Significance
While we age, our body accumulates random somatic mutations. These mutations spontaneously arise from endogenous and exogenous sources, such as DNA replication errors or environmental insults like smoking or sunlight. Direct measurement of rare mutations could help us understand the role of somatic mutations in human aging, normal biology, and disease processes. Here, we develop the bottleneck sequencing system (BotSeqS) as a simple genome-wide sequencing-based method that accurately quantitates nuclear and mitochondrial mutational load in normal human tissues. We demonstrate that mutation prevalence and spectrum vary depending on age, tissue type, DNA repair capacity, and carcinogen exposure. Our results suggest a varied landscape of rare mutations within the human body that has yet to be explored.
Keywords: next-generation sequencing, somatic mutation, aging, genomics
Abstract
We present the bottleneck sequencing system (BotSeqS), a next-generation sequencing method that simultaneously quantifies rare somatic point mutations across the mitochondrial and nuclear genomes. BotSeqS combines molecular barcoding with a simple dilution step immediately before library amplification. We use BotSeqS to show age- and tissue-dependent accumulations of rare mutations and demonstrate that somatic mutational burden in normal human tissues can vary by several orders of magnitude, depending on biologic and environmental factors. We further show major differences between the mutational patterns of the mitochondrial and nuclear genomes in normal tissues. Lastly, the mutation spectra of normal tissues were different from each other, but similar to those of the cancers that arose in them. This technology can provide insights into the number and nature of genetic alterations in normal tissues and can be used to address a variety of fundamental questions about the genomes of diseased tissues.
The accumulation of random somatic mutations in the nuclear and mitochondrial genome (mtDNA) over time underlies fundamental theories of carcinogenesis, neurodegeneration, and aging (1–3). Direct observation of these rare mutations in the human body with age therefore has the potential to enhance our understanding of human disease. Currently, no simple high-throughput method exists to directly and systematically quantify somatic mutational load in normal, nondiseased human tissues at a genome-wide level. Next-generation DNA sequencing (NGS) technologies are an ideal platform to address this issue, but their sequencing error rate limits the detection of rare mutations. For example, the Illumina platform has the lowest reported error rate, but, even with sophisticated postsequencing analysis, the sensitivity is at best 0.1% (4), far lower than required to detect rare mutations in normal human tissues (5, 6).
Two main NGS strategies have been developed for more sensitive detection of rare mutations: single cell genomic sequencing (7–9) and consensus sequencing with molecular barcodes (10–13). Single cell genomic sequencing has the potential to detect rare mutations in a genome-wide fashion, with sensitivity achieved through the isolation of single cells from the bulk population. However, point mutations are introduced during whole-genome amplification of the picograms of DNA isolated from single cells. To increase the specificity of point mutation calling with single cell methods, it is necessary to identify the same point mutation in at least two different cells (14). This approach, although useful for the evaluation of tumor heterogeneity and other purposes, cannot accurately call a point mutation that is private to a single cell. In contrast, consensus sequencing with molecular barcodes can accurately detect very rare point mutations (<10−6) by distinguishing individual DNA molecules in a population with a unique barcode. This unique molecule identifier (15) is used to group reads from the same DNA template; only mutations that are present in most or all of the reads from the same template are scored as mutations (10–13). Although sensitive and accurate, molecular barcoding methods are designed for targeted loci (16–18) or small, predefined genomic regions (19, 20) rather than unbiased detection across the human genome.
The bottleneck sequencing system (BotSeqS) technology described in this work was designed to accurately detect rare point mutations from a molecularly barcoded library in a completely unbiased fashion. In addition to describing the technology and demonstrating its high sensitivity, we report how we used it to gain insight into the accumulation of somatic mutations in normal human tissues.
Results
Principles Underlying BotSeqS.
The principal feature of BotSeqS is the dilution of a sequencing library before PCR amplification. Although the dilution could be performed before or after ligation of sequencing adapters, it is advantageous to dilute afterward; the whole procedure is simpler, and more reproducible when inputting nanograms than picograms of DNA. This dilution creates a bottleneck and permits an efficient random sampling of double-stranded template molecules with a minimal amount of sequencing. Rare mutations, which would normally be masked by an abundance of wild-type sequences in conventional libraries, account for much more of the signal at the corresponding genomic position in a bottlenecked library. Dilution also increases the likelihood that both the “Watson” and “Crick” strands of a DNA molecule will be sequenced redundantly, a feature critical for the high accuracy of BotSeqS and the relatively small amount of sequencing required to implement it. The presence of the same rare mutation on both strands can substantially decrease artifacts and increase specificity (12). Finally, the random nature of dilution allows DNA molecules from both nuclear and mitochondrial genomes to be assessed from one library.
Generation of BotSeqS Libraries.
A standard Illumina TruSeq PCR-Free kit was used to generate 44 BotSeqS libraries from the normal tissues of 34 individuals (Dataset S1, Table S1), which included 9 individuals with one or two technical replicates. In addition, 10 of our 12 cohorts had more than one biological replicate, each containing 2 to 6 individuals.
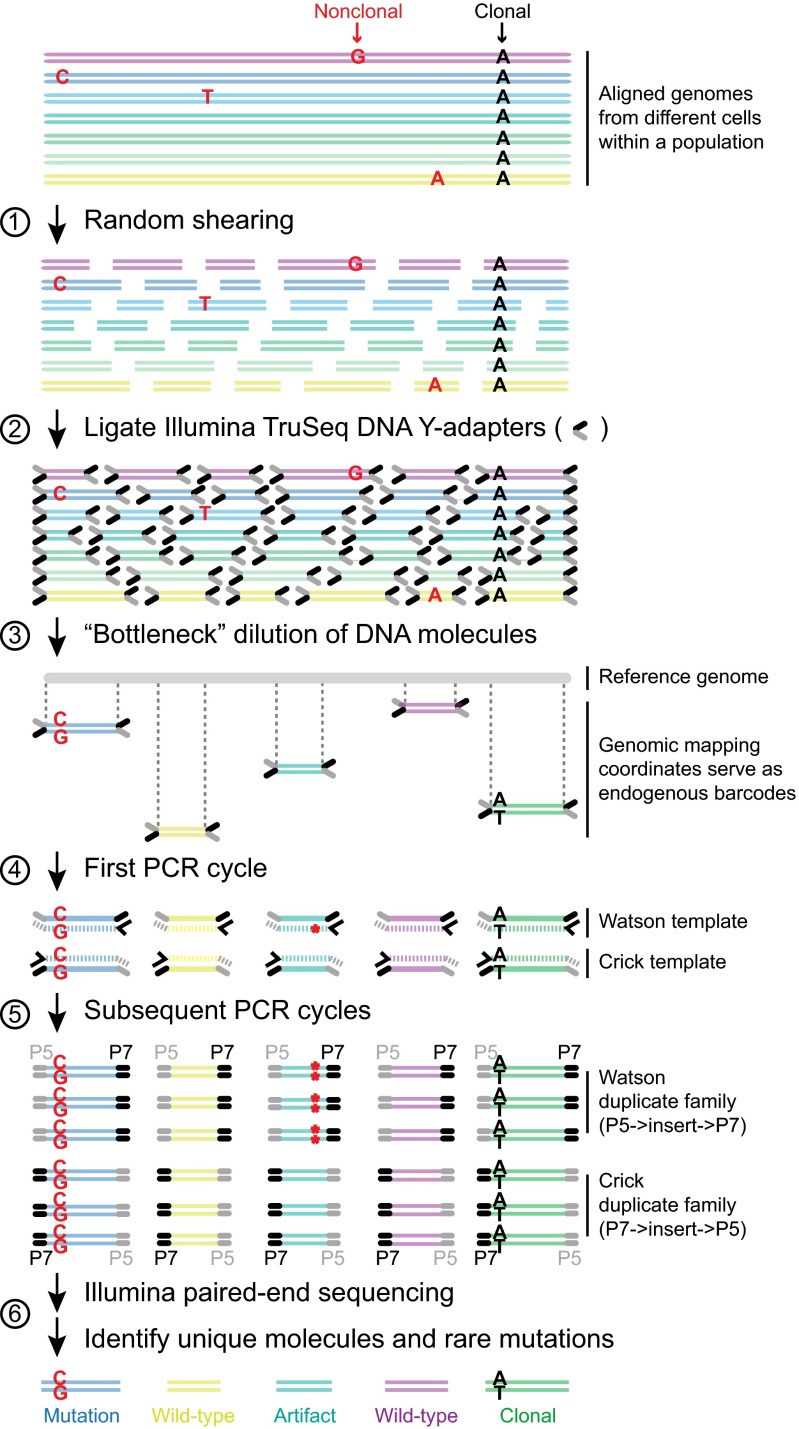
The preparation of BotSeqS libraries starts with the random shearing of genomic DNA (Fig. 1), which fragments the genomes into variably sized DNA molecules, each possessing unique end coordinates called endogenous barcodes (10). After ligation of standard sequencing adapters to the DNA molecules, the library was diluted to reduce the number of molecules in the population. To identify the correct dilution factor, a 10-fold dilution series was assessed on a MiSeq instrument (SI Appendix, Fig. S1). After dilution, PCR amplification of the library generated multiple copies (duplicates) of each DNA molecule. The endogenous barcodes enable the grouping of sequencing reads into families, also known as unique identifiers (UIDs) (10); each family represents the PCR-derived progeny of a single-stranded template and each member of a family represents the sequence from a single cluster on the Illumina instrument. In the following, we consider the Watson strand to be the sequence derived from the first read of the sequencing instrument (Illumina adapter P5) and the Crick strand to be the sequence derived from the second read (Illumina adapter P7) of each member of the family (Fig. 1). To be considered a potential mutation, BotSeqS required that the identical sequence change be observed in ≥90% of the Watson and in ≥90% of the Crick family members and that each family be composed of at least two members. BotSeqS libraries were analyzed using an Illumina HiSeq 2500 instrument on rapid run mode with paired-end reads of 100 bases each. A median of 70 million clusters per library passed the standard Illumina quality filters (range 37–190 million clusters per library) (Dataset S1, Table S1).
Fig. 1.
Bottleneck sequencing methodology. Each color at the top of the figure represents double-stranded DNA from a genome of one cell within a population. Random, nonclonal point mutations (red) are private to individual cells. In contrast, clonal reference changes (A in black) are present in all genomes within the cell population. (step 1) Random shearing generates variably sized DNA molecules. (step 2) Noncomplementary single-stranded regions of the Illumina Y-adapters (P5 in gray and P7 in black) are represented as forked structures ligated to both ends of each DNA molecule. (step 3) Dilution decreases the number of DNA molecules (five are shown) from the original population in a random manner. Ends of the DNA molecules align uniquely to the reference genome. Mapping coordinates are used as unique molecule “barcodes” during data processing. (step 4) PCR primer (black arrowhead) anneals and primer extends (hashed lines) the Watson and Crick template of the original DNA molecule independently. The red asterisk represents an error generated during PCR of the library. (step 5) Watson and Crick templates generate two families of PCR duplicates. Orientation of P5 (gray) and P7 (black) containing adapters to the DNA molecule (insert) distinguishes the two families. P5 and P7 sequences dictate which end will be sequenced in read 1 vs. read 2, respectively, on the Illumina flow cell. Red asterisks represent the PCR error propagated in the Watson but not the Crick family members. In contrast to artifacts, real mutations (C:G mutation in red) will be present in both the Watson and Crick family members. (step 6) The BotSeqS pipeline identifies and quantifies the number of unique DNA molecules and point mutations (C:G in red) in the sequencing data by eliminating artifacts and clonal changes (A:T in black).
BotSeqS Data Processing Pipeline.
The goal of the BotSeqS pipeline was to accurately identify rare, somatic point mutations and to calculate the prevalence of these mutations in the sample. To process the data for this purpose, raw sequencing data were inputted into Illumina’s secondary analysis package (CASAVA 1.8) with ELANDv2 mapping to the GRCh37/hg19 human reference genome. The BotSeqS pipeline begins by selecting high quality reads for analysis (SI Appendix, SI Materials and Methods and BotSeqS Pipeline Supplement). The data were then organized into two tables for each BotSeqS library: (i) a “change” table listed all differences from the reference sequence and (ii) a unique molecule table listed all families. Importantly, each table contained strand information.
Most BotSeqS libraries (37 of 44) had a median number of family members between 5 and 20 (SI Appendix, Fig. S2A), demonstrating that the libraries underwent successful bottlenecking. Almost half (median 45%, range 8–62%) of the unique molecules from each BotSeqS library had both the Watson and Crick duplicate families represented in the dataset, ensuring specificity in the subsequent mutation analysis (SI Appendix, Fig. S2A). Furthermore, a median of 60,640 (range 2,127–147,200) unique molecules from the nuclear genome was assessed per BotSeqS library, which resulted in ∼0.4% of the nuclear genome being covered, comparable with other genome-wide techniques such as exome sequencing.
To identify rare, somatic mutations, it was necessary to eliminate germ-line and clonal variants from the BotSeqS data (we defined clonal as those present in both strands of more than one template molecule). We performed whole genome sequencing (WGS) of the same DNA sample or the same libraries that had been diluted for BotSeqS for 32 of the 34 individuals in this study (Dataset S1, Table S1). For the remaining two individuals (COL238 and COL239), Sanger sequencing was performed to eliminate clonal variants, demonstrating that WGS was not necessary for BotSeqS. The vast majority (median 92%, range 88–94%) of variants were found to be germ-line, easily identifiable from the matched WGS dataset. In addition to clonality, we eliminated potential artifacts by considering only well-mapped positions and by using other filters (Dataset S1, Tables S2–S6 and SI Appendix, SI Materials and Methods). The requirement for mutations to be present on both strands was indeed necessary because, in the absence of this filter, there was ∼10-fold higher nuclear mutation prevalence associated with a large number of G→T transversions known to represent artifacts in NGS library preparations (SI Appendix, Fig. S2B) (21). Analysis of technical and biological replicates showed a similar average range (∼twofold) in mutation prevalence from both the mtDNA and nuclear genome (SI Appendix, Fig. S2C). We further performed a “spike-in” validation experiment by mixing one individual’s normal DNA (PEN93) into another individual’s normal DNA (PEN95) at two different ratios. Using BotSeqS, we were able to detect PEN93-specific SNPs in both samples, with a 7.4-fold difference in prevalence between the low and high spike-ins, within the expected error of the intended 10-fold difference (SI Appendix, SI Materials and Methods).
From the 44 BotSeqS libraries, we identified a total of 666 and 876 rare somatic point mutations in mtDNA and nuclear DNA, respectively (Dataset S1, Tables S7 and S8). All rare mutations passed visual inspection, and a subset was Sanger-sequenced to confirm that the mutations were not germ-line or highly prevalent in the samples evaluated (SI Appendix, SI Materials and Methods). As expected from previous studies, point mutation prevalence of mtDNA (1.4 ± 1.3 × 10−5 mutation per base pair, mean ± SD) were significantly higher than those of nuclear DNA (5.2 ± 3.5 × 10−7) in 25 control individuals (two-tailed t test, P < 0.0001) (Dataset S1, Table S9). We further determined the specificity of BotSeqS using discordant germ-line heterozygous calls to estimate a false positive rate of 2.6 × 10−12 (SI Appendix, SI Materials and Methods).
Mutation Prevalence Varies with DNA Repair Capacity and Carcinogen Exposure.
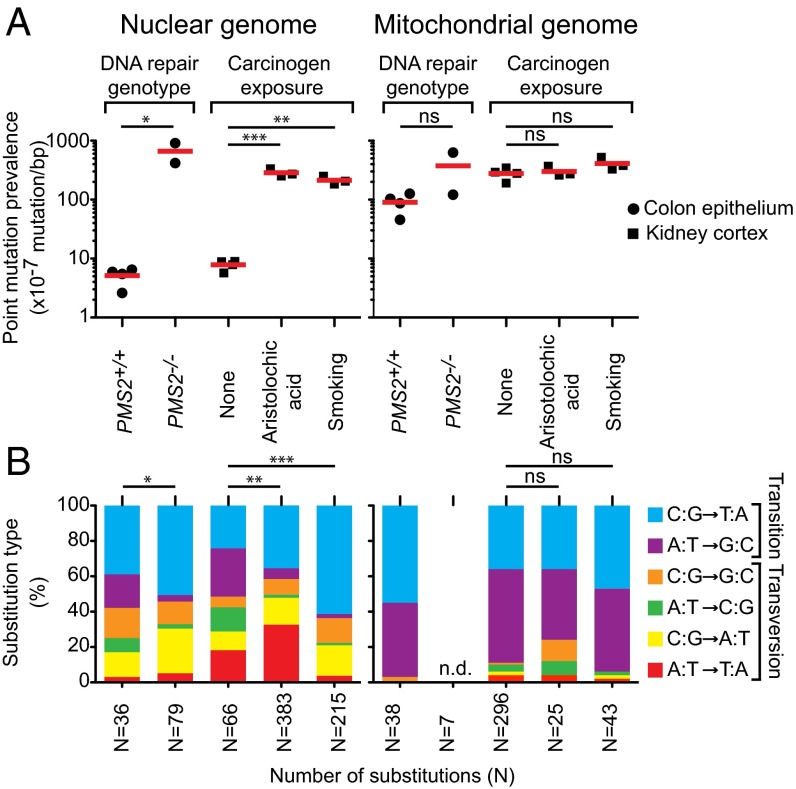
We first asked whether BotSeqS can detect the elevated levels of mutations in the normal tissues of mismatch repair deficient individuals. Individuals with biallelic inactivating germ-line mutations in mismatch repair machinery show higher levels of mutation in both normal and tumor tissues (22, 23). Therefore, we tested DNA from normal colon epithelium of individuals (COL238 and COL239) with biallelic germ-line inactivating mutations in the Post-Meiotic Segregation 2 (PMS2) gene. Using BotSeqS, we found that the average mutation prevalence of nuclear DNA in these two siblings (6.6 ± 3.5 × 10−5 mutation per base pair; ages 16 and 18 y) was significantly higher than that in similarly aged individuals (5.1 ± 1.7 × 10−7 for COL235, COL236, COL237, and COL374; average age 24 y) with proficient mismatch repair (two-tailed t test, P < 0.05) (Fig. 2A). This 130-fold increase in nuclear mutation prevalence was associated with a significant difference in the nuclear mutational spectrum between PMS2+/+ and PMS2−/− cohorts (Fisher’s exact test, P = 0.04, Fig. 2B).
Fig. 2.
Nuclear point mutations increase in normal tissues from individuals with defects in DNA repair or with exposure to environmental carcinogens compared with controls. (A) Comparison of point mutation prevalences in nuclear (Left) and mitochondrial (Right) genome in age-matched normal colon epithelium (filled circle) with different DNA mismatch repair genotypes (PMS2+/+ or PMS2−/−) or in age-matched normal kidney cortex (filled square) without (none) or with (aristolochic acid or smoking) carcinogen exposure. Red lines represent average. *P < 0.05, t test; **P < 0.001 and ***P < 0.0001, one-way ANOVA with Bonferroni multiple comparison posttest; ns, not significant, indicates P > 0.05. (B) Stacked columns representing the substitution frequencies (y axis) of each substitution out of the six possible types (see legend). Cohort labels are indicated in A directly above each column. Number of substitutions (N) generating each mutational spectrum is indicated on the x axis. n.d., not determined due to an insufficient number of mutations (N = 7) for mutational spectrum analysis. *P = 0.04, Fisher’s exact test; **P = 2.6 × 10−8 and ***P = 1.5 × 10−16, Fisher’s exact test with Bonferroni multiple comparison correction; ns, not significant, indicates P > 0.05. All statistical tests in this figure were two-tailed.
We also tested whether BotSeqS could identify a high number of mutations in the normal tissues of individuals exposed to environmental carcinogens. We previously performed genome-wide sequencing of upper tract urothelial carcinomas, representing a cancer type associated with exposure to aristolochic acid (AA) or smoking (24). Mutagens in tobacco smoke as well as AA are metabolized to form DNA-adducts in the normal kidney cortex (24, 25). We compared four age-matched normal kidney cortices from individuals (KID034, KID035, KID036, and KID037; average age 64 y) without known exposure to tobacco smoke or to AA with the normal kidney cortex of three heavy smokers (SA_117, SA_118, and SA_119; average age 65 y), as well as with three individuals who had been exposed to AA (AA_105, AA_124, and AA_126; average age 79 y). The nuclear point mutation prevalences in smokers and AA-exposed kidneys were significantly higher, by 27- and 36-fold, respectively, than in the nonexposed controls (one-way ANOVA with Bonferroni multiple comparison posttest, P < 0.0001 for AA and P < 0.001 for smoking) (Fig. 2A). This increased number of mutations in the nuclear genome was associated with a significantly altered nuclear mutational spectrum (Fisher’s exact test with Bonferroni multiple comparison correction, P = 2.6 × 10−8 for AA and P = 1.5 × 10−15 for smoking) (Fig. 2B). Interestingly, the mtDNA point mutation prevalences and spectra between the nonexposed and exposed groups were not significantly different, despite the dramatic difference in their nuclear genomes (Fig. 2 A and B).
Rare Mutations Accumulate with Age.
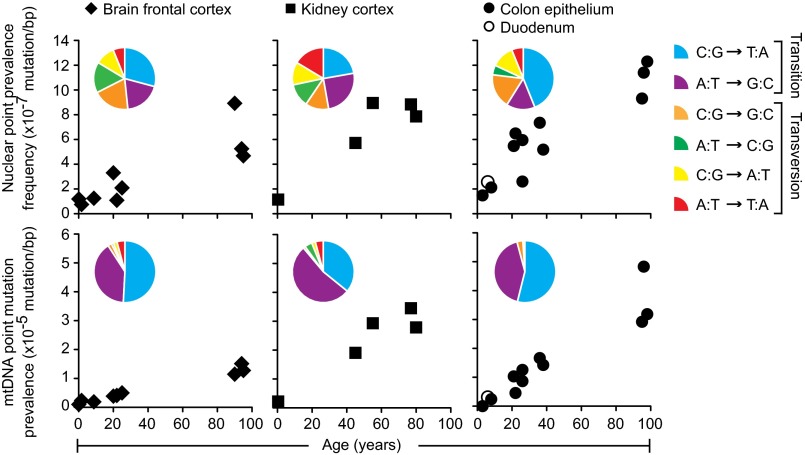
Many lines of evidence indicate that the human body accumulates random mutations with age. BotSeqS was designed to directly measure differences such as these, and we tested whether the prevalence of rare point mutations in the DNA of three normal human tissues was dependent upon age. Normal colonic epithelium from 11 individuals showed mutation prevalences that significantly increased with age, by an average of 30-fold in mtDNA and 6.1-fold in nuclear DNA, over 91 y (Fig. 3 and Table 1) (one-way ANOVA with Bonferroni multiple comparison posttest, P < 0.001 for both). Similarly, mutation prevalences increased by an average of 19-fold in mtDNA and 6.5-fold in nuclear DNA over 64 y in normal kidney cortices. The mutation prevalences in brain frontal cortex also significantly increased with age, albeit more slowly, by 7.3-fold in mtDNA and 5.7-fold in nuclear DNA over 90 y (one-way ANOVA with Bonferroni multiple comparison posttest, P < 0.001 for mtDNA and P < 0.05 for nuclear).
Fig. 3.
Normal human tissues accumulate point mutations over a lifetime with genome-specific and tissue-specific mutational patterns. Point mutation prevalences in nuclear (Top) and mitochondrial (Bottom) genome measured in four normal tissue types (brain frontal cortex of 9 individuals, kidney cortex of 5 individuals, colon epithelium of 11 individuals, and duodenum of 1 individual). Twenty-six total individuals were assessed, with each individual contributing to one normal tissue type. Pie chart Insets show the prevalences of each substitution out of the six possible substitution types (see pie chart legend, right side). Each pie chart was compiled from the individuals represented in their respective scatter plots, with the exception that duodenum was omitted. The number of substitutions generating the pie charts for the nuclear genome was n = 31 for brain, n = 73 for kidney, and n = 94 for colon, and for the mitochondrial genome was n = 181 for brain, n = 299 for kidney, and n = 116 for colon.
Table 1.
Summary of prevalence of rare mutations in normal human tissues in this study
| Genome | Normal human tissue | Average mutation prevalence ± SD (× 10−7) mutation per base pair | Average lifespan, y | Average lifespan fold-increase | ||
| Young child (<10 y) | Young adult (20–40 y) | Mid/old adult (>40 y) | ||||
| mtDNA | Brain frontal cortex | 18 ± 7 (n = 3) | 43 ± 6 (n = 3) | 130 ± 18 (n = 3) | 90 | 7.3 |
| Kidney cortex | 15 (n = 1) | n.d. | 280 ± 64 (n = 4) | 64 | 19 | |
| Colon epithelium | 12 ± 17 (n = 2) | 110 ± 43 (n = 3) | 370 ± 100 (n = 3) | 91 | 30 | |
| Nuclear | Brain frontal cortex | 1.1 ± 0.3 (n = 3) | 2.2 ± 1.1 (n = 3) | 6.3 ± 2.3 (n = 3) | 90 | 5.7 |
| Kidney cortex | 1.2 (n = 1) | n.d. | 7.8 ± 1.5 (n = 4) | 64 | 6.5 | |
| Colon epithelium | 1.8 ± 0.5 (n = 2) | 5.5 ± 1.6 (n = 3) | 11 ± 1.5 (n = 3) | 91 | 6.1 | |
n.d., not determined.
Within our dataset, we could directly compare point mutation prevalences in brain versus colonic tissues in three different age groups (children <10 y; adults between 20 and 40 y; and old adults ≥90 y). Interestingly, the nuclear mutation prevalence in the colon was not significantly different from that of the brain in children (1.8 ± 0.5 × 10−7 in colon vs. 1.1 ± 0.3 × 10−7 in brain, two-way ANOVA with Bonferroni multiple comparison posttest, P > 0.05). However, the mutation prevalence in the colon was significantly higher than that of the brain in young adults (5.5 ± 1.6 × 10−7 in colon vs. 2.2 ± 1.1 × 10−7 in brain, two-way ANOVA with Bonferroni multiple comparison posttest, P < 0.05) as well as in old adults (1.1 ± 0.2 × 10−6 in colon vs. 6.3 ± 2.3 × 10−7 in brain, two-way ANOVA with Bonferroni multiple comparison posttest, P < 0.01) (SI Appendix, Fig. S3). No significant differences were found between the mtDNA mutation prevalence of the colon versus that of the brain in relatively young individuals (children or young adults). However, the mtDNA mutation prevalence in the colon was significantly higher than that of the brain in old individuals (3.7 ± 1.0 × 10−5 in colon vs. 1.3 ± 0.2 × 10−5 in brain, two-way ANOVA with Bonferroni multiple comparison posttest, P < 0.0001) (SI Appendix, Fig. S3).
The Mutational Patterns in mtDNA Are Very Different from Those of Nuclear DNA.
We examined the spectra of the rare point mutations in each normal tissue studied. Mutations in mtDNA were dominated by transitions (97% in colon, 89% in kidney, and 91% in brain) with a heavy strand bias, as expected from previous studies (12) (Fig. 3 and Dataset S1, Table S7). The ratio of transitions-to-transversions was strikingly different in mtDNA (average of 15) compared with nuclear DNA (average of 1.1) in all three tissues.
To further assess the differences in mutation prevalence between the two genomes, we calculated the ratio between mtDNA-to-nuclear mutation prevalences for each individual (Dataset S1, Table S9). Point mutation prevalences in the mtDNA were on average 25-fold higher than the nuclear genome in normal tissues (SI Appendix, Fig. S4, control cohort). In patients with exposure histories or DNA repair defects, the ratios were significantly smaller due to the concomitantly greater number of nuclear (but not mitochondrial) DNA mutations in such individuals compared with those from the control cohort (one-way ANOVA with Bonferroni multiple comparison posttest, P < 0.05) (SI Appendix, Fig. S4).
Mutational Spectra Are Tissue Specific.
Although rare mutations in mtDNA are dominated by transitions, there are still tissue-specific mtDNA differences that can be appreciated from the pie charts in Fig. 3. For example, mitochondrial C:G-to-T:A transitions were more prominent, and A:T-to-G:C transitions less prominent, in normal colon (54% and 42%, respectively) and brain (51% and 40%, respectively) compared with normal kidney tissues (36% and 53%, respectively). The mutation spectra in the nuclear DNA of all three tissues were much more diverse. For example, C:G-to-T:A transitions predominated in normal colon (44% in colon compared with 22% in kidney and 29% in brain) whereas normal kidney and brain harbored a proportionately greater fraction of A:T-to-G:C transitions (25% in kidney and 19% in brain compared with 15% in colon) as well as A:T-to-C:G transversions (12% in kidney and 16% in brain compared with 5% in colon). Moreover, A:T-to-T:A transversions were more frequent in kidney (16%) compared with colon (6%) and brain (6%). Pairwise comparisons of the mutational spectra within each genome revealed significant differences between the substitution pattern of kidney and colon (Fisher’s exact test with Bonferroni multiple comparison correction, P = 0.0029 in mtDNA and P = 0.031 in nuclear DNA).
We compared the spectra of the rare mutations found in normal kidney and colon tissues to the clonal DNA mutations in cancers derived from the cells of these organs, using publicly available data for the latter (26, 27). Brain frontal cortex was excluded in this analysis because it was not clear what tumor type should be used for comparison. To search for similarities and differences among normal and tumor mutational spectra, principal component analysis was performed on the nuclear and mtDNA spectra derived from the data on normal kidney cortex, normal colon epithelium, clear cell renal carcinoma, and colorectal carcinoma. We found that the spectra of the rare mutations in normal colon and kidney tissues were very similar to those of the corresponding cancer type (SI Appendix, Fig. S5).
Discussion
BotSeqS is a straightforward NGS-based approach that can accurately measure rare point mutations in an unbiased, genome-wide manner. Using BotSeqS, we were able to achieve several important goals: (i) define estimates of rare mutation prevalences across the whole genome; (ii) simultaneously evaluate rare mutations in both the nuclear and mitochondrial genomes of the same population of cells; (iii) compare the prevalence of rare mutations among various normal tissues of individuals of different age, DNA repair capacity, or exposure histories; and (iv) identify the spectra of rare mutations in normal tissues, allowing their comparison with those of clonal mutations in cancers.
Our data show that mutations increase with age, a result that is broadly consistent with the literature (2, 3). The rate of increase of mutations is not as great in the brain as it is in the colon or kidney, presumably because the colon and kidney are both self-renewing tissues throughout adult life whereas the brain is not. On the other hand, the fact that the mutation prevalence increased at all after childhood was surprising, given that the major cell types in the prefrontal cortex are generally thought to be postmitotic (28). There are several potential explanations for this increase. A small number of cells that are replicating more actively than neurons or glia could be responsible for the increase. Such cells could include microglia or infiltrating lymphocytes or other inflammatory cells. Alternatively, these mutations could represent the results of spontaneous DNA damage independent of DNA replication. A recent single-cell sequencing study of human neurons suggested that spontaneous damage occurs during transcription (29). However, in contrast to single-cell sequencing, BotSeqS measures mutations that are found on both strands. Thus, for the explanation of spontaneous DNA damage to be plausible, the mutations identified by BotSeqS would have to have been subject to DNA repair. Consistent with this possibility, DNA repair processes are known to be active in postmitotic neurons and glia (30).
A third possibility is that these mutations are artifacts of the procedure we used to detect them. It is fascinating that this formal possibility is essentially impossible to exclude because many of the mutations we detected are likely found in only one cell of the tissue studied, and the DNA from that cell is no longer available for subsequent evaluation. Additionally, there is no other technique available to observe such mutations with the sensitivity achieved here. Our sensitivity is currently limited only by the amount of sequencing devoted to the project. We can easily detect mutations occurring at 6 ×10−8 per base pair using a small fraction of an Illumina HiSeq 2500 flow cell. We estimate that mutations could be detected at <10−9 per base pair using an entire flow cell. Supporting this sensitivity is BotSeqS’s high specificity, where strand-discordant germ-line SNPs yield a false positive rate of 2.6 × 10−12 errors per base pair. The only other method that approaches this sensitivity and specificity has been described by Loeb and coworkers (12, 31), but their method is applicable only to predefined regions of the genome. In the absence of direct confirmation, we are forced to use correlations and other approaches to support the accuracy of the technology described herein. These correlations include the following, as detailed in Dataset S1, Table S9: similar mutation prevalences and spectra identified in different DNA aliquots of the same samples; similar mutation prevalences and spectra identified in the same tissues of different individuals of similar age; expected increases in mutation prevalence with age; tissue-specific differences and age-dependent increases in mutation prevalence; higher mutation prevalences in normal tissues deficient in mismatch repair or exposed to environmental mutagens; and mutation spectra in normal tissues consistent with those previously observed in cancers from the same tissues. Other in silico and experimental approaches used to evaluate the accuracy of BotSeqS are described in the SI Appendix, SI Materials and Methods.
We also were able to compare mutation prevalence in the mitochondrial and nuclear genomes of the same tissues. In normal individuals, in the absence of exposures to mutagens, the mutation prevalence was much higher in the mitochondria than in the nuclear genome (median ratio of 26). This finding is consistent with the relatively poor efficiency of DNA repair in the mitochondria compared with the nuclear genome (32). Equally important, however, is that the ratio of mitochondrial to nuclear mutation prevalences was vastly lower (median of 1.3) in the normal kidneys of individuals exposed to either cigarette smoke or AA. This finding is not consistent with the known, less efficient repair of DNA in mitochondria. Moreover, there was a shift toward the AA mutational signature, A:T-to-T:A transversions, in the nuclear DNA of normal kidneys in individuals exposed to AA, but virtually none in the mtDNA. One possibility is that the higher mutation prevalence in the mtDNA could be masking the effect of environmental mutagens on the mitochondrial genome compared with its effect on the nuclear genome. Another possibility is that there are unexpected and pronounced differences in the ways through which these mutagens cause DNA damage in these two organelles.
Another original observation of our study is the finding that mutation spectra differed among normal human tissues, even in the absence of exposures to known mutagens. Whether such differences reflect varying exposures to as yet unidentified commonly encountered mutagens, or tissue-specific processes such as DNA repair, is not known. In some cases, the rare mutation spectra in normal tissues were found to be similar to the clonal mutations found in cancers. Although varying mutation spectra in cancers have often been attributed to cancer-specific processes, our data suggest that at least a subset of these mutations actually reflect tissue-specific processes. This concept is consistent with the idea that a substantial fraction of the mutations found in cancers occur in normal stem cells (33, 34). We envision that the straightforward approach described here, which can easily measure very rare mutations in any tissue or cell type of interest, will be applicable to questions of broad biomedical interest.
Materials and Methods
Human Tissue Samples.
Normal, nondiseased flash frozen tissues for this study were acquired from five sources (Dataset S1, Table S1). COL229, COL231, COL232, COL233, COL234, COL235, COL236, COL237, and SIN230 were obtained from consented patients at the Johns Hopkins Hospital with the approval of the Johns Hopkins Institutional Review Board. COL373, COL374, COL375, and BRA01-09 were requested from the NIH NeuroBioBank (https://neurobiobank.nih.gov/), with the request being approved and fulfilled by the University of Maryland Brain and Tissue Bank and the University of Miami Brain Endowment Bank. KID034-038 were purchased as 200-mg flash frozen blocks from the Windber Research Institute. COL238 and COL239 were previously reported (22, 35, 36). SA_117, SA_118, SA_119, AA_105, AA_124, and AA_126 were acquired by C.-H. Chen and Y.-S. Pu (Department of Urology, National Taiwan University Hospital and College of Medicine, Taipei, Taiwan) as previously reported (24).
Genomic DNA Purification.
Tissues were dissociated by pulverization, and genomic DNA was purified using Qiagen AllPrep. DNA was resuspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). DNA concentration was quantitated by Qubit (ThermoFisher) or TapeStation (Agilent). Assuming 6.6 pg of DNA per human cell, the median number of cell equivalents (before dilution for BotSeqS) was 1.2 million (range 0.2–5.3 million).
Preparation of Illumina Y-Adapter–Ligated Molecules.
Genomic DNA (34 ng to 1 µg) in 55 µL of TE buffer was fragmented using BioRuptor (Diagenode) at high intensity for 15 s on and 90 s off, using seven cycles at 3 °C. After random fragmentation, Illumina Y-adapters were ligated to the DNA fragments using a TruSeq DNA PCR-Free kit (Illumina) according to a standard low DNA input Illumina protocol with selection for 350-bp insert sizes. In general, with an input of 500 ng of genomic DNA into the library prep, a 105-fold dilution on Y-adaptor–ligated DNA molecules was performed prior to PCR. Further details on dilution of BotSeqS libraries, sequencing, and data analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We are grateful for our human tissue samples and sources. We thank Stephen Eacker, Surojit Sur, Yuxuan Wang, Austin Mattox, and Joshua Cohen for helpful comments on the project and manuscript. This work was supported by The Virginia and D. K. Ludwig Fund for Cancer Research; the Howard Hughes Medical Institute; National Institutes of Health Grants CA43460, CA57345, and CA62924; Henry and Marsha Laufer; and the Zickler Family Foundation (A.P.G.).
Footnotes
Conflict of interest statement: B.V. has no conflict of interest with respect to the new technology described in this manuscript, as defined by Johns Hopkins University's policy on conflict of interest. B.V. is a founder of PapGene and Personal Genome Diagnostics and a member of the Scientific Advisory Boards of Morphotek and Syxmex-Inostics. These companies and others have licensed patent applications on genetic technologies from Johns Hopkins, some of which result in royalty payments to B.V. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Genome-Phenome Archive (EGA) database (accession no. EGAS00001001838).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607794113/-/DCSupplemental.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133(4):118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijg J. Somatic mutations, genome mosaicism, cancer and aging. Curr Opin Genet Dev. 2014;26:141–149. doi: 10.1016/j.gde.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14(5):R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albertini RJ, Nicklas JA, O’Neill JP, Robison SH. In vivo somatic mutations in humans: Measurement and analysis. Annu Rev Genet. 1990;24:305–326. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 6.Cole J, Skopek TR. International Commission for Protection Against Environmental Mutagens and Carcinogens working paper no. 3: Somatic mutant frequency, mutation rates and mutational spectra in the human population in vivo. Mutat Res. 1994;304(1):33–105. doi: 10.1016/0027-5107(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 7.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150(2):402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108(23):9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci USA. 2011;108(50):20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA. 2012;109(36):14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiatt JB, Pritchard CC, Salipante SJ, O’Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23(5):843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baslan T, Hicks J. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev. 2014;26:59–65. doi: 10.1016/j.gde.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011;9(1):72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 16.Kinde I, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5(167):167ra4. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A, et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol. 2014;15(12):530. doi: 10.1186/s13059-014-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keys JR, et al. Primer ID informs next-generation sequencing platforms and reveals preexisting drug resistance mutations in the HIV-1 reverse transcriptase coding domain. AIDS Res Hum Retroviruses. 2015;31(6):658–668. doi: 10.1089/aid.2014.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9(9):e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt MW, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods. 2015;12(5):423–425. doi: 10.1038/nmeth.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41(6):e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons R, et al. Mismatch repair deficiency in phenotypically normal human cells. Science. 1995;268(5211):738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 23.Shlien A, et al. Biallelic Mismatch Repair Deficiency Consortium Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 24.Hoang ML, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5(197):197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randerath E, et al. Covalent DNA damage in tissues of cigarette smokers as determined by 32P-postlabeling assay. J Natl Cancer Inst. 1989;81(5):341–347. doi: 10.1093/jnci/81.5.341. [DOI] [PubMed] [Google Scholar]
- 26.Ju YS, et al. ICGC Breast Cancer Group; ICGC Chronic Myeloid Disorders Group; ICGC Prostate Cancer Group Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3:3. doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122(1):133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 29.Lodato MA, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350(6256):94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron. 2014;83(2):266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy SR, et al. Detecting ultralow-frequency mutations by duplex sequencing. Nat Protoc. 2014;9(11):2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM, 3rd, Bohr VA. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015;25(3):158–170. doi: 10.1016/j.tcb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci USA. 2013;110(6):1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasetti C, Vogelstein B. Cancer etiology: Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton SR, et al. The molecular basis of Turcot’s syndrome. N Engl J Med. 1995;332(13):839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 36.De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74(5):954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.