Significance
Our study helps answer the question of how G protein-coupled receptors bind an extracellular ligand and relay this signal through its transmembrane helices into an intracellular signaling event. This has been the central thesis that G protein-coupled receptor structural studies have sought to address through static snapshots of the crystallized proteins. Our functional approach using CXC chemokine receptor 4 as a model complements these structures by identifying the dynamic atomic pathway from chemokine engagement to G protein coupling.
Keywords: GPCR activation, G protein, chemokine receptor, hydrophobic bridge, shotgun mutagenesis
Abstract
The atomic-level mechanisms by which G protein-coupled receptors (GPCRs) transmit extracellular ligand binding events through their transmembrane helices to activate intracellular G proteins remain unclear. Using a comprehensive library of mutations covering all 352 residues of the GPCR CXC chemokine receptor 4 (CXCR4), we identified 41 amino acids that are required for signaling induced by the chemokine ligand CXCL12 (stromal cell-derived factor 1). CXCR4 variants with each of these mutations do not signal properly but remain folded, based on receptor surface trafficking, reactivity to conformationally sensitive monoclonal antibodies, and ligand binding. When visualized on the structure of CXCR4, the majority of these residues form a continuous intramolecular signaling chain through the transmembrane helices; this chain connects chemokine binding residues on the extracellular side of CXCR4 to G protein-coupling residues on its intracellular side. Integrated into a cohesive model of signal transmission, these CXCR4 residues cluster into five functional groups that mediate (i) chemokine engagement, (ii) signal initiation, (iii) signal propagation, (iv) microswitch activation, and (v) G protein coupling. Propagation of the signal passes through a “hydrophobic bridge” on helix VI that coordinates with nearly every known GPCR signaling motif. Our results agree with known conserved mechanisms of GPCR activation and significantly expand on understanding the structural principles of CXCR4 signaling.
The CXC chemokine receptor 4 (CXCR4) belongs to the G protein-coupled receptor (GPCR) superfamily of proteins, the largest class of integral membrane proteins encoded in the human genome, comprising greater than 30% of current drug targets. Deregulation of CXCR4 expression in multiple human cancers, its role in hematopoietic stem cell migration, and the utilization of CXCR4 by HIV-1 for T-cell entry, make this receptor an increasingly important therapeutic target (1). One FDA-approved drug against CXCR4 is currently on the market (Mozobil, for hematopoietic stem cell mobilization), and multiple additional drugs against this target are in development for oncology and other indications (2).
The crystal structures of class A GPCR superfamily members in their active and inactive conformations (reviewed in refs. 3 and 4) provide unprecedented insight into the structural basis of ligand binding, G protein coupling, and activation of GPCRs via rearrangements of transmembrane (TM) helices. GPCR helices V and VI in particular, and in some cases III and VII, are known to undergo significant conformational changes upon activation (5–7). However, static images alone have not been able to explain the residue-level mechanisms underlying the dynamic helical shifts that mediate GPCR signal transduction. Additionally, only inactive state structures have been solved for CXCR4 and most other GPCRs (8, 9). Over the last two decades, extensive mutagenesis studies of GPCRs in general [collectively describing >8,000 mutations (gpcrdb.org)] and of CXCR4 in particular (covering 81 primarily extracellular residues of 352 total) (10) have identified individual residues that are critical for receptor signaling. Whereas many of the individual critical residues and motifs have been described, the complete intramolecular signal transmission chain remains unclear.
Here we report a cohesive model for the mechanism by which CXCR4 transmits the signal induced by its extracellular chemokine ligand CXCL12 [also known as stromal cell-derived factor 1 (SDF-1)] to the intracellular G protein. Using a comprehensive library of 728 mutants covering all 352 residues of CXCR4, we experimentally identified 41 amino acids that are required for signal transmission. Our results complement structural studies of GPCRs and expand on previous mutagenesis studies from diverse laboratories to form a comprehensive functional model that explains how CXCR4 transmits an extracellular ligand binding event through its TM domains to dynamically affect helical shifts and intracellular G protein coupling.
Results
Comprehensive Mutagenesis Identifies Critical Residues for CXCR4 Signaling.
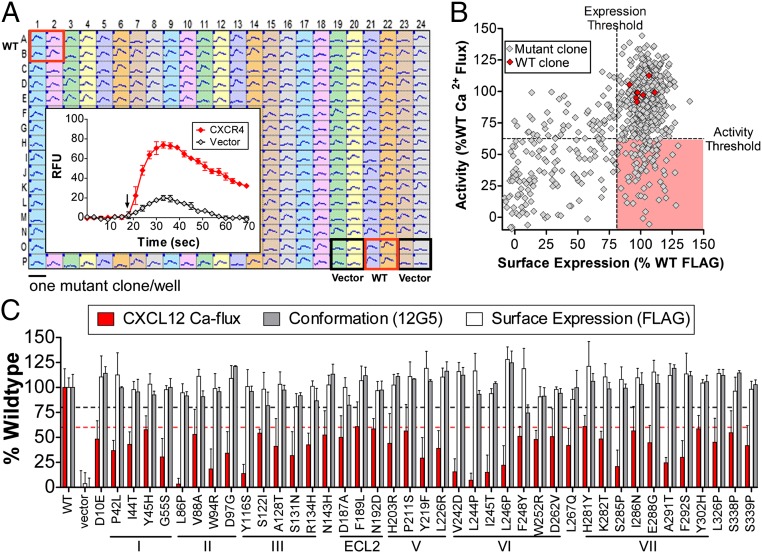
To identify critical residues required for CXCL12-dependent CXCR4 signal transmission, a comprehensive “shotgun mutagenesis” library of receptor variants was created with at least one mutation at each of the 352 residues of CXCR4 (11). The library contains a total of 728 mutant clones, representing an average of 2.7 substitutions at each amino acid position. The entire CXCR4 mutation library was transfected into mammalian cells in a 384-well array format (one clone per well) and evaluated in parallel for CXCL12-dependent activation as measured by a calcium flux assay (Fig. 1A). The addition of 20 nM CXCL12 to cells expressing WT CXCR4, but not mock-transfected cells, resulted in robust receptor activation, measured as an increase in cellular fluorescence. A high concentration of CXCL12 (20 nM, approximately three times the KD) was selected for stimulation to identify mutations that are the most important for (i.e., resulted in the most severe impairment of) CXCR4 signaling.
Fig. 1.
Critical residues for CXCR4 activation by CXCL12. (A) Representative 384-well mutation array plate showing calcium flux for CXCR4 variants upon stimulation with 20 nM CXCL12. Each plate contained eight positive and eight negative control wells used for normalization. Inset shows the mean normalized calcium flux curves for highlighted control wells. (B) Reactivity profiles of CXCR4 variants showing mean calcium flux (n = 5) and cell surface expression (FLAG reactivity, n = 5). Critical clones were chosen from the Lower Right (red) quadrant using thresholds outlined in Materials and Methods. (C) A total of 41 critical CXCR4 variants show reduced calcium flux (red bars), but near wild-type surface expression (FLAG reactivity, white bars) and correct conformation (12G5 reactivity, gray bars). The thresholds used for CXCL12 activity (62%, red dashed line) and for FLAG surface expression and 12G5 folding (80%, black dashed line) are shown. Bars represent the average and error bars show the SD (n = 3–5).
To differentiate nonsignaling mutant proteins from poorly trafficking or misfolded proteins, each CXCR4 variant was also independently tested for surface expression using an N-terminal FLAG epitope tag and for global folding using the conformation-dependent anti-CXCR4 monoclonal antibody 12G5 (Fig. 1B). We identified a total of 41 positions in CXCR4 where mutations resulted in significantly reduced CXCR4 activation (less than two SDs below wild-type, <62%) without disrupting its global structure (>80% 12G5 reactivity) or surface trafficking (>80% FLAG reactivity) (Fig. 1C and SI Appendix, Table S1). Each mutant was further tested for reactivity with three additional conformationally sensitive anti-CXCR4 MAbs, which confirmed that the selected proteins were correctly folded in nearly every case (SI Appendix, Fig. S1). Mutants with the greatest impairment of signaling (whose calcium flux value plus one SD <62%) were also tested for their ability to bind CXCL12 using a FRET assay (SI Appendix, Fig. S2). In this assay, a high concentration of chemokine (60 nM, vs. KD of 22 nM for the labeled CXCL12 used; CisBio) was used to identify mutations that most significantly impaired binding. Of these mutants, only W942.60R and D972.63G resulted in a significant loss of CXCL12 binding, consistent with previous reports (12, 13) (superscript nomenclature indicates Ballesteros–Weinstein numbering of GPCR residues).
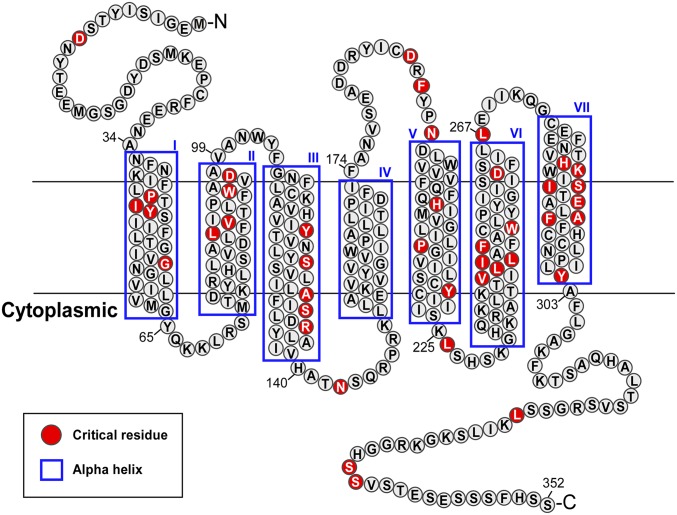
Mapping the 41 critical residues onto a snake plot of CXCR4 reveals that 33 of them localize within the TM helices, with half positioned in helices III, VI, and VII (Fig. 2), consistent with the known role of these helices in GPCR signal transduction (14, 15). No critical residues were identified in helix IV, which was previously also found to have little involvement in signal transduction (7, 15).
Fig. 2.
CXCR4 residues critical for CXCL12-mediated signaling. A total of 41 residues critical for CXCL12-dependent CXCR4 function are highlighted in red on a serpentine diagram of the receptor.
Functional Classification of Critical Residues.
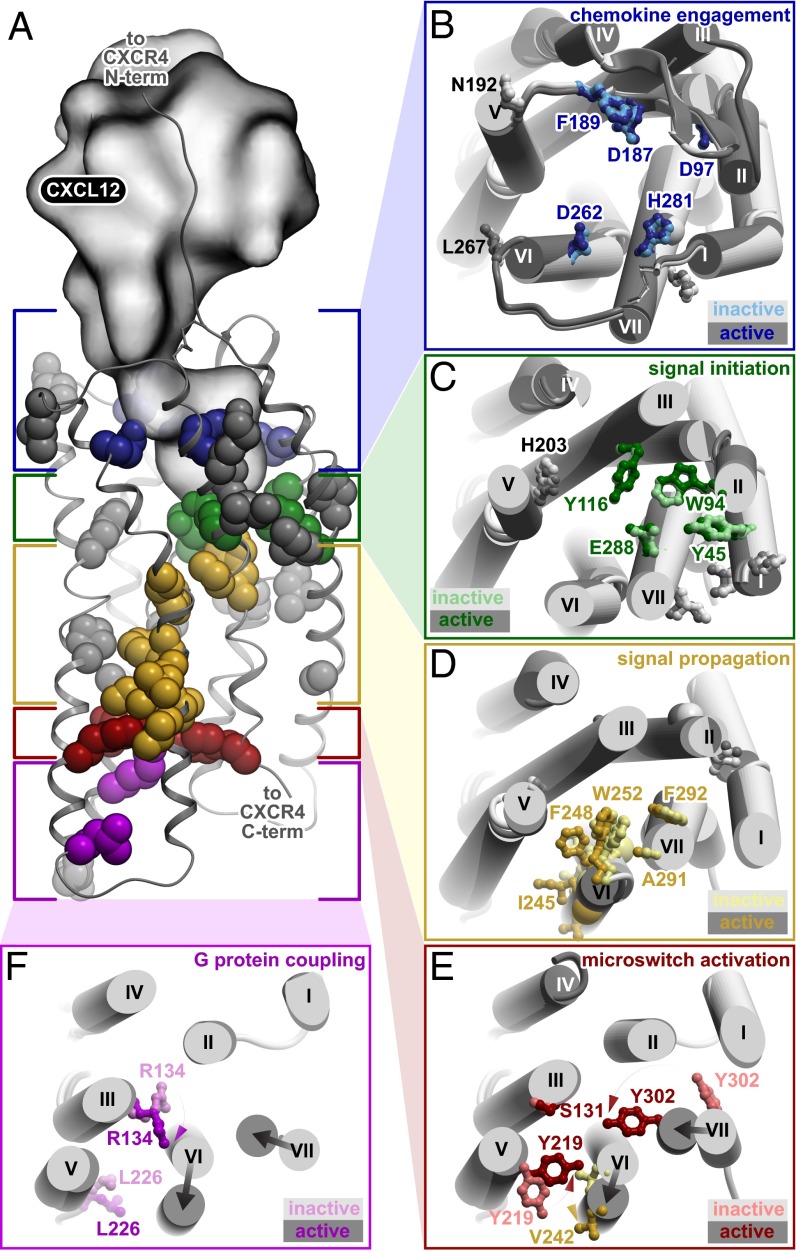
To better understand how each of the 41 identified residues contributes to CXCL12-induced signaling, we mapped them onto the crystal structures of CXCR4 (8, 9), as well as active and inactive state models of the CXCR4:CXCL12 complex derived by homology from the CXCR4:vMIP-II crystal structure (9) (Fig. 3A). We then measured the spatial distance of each critical residue to every other critical residue to identify their interactions (i.e., interatomic distances of <4.2 Å) (SI Appendix, Table S2). Remarkably, the majority of these critical residues form a continuous intramolecular signaling chain through the TM helices that connects the receptor’s extracellular residues to its intracellular residues (Fig. 4). Based on their function and location, critical residues identified were classified into one of five functional categories: (i) chemokine engagement, (ii) signal initiation, (iii) signal propagation, (iv) microswitch activation, and (v) G protein coupling. The following sections describe the composition of these functional categories and their role in signal transmission.
Fig. 3.
Functional classes of critical residues in CXCR4. (A) Critical residues of CXCR4 are divided into functional groups based on the CXCR4:CXCL12 model and CXCR4:vMIP-II structure. Critical residues that contact each other, CXCL12, or G protein are shown in color; other critical residues identified are shown in gray. The active state of the CXCR4:CXCL12 complex is shown. CXCR4 residues are involved in (B) chemokine engagement at the mouth of the orthosteric ligand binding pocket (blue), (C) signal initiation at the base of the ligand binding pocket (green), (D) signal propagation through the TM domain helices that include the hydrophobic bridge (yellow), (E) microswitch activation that transmits hydrophobic bridge conformational changes (red), and (F) G protein coupling (purple). Subpanels show top-down views of interactions in each group. In the subpanels, dark and light colors represent the active and inactive states of the CXCR4:CXCL12 complex model, respectively.
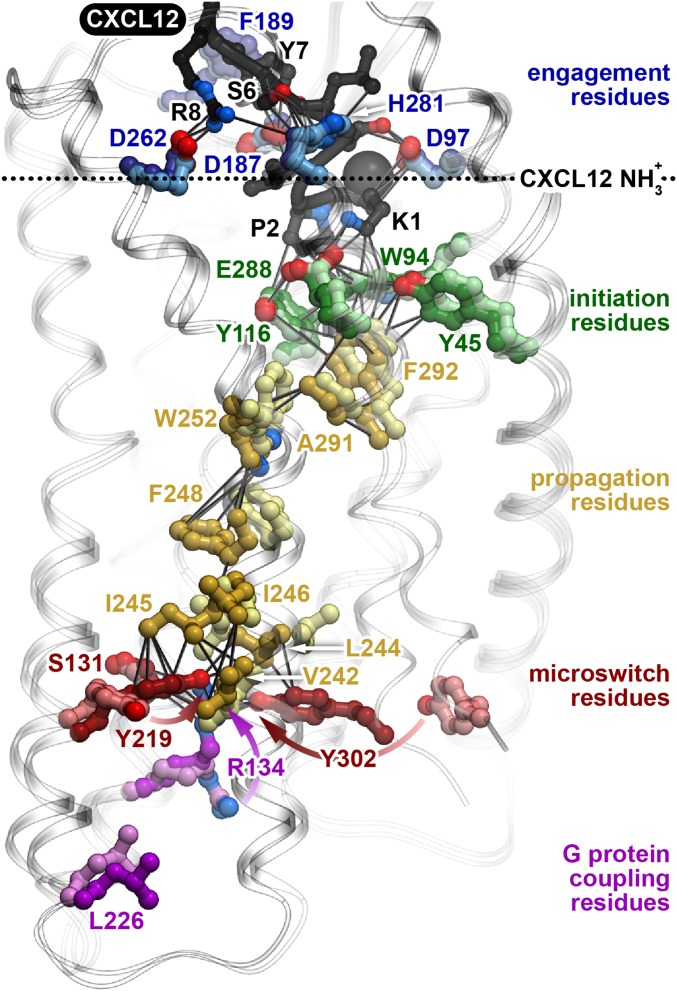
Fig. 4.
CXCR4 signal transmission through the TM helices. Critical CXCR4 residues identified are involved in chemokine engagement (blue), signal initiation (green), signal propagation (yellow), microswitch activation (red), and G protein coupling (purple). The dashed horizontal line highlights the N-terminal amine of CXCL12 (black). Oxygen atoms are shown in red, nitrogen atoms in blue. Solid connecting lines indicate interatomic distances that are <4.2 Å in the active state model of CXCR4:CXCL12. Dark and light colors represent the superimposed active and inactive state conformations of the CXCR4:CXCL12 models, respectively. R1343.50, Y2195.58, and Y3027.53 undergo particularly large conformational changes from the inactive to active state and their changes are highlighted with arrows.
CXCR4 residues involved in chemokine engagement.
The interaction of CXCL12 with CXCR4 is known to be mediated by two distinct epitopes (1, 8, 12, 16–22). In the first [chemokine recognition site 1 (CRS1)], the unstructured N terminus of CXCR4 interacts with the globular core of the chemokine, specifically with the N loop and the 40s loop of CXCL12. In the second site [chemokine recognition site 2 (CRS2)], the distal flexible N terminus of CXCL12 reaches into the TM binding pocket of CXCR4 to trigger signaling. Most of the critical residues that we identified on the extracellular side of the receptor cluster in the CRS2 binding pocket (blue and green layers in Fig. 3), consistent with the fact that CRS2 interactions are the primary driver for both affinity and signaling in CXCR4 (9).
Our study identified seven solvent-exposed critical residues on the extracellular face of the receptor that are positioned to mediate chemokine engagement (Fig. 3B, blue). These engagement residues include D972.63 at the top of helix II and D187ECL2 in ECL2, both of which have previously been implicated in binding CXCL12 (9, 17, 22, 23). On the other side of the pocket, D2626.58 toward the top of helix VI (consistent with previous studies, ref. 23) as well as H2817.32 at the top of helix VII were identified, both in direct proximity of the chemokine in the structure (9) and the models. Residues F189ECL2, N192ECL2, and L267ECL3 on the extracellular face of CXCR4 are also consistent with a potential role in chemokine engagement.
Transmembrane residues that initiate signal transmission.
Four of the 41 critical residues are solvent accessible but are located at the very bottom of the binding pocket and directly contact buried critical residues involved in signaling (Fig. 3C, green). This unique position suggests their role as signal initiators responsive to chemokine binding. One of these CXCR4 residues, W942.60, is highly conserved among chemokine receptors, has been implicated in binding the small molecule antagonist IT1t and vMIP-II (8, 9), and the equivalent residue in the chemokine receptor CCR5 (W862.60) has also been shown to play a role in ligand binding (24). Additional signal initiator residues identified in our screen include Y451.39, Y1163.32, and E2887.39, all previously reported as binding and/or signaling determinants in CXCR4 (13, 19, 22, 25). In the CXCR4:CXCL12 complex model, each of these four residues directly contacts or is in close proximity to the distal N terminus of CXCL12 (residues K1 and P2), which is widely recognized as the critical domain of the chemokine that initiates signaling (16, 26).
Transmembrane residues involved in signal propagation.
Eight critical residues from our dataset are located in the core of the receptor and form a continuous intramolecular chain between the residues involved in signal initiation on the extracellular side and the activation microswitches on the intracellular side, suggesting their role in signal propagation (Fig. 3D, yellow). Three of these critical residues are buried directly below the CXCR4 signal initiator residues discussed above. These residues are F2927.43, which has been previously shown to be important for CXCR4 activity (17, 19), A2917.42, which has a known role in introducing constitutive activity in CCR5 (27), and W2526.48, which is part of the well-characterized CWxP rotamer motif (28, 29). Homologous positions in the A2AR structure (H2787.43 and S2777.42) have been shown to move closer to helix III upon agonist binding (30), supporting an important role for these residues in GPCR signal transmission.
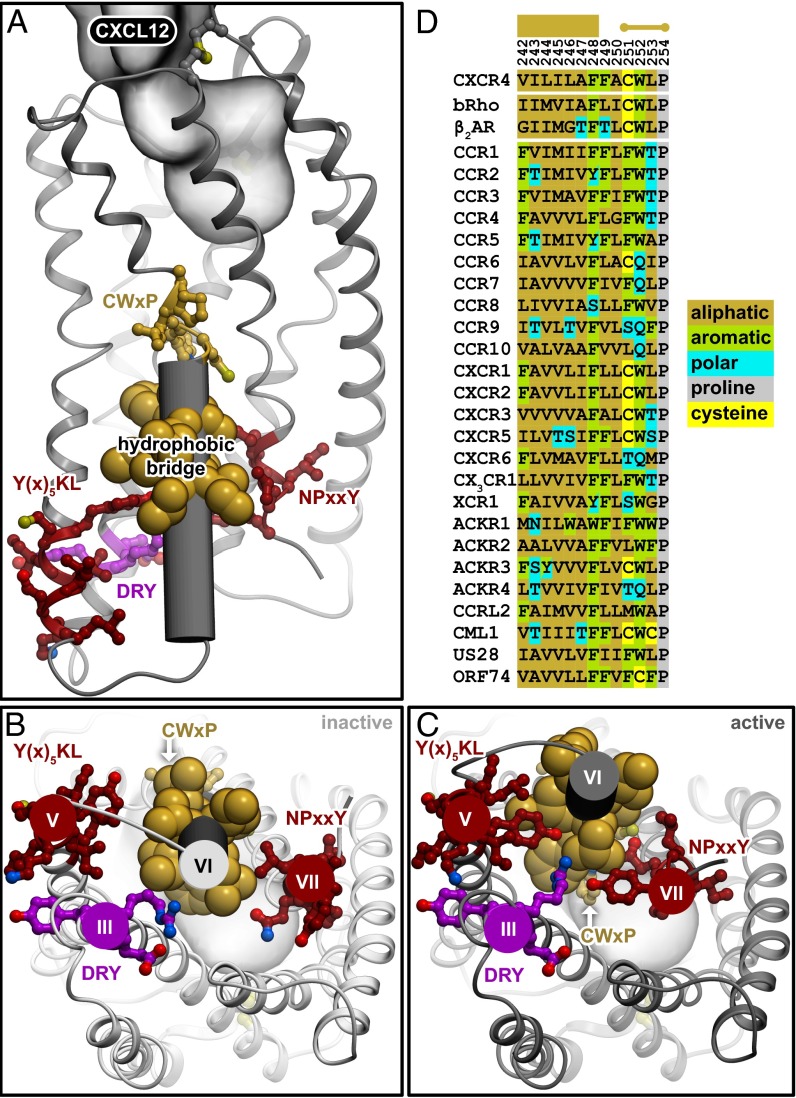
Directly below W2526.48, a string of five residues on helix VI (F2486.44, L2466.42, I2456.41, L2446.40, and V2426.38) were identified as critical. Interestingly, these five hydrophobic residues appear to form a “bridge” linking nearly every key signaling motif in GPCRs, including the CWxP rotamer, NPxxY, DRY, and Y(x)5KL microswitch motifs (Fig. 5A). Moreover, these motifs move closer to this hydrophobic bridge in the active state model of CXCR4 (Fig. 5 B and C), suggesting that the hydrophobic bridge enables helix and side chain repacking during the conformational transition. Previous mutation analyses at the 6.44 and 6.40 positions in different receptors support a role for these residues in mediating the transition between inactive and active GPCR states (3, 31–33). Whereas the identity of these residues is not absolutely conserved, the hydrophobic nature and helical compatibility of these residues is strictly conserved among all chemokine receptors (Fig. 5D) and other GPCRs (10). Two of these residues were deemed critical based on substitution to proline (L2446.40P and L2466.42P). Introducing a proline in this region of CXCR4 can eliminate signaling without altering extracellular structure or ligand binding (8), suggesting that the helical conformation of the intracellular half of TM6 is critically important for signal transmission.
Fig. 5.
Interactions of the hydrophobic bridge with GPCR signaling motifs. (A) An active-state model of the CXCR4:CXCL12 complex shows contacts of hydrophobic bridge residues (yellow) with motifs known to be important in GPCR activation. These include the CWxP motif (yellow) that is part of the signal propagation functional group, the Y(x)5KL and NPxxY motifs (red) that are part of the microswitch functional group, and the DRY box motif (purple) that is part of the G protein-coupling functional group. The lower half of helix VI is shown as a cylinder. (B and C) The hydrophobic bridge and known GPCR motifs are shown (bottom-up view) in the inactive and active states of CXCR4, respectively. The motifs move closer to the hydrophobic bridge in the active state. Oxygen atoms are shown in red, nitrogen atoms in blue. (D) Sequence alignments of homologous residues corresponding to the CXCR4 hydrophobic bridge V2426.38-ILILA-F2486.44 (highlighted at Top as wide horizontal bar) in bRho, β2AR, and the other chemokine receptors, showing that residues are not strictly conserved but need to be hydrophobic.
Activation microswitches that control G protein coupling.
Highly conserved microswitches within GPCRs are known to control the G protein interface during the inactive-to-active state transition. Our screen identified three amino acids that are critical microswitch residues in CXCR4, S1313.47, Y2195.58, and Y3027.53 (Fig. 3E, red). Residues Y2195.58 and Y3027.53 are predicted to significantly change position in the active state to form the structural support for the G protein interface (Fig. 3E, dark vs. light red). Residue Y2195.58 is the first position of the Y(x)5KL microswitch motif, whereas residue Y3027.53 is the last position of the conserved NPxxY motif, both recognized as critical determinants of GPCR activation (29). The importance of S1313.47 and Y2195.58 is also supported by studies of rhodopsin where the direct interaction of homologous residues has been reported to stabilize the activated state (34). Notably, hydrophobic bridge residue V2426.38 structurally resides directly in the center of the microswitch functional group, suggesting that it may serve as the trigger residue that activates the microswitches.
CXCR4 residues that directly couple to G protein.
Finally, our screen also identified two highly conserved critical residues on the intracellular side of the receptor, R1343.50 and L2265.65 (Fig. 3F, purple residues). R1343.50 is part of the well-known “DRY” box motif and L2265.65 is the last residue of the Y(x)5KL motif, consistent with both residues contributing directly to G protein coupling. The homologous positions are directly involved in binding the C terminus of G protein in the crystal structures of the active state ternary complex of the β2 adrenergic receptor (β2AR) (30) and bRho (35, 36). By homology with β2AR and bRho, 15 residues on the intracellular face of CXCR4 could be involved in G protein coupling, with 9 of them either identical or similar to the corresponding residues in β2AR (SI Appendix, Fig. S3). Our screen tested mutations at all 15 CXCR4 positions (SI Appendix, Table S3), and identified two of them as critical, likely representing hotspots within an otherwise large distributed interface. Mutations at the remaining 13 positions expressed well and were conformationally folded, but did not meet our threshold criteria for impaired signaling. However, the exact set of CXCR4 residues involved in G protein coupling remains unknown, and there are likely to be significant differences in how different GPCRs bind G proteins (37).
Discussion
A Functional Model for CXCR4 Signal Transmission.
Helical movements are well known to be essential in the GPCR signal transduction process, but static structural information alone has not been able to fully distinguish the functionally critical amino acids within these helices that mediate this dynamic process. The identification of 41 critical CXCR4 residues forming a continuous intramolecular signaling chain through the TM helices enables us to propose a comprehensive, hypothesis-based functional model for signal transmission. Although many previous mutagenesis studies have been conducted on CXCR4 and other GPCRs, our results provide unbiased, full-coverage characterization of all 352 residues in the receptor, with the results derived from a single laboratory using an internally consistent set of assays and protocols that account for expression and folding of receptor variants. Overall, the critical CXCR4 activation residues identified in our study agree with common activation mechanisms proposed for class A GPCRs, such as the prominent role of helix VI and the role of various conserved motifs and microswitches.
Critical residues of CXCR4 identified here also overlap with the water-mediated polar residue network that facilitates the inactive-to-active transition in the μ-opioid receptor (35, 38). All 17 homologous residue positions were tested in our screen, 13 expressed and folded well, and 8 were identified as critical signaling residues in our assays (Y1163.32, L1273.43, Y2195.58, L2446.40, W2526.48, E2887.39, A2917.42, and Y3027.53). These findings suggest that CXCR4 also features a polar residue network that may play a role in its activation.
Six crystal structures of CXCR4 have been reported to date, all featuring a common parallel homodimer interface (8, 9). Our data suggest that dimerization involving this interface may not be important for CXCL12-induced Ca+2 mobilization in our assay system; among 11 homodimer interface residues that were mutated in our screen, only N192ECL2 and L267ECL3 had any significant effect. All mutants of the homodimer interface were properly folded and trafficked to the cell surface (SI Appendix, Table S4), also suggesting that this interface does not play a role in folding or trafficking. However, we cannot exclude the role of this dimerization interface in other processes, such as internalization or other types of CXCR4 signaling (39) or of alternative oligomerization interfaces.
Unbiased High-Throughput Screening for Function.
The functional model described here integrates our comprehensive mutational dataset with crystal structures and models of CXCR4 complexes into a cohesive model of CXCL12-mediated CXCR4 activation. Although our final model of how each residue functions is hypothesis-driven, critical residues were identified using stringent criteria: critical mutants had to express, fold, and traffic similar to WT CXCR4 yet impair signaling in response to a saturating concentration of ligand (∼3× KD). It is likely that mutations that produce more modest effects on signaling escaped detection under these criteria. Our analysis does not distinguish effects of CXCR4 mutations on CXCL12 potency vs. efficacy, but it is unlikely that further increases in CXCL12 concentrations would identify additional residues more important to CXCR4 signal transmission.
Amino acids within all known GPCR signaling motifs were identified in our studies as critical signal transmission residues. Other positions within these motifs are also likely involved, but the tested mutations at these positions did not express or fold well enough, or did not significantly affect signaling (SI Appendix, Table S5). It is also possible that CXCR4 signal transmission pathways could be different when coupled to different G proteins, when signaling through different pathways, or for constitutive activation.
The use of random and unbiased mutagenesis across all 352 residues of CXCR4 led to the identification of a number of well-expressing but signaling-deficient mutants, such as W94R2.60, D97G2.63, and E288G7.39, that would not have been possible with alanine mutations. For example, W94A2.60, D97A2.63, and E288A7.39 have been previously shown to reduce receptor expression (12, 13, 22). Substitutions to proline can impact expression and folding, but many were well tolerated. For example, L244P6.40 and L246P6.42 support the critical helical nature of TM6. However, random mutagenesis also has limits; some of the positions tested had rather extreme substitutions that affected receptor folding, whereas others had only conservative substitutions that failed to impact signaling. To compensate for the random changes, we tested 2.7 different amino acid substitutions per position on average, but testing more substitutions would likely have revealed additional critical residues.
Conclusions
An interconnected chain of residues responsible for transmitting extracellular ligand signals to intracellular G proteins is likely a conserved mechanism across all GPCRs. Thus, we expect the results of our study to be largely applicable to other GPCRs, especially for the propagation, microswitch, and G protein-coupling groups. The chemokine engagement and signal initiation groups are likely to be more ligand specific, so we expect them to be most applicable to other chemokine receptors.
Understanding the activation dynamics of GPCRs has implications for studying the effects of mutations and naturally occurring variants, for structure determination efforts, and for the development of novel therapeutic targeting strategies using allosteric ligands. Modulation of CXCR4–CXCL12 signaling, in particular, has implications for controlling cancer metastasis, preventing HIV infection, and promoting immune and stem cell trafficking. The present study provides a missing link in understanding the dynamic multistep activation mechanisms of the GPCR CXCR4.
Experimental Methods
All experiments in this project were approved by the Institutional Biosafety Committee and management of Integral Molecular. No human subject materials were used.
Preparation of CXCR4 Shotgun Mutagenesis Mutation Library.
A shotgun mutagenesis mutation library was created as previously described (40). Briefly, a parental plasmid expressing full-length human CXCR4 cDNA was constructed with an N-terminal FLAG epitope tag and a C-terminal V5 epitope tag. Using the parental cDNA construct as a template, a library of random mutations was created using PCR-based mutagenesis (Diversify PCR Random Mutagenesis Kit, Clontech). Each mutant clone was sequence verified. A complete mutation library was assembled by selection of 2.7 mutant clones per residue, spanning the entire protein, and preferably representing a conserved and nonconserved substitution at each position. A total of 551 CXCR4 variants contained single mutations and the remaining 172 clones contained mutations at two or more positions.
Calcium Flux Assay.
The FLIPR Calcium 4 Assay (Molecular Devices) was performed according to the manufacturer’s protocol, with minor modifications. The CXCR4 mutation library and controls [WT (+) and vector alone (−)] were transfected and expressed in canine Cf2Th cells (selected because they lack endogenous CXCR4) in 384-well microplate format. Twenty-four hours posttransfection, cells were washed twice in HBSS/Hepes supplemented with 10 μM indomethacin, then incubated with 1× loading dye for 1.5 h at 37 °C. The plates were transferred to a FlexStation II-384 plate reader, and wells were injected (at t = 20 s) with 20 nM (final) human recombinant CXCL12 (PeproTech), and fluorescence was measured for 70 s, reading every 3 s (Ex485/Em525).
Immunodetection Assays.
The CXCR4 mutation library was expressed in HEK-293T cells. Twenty-four hours posttransfection, cells were washed and fixed in 4% (vol/vol) paraformaldehyde, incubated with anti-FLAG M2 monoclonal antibody (Stratagene, no. 200472), or anti-CXCR4 monoclonal antibodies 12G5 (a gift of James Hoxie, University of Pennsylvania, Philadelphia), 44708, 44712, or 44716 (R&D Systems), followed by goat anti-mouse Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Microplates were measured on a NovaRay imager (Alpha Innotech).
Ligand Binding FRET Assay.
Selected FLAG-tagged CXCR4 clones and controls were arrayed in duplicate in 96-well microplates and expressed in HEK-293T cells. Twenty-four hours posttransfection, cells were washed twice in HBSS supplemented with 10 mM Hepes, then incubated for 1 h at room temperature with the donor fluorophore, an anti-FLAG (M2)-Terbium-labeled antibody (Cisbio). Following five washes in PBS and one wash in 1× ligand binding buffer, cells were incubated for 3 h at room temperature with the acceptor fluorophore solution, 60 nM Tag-lite CXCR4 receptor red agonist (L0012RED). Fluorescent-positive agonist-bound cells were detected at 665-nm emission using a Perkin-Elmer Envision 2100.
Data Analyses.
The maximum calcium flux value for each clone was calculated from the peak flux value at t = 30 s minus the trace baseline value, then background subtracted using negative control values on the same plate and normalized to express each mutant activity as a percentage of wild-type. The average (n = 5) calcium flux value for each clone was compared with a 62% cut-off threshold [100 − (2 × [SD of wild-type controls])] to identify clones that signal significantly below wild-type levels. Similarly, the immunofluorescence values for each clone were calculated from raw plate data, background subtracted, and normalized to express each mutant as a percentage of wild-type. The average FLAG (n = 5) or 12G5 (n = 3) immunofluorescence value for each clone was compared with an 80% cut-off threshold to identify clones that react with each MAb at near wild-type levels.
Structural Modeling.
A model of the complex between wild-type CXCR4 in the inactive state and CXCL12 was previously published (9). A model of the active state for this complex was obtained using gradient minimization with restraints in Molsoft ICM molecular modeling package. For that, consensus intramolecular distance changes were first obtained by comparing the active and inactive state structures for each of β2AR, AA2AR, and bRho (7, 35, 36, 41). The calculated distance changes were converted into target distances by adding them to the homologous intramolecular distances measured within the inactive CXCR4 structure and then imposed onto the model as harmonic distance restraints. An additional set of restraints was derived from the inactive state intramolecular hydrogen bonds and used to maintain the receptor secondary structure during energy minimization. The 105 steps of gradient minimization were performed using a fully flexible representation of the receptor. The final model was visually inspected for the absence of steric conflicts and for consistency of the microswitch residue rotamers with the signature of an active state as described for crystallized active state GPCRs.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R44-GM076779 (to B.J.D.), R01-GM071872 (to I.K.), and R01-AI118985, R01-GM117424, R21-AI121918, and R21-AI122211 (to I.K. and T.M.H.).
Footnotes
Conflict of interest statement: B.J.D. and J.B.R. are shareholders of Integral Molecular.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601278113/-/DCSupplemental.
References
- 1.Kufareva I, Salanga CL, Handel TM. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol Cell Biol. 2015;93(4):372–383. doi: 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debnath B, Xu S, Grande F, Garofalo A, Neamati N. Small molecule inhibitors of CXCR4. Theranostics. 2013;3(1):47–75. doi: 10.7150/thno.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr Opin Struct Biol. 2011;21(4):541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33(1):17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274(5288):768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 6.Gether U, et al. Agonists induce conformational changes in transmembrane domains III and VI of the beta2 adrenoceptor. EMBO J. 1997;16(22):6737–6747. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin L, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347(6226):1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vroling B, et al. GPCRDB: Information system for G protein-coupled receptors. Nucleic Acids Res. 2011;39(Database issue):D309–D319. doi: 10.1093/nar/gkq1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson E, Doranz BJ. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143(1):13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275(31):23736–23744. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 13.Wong RS, et al. Comparison of the potential multiple binding modes of bicyclam, monocylam, and noncyclam small-molecule CXC chemokine receptor 4 inhibitors. Mol Pharmacol. 2008;74(6):1485–1495. doi: 10.1124/mol.108.049775. [DOI] [PubMed] [Google Scholar]
- 14.Deupi X, Kobilka B. Activation of G protein-coupled receptors. Adv Protein Chem. 2007;74:137–166. doi: 10.1016/S0065-3233(07)74004-4. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann KP, et al. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem Sci. 2009;34(11):540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Crump MP, et al. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16(23):6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi WT, et al. Unique ligand binding sites on CXCR4 probed by a chemical biology approach: Implications for the design of selective human immunodeficiency virus type 1 inhibitors. J Virol. 2005;79(24):15398–15404. doi: 10.1128/JVI.79.24.15398-15404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doranz BJ, et al. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73(4):2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian S, et al. Distinct functional sites for human immunodeficiency virus type 1 and stromal cell-derived factor 1alpha on CXCR4 transmembrane helical domains. J Virol. 2005;79(20):12667–12673. doi: 10.1128/JVI.79.20.12667-12673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou N, et al. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J Biol Chem. 2001;276(46):42826–42833. doi: 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]
- 21.Kofuku Y, et al. Structural basis of the interaction between chemokine stromal cell-derived factor-1/CXCL12 and its G-protein-coupled receptor CXCR4. J Biol Chem. 2009;284(50):35240–35250. doi: 10.1074/jbc.M109.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kufareva I, et al. Stoichiometry and geometry of the CXC chemokine receptor 4 complex with CXC ligand 12: Molecular modeling and experimental validation. Proc Natl Acad Sci USA. 2014;111(50):E5363–E5372. doi: 10.1073/pnas.1417037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71(6):4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341(6152):1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenkilde MM, et al. Molecular mechanism of AMD3100 antagonism in the CXCR4 receptor: Transfer of binding site to the CXCR3 receptor. J Biol Chem. 2004;279(4):3033–3041. doi: 10.1074/jbc.M309546200. [DOI] [PubMed] [Google Scholar]
- 26.Hanes MS, et al. Dual targeting of the chemokine receptors CXCR4 and ACKR3 with novel engineered chemokines. J Biol Chem. 2015;290(37):22385–22397. doi: 10.1074/jbc.M115.675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steen A, et al. Biased and constitutive signaling in the CC-chemokine receptor CCR5 by manipulating the interface between transmembrane helices 6 and 7. J Biol Chem. 2013;288(18):12511–12521. doi: 10.1074/jbc.M112.449587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holst B, et al. A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J Biol Chem. 2010;285(6):3973–3985. doi: 10.1074/jbc.M109.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469(7329):175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greasley PJ, Fanelli F, Rossier O, Abuin L, Cotecchia S. Mutagenesis and modelling of the alpha(1b)-adrenergic receptor highlight the role of the helix 3/helix 6 interface in receptor activation. Mol Pharmacol. 2002;61(5):1025–1032. doi: 10.1124/mol.61.5.1025. [DOI] [PubMed] [Google Scholar]
- 32.Deupi X, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA. 2012;109(1):119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han X, Tachado SD, Koziel H, Boisvert WA. Leu128(3.43) (l128) and Val247(6.40) (V247) of CXCR1 are critical amino acid residues for G protein coupling and receptor activation. PLoS One. 2012;7(8):e42765. doi: 10.1371/journal.pone.0042765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goncalves JA, et al. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc Natl Acad Sci USA. 2010;107(46):19861–19866. doi: 10.1073/pnas.1009405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471(7340):656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471(7340):651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 37.Huang CC, Tesmer JJ. Recognition in the face of diversity: Interactions of heterotrimeric G proteins and G protein-coupled receptor (GPCR) kinases with activated GPCRs. J Biol Chem. 2011;286(10):7715–7721. doi: 10.1074/jbc.R109.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W, et al. Structural insights into µ-opioid receptor activation. Nature. 2015;524(7565):315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, He L, Combs CA, Roderiquez G, Norcross MA. Dimerization of CXCR4 in living malignant cells: Control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Mol Cancer Ther. 2006;5(10):2474–2483. doi: 10.1158/1535-7163.MCT-05-0261. [DOI] [PubMed] [Google Scholar]
- 40.Paes C, et al. Atomic-level mapping of antibody epitopes on a GPCR. J Am Chem Soc. 2009;131(20):6952–6954. doi: 10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332(6027):322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.