Significance
Synapses functionally connect neurons in the brain and mediate information processing relevant to all aspects of life. Among others, synaptic connections are enabled by cell adhesion molecules, which connect presynaptic and postsynaptic membranes by binding to each other via the synaptic cleft. Mammalian genomes express hundreds of cell adhesion molecules whose combinatorial utilization is thought to contribute to the brain’s “connectivity code.” Such code could explain the versatility of synapses as well as the logic of connectivity between cell types. Here, we used single-cell RNA sequencing to analyze the expression of cell adhesion molecules and other signaling proteins in defined cell types, and found developmental patterns that potentially identify relevant elements of the connectivity code.
Keywords: synapse, cell adhesion, single cell, RNAseq
Abstract
In brain, signaling mediated by cell adhesion molecules defines the identity and functional properties of synapses. The specificity of presynaptic and postsynaptic interactions that is presumably mediated by cell adhesion molecules suggests that there exists a logic that could explain neuronal connectivity at the molecular level. Despite its importance, however, the nature of such logic is poorly understood, and even basic parameters, such as the number, identity, and single-cell expression profiles of candidate synaptic cell adhesion molecules, are not known. Here, we devised a comprehensive list of genes involved in cell adhesion, and used single-cell RNA sequencing (RNAseq) to analyze their expression in electrophysiologically defined interneurons and projection neurons. We compared the cell type-specific expression of these genes with that of genes involved in transmembrane ion conductances (i.e., channels), exocytosis, and rho/rac signaling, which regulates the actin cytoskeleton. Using these data, we identified two independent, developmentally regulated networks of interacting genes encoding molecules involved in cell adhesion, exocytosis, and signal transduction. Our approach provides a framework for a presumed cell adhesion and signaling code in neurons, enables correlating electrophysiological with molecular properties of neurons, and suggests avenues toward understanding synaptic specificity.
The brain’s “connectivity code” is thought to confer exquisite specificity to brain circuits by dictating connectivity between different types of neurons. Although its existence has not yet been conclusively demonstrated, synaptic cell adhesion molecules likely comprise a large part of such a code (1–4). Cell adhesion molecules are encoded by ∼2% of the genome and play central roles in all tissues. During brain development, precisely matching presynaptic and postsynaptic cell adhesion molecule interactions likely guide synapse formation and specify the properties of synapses by activating signal transduction cascades and recruiting scaffolding molecules, receptors, and active-zone proteins; in addition, such interactions could provide structural support to synapses. However, the molecular mechanisms involved are not understood. Because cell adhesion molecule-based interactions likely code for synapse specificity in a combinatorial fashion (2, 3), an important step toward gaining insight into these molecular mechanisms is to eliminate nonfunctional possibilities, which—to a great extent—can be done by examining cell type-specific expression of these molecules.
If cell adhesion molecules dictate synapse properties, then such differences must be clearly present in interneuron and pyramidal cell classes, which have diverse synaptic properties. For example, within the hippocampal circuit, CA1 pyramidal (CA1-PYR) neurons receive convergent inputs from multiple, electrophysiologically distinct inhibitory interneurons within the hippocampus (5). Inhibitory inputs include synapses formed by fast-spiking (FS) and regular-spiking (RS) interneurons (ref. 6; for reviews, see refs. 7 and 8). We previously showed using single-cell transcriptional profiling in FS parvalbumin (PV)- and RS cholecystokinin (CCK)-containing GABAergic interneurons that neurexin (Nrxn1 and Nrxn3; presynaptic cell adhesion molecules) isoforms were expressed cell type-specifically, with remarkable consistency in respective cell types (9). We also found that genetic deletion of neuroligin-3 (Nlgn3) (postsynaptic cell adhesion molecule) in PYR cells disabled tonic, cannabinoid type 1 receptor-mediated, endocannabinoid signaling in RS CCK synapses, but had no detectable phenotype in FS PV synapses (10). Thus, although no systematic assessment of cell adhesion molecules in GABAergic interneurons is available, previous studies established that cell adhesion molecules play a central role in controlling their properties.
Similar to their inputs, outputs of CA1-PYR cells display functional dichotomy: they primarily project to the subiculum, forming synapses on two, electrophysiologically different principal cell types: regular-spiking pyramidal (RS-PYR) and burst-spiking pyramidal (BS-PYR) cells. Analysis of these neurons is particularly difficult because, although RS-PYR and BS-PYR cells exhibit distinct electrophysiological signatures as well as dramatically different forms of long-term plasticity, no molecular markers are available to distinguish these neurons (11, 12). In examining synapse-specific mechanisms in these cells, we have shown that different forms of long-term plasticity were determined presynaptically by expression of specific neurexin isoforms in CA1-PYR cells (13–15). Together, these molecular and physiological analyses revealed specific control of synaptic properties (such as forms of LTP and endocannabinoid signaling) by cell adhesion molecules in CA1-PYR cell inputs and outputs, and raised the question whether other synaptic properties were also controlled by cell adhesion molecules. Because there are a large number of molecules with such potential, a logical step to this direction is the cell type-specific analysis of cell adhesion-related gene expression.
Here, we combined electrophysiology and single-cell RNA sequencing (RNAseq) to identify cells in the pathway involving hippocampal FS interneuron (FS-INT), RS-INT, CA1-PYR cells, and subiculum RS-PYR and BS-PYR cells, and to analyze their gene expression profiles. Our data represent an initial circuit-level single-cell RNAseq analysis from synaptically and electrophysiologically defined neurons. We find surprising differences in the total number of expressed genes among neuron types and show that hippocampal neurons can be characterized by the expression of two common, developmentally regulated gene networks comprising shared cell adhesion and signaling molecules. Moreover, we demonstrate that each type of electrophysiologically defined neuron expresses a separate set of candidate synaptic cell adhesion and signaling molecules. Finally, we extended these analyses to the two types of subiculum PYR neurons that feature highly similar transcriptomes but express a limited number of unique markers, including cell adhesion-related genes, which could potentially be used in future analyses. In this manner, the approaches and data developed here lay the foundation for a biologically relevant analysis of how cell type-specific neuronal gene expression may sculpt neural circuits during development and beyond.
Results
Cell Adhesion Molecules in the Hippocampus.
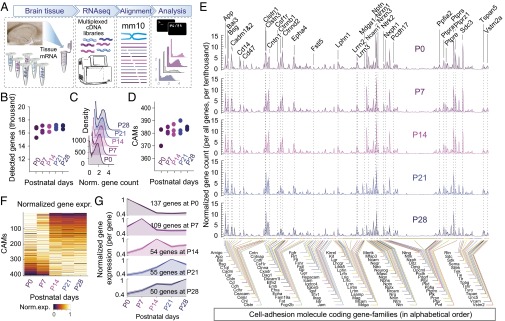
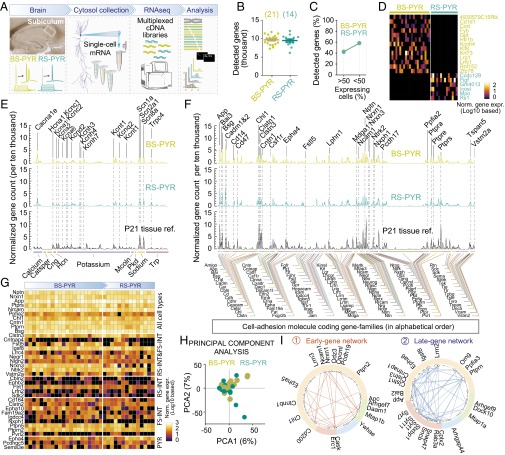
As a starting point, we examined the transcriptome of the entire hippocampus (Fig. 1A) at five developmental stages (in triplicate at postnatal days P0, P7, P14, P21, and P28). In these samples, the total number and distribution of expressed genes were similar (Fig. 1 B and C, and Fig. S1). To specifically analyze cell adhesion molecules, we created a comprehensive list of candidate genes involved in, or related to, transsynaptic cell adhesion (collectively referred to as CAMs, for candidate cell adhesion-related molecules). For this, we first considered all molecules with single transmembrane domains (a general, although not unique property of membrane surface adhesion molecules) and narrowed this list down to 406 genes based on preexisting data (Fig. S1C). This curated list of candidate cell adhesion molecules includes proteins implicated in cell–cell signaling but excludes, for example, intracellular signal transducers linked to cell adhesion signaling. We found these genes to be broadly represented in all replicates and developmental stages (Fig. 1D), highlighting diversity of CAMs in the hippocampal circuit and throughout development. Expression profiles (Fig. 1E) for each developmental stage included genes that were consistently highly expressed at all developmental stages [for example, amyloid precursor protein (App), contactins (Cntn), neurexins (Nrxn), and protein-tyrosine phosphatases (Ptpr); Fig. S1D], as well as genes with large changes during development (for example, Ncam1 and Sdc3). Normalization of individual gene expression levels also revealed an average change of 60% between P0 and P28 (Fig. 1 F and G).
Fig. 1.
RNAseq of the total hippocampus during development: analysis of the changing landscape of CAMs. (A) Experimental design and strategy for mRNA extraction from hippocampal tissue samples (represented in photograph) and RNAseq analyses. (B and C) Similar number of detected genes in our analyses of three sample replicates at five developmental stages (from P0 to P28) suggested replicability, and similar normalized gene count distributions (i.e., histogram of gene numbers with given normalized gene count) suggested comparability between samples. (D) Number of detected CAMs was consistent across developmental stages. (E) Averaged expression of CAMs in five developmental stages (in the plot, each gene is spaced equidistant and represented by mean ± SEM; expression values are connected with lines for visibility; for numerical values, see Dataset S1). (F) Heat plot of normalized gene expression shows developmental regulation of CAMs, which were ranked by linear fit of developmental expression values. (G) Average gene expression levels grouped by peak expression throughout development. Label Insets detail number of genes that belong to each group. Averaged data represent mean ± SEM.
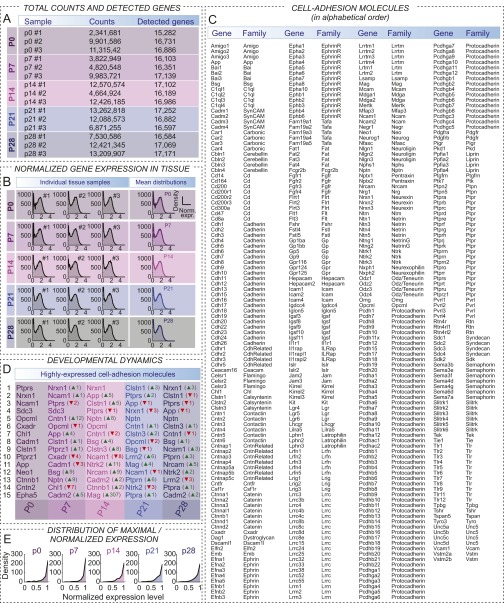
Fig. S1.
Cell adhesion molecules in hippocampal tissue RNAseq samples. (A) To assay comprehensive, tissue-level expression of CAMs, we analyzed hippocampal tissue samples from multiple developmental age groups, including newborn (P0) as well as postnatal day 7, 14, 21, and 28 (labeled as P7, P14, P21, and P28, respectively) mice. For each age group, we processed three independent samples, from littermate mice. We found that the number of gene-aligned reads ranged from 2.3 to 13.2 million; however, the number of detected genes was consistent in all samples and age groups. (B) Normalized gene expression was comparable in all samples, reflecting similar postsequencing sample quality, and therefore gene expression in these samples could be directly compared, after sample-to-sample normalization. Right panels show average distribution for each age group, respectively. (C) Comprehensive list of known and potential CAMs. Central to this study, we analyzed expression of these molecules at the whole hippocampus tissue level as well as their cell type-specific expression in five different cell types, including RS-INT and FS-INT interneurons and CA1 PYR cells, and two types of subiculum PYR neuron cell types, including BS-PYR and RS-PYR cells. (D) Table depicts developmental dynamics of highly expressed CAMs. By rank-ordering mean CAM levels in each developmental stage, we tracked whether a gene was up- or down-regulated or displayed stable expression over development: up/down arrows and ranking numbers indicate changes compared with previous developmental stages. (E) Plots show distributions of normalized gene expression, including all CAMs. These distributions were similar across developmental stages, as shown in Fig. 1C.
Single-Cell RNAseq from Electrophysiologically Identified Cells.
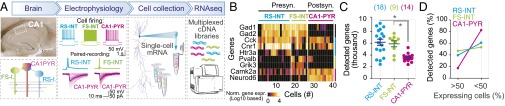
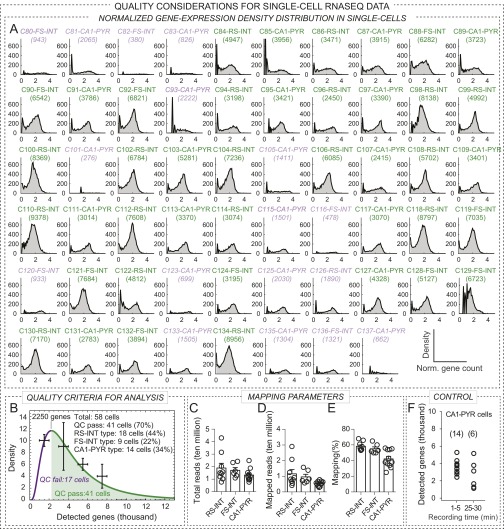
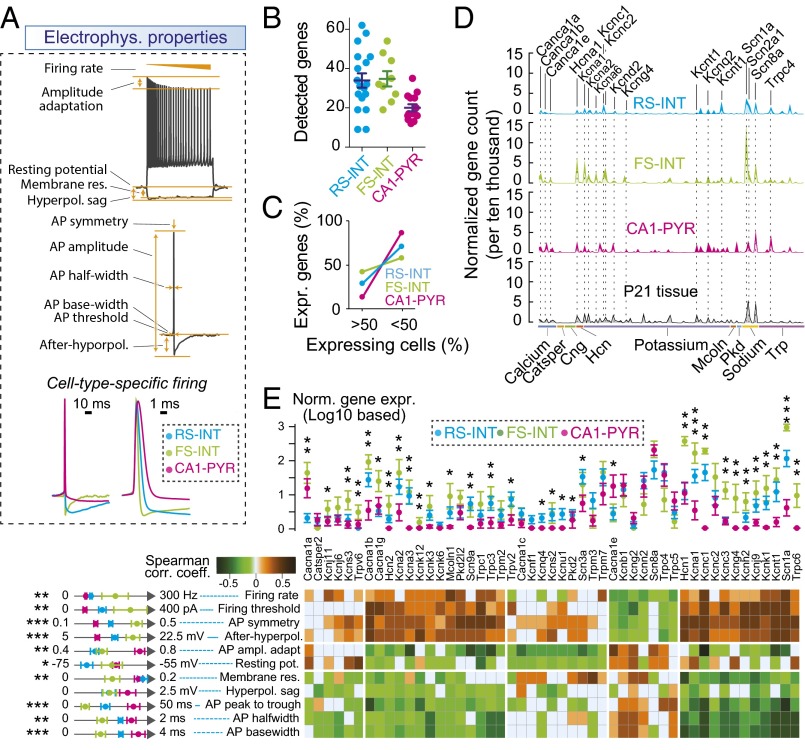
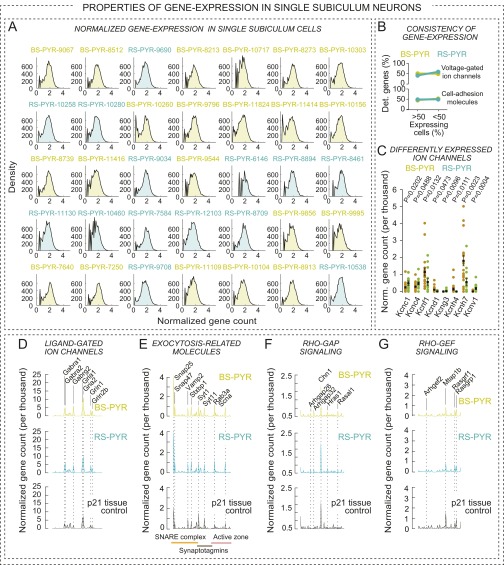
To analyze gene expression in specific cell types in a defined circuit, we developed a method—which was also recently introduced by others (16, 17)—with which we first characterized neurons electrophysiologically, and subsequently analyzed their transcriptome by aspiration of cytosol followed by single-cell RNAseq. Using this approach, we examined FS and RS interneurons that directly inhibit CA1-PYR cells, as well as the target CA1-PYR cells for these interneurons. We patched FS and regular-firing interneurons located within, or in close proximity to, the pyramidal cell layer in wild-type mice, and then used paired recordings by simultaneously patching PYR cells to test whether presynaptic action potentials (APs) evoked inhibitory postsynaptic responses, characterizing these cells as FS or RS inhibitory interneurons (Fig. 2A). Note that such distinction did not necessarily identify individual cell types. For instance, FS cells could include PV+ basket, bistratified, and axoaxonic cells, whereas RS cells could include CCK+ basket or bistratified cells as well. At the end of the recordings, we collected the neuronal cytosol via pipette aspiration for RNAseq (Fig. 2A). Upon alignment of sequencing reads and assignment of gene counts for each gene, we applied stringent quality control metrics to each cell (Fig. S2 A and B). RNAseq data obtained in this manner from 18 RS-INT, 9 FS-INT, and 14 CA1-PYR cells passed quality control.
Fig. 2.
Single-cell RNAseq in situ of electrophysiologically characterized neurons in identified microcircuits. (A) Experimental design and strategy. Single-cell mRNA samples were collected after paired electrophysiological recordings from RS-INT, FS-INT, and CA1-PYR cells. White box indicates the CA1 region. After sequencing, transcripts were aligned to the mm10 assembly. (B) Representative heat map shows examples of gene expression in each cell type. For example, Gad1 and Gad2 expression in most presynaptic FS-INT and RS-INT cells confirmed that they were GABAergic, whereas Neuro6d was specific to postsynaptic PYR cells. (C) Consistency of gene expression within and between cell types. On average, RS-INT and FS-INT cells expressed twice as many genes as CA1-PYR cells. (D) Plot of the percentage of genes that were detected in less (1–50%) and more (50–100%) than one-half of the cells (data points were connected for visualization) in respective cell types. RS-INT and FS-INT cells had more consistent gene expression profiles than CA1-PYR cells. Averaged data in C represent means ± SEM.
Fig. S2.
Sample quality in single-cell RNAseq data. (A) Plots show individual normalized gene count density for each single cell (i.e., histogram of gene numbers with given normalized gene counts), in order of data collection. The shape of these distributions reflect sample quality, where distinct peaks around 0 and 2 represent nonexpressed vs. expressed genes. Title of each plot displays sample id, cell type, and the number of detected genes. (B) To establish an adequate quality control (QC) measure, we counted all genes with expression level >1 (reflecting reliably detected genes) in each cell, and binned cells by these cell-specific gene count values, using a bin size of 1,000. To derive statistical inferences, we averaged neighboring data points and fitted these results with a log-normal distribution. The resulting fit served as quality assessment of the complete dataset, and we used the peak of the fit as a QC cutoff threshold. Therefore, for these analysis, we included cells with at least 2,250 detected genes; 41 cells of the total of 51 cells processed. In A, green title colors refer to cells further analyzed, whereas faded colors refer to excluded cells. (C–E) Plots show mapping parameters for each cell type, including total reads, mapped reads, and mapping efficacy. (F) Because of the sequential nature of paired recordings, PYR cells were collected 1–5 min, and RS-INT and FS-INT cells were collected 20–30 min after forming whole-cell break in. Therefore, differences in recording time could have significantly impacted the number of detected genes, resulting in the reported differences in Fig. 2C. In this control experiment, we recorded six CA1 PYR cells for 25–30 min and performed RNAseq. Plot shows that the number of detected genes after 25- to 30-min recording were not different from those collected within 1–5 min. For the later group, only QC passed gene count values were included (A), and for control group, all cells were plotted.
To gain insight into the molecular identity of recorded cells, we examined expression of genes that had been previously associated with RS-INT, FS-INT, or PYR cells (Fig. 2B). As expected, Gad1 and Gad2 were highly expressed in interneurons but not in PYR cells, consistent with their respective neurotransmitters. Although CCK peptide is a unique marker for RS CCK cells, we found that the CCK gene was nonspecifically expressed in all three cell types. Similarly, although cannabinoid type-1 receptor (Cnr1) is most highly expressed in RS CCK cells (for specific examples, see refs. 10, 18, and 19; for review, see ref. 7), it was also produced in FS-PV and PYR cells. In contrast, Htr3a and PV (parvalbumin) were more specific to RS-INT and FS-INT cells, respectively. Although the latter is a unique marker for FS subtypes at the protein level, the corresponding mRNA was detected in two RS-INT and three PYR cells as well. To identify PYR cells, we examined Camk2a and Neurod6 expression: although the former’s expression was rather nonspecific, Neurod6 was a reliable predictor of PYR cell identity. We also found that Grik3 (kainate receptor GluR7) uniquely, but not exclusively, identified FS-INT cells.
Examining overall gene expression, we found that interneurons consistently expressed almost twice as many genes as CA1-PYR cells (Fig. 2C; see Fig. S2F for controls), and that detection of individual genes was also more consistent in RS- and FS-INT cells than in CA1-PYR cells (Fig. 2D). A potential explanation for the latter observation involves spatial gene expression gradients in the hippocampus that likely underlie heterogeneity of CA1 pyramidal neurons (20).
Molecular Correlates of Electrophysiological Properties.
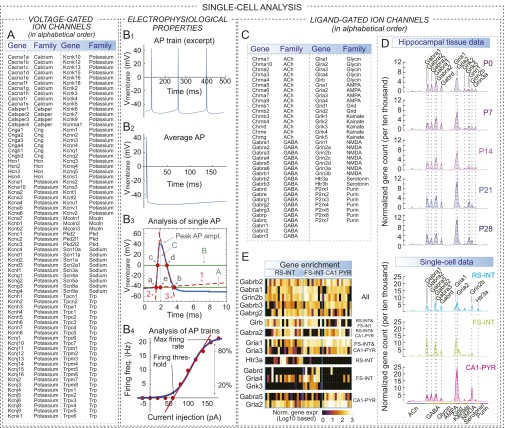
To link single-cell transcriptional profiles to functional properties, we examined expression of 140 voltage-gated ion channels (see Materials and Methods for gene set assembly; Fig. 3A, Fig. S3A, and Dataset S1). Because RS-, FS-, and CA1-PYR cells have distinct electrophysiological properties (Fig. 3A), we asked whether ion channel expression could account for their physiological differences. On average, we detected more ion channels and more consistent gene expression in interneurons than in PYR cells (Fig. 3 B and C), following total transcript levels. As a pivotal example of cell type-specific gene expression, we found enrichment of Na+ and K+ channels in FS cells (Fig. 3 D and E, Upper).
Fig. 3.
Combined electrophysiological and transcriptional analysis. (A) Ion channels define neuronal excitability that can be quantified using active (AP-related) and passive electrophysiological properties (e.g., voltage response for hyperpolarizing and depolarizing current injections). These parameters differ between RS-INT, FS-INT, and CA1-PYR cells, and the lower panel shows differences in AP firing in the three cell types. (B) Number of detected ion channels was similar in RS-INT and FS-INT cells, but lower in CA1-PYR cells. (C) Consistency of ion channel expression was similar in all three cell types. (D) Averaged, single-cell ion channel expression in RS-INT (n = 14), FS-INT (n = 9), and CA1-PYR (n = 14) cells, and in aged-matched P21 tissue control (for numerical values, see Dataset S1). (E) Correlation analysis of electrophysiological parameters and ion channel expression in RS-INT, FS-INT, and CA1-PYR cells. (Upper) Normalized gene expression data and (Left) electrophysiology parameter measurements for RS-INT, FS-INT, and CA1-PYR cells. Averaged data represent mean ± SEM. Asterisks represent significant differences between cell types (*difference between two cell types; **difference between two–two cell types; ***difference between all three cell types).
Fig. S3.
Comprehensive molecular and electrophysiological analyses of single cells. (A) Table shows a comprehensive list of voltage-gated ion channel coding genes. We specifically examined this gene category, because ion channels in the plasma membrane render complex electrophysiological properties to neurons. See Materials and Methods for more information on assembly of this list. (B1–B4) These plots depict step-by-step analysis of electrophysiological properties. (B1) Excerpt from of an example AP train, which, in this case, depicts typical PYR cell firing. (B2) APs from each train were peak aligned and averaged for subsequent analyses. (B3) Analysis of single AP properties: baseline, ascending, and descending phases of APs were linearly fit (red, dashed lines; labeled “1,” “2,” and “3,” respectively). Intersection of lines 1 and 2 marked onset of APs, labeled “a.” Then, a horizontal line (green line, labeled “A”) through point “a” determined baseline width, between points “a” and “b.” Next, we determined AP half-amplitude (green line, labeled “B”) between peak amplitude and line “A”; AP half-width was determined as distance between “d” and “c.” Finally, intersection of “A” and a vertical line through the peak was used to determine point “e,” which (together with “a” and “b”) was used to quantify AP symmetry. (B4) Analysis of AP trains: red points demonstrate input/output curve of the neuron. Blue curve demonstrates a sigmoid curve that was used to fit data. Linear fit through 20–80% of the sigmoid (red line) was used to determine firing threshold, and amplitude of the sigmoid fit was used to determine maximal firing rate. (C) List of ligand-gated ion channels included in our analyses. See Materials and Methods for more information on assembly of this list. (D) Expression of ligand-gated ion channels in hippocampal tissue (Upper) and single-cell data (Lower; for numerical values, see Dataset S1). (E) Heat map shows single-cell mRNA expression of ligand-gated ion channels. Cells were grouped according to cell types (labeled on Top), and genes were grouped according to cell type-specific enrichment (labeled on Right). Statistical significance (i.e., cell type-specific enrichment of gene expression) was assessed using Mann–Whitney test (P < 0.05). Ubiquitously enriched genes (in “All” category) showed no significant difference in any of the cell types. However, cell type-specific gene enrichment suggested further clues of cell type identity: in particular, selective expression of serotonin receptor Htr3a [together with high expression of cannabinoid type 1 receptor (Cnr1); Fig. 2B] suggested that most RS-INT cells were “CCK-containing interneurons,” which as a peptide uniquely identifies these cells at posttranslational levels (5, 7). Enrichment of Gabrd and coexpression of Gria1, Gria2, Gria3, and Gria4 mRNA in FS-INT [together with expression of parvalbumin (Pvalb); Fig. 2B] cells suggested that these cells were “PV-containing interneurons” (49–51), in which cells moderate-to-weak presence of Gabra2 in FS-INT (50) has been also described. We also consistently detected Gabra1 in RS-INT cells, although the gene product itself is not present in related CCK-containing interneuron cell types (52). Furthermore, we detected Grik3 exclusively in FS-INT cells, which may identify these cells as kainate-responsive interneurons (53).
Next, we quantified electrophysiological parameters (n = 11; Fig. 3 A and E, Left) because their correlation with gene expression patterns could reflect characteristic, cell type-specific properties in these cells. We used unsupervised clustering and plotted genes/electrophysiology parameters with at least one correlation value of greater than 0.35 or less than −0.35 (arbitrary threshold to eliminate no or minimal correlations; Fig. 3E). Here, we found that AP firing rate, threshold, symmetry, and afterhyperpolarization, parameters that typically exhibit high values in FS cells, correlated with high expression of Na+ and K+ channels (Fig. 3E), including Kcnc1 (Kv3.1) and Kcnc2 (Kv3.2). Furthermore, we found that expression of these genes inversely correlated with AP peak-to-trough and width times (i.e., larger gene expression values correspond to lower electrophysiological property values, and vice versa), which as properties also correspond to the fast signaling characteristics of FS-INT cells. These results are not unexpected as expression of Kcnc1 and Kcnc2 is a hallmark of FS PV interneurons (refs. 21 and 22; for review, see ref. 23), validating the single-cell RNAseq analysis.
Surprisingly, unbiased correlation analysis can also deliver seemingly contradictory results. For example, the observation of high Hcn1 expression but a low hyperpolarization sag in FS-INT is puzzling because a primary readout for Hcn-mediated Ih currents is the sag amplitude. However, the correlation between hyperpolarization sag and Ih current is not absolute, nor the involvement of Hcn1 in modulating active membrane properties (24). In addition, we found significantly higher expression of Kcnc3, Scn1a, and Trpc6 (Fig. 3E) in FS cells (described for PV cells in refs. 25–27, respectively), whereas specific expression of Kcng4 (Kv6.3, Kv6.4) identifies a thus far uncharacterized component of the ion channel repertoire of FS cells. For further clues about molecular architecture of these cell types, see expression of ligand-gated ion channels in Fig. S3 C–E.
Together, these combined analyses extend previous single-cell studies (including refs. 22 and 26–29) that used probe-based gene expression analysis, and enable comprehensive and unbiased RNAseq-based functional inferences.
CAMs in Single Cells.
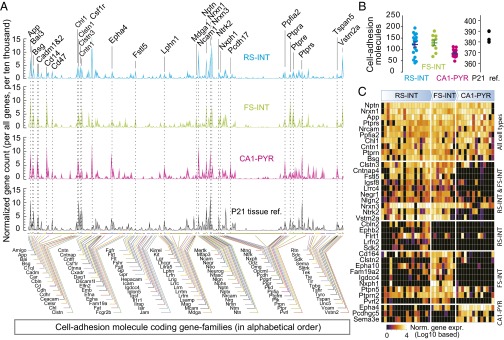
Our analyses in whole-tissue preparations (Fig. 1) consistently detected transcripts encoding 383.2 ± 1.1 (mean ± SEM) different CAMs in the hippocampus, which comprises a large variety of neuronal and nonneuronal cell types. In examining expression of CAMs at the single-cell level, we focused on how many CAMs were expressed in a neuron, and whether these were expressed with cell type specificity. In single RS-INT, FS-INT, and CA1-PYR cells, we detected NRS-INT = 121.6 ± 11.2, NFS-INT = 128.7 ± 12.7, and NCA1-PYR = 82.7 ± 4.8 different CAM transcripts, respectively [PRS-INT vs. FS-INT = 0.73, PRS-INT vs. CA1-PYR = 0.02, PFS-INT vs. CA1-PYR = 0.006; pairwise Mann–Whitney (MW) test; Fig. 4 A and B and Dataset S1]. Examining these genes revealed that expression of CAMs partially overlapped between different cell types. There were NRS-INT vs. FS-INT = 48, NRS-INT vs. CA1-PYR = 58, and NFS-INT vs. CA1-PYR = 62 differentially expressed CAMs between cell types (pairwise MW test, P < 0.05 in all cases). Thirty CAMs were expressed higher (n = 26; MW test, P < 0.05 in all cases) or lower (n = 4; Epha4, Mdga1, Pcdhgc5, Sema3e; MW test, P < 0.05 in all cases) in both interneuron types compared with PYRs. We found that genes that were highly expressed in tissue samples were also highly expressed in the RS-INT, FS-INT, and CA1-PYR cells (note that for sequencing libraries, tissue samples were diluted and processed using same conditions and reagents as for single cells, albeit with larger amounts of starting mRNAs). Such genes included App, Chl1, Cntn1, Nptn, Nrxn1, and Ptprs that were consistently detected in all three cell types (Fig. 4C, “All cell types”).
Fig. 4.
Hierarchy of CAMs in single neurons. (A) Averaged, single-cell expression of CAMs in RS-INT, FS-INT, and CA1-PYR cells, and in aged-matched P21 tissue control (for numerical values, see Dataset S1). (B) Number of CAMs detected in the three cell types and in age-matched tissue control. (C) Heat plot shows single-cell expression for ubiquitously (“All cell types”) as well as cell type-specifically expressed CAMs. Statistical differences were determined using MW test, with P < 0.05. Averaged data in B represent mean ± SEM.
The 26 CAMs that were expressed higher in interneurons than in CA1-PYR cells included Clstn3, Cntnap4, and Lrrc4, which have been implicated in specification of inhibitory synapses (30–32), as well as Nrxn3, a presumed presynaptic organizer of synapse function (13, 14, 33). Note that, although expression of Nrxn3 was not specific for interneurons, it was consistently enriched in interneurons, which we also considered here as a cell type-specific feature. Other examples for cell type specificity included Cbln2 and Ephb2 for RS-INT cells, Nphx1 for FS-INT cells, and Epha4 for CA1-PYR cells. Although some of these molecules have been directly implicated in transsynaptic signaling (e.g., Nrxn1, Nrxn3, Ptprs) or in synapse specialization (see examples above), others have not been studied in detail. Nevertheless, these data suggest cell type-specific expression of CAMs can potentially explain neuronal connectivity at the molecular level. Such cell type-specific features were further highlighted by principal-component axes (PCA) analysis, which revealed two main components, including overall expression as well as differential expression in interneurons vs. PYR cells (Fig. S4 A–C).
Fig. S4.
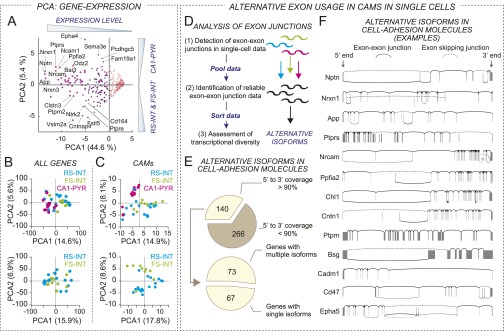
Single-cell analysis of CAMs. (A) We performed principal-component axes (PCA) analysis on expression of CAMs in RS-INT, FS-INT, and CA1 PYR cells. Two main components that were revealed by these analyses involved overall expression level of genes (PCA1), and separation of gene expression between CA1 PYR cells, and RS-INT and FS-INT cells combined (PCA2). (B and C) Because of the cell type-specific differences in gene expression between RS-INT, FS-INT, and CA1 PYR cells, we further examined the extent to which gene expression could identify cell types. For this, we used PCA analysis, using “All genes” and “Cell adhesion molecules” as selection criteria. The upper panels display analyses of all three cell types, whereas the lower panels include analyses of interneurons (RS-INT and FS-INT cells) only. (D) Alternative splicing can prominently alter cell adhesion function, and therefore we examined the potential extent of alternative exon use in single cells. Diagram depicts our strategy to determine the presence of alternative exon use in single gene transcripts in a “yes-or-no” fashion, independent of cell type-specific expression. For this, we pooled data from each cell and examined whether (i) there existed any overlapping exon–exon junction (i.e., where an exon was linked to multiple other exons) and (ii) there existed any neighboring exon pair without common exon–exon junction that was bridged over by other exon–exon junction in the mRNA transcript. The presence of either of these implicated alternative exon choices during transcription. Although the amplification method used in this study allowed detection of virtually any poly-A–tailed mRNA sequence, its inherent 3′-biased nature limited efforts to draw statistically relevant, cell type-specific conclusions about splice isoforms. Therefore, to estimate overall mRNA isoform diversity in single-cell samples, we only analyzed reliably detected exon–exon junctions (i.e., at least 90% coverage in more than three single-cell samples; reasons for less coverage could include low/none expression as well as potentially GC-rich nucleotide structure, making specific amplification less efficient) and used these data to reconstruct transcriptional variants. (E) To quantify the overall diversity of alternatively spliced exons, we screened all CAMs for alternative exon use, as explained above. Using this approach, we examined 139 CAMs and found that ∼50% of these molecules had multiple isoforms. (F) Example exon structure reconstruction of highly and ubiquitously (present in all three cell types, including RS-INT, FS-INT, and CA1 PYR cells) expressed CAMs. Each vertical line represents an exon (for these, genetic coordinates were obtained from the mm10 assembly), solid lines represent exon–exon junctions between neighboring exons, whereas dashed lines represent junctions that skipped one or more intermediate exons. In these analyses, we found transcripts that lacked any alterative exon use (such as Ptprn and Cadm1), and transcripts with consistently skipped exons in all single-cell samples (such as Ptprs). However, analyses of most transcripts revealed presence of multiple, alternatively used exon–exon junctions.
Beyond cell type-specific control of gene expression levels, diversity in CAM signaling could have been generated by other cellular mechanisms, especially by alternative splicing. Some of the most notable examples for alternative splicing include CAMs, such as Dscam in Drosophila and neurexins in mammals, which can express thousands of alternatively spliced isoforms (34–37). Therefore, we examined alternative exon use of CAMs at the single-cell level. We analyzed genes with reliable end-to-end exon junction coverage, which was applicable to n = 139 CAM genes. Out of these, we identified a single isoform in n = 67 and multiple isoforms in n = 72 genes (Fig. S4 D and E). Because of their potential cell type specificity, presence of different isoforms in the latter group would be especially interesting. However, currently available single-cell RNAseq platforms—including ours—require library fragmentation. As a result, precise exon use of full-length mRNAs can be inferred only indirectly, with sample sizes typically larger than those of single cells, hindering further analyses of cell type-specific splice variants. Nevertheless, we examined whether any splice variants of the highly expressed ubiquitous CAMs had the potential to be expressed with cell type specificity. We found that some of these molecules had only single isoforms that may or may not involve alternative splicing (for example, in Ptprs, we consistently detected exon skipping, whereas in Ptprn and Cadm, we only detected canonical isoforms), whereas others had multiple isoforms (for example, in Nptn, Nrxn1, and Cd47; Fig. S4F). Together, these exon junction observations suggested further diversification of cell adhesion signaling in single cells, which—at least regarding neurexins—was also suggested in our previous findings (9).
Synaptic Vesicle Exocytosis and Intracellular Signal Transduction.
One presumed role of CAMs is to convey transsynaptic information and thereby establish a functional match between presynaptic and postsynaptic specializations. However, neither the precise molecular targets nor the underlying mechanisms have been described for most CAMs. Thus, we analyzed expression of two broadly defined groups of genes that are likely linked to cell adhesion function, namely genes related to vesicle exocytosis and to RhoGAP/RhoGEF signaling (see detailed list in Fig. S5A and Dataset S1). Exocytosis-related molecules (38) were expressed broadly in all single cells and at all developmental stages (Fig. 5 A and B, and Fig. S5B). In single cells, we detected both ubiquitous and cell type-specific features, where the former included Snap25, Synaptobrevin-2 (Vamp2), Snap47, and Syt11 (for which no function has been uncovered yet, despite its high abundance), whereas Cplx1, Stx1a, Syt13, and Synaptobrevin-1 (Vamp1) were expressed higher in interneurons (MW test, P < 0.05, for all genes; expression of Synaptobrevin-1 also appeared to be exclusive, as it was not detected in any PYRs; Fig. 5 A and B, and Dataset S1). In addition, we found exclusive expression of Sncg and Syt6 genes in RS-INT cells, with unknown functional consequences.
Fig. S5.
Exocytosis-, RhoGAP-, and RhoGEF-related gene expression in whole hippocampal RNAseq data. (A) Panels show comprehensive lists of genes, which were included in our analyses (see Materials and Methods for information on assembly of these lists). (B–D) Graphs show normalized expression levels of exocytosis-, RhoGAP-, and RhoGEF-related genes, respectively, in five developmental stages (for numerical values, see Dataset S1). We found an overall monotonic increase in enrichment of exocytosis-related gene expression, which may directly correspond to synapse maturation and increase in the numbers of synapses in the developing circuit. Such tendency was mirrored by Chn1 (RhoGAP), which was, however, the direct opposite of marked decrease in Mtap1b (RhoGEF) expression. These data provide developmental context for single-cell data presented for RS-INT, FS-INT, and CA1 PYR cells in Fig. 5. Averaged data represent mean ± SEM.
Fig. 5.
Cell-specific expression of exocytosis and RhoGAP/RhoGEF signal transducer molecules. (A) Schematic drawing depicting the exocytotic machinery of synapses. (B–D) Averaged single-cell expression of exocytosis- (B), RhoGAP- (C), and RhoGEF-related genes (D) in RS-INT, FS-INT, and CA1-PYR cells, and in aged-matched P21 tissue control (for numerical values, see Dataset S1). RhoGAP- and RhoGEF-related molecules are implicated in transmembrane signal transduction and intracellular cytoskeleton reorganization. Averaged data represent mean ± SEM.
Next, we analyzed the expression of RhoGAP- and RhoGEF-domain–containing and related proteins. These are membrane-associated proteins that are thought to transform extracellular receptor-mediated signals into an intracellular response, prominently the organization of the actin cytoskeleton. Among others, the Rho/rac/CDC42 signaling machinery is thought to be involved in activity-dependent changes in postsynaptic spines (39–41). Therefore, it is plausible that RhoGAPs and RhoGEFs act as downstream effectors for cell adhesion signals. We observed diverse expression patterns for RhoGAPs and RhoGEFs between RS-INT, FS-INT, and CA1-PYR cells (Fig. 5 C and D, and Fig. S5 C and D, and Dataset S1). Particularly striking was the selectively high expression of chimaerin-1 (Chn1) in CA1-PYR cells, consistent with a possible spine-specific function (42).
Coregulation and Coexpression of Synaptic Molecules.
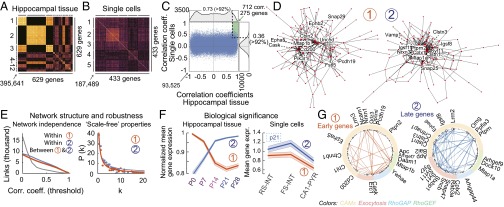
Thus, far, our data offer a comprehensive view of gene expression and identified multiple cell type-specific differences. Next, we aimed to evaluate correlations in gene expression, because our data could potentially reveal genes that were developmentally coregulated and coexpressed at the single-cell level, and because such genes may contribute to core synaptic features. For this analysis, we included data on genes related to CAMs, exocytosis, and RhoGAPs/RhoGEFs, collected from five developmental stages (hippocampal tissue) and three cell types (single hippocampal cells monitored at ∼P21). To avoid inferences from inconsistently detected molecules, we excluded genes that were detected in less than five tissue or less than five single-cell samples, narrowing our analyses to 632 and 428 genes, respectively.
First, we independently examined gene correlations in tissue and single-cell data, and used unsupervised clustering to consolidate these results (Fig. 6 A and B), which reflected developmental and cell type-specific characteristics, respectively (Fig. S6A). We reasoned that functionally relevant correlations may be present in both sets with high correlation coefficients, and therefore selected gene pair correlations that were in the tail of distributions (>92nd percentile; note that this threshold was chosen arbitrarily but that consequences of this analysis were thoroughly tested at multiple threshold values; Fig. 6E) both for tissue and single-cell data (Fig. 6C). This step narrowed our focus to 269 genes and 692 correlations. Genes were outnumbered by correlations because some genes correlated with multiple others (i.e., they represented correlation “hubs”). We used graph analysis to test such possibility. In such graphs, each node represents a gene and each vertex represents correlation between two genes, if any, with a length inversely proportional to the respective coefficient; genes with more correlations simply had more vertices connecting them.
Fig. 6.
Coregulation and coexpression of synaptic molecules. (A) Heat plot of correlations between genes representing cell adhesion-, exocytosis-, RhoGAP-, and RhoGEF-related molecules in hippocampal tissue. Coefficients were computed based on gene expression values in five developmental stages in tissue samples. Unsupervised clustering revealed 12 clusters (labeled on the Right and Top) based on 395,641 gene correlations. (B) Same analysis as in A, but for single-cell data. Coefficients were computed based on single-cell gene expression values independent of cell type identity. Unsupervised clustering revealed five clusters (labeled on the Right and Top) based on the 187,489 gene correlations. (C) Combined data of correlation coefficients from tissue and single-cell samples. Gene pairs with both coefficients larger than the 92nd percentile of the respective distributions were further analyzed (green shaded area; see text for further information). (D) Graph representation of correlation coefficients displayed in the green shaded area of C. Each vertex connects two genes according to the correlation coefficient between the two. Graph representation of correlations in C revealed two independent, uncorrelated subgraphs. (E, Left) Subgraph 1 and subgraph 2 remained independent when relaxing gene inclusion criteria (i.e., when green area in C was gradually increased). (Right) Density of vertex distribution (number of gene–gene correlations) followed power law distribution, indicating that both subgraphs were scale-free and nonrandom in nature. (F, Left) Mean normalized gene expression values in the two subgraphs showed diametrically opposite developmental trajectories, and suggest that genes in subgraph were developmentally coregulated. Because single-cell expression was a selection criterion in C, these genes were also coexpressed at the single-cell level. (Right) As suggested by the developmental differences, genes in subgraph 1 and subgraph 2 had different expression values in single cells of all three cell types, collected at ∼P21. (G) Core early- and late-gene networks (subgraph 1 and subgraph 2, respectively) were identified by common motifs found in cell type-specific analyses (i.e., these correlations were present in pooled data from the three cell types, in pooled data from RS-INT and FS-INT cells, representing interneurons, and RS-INT, FS-INT, and CA1-PYR data, analyzed independently). Averaged data represent mean ± SEM.
Fig. S6.
Coregulation and coexpression of cell adhesion-, exocytosis-, RhoGAP-, and RhoGEF-related genes. (A) Analyses of clustering properties revealed that developmental stages (“tissue data” at Left; corresponding to Fig. 6A) and cell type-specific gene expression (“single-cell data” at Right; corresponding to Fig. 6B) explained respective clustering patterns. Cluster numbers in the plot correspond to those in Fig. 6 A and B. Averaged data represent mean ± SEM. (B, Upper) Diagram outlines combined analyses of cell adhesion-, exocytosis-, RhoGAP-, and RhoGEF-related genes that were performed on multiple, cell type-specific datasets. To examine the size of network in each data, we quantified the number of genes involved in these networks (Lower Left), and found a decreasing trend in network size, corresponding to smaller datasets (i.e., individual cell types) and/or less consistent gene expression (e.g., CA1-PYR cells). Next, we examined whether there was any overlap between the different cell type-specific networks (Lower Right). For this, we compared genes in each network to those that were determined using all three cell types (these were performed for early- and late-gene networks separately) and found that more than 80% of these genes in individual cell type-specific data (e.g., RS-INT and FS-INT) were also present in the three cell type data, when analyzed together (represented by “100%”). These suggested the existence of core networks independent of cell type identity. We identified these two core networks, as shown in Fig. 6G, by determining common gene correlations in each cell type-specific data. (C) These graphs represent the complete early- and late-gene networks (subgraph 1 and subgraph 2, respectively), as shown in Fig. 6D, such that the name of each gene is displayed in alphabetical order, grouped by gene families. Larger font sizes symbolize higher expression values, and two genes are connected if their correlation value was in >92nd percentile, in this case (see Coregulation and Coexpression of Synaptic Molecules for more information on percentile threshold). (D) Insets depict molecular composition of the two subgraphs, displaying gene families and respective gene numbers (lines and circles represent cumulative vertices between the respective groups, such that the numbers of such vertices are labeled adjacently).
The structure of the resulting graph was surprising because two independent subgraphs emerged that exhibited dense correlations within, but no correlations between them (Fig. 6D; note the complete lack of interconnecting vertices; see Fig. S6C for complete gene listings in this graph). A biologically relevant explanation for these subgraphs was not immediately obvious, prompting us to perform additional analyses. First, we examined whether the two subgraphs were defined by nonoverlapping gene families (for example, they only include exocytosis or CAM genes). However, this was not the case, as both subgraphs included genes related to CAMs, exocytosis, and RhoGAP/RhoGEF (Fig. S6C). Second, we examined robustness of their independence by quantifying vertices between the two subgraphs while relaxing criteria for gene inclusion (from >90th to 0 percentile, i.e., all inclusion, in steps of 10 percentile). These analyses consistently revealed more intranetwork than internetwork correlations, which reversed when lowering inclusion threshold to 20 percentiles (0.2 correlation coefficient threshold in Fig. 6E, Left), suggesting robust independence (note that with “all inclusion,” 0 percentile, the graph is perfectly connected). Third, we examined randomness in graph connectivity. In a random graph, each gene would have a similar number of correlations, or vertices in the graph. However, both subgraphs displayed a nonrandom, “scale-free” nature where some genes were more interconnected than others, following a power law distribution and thus suggesting a biological origin (Fig. 6E, Right) (43). Fourth, we analyzed gene expression levels in each subgraph. Here, we found that developmental gene expression profiles were very similar within each, but not when the two sets were compared with each other. Specifically, subgraph 1 included genes that were highly expressed during early development, whereas subgraph 2 contained genes that exhibited later expression onsets, with peak expressions occurring >2 wk after birth (Fig. 6F, Left). In agreement with this finding, late expressing genes (subgraph 2) displayed higher expression than early expressing genes (subgraph 1) in single cells of all three types (Fig. 6F, Right), which were collected at ∼P21. Together, these analyses suggested developmental coregulation as a primary determinant for separation of the two graphs.
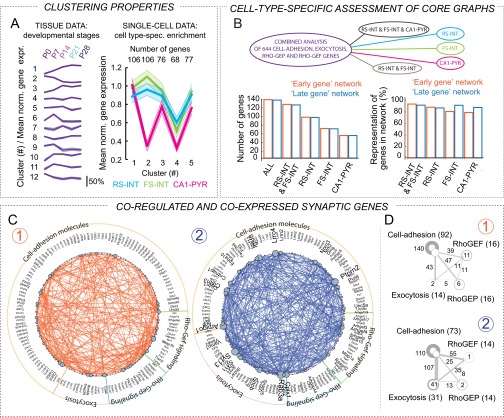
Next, we sought to further examine graph structures using added knowledge about identity of single-cell samples. We reasoned that robust gene expression correlations should be consistently present if cell types were analyzed independently. Therefore, we repeated analyses such that Fig. 6B only included RS-INT, or FS-INT, or CA1-PYR cells separately, or RS-INT and FS-INT cells combined, because they both represented GABAergic interneurons. We found that each of these analyses confirmed the independence of subgraphs 1 and 2 (Fig. S6B), and that the gene representation in each cell type-specific dataset was at least 80% identical to that in Fig. 6D (Fig. S6B), suggesting that coexpression of these genes occurs at the single-cell level. Moreover, we identified core parts of these subgraphs (i.e., parts that can be consistently detected in each cell type) by taking the intersection of the cell type-specific analyses (Fig. 6G). Because of their robust presence in each cell type, these correlations conceivably represented ubiquitous features, at different developmental stages of synapse maturation. This hypothesis was supported by the existence of correlations between structurally relevant RhoGEF and RhoGAP molecules in early development, when synapses are formed (Fig. 6G, Left, and Fig. S6D, Upper). Conversely, emerging correlations between cell adhesion and exocytosis in the late-gene network may reflect synapse specialization (Fig. 6G, Right, and Fig. S6D, Lower). Although our analyses did not reveal a mechanistic explanation for the observed correlations, they identified synaptic genes that were both developmentally coregulated and coexpressed at the single-cell level.
CAMs in BS-PYR and RS-PYR Cells of the Subiculum.
Our findings thus far revealed ubiquitous and cell type-specific features of CAM and signaling molecule expression as well as the contextual determinants of their expression, involving molecules related to exocytosis and Rho signaling. Next, we wanted to examine the generality of our central findings, including that of (i) the identity of ubiquitously expressed CAMs and of (ii) the existence and identity of synaptic early- (subgraph 1) and late-gene interaction networks (subgraph 2). To this end, we performed single-cell RNAseq experiments to analyze electrophysiologically defined BS-PYR and RS-PYR neurons in the subiculum (Fig. 7A), which are the major projection targets of CA1-PYR cells. We hypothesized that their gene expression profiles are different, because they exhibit distinct excitability and synaptic properties (12, 13, 15).
Fig. 7.
Molecular identity of BS-PYR and RS-PYR neurons in the subiculum. (A) Experimental design shows electrophysiological identification of BS and RS neurons as well as sample collections, and processing of single-cell RNAseq data. White box indicates the subiculum region. (B) The number of detected genes were not different in burst- (n = 21) and regular-firing (n = 14) neurons. (C) Plot shows consistency of gene expression in the two cell types. (D) Heat map shows exclusively expressed genes in burst- and regular-firing neurons. (E and F) Expression of voltage-gated ion channels and CAMs in BS-PYR and RS-PYR cells as well as in age-matched tissue controls (P21; for numerical values, see Dataset S1). (G) Heat map shows single-cell expression of CAMs respective to the order of their expression in the CA1 region, as shown in Fig. 4C. Expression of these molecules in BS-PYR and RS-PYR cells were not distinguishable. (H) Whole-transcriptome PCA was also unable to distinguish between BS-PYR and RS-PYR cells. (I) Analyses of gene expression in BS-PYR and RS-PYR cells confirmed existence of early- and late-gene networks (shown in Fig. 6). These plots show core consensus networks defined by independent analyses in five cell types examined, including RS-INT, FS-INT, CA1-PYR, BS-PYR, and RS-PYR neurons.
We detected 9,671 ± 278 and 9,499 ± 405 genes in BS-PYR and RS-PYR cells, respectively (P = 0.85; Fig. 7B and Fig. S7A), with more consistent gene expression patterns than among CA1-PYRs (Fig. 7C and Fig. S7B). Next, we examined the molecular profiles of these cells, especially seeking exclusive markers that would unequivocally identify these functionally different cell types. However, we found that they were largely identical and detected only a surprisingly small number of exclusively expressed genes (Fig. 7D). Such genes included, for example, the neuropeptide cortistatin (Cort) and the c-Fos–induced growth factor (Figf) (a member of the VEGF family). Apart from these genes, BS-PYR and RS-PYR neurons expressed similar patterns of voltage-gated ion channels (Fig. 7E; see differently expressed ion channels in Fig. S7C), CAMs (Fig. 7F), ligand-gated ion channels, and exocytosis- and RhoGAP-/RhoGEF-related molecules (Fig. S7 C–G and Dataset S1).
Fig. S7.
Single-cell gene expression analyses in subiculum BS-PYR and RS-PYR neurons. (A) Plots show individual normalized gene expression density (i.e., histogram of gene numbers with given normalized gene counts) for each single cells; plot titles include cell type and number of detected genes, respectively. (B) Consistency of gene expression for voltage-gated ion channels (Upper) and CAMs (Lower). We found that, in both burst and regular firing cells, ∼50% of expressed genes were consistently detected in more than one-half of the cells, rendering more pronounced homogeneity in these samples compared those in the CA1 region (Fig. 2 C and D). (C) We found eight ion channels, all K+ channels, that were differently expressed in BS-PYR and FS-PYR cells. This plot shows single-cell expression values for these genes, mean ± SEM values, and values of significance when using pairwise Mann–Whitney test. It is currently not clear which of these gene or genes are responsible for electrophysiological differences between BS-PYR and RS-PYR cells. (D–G) Single-cell expression of ligand-gated ion channels, exocytosis-related molecules, as well as RhoGAP and Rho-GEF signaling molecules in subiculum BS-PYR and RS-PYR neurons, and age-matched tissue control data (P21). For numerical values, see Dataset S1.
We next examined expression of ubiquitously present CAMs (as identified in CA1 interneurons and PYR cells), which we reliably detected in subiculum PYR cells similar to CA1-PYRs (Figs. 4C and 7G), further strengthening the notion of general importance of these genes in neuron and synapse function. Although 42 CAMs had different expression levels between the two subiculum PYR cell types (P < 0.05; pairwise MW test; Dataset S1), none of these CAMs was exclusively expressed in one or the other cell type. However, their expression differed markedly from that of cell type-specifically expressed CAMs in CA1 PYR cells. Subiculum PYR cells consistently used CAMs that were not observed in CA1-PYR cells. For example, Clstn3 (significantly enriched only in interneurons in CA1), Ptpn5 and Ptprn2 (both were enriched in FS-PV cells), as well as Lrrc4 (not present in any CA1-PYR) were detected and highly expressed in subiculum PYR cells. Further cell-to-cell analysis corroborated molecular similarities between BS-PYR and RS-PYR cells, as confirmed by the inability of PCA (used on the complete transcriptome) to distinguish between these cells (Fig. 7H).
Finally, the subiculum neuron data allowed us to test the validity of synaptic gene correlation networks described in Fig. 6. We repeated the combined analysis of tissue and single-cell RNAseq data on subiculum PYR cells, independent from CA1 cells. In these analyses, we also identified two independent subgraphs, which were essentially identical to those found in CA1 RS, FS, and PYR cells. To reexamine the identity of the two networks, we singled out overlapping gene correlations in all five cell types (Fig. 7I) and found that they were nearly identical to subgraph 1 and subgraph 2, identified based on the three CA1 cell types (Fig. 6G). Therefore, these analyses independently confirmed the existence of coregulated and coexpressed synaptic gene networks, which may be generally present in cells in the brain independent of cell type identity.
Discussion
A molecular description of gene expression in individual, independently identified neurons in combination with knowledge of their physiological properties and synaptic connectivity is a prerequisite for insight into how synaptic connections are formed and maintained in brain. Moreover, such data are essential for understanding why disease-associated gene mutations impair brain function. RNAseq of single neurons can be routinely obtained from cells isolated from tissue lysates (44–47) and has recently been achieved from electrophysiologically recorded cells (the current study; also see refs. 16 and 17). Here, we used single-cell RNAseq of individual synaptically and electrophysiologically characterized neurons, and cross-referenced their gene expression profiles to that of total brain tissue to explore cell adhesion signaling in a well-defined hippocampal circuit. In particular, we examined inhibitory projections from RS and FS interneurons to CA1-PYR cells (6, 7, 10) and excitatory projections from CA1-PYR cells to subiculum BS-PYR and RS-PYR cells (13, 15). Our results show that capture of mRNAs by aspiration of cytosol allows production of high-quality transcriptome data (Figs. S2 and S7), opening up identification of the precise expression profiles of single neurons as a molecular portal for a detailed functional understanding.
Functional implications of gene expression can be most directly examined by relating gene expression to electrophysiological properties (16, 17, 26–29, 48). Therefore, we first tested whether the electrophysiological properties of a neuron could be explained by its ion channel expression pattern. Previous studies established that expression of Kv3 K+ channels and a high Na+-channel density enable the FS properties of PV cells (23). In line with this conclusion, we found specific enrichment of Kcnc1 (Kv3.1), Kcnc2 (Kv3.2), and Scn1a (NaV1.1) in FS-INT cells. By taking advantage of our comprehensive gene expression data, we extended these findings and correlated 11 electrophysiological properties with 104 ion channel genes. We found strong correlations between specific sets of genes and physiological properties, consistent with the different cell types. Although these correlations may mostly reflect group-level differences, an added value of these analyses is the conclusion that electrophysiological properties can be identified using single-cell transcriptome information. More detailed quantification of electrophysiological properties in combination with extended sample sets and morphological analyses should, for example, allow differentiation of interneuron subtypes (5) on a molecular basis. In addition, these results suggest that the validity of RNAseq data in electrophysiologically defined cells extends to genes whose functional readouts were not readily available, as is the case for CAMs.
We hypothesized that combinatorial expression of CAMs in single cells may define connectivity in a neuronal network. A corollary of this hypothesis is that CAMs in a presynaptic terminal must interact with complementary CAMs in the postsynaptic compartment; this interaction is—through reciprocal ligand–receptor coupling—enabled via the synaptic cleft. Abundant presence of CAMs in hippocampal tissue (Fig. 1) supports this hypothesis, because the diversity of CAMs likely permits a large number of combinations, possibly defining synaptic connectivity between the more than 20 cell types in the hippocampus and beyond (5). Our data present the most current account of CAMs in the brain, described by expression analysis of 406 genes. A limitation of our RNAseq results is that a transcriptome profile represents the abundance of mRNAs in a given cell type but does not provide information regarding protein abundance or localization. A given neuron receives heterogeneous inputs (excitatory and inhibitory synapses from multiple sources) and can innervate multiple postsynaptic cells of differing identities. Thus, it is plausible that CAMs are differentially localized and combinatorially used in a synapse-specific manner. To understand the organization of these molecules and possibly interpret them as a code for connectivity, we need to clarify which of these molecules are expressed in specific synapses and physically interact with each other. An important step to this direction is to examine cell type-specific expression of these molecules.
Our hypothesis suggests that expression of CAMs should differ among cell types, including RS-INT, FS-INT, CA1-PYR, BS-PYR, and RS-PYR cells, in which we previously established functionally relevant differences in cell adhesion signaling (9, 10, 13, 15). Thus, we examined candidate synaptic CAMs in single cells from these types. Presynaptic and postsynaptic CAMs presumably engage in homophilic and heterophilic interactions in the synaptic cleft, but the rules that associate particular CAMs to specific synapses and their transcriptional regulation in different cell types were not known. In examining the cell type-specific expression of CAMs, we made two fundamental observations (Figs. 4 and 7).
First, CAMs are hierarchically expressed. This conclusion is based on the observation that some CAMs were consistently and highly expressed in all five cell types examined (these included Nptn, Nrxn1, App, Ptprs, Nrcam, Ppfia2, Chl1, Cntn1, Ptprn, and Bsg), whereas others were expressed cell type-specifically (these included, for example, Cntnap4, Fstl5, Igsf8, Lrrc4, Cbln2, Eph2b, Idgcc4, Nxph1, Epha4, and Sema3e). The most likely explanation for such hierarchy is that commonly expressed molecules are required for core synaptic features as well as other neuronal cell interactions, whereas cell type-specific molecules may enable functional specialization. For example, it is intriguing to speculate which, if any, of these molecules are transsynaptically involved in sculpting endocannabinoid signaling in RS CCK or opioid signaling in FS PV synapses, respectively (6, 7, 10). Overall, these data support our hypothesis that CAMs are combinatorially expressed at the single-cell level.
Second, we identified synaptic genes that were both developmentally coregulated as well as coexpressed in single cells (Figs. 5–7). This was made possible by a combined analysis of tissue and single-cell data, and revealed two independent gene networks: an “early-gene” and a “late-gene” network. Beyond developmental differences, molecular composition of the two networks implicated distinct functional relevance. Specifically, interactions between intracellular signal transducer molecules were enriched in the early-gene cluster, whereas representation of exocytosis-related molecules as well as their interactions with CAMs were more apparent in the late-gene cluster. Such findings are consistent with the developmental sequels of structural formation and functional maturation of synapses and neuron–glia interactions. Because they are comprehensive in nature, experimental probing of these networks is challenging as it may require simultaneous manipulation of multiple genes. Such approach, targeting multiple heavily connected genes or hubs (e.g., Clstn1, Clstn3, or Igsf8), could reveal susceptibility to genetic conditions with especially disruptive pathophysiological consequences.
Together, our results provide a description of which electrophysiologically and synaptically characterized neurons express which specific CAMs. Some of these molecules are already known to play important synaptic roles, as their genetic disruptions have been linked to neuropsychiatric disorders in humans and have been shown to be lethal in mice (33). However, the function of the majority of these molecules as well as the logic that integrates them into assembly and specification of synapses or other intercellular junctions remain unknown. Here, we made progress toward understanding such logic by measuring developmental and single-cell expression of CAMs. When revealed, underlying principles can help us to use the brain’s cell adhesion code to formally describe connectivity in neuronal circuits. Such applications would lead to developments in molecular diagnostics allowing comprehensive analysis of brain circuits from single-cell samples.
Materials and Methods
Electrophysiology and Single-Cell Sample Collection.
All animal protocols and husbandry practices were approved by the Institutional Animal Care and Use Committee at Stanford University. Hippocampal slices (300 µm) were prepared from 3- to 4-wk-old wild-type CD1 mice, as described in ref. 10 and SI Materials and Methods.
cDNA Preparation and Library Preparation for Next-Generation Sequencing.
Single-cell mRNA was performed using the Clontech’s SMARTer Ultra Low Input RNA Kit. As a first step, cells were collected via pipette aspiration into sample collection buffer, were spun briefly, and were snap frozen on dry ice. Samples were stored at −80 °C until further processing, which was performed according to manufacturer’s protocol. Library preparation was performed using Nextera XT DNA Sample Preparation Kit (Illumina) as described in the protocol. Then, cells were pooled and sequenced in an Illumina NextSeq500 instrument using 2 × 75 paired end reads on a NextSeq high-output kit (Illumina; see SI Materials and Methods for details).
Processing of mRNA Sequencing Data.
After de-multiplexing the raw reads to single-cell datasets, we used Prinseq to remove short reads. After trimming and removal of overrepresented sequences and adapters, remaining reads were aligned to the mm10 genome with STAR aligner. Aligned read were converted to gene counts using HTSeq (see SI Materials and Methods for detailed parameter descriptions).
Gene Categories.
For gene expression analysis, we examined six functionally related categories: CAMs, voltage-gated ion channels, ligand-gated ion channels, synaptic exocytosis-related molecules, as well as RhoGAP and RhoGEF signaling-related molecules. For each of these, we assembled a comprehensive list, which included all genes examined (see SI Materials and Methods for a detailed description of categories).
Data Analysis.
All data analyses were performed using Mathematica10 (Wolfram Research). These analyses included (i) normalization of gene expression data, (ii) quality control, (iii) analysis of physiological properties, (iv) analysis of exon junctions, and (v) correlation and graph analyses of expression data (see SI Materials and Methods for more details).
SI Materials and Methods
Electrophysiology and Single-Cell Sample Collection.
Hippocampal slices (300 µm) were prepared from 3- to 4-wk-old wild-type CD1 mice incubated at 33 °C in sucrose-containing artificial cerebrospinal fluid (sucrose-ACSF) (85 mM NaCl, 75 mM sucrose, 2.5 mM KCl, 25 mM glucose, 1.25 mM NaH2PO4, 4 mM MgCl2, 0.5 mM CaCl2, and 24 mM NaHCO3) for 1 h, and then held at room temperature until recording. Cells were visualized by infrared differential interference contrast optics in an upright microscope (Olympus; BX-61WI). Recordings were performed using borosilicate glass pipettes with filament (Sutter Instruments; BF100-58-10; o.d., 1.0 mm; i.d., 0.58 mm; 10-cm length) at 33 °C in ACSF (126 mM NaCl, 2.5 mM KCl, 10 mM glucose, 1.25 mM NaH2PO4, 2 mM MgCl2, 2 mM CaCl2, and 26 mM NaHCO3) with a standard intracellular solution (95 mM K-gluconate, 50 mM KCl, 10 mM Hepes, 4 mM Mg-ATP, 0.3 Na-GTP, 10 mM phosphocreatine; pH 7.2, 300 mOsm).
Identification of cell types.
Cells were identified based on soma location, action potential (AP) firing properties, and/or synaptic connectivity. First, we examined FS and RS interneurons that directly inhibit CA1-PYR cells, as described in the main text. Because these combined electrophysiology and single-cell RNAseq were first of the kind, we did not use any labeling method that could potentially interfere with RNA quality, and therefore post hoc morphological or immunocytochemical analyses were not performed on these cells. After recording, cytosol samples were collected from both presynaptic and postsynaptic cells. Because of the sequential nature of paired-recording experiments, typical recording time was 20–30 min for FS and RS interneurons and 1–5 min for CA1-PYR cells. Because such time difference could have impacted the number of detected genes, we conducted additional control experiments, where we recorded from CA1-PYR cells for 25–30 min and performed single-cell RNAseq: however, the number of detected genes was not different (Fig. S2F). Second, we recorded from subiculum burst-spiking and regular-spiking PYR neurons (BS-PYR and RS-PYR, respectively), which were identified based on their soma locations and electrophysiological properties. Specifically, these cells were localized in the middle third of the subiculum in 300-µm-thick horizontal slices from the intermediate hippocampus. To identify cells, depolarizing current steps were applied in current clamp until APs were evoked. Cells that displayed multiple (typically two to three) APs within 3 ms were classified as BS-PYR cells, and cells that fired only one AP, or fired multiple, but with a delay of >10 ms after the first, were classified as RS-PYR cells. The typical recording time was 1–5 min for these neurons. After recordings, neuronal cytosol was aspirated via the glass pipette used for recording.
Sample collection.
To minimize interference with subsequent molecular experiments (such as sensitivity to different salts), we minimized the amount of intracellular solution used in the glass pipette to ∼0.5 µL. The intracellular solution used for recordings was not autoclaved or treated with RNase inhibitors otherwise. Before and during recording, all surface areas that the experimenter needed to contact during experiments (such as manipulators, microscope knobs, computer keyboard, etc.) were thoroughly cleaned with RNase Away solution (Molecular BioProducts). After recordings, neuronal cytosol was aspirated via the glass pipette used for recording. Although the aspirated cytosol may have contained some of the genomic DNA, our choice of cDNA preparation, which involved poly-A selection virtually eliminated the possibility of genomic contamination in the RNAseq data. For sample collection, we quickly removed the pipette holder from amplifier head stage, and used positive pressure to expel samples into microtubes containing cell collection buffer while gently breaking the glass pipette tip. Cell collection microtubes containing cell collection buffer were stored on ice until they were used. All recordings were made using MultiClamp700B amplifier (Molecular Devices), and signals were filtered at 4 kHz (Bessel filter) and digitized (10 kHz) with a Digidata1440A (Molecular Devices); signals were analyzed with custom-written Mathematica10 (Wolfram Research) scripts.
cDNA Preparation and Library Preparation for Next-Generation Sequencing.
Single-cell mRNA was performed using the Clontech’s SMARTer Ultra Low Input RNA Kit for Sequencing, version 3 (for tissue/RS-INT/FS-INT/CA1-PYR samples) and version 4 (for BS-PYR/RS-PYR samples). As first step, cells were collected via pipette aspiration into 1.1 µL of 10× collection buffer, were spun briefly, and were snap frozen on dry ice. Samples were stored at −80 °C until further processing, which was performed according to manufacturer’s protocol. Resulting cDNA was harvested and analyzed on the Fragment analyzer automated fragment analyzer by Advanced Analytical. Only cells that had a concentration higher than 0.05 ng/µL were selected for library preparation. Library preparation was performed using Nextera XT DNA Sample Preparation Kit (Illumina) as described in the protocol. Following library preparation, cells were pooled and sequenced in an Illumina NextSeq500 instrument using 2 × 75 paired end reads on a NextSeq high-output kit (Illumina).
Processing of mRNA Sequencing Data.
After demultiplexing the raw reads to single-cell datasets, we used Prinseq to remove short reads (-min_len 30), to trim the first 10 bp on the 5′-end (-trim_left 10), to trim reads with low quality on the 3′-end (-trim_qual_right 25), and to filter low complexity reads (-lc_method entropy \-lc_threshold 65). We used FASTQC to determine overrepresented sequences and removed those using cutadapt (-e 0.15 –m 30). We then used Prinseq to remove orphan pairs less than 30 bp in length followed by removal of Nextera adapters using Trim Galore (–stringency 1). Remaining reads were aligned to the mm10 genome with STAR using the following options (–outFilterType BySJout \–outFilterMultimapNmax 20 \–alignSJoverhangMin 8 \–alignSJDBoverhangMin 1 \–outFilterMismatchNmax 999 \–outFilterMismatchNoverLmax 0.04 \–alignIntronMin 20 \–alignIntronMax 1000000 \–alignMatesGapMax 1000000 \–outSAMstrandField intronMotif). Then, in each sample, aligned reads were converted to counts for every gene using HTSeq (-m intersection-nonempty \-s no).
Gene Categories.
For gene expression analysis, we examined six functionally related categories: CAMs, voltage-gated ion channels, ligand-gated ion channels, synaptic exocytosis-related molecules, as well as RhoGAP and RhoGEF signaling-related molecules. For each of these, we assembled a comprehensive list, which included all genes examined (see Dataset S1 for more details).
CAMs.
A major characteristics of CAMs is that they contain a single transmembrane domain (STM domain) (also called “single-pass” molecules), which links the intracellular and extracellular signaling domains. However, the presence of an STM domain does not necessarily render cell adhesion function. To narrow our focus from over 1,000 STM domain-containing molecules, we examined cell type-specific expression of 406 molecules, which were previously implicated in cell adhesion function because they established homophilic or heterophilic biochemical interactions with other molecules within this list (Fig. S1C).
Voltage- and ligand-gated ion channels.
These gene listings were adopted from the NC-IUPHAR guide (the International Union of Basic and Clinical Pharmacology Committee on Receptor Nomenclature and Drug Classification; see guidetopharmacology.org), including 140 and 67 genes, respectively (Fig. S3A and Fig. 3C).
Synaptic exocytosis-related molecules.
For comprehensive analysis of this category, we enlisted SNARE complex-associated genes, synaptotagmins, and active-zone proteins, including a total of 103 genes (based on ref. 38; Fig. S5A).
RhoGAP and RhoGEF signaling-related molecules.
These gene listings were assembled using “RhoGAP” and “RhoGEF” search criteria in the UniProtKB database at uniprot.org and include 78 and 58 genes, respectively (Fig. S5A).
Data Analysis.
All data analyses were performed using Mathematica10 (Wolfram Research). These analyses included (i) normalization of gene expression data, (ii) quality control (QC), (iii) analysis of physiological properties, (iv) analysis of exon junctions, and (v) correlation and graph analyses of expression data.
Normalization of gene expression.
The following procedures applied for tissue as well as single-cell data. First, total gene counts were determined in each sample, independently. Then, in each sample, all gene count values were divided by the total gene count value in that sample (“Normalized gene count”; for example, in Fig. 1E). The resulting normalized gene counts thus allowed comparison between different samples irrespectively of sequencing depth [for additional information, see QC in single cells (related to Fig. 2 and Fig. S2)]. For heat maps (for example, in Fig. 1F), normalized gene count values were log-normalized by multiplication with 106 [thus converting them to counts per million (cpm)], addition of 1 (to avoid values smaller than 1 or equal to zero upon calculation of logarithmic values), and derivation of logarithm based 10. Note that the sole purpose of this log-normalization was to accommodate visualization in heat map plots.
QC in single cells (related to Fig. 2 and Fig. S2).
Although the above-described steps ensured normalization of gene expression, we sought additional measures to ensure identification of samples where sample collection and/or preparation may have been insufficient to match quality of the rest of sample population. Therefore, we analyzed distribution of normalized gene counts in single-cells samples (Fig. S2). In these plots, peak of nonexpressing genes (which would appear at 0) were omitted for clarity. The remaining two peaks characteristically represented those genes with low expression (“first peak,” between 0 and 1) and those with high expression (“second peak,” above 2). Then, in each single cell, we counted the number of genes with expression level >1, binned genes by these numbers (the bin size was 1,000), and grouped neighboring gene counts pairwise to determine their averaged values, respective to both axes. The resulting data followed log-normal distribution, and we used the following log-normal equation to fit the data: . Our data analysis resulted in the following fit parameters: A = 59,705.1, µ = 8.22, and σ = 0.7, which enabled us to plot the fitting distribution curve on the data (Fig. S2B). Last, we determined the maximum value of this curve, 2,249.59 genes, which we used as a lower cutoff threshold for including single-cell samples for further analysis.
Analysis of electrophysiological properties (related to Fig. 3 and Fig. S3).
To examine neuronal excitability, we quantified several electrophysiological properties, including passive membrane properties, and properties of single and trains of APs (for graphical illustration, see Fig. S3 B1–B4). Data were collected with standardized protocols, and cell classifications were determined during recording. After recordings, no post hoc adjustments, such as cell exclusion or reclassification, were made based on individual electrophysiological properties. However, only those cells were used for further analyses (NRS-INT = 18, NCA1-PYR = 12, NFS-INT = 8), in which all parameters were adequately quantified, including convergent parameter fitting in all mathematical models used (see below).
Passive membrane properties.
We injected a series of current (I) steps through the patch pipette, starting from negative (hyperpolarizing) steps to positive (depolarizing) current steps. For determining passive membrane properties, we used those voltage response (V) measurements, which did not elicit AP firing. From these, we calculated (i) membrane resistance (Rm; as V/I ratio), (ii) hyperpolarization sag, as well as (iii) resting membrane potential (V0).
Single APs.
We analyzed single AP properties in 39 cells. In the majority of the cells (36 of 39), we averaged two to five individual, peak-aligned APs for analysis, whereas in three cells only single APs were analyzed. These were fitted with three linear regressions, which included baseline fit before AP as well as ascending and descending phases of AP. In addition, we determined (iv) AP peak (max) and trough (min) times and voltage amplitudes. Furthermore, using these readouts, we determined (v) time from AP peak to trough, (vi) AP base width, (vii) AP half-width, as well as (viii) AP symmetry (i.e., temporal position of peak between ascending and descending phases of AP).
AP trains.
We collected all peak AP amplitude and timing in response, if any, to current injections (including negative and positive) delivered in each cell. These together represent the input/output function of the cell, which typically do not display AP firing at low current steps, and gradually increasing firing rates at increasing current steps. Then, we fitted each cell’s input/output curve with a sigmoid function , and subsequently fitted 20–80% (amplitude) of the resulting sigmoid with a linear function. Using these, (ix) firing threshold was determined where the linear fit intersected the “x axis,” representing current injection steps, whereas amplitude of the sigmoid was used to quantify (x) maximal firing frequency. Next, we determined (xi) the rate of AP amplitude adaptation for each cell. For this, we considered AP trains consisting at least five APs and determined all AP amplitudes within these trains. Then, from each AP amplitude value (starting from the second), we subtracted amplitude of its preceding neighbor. Elements of these newly created lists were positive if amplitude increased and negative if decreased. In each cell, we analyzed all available AP trains, independent of the current injection used, and pooled these values. Finally, we determined the proportion of negative values, resulting in a number between 0 and 1, for each cell. In case of “1,” all APs were smaller than the preceding one (AP depression); in case of “0.5,” AP amplitudes did not change; in case of “0,” all APs were larger than the preceding ones (AP facilitation). Values in between suggested respective proportions.
Correlation between electrophysiological properties and gene expression.
We used Mathematica’s SpearmanRho function to identify correlations between electrophysiological properties and voltage-gated ion channel expression, and used Mathematica’s FindClusters function to identify clusters in the correlation data (Fig. 3E).
Analysis of exon junctions (related to Fig. S4).
Exon junction were only analyzed in single-cell samples. For these analyses, we used the STAR aligner’s SJ.out.tab output, which contained exon junction-specific information. Specifically, these files contained count numbers for each detected exon–exon junction together with genomic coordinates of their respective “start” and “end” exons. To increase robustness of detection, we analyzed exon–exon junctions that were detected in at least three single cells, independent of their cell types. Then, for each detected exon–exon junction, we determined (i) if there was any other annotated exon(s) between the start and end coordinates, implicating “exon skipping,” and (ii) if there was any other exon–exon junction with overlapping genomic coordinates, conferring presence of “alternative splicing.” Finally, we used this information to reconstruct the exon use map of highly expressed CAMs and determined the proportion of CAMs with single vs. multiple isoforms (Fig. S4 D–F).
Correlation and graph analyses of expression data (related to Fig. 6 and Fig. S6).
We analyzed expression of cell adhesion, synaptic vesicle exocytosis, as well as RhoGAP- and RhoGEF-related molecules. To increase dimensionality of analysis, we combined tissue (representing five developmental stages) and single-cell (first, including three cell types, and then extending to five cell types) data. To eliminate correlations reflecting coordinated gene expression in a very low fraction of cells, we only considered genes that were detected in at least five tissue or single-cell samples, and evaluated their correlations using Mathematica’s SpearmanRho function. To examine graph structure, we used Mathematica’s FindGraphPartition function, which found a partition of vertices such that the number of edges having endpoints in different parts was minimized. To ensure robustness of analysis, graph analysis was repeated at multiple gene correlation including thresholds, as indicated in the main text. To examine the scale-free vs. random nature of the two networks, we plotted the number of vertices with k edges, P(k), and fit these data with , where α and γ represented amplitude and power law parameter, respectively. Core consensus networks were determined by independently analyzing (RS-INT, FS-INT, CA1-PYR), (RS-INT, FS-INT), (RS-INT), (FS-INT), and (CA1-PYR) cell type-containing datasets and by subsequently determining shared gene correlation networks within these datasets.
Supplementary Material
Acknowledgments
This work was supported by National Institute on Drug Abuse Grant K99 DA034029 (to C.F.), Swiss National Science Foundation Grant CRETP3_166815 (to C.F.), and the National Institute of Mental Health Grants R37 MH52804 (to T.C.S.) and K99 MH103531 (to J.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE75386).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610155113/-/DCSupplemental.
References
- 1.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch C, Laurent G. Complexity and the nervous system. Science. 1999;284(5411):96–98. doi: 10.1126/science.284.5411.96. [DOI] [PubMed] [Google Scholar]
- 3.Itzkovitz S, Baruch L, Shapiro E, Segal E. Geometric constraints on neuronal connectivity facilitate a concise synaptic adhesive code. Proc Natl Acad Sci USA. 2008;105(27):9278–9283. doi: 10.1073/pnas.0712207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17(1):22–35. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
- 5.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: The unity of hippocampal circuit operations. Science. 2008;321(5885):53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glickfeld LL, Atallah BV, Scanziani M. Complementary modulation of somatic inhibition by opioids and cannabinoids. J Neurosci. 2008;28(8):1824–1832. doi: 10.1523/JNEUROSCI.4700-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freund TF, Katona I. Perisomatic inhibition. Neuron. 2007;56(1):33–42. doi: 10.1016/j.neuron.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Caroni P. Inhibitory microcircuit modules in hippocampal learning. Curr Opin Neurobiol. 2015;35:66–73. doi: 10.1016/j.conb.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Fuccillo MV, et al. Single-cell mRNA profiling reveals cell-type specific expression of neurexin isoforms. Neuron. 2015;87(2):326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Földy C, Malenka RC, Südhof TC. Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron. 2013;78(3):498–509. doi: 10.1016/j.neuron.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staff NP, Jung HY, Thiagarajan T, Yao M, Spruston N. Resting and active properties of pyramidal neurons in subiculum and CA1 of rat hippocampus. J Neurophysiol. 2000;84(5):2398–2408. doi: 10.1152/jn.2000.84.5.2398. [DOI] [PubMed] [Google Scholar]
- 12.Wozny C, Maier N, Schmitz D, Behr J. Two different forms of long-term potentiation at CA1-subiculum synapses. J Physiol. 2008;586(11):2725–2734. doi: 10.1113/jphysiol.2007.149203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154(1):75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aoto J, Földy C, Ilcus SM, Tabuchi K, Südhof TC. Distinct circuit-dependent functions of presynaptic neurexin-3 at GABAergic and glutamatergic synapses. Nat Neurosci. 2015;18(7):997–1007. doi: 10.1038/nn.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson GR, et al. β-Neurexins control neural circuits by regulating synaptic endocannabinoid signaling. Cell. 2015;162(3):593–606. doi: 10.1016/j.cell.2015.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadwell CR, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol. 2016;34(2):199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuzik J, et al. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol. 2016;34(2):175–183. doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neu A, Földy C, Soltesz I. Postsynaptic origin of CB1-dependent tonic inhibition of GABA release at cholecystokinin-positive basket cell to pyramidal cell synapses in the CA1 region of the rat hippocampus. J Physiol. 2007;578(Pt 1):233–247. doi: 10.1113/jphysiol.2006.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenkey N, et al. Tonic endocannabinoid-mediated modulation of GABA release is independent of the CB1 content of axon terminals. Nat Commun. 2015;6:6557. doi: 10.1038/ncomms7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cembrowski MS, et al. Spatial gene-expression gradients underlie prominent heterogeneity of CA1 pyramidal neurons. Neuron. 2016;89(2):351–368. doi: 10.1016/j.neuron.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Du J, Zhang L, Weiser M, Rudy B, McBain CJ. Developmental expression and functional characterization of the potassium-channel subunit Kv3.1b in parvalbumin-containing interneurons of the rat hippocampus. J Neurosci. 1996;16(2):506–518. doi: 10.1523/JNEUROSCI.16-02-00506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martina M, Schultz JH, Ehmke H, Monyer H, Jonas P. Functional and molecular differences between voltage-gated K+ channels of fast-spiking interneurons and pyramidal neurons of rat hippocampus. J Neurosci. 1998;18(20):8111–8125. doi: 10.1523/JNEUROSCI.18-20-08111.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu H, Gan J, Jonas P. Interneurons. Fast-spiking, parvalbumin+ GABAergic interneurons: From cellular design to microcircuit function. Science. 2014;345(6196):1255263. doi: 10.1126/science.1255263. [DOI] [PubMed] [Google Scholar]
- 24.Aponte Y, Lien C-C, Reisinger E, Jonas P. Hyperpolarization-activated cation channels in fast-spiking interneurons of rat hippocampus. J Physiol. 2006;574(Pt 1):229–243. doi: 10.1113/jphysiol.2005.104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang SY, et al. Distribution of Kv3.3 potassium channel subunits in distinct neuronal populations of mouse brain. J Comp Neurol. 2007;502(6):953–972. doi: 10.1002/cne.21353. [DOI] [PubMed] [Google Scholar]
- 26.Ogiwara I, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: A circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27(22):5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy GA, et al. DAG-sensitive and Ca2+ permeable TRPC6 channels are expressed in dentate granule cells and interneurons in the hippocampal formation. Hippocampus. 2013;23(3):221–232. doi: 10.1002/hipo.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambolez B, Audinat E, Bochet P, Crépel F, Rossier J. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9(2):247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 29.Toledo-Rodriguez M, et al. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14(12):1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z, et al. Calsyntenin-3 molecular architecture and interaction with neurexin 1α. J Biol Chem. 2014;289(50):34530–34542. doi: 10.1074/jbc.M114.606806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettem KL, et al. The specific α-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron. 2013;80(1):113–128. doi: 10.1016/j.neuron.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karayannis T, et al. Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature. 2014;511(7508):236–240. doi: 10.1038/nature13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14(3):497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 35.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101(6):671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 36.Treutlein B, Gokce O, Quake SR, Südhof TC. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA. 2014;111(13):E1291–E1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiner D, et al. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 2014;84(2):386–398. doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Südhof TC. Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron. 2013;80(3):675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaudon F, et al. The RhoGEF DOCK10 is essential for dendritic spine morphogenesis. Mol Biol Cell. 2015;26(11):2112–2127. doi: 10.1091/mbc.E14-08-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai KO, Ip NY. Structural plasticity of dendritic spines: The underlying mechanisms and its dysregulation in brain disorders. Biochim Biophys Acta. 2013;1832(12):2257–2263. doi: 10.1016/j.bbadis.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Cheadle L, Biederer T. The novel synaptogenic protein Farp1 links postsynaptic cytoskeletal dynamics and transsynaptic organization. J Cell Biol. 2012;199(6):985–1001. doi: 10.1083/jcb.201205041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van de Ven TJ, VanDongen HM, VanDongen AM. The nonkinase phorbol ester receptor alpha 1-chimerin binds the NMDA receptor NR2A subunit and regulates dendritic spine density. J Neurosci. 2005;25(41):9488–9496. doi: 10.1523/JNEUROSCI.2450-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 45.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 46.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112(23):7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanchate NK, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350(6265):1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geiger JR, et al. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15(1):193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 49.Catania MV, et al. AMPA receptor subunits are differentially expressed in parvalbumin- and calretinin-positive neurons of the rat hippocampus. Eur J Neurosci. 1998;10(11):3479–3490. doi: 10.1046/j.1460-9568.1998.00356.x. [DOI] [PubMed] [Google Scholar]
- 50.Milenkovic I, et al. The parvalbumin-positive interneurons in the mouse dentate gyrus express GABAA receptor subunits α1, β2, and δ along their extrasynaptic cell membrane. Neuroscience. 2013;254:80–96. doi: 10.1016/j.neuroscience.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 51.Ferando I, Mody I. In vitro gamma oscillations following partial and complete ablation of δ subunit-containing GABAA receptors from parvalbumin interneurons. Neuropharmacology. 2015;88:91–98. doi: 10.1016/j.neuropharm.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao B, Fritschy JM. Selective allocation of GABAA receptors containing the alpha 1 subunit to neurochemically distinct subpopulations of rat hippocampal interneurons. Eur J Neurosci. 1994;6(5):837–853. doi: 10.1111/j.1460-9568.1994.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 53.Frerking M, Malenka RC, Nicoll RA. Synaptic activation of kainate receptors on hippocampal interneurons. Nat Neurosci. 1998;1(6):479–486. doi: 10.1038/2194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.