Significance
A key challenge in understanding cell communication is to characterize the coordinated activity of signaling pathways. A number of studies suggest that signaling pathways can regulate each other by direct control of ligand and receptor expression levels, triggering sequential signaling events in cells. To address the extent of sequential signaling, we profiled the transcriptional responses of ligand and receptor genes to single and combinatorial signaling pathway inductions in cell lines. Our analysis revealed that transcriptional regulation of genes encoding pathway components is a major level of signaling cross-talk, especially in the context of costimulation of signaling pathways.
Keywords: Drosophila, signaling pathways, signaling cross-talk, signaling networks, transcriptional regulation
Abstract
During development and homeostasis, cells integrate multiple signals originating either from neighboring cells or systemically. In turn, responding cells can produce signals that act in an autocrine, paracrine, or endocrine manner. Although the nature of the signals and pathways used in cell–cell communication are well characterized, we lack, in most cases, an integrative view of signaling describing the spatial and temporal interactions between pathways (e.g., whether the signals are processed sequentially or concomitantly when two pathways are required for a specific outcome). To address the extent of cross-talk between the major metazoan signaling pathways, we characterized immediate transcriptional responses to either single- or multiple pathway stimulations in homogeneous Drosophila cell lines. Our study, focusing on seven core pathways, epidermal growth factor receptor (EGFR), bone morphogenetic protein (BMP), Jun kinase (JNK), JAK/STAT, Notch, Insulin, and Wnt, revealed that many ligands and receptors are primary targets of signaling pathways, highlighting that transcriptional regulation of genes encoding pathway components is a major level of signaling cross-talk. In addition, we found that ligands and receptors can integrate multiple pathway activities and adjust their transcriptional responses accordingly.
Cellular communication is fundamental to all organisms and mediates numerous processes, such as cell fate decisions, proliferation, migration, and homeostasis. Remarkably, based on the types of ligands and signal transducers, only a limited number of pathways have been identified (1). These pathways include Notch, receptor tyrosine kinases, Wnt, Hedgehog, TGF-β, Hippo, Jun kinase (JNK), NF-κB, and JAK/STAT pathways. This limited set of pathways underscores that intricate signaling cross-talk must underlie most cellular interactions.
Signaling pathways are highly interconnected, and their cellular outcomes are context-dependent (2–6). Pathways can antagonize, enhance, or complement each other, and synergistic or inhibitory effects can dramatically change the nature of responses to a stimulus (7–9). Therefore, cells generate responses that are extremely diverse, explaining the emergence of a vast array of cell fate decisions and biological processes controlled by relatively few signaling pathways. In addition, pathway interactions are influenced by the signaling state of cells, and their specific outcomes can depend on previously acting signals (4, 10).
Although in vivo and in vitro examples of cross-talk between pathways have been well documented, we lack an integrative view of signaling incorporating the spatial and temporal interactions between pathways. Indeed, in most biological systems, we may know which pathways are required for a specific process, but we may not understand whether signals are processed sequentially or concomitantly and how pathways interact to produce the final outcome. For example, studies of the adult Drosophila midgut have revealed that most of the main signaling pathways are involved in homeostasis and/or regeneration (11). However, how they interact to promote proliferation or differentiation is poorly understood, because, in most cases, experiments lack the spatial and temporal information necessary to decipher whether signals are received concomitantly or whether, for example, one pathway regulates the activity of other pathways. Furthermore, in many cases, signaling pathway readouts are based on fluorescent reporters with low temporal resolution, making it difficult to evaluate whether a response is primary or secondary (12). Finally, most previous studies have been performed by activating signaling pathways to levels unlikely to be achieved physiologically, making it difficult to determine the biological significance of the regulated genes.
To obtain a systems level understanding of cross-talk mechanisms between the major metazoan transduction pathways, we examined the regulation of central components of signaling pathways, mostly ligands and receptors, in response to pathway activation. Specifically, we examined changes in gene expression in homogeneous cultured cells at a time scale allowing us to detect primary transcriptional targets (12–16). We selected Drosophila cell lines for these studies, because the signaling systems are simpler than in mammals. Importantly, in fly cell lines, most signaling pathways are not active under normal culture conditions (17). However, because these pathways can be activated by the simple addition of ligands or specific agonists (Materials and Methods), the transcriptional responses to various pathway activations can be readily studied with high temporal resolution.
Our analysis, focusing on the main limiting components for signal activation, demonstrates that activation of specific signaling pathways in cells often directly regulates the expression of ligand and receptor pathway components, highlighting sequential activation of pathways as a major mechanism of signaling cross-talk.
Results
Pathway Stimulation Methods and Profiling Transcriptional Outputs.
To characterize the regulation of key components of signaling pathways in response to various signal stimulations [JNK, JAK/STAT, epidermal growth factor receptor (EGFR), bone morphogenetic protein (BMP), Notch, Insulin, and Wnt], we selected Drosophila cell lines expressing the relevant receptors (17) (Fig. 1A). Then, we analyzed changes in transcript levels of canonical target genes for each pathway identified from the literature (18–24) using quantitative PCR (qPCR), 30 min and 1 h after stimulation. In all cases, the reporter genes showed the expected transcriptional responses, validating the pathway stimulation methods (Fig. 1B).
Fig. 1.
Genes regulated by single-pathway inductions. (A) Reagents used to stimulate each signaling pathway. (B) Expression changes of primary target genes at 30 min (light gray) and 1 h (dark gray). Bars represent the log2 fold changes of induced versus control samples from two biological replicates. Error bars indicate the minimum and maximum values. (C) Heat map showing expression changes of all genes included in the Nanostring code set at five time points following pathway stimulations (15 min, 30 min, 1 h, 2 h, and 6 h). Transcript levels are displayed as log2 fold change of normalized counts compared with nontreated cells. (D) Volcano plot representing statistical significance as a function of average fold change in gene expression for the pathway stimulations indicated. Averages of 30-min and 1-h time points are displayed in the graph to simplify visualization. Enlarged versions of single assay plots are shown in Fig. S3.
Next, to determine the optimal time points at which to analyze the transcriptional response to pathway activation, we performed a time course analysis (0 min, 15 min, 30 min, 1 h, 2 h, and 6 h) following stimulation of each pathway. We used the Nanostring nCounter platform to analyze changes in gene expression, because it is a robust and sensitive method effective for measurement of low-abundance mRNAs, such as ligands (Dataset S1), for a relatively large cohort of genes (25, 26). The method detects transcripts with a high level of linearity, reproducibility, and accuracy, avoiding biases introduced by amplification steps, such as for qPCR or RNA-sequencing (RNA-seq) (Materials and Methods and Figs. S1 and S2). Although signaling cross-talk could potentially be mediated by transcriptional regulation of any pathway component, we focused our analysis on ligand and receptor expression, because such expression is often the rate-limiting component of signaling pathways (Dataset S1 and Nanostring probe sets in Dataset S2). Gene lists were compiled based on previously reported pathway ligands, receptors, and reporters (1, 4).
Analysis of the time courses revealed that the majority of changes in ligand and receptor transcript levels were detectable at 30 min or 1 h (Fig. 1C). In addition, these time points are likely optimal for detecting the expression of primary target genes (12, 13, 16, 27).
Direct Transcriptional Regulation of Pathway Components by Signaling Pathways.
To identify direct targets of the seven signaling pathways, we compared data from stimulated samples at 30-min and 1-h time points with data from unstimulated control samples. We selected differentially expressed genes based on fold changes and significance. The selected hits were divided into three categories based on confidence (high confidence, mean fold ≥ 1.5 and P ≤ 0.05; medium confidence, mean fold ≥ 2 in two-thirds of replicates; and low confidence, mean fold ≥ 1.5 in two-thirds replicates; hits were called at 30 min and 1 h or using the combination of both time points (Fig. 1D and Dataset S3).
For each pathway, we identified reporter, receptor, and ligand genes that scored as hits (Dataset S4). With the exception of Thor (Insulin pathway), all seven canonical reporters were identified as targets of the expected pathways. The transcriptional response following insulin stimulation was relatively weak, most likely due to the presence of insulin in culture media that causes background pathway activation in control samples. Nevertheless, although Thor did not score as a hit due to our stringent criteria, transcript levels were down-regulated as expected at both time points (0.76- and 0.70-fold, respectively). Fourteen other pathway reporters were also included in the dataset, yet relatively few of these reporters were responsive in the cell lines tested (Dataset S3), likely due to contextual differences between the cell lines tested and the tissues in which each of these reporters were originally identified.
Each pathway regulated at least one ligand gene, demonstrating that ligand transcriptional regulation is likely a major mechanism of pathway cross-talk (Fig. 1D and Dataset S4). In addition, there were considerable differences between pathways in the number of ligand and receptor genes regulated (Fig. 1D and Dataset S4), with JNK (14 genes) being most prominent, followed by JAK/STAT (6 genes), Notch (5 genes), EGFR (5 genes), BMP (2 genes), Insulin (2 genes) and, finally, Wnt with only one ligand regulated. Some of these differences in response may be due to lower sensitivity in detecting expression changes in genes that are already expressed under control conditions. However, we found no correlation between fold induction of target genes and their basal expression (Fig. S2), indicating that expression level is unlikely to explain the differences in response between pathways completely. Alternatively, differences in the number of target genes identified could reflect the methods used to activate the pathways, which may result in different levels of activation, although it is also possible that the observed differences are inherent to the pathways analyzed.
Among the responsive pathway components for each assay were often ligands of the stimulated pathway, suggesting that feedback signal amplification may be a common output of signaling. For instance, the strongest responding gene to EGFR pathway stimulation was the ligand spitz (spi). Likewise, activation of the JAK/STAT, BMP, and Wnt pathways increases the expression of their own ligands [unpaired 1 (upd), decapentaplegic (dpp), and wingless (wg), respectively].
As in the case of ligands, several signaling receptors were primary targets of pathways (Dataset S4). For example, fz, Tollo, and Pvr are all targets of JNK signaling (Fig. S3). However, receptor inductions were generally weaker than ligand genes, and we did not find any receptor genes involved in feedback regulation following single-pathway stimulations.
The hits also included examples of incoherent feedback motifs, where both positive and negative regulators of the stimulated pathway were simultaneously activated (Dataset S4). For example, BMP signaling activated both the BMP ligand dpp and a negative regulator of BMP signaling, Dad. Similarly, EGFR pathway stimulation results in transcription of ligands and inhibitors of EGFR, spi and aos, respectively. Further, activation of JAK/STAT signaling stimulated the upd and upd3 ligands, as well as the negative regulator Socs36E. Incoherent feedforward loops were also identified, where positive and negative regulators of other pathways were simultaneously regulated. For example, stimulation of the Insulin, EGFR, or JNK pathway caused activation of both JAK/STAT ligands and the JAK/STAT repressor Socs36E (Fig. 1D and Dataset S4). Indeed, at least one incoherent feedforward or feedback loop was present among the targets of each pathway, with the exception of Notch and Wnt. We note, however, that incoherent feedforward loops have previously been identified downstream of Notch in another cell type (13, 14).
Responses to Combinatorial Stimulations.
Because cells in tissues are often exposed to multiple ligands, we investigated the effects of combinatorial pathway stimulations on the expression of ligand and receptor genes. We used the same experimental setup as for single-pathway assays and performed combinatorial stimulations of five pairs of pathways: Insulin + JNK, JNK + JAK/STAT, Insulin + JAK/STAT, BMP + JNK, and BMP + JAK/STAT.
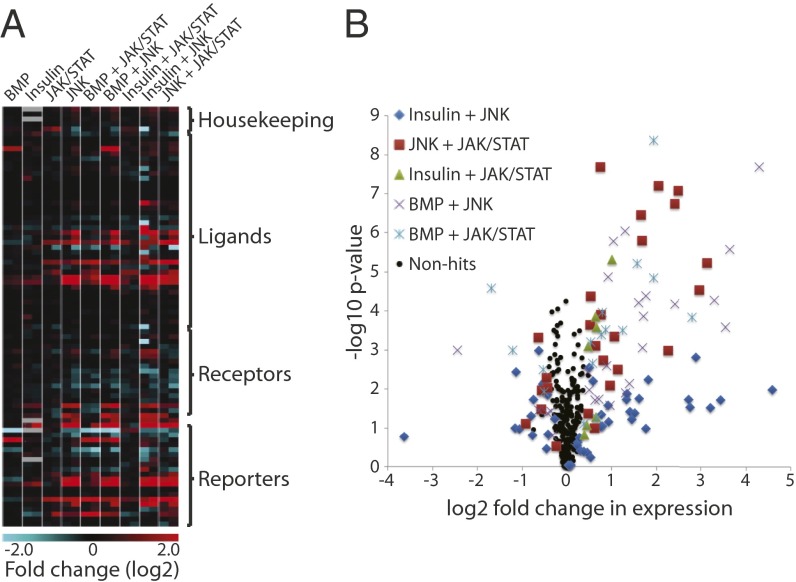
For all combinatorial stimulations, at least one known reporter gene for each activated pathway was transcriptionally affected (with the exception of Thor following JAK/STAT + Insulin stimulation, which was reduced but did not meet the criteria to be classed as a hit), confirming the simultaneous activation of both pathways (Fig. 2 and Datasets S3 and S4). The majority of hits identified in single-pathway assays were also detected in combinatorial assays (Fig. 1D and Table 1). Interestingly, combinatorial stimulation of JAK/STAT and Insulin only regulated half of the hits identified from individual assays, indicating a possible inhibitory interaction between these pathways on the transcriptional induction of their respective target genes (Table 1 and Dataset S4).
Fig. 2.
Genes regulated by combinatorial pathway inductions. (A) Expression heat map for all genes at two time points (30 min and 1 h) following either single- or combinatorial pathway stimulations. Transcript levels are displayed as the average log2 fold change of normalized counts in induced versus control samples from three biological replicates. (B) Volcano plot of combinatorial stimulation assays as described in Fig. 1D. Enlarged versions are shown in Fig. S3.
Table 1.
Comparison of hits between single and combinatorial stimulation assays
| Pathway combination | Additional hits in combinatorial assays | Missing hits in combinatorial assays |
| JNK + BMP | ds | ptc aos E(spl)mβ fz |
| JAK/STAT + BMP | E(spl)m3 PvR Rel upd2 AttA | aos dpp |
| JNK + Insulin | Alk Dad dally Egfr fz2 gbb grk dilp1 dilp6 InR numb Ptth Pvf1 sgg spi spz tkv Tl trk Wnt5 N | ptc PvR Act5C |
| JAK/STAT + Insulin | brk | Ptp61F Thor Tollo Ser Rel E(spl)mβ |
| JAK/STAT + JNK | bnl brk numb th | aos ptc vn dpp fz E(spl)mβ |
Overall, combinatorial stimulations resulted in regulation of more genes than expected based on single stimulations, suggesting that several pathways have cooperative interactions (Table 1 and Dataset S4).
Validation of Target Genes by qPCR and RNA-Seq.
To determine whether the target genes identified by Nanostring are robust, we used two orthogonal technologies to validate target genes from JNK and JAK/STAT single stimulations. For the JNK pathway, we selected six targets with varying levels of induction (dpp, upd2, upd3, pvf2, pyr, and ths) and measured changes in expression following 30-min and 1-h stimulations using qPCR. The results correlated well with results from the Nanostring assays (correlation coefficient = 0.90), and all target genes were regulated by at least 1.5-fold in qPCR assays at one or both time points (Fig. 3A).
Fig. 3.
Validation of target genes by qPCR and RNA-seq. (A) Graph comparing fold changes determined by qPCR and Nanostring for JNK stimulations at 30 min and 1 h. Black dots represent average fold change determined from two replicates of qPCR and three replicates of Nanostring assays. Correlation coefficient = 0.90. (B) Graph comparing expression levels under control or Upd-stimulated conditions measured by Nanostring or RNA-seq. Black dots represent log2 average normalized counts from three biological replicates for Nanostring and two biological replicates for RNA-seq. Correlation coefficient = 0.86. (C) Table showing fold change of JAK/STAT target genes measured using Nanostring or RNA-seq under the conditions indicated. P values are shown in parentheses. Max expression indicates the maximum number of reads under any condition measured by RNA-seq. NA, not applicable. (D) Relationships (lines) between signaling pathways, ligands (pink circles), and receptors (green polygons) identified in the single-pathway stimulation assays. Pathway activation can induce (arrows) or repress (bars) the expression of pathway components (yellow rectangles).
Next, we used RNA-seq to identify genes regulated following JAK/STAT pathway stimulation. Wild-type or STAT92E mutant cells (28) were treated with control or Upd-conditioned media for 1 h before assessing changes in gene expression by RNA-seq. First, we combined results for the experimental and control treatments in wild-type cells and compared gene expression levels between RNA-seq and Nanostring assays performed under similar conditions. Results were highly reproducible, with a correlation coefficient of 0.86 (Fig. 3B), indicating that results from both approaches are robust. Next, we considered whether JAK/STAT target genes identified by Nanostring were also differentially expressed in the RNA-seq data. Of the nine target genes, four (ths, pyr, Ser, and upd) could not be reliably detected under at least one condition due to low expression levels. Of the remaining five genes, three (Socs36E, Ptp61F, and Thor) showed significant differential expression (P < 0.1) in the expected direction (Fig. 3C). Finally, we compared wild-type and STAT92E mutant cells under control conditions and found that upd3 was significantly differentially expressed (Fig. 3C), suggesting that it is either an indirect target of the pathway or requires stronger stimulation to be regulated, consistent with its weak signal at early time points (Fig. S3).
Overall, the qPCR and RNA-seq results indicate that the Nanostring data are robust and the majority of target genes can be detected with multiple approaches.
Discussion
Signaling pathways are highly interconnected, and understanding how they cooperate to achieve specific biological outcomes is complicated by the diversity of mechanisms by which they can regulate each other (1, 4, 5, 29, 30). Secreted ligands and their receptors are key to the activity of signaling pathways, and the extent to which their transcriptional regulation is used to connect signaling pathway activities is not fully appreciated. Here, we analyzed the immediate transcriptional response to pathway activation in homogeneous cell lines to address three basic questions in cell signaling with regard to ligand and receptor gene expression. First, what is the extent of transcriptional feedback regulation within a signaling pathway occurring at the level of ligands and receptors? Second, to what extent does one specific signaling pathway regulate the expression of ligands and receptors of other pathways? Third, how is the regulation of ligand and receptor expression modulated by costimulation of multiple signaling pathways? Our study, focusing on evolutionarily conserved metazoan signaling pathways, revealed intricate relationships between pathway activities and expression of ligand and receptor genes, suggesting that coordination of signaling pathway activities is more pervasive than previously thought. In addition, we identified many interactions that have previously been shown in in vivo systems or mammalian cells, suggesting that this level of signaling cross-talk is not specific to cultured Drosophila cells.
Transcriptional Feedback Regulation Within a Signaling Pathway.
Examination of the transcriptional regulation of ligands and receptors by their own pathways revealed that half of the pathways tested have positive feedback loops at the level of ligands. Specifically, induction of the BMP, EGFR, JAK/STAT, and Wnt pathways up-regulated the expression of their respective ligand genes dpp, spi, upd-upd3, and wg (Fig. 1D and Dataset S4). These observations from cell culture correlate well with previous in vivo studies, where positive feedback loops through ligand expression have been observed in a number of tissues for the JAK/STAT, Wnt, and EGFR pathways (23, 31, 32). For instance, the regulation of spi by Ras signaling (a downstream component of the EGFR pathway) has been shown in vivo during embryonic stages (32). Interestingly, Spi is produced as an inactive precursor and requires processing by Rhomboid (Rho) to generate the active ligand (33). Expression of rho is generally the limiting step in Spi-mediated signaling. However, the transcriptional regulation of spi by EGFR signaling suggests that there may be situations in which Spi expression is limiting. Alternatively, Spi expression levels may be important to modify the range or strength of signaling rather than inducing the signaling event itself. Signaling feedback via regulation of ligands has also been shown to play a central role in mammalian cells (8).
Remarkably, transcriptional feedback loops were present in all combinatorial pathway inductions performed, and these feedback loops were mediated by multiple ligands in some cases (Dataset S4). For instance JAK/STAT + BMP stimulation up-regulated the three upd genes, although only two of them were induced by JAK/STAT stimulation alone.
Furthermore, incoherent feedback loops were also identified in our study as exemplified by the EGFR assay, where both spi and its antagonist aos were induced. Another case is in the context of JAK/STAT pathway activation, where both upd ligand and the Socs36E repressor were activated (Fig. 1D and Dataset S4).
Together, the results from our study indicate that autoregulation of signaling pathways via ligand transcription is a common mechanism found downstream of multiple pathways (Figs. 1D and 3D). In addition, feedback loops appear to be more common following combinatorial pathway stimulations (Fig. 2), suggesting that these motifs may play an important role in the integration of signals by altering the strength, duration, and extent of signaling (34, 35).
Transcriptional Regulation Between Pathways.
With the exception of the Wnt pathway, all individual and combinatorial pathway stimulations that we tested resulted in the regulation of ligands and/or receptors of other pathways, indicating that the transcriptional regulation of these components is a common mechanism by which pathways cross-regulate each other (Dataset S4).
The extent to which different pathways regulate ligands and receptors was variable. For example, JNK activation regulated 10 ligands and four receptors, whereas Wnt activation regulated only one ligand and no receptors. The response to JNK activation is consistent with its role in the Drosophila midgut, where JNK is thought to be stimulated immediately following tissue damage and multiple other pathways are regulated downstream of this initial signaling event (11, 36, 37). This variability in the response to stimulation may indicate inherent differences in the ability of each pathway to regulate other pathways sequentially. For example, the JNK pathway may act as a mediator signal that coordinates downstream signaling events, whereas pathways with lower connectivity may act as effectors rather than mediators. However, more extensive studies in a greater range of cellular contexts will be needed to investigate this possibility.
Finally, it has previously been suggested that pathways with more complex intracellular transduction cascades may have an increased potential for cross-talk (4). Whereas this possibility may be true for direct cross-talk between transduction cascades, we saw no such correlation at the transcriptional level.
Integration of Multiple Pathway Activities.
In in vivo situations, cells are likely to be simultaneously exposed to ligands from multiple different pathways. For example, almost every core pathway described has been implicated in the response to tissue damage in the Drosophila midgut. However, it is currently unclear how these signals are integrated to produce the correct response.
The target genes regulated by combinatorial stimulations are not simply the sum of those target genes regulated by each pathway alone. Indeed, for all pathway combinations tested, there were examples of target genes that were absent from either pathway stimulation alone or genes regulated by one or both pathways alone that were not identified as hits following costimulation (Table 1). For example, following JNK and Insulin costimulation, 21 of the 45 targets of costimulation were not regulated by either JNK or Insulin alone. This result indicates that integration of signals from multiple pathways is not simply additive and more complex processing of these signals must be occurring.
Relevance of the Cell Line Interactions to the Study of Cell Signaling.
The pathway interactions that we have identified from signaling pathway stimulation assays were summarized as a network of pathways to ligand and receptor links (Fig. 3D). Importantly, many of these connections have been previously identified in in vivo experiments. For example, studies in the Drosophila adult gut have documented that most signaling pathways are required for either regeneration or/and homeostasis (11, 36, 37). Activation of the JNK pathway in the injured or stressed midgut epithelium leads to expression of the ligands vein, decapentaplegic (dpp), Pvf2, upd2, and upd3 from the enterocytes, enteroblasts, and visceral muscle, and subsequent activation of the EGFR, BMP, PVR, and JAK/STAT pathways. All of these ligands were identified by our JNK activation assay, indicating that our data are applicable to in vivo contexts.
In addition to the gut system, our results are reminiscent of the pathway interactions observed in tumor systems. For example, activation of the EGFR pathway leads to overproliferation of tissues in Drosophila and mammals, but the combinatorial activation of the JAK/STAT, EGFR, and JNK pathways has been shown to result in more aggressive tumors than EGFR alone (38). This conservation of signal integration suggests that studies of cross-talk in Drosophila may be applicable to understanding human diseases such as cancers. In particular, knowledge of interactions between signaling pathways will likely be important for understanding mechanisms of drug resistance in cancers.
In addition to the known connections, we have identified many previously unidentified links between signaling pathways. This dataset can therefore be used to guide future studies into signaling pathway connectivity in Drosophila and, due to the highly conserved nature of signaling pathways, other organisms as well. However, cross-talk between signaling pathways is likely to be context-dependent; therefore, the connections we have identified represent a network of possible interactions that may not be used in all situations.
Finally, although we have identified many transcriptional links between pathways, it is not clear from this study which of these events will lead to a change in signaling activity. Many of the transcriptional changes observed were weak (Dataset S3), and more sustained signaling or integration of other signals may be required to produce a biologically meaningful effect on the target pathway.
Materials and Methods
Pathway Stimulation Assays.
Cell lines and cell culture.
S2R+ (39), S2 (40), Kc167 (41), and STAT92E mutant (28) Drosophila cell lines were cultured using standard procedures. STAT92E mutant cells contain a frameshift deletion within the STAT92E gene and were previously shown to result in complete loss of detectible transcription factor activity (28).
Cell induction.
To activate signaling pathways, we used the following: Insulin at 0.5 mg/mL (Sigma I6634) for the Insulin pathway, LPS from Escherichia coli at 25 mg/mL (O55:B5; Sigma) for the JNK pathway, recombinant Dpp protein at 33 μL/mL (159-dp-020/CF; R&D Systems) for the BMP pathway, and 2 mM EDTA in PBS solution for the Notch pathway (42). For the Wnt, JAK/STAT, and EGFR pathways, conditioned media expressing Wg, Upd (Upd cDNA cloned into the pMK33 vector), or Spi protein were produced, respectively, from S2R+ cells transfected with the relevant plasmid, as previously reported (40, 43). Protein expression was induced with 0.5 mM CuSO4, and media were harvested after overnight incubation. For the control media, 0.5 mM CuSO4 was added overnight to untransfected cells. Supernatants from induced and control cells were collected, sterilized, and concentrated to optimize their efficiency for stimulation assays. For EGFR pathway assays, experiments were performed in S2 cells stably transfected with an inducible transgene encoding EGFR (43).
Analysis of Target Gene Expression Levels.
Nanostring nCounter gene expression assays.
To profile gene expression levels following signaling pathway induction, we used the Nanostring nCounter method. We designed a specific code set targeting 86 genes, including 40 ligands belonging to the main signaling pathways, 18 corresponding signaling pathway receptors, 21 previously characterized known transcriptional targets of the pathways, 3 additional components of signaling pathways (shotgun/E cadherin, rho, and Diap1), and 4 housekeeping genes. Genes encoding pathway components and reporters were identified using recent literature reviews (1). Nanostring probe sequences are listed in Dataset S2. Total RNA extracts were processed according to Nanostring instructions (nCounter Expression Data Analysis Guide, 2001; Nanostring Technologies, Inc.; www.nanostring.com/media/pdf/MAN_nCounter_Gene_Expression_Data_Analysis_Guidelines.pdf).
Data normalization and fold change calculation.
Technical normalization of data was performed as described by Nanostring (nCounter Expression Data Analysis Guide, 2001), whereby raw counts were normalized by the geometric mean of six different positive control probes. Next, for biological normalization, 10 genes showing the lowest coefficient of variation across all experiments were selected as references (Dataset S5). This reference set was selected separately for each cell line to account for differences in background expression levels. To account for variation in mRNA load per cartridge, counts were normalized by the geometric mean of the reference genes across each cell line.
To evaluate background noise, we calculated the average counts plus 2 SDs from eight negative control probes (which have no complementary sequences in the Drosophila genome) in each assay. In addition, we determined the lowest concentration of positive control probes within the linear detection range, representing the threshold of accurate transcript measurement. The higher value of these two measurements was selected as the sensitivity cutoff, and counts below the cutoff were adjusted to the cutoff for fold induction calculations. The ratios of treated versus control samples were calculated for each biological replicate.
Determination of hits.
Responsive genes for single and combinatorial treatments were determined using different criteria for high-, medium-, and low-confidence hits. For the high-confidence hits, genes with average fold change ≥ 1.5 and P ≤ 0.05 determined using t tests were selected. For the medium- and low-confidence hits, genes having a minimum of twofold and 1.5-fold similar trend expression changes in at least two of three biological replicates were selected, respectively.
RNA-Seq.
RNA-seq library preparation was performed on total RNA by poly-A selection using the Wafergen PrepX RNA-Seq Library Kit for Illumina and the Apollo 343 NGS Library Prep System (Wafergen). Single end-sequencing reads (75 nt) were collected using the Illumina NextSeq sequencer. RNA-seq reads were mapped to the dm6.05 genome sequence using the TopHat 2.1.0 mapper (https://ccb.jhu.edu/software/tophat/index.shtml) and Bowtie 1.1.2 aligner (bowtie-bio.sourceforge.net/index.shtml). Expression was measured by raw count using HTSeq (www.huber.embl.de/users/anders/HTSeq/doc/overview.html). Initial normalization was performed using the DESeq2 Bioconductor package in R (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) with the count() function. Batch correction was calculated using the limma Bioconductor package in R (https://bioconductor.org/packages/release/bioc/html/limma.html) with the removeBatchEffect() function. A Student t test was then used to determine the significance of fold change between experimental groups for the genes of interest with the multtest Bioconductor package in R (bioconductor.org/packages/release/bioc/html/multtest.html).
Supplementary Material
Acknowledgments
We thank Valentine Ammeux for the design of Fig. 3D, Vinayagam Arunachalam for help with YEd Graph Editor, Young Kwon for providing the upd-pMK33 expression plasmid, and Richelle Sopko for helpful comments. We thank the Nanostring company, particularly Erroll Rueckert, for thoughtful support. N.A. was supported by the Boehringer Ingelheim Fonds PhD fellowships. Work in the laboratory of N.P. is supported by the Howard Hughes Medical Institute and the NIH.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610432113/-/DCSupplemental.
References
- 1.Perrimon N, Pitsouli C, Shilo B-Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol. 2012;4(8):a005975. doi: 10.1101/cshperspect.a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128(2):225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Doroquez DB, Rebay I. Signal integration during development: Mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41(6):339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- 4.Housden BE, Perrimon N. Spatial and temporal organization of signaling pathways. Trends Biochem Sci. 2014;39(10):457–464. doi: 10.1016/j.tibs.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vivekanand P, Rebay I. Intersection of signal transduction pathways and development. Annu Rev Genet. 2006;40:139–157. doi: 10.1146/annurev.genet.40.110405.090555. [DOI] [PubMed] [Google Scholar]
- 6.Bray SJ. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 7.Hsueh RC, et al. Deciphering signaling outcomes from a system of complex networks. Sci Signal. 2009;2(71):ra22. doi: 10.1126/scisignal.2000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janes KA, et al. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124(6):1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 9.Natarajan M, Lin K-M, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8(6):571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 10.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11(6):414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang H, Edgar BA. Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res. 2011;317(19):2780–2788. doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doupé DP, Perrimon N. Visualizing and manipulating temporal signaling dynamics with fluorescence-based tools. Sci Signal. 2014;7(319):re1. doi: 10.1126/scisignal.2005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Housden BE, et al. Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes. PLoS Genet. 2013;9(1):e1003162. doi: 10.1371/journal.pgen.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krejcí A, Bernard F, Housden BE, Collins S, Bray SJ. Direct response to Notch activation: Signaling crosstalk and incoherent logic. Sci Signal. 2009;2(55):ra1. doi: 10.1126/scisignal.2000140. [DOI] [PubMed] [Google Scholar]
- 15.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3(5):711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 16.Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 2010;11(3):201–207. doi: 10.1038/embor.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherbas L, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21(2):301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122(1):223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- 19.Karsten P, Häder S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117(1-2):343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 20.Martín-Blanco E, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12(4):557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17(16):2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuneizumi K, et al. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389(6651):627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Noll M. Role of the gooseberry gene in Drosophila embryos: Maintenance of wingless expression by a wingless--gooseberry autoregulatory loop. EMBO J. 1993;12(12):4499–4509. doi: 10.1002/j.1460-2075.1993.tb06139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper MT, et al. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev Biol. 2000;221(2):390–403. doi: 10.1006/dbio.2000.9691. [DOI] [PubMed] [Google Scholar]
- 25.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 26.Kulkarni MM. 2011. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol Chapter 25:Unit25B.10.
- 27.Lee REC, Walker SR, Savery K, Frank DA, Gaudet S. Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol Cell. 2014;53(6):867–879. doi: 10.1016/j.molcel.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Housden BE, et al. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal. 2015;8(393):rs9. doi: 10.1126/scisignal.aab3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hancock JT. Cell signalling is the music of life. Br J Biomed Sci. 2008;65(4):205–208. doi: 10.1080/09674845.2008.11732830. [DOI] [PubMed] [Google Scholar]
- 30.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16(10):1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 31.Osman D, et al. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J Cell Sci. 2012;125(Pt 24):5944–5949. doi: 10.1242/jcs.113100. [DOI] [PubMed] [Google Scholar]
- 32.Hurlbut GD, Kankel MW, Artavanis-Tsakonas S. Nodal points and complexity of Notch-Ras signal integration. Proc Natl Acad Sci USA. 2009;106(7):2218–2223. doi: 10.1073/pnas.0812024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bang AG, Kintner C. Rhomboid and Star facilitate presentation and processing of the Drosophila TGF-alpha homolog Spitz. Genes Dev. 2000;14(2):177–186. [PMC free article] [PubMed] [Google Scholar]
- 34.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152(5):945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 36.Lucchetta EM, Ohlstein B. The Drosophila midgut: A model for stem cell driven tissue regeneration. Wiley Interdiscip Rev Dev Biol. 2012;1(5):781–788. doi: 10.1002/wdev.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kux K, Pitsouli C. Tissue communication in regenerative inflammatory signaling: lessons from the fly gut. Front Cell Infect Microbiol. 2014;4:49. doi: 10.3389/fcimb.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hombría JC-G, Serras F. Why should we care about fly tumors?: The case of JAK-STAT and EGFR cooperation in oncogenesis. JAK-STAT. 2013;2(2):e23203. doi: 10.4161/jkst.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273(48):32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- 40.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27(2):353–365. [PubMed] [Google Scholar]
- 41.Echalier G, Ohanessian A. In vitro culture of Drosophila melanogaster embryonic cells. In Vitro. 1970;6(3):162–172. doi: 10.1007/BF02617759. [DOI] [PubMed] [Google Scholar]
- 42.Krejcí A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21(11):1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 1995;9(12):1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.