Significance
Understanding the evolutionary history of Haemosporidian parasites would help to understand human malaria evolution. Nevertheless, Haemosporidia parasite diversity in bats remains largely unstudied. In addition, some cases of unsuccessful PCR amplification of cytochrome b, the most widely used molecular marker to characterize Haemosporidia parasites, have recently been reported for Nycteria parasites infecting Nycteridae bats. Here we used next-generation sequencing to characterize mitochondrial genomes of parasites from the Nycteria genus. Our results have revealed an unsuspected mitochondrial genome rearrangement within Haemosporidia.
Keywords: cytochrome b, evolution, phylogeny, Nycteris
Abstract
Haemosporidia parasites have mostly and abundantly been described using mitochondrial genes, and in particular cytochrome b (cytb). Failure to amplify the mitochondrial cytb gene of Nycteria parasites isolated from Nycteridae bats has been recently reported. Bats are hosts to a diverse and profuse array of Haemosporidia parasites that remain largely unstudied. There is a need to obtain more molecular data from chiropteran parasites. Such data would help to better understand the evolutionary history of Haemosporidia, which notably include the Plasmodium parasites, malaria’s agents. We use next-generation sequencing to obtain the complete mitochondrial genome of Nycteria parasites from African Nycteris grandis (Nycteridae) and Rhinolophus alcyone (Rhinolophidae) and Asian Megaderma spasma (Megadermatidae). We report four complete mitochondrial genomes, including two rearranged mitochondrial genomes within Haemosporidia. Our results open outlooks into potentially undiscovered Haemosporidian diversity.
Haemosporidia blood parasites (phylum Apicomplexa) are vector-borne protozoan parasites that infect multiple vertebrate hosts such as squamates, chelonians, birds, and mammals, including humans, in which five species of the genus Plasmodium are known to cause malaria (1, 2). The diversity of Haemosporidia has been mostly investigated using mitochondrial genes. In particular, the mitochondrial (mt) cytochrome b (cytb) gene is one of the most widely used genetic markers to characterize the diversity of Haemosporidia parasites, and has given rise to many phylogenetic reconstructions to document the evolutionary history of the Haemosporidia (3–5). All Haemosporidia mt genomes previously sequenced share the same structure, characterized by a tandemly repeated linear element of ∼6 kb containing the three protein-coding genes cytb, cytochrome c oxidase subunit 1 (cox1), and cytochrome c oxidase subunit 3 (cox3) and two highly fragmented (small and large subunits) ribosomal RNA (rRNA) genes (6). In the Plasmodium species, copy numbers of the mt genome are dozens to hundreds of times more numerous than the nuclear genomes (7, 8). Consequently, mt genes are generally easily amplified using traditional PCR techniques.
Nevertheless, several cases of unsuccessful PCR amplification of mt DNA markers have been recently reported, in particular for Nycteria and Plasmodium parasites infecting, respectively, Nycteridae bats and Alaudidae birds (9, 10). Three alternative hypotheses have been proposed to explain these negative results: (i) loss of the mitochondrial genome during the evolution of these parasites; (ii) transfer of some mitochondrial genes into the nuclear genome; and (iii) technical problems due to the use of inefficient or misdesigned primer sets.
Bats are hosts to a large, diverse, and specific array of Haemosporidia parasites (11). However, this parasite diversity remains largely unstudied at the molecular and genomic levels, and this lack of knowledge limits the understanding of the evolutionary history of Haemosporidia, in particular their basal diversification.
Nycteria parasites have been primarily described, based on traditional taxonomy, in African insectivorous bats of two families, Nycteridae and Rhinolophidae (12). The type species of the genus Nycteria is Nycteria medusiformis, first described from Kenya by Garnham and Heisch in 1953 (13) from the Egyptian slit-faced bat (Nycteris thebaica). Later, this species, N. medusiformis, was reported in other species of Nycteris, including the large slit-faced bat, Nycteris grandis (14). A total of six species have so far been described in the genus Nycteria (14).
To contribute to the identification and characterization of Nycteria parasites in bats and to ascertain the reasons for the failure of PCR amplification in previous studies, we have used deep next-generation sequencing (NGS) to assemble the complete mitochondrial genome of four different Nycteria species: N. medusiformis and Nycteria sp. (mixed infection) from N. grandis (Nycteridae); Nycteria gabonensis from Rhinolophus alcyone (Rhinolophidae); and a species of Nycteria, herein described and named Nycteria heischi n. sp., from Megaderma spasma (Megadermatidae). Our results have revealed an unsuspected mitochondrial genome rearrangement within the Haemosporidia.
Results
Three out of 13 N. grandis and four out of four R. alcyone from a total of 195 bats (six families) were found to be infected with Nycteria parasites in the Democratic Republic of Congo. Four out of five M. spasma from a total of 90 bats (six families) were found to be infected with Nycteria parasites in Cambodia.
All of the three N. grandis were coinfected with N. medusiformis (13) and an unidentified Nycteria sp. The four R. alcyone were infected with N. gabonensis (14). N. medusiformis and N. gabonensis were identified according to their morphological traits, from N. grandis and R. alcyone, respectively (Fig. 1). Thin blood smears of N. medusiformis from N. grandis [Museum National d’Histoire Naturelle (MNHN) registration nos. 178PBP, 225PB, and 226PB] and N. gabonensis from Rhinolophus sylvestris (MNHN registration nos. 155GG, 376GG, and 377GG), collected respectively in 1977 and 1976–1977 in Gabon, from the Landau collection stored at the Museum National d’Histoire Naturelle, were included in the study for taxonomic examinations. The Nycteria parasites from M. spasma were characterized and described morphologically and named N. heischi (MNHN registration no. 206YZ; hapantotype collection no. PXX99).
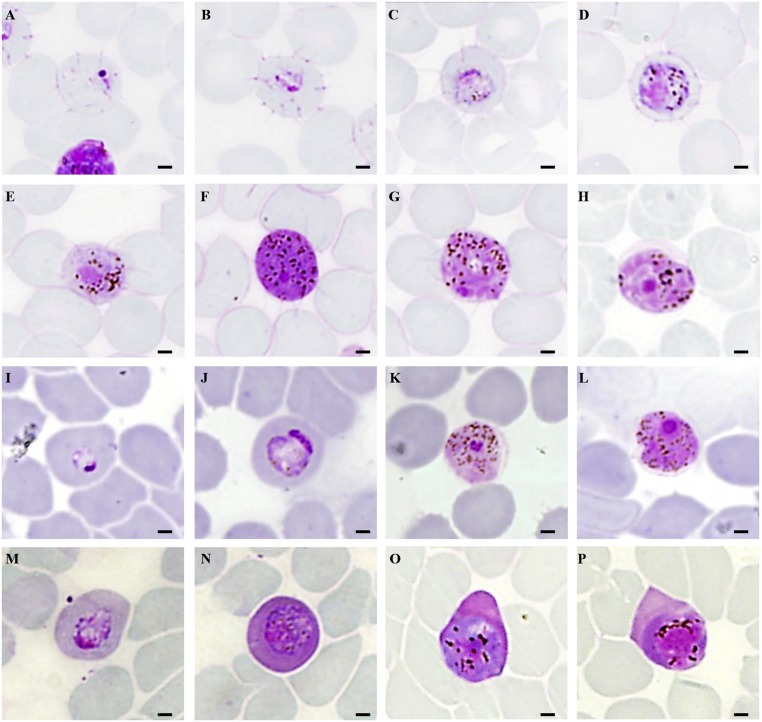
Fig. 1.
Microphotographs of gametocytes of Nycteria parasites from N. grandis (A–H), M. spasma (I–L), and R. alycone (M–P). (A and B) Very young gametocytes. (C–E) Immature gametocytes. (F and G) Macro- and microgametocytes. (H) Old gametocyte of N. medusiformis from N. grandis (273VI). (I and J) Immature gametocytes. (K and L) Macro- and microgametocytes of N. heischi n. sp. from M. spasma (C289). (M and N) Immature gametocytes. (O and P) Macro- and microgametocytes of N. gabonensis from R. alcyone (289VI). (Scale bars, 1.5 μm.)
The morphological characteristics of N. medusiformis gametocytes were consistent with the original description (12, 13). Mature gametocytes completely filled the red blood cells (RBCs), which were slightly enlarged (8 µm instead of 6 µm for uninfected RBCs; n = 10). In most stages of gametocytes, filaments could be observed at the surface of infected erythrocytes. These filaments became fewer and shorter as the gametocytes matured, and sometimes disappeared at the later stages. Pigment grains were fine and irregularly scattered throughout the cytoplasm. Macrogametocytes were highly chromophilic and appeared dark blue-purple with small and condensed nuclei. Microgametocytes had a pale pink color and the nuclei displayed a small central condensation of chromatin reminiscent of a “cocarde” (rosette) (Fig. 1 A–H). The Nycteria sp. morphological type, observed in N. grandis in a mixed infection, was similar to N. medusiformis. Nevertheless, it did not present the highly characteristic filaments observed in N. medusiformis. This morphotype differed mostly in size, in the nucleus of the microgametocytes, and in pigment features. However, the number of gametocytes of this Nycteria sp. was insufficient for a full description. N. medusiformis was highly predominant, whereas Nycteria sp. was less frequent.
With NGS sequencing, we obtained two complete mitochondrial genomes of Nycteria from N. grandis (273VI), using iSeGWalker. No other Haemosporidia mt genomes were detected in this bat sample by NGS sequencing. Both mt genomes were 6,091-bp-long, with a total of 113,661 reads and a mean coverage of 1,006. These two Nycteria parasites share the same genome organization, and their nucleotide sequences differ in only 83 out of the 6,091 positions (about 1.4%). Read coverages of 80% and 20% were obtained for the two Nycteria mt genomes. Based on the morphological observation frequencies of both Nycteria morphotypes, we propose assigning the predominant mt genome to N. medusiformis and the less frequent to Nycteria sp.
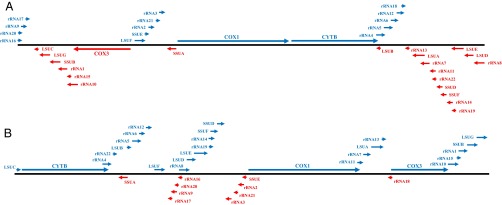
Both N. medusiformis and Nycteria sp. mt genomes contained the three protein-coding genes cytb, cox1, and cox3 as well as the two highly fragmented rRNA genes (Fig. 2B). They displayed a highly rearranged mt genome organization compared with other Nycteria parasites and all other Haemosporidia mt genomes sequenced until now. These rearrangements concerned gene order, transcriptional direction (gene inversion at cox3), as well as the pattern of high fragmentations of the large- and small-subunit rRNA genes (Fig. 2B). However, the rRNA sequences were well-conserved between the Nycteria parasites from N. grandis and all other known Haemosporidia mt genome parasites. The three protein-coding genes, cox1, cytb, and cox3, were divergent from those of N. gabonensis and N. heischi and from all other Haemosporidia parasites.
Fig. 2.
Two schematic types of mitochondrial genome organization. (A) Typical Haemosporidia mt genome organization, as in N. gabonensis and N. heischi. (B) N. medusiformis and Nycteria sp. mt genome organization. LSU, large subunit; SSU, small subunit.
N. medusiformis mt genome reconstruction was successfully validated by the amplification of a 1,439-bp mt fragment using classical PCR tools with specifically designed primers followed by Sanger sequencing. However, we did not succeed in amplifying and sequencing the mt fragment of Nycteria sp. using classical PCR and Sanger sequencing. This classical method favored the amplification and/or sequencing of only the N. medusiformis parasite in the case of the N. grandis mixed infection (15).
Only one species of Nycteria was observed in thin blood smears from R. alcyone. Morphological characteristics were consistent with the original description of N. gabonensis gametocytes (14) (Fig. 1 M–P). Briefly, the erythrocyte was deformed and enlarged (mean measurements of 9 µm; n = 10). It displayed a more or less dark pink color. Mature gametocytes (mean measurements of 7.65 µm; n = 10) were roundish and did not fill the erythrocyte. Microgametocytes had a large well-limited nucleus with a diffuse chromatin and dark pigments, which were mostly seen in the periphery of the nucleus. The macrogametocyte nuclei were smaller and irregular in shape. Only one mitochondrial genome of 5,999 bp was reconstructed from the R. alcyone (289VI) sample, with a total of 16,361 reads and a mean coverage of 272. This mt genome could confidently be assigned to N. gabonensis.
N. heischi from M. spasma.
The shape and staining affinities of the M. spasma RBCs were not modified by the presence of the N. heischi gametocytes (Fig. 1 I–L). Young gametocytes were ovals and occupied about a quarter of the RBCs. The nucleus was made of two or three chromatin grains of variable size, often located between a bluish relatively large vacuole and a smaller white one. A few very fine grains of pigment could be observed in the cytoplasm. As gametocytes matured, they became rounded. The cytoplasm was light blue or grayish and the nucleus was granular and peripheral, either half-moon–shaped in younger stages or larger in older stages. Mature gametocytes of both sexes were pink, of a light color in macrogametocytes and much darker in microgametocytes. They filled the RBCs almost completely, which were slightly enlarged (mean measurements of about 7 µm; n = 10). There was no “accessory chromatin dot.” The nucleus of the microgametocytes was arranged as a rosette (en cocarde) comprising a pink more or less homogeneous area devoid of pigment and a central condensed granular zone. The pigment (about 30 grains) was fine, of variable shape and size, and dispersed in the cytoplasm. The nucleus of the macrogametocytes was smaller, dense, and granular. The cytoplasm was slightly foamy and the pigment was coarser and more abundant than in the microgametocytes.
N. heischi was morphologically related to the group of African Nycteria from Nycteris bats with the nucleus of the gametocytes en cocarde: N. medusiformis (13), Nycteria houini, and Nycteria erardi (14). The schizogonic stages of N. heischi are unknown. Gametocytes differ morphologically from (i) N. medusiformis by the absence of filaments on the erythrocyte; (ii) N. houini by their effect on RBCs, which stain a dark-pink color in the latter (i.e., N. houini) but are colorless when seen in N. heischi, and by the “rice grains” aspect of the pigment, scarcer and irregularly disposed in N. houini and abundant, dispersed, and in small grains in N. heischi; and (iii) N. erardi by the “pine needle” pigment, scarcer, irregularly disposed, and abundant, yet dispersed and in small grains in N. heischi.
Taxonomic Summary.
Taxon: N. heischi n. sp. (Karadjian, Landau, and Duval, 2016).
Type host: M. spasma (Linnaeus, 1758).
Type locality: Mondolkiri, Cambodia.
Etymology: Named in honor of R. B. Heisch, codiscoverer with P. C. C. Garnham of N. medusiformis, the type species of the Nycteria genus.
Hapantotype: One blood film deposited at the collection of the Museum National d’Histoire Naturelle de Paris (collection no. PXX99).
The complete mitochondrial genome of 5,989 bp from N. heischi (C289) was reconstructed with a total of 64,660 reads and a mean coverage of 734. Both mt genomes of N. gabonensis and N. heischi, obtained respectively from R. alcyone (289VI) and M. spasma (C289), show the same genome organization as that of all other species of Haemosporidia parasites currently known (Fig. 2A).
GenBank accession numbers of the four Nycteria mt genomes are N. medusiformis (273VI), KX090645; Nycteria sp. (273VI), KX090646; N. gabonensis (289VI), KX090647; and N. heischi (C289), KX090648.
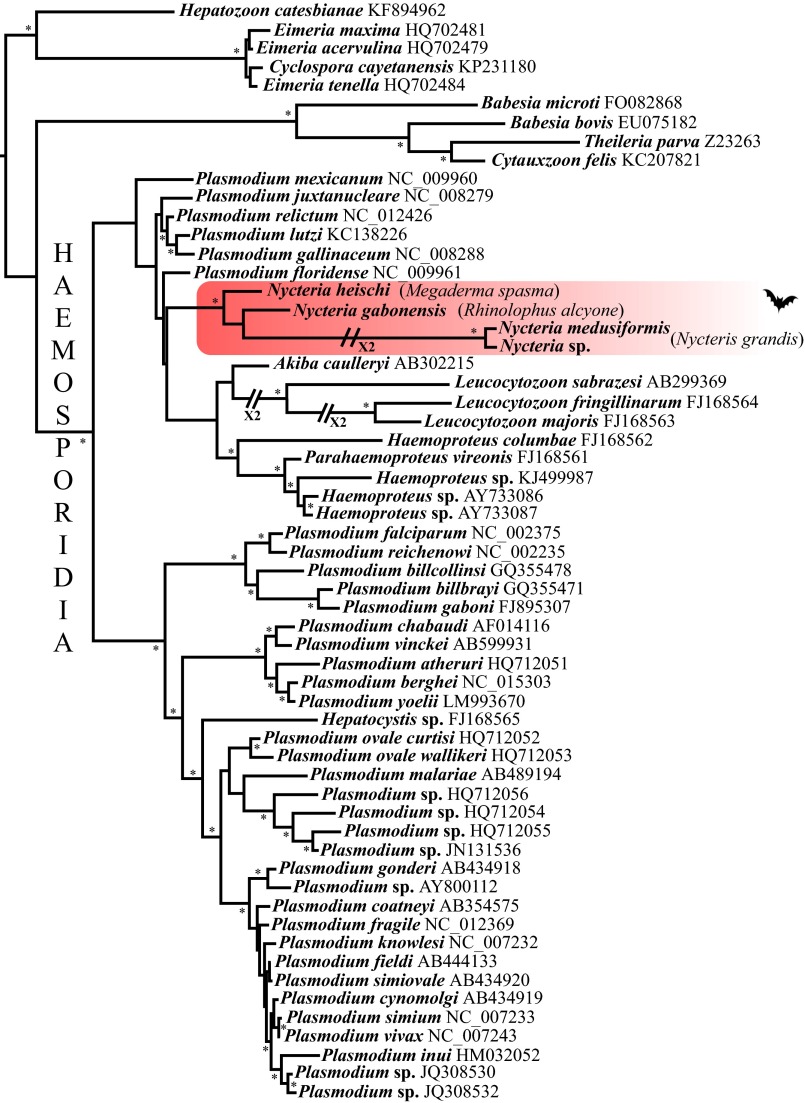
Phylogenetic analyses based on the datasets (3PCG and cytb) supported the monophyly of the genus Nycteria [posterior probabilities (PP) > 0.90] as well as a sister-group relationship between N. medusiformis and Nycteria sp. from N. grandis (PP = 1) (Fig. 3 and Fig. S1). The branch leading to the two latter species is much longer than other branches of the tree.
Fig. 3.
Mitochondrial genome phylogeny (3PCG) of Haemosporidia parasites including the four Nycteria species obtained by MrBayes. The Nycteria species group is highlighted in red. Splits with a posterior probability >0.90 are indicated with asterisks. The tree was rooted with Eucoccidiorida and Piroplasmida.
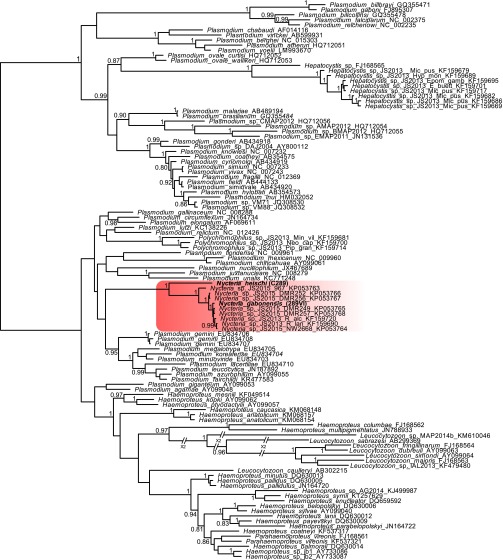
Fig. S1.
cytb phylogeny of the Haemosporidia parasites, including Nycteria sp. sequences from ref. 9. Splits with a posterior probability >0.80 are indicated. The Nycteria group is highlighted in red. The tree is unrooted.
The placement of the genus Nycteria in deep-level relationships within the Haemosporidia could not be confidently determined.
Discussion
The use of the deep NGS approach has allowed us to discover an unsuspected and highly rearranged mitochondrial genome in Nycteria parasites infecting African bats of the family Nycteridae, which had not been successfully amplified and sequenced using classical PCR and Sanger sequencing approaches. Our results reveal that neither the order nor the transcriptional direction of the three protein-coding mitochondrial genes (cytb, cox1, and cox3) is conserved within the Nycteria genus and Haemosporidia parasites. The large and small subunits of the rRNA gene sequences are well-conserved, but their highly fragmented patterns in N. medusiformis and Nycteria sp. are remarkably different from all other Haemosporidia mt genomes sequenced until now (8). This rearrangement of the mt genomes of N. medusiformis and Nycteria sp. was discovered by means of deep NGS sequencing and primer sets. In other Apicomplexan parasites outside the order Haemosporidia, such as the order Piroplasma (genera Babesia and genera Theileria), the mt genomes exhibit remarkably diverse structures (6). However, the biological and evolutionary significance of this mt genome diversity remains to be explored (6).
The use of deep NGS sequencing associated with morphological descriptions might also be an alternative and reliable method to determine the occurrence of coinfections within single hosts (16). Coinfections are often undetectable using standard PCR protocols, resulting frequently in the amplification of a single cytb sequence and thus to underestimation of Haemosporidian diversity (15). The PCR underestimation in mixed infection cases could, in turn, lead to unreliable morphomolecular parasite identifications and phylogenetic analyses, resulting in erroneous interpretations and conclusions. In the present investigation, using NGS, we have confidently identified the occurrence of two distinct Nycteria mt genomes present in mixed infection from N. grandis (273VI) and two Nycteria monoinfections from R. alcyone (289VI) and M. spasma (C289). Based on morphological observations of the two Nycteria morphotypes and on the large NGS read coverage of the two mt genomes in N. grandis, we associated each Nycteria species with its corresponding mitochondrial genome. Thus, the mt genome predominantly found in N. grandis could consistently be ascribed to N. medusiformis, and the minority mt genome could belong to Nycteria sp. In the same way, we confidently associated N. gabonensis and N. heischi with their corresponding mt genomes.
Our phylogenetic analyses support the classification of Nycteria parasites as a monophyletic group composed of at least two distinct but closely related parasitic lineages associated with different bat families (12). The distinction between Nycteria parasites infecting Nycteris and Rhinolophus host bats has been proposed previously based on traditional taxonomy (14). Thus, six species have been described and divided into two distinct groups based on the following morphological sexual-stage characteristics in blood (gametocytes) and schizont characteristics: (i) Nycteris sp. (Nycteridae) parasites with gametocytes arranged as a rosette (en cocarde) (N. medusiformis, N. houini, and N. erardi), and (ii) Rhinolophus sp. (Rhinolophidae) parasites with gametocytes of the dispersed (“diffus”) type (N. gabonensis, Nycteria krampitzi, and Nycteria congolensis) (14). The N. heischi parasites from Southeast Asian M. spasma were morphologically related to the Nycteria from African Nycteris, with a nucleus of gametocytes en cocarde.
Two phylogenetic placements among all major Haemosporidia parasites and evolutionary scenarios based on multigene phylogenetic analyses (mostly inferred from cytb phylogenetic information) have been previously proposed for the Nycteria parasite group. Nycteria parasites were placed as a sister clade closely related to the mammalian Plasmodium/Hepatocystis parasite clade, suggesting that the origin of mammalian Haemosporidia may have been in bats (11). Alternatively, these parasites were placed closer to Plasmodium species from lizards, suggesting an alternative scenario such as a host-switch event between bat and reptile Haemosporidia parasites in their evolutionary history (9). Such a host-switch event between these terrestrial vertebrate host classes has also been assumed for Polychromophilus parasites (infecting Miniopteridae and Vespertilionidae bats) based on close phylogenetic relationships and shared biological traits (schizogony present in the reticuloendothelial system) (17–20). The mitochondrial genomic phylogenetic analyses presented in this study, using two genetically distantly related Apicomplexan outgroups of Haemosporidia parasites to root phylogenetic reconstructions, did not produce a robust placement of Nycteria parasites among the Haemosporidia parasites (21, 22). However, the Nycteria parasites that have a hepatic schizogony shared this biological trait with mammalian Plasmodium and Hepatocystis parasites (12, 14).
Next-generation sequencing would further open new outlooks into unsuspected and unexplored Haemosporidia parasite diversity, and would help to understand the systematics and evolution of Haemosporidia parasites.
Materials and Methods
Specimens.
All animal experiments were approved by the ethical committee of the Museum National d’Histoire Naturelle (Comité Cuvier). A total of 195 bats (six families) were caught using mist nets and harp traps during field expeditions in the Orientale province of the Democratic Republic of Congo (November–December 2013), as well as 90 bats (six families) in Cambodia (July 2005 and January 2006). For morphological parasite identification, thin blood smears were used and blood spots were made on Whatman filter paper to be used later for genome analyses.
One sample each of N. grandis (273VI), R. alcyone (289VI), and M. spasma (C289), infected respectively with N. medusiformis and N. sp., N. gabonensis, and N. heischi, were used for whole mt genome sequencing.
Microscopy.
Thin blood smears were fixed with absolute methanol, stained with 10% (vol/vol) Giemsa solution (in phosphate-buffered solution, pH 7.4) for 45 min, and then preserved with mounting medium (Eukitt; Polylabo) and a coverslip. Smears were then carefully screened for morphological examinations with a motorized BX63 upright Olympus microscope at a magnification of 100× and photographed with an Olympus DP72 camera (High-Speed 12.8 Megapixel Image Capture).
DNA Extraction.
Whole genomic DNA from N. grandis, R. alcyone, and M. spasma dried blood spots was extracted using a QIAamp DNA Micro Kit (Qiagen) following the manufacturer’s instruction handbook. Samples were eluted in 20 µL of molecular biology-grade water. Whole genomic DNAs were quantified using a Qubit 2.0 fluorometer (Life Technologies), and concentrations of 0.997 ng/µL, 0.74 ng/µL, and 0.15 ng/µL were respectively obtained for the N. grandis (including N. medusiformis and N. sp. parasites), R. alcyone (including N. gabonensis), and M. spasma samples (including N. heischi).
Whole-Genome Sequencing of Nycteria Parasites.
Whole mitochondrial genome sequencing was performed on Nycteria parasites using HiSeq 2000 Illumina paired-read sequencing technology at the Genomics Platform at the Pasteur Institute. The Illumina library preparations were produced using Nextera XT DNA Library Preparation Kits following standard protocols developed by the supplier (Illumina). Briefly, genomic DNA was sheared by nebulization, and sheared fragments were end-repaired and phosphorylated. Blunt-end fragments were A-tailed, and sequencing adapters were ligated to the fragments. Inserts were sized using Agencourt AMPure XP beads (±500 bp; Beckman Coulter Genomics) and enriched using 10 cycles of PCR before library quantification and validation. Hybridization of the Nextera DNA libraries to the flow cell and bridge amplification were performed to generate clusters; paired-end reads of 100 cycles were collected on a HiSeq 2000 (Illumina). Raw sequence files were filtered using the Fqquality tool, read-quality filtering software developed by the gensoft team at the Pasteur Institute, which enables the trimming of the first and last low-quality bases in reads.
Perl software named iSeGWalker was developed to accomplish a de novo genome reconstruction from the reads file in fastq format provided by next-generation sequencing data (Illumina). The exact-matching algorithm removed all variations due to sequencing errors. iSeGWalker was based on a unique and specific seed to Haemosporidia parasites. The sequence TCTTGAGGCAGTTTGTTCCCTATCTACC, conserved throughout 72 Haemosporidia species, was chosen as initial seed. The search step is an exact-matching read selection using a regular expression and a very simple Perl script, reading each sequence one by one. Once all matching reads have been selected, a consensus sequence is determined. Once consensus is determined, a new seed, composed by the 30 last consecutive nucleotides, is obtained and a new search is performed. Reads are then mapped using the new sequence as a reference (BWA software) for all samples to identify the parasitic variant present in mixed infection (N. grandis).
Assessment of de Novo Reconstruction.
The accuracy of the de novo iSeGWalker software was first validated by reconstructing the complete mitochondrial genome of the Plasmodium falciparum 3D7 reference strain from NGS data. PCR and nested PCR, using designed specific primers, were also performed to amplify an mt fragment of 1,403 bp from Nycteria parasites from N. grandis (including rRNA fragments and a portion of the cox1 sequence). PCR was carried out in a total volume of 25 µL consisting of 3 µL DNA, 5 µL 5× PCR buffer, 1.5 mM MgCl2, 0.2 mM each dNTP, 1 U Taq DNA polymerase (Promega), and 0.2 mM primers P5 (5′-AACGCCTGABATGRATGGATAA-3′) and P5R (5′-TTCDGGATGWCCAAARAACCAG-3′). PCR cycling conditions were 2 min 30 s denaturation at 95 °C followed by 30 s at 53 °C and 2 min 30 s at 72 °C for 40 cycles, and a final 5-min extension at 72 °C. Nested PCR was performed using 5 µL PCR products (diluted 1/10), 5 µL 5× PCR buffer, 2.5 mM MgCl2, 0.2 mM each dNTP, 1 U Taq DNA polymerase (Invitrogen), and 0.2 mM primers P4 (5′-TCCATGTCKTCTCATCGCAG-3′) and P6iN (5′-ATAATGTCCATCCAGTTCCACC-3′). PCR cycling conditions were 2 min 30 s denaturation at 95 °C followed by 30 s at 54 °C and 1 min 30 s at 72 °C for 40 cycles, and a final 5-min extension at 72 °C. The PCR products were sent to Eurofins Genomics for sequencing using nested-PCR primers.
Phylogenetic Analyses.
The four complete mitochondrial genomes of Nycteria specifically produced for this study were compared with the mitochondrial sequences available in the nucleotide databases for Haemosporidia parasites. Phylogenetic analyses were performed using two different datasets, named 3PCG for the protein alignment of three protein-coding genes (1,014 amino acids; 59 taxa, including 50 Haemosporidians and 9 outgroups) and cytb for the DNA alignment of the cytb gene (1,054 nt; 110 Haemosporidians). For protein analyses, the Haemosporidian tree was rooted using nine outgroup species belonging to Eucoccidiorida (Cyclospora cayetanensis, Eimeria acervulina, Eimeria maxima, Eimeria tenella, and Hepatozoon catesbianae) or Piroplasmida (Babesia bovis, Babesia microti, Cytauxzoon felis, and Theileria parva). GenBank accession numbers of all the genomic and cytb sequences are provided in the trees in Fig. 3 and Fig. S1. Complete mitochondrial genomes were aligned using MUSCLE (23) and then further adjusted by eye with Se-Al version 2.0a11 (tree.bio.ed.ac.uk/software/seal/). For cytb datasets, the best-fitting model of sequence evolution (GTR+I+G) was selected under jModelTest 2.1.4 (24) using the Akaike information criterion. Phylogenetic relationships were inferred using Bayesian analyses with MrBayes version 3.2.1 (25), and posterior probabilities were calculated using four independent Markov chains run for 10 million Metropolis-coupled Markov chain Monte Carlo generations, with tree sampling every 1,000 generations and a burn-in of 25%. For the protein alignment (3PCG dataset), phylogenetic relationships were reconstructed with the same Bayesian approach by applying the mixed model (25).
Acknowledgments
We thank the individuals who helped in collecting tissue samples: Raphaël Colombo, Laurent Daudet, Tamas Görföl, Ros Kiri Ing, Jean-François Julien, Prescott Musaba Akawa, and Vuong Tan Tu. We also thank Faustin Toengaho Lokundo, the Rector of the University of Kisangani, for his invitation, and Benjamin Dudu Akaibe, Keunen Hilde, Erick Verheyen, and all other members of the Centre de Surveillance de la Biodiversité for technical support and administrative assistance. This research study was funded by Labex BCDiv (Biological and Cultural Diversities) and ATM (Actions Thématiques Muséum) Génomique et Collections, Muséum National d’Histoire Naturelle de Paris.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. KX090645, KX090646, KX090647, and KX090648).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610643113/-/DCSupplemental.
References
- 1.Santiago-Alarcon D, Rodríguez-Ferraro A, Parker PG, Ricklefs RE. Different meal, same flavor: Cospeciation and host switching of Haemosporidian parasites in some non-passerine birds. Parasit Vectors. 2014;7:286. doi: 10.1186/1756-3305-7-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Outlaw DC, Ricklefs RE. Comparative gene evolution in Haemosporidian (Apicomplexa) parasites of birds and mammals. Mol Biol Evol. 2010;27(3):537–542. doi: 10.1093/molbev/msp283. [DOI] [PubMed] [Google Scholar]
- 4.Lacorte GA, et al. Exploring the diversity and distribution of neotropical avian malaria parasites—A molecular survey from southeast Brazil. PLoS One. 2013;8(3):e57770. doi: 10.1371/journal.pone.0057770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark NJ, Clegg SM, Lima MR. A review of global diversity in avian Haemosporidians (Plasmodium and Haemoproteus: Haemosporida): New insights from molecular data. Int J Parasitol. 2014;44(5):329–338. doi: 10.1016/j.ijpara.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Hikosaka K, Kita K, Tanabe K. Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol. 2013;188(1):26–33. doi: 10.1016/j.molbiopara.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- 8.Feagin JE, et al. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One. 2012;7(6):e38320. doi: 10.1371/journal.pone.0038320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaer J, et al. Nycteria parasites of Afrotropical insectivorous bats. Int J Parasitol. 2015;45(6):375–384. doi: 10.1016/j.ijpara.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Zehtindjiev P, et al. A new morphologically distinct avian malaria parasite that fails detection by established polymerase chain reaction-based protocols for amplification of the cytochrome b gene. J Parasitol. 2012;98(3):657–665. doi: 10.1645/GE-3006.1. [DOI] [PubMed] [Google Scholar]
- 11.Schaer J, et al. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc Natl Acad Sci USA. 2013;110(43):17415–17419. doi: 10.1073/pnas.1311016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garnham PCC. 1966. Malaria Parasites and Other Haemosporidia (Blackwell Scientific, Oxford)
- 13.Garnham PCC, Heisch RB. On a new blood parasite of insectivorous bats. Trans R Soc Trop Med Hyg. 1953;47(5):357–363. doi: 10.1016/s0035-9203(53)80016-7. [DOI] [PubMed] [Google Scholar]
- 14.Rosin G, Landau I, Hugot JP. Considérations sur le genre Nycteria (Haemoproteidae) parasite de Microchiroptères africains avec description de quatre espèces nouvelles [Considerations on the genus Nycteria (Haemoproteidae) parasite of African Microchiroptera with the description of four new species] Ann Parasitol Hum Comp. 1978;53(5):447–459. French. [PubMed] [Google Scholar]
- 15.Bernotienė R, Palinauskas V, Iezhova T, Murauskaitė D, Valkiūnas G. Avian Haemosporidian parasites (Haemosporida): A comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp Parasitol. 2016;163(163):31–37. doi: 10.1016/j.exppara.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Juliano JJ, et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Natl Acad Sci USA. 2010;107(46):20138–20143. doi: 10.1073/pnas.1007068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duval L, et al. Multiple host-switching of Haemosporidia parasites in bats. Malar J. 2007;6:157. doi: 10.1186/1475-2875-6-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landau I, Rosin G, Miltgen F, Léger N, Mialhe E. [The schizogony of Polychromophilus (Haemoproteidae), cosmopolitan parasite of Microchiroptera] C R Acad Sci Hebd Seances Acad Sci D. 1977;285(15):1311–1313. French. [PubMed] [Google Scholar]
- 19.Landau I, et al. Sur le genre Polychromophilus (Haemoproteidae, parasite de Microchiropteres) Ann Parasitol. 1980;55(1):13–32. French. [Google Scholar]
- 20.Witsenburg F, Salamin N, Christe P. The evolutionary host switches of Polychromophilus: A multi-gene phylogeny of the bat malaria genus suggests a second invasion of mammals by a Haemosporidian parasite. Malar J. 2012;11:53. doi: 10.1186/1475-2875-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Outlaw DC, Ricklefs RE. Rerooting the evolutionary tree of malaria parasites. Proc Natl Acad Sci USA. 2011;108(32):13183–13187. doi: 10.1073/pnas.1109153108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borner J, et al. Phylogeny of Haemosporidian blood parasites revealed by a multi-gene approach. Mol Phylogenet Evol. 2016;94(Pt A):221–231. doi: 10.1016/j.ympev.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]