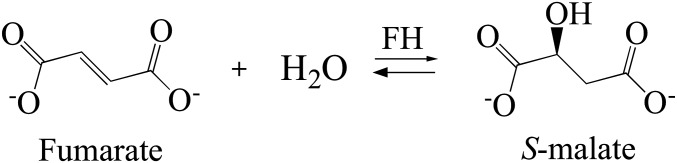Significance
Leishmaniases, Chagas disease, and sleeping sickness are parasitic diseases classified as neglected tropical diseases, affecting approximately one-sixth of the world’s population. Because of the absence of effective medicines to treat these diseases, there is a substantial interest in the identification of new targets for the development of therapeutic strategies to combat neglected tropical diseases. We have determined the crystal structure of a class I fumarate hydratase (FH) from Leishmania major, the parasite responsible for cutaneous leishmaniasis, and find that the structure is distinct from class II human FH. Thus, Leishmania class I FH, which is an essential metabolic enzyme, offers a new perspective for the development of antileishmaniases therapies.
Keywords: leishmaniases, fumarate hydratase, Fe-S cluster, X-ray crystallography
Abstract
Fumarate hydratases (FHs) are essential metabolic enzymes grouped into two classes. Here, we present the crystal structure of a class I FH, the cytosolic FH from Leishmania major, which reveals a previously undiscovered protein fold that coordinates a catalytically essential [4Fe-4S] cluster. Our 2.05 Å resolution data further reveal a dimeric architecture for this FH that resembles a heart, with each lobe comprised of two domains that are arranged around the active site. Besides the active site, where the substrate S-malate is bound bidentate to the unique iron of the [4Fe-4S] cluster, other binding pockets are found near the dimeric enzyme interface, some of which are occupied by malonate, shown here to be a weak inhibitor of this enzyme. Taken together, these data provide a framework both for investigations of the class I FH catalytic mechanism and for drug design aimed at fighting neglected tropical diseases.
Fumarate hydratase (FH; also known as fumarase; EC 4.2.1.2) catalyzes the stereospecific reversible hydration/dehydration of fumarate to S-malate (Scheme 1). Eukaryotic cells express both mitochondrial and cytosolic FH isoforms. The mitochondrial FH is well known for its participation in the tricarboxylic acid cycle and may also take part in the succinic fermentation pathway (1). The cytosolic FH is thought to produce the fumarate substrate for dihydroorotate dehydrogenase (2), an enzyme involved in the de novo pyrimidine biosynthetic pathway. In addition, cytosolic FH can migrate from the cytosol to the nucleus, where it plays a key role in DNA repair (3). Because of these vital roles, FHs are potential therapeutic targets. FHs have been grouped into two classes with no obvious amino acid sequence similarity. Class I FHs include trypanosomatids FHs (4, 5) and are [4Fe-4S] cluster-containing homodimeric enzymes. Class II FHs include human FH (6) and are Fe-independent homotetrameric enzymes. Although the FHs from class II have been structurally characterized (7), a structure of an intact class I FH has been elusive until now.
Scheme 1.
Here, we report the 2.05 Å resolution structure of a FH from Leishmania major (LmFH-2). LmFH-2, encoded by the LmjF29.1960 gene, was previously shown to be a cytosolic enzyme able to reversibly convert fumarate to S-malate with preference for S-malate production (5). Consistent with the classification as a class I FH, LmFH-2 activity assays reveal the presence of an oxygen-sensitive cofactor (5). Only a few other class I FH enzymes have been biochemically characterized, including class I FHs from Euglena gracilis var. bacillaris (8), Escherichia coli (9, 10), bacterium strain MPOB (11), and Burkholderia xenovorans (12), but the L. major enzymes LmFH-2 and LmFH-1 (the mitochondrial isoform) are the only class I FH characterized from a parasite in the Trypanosomatidae family. Trypanosomatids are responsible for neglected tropical diseases (NTD), such as leishmaniases, Chagas disease, and sleeping sickness, which infect millions of people and represent a substantial health and economic burden on developing countries. The high sequence identity shared (60–75%) by trypanosomatid FHs suggests not only structural similarity but also a similar mechanism of action. Our data reveal that LmFH-2 has a previously undiscovered protein fold, providing a structural framework both for investigations of the class I FH catalytic mechanism as well as for drug design aimed at fighting NTD.
Results and Discussion
Overall Structure of LmFH-2.
The crystal structure of LmFH-2 was solved by single-wavelength anomalous dispersion (SAD) using iron as the anomalous scatterer and refined to 2.05 Å resolution (Table S1). The asymmetric unit contains one copy of the functional homodimeric enzyme, and the monomers are related by a noncrystallographic twofold axis (Fig. 1A). The superposition of Cα atoms between chains A and B shows a high level of structural similarity with an rmsd of 0.22 Å. The quaternary structure of LmFH-2 resembles an upside-down heart (Fig. 1A), with each monomer consisting of two structural domains arranged around the catalytic [4Fe-4S] cluster (Fig. 1B).
Table S1.
Data collection and refinement statistics
| Data name | LmFH-2–SAD* | LmFH-2–native |
| Ligands | S-malate and malonate | |
| Data collection | ||
| Wavelength (Å) | 1.7399 | 0.9795 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 65.75, 85.66, 241.62 | 65.67, 85.13, 241.48 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–2.7 (2.75–2.70)† | 200–2.05 (2.09–2.05)† |
| No. of unique reflections | 36,042 | 83,691 |
| Rsym | 0.091 (0.367)† | 0.114 (0.41)† |
| I/σ(I) | 21 (3.7)† | 16.5 (2.8)† |
| Completeness (%) | 94.9 (70.2)† | 98.1 (97.9)† |
| Redundancy | 6.6 (5.1)† | 5.5 (5.4)† |
| Refinement | ||
| Resolution (Å) | 200–2.05 | |
| No. reflections | 83,691 | |
| Rwork/Rfree‡ | 0.144/0.177 | |
| No. atoms | 9,103 | |
| Protein | 8,196 | |
| Ligand | 135 | |
| Water | 772 | |
| B-factors (Å2) | 29.8 | |
| Protein | 28.9 | |
| Ligand | 36.8 | |
| Water | 38.2 | |
| Rmsd | ||
| Bond lengths (Å) | 0.010 | |
| Bond angles (°) | 1.066 |
LmFH-2–SAD and LmFH-2–native datasets were collected using two and one crystals, respectively.
Statistics are for data that were scaled anomalously.
Highest resolution shell is shown in parenthesis.
Rfree was calculated with 5% of the data.
Fig. 1.
Crystal structure of LmFH-2. (A) Overall structure of the LmFH-2 functional dimer. The Upper and Lower panels represent two orthogonal views of the structure with two domains: N terminal (blue and green) and C terminal (yellow). The Left and Right panels show the cartoon and electrostatic surface potential representation of the LmFH-2 dimer, respectively. The [4Fe-4S] clusters are shown in magenta. (B) Ribbon diagram of LmFH-2 monomer. The N-terminal domain is divided into two nonsequential subdomains 1 (light blue and dark blue) and 2 (green), and is connected to C-terminal domain (yellow) by a linker (black arrow), as indicated in the linear schematic. (C) Ribbon diagram of LmFH-2 N-terminal subdomains 1 and 2. (D) Ribbon diagram of LmFH-2 C-terminal domain.
The LmFH-2 monomer contains 23 β-strands (β1 to β23) and 18 α-helices (α1 to α18), and can be described as being composed of two domains: an N-terminal domain (Asp-28 to Pro-375) and a C-terminal domain (Thr-385 to Ala-568), connected by a flexible linker (Asp-376 to Thr-384) (Fig. 1B and Fig. S1). The first 27 residues and the flexible linker were excluded from the structure because of the lack of interpretable electron density. The N-terminal domain can be divided in subdomains 1 (Asp-28 to Lys-213 and Asp-349 to Pro-375) and 2 (Gly-214 to Ala-348) (Fig. 1C). Subdomain 1 contains 11 β-strands (β1 to β9, β15, and β16) and six α-helices (α1 to α5 and α11). Subdomain 2 is located between the β9 and β15 of subdomain 1 and contains five β-strands (β10 to β14) and five α-helices (α6 to α10). The C-terminal domain contains seven α-helices (α12 to α18) and seven β-strands (β17 to β23) organized as independent β-barrel and α-helical motifs (Fig. 1D). The monomer has a [4Fe-4S] cluster coordinated by three cysteine residues (Cys-133, Cys-252, and Cys-346; the motif is C-X118-C-X93-C) from the N-terminal domain (Fig. 2A). The fourth iron of the cluster has no protein ligand.
Fig. S1.
Topology diagram of LmFH-2. LmFH-2 is described in two domains: N- and C-terminal domains. The N-terminal domain is divided into subdomains 1 and 2 that are shown in blue and green, respectively. The C-terminal domain is shown in yellow. The topology was performed using PDBsum, a pictorial database of 3D structures in the Protein Data Bank (www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html).
Fig. 2.
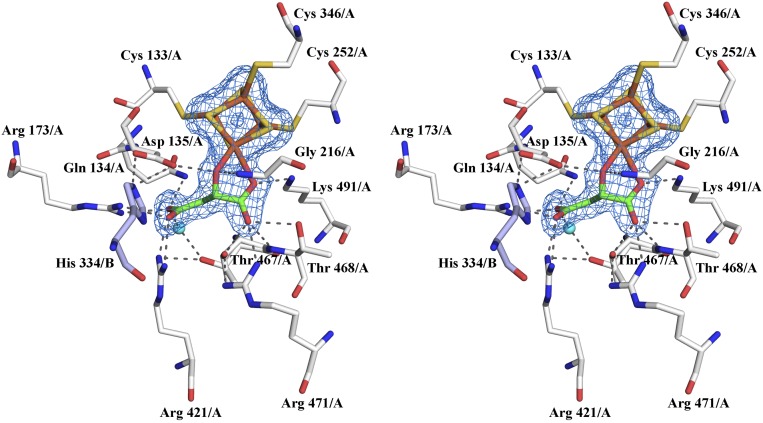
LmFH-2 active site. (A) The residues of chains A and B are shown in white and light blue, respectively. The substrate S-malate, [4Fe-4S] cluster and water molecule are shown in green, yellow (S) and orange (Fe), and cyan, respectively. Mesh represents the final 2Fo–Fc electron density map contoured at 1.5 σ level (blue) for S-malate and the [4Fe-4S] cluster. A stereoview is shown in Fig. S4. (B) Interactions between S-malate and the active site residues in LmFH-2. The water molecule, C, N, O, Fe, and S atoms are shown in cyan, black, blue, red, orange, and yellow, respectively. The hydrogen bonds are shown as green dashed lines. The distance between the OH group of Thr-467 to the S-malate C3 carbon is shown as a black dashed line. Image created with LigPlot (30). (C) Proposed mechanism for class I FHs to catalyze the dehydration of S-malate to fumarate. The first step is thought to be deprotonation of S-malate C3 by a catalytic base (B:). The closest residue to C3 is Thr-467 (3.34 Å), which is near a water molecule and two Arg residues (Arg-421 and Arg-471), any of which could accept the proton. The residue Asp-135, and its hydrogen-bonding partner His-334/B, are ideally positioned to play a role as a catalytic acid to protonate the C2 hydroxyl group of S-malate for elimination as H2O and subsequent formation of fumarate.
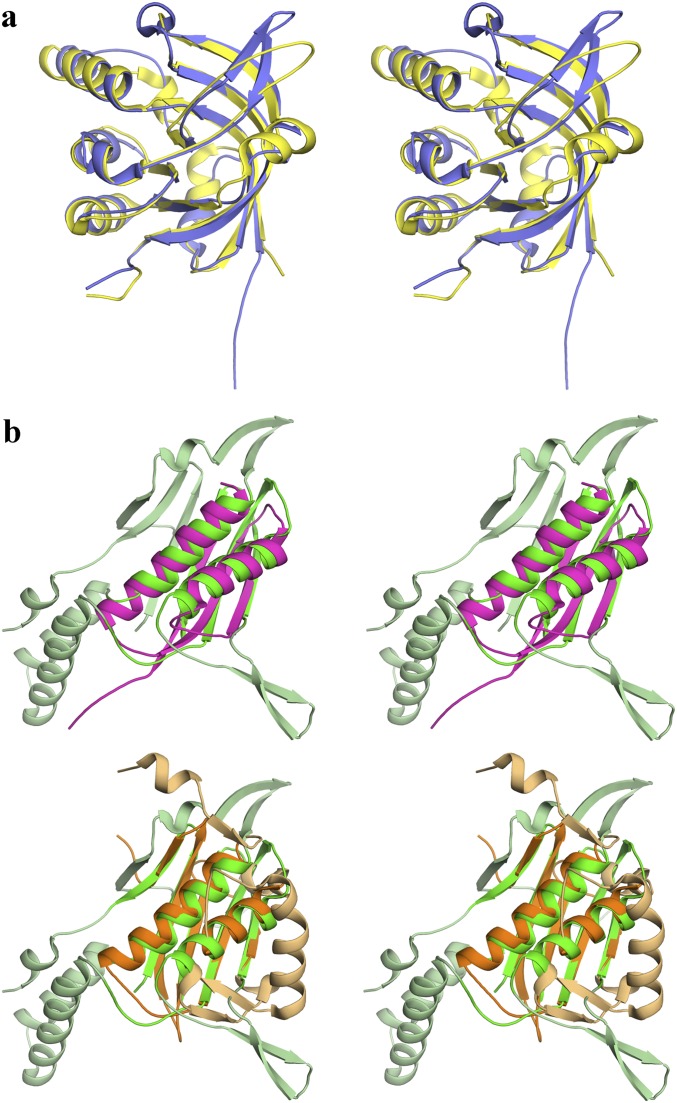
The DALI server (13) was used to search the Protein Data Bank to identify known structures with similar folds to LmFH-2. Unsurprisingly, DALI finds the β-subunit of a putative class I FH from Archaeoglobus fulgidus (PDB ID code 2ISB) as the strongest match to the C-terminal domain of LmFH-2 (z-score of 22.9). The high structural similarity is evident in the structural superposition, which has an rmsd of 2.2 Å for 172 Cα atoms (of the 183 residues comprising the C-terminal domain) (Fig. S2A). In fact, this conserved region is classified by Structural Classification of Proteins database (SCOP) (14) as a swiveling β/β/α domain, known to be a mobile motif in multidomain proteins. Although the structure of the α-subunit of this putative class I FH from A. fulgidus may turn out to be similar to the N-terminal domain of LmFH-2, it has not been determined. Moreover, the DALI server finds no close matches to the full N-terminal domain of LmFH-2. The best matches are to the Ni-binding domain of HypA from Thermococcus kodakaraensis KOD1 (PDB ID code 3A43) (15) and the N-terminal β-domain of l-serine dehydratase from Legionella pneumophila (PDB ID code 4RQO) (16), but in both cases the z-scores are low: 6 and 5.7, respectively. Z-scores below 2 indicate structural dissimilarity. Structural comparisons to the identified proteins reveal that only a fragment of LmFH-2 subdomain 1, comprised of residues from one β-sheet (β4, β8, β9, β15, and β16) and two helices (α2 and α5), is structurally similar (Fig. S2B). The rmsds between this fragment of LmFH-2 and PDB ID codes 3A43 and 4RQO models are 2.7 and 2.7 Å for 74 and 100 aligned Cα atoms, respectively. Thus, we can interpret the relatively low z-scores as being a reasonably good alignment with a small portion of the fold. According to the DALI server, no protein with a known structure shares the full α + β-fold that we find here for LmFH-2.
Fig. S2.
Structural comparison of the proteins that share similarity to N- and C-terminal domains of LmFH-2 according to the DALI server. (A) Stereoview of the superposition between LmFH-2 C-terminal domain (residues 385–568) (yellow) and full β-subunit of FH from Archaeoglobus fulgidus (blue; PDB ID code 2ISB), showing a high level of similarity. Orientation is the same as in Fig. 1D. (B) Stereoview of the superposition between subdomain 1 (residues 28–213 and 349–375) of the LmFH-2 N-terminal domain (green and light green), with (Upper) Ni-binding domain (residues 1–68) of HypA from Thermococcus kodakaraensis KOD1 (pink; PDB ID code 3A43) and (Lower) the N-terminal β-domain (residues 2–161) of l-serine dehydratase from Legionella pneumophila (orange and light orange; PDB ID code 4RQO). Secondary structure in light orange does not align with LmFH-2. Orientation is the same as in Fig. 1C. Of the 11 β-strands of the subdomain 1 of the LmFH-2 N-terminal domain, only 3 β-strands (HypA; pink) or 5 β-strands (l-serine dehydratase; orange) are similar, and of 6 helices of the subdomain 1, 2 helices from HypA (pink), and l-serine dehydratase (orange) are similar. No matches to the subdomain 2 of the LmFH-2 N-terminal domain were identified.
The dimer interface of LmFH-2 involves 32 residues of the N-terminal domain from chains A and B, and is stabilized by 42 hydrogen bonds (Table S2) and two cation–π interactions between Phe-63 and Lys-69. The great majority of residues at the interface are completely conserved in class I FHs (Fig. S3). Phe-63 and Lys-69 are not fully conserved, showing that the cation–π interaction is a unique feature of cytosolic FHs from Leishmania spp.
Table S2.
Hydrogen bonds formed between residues of chains A and B from LmFH-2
| Chain A | Chain B | ||||||||
| Hydrogen bond no. | Atom name | Residue name | Residue number | Chain | Atom name | Residue name | Residue number | Chain | Distance (Å) |
| 1 | OD1 | ASP | 135 | A | NE2 | HIS | 334 | B | 2.87 |
| 2 | NH2 | ARG | 145 | A | O | VAL | 201 | B | 3.03 |
| 3 | OE2 | GLU | 147 | A | N | THR | 151 | B | 2.77 |
| 4 | OE2 | GLU | 147 | A | N | GLY | 152 | B | 2.79 |
| 5 | N | THR | 151 | A | OE2 | GLU | 147 | B | 2.89 |
| 6 | N | GLY | 152 | A | OE2 | GLU | 147 | B | 2.81 |
| 7 | O | SER | 175 | A | N | GLY | 335 | B | 2.84 |
| 8 | N | THR | 177 | A | O | ARG | 333 | B | 2.71 |
| 9 | O | THR | 177 | A | N | ARG | 333 | B | 3.21 |
| 10 | O | MET | 182 | A | NE | ARG | 330 | B | 3.00 |
| 11 | O | MET | 182 | A | NH2 | ARG | 330 | B | 3.01 |
| 12 | OE1 | GLU | 185 | A | NZ | LYS | 228 | B | 3.30 |
| 13 | OE1 | GLU | 185 | A | NH2 | ARG | 333 | B | 2.60 |
| 14 | OE2 | GLU | 185 | A | NH2 | ARG | 330 | B | 3.00 |
| 15 | OE2 | GLU | 185 | A | NE | ARG | 333 | B | 2.73 |
| 16 | OE1 | GLN | 195 | A | OG1 | THR | 264 | B | 2.66 |
| 17 | OE1 | GLN | 195 | A | OG | SER | 265 | B | 2.64 |
| 18 | NE2 | GLN | 195 | A | OG | SER | 265 | B | 3.35 |
| 19 | O | LEU | 198 | A | N | ALA | 200 | B | 2.89 |
| 20 | N | ALA | 200 | A | O | LEU | 198 | B | 2.81 |
| 21 | O | VAL | 201 | A | NH2 | ARG | 145 | B | 3.03 |
| 22 | OD1 | ASN | 219 | A | OG | SER | 337 | B | 2.82 |
| 23 | ND2 | ASN | 219 | A | O | ALA | 336 | B | 2.86 |
| 24 | O | ALA | 221 | A | NE2 | GLN | 225 | B | 2.98 |
| 25 | NE2 | GLN | 225 | A | O | ALA | 221 | B | 2.87 |
| 26 | OG1 | THR | 264 | A | OE1 | GLN | 195 | B | 2.61 |
| 27 | OG | SER | 265 | A | OE1 | GLN | 195 | B | 2.74 |
| 28 | OE2 | GLU | 267 | A | NZ | LYS | 274 | B | 2.84 |
| 29 | NZ | LYS | 274 | A | OE2 | GLU | 267 | B | 2.88 |
| 30 | N | GLY | 288 | A | O | LEU | 180 | B | 3.28 |
| 31 | NE | ARG | 330 | A | O | MET | 182 | B | 2.99 |
| 32 | NH2 | ARG | 330 | A | O | MET | 182 | B | 3.18 |
| 33 | NH2 | ARG | 330 | A | OE2 | GLU | 185 | B | 2.79 |
| 34 | N | ARG | 333 | A | O | THR | 177 | B | 3.23 |
| 35 | O | ARG | 333 | A | N | THR | 177 | B | 2.79 |
| 36 | NE | ARG | 333 | A | OE2 | GLU | 185 | B | 2.88 |
| 37 | NH2 | ARG | 333 | A | OE1 | GLU | 185 | B | 2.85 |
| 38 | NE2 | HIS | 334 | A | OD1 | ASP | 135 | B | 2.70 |
| 39 | N | GLY | 335 | A | O | SER | 175 | B | 2.89 |
| 40 | O | ALA | 336 | A | ND2 | ASN | 219 | B | 2.76 |
| 41 | OG | SER | 337 | A | O | GLY | 215 | B | 3.27 |
| 42 | OG | SER | 337 | A | OD1 | ASN | 219 | B | 2.72 |
Fig. S3.
Sequence alignment of class I FHs. LmFH-2 (cytosolic isoform of L. major), LtFH-2 (cytosolic isoform of Leishmania tropica), LdFH-2 (cytosolic isoform of Leishmania donovani), LiFH-2 (cytosolic isoform of Leishmania infantum), LbFH-2 (cytosolic isoform of Leishmania braziliensis), TcFH-2 (cytosolic isoform of Trypanosoma cruzi), TbFH-2 (cytosolic isoform of Trypanosoma brucei), TvFH-2 (cytosolic isoform of Trypanosoma vivax), SpFH (Strongylocentrotus purpuratus), SmFH (Schistosoma mansoni), SjFH (Schistosoma japonicum), AcFH (Acanthamoeba castellanii str. Neff), EcFH-A (fum A of E. coli), SeFH (Salmonella enterica), EnterobacterFH (Enterobacter sp. SST3), SbFH (Shigella boydii), YpFH (Yersinia pseudotuberculosis IP 31758), RhodococcusFH (Rhodococcus sp. P14), LmFH-1 (mitochondrial isoform of L. major), LiFH-1 (mitochondrial isoform of L. infantum), LmxFH-1 (mitochondrial isoform of Leishmania mexicana), LbFH-1 (mitochondrial isoform of L. braziliensis), TbFH-1 (mitochondrial isoform of T. brucei), TcongoFH-1 (mitochondrial isoform of Trypanosoma congolense), TvFH-1 (mitochondrial isoform of T. vivax), TcFH-1 (mitochondrial isoform of T. cruzi). The conserved residues are indicated in the blue boxes. The similar residues are indicated in blue texts. The conserved active site residues among class I FHs are indicated in the pink boxes. The three conserved cysteine residues, which are shown to bind a [4Fe-4S] cluster, are indicated in the yellow boxes. The conserved dimer interface residues among class I FHs are indicated in green circles. The alignment was performed using MULTALIN (31) and graphically displayed using ESPript (32).
Active Site of LmFH-2.
The substrate S-malate is clearly identified in both active sites of the LmFH-2 structure and it is found coordinated to the unique iron (Fe4) of the [4Fe-4S] cluster via C2 carboxyl and hydroxyl oxygen atoms, as observed by the final σA-weighted 2Fo–Fc map (Fig. 2A and Fig. S4). Notably, aconitase, a [4Fe-4S] cluster-containing enzyme that catalyzes the stereospecific dehydration/rehydration of citrate to isocitrate via cis-aconitate, also coordinates its substrate isocitrate via a carboxyl and hydroxyl oxygen to the unique Fe atom of its [4Fe-4S] (17). The active site of LmFH-2 is located in a deep cleft formed between the N- and C-terminal domains, and comprises the [4Fe-4S] cluster, a water molecule, and 12 residues from chain A and 1 residue from chain B, suggesting that the dimerization can be important for enzyme activity (Fig. 2 A and B). The N-terminal domain of chain A provides seven residues (Cys-133, Gln-134, Asp-135, Arg-173, Gly-216, Cys-252, Cys-346), with only Gly-216 located in a helix (α6), whereas all of the other residues are found in loops. The C-terminal domain of chain A provides five residues (Arg-421, Thr-467, Thr-468, Arg-471, Lys-491), with Arg-421 and Arg-471 located in α13 and α15, respectively, and other residues in loops. The N-terminal domain of chain B provides the His-334 that is located in a loop and is not directly involved in substrate binding. Sequence comparisons with 26 members of the class I FHs indicate that active site residues are fully conserved (Fig. S3).
Fig. S4.
Stereoview of LmFH-2 active site. The residues of chains A and B are shown in white and light blue, respectively. The substrate S-malate, [4Fe-4S] cluster, and water molecule are shown in green, yellow (S) and orange (Fe), and cyan, respectively. Mesh represents the final 2Fo–Fc electron density map contoured at 1.5 σ level (blue) for S-malate and [4Fe-4S] cluster.
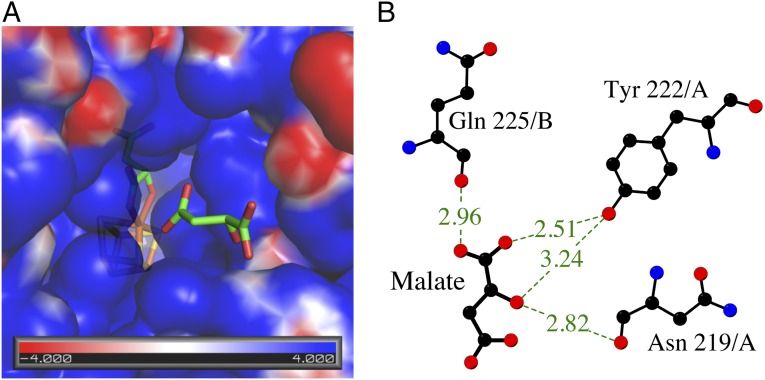
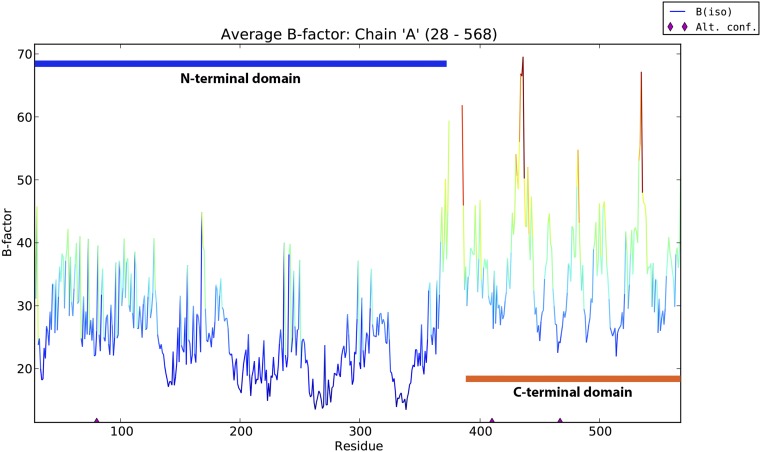
The electrostatic surface potential of LmFH-2 dimer reveals a positively charged cavity located at the interface between N- and C-terminal domains from each monomer (Fig. 1A). This cavity, which leads to the active site, contains two S-malate molecules (Fig. 3A), suggesting that access to the active site is favored by the charge distribution within this region. The residues Asn-219, Gln-225, and Tyr-222 coordinate to S-malate in this positive cavity (Fig. 3B), and sequence comparison indicates that only Tyr-222 is not conserved in class I FHs (Fig. S3). Movement of the “swiveling” C-terminal domain, which has higher B-factors than average for the rest of the structure (Fig. S5), may also regulate access to the active site.
Fig. 3.
Substrate access to the active site of LmFH-2. (A) Electrostatic surface potential of the substrate-binding pocket showing the access of another S-malate (green) to the active site. The [4Fe-4S] cluster is shown in yellow (S) and orange (Fe). (B) Interactions between the second molecule of S-malate found in the positive cavity and N-terminal domain residues. The hydrogen bonds are shown as dashed lines. Image created with LigPlot (30).
Fig. S5.
B-factor versus number of residues of LmFH-2 chain A. The blue and orange bars represent the residues of N- and C-terminal domains, respectively. The graphic was generated by phenix.pdbtools (26).
Experimental evidence suggests that class I FHs catalyze the dehydration of S-malate to fumarate by a carbanion intermediate (E1cB) mechanism (Fig. 2C), with the [4Fe-4S] cluster acting as a Lewis acid (9). The first step is thought to be deprotonation at C3 to form the carbanion, and surprisingly we find that a Thr (Thr-467) is the closest residue to C3, with the distance from the OH group of Thr to the substrate C3 carbon of 3.34 Å (Fig. 2B). Although the hydroxide group of Thr would not be expected to be deprotonated, this OH group is close to a water molecule and to two Arg residues (Arg-421 and Arg-471), any of which could accept the proton. The reaction also requires a catalytic acid to protonate the C2 hydroxyl group of S-malate for elimination as H2O and subsequent formation of fumarate. Here, Asp-135 and its hydrogen-bonding partner, His-334/B, are ideally positioned to play this role.
The structure of class II FHs is well known to be a homotetramer, with each monomer consisting of three domains, and its active site is formed by residues of three monomers (7), which is different from that found in LmFH-2. In addition, class II FH performs the reaction without a [4Fe-4S] cluster as a cofactor. Because of these differences in structure and cofactor use, along with their essential metabolic roles, class I FHs appear to represent an excellent target for structure-based drug design against NTD.
LmFH-2 Tunnel as a Ligand-Binding Site.
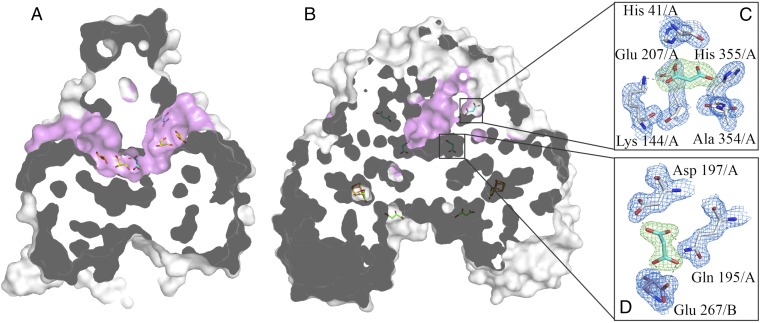
Interestingly, the dimerization of LmFH-2 reveals a deep cavity on the top of the protein formed between N-terminal domains from both chains (Fig. 4 and Movie S1). This cavity is ∼15 Å from the [4Fe-4S] cluster (Fig. 4B), and does not connect to the active site cavity described above, which is at the bottom of the structure. This “top” cavity generates a tunnel that goes through the entire protein and exhibits a volume of 1,168.5 Å3 (Fig. 4A). Although tunnels have been observed in a number of enzyme structures, they commonly provide passage from the protein surface to a buried active site (18) or provide a route from one active site to another to protect a highly reactive (19) or very valuable (20) reaction intermediate. None of these functions would seem to apply here. Interestingly, analysis of the difference electron density map in this tunnel reveals the presence of ligands from the crystallization condition, such as polyethylene glycol and malonate, as well as glycerol.
Fig. 4.
LmFH-2 cavities. (A) View of the tunnel (pink), formed between the N-terminal domains, which goes through the entire protein. The ligands polyethylene glycol (orange), glycerol (yellow), and malonate (cyan) are found in this tunnel. (B) View of the dimer surface, showing the entrance of the tunnel (pink) and the binding pockets of malonate (cyan), S-malate (green), and [4Fe-4S] cluster [yellow (S) and orange (Fe)]. (C and D) The interactions between malonate (cyan) and the residues of the binding pocket on the top of protein and in the dimer interface, respectively. Blue mesh represents the final 2Fo–Fc electron density map contoured at 1.5 σ level for residues of LmFH-2. Green mesh represents the Fo–Fc difference electron density map contoured at 3.0 σ level for malonate.
Malonate is a known inhibitor of porcine class II FH, although not a strong one, with a Ki of 40 mM (21), and has been observed bound in the active site of class II FH from Rickettsia prowazekii (22). Given that inhibition of class I FHs by malonate has not been examined previously, we investigated its ability to inhibit the L. major class I FH. Our results demonstrate that malonate is a weak inhibitor of LmFH-2, with an IC50 of 9.8 ± 0.3 mM against S-malate and 5.6 ± 0.3 mM against fumarate (Fig. S6). Consistent with this finding, we observe S-malate and not malonate bound in the active site when both are present in the crystallization buffer. Instead of being bound in the active site, malonate occupies two different types of pockets within the “top” cavity (Fig. 4B), one near the tunnel entrance (Fig. 4C) and the other at the tunnel center near the dimer interface (Fig. 4D). The residues His-41, Lys-144, Glu-207, Ala-354, and His-355 coordinate to malonate at a pocket near the tunnel entrance, and sequence comparison indicates that those residues are not conserved within class I FHs. However, the malonate bound near the dimer interface is coordinated by residues Gln-195/A, Asp-197/A, and Glu-267/B, where Gln-195 and Glu-267 are conserved and Asp-197 is either Asp or Glu. Although it is unclear if this interface binding site is the source of the weak inhibition by malonate, the interface site does seem to be a conserved small molecule binding site at an interesting position in the structure.
Fig. S6.
Determination of IC50 values for malonate. The enzymatic activity was determined under anaerobic conditions in an MBraun glovebox by measuring the fumarate production or consumption at 250 nm, using 0–120 mM malonate in 50 mM Tris, pH 9, 150 mM NaCl, 0.5 mM fumarate (A) or 2 mM S-malate (B). The reaction was initiated by the addition of the LmFH-2 at 10 μg/mL. IC50 values were determined by fitting the data to the equation v = V0/[1 + (I/IC50)], where v is the observed velocity, V0 is the uninhibited velocity, I is the inhibitor concentration, and IC50 is the 50% inhibitory concentration.
Conclusion
The crystal structure of LmFH-2 represents an important step toward the validation of the metabolically essential class I FH enzymes as targets against leishmaniasis, Chagas disease, and sleeping sickness. We are excited to find that this class I FH has a fold that shares no resemblance to mammalian class II FHs or to any other protein of known structure, thus representing a structurally unique drug target. With a structure of class I FH finally in hand, the catalytic mechanism can be interrogated and designing a structure-based drug to combat these NTDs can begin.
Materials and Methods
Crystallization and Data Collection.
Recombinant LmFH-2 was expressed in E. coli T7 express and purified by nickel affinity chromatography, as described previously (5). The purification of LmFH-2 was performed with 1 mM DTT in all buffers at 4 °C in an MBraun anaerobic glovebox. Initial LmFH-2 crystallization conditions were identified using a Mosquito robot (TTP Labtech) in a room-temperature MBraun anaerobic glovebox, and optimized using a hanging-drop vapor-diffusion method at room temperature in a Coy anaerobic chamber. Drops were prepared by mixing 1 μL of protein solution (8.7–10 mg/mL in 50 mM Tris, pH 8.5, 150 mM NaCl, 1 mM DTT) and 1 μL of reservoir solution [2–4% (vol/vol) tacsimate, pH 5 (Hampton Research), 12–14% (vol/vol) polyethylene glycol (PEG) 3,350 (Hampton Research)], equilibrated against 400 μL of reservoir solution. Tacsimate is composed of a mixture of titrated organic acid salts (23), containing the substrate S-malate and the inhibitor malonate, which resulted in a structure with both molecules bound without further addition of these molecules to the crystallization buffer. After 1 d, brownish needle cluster-like crystals were obtained. The optimization of the crystals was performed using microseeding techniques (24) and ethanol [2.7% (vol/vol)] as an additive. The crystals were transferred to a cryoprotectant solution [8% (vol/vol) tacsimate, pH 5, 18% (vol/vol) PEG 3,350, 25% (vol/vol) glycerol], and flash-cooled in liquid nitrogen in the Coy chamber. Data collection was performed by an inverse-beam method (Friedel mates were measured rotating the crystal 180° every 15 frames with 1° oscillation and exposure time of 1 s) at the 24-ID-C beamline of the Advanced Photon Source. Diffraction data were processed and scaled using HKL2000 (25). The statistics are summarized in Table S1.
Structure Determination and Refinement.
The crystal structure of LmFH-2 was solved by iron-SAD. The positions of the one sulfur and eight iron sites per functional dimer were determined and refined using phenix.autosol (26). A partial model was built in Coot (27) using a 4 Å resolution experimental map with a figure of merit of 0.488, followed by model building using phenix.autosol to 2.7 Å resolution and figure of merit of 0.368. Native data from 200 to 2.05 Å were used for structure refinement and iterative rounds of model building and addition of water molecules in Coot (27). Refinement in phenix.refine (26) used noncrystallographic symmetry restraints, TLS (translation, libration, and screw), and positional and individual B-factor refinement, with S-malate and malonate geometry restraints generated by phenix.elbow (26). The Ramachandran statistics are 97.4% in the most favored region and 2.6% in the allowed region. The final model contains 2 polypeptide chains, 2 [4Fe-4S] clusters, 4 S-malate molecules, 4 malonate molecules, 8 glycerol molecules, 1 PEG molecule, and 772 water molecules (Table S1). Figures were created with PyMol Software (28). The electrostatic surface potentials were calculated using the Adaptive Poisson-Boltzmann Solver (29) plugin implemented in PyMol, using default parameters.
Supplementary Material
Acknowledgments
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by NIH-Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. This study was supported in part by Fundação de Amparo a Pesquisa do Estado de São Paulo Grants 2009/10454-3 and 2011/19674-6 (to P.R.F.) and Grant 2008/08262-6 (to M.C.N.). C.L.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5L2R).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605031113/-/DCSupplemental.
References
- 1.Coustou V, et al. A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J Biol Chem. 2005;280(17):16559–16570. doi: 10.1074/jbc.M500343200. [DOI] [PubMed] [Google Scholar]
- 2.Cordeiro AT, Feliciano PR, Pinheiro MP, Nonato MC. Crystal structure of dihydroorotate dehydrogenase from Leishmania major. Biochimie. 2012;94(8):1739–1748. doi: 10.1016/j.biochi.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Yogev O, et al. Fumarase: A mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8(3):e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coustou V, et al. Fumarate is an essential intermediary metabolite produced by the procyclic Trypanosoma brucei. J Biol Chem. 2006;281(37):26832–26846. doi: 10.1074/jbc.M601377200. [DOI] [PubMed] [Google Scholar]
- 5.Feliciano PR, et al. Fumarate hydratase isoforms of Leishmania major: Subcellular localization, structural and kinetic properties. Int J Biol Macromol. 2012;51(1-2):25–31. doi: 10.1016/j.ijbiomac.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Kinsella BT, Doonan S. Nucleotide sequence of a cDNA coding for mitochondrial fumarase from human liver. Biosci Rep. 1986;6(10):921–929. doi: 10.1007/BF01116247. [DOI] [PubMed] [Google Scholar]
- 7.Weaver TM, Levitt DG, Donnelly MI, Stevens PPW, Banaszak LJ. The multisubunit active site of fumarase C from Escherichia coli. Nat Struct Biol. 1995;2(8):654–662. doi: 10.1038/nsb0895-654. [DOI] [PubMed] [Google Scholar]
- 8.Shibata H, Gardiner WE, Schwartzbach SD. Purification, characterization, and immunological properties of fumarase from Euglena gracilis var. bacillaris. J Bacteriol. 1985;164(2):762–768. doi: 10.1128/jb.164.2.762-768.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint DH. Initial kinetic and mechanistic characterization of Escherichia coli fumarase A. Arch Biochem Biophys. 1994;311(2):509–516. doi: 10.1006/abbi.1994.1269. [DOI] [PubMed] [Google Scholar]
- 10.van Vugt-Lussenburg BMA, van der Weel L, Hagen WR, Hagedoorn P-L. Biochemical similarities and differences between the catalytic [4Fe-4S] cluster containing fumarases FumA and FumB from Escherichia coli. PLoS One. 2013;8(2):e55549. doi: 10.1371/journal.pone.0055549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Kuijk BLM, Van Loo ND, Arendsen AF, Hagen WR, Stams AJM. Purification and characterization of fumarase from the syntrophic propionate-oxidizing bacterium strain MPOB. Arch Microbiol. 1996;165(2):126–131. doi: 10.1007/s002030050307. [DOI] [PubMed] [Google Scholar]
- 12.Kronen M, Sasikaran J, Berg IA. Mesaconase activity of class I fumarase contributes to mesaconate utilization by Burkholderia xenovorans. Appl Environ Microbiol. 2015;81(16):5632–5638. doi: 10.1128/AEM.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe S, et al. Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation. J Mol Biol. 2009;394(3):448–459. doi: 10.1016/j.jmb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 16.Thoden JB, Holden HM, Grant GA. Structure of L-serine dehydratase from Legionella pneumophila: Novel use of the C-terminal cysteine as an intrinsic competitive inhibitor. Biochemistry. 2014;53(48):7615–7624. doi: 10.1021/bi501253w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauble H, Kennedy MC, Beinert H, Stout CD. Crystal structures of aconitase with isocitrate and nitroisocitrate bound. Biochemistry. 1992;31(10):2735–2748. doi: 10.1021/bi00125a014. [DOI] [PubMed] [Google Scholar]
- 18.Pravda L, et al. Anatomy of enzyme channels. BMC Bioinformatics. 2014;15:379. doi: 10.1186/s12859-014-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raushel FM, Thoden JB, Holden HM. Enzymes with molecular tunnels. Acc Chem Res. 2003;36(7):539–548. doi: 10.1021/ar020047k. [DOI] [PubMed] [Google Scholar]
- 20.Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL. Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry. 2008;47(11):3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massey V. Studies on fumarase. 4. The effects of inhibitors on fumarase activity. Biochem J. 1953;55(1):172–177. doi: 10.1042/bj0550172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan I, et al. Structure of fumarate hydratase from Rickettsia prowazekii, the agent of typhus and suspected relative of the mitochondria. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 9):1123–1128. doi: 10.1107/S174430911102690X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McPherson A, Cudney B. Searching for silver bullets: An alternative strategy for crystallizing macromolecules. J Struct Biol. 2006;156(3):387–406. doi: 10.1016/j.jsb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Luft JR, DeTitta GT. A method to produce microseed stock for use in the crystallization of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 5):988–993. doi: 10.1107/s0907444999002085. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schrödinger, LCC (2016) The PyMOL Molecular Graphics System, Version 1.7.6. Available at www.pymol.org.
- 29.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 31.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouet P, Courcelle E, Stuart DI, Métoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.