computationally linking the overlapping ends of the P2a1a model structure, the P2ab solution NMR structure, and the P2b–P3 pseudoknot model structure (Fig. 8A) and minimizing the final model using Rosetta to eliminate steric clashes. This approach was similar to that used to make a model of hFL-PK (31), except in that case the model was refined with Amber. The P2b–P3 pseudoknot and P2a.1–P2a domains are located on either side of the J2a/b bulge, which induces a 79° bend between P2a and P2b, thereby defining the overall topology of the FL-PK. The conserved nucleotides at the 3′ end of the J2a/3 loop interact in the minor groove of the pseudoknot stem P2b. At the 5′ end of the J2a/3 loop defined by the P2a.1 hairpin the conserved UU nucleotides (109 and 110) interact with the major groove. Therefore the J2a/3 loop must cross from the major groove to the minor groove at some point. Although not modeled, the J2a/3 loop could cross into the minor groove at the J2a/b bulge (shown as a gray line in Fig. 8A). hFL-PK has helical regions of ∼50 Å in length on either side of the J2a/b bulge, while mdFL-PK helical regions are ∼60 Å (P2b–P3) and ∼45 Å (P2a–P2a.1), where P2a is ∼30 Å (Fig. 8A). The bend induced by the medaka J2a/b bulge creates an ∼72-Å end–end distance between P2b/P3 and P2a. The template strand would connect to the G34 at the 5′ end of P2a, at the junction of P2a and P2a.1 (Fig. 8A). Although the P2ab bend is somewhat larger for hTR (90°) than for mdTR (79°), medaka has a longer single strand (27 nt) containing the template region than human (24 nt). Overall, the mdFL-PK model structure is similar to the model of hFL-PK (Fig. 8B), indicating that the vertebrate telomerase t/PK is structurally conserved.
Fig. 8.
Model structure of mdFL-PK and hFL-PK. (A) Model structure of mdFL-PK. (B) Superimposition of the model structures of mdFL-PK (red) and hFL-PK (green).
Model Structure of Medaka t/PK on TERT.
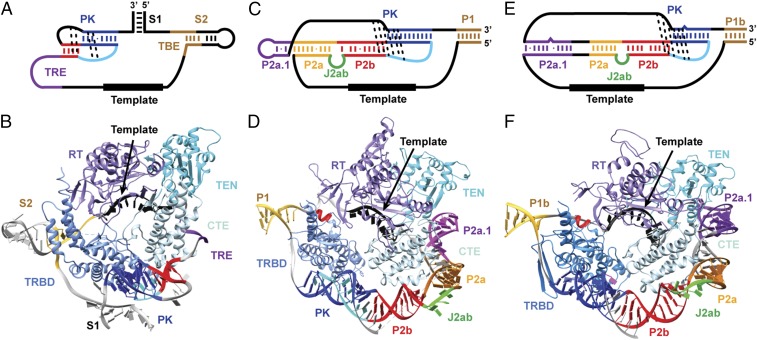
The recently reported ∼9-Å–resolution cryoEM structure of Tetrahymena holoenzyme showed that the t/PK encircles the TERT ring, approximately perpendicular to the plane of the ring, with the pseudoknot on one side and the template on the other (Fig. 9 A and B) (32, 45). The Tetrahymena TR is smaller than the mdTR (159 vs. 312 nt) overall and has a significantly smaller pseudoknot, approximately equivalent in size to the vertebrate minimal pseudoknot. In Tetrahymena, the t/PK circle is closed by S1, and the TBE is S2 and flanking single-stranded nucleotides, located 5′ to the template (Fig. 9A). In medaka, P1 both closes the circle and is the stem of the TBE (based on homology to hTR) (Fig. 9C) (24, 46).
Fig. 9.
Models of medaka and human TERT–t/PK. Secondary structure of the t/PK (A, C, and E) and structural models of TERT–t/PK (B, D, and F) of Tetrahymena (A and B), medaka (C and D), and human (E and F). The structural model of Tetrahymena t/PK is based on the cryoEM structure. The color scheme for TR is same as in Fig. 1. TEN is cyan, TRBD is blue, RT is purple, and CTE is light blue. The TLFY motif within TRBD is red.
We used the position of the Tetrahymena pseudoknot, template, TBE, and the general location of the TR on TERT in the cryoEM structure of Tetrahymena telomerase to model the medaka t/PK on a homology model of medaka TERT (Fig. 9 C and D). In Tetrahymena, the bottom of S2 and the flanking single-stranded residues of the TBE wrap on either side of the TRBD, serving as an anchor to prevent the use of nontemplate nucleotides for replication (47). In the medaka model, the bottom of P1 and flanking single strands of the TBE are positioned similarly. The 3′ single strand (nucleotides 7–14) of the TBE contacts the TFLY motif of TRBD, as proposed for the teleost fish Takifugu rubripes (48). In the medaka model the position of the minimal P2b–P3 pseudoknot is slightly different from the Tetrahymena pseudoknot because of the short linker between P1 and the pseudoknot. However, both are near the TRBD–CTE interface. In Tetrahymena TR, the nucleotides between the TBE and pseudoknot include two single-stranded regions connected by stem S1, whereas in mdTR the TBE is connected directly to the (larger) pseudoknot. In medaka, P1 both closes the t/PK circle and participates in template boundary definition (24, 46), whereas in Tetrahymena S2 participates in template boundary definition and S1 closes the t/PK circle. hTR circular permutation experiments show that breaking the t/PK circle in the P1b (equivalent to medaka P1) region inhibits or abolishes telomerase activity (49), indicating the importance of P1 for closing the t/PK circle. In Tetrahymena, nucleotides between the 3′ end of the template and the pseudoknot are single stranded. This region is called the “template recognition element,” which is proposed to position the 3′ end of the template in the active site (50) and to interact with TERT TEN domain (51). In medaka, there is a shorter region of single-strand nucleotides adjacent to the template which connects to P2ab. The large bend between P2a and P2b induced by the J2a/b bulge allows the TR to fold around TERT, with the nucleotides of the J2a/b bulge facing away from TERT. The model explains why mutations on P2ab that decrease or abolish the bend decrease activity and why it is the bend, not the exact sequence, that is important. The outward-facing position of the bulge nucleotides is consistent with the low sequence conservation of the J2a/b bulge and the proposal that J2a/b flexibility is important for activity (31). The TERT–t/PK model was made using the bend angle found in the NMR structure of P2ab, but it is possible that this bend may be different in the complex with TERT.
Summary and Comparison with Human Telomerase.
Telomerase RNA in general, and the t/PK in particular, contains structured subdomains and single-stranded regions that assemble with TERT to form an active TERT–TR complex. We find that mdPK has the same tertiary interactions as hPK but has a longer P3 that lacks a bulge base and has an additional J2b/3–P3 G–G–C triple. mdP2ab, predicted to have a 6-nt internal loop, has a 5-nt internal loop with a bend angle and dynamics similar to those in hP2ab. Medaka was not predicted to have a P2a.1 extension, but we find that the 5′ end of what was predicted to be J2a/3 forms an mdP2a.1 hairpin that stacks on mdP2ab. By separately determining models or structures of the vertebrate PK, P2ab, and P2a.1 hairpin and combining them computationally, we were able to generate a model of the mdFL-PK and compare it with the previously determined model of hFL-PK (Fig. 8) (31). The 9-Å cryoEM structure of Tetrahymena telomerase provided clear evidence that the pseudoknot is fully folded in the complex with TERT (32), in contrast to a pseudoknot–hairpin equilibrium for the free RNA, as observed by NMR studies of human and medaka PK and FRET studies of hFL-PK (28, 39, 43). Using the positions of the pseudoknot, TBE, and template from the Tetrahymena telomerase cryoEM model structure (32), we were able to model the medaka t/PK on TERT. This information also allowed us to refine previous models of the human TERT–t/PK structure (31). We generated a human telomerase TERT–t/PK model structure using the previously determined model of hFL-PK (31) and a homology model of human TERT (Fig. 9 E and F). The human TERT–t/PK model is very similar to medaka TERT–t/PK, with identical locations of P1, the P2b–P3 pseudoknot, and the P2a.1 extension. As in medaka, the hP2ab subdomain appears to provide a flexible bend to allow the t/PK to encircle the TERT ring. The human and medaka FL-PKs are much larger than that in Tetrahymena, and the position of the minimal pseudoknot in the models is slightly different among the three. In hTR, P2a.1 is an extension of P2a, and in medaka the putative P2a.1 hairpin is formed by J2a/3 loop residues. In both models the P2a.1 is close to or is in contact with the TEN domain, as suggested for human telomerase (12, 52). In addition, in the model human TBE (P1 and its flanking single strands) is positioned in the groove formed by the T-CP motif of the TRBD so that the TBE can contact the TFLY motif of the TRBD directly (48). Although not modeled, the J2a/3 loop is proposed here to wrap around human P2b–P2a–P2a.1 and to have no specific interaction with TERT, consistent with studies that show that deletions and mutations of the J2a/3 loop (except for the CAAA at the 3′ end) have no detectable effect on telomerase activity (26). Comparisons among these models suggest that the architecture and a set of TERT interactions for the vertebrate t/PK domain are conserved from teleost fish to human. Moreover, our human TERT–t/PK model offers a starting point for further investigations into the structure and function of the human telomerase.
SI Materials and Methods
NMR Sample Preparation.
All RNAs were prepared by in vitro transcription using T7 RNA polymerase (P266L mutant) (53) with synthetic DNA templates as described previously (31). RNAs for mdPK (nucleotides 52–82 and 120–140), mdPKΔA131, mdPKA131U, mdPK-0 (nucleotides 52–82 and 121–140), mdP2ab [nucleotides GG;39–55;UUCG;79–89;CC (semicolons are between connected nucleotides)], mdP2a1a (nucleotides 34–38;UUCG;90–112), mdFL-PK (nucleotides 34–140), mdP2b (nucleotides 52–82), mdP3 (nucleotides GG;67–82;120–138;CC), and mdP2a.1 hairpin (nucleotides 95–108) were made unlabeled. In addition, for NMR structure determination of mdP2ab, uniformly 13C,15N-labeled and base-specifically (A, U, G, or C) 13C,15N-labeled RNAs were synthesized. For assignments of mdPK and mdP2a1a, base-specifically 13C,15N-A,U–labeled mdPK and 13C,15N-G–labeled mdP2a1a were synthesized. All transcribed RNAs were ethanol precipitated and purified by 20% denaturing PAGE. RNA was eluted from the sliced gel pieces using an Elutrap apparatus (Whatman) and was further purified from the collected solution by anion-exchange chromatography on a 5-mL Hi-Trap Q column (GE Healthcare). All purified RNA was desalted and exchanged into nanopure water using an Amicon filtration system with 3,000 molecular weight cutoff membranes (Millipore). RNA samples were diluted to concentrations of ∼10 μM in the desired buffer containing 10 mM potassium phosphate, heated at 95 °C for 3 min, snap-cooled on ice for 30 min, and then concentrated to 1–1.5 mM. For each RNA construct, a variety of different KCl concentrations, Mg2+ concentrations, and pH were tested to find solution conditions that gave spectra with the best spectral dispersion and narrowest linewidths. Final conditions for all NMR samples were 10 mM potassium phosphate (pH 6.3), 100 mM KCl except for mdP2ab, which was at pH 6.4. For H2O samples, 10% D2O was included. For D2O samples, the RNA solutions were lyophilized, and the sample was redissolved in the same volume of 99.996% D2O (Sigma); this procedure was repeated at least twice.
NMR Spectroscopy.
NMR spectra were recorded on Bruker DRX 500-MHz and 600-MHz and Avance 800-MHz spectrometers equipped with HCN cryogenic probes. NMR spectra were processed and analyzed with XWINNMR 3.5 (Bruker), NMRPipe (62), and Sparky 3.110 (University of California, San Francisco). For secondary structure determination, the exchangeable-proton NOESY spectra were recorded using H2O samples at 283 K, and nonexchangeable-proton NOESY spectra were recorded on D2O samples at 298 K. The mixing time for mdP2ab and mdP2a1a was 200 ms. For mdPK and mdFL-PK, the mixing time was 150 ms and 100 ms, respectively. For all RNAs except mdFL-PK, base pairs were assigned from 2D NOESY spectra in H2O, and the nonexchangeable aromatic and H1′ base protons were assigned using 2D NOESY in D2O, TOCSY, and natural-abundance 1H-15N HSQC. For mdPK, the imino hydrogen bonds in base pairs were confirmed with 2D H5(C5C4N)H and JNN-COSY experiments (63, 64). For mdFL-PK, the imino proton assignments were assisted by the assignments from the subdomains mdP2ab, mdPK, mdP2b, and mdP3.
For structure determination of mdP2ab, the assignments for all nonexchangeable protons were achieved from analysis of 2D NOESY, 1H-13C HSQC, TOCSY, HCCH-COSY, and 3D HCCH-TOCSY of base-specifically 13C,15N-labeled RNA samples (54, 65, 66). A suite of 2D-filtered/edited proton NOESY (F2f, F1fF2e, F1fF2f, and F1eF2e) experiments on base-specific 13C,15N-labeled RNAs was used to resolve ambiguous assignments in overlapped regions and to obtain NOE restraints, as previously described (55). For structure refinement and characterization of interhelical dynamics of mdP2ab, RDCs were obtained on uniformly 13C,15N-labeled samples in the absence and presence of ∼15 mg/mL Pf1 phage (ASLA Biotech, Ltd.) at 298 K on the 800-MHz spectrometer. One-bond C–H (C1′H1′, C2H2, C5H5, C6H6, and C8H8) RDCs were measured from the splitting of 1H-13C doublets along the 1H dimension using the transverse relaxation-optimized spectroscopy (TROSY)-based 2D 1H-13C S3CT-HSQC (67). One-bond H–N (N1H1 and N3H3) RDCs were determined by using standard 1H-15N HSQC experiments without decoupling in the indirect or direct dimension (68), as described previously (69), and are listed in Table S2. A total of 95 one-bond C–H and N–H RDCs were obtained for mdP2ab. NMR spectra for RDCs were processed and analyzed using NMRPipe/NMRDraw (62). For structural refinement, all C–H and N–H RDCs were input as normalized C–H RDCs with a fixed bond length of 1.0 Å as previously described (31).
Structure Calculation of mdP2ab.
NOE restraints for interproton distances were obtained from the 2D-filtered/edited NOESY, 2D NOESY, and 2D Watergate NOESY spectra acquired in D2O and H2O samples. NOE distance restraints were classified as very strong (2.5 + 1.0/−0.7 Å), strong (3.5 ± 1 Å), medium (4.5 ± 1 Å), weak (5.5 ± 1 Å), and very weak (6.5 ± 1 Å), normalized using the average stem pyrimidine H5–H6 cross-peak intensity (2.45 Å) as a reference. For dihedral angle restraints, stem residues were given A-form restraints (−62.1 ± 30.0° for α, 180.1 ± 30.0° for β, 47.4 ± 30.0° for γ, 37.3 ± 30.0° for ν2, and −73.6 ± 30.0° for ζ); the J2a/b bulge and UUCG tetraloop were left unrestrained. The ribose sugar puckers and δ values were determined according to the intensities of H1′–H2′ cross-peaks in a double-quantum-filtered COSY, i.e., strong (C2′-endo, δ = 145 ± 30°), intermediate (C2′-endo, δ = 120 ± 30°), or no cross-peak (C3′-endo, δ = 82 ± 30°). Hydrogen bond distance and weak planarity restraints were used for the 12 Watson–Crick base pairs and two G–U base pairs. There is only one syn base (G78) in the UUCG tetraloop according to NOE analysis, and the χ angles were restrained to syn (60 ± 20°) for G78 and anti (−160 ± 20°) for the other nucleotides.
An initial 200 structures of mdP2ab were calculated using Xplor-NIH 2.42 (56), starting from an extended unfolded RNA generated with nucleic-3.1, using 802 NOEs and 459 dihedral angle restraints following standard Xplor protocols. The 200 lowest-energy structures were further refined with RDCs. To account for different degrees of alignment caused by interhelical motions, the values for the magnitude (Da) and asymmetry (R) of the alignment tensor for each stem (P2a and P2b) of mdP2ab were first evaluated by analyzing their RDCs using the RAMAH program (57), with the lowest-energy NOE-refined solution structure used as input coordinates. Residues at the end of helices (nucleotides 37, 43–46, and 91) or determined to be flexible from relaxation experiments (Fig. S4) were excluded from the order tensor analysis. During this initial-order tensor analysis (70) idealized A-form helices corresponding to P2a and P2b were constructed using Insight II (Molecular Simulations, Inc.) with propeller twist angles corrected from +14.5° to −14.5° and were used as input coordinates (71). These initial values then were used as the starting values and were further optimized using the structure calculated without RDCs. The optimal values for the magnitude and asymmetry of the alignment tensor are Da = −18.2 Hz and −27.6 Hz and R = 0.15 and 0.26, for P2a and P2b, respectively. During the final structure calculations with RDCs, the force constants for RDCs were gradually increased from 0.001 to 1.0 and 0.5 kcal·mol−1·Hz−2 for P2a and P2b, respectively to account for the difference in the Da values. In the final step of the refinement, database potential was applied with a low weighting. The 20 lowest-energy structures from the final refinement are reported. Restraints and structural statistics are summarized in Table S1. Structures were viewed and analyzed with MOLMOL (58) and PyMOL (PyMOL Molecular Graphics System, version 1.7; Schrödinger, LLC).
The interhelical dynamics were characterized by the resultant generalized degree of orders (GDO): ], where Sxx, Syy, and Szz are the order tensor frames that describe the degree of alignment of each stem in the alignment medium. The resultant Euler angles (α, β, γ) were used to generate a structural model that represents the average relative orientation between the two stems by superimposing their order tensor frames (70, 72). The average interhelical bend, defined as the internal bend between P2a and P2b, was obtained from analyzing these structural models using EularRNA (72).
Model Structure of the mdP2a.1 Hairpin and the mdP2a1a Domain.
The P2a.1 hairpin models were generated using an SWM method, which is based on the Stepwise Assembly method (44) but is far less computation intensive. Although not formally released, extensive documentation for SWM is available on the Rosetta Wikipedia page (Git commit: cb7057ce678691015e3cac64f83eefa4b472d2f9). More general information is available at https://www.rosettacommons.org/docs/latest/application_documentation/stepwise/stepwise_monte_carlo/stepwise.
We initially generated a starting structure using the rna_helix application, which built an idealized helix from our starting sequence, and the command displayed below. This structure first contains the helical stem that will not be built during the SWM modeling to save computational resources. Second, the version of SWM that we used required a native structure to be supplied to do rmsd calculations; this requirement has since been removed, but for backward compatibility we included it:
rna_helix -seq guugggcgugaagc -o octoloop.pdb.
Finally, we ran stepwise the application to run the SWM algorithm. The “–fasta” flag specifies the fasta file containing the full sequence of the target structure. The “-s” flag specifies the template structure, in this case the structure previously generated by rna_helix. The “-extra_min_res” specifies that residues other than those being built should be minimized; in this case a base pair right under the loop should be minimized on the addition of newly built residues. “-nstruct” specifies how many final structures should be generated or how many times the full algorithm should be run. Last, “-cycles” controls the number of Monte Carlo steps that should be used.
The command
stepwise -s octoloop.pdb -native octoloop.pdb -terminal_res 1 14 -extra_min_res 3 12 –fasta octoloop.fasta -nstruct 10 -cycles 5000 -out:file:silent swm_rebuild.out
was run 256 times for a total of ∼2,500 structures (some jobs ended prematurely because wall time ran out). The corresponding commands used to generate the mutant loop structure are shown below:
rna_helix -seq cgagggcgugauug -o mutant_octoloop.pdb
stepwise -s mutant_octoloop.pdb -native mutant_octoloop.pdb -terminal_res 1 14 -extra_min_res 3 12 –fasta mutant_octoloop.fasta -nstruct 10 -cycles 5000 -out:file:silent swm_rebuild.out
The lowest-energy WT hairpin structure was used for the mdP2a1a and mdFL-PK modeling. The stem of mdP2a (nucleotides 90–94 and 34–38) was generated as an A-form helix by the online server RNAComposer (59). mdP2a1 and mdP2a were assembled together by PyMOL, and the 5′ end of the J2a/3 loop was placed and optimized on the major groove of mdP2a using Coot. Both mdPK and mdP2a1a models were energy optimized by Rosetta (73).
Model Structures of mdPK and mdFL-PK.
A model structure of mdPK (nucleotides 52–82 and 120–140) was created based on the solution NMR structure of hPKΔU (PDB ID code 2K96) by substitution. Nucleotides that differed from hPKΔU were replaced, the extra C nucleotide was added to the beginning of the J2a/3 loop, and the two additional U–A base pairs were added to the end of P3. The J2a/3 and J2b/3 loops were placed in the minor groove of P2b and the major groove of P3, respectively. Standard bond angles and lengths of backbone of the nucleotides in the loops were optimized by using the Regularize Zone tool in Coot (74). A model structure of mdFL-PK was generated by linking three subdomains (mdP2ab, mdPK, and mdP2a1a) using overlapping nucleotides (nucleotides 53–55 and 79–81 between mdP2ab and mdPK; nucleotides 37–38 and 90–91 between mdP2ab and mdP2a1a) by PyMOL and Coot. The model structure of hFL-PK was generated by RDC–MC-Sym in our previous study (31).
Model Structure of Medaka TERT-t/PK.
A model structure of medaka t/PK in complex with TERT was generated as follows. For TERT, medaka RT and CTE domains (residues 562–1089) were modeled using the online server SWISS-MODEL (60) based on the crystal structure of Tribolium TERT in complex with an RNA–DNA hybrid helix (PDB ID code 3KYL). The TEN domain (residues 1–190) was modeled using the online server HHpred (61) based on the Tetrahymena TEN crystal structure (PDB ID code 2B2A). The insertion in fingers domain (residues 711–780) was removed because its structure was poorly predicted. The medaka TRBD (residues 310–561) model is a combination of a crystal structure of medaka TRBD lacking the TFLY motif (PDB ID code 4O26) and a homology model of the TFLY motif from the Takifugu TRBD crystal structure (PDB ID code 4LMO) generated by the Swiss-MODEL server (60). The TRBD and the RT and CTE domains were assembled together to form the TERT ring. The position of the TEN domain relative to the TERT ring (TRBD–RT–CTE) was based on its position in the Tetrahymena cryoEM holoenzyme model structure (32) and was placed using Chimera. The t/PK domain was generated by connecting P1 (modeled as ideal A-form RNA) and the mdFL-PK model with the single-stranded regions between these domains using PyMOL and Coot. Briefly, ideal A-form single-stranded RNA fragments were approximately placed between their flanking TR subdomains using PyMOL, and then the bond angles were adjusted by Coot. The TERT and t/PK models were assembled together using Chimera. First, the template and region of the pseudoknot corresponding to mdPK were placed based on their respective locations in the Tetrahymena telomerase cryoEM model structure. P1 was placed based on the location of Tetrahymena telomerase SL2. Second, the position of mdFL-PK was adjusted to satisfy the linker length between P1 and the pseudoknot. Finally, the bond angles of single-stranded regions were optimized by Coot to avoid steric clashes between TR and TERT.
A model of human TERT–t/PK was generated similarly. For human TERT, residues 595–1111 were used for the RT and CTE domains, residues 322–594 were used for the TRBD, and residues 1–200 were used for the TEN domain. A Swiss homology model of the TRBD was generated based on the Takifugu TRBD crystal structure.
Preparation of WT and Mutant Full-Length mdTRs.
Full-length (312-nt) WT mdTR (a gift from Julian J. L. Chen, Department of Chemistry & Biochemistry, Arizona State University, Tempe, AZ) was prepared from the pUC19 plasmid (pUC19mdTR). All J2a/b substitutions were carried out directly on the pUC19mdTR plasmid using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Both WT and mutant RNAs were transcribed in vitro using linearized DNA templates obtained from PCR amplification of the pUC19mdTR using a 5′ primer that contained the T7 promoter added to the 5′ end of the full-length mdTR sequence and a 3′ primer that is complementary to the 3′ end of the full-length mdTR. RNA products were purified by 5% PAGE and electroelution as described above for NMR samples.
Materials and Methods
NMR Sample Preparation and Spectroscopy.
All RNAs were prepared by in vitro transcription using T7 RNA polymerase (P266L mutant) with synthetic DNA templates (53). NMR experiments were carried out on Bruker DRX 500- and 600- and Avance 800-MHz spectrometers equipped with HCN cryogenic probes. All NMR samples were ∼1 mM in 10 mM phosphate buffer, pH 6.3, 100 mM KCl, except for mdP2ab which was at pH 6.4. For secondary structure determination, the exchangeable-proton NOESY spectra were recorded using 90% H2O/10% D2O samples at 283 K, and nonexchangeable proton NOESY spectra were recorded on 100% D2O samples at 298 K. For structure determination of mdP2ab, the assignments for all nonexchangeable protons were achieved from analysis of 2D NOESY, 1H-13C HSQC, TOCSY, HCCH-COSY, and 3D HCCH-TOCSY of base-specifically 13C,15N-labeled RNA samples (54). A suite of 2D-filtered/edited proton NOESY (F2f, F1fF2e, F1fF2f, and F1eF2e) experiments on base-specifically 13C,15N-labeled RNAs was used to resolve ambiguous assignments in overlapped regions and to obtain NOE restraints, as previously described (55). For structure refinement and characterization of interhelical dynamics of mdP2ab, RDCs were obtained on uniformly 13C,15N-labeled samples in the absence and presence of ∼15 mg/mL Pf1 phage (ASLA Biotech) at 298 K on the 800-MHz spectrometer. Detailed descriptions of sample preparation, NMR experiments, and assignment methodology are given in SI Materials and Methods.
Structure Calculations.
The structure of mdP2ab was calculated from an extended unfolded RNA using 802 NOEs and 459 dihedral angle restraints following standard Xplor protocols as described previously (31) but modified to use the updated NIH-Xplor 2.42 (56). The 200 lowest-energy structures were further refined with 95 RDCs. To account for different degrees of alignment resulting from interhelical motions, the values for the magnitude (Da) and asymmetry (R) of the alignment tensor for each stem (P2a and P2b) of mdP2ab were first evaluated by analyzing their RDCs using the RAMAH program (57), with the lowest-energy NOE-refined solution structure used as input coordinates. The residues at the end of helices (37, 43–46, and 91) or determined to be flexible from relaxation experiments (Fig. S4) were excluded from the order tensor analysis. The optimal values for the magnitude and asymmetry of the alignment tensor are Da = −18.2 Hz and −27.6 Hz and R = 0.15 and 0.26, for P2a and P2b, respectively. The 20 lowest-energy structures from the final refinement are reported. Restraints and structural statistics are summarized in Table S1. Structures were viewed and analyzed with MOLMOL (58) and PyMOL (PyMOL Molecular Graphics System, version 1.7; Schrödinger).
Model Structure of TERT–t/PK.
A model structure of mdPK was created based on the solution NMR structure of hPKΔU (PDB ID code 2K96) by substitution. A model structure of the mdP2a1 hairpin was calculated using the SWM method (44) and was incorporated into the Rosetta codebase written in C++. A description of the SWM application and detailed command-line examples are provided in SI Materials and Methods. The lowest-energy WT hairpin structure was used for the mdP2a1a and mdFL-PK modeling. The stem of mdP2a was generated as an A-form helix by the online server RNAComposer (59). mdP2a1 and mdP2a were assembled together by PyMOL, and the 5′ end of the J2a/3 loop was placed and optimized on the major groove of mdP2a using Coot. A model structure of mdFL-PK was generated by PyMOL and Coot by linking three subdomains (mdP2ab, mdPK, and mdP2a1a) using overlapping nucleotides. For Medaka TERT, the medaka RT and CTE domains were modeled using the online server SWISS-MODEL (60) based on the crystal structure of Tribolium TERT in complex with an RNA–DNA hybrid helix (PDB ID code 3KYL). The TEN domain was modeled using the online server HHpred (61) based on the Tetrahymena TEN crystal structure (PDB ID code 2B2A). The medaka TRBD model is a combination of a crystal structure of medaka TRBD lacking the TFLY motif (PDB ID code 4O26) and a homology model of the TFLY motif from the Takifugu TRBD crystal structure (PDB ID code 4LMO) generated by the Swiss-MODEL server (60). A model structure of medaka t/PK in complex with TERT was generated based on its position in the Tetrahymena cryoEM holoenzyme model structure (32) and was placed using Chimera. A model of human TERT–t/PK was generated similarly; detailed procedures of the modeling are described in SI Materials and Methods.
In Vitro Reconstitution of Telomerase and Direct Telomerase Activity Assays.
TERT and TR (WT and mutant) were reconstituted in RRL using the TNT quick-coupled transcription/translation system (Promega), and direct telomerase activity assays were performed as previously described (35). The relative activity was determined by measuring the total intensity of extended telomere primers after background correction and normalizing against the recovery control (33). The activity with WT mdTR was set to be 100. Each activity assay was repeated a minimum of two times.
Acknowledgments
This work was supported by NIH Grant GM48123 (to J.F.); National Science Foundation Grant MCB1517625 (to J.F.); and Ruth L. Kirschstein National Research Service Award Postdoctoral Fellowships GM112503 (to Y.W.) and GM112294 (to J.D.Y.). The NMR facility was supported in part by US Department of Energy Grant DE-FC03-02ER63421.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5KQE). The NMR chemical shifts have been deposited in the Biological Magnetic Resonance Bank, www.bmrb.wisc.edu (accession no. 30132).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607411113/-/DCSupplemental.
References
- 1.Nandakumar J, Cech TR. Finding the end: Recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14(2):69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capper R, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21(19):2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol. 2000;1(1):72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8(3):167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 5.Shay JW, Wright WE. Telomerase therapeutics for cancer: Challenges and new directions. Nat Rev Drug Discov. 2006;5(7):577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 6.Kirwan M, Dokal I. Dyskeratosis congenita: A genetic disorder of many faces. Clin Genet. 2008;73(2):103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 7.Calado RT, Young NS. Telomere maintenance and human bone marrow failure. Blood. 2008;111(9):4446–4455. doi: 10.1182/blood-2007-08-019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13(10):693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JL, Greider CW. Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends Biochem Sci. 2004;29(4):183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi H, et al. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102(3):916–918. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 11.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7(7):484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014;33(8):921–935. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287(16):12805–12814. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13(3):218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 15.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21(4):990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 17.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38(17):5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455(7213):633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17(4):513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 20.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100(5):503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 21.Kachouri-Lafond R, et al. Large telomerase RNA, telomere length heterogeneity and escape from senescence in Candida glabrata. FEBS Lett. 2009;583(22):3605–3610. doi: 10.1016/j.febslet.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Kuprys PV, et al. Identification of telomerase RNAs from filamentous fungi reveals conservation with vertebrates and yeasts. PLoS One. 2013;8(3):e58661. doi: 10.1371/journal.pone.0058661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6(2):361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 24.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17(22):2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc Natl Acad Sci USA. 2004;101(41):14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ly H, Blackburn EH, Parslow TG. Comprehensive structure-function analysis of the core domain of human telomerase RNA. Mol Cell Biol. 2003;23(19):6849–6856. doi: 10.1128/MCB.23.19.6849-6856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JL, Greider CW. Functional analysis of the pseudoknot structure in human telomerase RNA. Proc Natl Acad Sci USA. 2005;102(23):8080–8085, discussion 8077–8079. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17(5):671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Cash DD, et al. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci USA. 2013;110(27):10970–10975. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15(6):634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci USA. 2010;107(44):18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350(6260):aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie M, et al. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem. 2008;283(4):2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- 34.Huang J, et al. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21(6):507–512. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim NK, Zhang Q, Feigon J. Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic Acids Res. 2014;42(5):3395–3408. doi: 10.1093/nar/gkt1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim NK, et al. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol. 2008;384(5):1249–1261. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luy B, Marino JP. Direct evidence for Watson-Crick base pairs in a dynamic region of RNA structure. J Am Chem Soc. 2000;122(33):8095–8096. [Google Scholar]
- 38.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. A molecular switch underlies a human telomerase disease. Proc Natl Acad Sci USA. 2002;99(26):16998–17003. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theimer CA, Finger LD, Trantirek L, Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc Natl Acad Sci USA. 2003;100(2):449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36(Database issue):D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman RA, Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys J. 1995;69(4):1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22(2):304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hengesbach M, Kim NK, Feigon J, Stone MD. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew Chem Int Ed Engl. 2012;51(24):5876–5879. doi: 10.1002/anie.201200526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sripakdeevong P, Kladwang W, Das R. An enumerative stepwise ansatz enables atomic-accuracy RNA loop modeling. Proc Natl Acad Sci USA. 2011;108(51):20573–20578. doi: 10.1073/pnas.1106516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang J, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496(7444):187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown AF, et al. A self-regulating template in human telomerase. Proc Natl Acad Sci USA. 2014;111(31):11311–11316. doi: 10.1073/pnas.1402531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jansson LI, et al. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22(11):883–888. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure. 2013;21(10):1870–1878. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mefford MA, Zappulla DC. Physical connectivity mapping by circular permutation of human telomerase RNA reveals new regions critical for activity and processivity. Mol Cell Biol. 2015;36(2):251–261. doi: 10.1128/MCB.00794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci USA. 2002;99(10):6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280(17):17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015;43(11):5537–5549. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA. 2005;102(17):5958–5963. doi: 10.1073/pnas.0407141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dieckmann T, Feigon J. Assignment methodology for larger RNA oligonucleotides: Application to an ATP-binding RNA aptamer. J Biomol NMR. 1997;9(3):259–272. doi: 10.1023/a:1018622708674. [DOI] [PubMed] [Google Scholar]
- 55.Peterson RD, Theimer CA, Wu H, Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J Biomol NMR. 2004;28(1):59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- 56.Bermejo GA, Clore GM, Schwieters CD. Improving NMR Structures of RNA. Structure. 2016;24(5):806–815. doi: 10.1016/j.str.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen AL, Al-Hashimi HM. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: Towards new long-range orientational constraints. J Magn Reson. 2006;179(2):299–307. doi: 10.1016/j.jmr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Molecular Graphics. 1996;14(1):51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 59.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40(14):e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 63.Dingley AJ, Nisius L, Cordier F, Grzesiek S. Direct detection of N-H[...]N hydrogen bonds in biomolecules by NMR spectroscopy. Nat Protoc. 2008;3(2):242–248. doi: 10.1038/nprot.2007.497. [DOI] [PubMed] [Google Scholar]
- 64.Wöhnert J, Ramachandran R, Görlach M, Brown LR. Triple-resonance experiments for correlation of H5 and exchangeable pyrimidine base hydrogens in (13)C,(15)N-labeled RNA. J Magn Reson. 1999;139(2):430–433. doi: 10.1006/jmre.1999.1797. [DOI] [PubMed] [Google Scholar]
- 65.Cavanagh J, Rance M. Sensitivity improvement in isotropic mixing (TOCSY) experiments. J Magn Reson. 1990;88(1):72–85. [Google Scholar]
- 66.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 67.Meissner A, Sørensen OW. The role of coherence transfer efficiency in design of TROSY-type multidimensional NMR experiments. J Magn Reson. 1999;139(2):439–442. doi: 10.1006/jmre.1999.1788. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450(7173):1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 69.Shen K, et al. Molecular mechanism of GTPase activation at the signal recognition particle (SRP) RNA distal end. J Biol Chem. 2013;288(51):36385–36397. doi: 10.1074/jbc.M113.513614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33(4):371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 71.Musselman C, et al. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J Biomol NMR. 2006;36(4):235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 72.Bailor MH, et al. Characterizing the relative orientation and dynamics of RNA A-form helices using NMR residual dipolar couplings. Nat Protoc. 2007;2(6):1536–1546. doi: 10.1038/nprot.2007.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 74.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]