In this study, Reid et al. investigate how cancer cells adapt to low glutamine conditions, which is needed for cancer cell proliferation and survival. They show that IKKβ directly interacts with and phosphorylates PFKFB3, a major driver of aerobic glycolysis, at Ser269 upon glutamine deprivation to inhibit its activity, thereby down-regulating aerobic glycolysis when glutamine levels are low and thus providing new insights into cancer cell adaptation.
Keywords: metabolic stress, aerobic glycolysis, glutamine, IKKβ, PFKFB3
Abstract
Glutamine is an essential nutrient for cancer cell survival and proliferation. Enhanced utilization of glutamine often depletes its local supply, yet how cancer cells adapt to low glutamine conditions is largely unknown. Here, we report that IκB kinase β (IKKβ) is activated upon glutamine deprivation and is required for cell survival independently of NF-κB transcription. We demonstrate that IKKβ directly interacts with and phosphorylates 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase isoform 3 (PFKFB3), a major driver of aerobic glycolysis, at Ser269 upon glutamine deprivation to inhibit its activity, thereby down-regulating aerobic glycolysis when glutamine levels are low. Thus, due to lack of inhibition of PFKFB3, IKKβ-deficient cells exhibit elevated aerobic glycolysis and lactate production, leading to less glucose carbons contributing to tricarboxylic acid (TCA) cycle intermediates and the pentose phosphate pathway, which results in increased glutamine dependence for both TCA cycle intermediates and reactive oxygen species suppression. Therefore, coinhibition of IKKβ and glutamine metabolism results in dramatic synergistic killing of cancer cells both in vitro and in vivo. In all, our results uncover a previously unidentified role of IKKβ in regulating glycolysis, sensing low-glutamine-induced metabolic stress, and promoting cellular adaptation to nutrient availability.
Driven by oncogenic signaling, cancer cells rapidly take up glucose and glutamine to support the bioenergetic demands of proliferation (Gao et al. 2009; Boroughs and DeBerardinis 2015). In contrast to normal cells, cancer cells divert glucose away from the tricarboxylic acid (TCA) cycle by converting a majority of their glucose-derived pyruvate to lactate, a phenomenon known as the Warburg effect or aerobic glycolysis (Lunt and Vander Heiden 2011). Because of this, cancer cells rely on glutamine to replenish TCA cycle intermediates through anaplerosis. Glutamine is metabolized to glutamate by glutaminase, which enters the mitochondria and is converted to α-ketoglutarate before continuing into a truncated TCA cycle (DeBerardinis et al. 2008). Besides replenishing TCA cycle intermediates, glutamine is important for highly proliferative cells, as it donates nitrogen to maintain pools of nonessential amino acids and also is a precursor for glutathione (GSH), a major cellular antioxidant (DeBerardinis et al. 2007). As aerobic glycolysis increases, more glutamine-derived GSH is produced to combat reactive oxygen species (ROS) generated by enhanced metabolism and proliferation (Le et al. 2012).
As tumors grow, increased glutamine demand and glutamine metabolism often leads to depletion of local supplies (Roberts and Frankel 1949; Kamphorst et al. 2015). This causes periods of glutamine deprivation as cancer cells wait for new vasculature to develop. Surprisingly, cancer cells are able to survive glutamine-restricted environments, which suggests that tumors have developed adaptations to this metabolic stress. We recently reported a cell cycle arrest checkpoint used by tumor cells in response to low glutamine levels, but other mechanisms may also contribute to cell survival in this context, such as alternative metabolism of available nutrients (Reid et al. 2013). For example, a recent report showed that, in highly glycolytic cells, the majority of TCA cycle carbons is derived from glutamine, but, when aerobic glycolysis was reduced, less glutamine contributed to TCA cycle intermediates, which decreased glutamine dependence (Le et al. 2012). Of particular interest, glutamine starvation reduces glycolytic flux; however, the molecular mechanisms mediating this remain unknown (Wong et al. 2004). Thus, inhibition of glycolytic flux may decrease glutamine dependence and contribute to cell survival under low-glutamine conditions.
The IκB kinase β (IKKβ) is well studied as a master regulator of cell survival and the inflammatory response (Comb et al. 2011, 2012; Oeckinghaus et al. 2011). Upon stimulation, IKKβ phosphorylates the IκB proteins, leading to their degradation and subsequent activation of NF-κB transcription factors (Oeckinghaus et al. 2011). Besides initiating NF-κB transcriptional activity, IKKβ directly phosphorylates numerous other substrates, such as TSC1, BAD, p85α, β-Catenin, and IRF5 kinase (Lamberti et al. 2001; Lee et al. 2007; Comb et al. 2012; Yan et al. 2013; Ren et al. 2014). Recently, IKKβ has emerged as a sensor of oxidative and metabolic stress. For example, IKKβ is activated by ROS and contributes to cell survival (Storz and Toker 2003). In addition, IKKβ–NF-κB promotes cell survival upon glucose starvation via up-regulation of mitochondrial respiration (Mauro et al. 2011) and also modulates glucose metabolism through expression of GLUT3 (Kawauchi et al. 2008). Furthermore, acute leucine starvation activates IKKβ, which phosphorylates and inhibits the p85α subunit of PI3K, leading to a downstream adaptation that is independent of NF-κB transcription (Comb et al. 2012). Moreover, multiple inducers of autophagy, including nutrient depletion, trigger the activation of IKKβ (Criollo et al. 2010). However, whether IKKβ is activated in response to prolonged glutamine deprivation, which is a major metabolic stress that elevates cellular ROS levels, has not been explored.
Isoform 3 of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB3) is overexpressed in numerous cancers and has emerged recently as a driving force of aerobic glycolysis (Atsumi et al. 2002; Ros et al. 2012). PFKFBs facilitate the conversion of fructose-6-phosphate (F-6-P) to fructose-2,6-bisphosphate (F-2,6-BP) as well as the reverse reaction. PFKFB3 has the highest kinase to phosphatase ratio of the PFKFB family, favoring F-2,6-BP production 700-fold (Sakakibara et al. 1997). Mechanistically, F-2,6-BP allosterically activates phosphofructokinase 1 (PFK1), which promotes the conversion of F-6-P to fructose-1,6-bisphosphate (F-1,6-BP), a rate-limiting step in glycolysis (Van Schaftingen et al. 1980). PFK1 is allosterically inhibited by downstream glycolytic products phosphoenolpyruvate and ATP, but PFKFB3-produced F-2,6-BP can override this inhibition, thus increasing the glycolytic capacity of the cell (Van Schaftingen et al. 1981). Consistent with the fact that PFKFB3 promotes the Warburg effect, inhibition of PFKFB3 results in decreased glycolytic activity and lactate production (Calvo et al. 2006; Schoors et al. 2014).
In this study, we found that IKKβ activity is enhanced under glutamine starvation, and its activation is essential for cell survival under low glutamine. Using a proteomics and biochemical approach, we found that IKKβ phosphorylates PFKFB3 at Ser269, which decreases its activity. Consistently, IKKβ knockdown leads to increased PFKFB3 activity, which causes enhanced aerobic glycolysis, lactate production, and glutamine dependence. Using tracer-labeling experiments, we demonstrated that IKKβ-deficient cells convert less of their glucose into TCA cycle intermediates yet have more overall intermediates due to increased glutamine uptake. Consequently, IKKβ-deficient cells are more sensitive to glutamine depletion, and combinatorial targeting of IKKβ and glutamine metabolism dramatically sensitizes cancer cells to glutaminase inhibitor treatment both in vitro and in vivo.
Results
IKKβ is required for cell survival upon glutamine deprivation, independent of NF-κB transcription
Recently, IKKβ has emerged as a sensor of oxidative and metabolic stress (Storz and Toker 2003; Criollo et al. 2010). To investigate whether glutamine deprivation, which is a major metabolic stress that elevates cellular ROS levels (Reid et al. 2013), can lead to the activation of IKKβ, 293T cells were transiently transfected with Flag-tagged IKKβ and cultured in complete medium or glutamine-free medium for 24 h followed by immunoprecipitation with anti-Flag conjugated agarose. TNFα treatment (15 min) was used as a positive control. We found that IKKβ is phosphorylated at Ser177 in complete medium conditions, and glutamine starvation led to robust IKKβ Ser177 phosphorylation, similar to TNFα treatment (Fig. 1A). Moreover, we observed similar IKKβ phosphorylation in HeLa cells upon glutamine deprivation (Supplemental Fig. S1A). Interestingly, no phosphorylation was found in IKKα purified by anti-Flag-conjugated beads when glutamine levels were depleted (Supplemental Fig. S1B). Because IKKβ transautophosphorylates (Polley et al. 2013), we used the same conditions and subjected Flag-IKKβ immunoprecipitates to a 32P-ATP in vitro kinase assay and again found that IKKβ is phosphorylated in complete medium and is further activated in response to glutamine deprivation (Fig. 1B). It is well established that IKKβ is activated in response to ROS accumulation (Schoonbroodt et al. 2000; Storz and Toker 2003; Takada et al. 2003; Storz et al. 2004; Gloire et al. 2006), and we reported that glutamine deprivation leads to increased cellular ROS (Reid et al. 2013). Thus, in order to determine whether low-glutamine-induced IKKβ activation was ROS-dependent, cells cultured in low glutamine were supplemented with antioxidant N-acetyl-L-cysteine (NAC), which was sufficient to prevent IKKβ phosphorylation upon glutamine starvation (Fig. 1C). We next examined the activation of the IKKβ signaling pathway upon glutamine limitation in mouse embryonic fibroblasts (MEFs) and HT1080 cells. In addition to IKKβ phosphorylation, we observed p65 phosphorylation and IκB degradation, demonstrating activation of IKKβ under glutamine starvation and confirming the results found using our ectopic system (Fig. 1D; Supplemental Fig. S1C). Because tumor cells often experience periods of low glutamine in vivo, we next asked whether IKKβ was important for cell survival under this metabolic stress condition. To test this, we starved wild-type, Ikkα−/−, and Ikkβ−/− MEFs of glutamine and measured cell viability over time using propidium iodide exclusion flow cytometry. Strikingly, Ikkβ−/− MEFs were much more sensitive to glutamine deprivation (Fig. 1E). Similarly, we transiently knocked down IKKβ using siRNA in human sarcoma HT1080 cells and found that IKKβ-deficient cells were also more sensitive to low glutamine (Fig. 1F). Because IKKβ is a master regulator of NF-κB and because p65 plays an important role in low-glucose cell survival (Mauro et al. 2011), we examined whether NF-κB-deficient cells displayed sensitivity to glutamine starvation but, surprisingly, found that p65−/− MEFs and p65 knockdown HT1080 cells displayed no difference in viability upon glutamine starvation compared with controls (Fig. 1G,H). To further investigate the activity of p65 under low-glutamine conditions, we used a luciferase-based p65 response element reporter construct. To our surprise, even though p65 was phosphorylated, we found no significant increase in p65 transactivation in 293T cells or MEFs upon glutamine deprivation but did observe p65 transcriptional activity upon glucose starvation and TNFα treatment (Supplemental Fig. S1D). Since p65 is not the only NF-κB transcription factor, we overexpressed the superrepressive form of IκBα, a dominant-negative mutant that prevents activity of all NF-κB family members, in wild-type MEFs using retrovirus (Lin et al. 1996) and found that these cells displayed viability similar to that of wild-type MEFs under glutamine starvation (Fig. 1I). Taken together, these data suggest that low-glutamine-induced ROS accumulation activates IKKβ to promote cell survival independent of NF-κB transcription.
Figure 1.
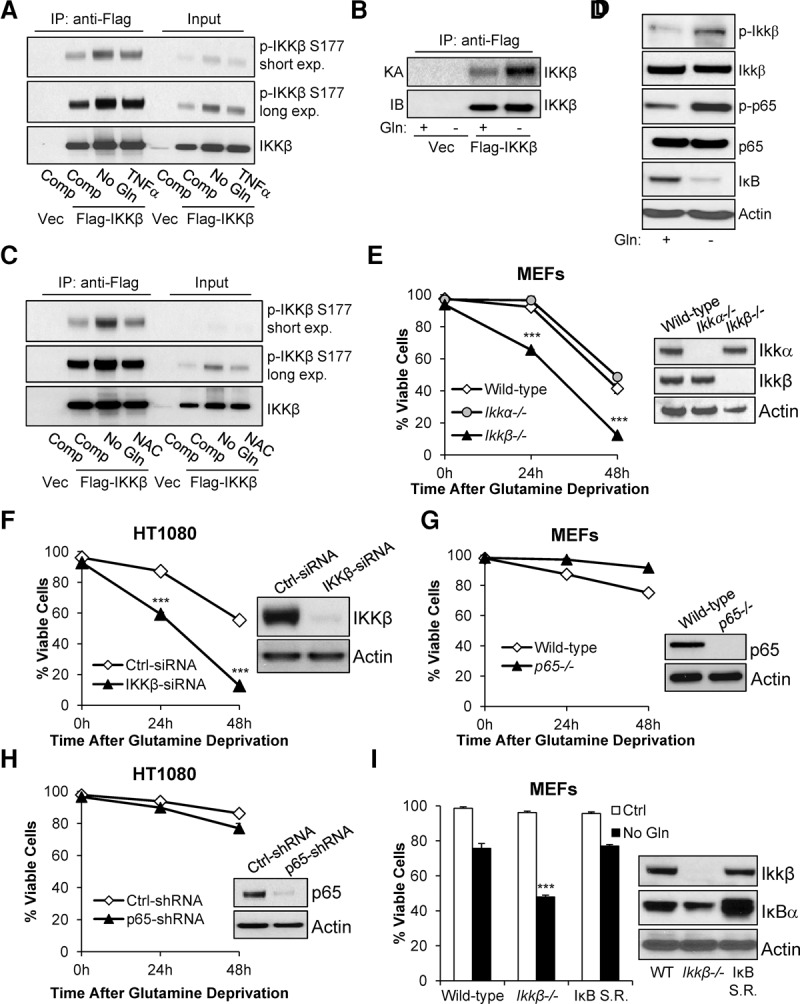
IKKβ is required for cell survival upon glutamine deprivation independent of NF-κB transcription. (A) 293T cells ectopically expressing Flag-IKKβ were cultured for 24 h in complete medium or glutamine-free medium or treated for 15 min with TNFα followed by immunoprecipitation with anti-Flag-conjugated agarose; immunoblotting was performed with the indicated antibodies (15% input). (B) 293T cells ectopically expressing vector (Vec) or Flag-IKKβ were cultured for 24 h in complete medium or glutamine-free medium followed by 32P-ATP in vitro kinase assays. Kinase assay reactions were quenched with SDS loading dye before SDS-PAGE separation and autoradiograph exposure. (C) 293T cells ectopically expressing Flag-IKKβ were cultured for 24 h in complete medium, glutamine-free medium, or glutamine-free medium supplemented with 5 mM NAC followed by immunoprecipitation with anti-Flag-conjugated agarose; immunoblotting was performed with the indicated antibodies (15% input). (D) Wild-type MEFs were cultured in complete or glutamine-free medium for 24 h, cells were lysed, and immunoblotting was performed with the indicated antibodies. (E, left panel) Wild-type, Ikkα−/−, and Ikkβ−/− MEFs were cultured in glutamine-free medium, and viability was assessed by propidium iodide exclusion at the indicated time points. Data presented are mean ± SEM of three independent experiments performed in duplicate. (Right panel) Cells were lysed, and immunoblotting was performed with the indicated antibodies. (F) HT1080 cells were transiently transfected with 20 nM control (Ctrl) siRNA or IKKβ siRNA. (Left panel) Forty-eight hours after transfection, cells were cultured in glutamine-free medium, and viability was assessed by propodium iodide exclusion at the indicated time points. Data presented are mean ± SEM of three independent experiments performed in duplicate. (Right panel) Forty-eight hours after transfection, cells were lysed, and immunoblotting was performed with the indicated antibodies. (G, left panel) Wild-type and p65−/− MEFs were cultured in complete or glutamine-free medium, and viability was assessed by propidium iodide exclusion at the indicated time points. (Right panel) Cells were lysed, and immunoblotting was performed with the indicated antibodies. Data presented are mean ± SEM of three independent experiments performed in duplicate. (H, left panel) HT1080 cells were transduced with control or p65 shRNA followed by puromycin selection, cells were cultured in complete or glutamine-free medium, and viability was assessed by propidium iodide exclusion at the indicated time points. Data presented are mean ± SEM of three independent experiments performed in duplicate. (Right panel) After puromycin selection, cells were lysed, and immunoblotting was performed with the indicated antibodies. (I, left panel) Wild-type, Ikkβ−/−, and wild-type MEFs stably expressing superrepressive IκBα (IκB S.R.) were cultured in complete or glutamine-free medium, and viability was assessed by propidium iodide exclusion at 24 h. Data presented are mean ± SEM of three independent experiments performed in duplicate. (Right panel) Cells were lysed, and immunoblotting was performed with the indicated antibodies. (***) P < 0.005, Student's t-test.
IKKβ is a PFKFB3 kinase
To identify the downstream substrate of IKKβ under low-glutamine conditions, 293T cells were transiently transfected with Flag-tagged IKKβ and starved of glutamine for 24 h followed by immunoprecipitation using anti-Flag-conjugated agarose to purify proteins interacting with IKKβ. The potential binding partners were further eluted with Flag peptide and identified by protein mass spectrometric peptide sequencing. Known IKKβ-interacting proteins such as IKKα, NEMO, CDC37, and PP1 were identified, thus validating the approach (Fig. 2A). Of particular interest, we found that PFKFB3 interacted with IKKβ (Fig. 2A). To confirm our mass spectrometry (MS) findings, we transiently coexpressed Flag-tagged IKKβ and V5-tagged PFKFB3 in 293T cells followed by immunoprecipitation with anti-Flag or anti-V5 antibody and found that IKKβ and PFKFB3 bind together (Fig. 2B,C). Importantly, we found increased association between IKKβ and PFKFB3 upon glutamine starvation (Fig. 2D). Although glycolysis occurs mostly in the cytosol and although PFKFB3 is a major regulator of glycolysis, recent reports have indicated cytosolic and nuclear roles for PFKFB3 (Yalcin et al. 2014; Domenech et al. 2015). This prompted us to examine the colocalization of IKKβ and PFKFB3 using immunofluorescence on the endogenous proteins. We indeed found a robust amount of PFKFB3 in the cytosol and nucleus and a strong colocalization of IKKβ and PFKFB3 in both compartments (Supplemental Fig. S2A). Moreover, we fractionated HT1080 cells and confirmed our immunofluorescence findings that both IKKβ and PFKFB3 are in the nucleus and cytosol (Supplemental Fig. S2B). In order to gain insight into how these proteins interact, we used protein interactions by structural matching (PRISM) (Tuncbag et al. 2011; Baspinar et al. 2014) and found that PFKFB3 is predicted to interact with the catalytic region of IKKβ in a fashion similar to IκBα, a well-known IKKβ substrate (Fig. 2E). Therefore, we hypothesized that PFKFB3 is phosphorylated by IKKβ. To test this hypothesis, we performed a 32P-ATP in vitro kinase assay using recombinant GST-PFKFB3 with and without constitutively active GST-IKKβ. Indeed, active IKKβ was sufficient to phosphorylate PFKFB3 but not the JNK substrate c-JUN, as expected (Fig. 2F). Furthermore, we tested whether immunoprecipitated Flag-IKKβ from cells cultured in complete or glutamine-free medium could phosphorylate GST-PFKFB3 in a 32P-ATP in vitro kinase assay and found that, consistent with IKKβ phosphorylation levels (Fig. 1A), IKKβ phosphorylated PFKFB3 at low levels in complete medium conditions and robustly when cells are starved of glutamine (Fig. 2G). To exclude the possibility that we copurified a contaminating kinase, we performed the same experiment using a kinase-inactive Flag-IKKβ and found no PFKFB3 phosphorylation (Fig. 2H). Moreover, we used IKKβ-specific inhibitor TPCA-1 and found that this greatly diminished phosphorylation of PFKFB3 (Supplemental Fig. S2C). Taken together, we identified PFKFB3 as a putative IKKβ substrate that is phosphorylated at low levels in proliferating cells and higher levels when cells are deprived of glutamine.
Figure 2.
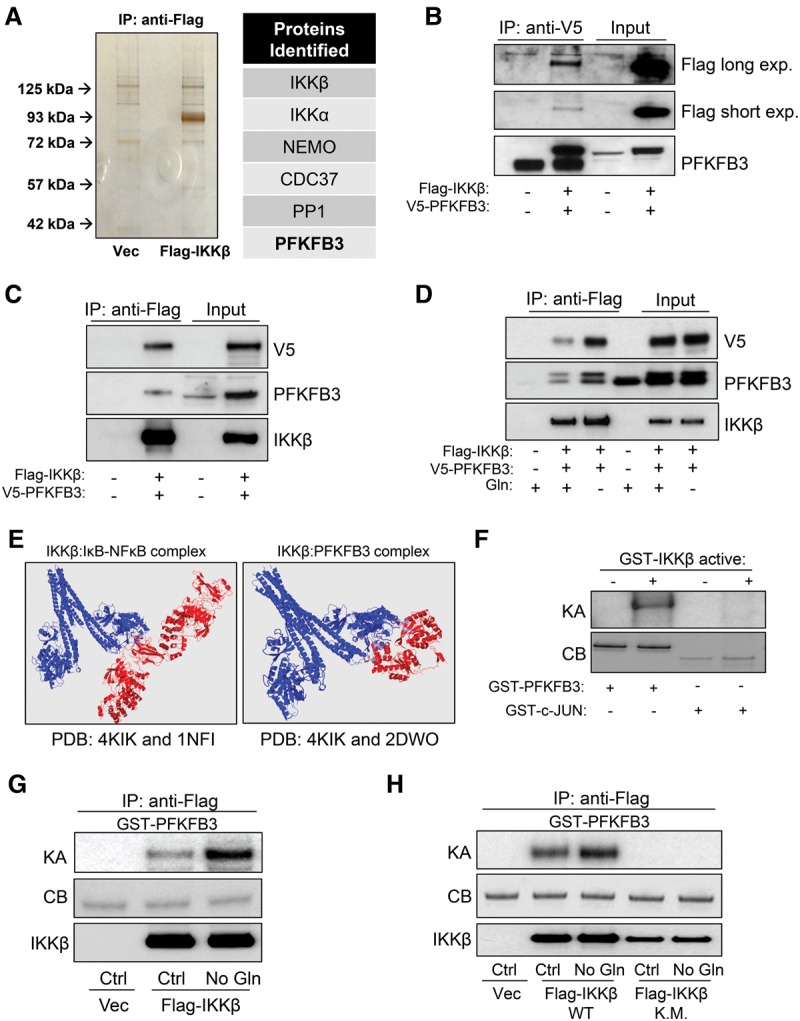
IKKβ is a PFKFB3 kinase. (A) Silver-stained gel of eluates from 24-h glutamine-starved 293T cells expressing vector (Vec) or Flag-IKKβ. The bands unique to the Flag-IKKβ eluates were identified by mass spectrometric analysis. (B,C) 293T cells ectopically coexpressing Flag-IKKβ and V5-PFKFB3 were immunoprecipitated with anti-V5 (B) or anti-Flag (C) antibody followed by immunoblotting with the indicated antibodies (15% input). (D) 293T cells ectopically coexpressing Flag-IKKβ and V5-PFKFB3 were cultured in complete or glutamine-free medium for 24 h followed by immunoprecipitation with anti-Flag antibody and immunoblotting with the indicated antibodies (15% input). (E, left panel) PRISM structure analysis for the predicted interaction between IKKβ (Protein Data Bank [PDB]: 4KIK) and the IκBα–NFκB complex (PDB: 1NFI). (Right panel) PRISM structure analysis for the predicted interaction between IKKβ (PDB: 4KIK) and PFKFB3 (PDB: 2DWO). (F) 32P-ATP in vitro kinase assay (KA) was performed using 0.2 µg of recombinant active GST-IKKβ with 2 µg of recombinant GST-PFKFB3 or 2 µg of recombinant GST-c-JUN followed by SDS-PAGE separation and autoradiograph exposure. Coomassie blue (CB) staining confirmed loading in the absence of the kinase assay band. (G) 293T cells ectopically expressing Flag-IKKβ were cultured for 24 h in complete medium or glutamine-free medium followed by immunoprecipitation with anti-Flag-conjugated agarose. 32P-ATP in vitro kinase assay was performed on immunoprecipitates incubated with 2 µg of recombinant GST-PFKFB3 followed by SDS-PAGE separation and autoradiograph exposure. (H) 293T cells ectopically expressing Flag-IKKβ (wild type) or the kinase-inactive mutant Flag-IKKβ (K.M.) were cultured for 24 h in complete medium or glutamine-free medium followed by immunoprecipitation with anti-Flag-conjugated agarose. 32P-ATP in vitro kinase assay was performed on immunoprecipitates incubated with 2 µg of recombinant GST-PFKFB3 followed by SDS-PAGE separation and autoradiograph exposure.
IKKβ phosphorylates PFKFB3 at Ser269
Next, we wanted to identify which residue on PFKFB3 is phosphorylated by IKKβ. To do this, we performed an in vitro kinase assay using recombinant constitutively active GST-IKKβ and GST-PFKFB3 followed by protein mass spectrometric phosphopeptide mapping and found that PFKFB3 was phosphorylated on Ser269 (Fig. 3A). Strikingly, the phosphopeptide that we identified matched the consensus IKKβ phospho-motif and shared high sequence similarity with other IKKβ substrates such as IκBα, p105, β-Catenin, FOXO3a, and TSC1 (Fig. 3B). To further investigate Ser269 phosphorylation, we raised an antibody against this site and performed an in vitro kinase assay followed by immunoblotting and found that Ser269 was indeed phosphorylated by active IKKβ (Fig. 3C). We further validated this antibody by performing the same experiment on S269A mutant PFKFB3 (Supplemental Fig. S3A). Intriguingly, PFKFB4 has a proline in place of serine in the corresponding sequence homology with PFKFB3 (Fig. 3D). With a 32P-ATP in vitro kinase assay, we found that active IKKβ phosphorylated GST-IκBα and GST-PFKFB3 but was unable to phosphorylate recombinant GST-PFKFB4 (Fig. 3E). To conclusively validate the specificity of the PFKFB3 Ser269 phosphosite, we performed a 32P-ATP in vitro kinase assay using wild-type and S269A mutant GST-PFKFB3. We found that only wild-type PFKFB3 was highly phosphorylated by active IKKβ, while phosphorylation of S269A was significantly diminished (Fig. 3F), suggesting that Ser269 is the major site that is phosphorylated by IKKβ. To test whether endogenous PFKFB3 is phosphorylated at Ser269, PFKFB3 was immunoprecipitated from HT1080 cells cultured in complete or glutamine-free medium followed by immunoblotting. Indeed, PFKFB3 is phosphorylated at Ser269 endogenously at low levels in proliferating cells in complete medium and at high levels in cells starved of glutamine (Fig. 3G). Interestingly, we further found that other IKKβ stimuli, including LPS and TNFα, also led to PFKFB3 Ser269 phosphorylation (Supplemental Fig. S3B,C). To examine whether PFKFB3 Ser269 occurs in vivo, we treated HT1080 mouse xenograft tumors with glutaminase inhibitor 6-diazo-5-oxo-L-norleucine (DON) and found that, compared with PBS control, DON-treated tumors showed significant PFKFB3 Ser269 phosphorylation (Fig. 3H). Importantly, using HT1080 cells stably expressing control or IKKβ-shRNA, we found that PFKFB3 Ser269 phosphorylation is IKKβ-dependent (Fig. 3I).
Figure 3.
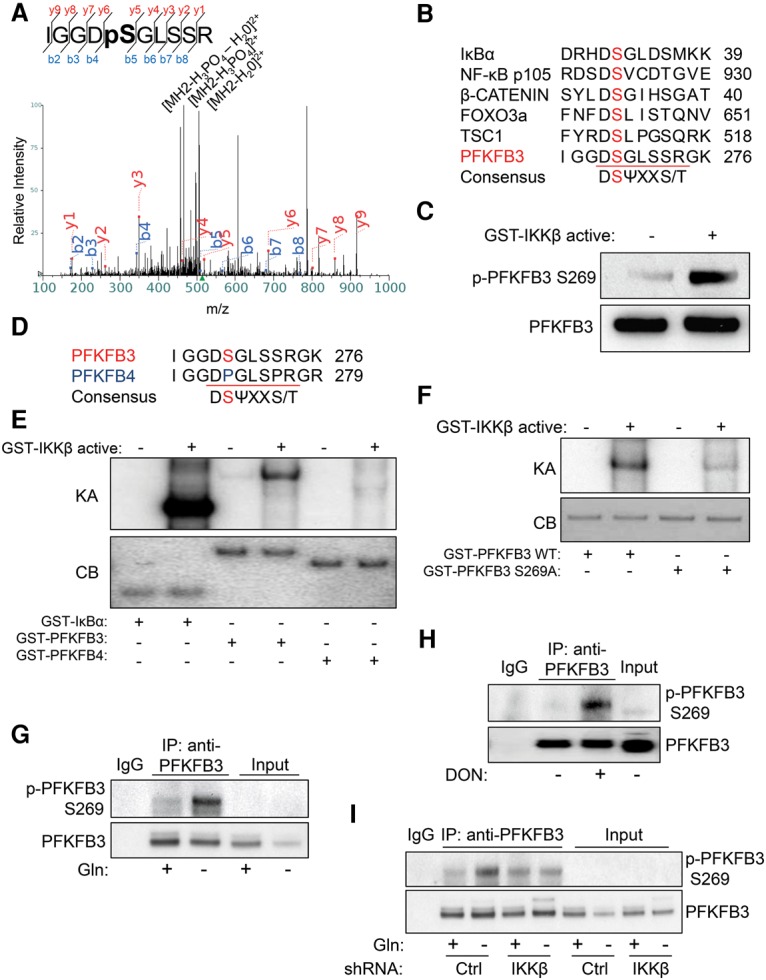
IKKβ phosphorylates PFKFB3 at Ser269. (A) Cold kinase assay using recombinant active GST-IKKβ and recombinant GST-PFKFB3 was performed followed by SDS-PAGE separation and phosphopeptide mapping. Mass spectra of identified phosphopeptide IGGDpSGLSSR are presented. (B) Sequence homology between the consensus IKKβ phospho-motif, known IKKβ substrates, and PFKFB3. (C) Cold kinase assay using recombinant active GST-IKKβ and recombinant GST-PFKFB3 was performed followed by immunoblotting with the indicated antibodies. PFKFB3 phospho-Ser269 antibody was raised against the antigen LGGRIGGD-pS-GLSSR. (D) Sequence homology between the consensus IKKβ phospho-motif, PFKFB3, and PFKFB4. (E) 32P-ATP in vitro kinase assay (KA) was performed using 0.2 µg of recombinant active GST-IKKβ with 0.5 µg of recombinant GST-IκBα, 2 µg of recombinant GST-PFKFB3, or 2 µg of recombinant GST-PFKFB4 followed by SDS-PAGE separation and autoradiograph exposure. Coomassie blue (CB) staining confirmed loading in the absence of the kinase assay band. (F) 32P-ATP in vitro kinase assay (KA) was performed using 0.2 µg of recombinant active GST-IKKβ with 2 µg of recombinant GST-PFKFB3 wild type or 2 µg of recombinant GST-PFKFB3 S269A followed by SDS-PAGE separation and autoradiograph exposure. Coomassie blue (CB) staining confirmed loading in the absence of the kinase assay band. (G) HT1080 cells were cultured for 48 h in complete medium or glutamine-free medium followed by immunoprecipitation with anti-PFKFB3 antibody; immunoblotting was performed with the indicated antibodies (15% input). (H) Athymic (nude) mice were injected subcutaneously with 2 × 106 HT1080 cells. Mice were treated with 10 mg/kg PBS or DON three times per week for 2 wk; tumors were harvested, followed by lysis and immunoprecipitation with anti-PFKFB3 antibody and immunoblotting with the indicated antibodies (25% input). (I) HT1080 cells transduced with either control (Ctrl) or IKKβ shRNA were cultured for 24 h in complete medium or glutamine-free medium followed by immunoprecipitation with anti-PFKFB3 antibody; immunoblotting was performed with the indicated antibodies (15% input).
PFKFB3 Ser269 phosphorylation reduces its activity
We then sought to determine how IKKβ phosphorylation affected PFKFB3. We first examined PFKFB3 function by performing a PFKFB3 activity assay on HT1080 cells expressing control or IKKβ shRNA and found that IKKβ knockdown cells have a twofold increase in PFKFB3 activity, suggesting that IKKβ negatively regulates PFKFB3 (Fig. 4A). Moreover, using liquid chromatography high-resolution MS (LC-HR-MS), we found that, compared with control cells, IKKβ knockdown cells exhibit threefold higher levels of F-1,6-BP, the major product of PFKFB3-induced PFK1 activity (Fig. 4B). To investigate how Ser269 phosphorylation affects PFKFB3 activity, we first depleted endogenous PFKFB3 from HT1080 cells using shRNA (Fig. 4C, lanes 1,2). We then ectopically expressed V5-tagged wild-type, Ser269Ala (S269A) mutant, or Ser269Asp (S269D) mutant PFKFB3 in PFKFB3-shRNA cells (Fig. 4C, lanes 3–5). Using these cells for a PFKFB3 activity assay, we found that cells expressing S269A PFKFB3 had 65% more activity, while S269D PFKFB3 displayed 80% less activity than cells expressing wild-type PFKFB3 (Fig. 4D), providing strong evidence that IKKβ-dependent PFKFB3 Ser269 phosphorylation inhibits its activity. Furthermore, we characterized the effect that S269A and S269D mutant PFKFB3 had on aerobic glycolysis and found that S269A mutant PFKFB3-expressing cells had significantly increased glucose uptake and lactate production compared with wild type, while S269D mutant PFKFB3 showed much less (Fig. 4E). Intriguingly, we found that glutamine starvation reduced the total protein level of PFKFB3 in an IKKβ-dependent manner (Fig. 3G,I, inputs). Moreover, it has been reported that PFKFB3 is regulated by the SCFβ–TRCP–ubiquitin ligase complex (Colombo et al. 2011; Duan and Pagano 2011), which often targets IKKβ phosphorylation substrates (Winston et al. 1999). To further investigate whether Ser269 played a role in PFKFB3 protein stability, we ectopically expressed wild type or the S269A mutant in HT1080 and 293T cells overnight and found a striking increase in protein level of the S269A mutant, suggesting that Ser269 phosphorylation reduces PFKFB3 stability (Fig. 4F). Taken together, these data demonstrate that PFKFB3 Ser269 phosphorylation reduces enzymatic activity and protein stability.
Figure 4.
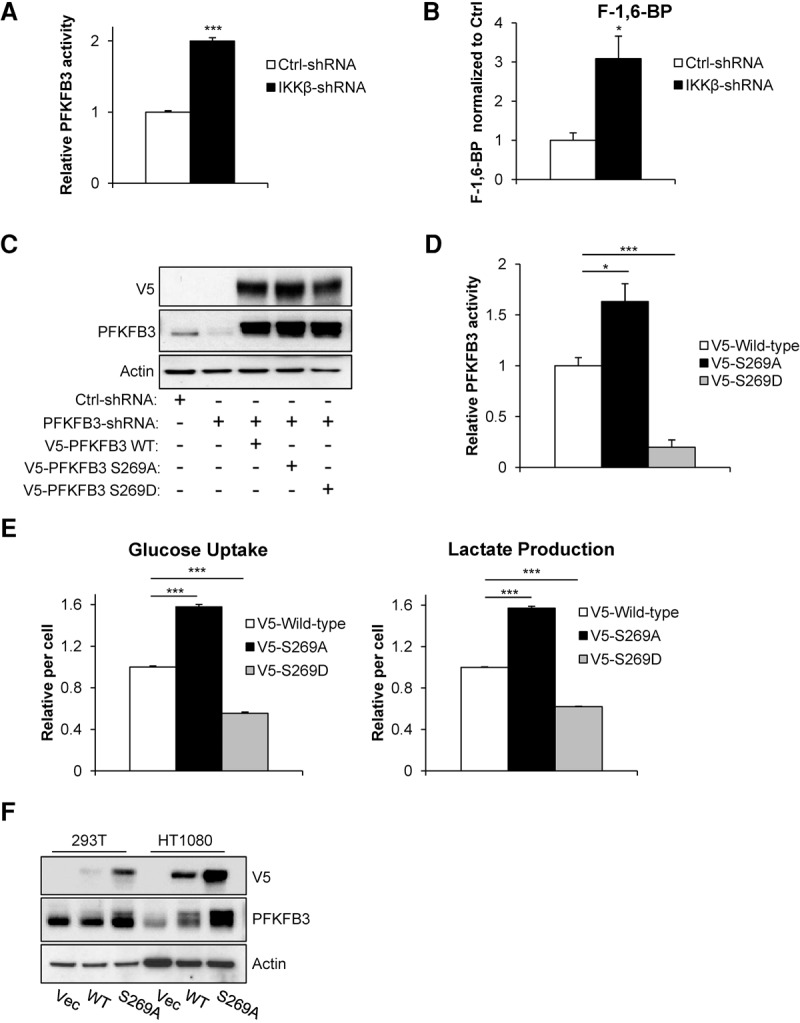
PFKFB3 Ser269 phosphorylation reduces its activity. (A) PFKFB3 activity was measured in HT1080 cells transduced with either control (Ctrl) or IKKβ shRNA. OD values were normalized to the control shRNA reading. Data presented are mean ± SEM of three independent experiments performed in duplicate. (B) Metabolites were extracted from HT1080 cells transduced with either control (Ctrl) or IKKβ shRNA, and F-1,6-BP levels were measured by LC-HR-MS. Data presented are mean ± SEM of three independent experiments performed in duplicate. (C) HT1080 cells were transduced with PFKFB3 shRNA to deplete endogenous PFKFB3 followed by ectopic expression of V5-tagged wild-type, S269A mutant, or S269D mutant PFKFB3 and blasticidin selection. Immunoblotting was performed with the indicated antibodies. (D) PFKFB3 activity was measured in HT1080 cells depleted of endogenous PFKFB3 and ectopically expressing wild-type, S269A, or S269D PFKFB3. OD values from PFKFB3 shRNA cells expressing V5 vector control were subtracted from wild-type, S269A, and S269D OD values before being normalized to wild type. Data presented are mean ± SEM of three independent experiments performed in duplicate. (E) HT1080 cells depleted of endogenous PFKFB3 and ectopically expressing wild-type, S269A, or S269D PFKFB3 were cultured for 48 h in complete medium followed by glucose uptake and lactate production measurements using the Nova Bioprofile 100 analyzer. Metabolite levels per cell were normalized to wild type. Data presented are mean ± SEM of three independent experiments performed in duplicate. (F) 293T and HT1080 cells were transduced with V5-tagged wild-type or mutant (S269A) PFKFB3. Forty-eight hours after transduction, cells were lysed, and immunoblotting was performed with the indicated antibodies. (*) P < 0.05; (***) P < 0.005, Student's t-test.
IKKβ down-regulates aerobic glycolysis through PFKFB3 inhibition
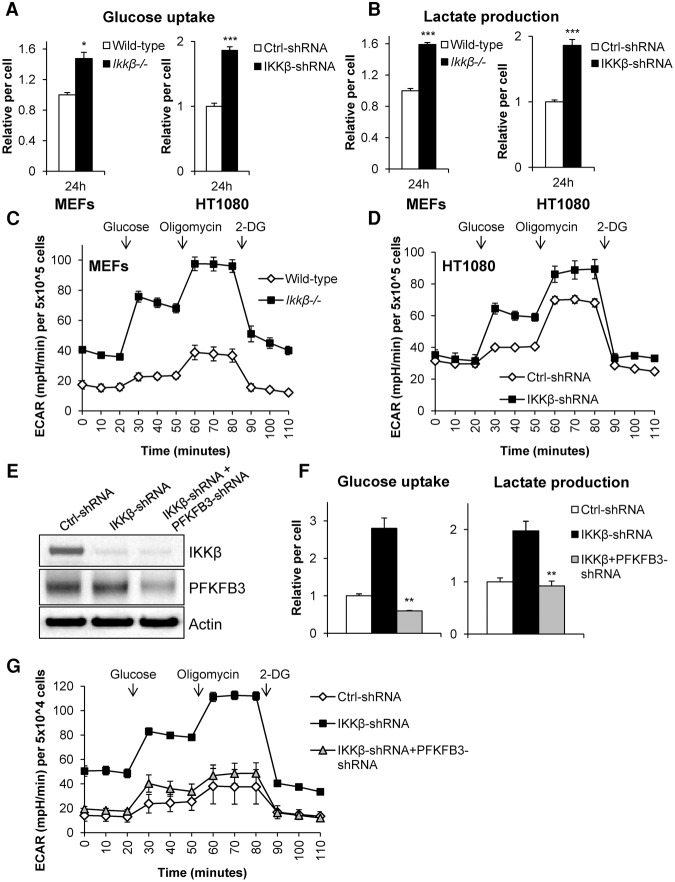
Because PFKFB3 is a major regulator of the Warburg effect (Almeida et al. 2010), we hypothesized that IKKβ-deficient cells, which have hyperactive PFKFB3, would have increased aerobic glycolysis. To test this, we cultured wild-type and Ikkβ−/− MEFs and control and IKKβ shRNA HT1080 cells in complete medium for 24 h to allow sufficient growth and nutrient use. Consistently, we found that genetic ablation of IKKβ led to increased glucose uptake and lactate production (Fig. 5A,B). To further evaluate the glycolytic capacity, we measured the extracellular acidification rate (ECAR) and found that IKKβ-deficient cells have increased glycolytic flux, consistent with the observed increased glucose uptake and lactate production (Fig. 5C,D). In order to determine whether the increased aerobic glycolysis observed in IKKβ-deficient cells was due to increased PFKFB3 activity, we stably knocked down both IKKβ and PFKFB3 in HT1080 cells (Fig. 5E). We found that knockdown of PFKFB3 was sufficient to rescue the increased aerobic glycolysis found in IKKβ-deficient cells as glucose uptake, lactate production, and glycolytic flux were restored (Fig. 5F,G). It was reported that NF-κB-dependent GLUT3 expression plays an important role in glycolysis (Kawauchi et al. 2008). To further test whether other potential mechanisms, such as GLUT3, could contribute to IKKβ-regulated aerobic glycolysis, we knocked down GLUT3 in HT1080 cells; however, no significant difference in cell viability upon glutamine withdrawal was observed (Supplemental Fig. S4A). This was consistent with our observation that IKKβ, but not NF-κB, plays a role in cell adaptation to low-glutamine conditions (Fig. 1E–I). These data demonstrate that IKKβ antagonizes aerobic glycolysis mainly through inhibition of PFKFB3.
Figure 5.
IKKβ down-regulates aerobic glycolysis through PFKFB3 inhibition. (A,B) MEFs (wild type and Ikkβ−/−) and HT1080 cells (control [Ctrl] and IKKβ shRNA) were cultured for 24 h in complete medium followed by glucose uptake and lactate production measurements using the Nova Bioprofile 100 analyzer. Metabolite levels per cell were normalized to wild-type or control shRNA. Data presented are mean ± SEM of three independent experiments performed in duplicate. (C,D) ECAR was measured in MEFs (wild type and Ikkβ−/−) and HT1080 cells (control [Ctrl] and IKKβ shRNA) using the XF-24 Seahorse system. Data presented are mean ± SEM of five independent experiments. (E) HT1080 cells expressing IKKβ shRNA were transduced with PFKFB shRNA followed by cell lysis and immunoblotting with the indicated antibodies. (F) HT1080 cells transduced with control (Ctrl), IKKβ, or IKKβ and PFKFB3 shRNA were cultured for 24 h in complete medium followed by glucose uptake and lactate production measurements using the Nova Bioprofile 100 analyzer. Metabolite levels per cell were normalized to control shRNA. Data presented are mean ± SEM of three independent experiments performed in duplicate. (G) ECAR was measured in cells transduced with control (Ctrl), IKKβ, or IKKβ and PFKFB3 shRNA using the XF-24 Seahorse system. Data presented are mean ± SEM of five independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.005, Student's t-test.
Enhanced aerobic glycolysis in IKKβ-deficient cells leads to increased glutamine dependence for TCA cycle intermediates
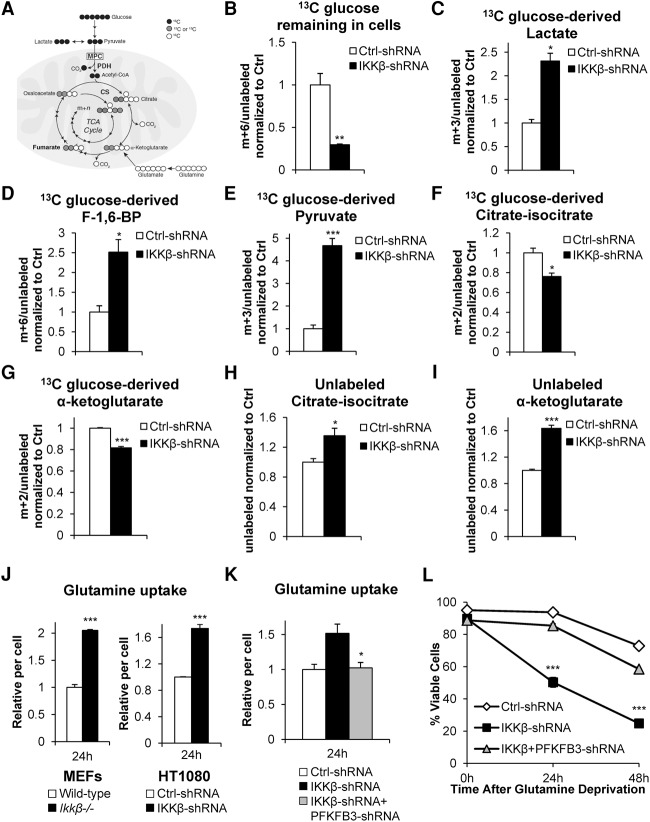
To further elucidate how IKKβ affects glucose metabolism beyond inducing lactate production, we performed stable isotope tracer-labeling experiments by culturing control or IKKβ shRNA HT1080 cells for 6 h in medium containing universally labeled 13C-glucose (Fig. 6A). Metabolites were extracted and analyzed by LC-HR-MS. Consistent with our uptake experiments, these labeling experiments revealed a significant decrease in 13C-glucose remaining in IKKβ shRNA cells compared with control, which signifies that these cells used more glucose (Fig. 6B). Moreover, we observed increased glucose-derived lactate when IKKβ was knocked down (Fig. 6C). Consistent with PFKFB3 activity assays, we found that IKKβ knockdown cells have increased glucose-derived F-1,6-BP, providing further evidence that PFKFB3 activity is up-regulated in these cells (Fig. 6D). Strikingly, IKKβ-deficient cells produced almost fivefold more pyruvate, the end product of glycolysis (Fig. 6E). Interestingly, very little of this glucose-derived pyruvate enters into the TCA cycle, as IKKβ knockdown cells have ∼20% less glucose-derived citrate and α-ketoglutarate (Fig. 6F,G). However, IKKβ-deficient cells have increased total levels of unlabeled citrate and α-ketoglutarate, which suggests that these cells may use glutamine for anaplerosis (Fig. 6H,I). Indeed, cells lacking IKKβ have increased glutamine uptake (Fig. 6J). Importantly, the increased glutamine uptake is PFKFB3-dependent because cells with knockdown of both IKKβ and PFKFB3 uptake glutamine at the same level as control shRNA cells (Fig. 6K). We reasoned that the increased glutamine uptake could explain why IKKβ-deficient cells are more sensitive to glutamine starvation. To confirm this, we performed propidium iodide exclusion flow cytometry to measure the viability of control, IKKβ, and IKKβ–PFKFB3 shRNA cells and found that IKKβ knockdown cell viability in low glutamine is almost completely rescued by PFKFB3 ablation (Fig. 6L). Similarly, cells depleted of endogenous PFKFB3 and ectopically expressing S269A mutant PFKFB3 displayed increased glutamine uptake and sensitivity to glutamine starvation, demonstrating that the IKKβ-deficient phenotype is largely driven by PFKFB3 Ser269 phosphorylation and not other IKKβ-regulated metabolic processes (Supplemental Fig. S5A,B). Because increased aerobic glycolysis driven by hyperactive PFKFB3 in IKKβ-deficient cells results in less glucose-derived carbons used by both the TCA cycle and the pentose phosphate pathway (Yamamoto et al. 2014), it is possible that the loss of glucose-derived carbons from both the pentose phosphate pathway and TCA cycle contributes to the loss of cell viability upon glutamine deprivation in IKKβ-deficient cells. Indeed, IKKβ knockdown reduced cellular NADPH levels (Supplemental Fig. S5C), and while cell-permeable TCA cycle intermediate succinate or antioxidant GSH rescued cell viability at early time points, the combination of both agents synergistically rescued IKKβ-deficient cell viability upon glutamine starvation at later time points (the viability of the combination group was 40%, while the sum of both agents alone was 22%) (Supplemental Fig. S5D). Taken together, these data support the hypothesis that highly glycolytic cells rely on glutamine to replenish TCA intermediates and antioxidants, and IKKβ acts to dampen aerobic glycolysis when cells experience periods of glutamine deprivation.
Figure 6.
Enhanced aerobic glycolysis in IKKβ-deficient cells leads to increased glutamine dependence for TCA cycle intermediates. (A) Schematic representation of 13C-glucose metabolism. M+n refers to the number of 13C found in each metabolite. (B–G) HT1080 cells transduced with either control (Ctrl) or IKKβ shRNA were cultured for 6 h in medium containing universally labeled 13C-glucose followed by metabolite extraction and analysis using LC-HR-MS. Labeled metabolites were divided by unlabeled metabolites and then normalized to control shRNA levels. Data presented are mean ± SEM of three independent experiments. (H,I) HT1080 cells transduced with either control (Ctrl) or IKKβ shRNA were cultured for 6 h in medium containing universally labeled 13C-glucose followed by metabolite extraction and analysis using LC-HR-MS. Unlabeled metabolites (i.e., metabolites not derived from glucose) were normalized to control shRNA levels. Data presented are mean ± SEM of three individual experiments. (J) MEFs (wild type and Ikkβ−/−) and HT1080 cells (control [Ctrl] and IKKβ shRNA) were cultured in complete medium for 24 h followed by glutamine uptake measurement using the Nova Bioprofile 100 analyzer. Metabolite levels per cell were normalized to wild-type or control shRNA. Data presented are mean ± SEM of three independent experiments performed in duplicate. (K) HT1080 cells transduced with control (Ctrl), IKKβ, or IKKβ and PFKFB3 shRNA were cultured in complete medium for 24 h followed by glutamine uptake measurement using the Nova Bioprofile 100 analyzer. Metabolite levels per cell were normalized to control shRNA. Data presented are mean ± SEM of three independent experiments performed in duplicate. (L) HT1080 cells transduced with control (Ctrl), IKKβ, or IKKβ and PFKFB3 shRNA were cultured in glutamine-free medium, and viability was assessed by propidium iodide exclusion at the indicated time points. Data presented are mean ± SEM of three independent experiments performed in duplicate. (*) P < 0.05; (**) P < 0.01; (***) P < 0.005, Student's t-test.
Inhibition of IKKβ sensitizes cancer cells and tumors to glutaminase inhibitor treatment
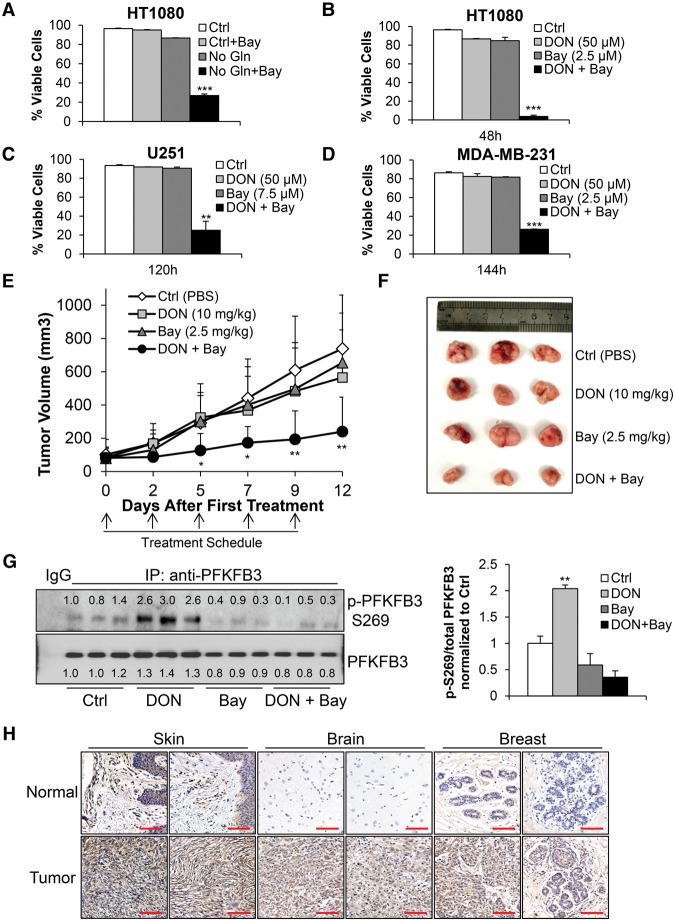
Targeting glutamine metabolism is quickly emerging as a viable option for numerous cancer types (Wise and Thompson 2010). One strategy for targeting glutamine metabolism is treating patients with glutaminase inhibitors such as DON, and these inhibitors have been used in clinical trials over the years; however, the trials have had little success due to cytotoxicity and adverse side effects (Sullivan et al. 1988; Earhart et al. 1990). Another explanation for the low efficacy of glutaminase inhibitors is adaptive pathways used by tumors to survive periods of glutamine starvation, suggesting that ablation of these adaptive pathways may enhance the effect, which could reduce the dose of glutaminase inhibitor required, thereby potentially alleviating off-target effects. To test whether disruption of the IKKβ–PFKFB3 signaling axis could sensitize cancer cells to glutamine metabolism inhibition, we first determined whether treatment with the IKKβ inhibitor Bay 11-7082 (Bay) had any effect on cells under glutamine deprivation. Indeed, much like genetic ablation of IKKβ, chemical inhibition using Bay dramatically sensitized HT1080 cells to low glutamine deprivation-induced cell death, but Bay had little effect on cells in complete medium (Fig. 7A). To investigate the possibility of Bay off-target effects, we treated IKKβ-deficient HT1080 cells with the same dose as our previous experiment and observed no change in cell death in IKKβ knockdown cells under glutamine withdrawal, while the control cells were dramatically sensitized (Supplemental Fig. S6A). We then proceeded with the combination of Bay and DON and found striking synergistic killing of HT1080, glioblastoma U251, and triple-negative breast cancer MD-MBA-231 cells, suggesting that this combinational therapy can be applied to a broad range of cancer types (Fig. 7B–D). To test the combinational therapy in vivo, athymic (nude) mice were subcutaneously injected with 2 × 106 HT1080 cells. Once tumors reached an average volume of 100 mm3, mice were placed in four groups and treated (intraperitoneally) three times per week with PBS control, 2.5 mg/kg Bay, 10 mg/kg DON, or 2.5 mg/kg Bay and 10 mg/kg DON combination, and a significant reduction in tumor growth was observed when the two drugs were administered together (Fig. 7E,F). Following completion of the study, tumors were harvested and homogenized followed by immunoprecipitation using anti-PFKFB3 antibody. Immunoblotting using anti-PFKFB3 p-Ser269 antibody revealed that animals treated with DON showed strong PFKFB3 Ser269 phosphorylation, while this phosphorylation was blocked in tumors of animals that received the combination of Bay and DON (Fig. 7G). Finally, we examined PFKFB3 Ser269 phosphorylation in human tissues using immunohistochemistry and found striking increases in the malignant tissues compared with healthy tissues, including breast, brain, and skin (Fig. 7H). These data demonstrate that PFKFB3 phosphorylation at Ser269 is important for maintaining tumor growth when glutamine metabolism is inhibited or when local supplies are depleted, and disruption of PFKFB3 phosphorylation via IKKβ inhibition could be a strong adjuvant for established glutaminase inhibitor treatments.
Figure 7.
Inhibition of IKKβ sensitizes cancer cells and tumors to glutaminase inhibitor treatment. (A) HT1080 cells were cultured for 24 h in complete medium, complete medium supplemented with 2.5 µM Bay, glutamine-free medium, or glutamine-free medium supplemented with 2.5 µM Bay, and viability was assessed by propidium iodide exclusion. Data presented are mean ± SEM of three independent experiments performed in duplicate. (B–D) HT1080, U251, or MDA-MB-231 cells were cultured complete medium supplemented with PBS (control [Ctrl]), 50 µM DON, Bay (as indicated), or the combination of DON and Bay for indicated time points, and viability was assessed by propidium iodide exclusion. Data presented are mean ± SEM of three independent experiments performed in duplicate. (E) Athymic (nude) mice were injected subcutaneously with 2 × 106 HT1080 cells. Mice were then placed into four groups and treated with control (PBS), 10 mg/kg DON, 2.5 mg/kg Bay, or the combination of 10 mg/kg DON and 2.5 mg/kg Bay three times per week for 2 wk, and tumor growth was measured over time. Data presented are mean ± SD of eight tumors per group. (F,G) HT1080 xenograft tumors were harvested and photographed, followed by lysis and immunoprecipitation with anti-PFKFB3 antibody and immunoblotting with the indicated antibodies. Immunoblots were quantified using ImageJ, and the graph represents the mean of each group p-Ser269 divided by the total PFKFB3 normalized to the control treatment values. (H) Paraffin-embedded tissues were deparaffinized and stained with p-Ser269 PFKFB3 antibody followed by counterstaining with hematoxylin. Images were acquired using a Zeiss Observer II microscope. Bar, 100 µm. (*) P < 0.05; (**) P < 0.01; (***) P < 0.005, Student's t-test.
Discussion
In this study, we uncovered a previously unidentified pathway by which IKKβ is activated as glutamine levels decrease and directly phosphorylates PFKFB3 to inhibit its activity, leading to decreased aerobic glycolysis and glutamine dependency. Thus, IKKβ deficiency and subsequent hyperactivation of PFKFB3 led to increased aerobic glycolysis and glutamine uptake (Figs. 5, 6), and the inability to adapt to low glutamine levels led to cell death (Figs. 1, 7). The relationship between highly glycolytic cells and their reliance on glutamine is quickly becoming well established (Vander Heiden et al. 2009; Le et al. 2012; Qing et al. 2012). For example, a recent report demonstrated that, in highly glycolytic cells, the majority of TCA cycle carbons are derived from glutamine, but, when aerobic glycolysis was reduced, less glutamine contributed to TCA cycle intermediates, which decreased glutamine dependence (Le et al. 2012). Intriguingly, it has been shown that high glutamine concentrations and glutamine-dependent anaplerosis led to increased glucose uptake and aerobic glycolysis (Kaadige et al. 2009). Moreover, glutamine depletion/repletion studies found that aerobic glycolysis is decreased in response to glutamine starvation (Wong et al. 2004). Our data are in agreement with these reports in that glutamine availability and aerobic glycolysis are tightly correlated, and a disconnect results in loss of cell viability.
Although IKKβ is most well known as an initiator of NF-κB transcription, we found little effect of NF-κB on low-glutamine-induced cell survival. Although we did observe p65 Ser536 phosphorylation in response to glutamine withdrawal, we found no significant increase in p65 trans-activity (Fig. 1; Supplemental Fig. S1). This is not entirely surprising, as p65 Ser536 phosphorylation does not necessarily affect p65 transcriptional activity (Okazaki et al. 2003). Interestingly, previous reports have indicated that glutamine level affects LPS or interleukin 1-β function via modulation of p65 nuclear activity (Liboni et al. 2005; Brasse-Lagnel et al. 2007), indicating that glutamine level may play a role in coordinating cytokine-regulated NF-κB activity and cell survival.
In recent years, several reports have demonstrated that IKKβ is a diverse kinase that phosphorylates numerous other substrates, such as the p85α subunit of PI3K, TSC1, BAD, and β-Catenin (Lamberti et al. 2001; Lee et al. 2007; Comb et al. 2012; Yan et al. 2013). More recently, a high-throughput screen combining quantitative MS with random forest bioinformatics to dissect the TNFα–IKKβ-induced phosphoproteome in MCF-7 breast cancer cells revealed dozens of potential IKKβ substrates (Krishnan et al. 2015). Of significance, PFKFB3 phosphorylation was identified in this study, providing independent confirmation that PFKFB3 is a putative IKKβ substrate. Interestingly, IKKβ phosphorylation of its substrates often marks them for degradation via the SCFβ–TRCP–ubiquitin ligase complex (Winston et al. 1999), and PFKFB3 is reportedly regulated by the SCFβ–TRCP–ubiquitin ligase complex, which requires phosphorylation at Ser269 (Colombo et al. 2011; Duan and Pagano 2011). Consistently, we found that the PFKFB3 protein level is reduced upon glutamine deprivation in an IKKβ-dependent manner (Fig. 3I, input). Moreover, we found the unphosphorylateable form of PFKFB3 (S269A mutant) to be much more stable in cells (Fig. 4F). Interestingly, we found that PFKFB3 Ser269 phosphorylation also affects its activity (Fig. 4D). Although the observed increase in PFKFB3 activity of S269A mutant could be explained by protein level, the expression of wild-type and S269D mutant PFKFB3 is similar, yet the activity of the S269D mutant is 80% reduced. Ser269 is located in a highly disordered region critical for PFKFB3 enzyme function, as the nearby residues must remain in contact with two-phosphate oxygens and help stabilize a tele-phosphohistidine intermediate (Cavalier et al. 2012; Boyd et al. 2015); therefore, phosphorylation could disrupt these interactions and normal function, which could ultimately decrease the activity. Taken together, our data suggest a dual consequence of IKKβ-dependent phosphorylation of PFKFB3 Ser269: (1) acutely reducing its activity and (2) marking it for ubiquitin-mediated degradation. Further studies will examine whether the SCFβ–TRCP–ubiquitin ligase complex is active in response to low-glutamine-induced metabolic stress. It is worth mentioning that, although other PFKFB3 phosphosites were identified using MS, we believe that Ser269 is the major site that is regulated by IKKβ due to the highly conserved IKKβ phospho-motif and the almost complete reduction in phosphorylation of the PFKFB3 S269A mutant (Fig. 3F).
The finding of the IKKβ–PFKFB3 signaling axis may be important in many other biological contexts. For example, it has been reported that constitutively active IKKβ in hepatocytes leads to hyperglycemia and insulin resistance, and inhibition of PFKFB3 also leads to insulin resistance in mice (Cai et al. 2005; Huo et al. 2010). These studies support our model in that active IKKβ will inhibit PFKFB3, which will decrease glucose uptake and glycolysis that can lead to hyperglycemia and insulin resistance. In addition, a dramatic shift toward aerobic glycolysis is observed when CD4+ T cells are proliferating, and a return to high oxidative phosphorylation (low glycolysis) occurs when activated CD4+ T cells become memory T cells (Gerriets et al. 2015), and T-reg cells rarely exhibit high aerobic glycolysis and generally maintain high oxidative phosphorylation (Beier et al. 2015). Of particular interest, a study conducted using CD4+ T-cell-specific IKKβ knockout mice found that IKKβ-deficient mice had dramatically less memory T cells and almost no T-reg cells, suggesting an inability to down-regulate aerobic glycolysis (Schmidt-Supprian et al. 2003). Therefore, it will be interesting to examine the IKKβ–PFKFB3 axis in these contexts as well as others where IKKβ activation occurs.
In this study, we also demonstrated the potential for targeting IKKβ in combination with glutamine metabolism inhibition. The finding that cancer cells are addicted to glutamine led to therapeutic approaches aimed at impairing glutamine metabolism. For example, glutaminase inhibitors DON or bis-2-[5-(phenylacetamido)-1,3,4-thiadiazol-2-yl]ethyl sulfide (BPTES) significantly diminish tumor growth in vivo (Kisner et al. 1980; Beier et al. 2015; Hernandez-Davies et al. 2015; Xiang et al. 2015). However, this approach will inevitably trigger metabolic stress and the adaptive response in cancer cells. We found that PFKFB3 is phosphorylated at Ser269 in tumors and even higher in tumors treated with glutamine deprivation-mimicking glutaminase inhibitor (DON) (Fig. 7C). Interestingly, DON-induced PFKFB3 Ser269 phosphorylation was blocked when coadministered with Bay, and these tumors grew at a significantly slower rate than those treated with DON or Bay alone, suggesting that PFKFB3 Ser269 phosphorylation is important for tumorigenesis (Fig. 7E). Thus, by eliminating IKKβ-dependent PFKFB3 inhibition, cancer cells are unable to adapt and are more susceptible to glutaminase inhibitor treatment. Strikingly, we found that multiple types of cancer cells are synergistically killed when IKKβ and glutaminase inhibitors are combined and observed PFKFB3 Ser269 phosphorylation in several human malignancies, providing proof of concept that combinational targeting could provide a new therapeutic option for cancer patients.
Materials and methods
Reagents and plasmids
Recombinant active GST- IKKβ (31176) was purchased from Active Motif. Recombinant GST-IκBα (I20-30G), GST-PFKFB3 (P323-30G), GST-PFKFB3 S269A (custom order), and GST-PFKFB4 (P324-30G) were purchased from SignalChem. Recombinant GST-c-JUN (GLO127G) was purchased from GloboZymes. and adenosine 5′-triphosphate, [γ-32P]-(BLU5024) was purchased from PerkinElmer. Bay (10010266) and TPCA-1 (15115) were purchased from Cayman Chemical. 6-dizao-5-oxo-L-norleucine (D2141), dimethyl succinate (W239607CSBOLDSTART),CSBOLDEND and NAC (138061) were purchased from Sigma. LPS serotype 0111:B4 (ALX-581-012-L001) was purchased from Enzo Life Sciences. GSH monoethyl ester (353905) was purchased from Calbiochem. pLX304 (25890), pLPC-N-Flag (12521), pLKO.1-Scramble (1864), pCMV2-Flag-IKKβ (11103), pCMV2-Flag-IKKβ kinase inactive (11104), and pBabe-Puro-IKBα-mut (15291) were purchased from Addgene. pLX304-PFKFB3 was purchased from Thermo-Dharmacon. pLX304-PFKFB3 S269A and S269D mutants were generated by MCLab. TRC human p65 shRNA (TRCN0000014686), TRC human IKKβ shRNA (TRCN0000 018917), and TRC human PFKFB3 shRNA (TRCN0000007338) plasmids were purchased from Thermo-Dharmacon.
Cell lines and cell culture
293T, HT1080, MCF-7, HeLa, and MDA-MB-231 cells were purchased from American Type Culture Collection (ATCC). Wild-type, Ikkα−/−, and Ikkβ−/− MEFs were kindly provided by Dr. Michael Karin's laboratory (University of California at San Diego). Wild-type and p65−/− MEFs were a gift from Dr. Hua Yu's laboratory (City of Hope Cancer Center). U251 cells were kindly provided by Dr. Craig Thompson's laboratory (Sloan Kettering Cancer Center). HT1080 cells expressing shRNA against p65, IKKβ, and/or PFKFB3 were generated via lentiviral-mediated gene transfer followed by puromycin selection and limited dilution to generate clonal cell lines. Wild-type MEFs ectopically expressing superrepressive IκBα were generated via retroviral-mediated gene transfer followed by puromycin selection. HT1080 cells ectopically expressing V5 wild-type, V5-S269A, or V5-S269D PFKFB3 were generated via lentiviral-mediated gene transfer followed by blasticidin selection. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 25 mM glucose and 4 mM L-glutamine supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products), 100 U/mL penicillin, and 100 µg/mL streptomycin. For glutamine starvation experiments, DMEM without glutamine (Invitrogen, 11960) was supplemented with 10% dialyzed FBS (Gemini Bio-Products). For starvation experiments, cells cultured in complete medium were supplemented with 10% dialyzed FBS (Gemini Bio-Products). Cell death was determined using propidium iodide exclusion flow cytometry.
siRNA transfection
On-target Smart Pool control siRNA (D-001810-10-20) and siRNA oligonucleotides targeting human p65 (L-003533-00-0005), human IKKβ (L-003503-00-0005), and human GLUT3 (L-007516-02-0005) were obtained from Thermo-Dharmacon. siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol.
Immunoblotting
Cells were lysed in buffer (50 mM Tris-HCl at pH 7.4, 5 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM EDTA, 1 mM EGTA, 250 mM mannitol, 1% [v/v] Triton X-100) containing freshly added protease inhibitor complex (Roche) and phosphatase inhibitor (Thermo Scientific). Equal amounts of protein (10–30 µg) were loaded onto precast NuPAGE Bis-Tris gels (Life Technologies) followed by transfer onto nitrocellulose. The Thermo Scientific NE-PER nuclear and cytoplasmic extraction kit was used for cellular fractionation according to the manufacturer's guidelines. Immunoblotting was performed with the following antibodies: IKKβ (Cell Signaling), IKKα (Cell Signaling), p-IKKβ Ser177 and p-IKKα Ser176 (Cell Signaling), p65 (Cell Signaling), p-p65 (Cell Signaling), IκB (Cell Signaling), V5 (Cell Signaling), β-Actin (Sigma), Flag M2 (Sigma), Tubulin (Sigma), Lamin B1 (Novus Biologicals), and PFKFB3 (Proteintech). p-PFKFB3 Ser269 was generated by Abiocode using the antigen “LGGRIGGD-pS-GLSSR” followed by affinity purification of crude antiserum.
Immunoprecipitation
For coimmunoprecipitation, one 10-cm plate containing cells grown to 80% confluence was used for each treatment condition and divided into aliquots for different immunoprecipitation conditions or antibodies. To harvest, cells were washed twice on the plate with PBS and lysed on ice using a cell scraper with lysis buffer (150 mM KCl, 0.2% [v/v] NP-40, 10% [v/v] glycerol, 20 mM Tris at pH 7.5, 0.5 mM DTT) containing freshly added protease inhibitor complex (Roche) and phosphatase inhibitor (Thermo Scientific), and protein quantification was performed using a BCA assay (Pierce). Equal amounts of lysate (500 µg to 1 mg) were immunoprecipitated while rotating at 4°C with 4 µg of one of the following antibodies: V5 (Cell Signaling), PFKFB3 (anti-mouse; Abnova), or control IgG mouse (Sigma). Protein G agarose (20 µL) or anti-Flag-conjugated agarose (20 µL) beads were added to each immunoprecipitate and rotated for 24 h. Beads were washed four times with lysis buffer, resuspended in SDS loading dye, and boiled prior to immunoblotting. For PFKFB3 immunoprecipitation followed by p-Ser269 immunoblotting, cells were lysed in the immunoblotting buffer.
Immunofluorescence and immunohistochemistry
Immunofluorescence staining was performed as described previously (Rosales et al. 2015). Cells were starved of glutamine for 24 h prior to staining with 1:75 diluted anti-IKKβ (Novus Biologicals 10AG2) and 1:300 diluted anti-PFKFB3 (OriGene TA590185) antibodies. Nuclei were visualized by staining with 1 µg/mL DAPI. Images were captured using a Zeiss LSM 700 confocal (immunofluorescence) or Zeiss Observer II (immunohistochemistry) microscope at 20× objective and analyzed using Zen 2012 software. Immunohistochemistry was performed as described previously (Fong et al. 2015) using 10 mM Tris-HCl, 1 mM EDTA, and 0.05% Tween-20 (pH 9.0) retrieval buffer. p-PFKFB3 Ser 269 antibody was used at 1:100 dilution. Tissue arrays were purchased from US Biomax (brain BS17016a, breast BC08013a, and skin BC21014)
Quantitative real-time PCR
Total RNA was extracted using Trizol (Invitrogen) and purified according to the manufacturer's guidelines. One microgram of RNA was added as a template to reverse transcriptase reactions carried out using the qScript cDNA synthesis kit (Quanta Biosciences). Quantitative real-time PCRs were carried out with the resulting cDNAs using SYBR Green PCR master mix (Quanta Biosciences) using an iQ5 real-time PCR machine (Biorad) with the following primers: 18S-F (5′-CCCGTTGAACCCCATTCGTGA-3′), 18S-R (5′-GCCTCACTAAACCATCCAATCGG-3′), GLUT3-F (5′-CAACTTCATGTCAACTTCTGGCT-3′), and GLUT3-R (5′-CTCAGTGAGAAATGGGACCCT-3′).
PFKFB3 activity assays
PFKFB3 activity was assayed using the Biovision, Inc. (K776-100), phosphofructokinase activity colorimetric assay kit according to the manufacturer's guidelines. Briefly, cells were seeded to 80% confluence followed by lysis using the provided lysis buffer. Cells were then spun down, and the supernatant was subjected to a 10-KD spin column to remove interfering small molecules. Lysates were the quantified, and 50 µg of protein was used per reaction.
Glucose/glutamine uptake and lactate production measurements
Cells were seeded at 1 × 105 cells per well into a six-well plate, and, the following day, medium was aspirated and replaced with 1 mL of fresh DMEM. Twenty-four hours later, medium was collected and measured using the Nova Biomedical BioProfile 100 plus machine. Cells were counted, and all metabolite measurements were then normalized per cell.
Seahorse assays
ECAR of cells was measured with the Searhorse Bioscience XF24 extracellular flux analyzer. Briefly, 5 × 104 cells per well were seeded in 525 µL of XF assay medium supplemented with 2 mM glutamine. Cells were incubated in a CO2-free incubator for 1 h at 37°C to allow for temperature and pH equilibration prior to loading into the XF24 apparatus. XF assays consisted of sequential mix (3 min), pause (3 min), and measurement (5 min) cycles, allowing for determination of ECAR every 10 min.
Kinase assays
Recombinant active GST-IKKβ or immunoprecipitated Flag-IKKβ was incubated for 30 min at 30°C with kinase assay buffer (Cell Signaling), 200 µM ATP (10 mM stock; Cell Signaling), the specified recombinant GST-tagged proteins, and 1 µL of [γ-32P]-ATP (10 mCi per milliliter of stock) in a final volume of 30 µL. The reaction was quenched by adding 10 µL of SDS loading dye, and reactions were separated by SDS-PAGE and exposed to autoradiograph film for 15 min to 1 h. For cold kinase assays, all reaction conditions were the same except for the addition of [γ-32P]-ATP.
13C-glucose tracing
Cells were seeded in a 6-cm plate at a density of 2 × 105 cells per dish. After overnight incubation, medium was removed, and cells were washed with 3 mL of PBS before the addition of glucose-free DMEM supplemented with 10% dialyzed and heat-inactivated FBS and 25 mM 13C-glucose (Cambridge Isotope Laboratory). After 6 h of incubation, the cells were harvested, and the metabolites were extracted as described previously (Liu et al. 2015). 13C metabolite measurements were determined using LC-HR-MS as described previously (Liu et al. 2015).
NADPH measurement
Cells were cultured for 24 h in complete medium and then measured using the NADP/NADPH-Glo kit (Promega, G9081) according to the manufacturer's guidelines.
Mouse xenografts
All studies involving animals were performed according to approved IACUC protocols at the City of Hope Cancer Center. In brief, 2 × 106 HT1080 cells were resuspended in 200 µL of DMEM, and tumors were established subcutaneously in 10-wk-old male athymic mice (Taconic). Mice were then placed into four groups and treated with PBS, 10 mg/kg DON, 2.5 mg/kg Bay, or the combination of 10 mg/kg DON and 2.5 mg/kg Bay three times per week for 2 wk, and tumor growth was measured over time using the formula 1/2(length × width2).
Statistics
Results are shown as means; error bars represent the standard error of the mean or standard deviation as indicated. The unpaired Student's t-test was used to determine the statistical significance of differences between means (P < 0.05 [*], P < 0.01 [**], and P < 0.005 [***]).
Supplementary Material
Acknowledgments
We thank members of the Kong laboratory for helpful comments on the manuscript. We thank Ross Tomaino at the Harvard Mass Spectrometry Core for assisting in the protein mass spectrometric peptide sequencing and phosphopeptide mapping experiments. Research is supported by National Institutes of Health/National Cancer Institute 1R01CA183989-01A1. M.A.R. is supported by the Ralph M. Parsons Foundation. M.K. is the Pew Scholar in the Biomedical Sciences, V Scholar in Cancer Research, and American Cancer Society Research Scholar. X.H.L. is supported by DNA Damage Response and Oncogenic Signaling (DDROS) Training Program at City of Hope. Research reported here includes work performed in Core Facilities supported by the National Institutes of Health/National Cancer Institute under grant number P30CA33572.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.287235.116.
References
- Almeida A, Bolanos JP, Moncada S. 2010. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci 107: 738–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. 2002. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res 62: 5881–5887. [PubMed] [Google Scholar]
- Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A. 2014. PRISM: a Web server and repository for prediction of protein–protein interactions and modeling their 3D complexes. Nucleic Acids Res 42: W285–W289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, Koike MA, Hancock SA, Bhatti TR, Han R, et al. 2015. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J 29: 2315–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroughs LK, DeBerardinis RJ. 2015. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S, Brookfield JL, Critchlow SE, Cumming IA, Curtis NJ, Debreczeni J, Degorce SL, Donald C, Evans NJ, Groombridge S, et al. 2015. Structure-based design of potent and selective inhibitors of the metabolic kinase PFKFB3. J Med Chem 58: 3611–3625. [DOI] [PubMed] [Google Scholar]
- Brasse-Lagnel C, Lavoinne A, Loeber D, Fairand A, Bole-Feysot C, Deniel N, Husson A. 2007. Glutamine and interleukin-1β interact at the level of Sp1 and nuclear factor-κB to regulate argininosuccinate synthetase gene expression. FEBS J 274: 5250–5262. [DOI] [PubMed] [Google Scholar]
- Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, Shoelson SE. 2005. Local and systemic insulin resistance resulting from hepatic activation of IKK-β and NF-κB. Nat Med 11: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo MN, Bartrons R, Castano E, Perales JC, Navarro-Sabate A, Manzano A. 2006. PFKFB3 gene silencing decreases glycolysis, induces cell-cycle delay and inhibits anchorage-independent growth in HeLa cells. FEBS Lett 580: 3308–3314. [DOI] [PubMed] [Google Scholar]
- Cavalier MC, Kim SG, Neau D, Lee YH. 2012. Molecular basis of the fructose-2,6-bisphosphatase reaction of PFKFB3: transition state and the C-terminal function. Proteins 80: 1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, Moncada S. 2011. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci 108: 21069–21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb WC, Cogswell P, Sitcheran R, Baldwin AS. 2011. IKK-dependent, NF-κB-independent control of autophagic gene expression. Oncogene 30: 1727–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb WC, Hutti JE, Cogswell P, Cantley LC, Baldwin AS. 2012. p85α SH2 domain phosphorylation by IKK promotes feedback inhibition of PI3K and Akt in response to cellular starvation. Mol Cell 45: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. 2010. The IKK complex contributes to the induction of autophagy. EMBO J 29: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. 2007. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. 2008. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20. [DOI] [PubMed] [Google Scholar]
- Domenech E, Maestre C, Esteban-Martinez L, Partida D, Pascual R, Fernandez-Miranda G, Seco E, Campos-Olivas R, Perez M, Megias D, et al. 2015. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nat Cell Biol 17: 1304–1316. [DOI] [PubMed] [Google Scholar]
- Duan S, Pagano M. 2011. Linking metabolism and cell cycle progression via the APC/CCdh1 and SCFβTrCP ubiquitin ligases. Proc Natl Acad Sci 108: 20857–20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earhart RH, Amato DJ, Chang AY, Borden EC, Shiraki M, Dowd ME, Comis RL, Davis TE, Smith TJ. 1990. Phase II trial of 6-diazo-5-oxo-L-norleucine versus aclacinomycin-A in advanced sarcomas and mesotheliomas. Invest New Drugs 8: 113–119. [DOI] [PubMed] [Google Scholar]
- Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, et al. 2015. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 17: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, Winter PS, Liu X, Priyadharshini B, Slawinska ME, et al. 2015. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest 125: 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloire G, Charlier E, Rahmouni S, Volanti C, Chariot A, Erneux C, Piette J. 2006. Restoration of SHIP-1 activity in human leukemic cells modifies NF-κB activation pathway and cellular survival upon oxidative stress. Oncogene 25: 5485–5494. [DOI] [PubMed] [Google Scholar]
- Hernandez-Davies JE, Tran TQ, Reid MA, Rosales KR, Lowman XH, Pan M, Moriceau G, Yang Y, Wu J, Lo RS, et al. 2015. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. J Transl Med 13: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Guo X, Li H, Wang H, Zhang W, Wang Y, Zhou H, Gao Z, Telang S, Chesney J, et al. 2010. Disruption of inducible 6-phosphofructo-2-kinase ameliorates diet-induced adiposity but exacerbates systemic insulin resistance and adipose tissue inflammatory response. J Biol Chem 285: 3713–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaadige MR, Looper RE, Kamalanaadhan S, Ayer DE. 2009. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci 106: 14878–14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. 2015. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res 75: 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi K, Araki K, Tobiume K, Tanaka N. 2008. p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat Cell Biol 10: 611–618. [DOI] [PubMed] [Google Scholar]
- Kisner DL, Catane R, Muggia FM. 1980. The rediscovery of DON (6-diazo-5-oxo-L-norleucine). Recent Results Cancer Res 74: 258–263. [DOI] [PubMed] [Google Scholar]
- Krishnan RK, Nolte H, Sun T, Kaur H, Sreenivasan K, Looso M, Offermanns S, Kruger M, Swiercz JM. 2015. Quantitative analysis of the TNF-α-induced phosphoproteome reveals AEG-1/MTDH/LYRIC as an IKKβ substrate. Nat Commun 6: 6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti C, Lin KM, Yamamoto Y, Verma U, Verma IM, Byers S, Gaynor RB. 2001. Regulation of β-catenin function by the IκB kinases. J Biol Chem 276: 42276–42286. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab 15: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, et al. 2007. IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 130: 440–455. [DOI] [PubMed] [Google Scholar]
- Liboni KC, Li N, Scumpia PO, Neu J. 2005. Glutamine modulates LPS-induced IL-8 production through IκB/NF-κB in human fetal and adult intestinal epithelium. J Nutr 135: 245–251. [DOI] [PubMed] [Google Scholar]
- Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. 1996. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol 16: 1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sadhukhan S, Sun S, Wagner GR, Hirschey MD, Qi L, Lin H, Locasale JW. 2015. High-resolution metabolomics with Acyl-CoA profiling reveals widespread remodeling in response to diet. Mol Cell Proteomics 14: 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27: 441–464. [DOI] [PubMed] [Google Scholar]
- Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, Moretti M, De Smaele E, Beg AA, Tergaonkar V, et al. 2011. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol 13: 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Hayden MS, Ghosh S. 2011. Crosstalk in NF-κB signaling pathways. Nat Immunol 12: 695–708. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Sakon S, Sasazuki T, Sakurai H, Doi T, Yagita H, Okumura K, Nakano H. 2003. Phosphorylation of serine 276 is essential for p65 NF-κB subunit-dependent cellular responses. Biochem Biophys Res Commun 300: 807–812. [DOI] [PubMed] [Google Scholar]
- Polley S, Huang DB, Hauenstein AV, Fusco AJ, Zhong X, Vu D, Schrofelbauer B, Kim Y, Hoffmann A, Verma IM, et al. 2013. A structural basis for IκB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS Biol 11: e1001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, Mayes PA, Wise DR, Thompson CB, Maris JM, et al. 2012. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 22: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MA, Wang WI, Rosales KR, Welliver MX, Pan M, Kong M. 2013. The B55α subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol Cell 50: 200–211. [DOI] [PubMed] [Google Scholar]
- Ren J, Chen X, Chen ZJ. 2014. IKKβ is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci 111: 17438–17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Frankel S. 1949. Free amino acids in normal and neoplastic tissues of mice as studied by paper chromatography. Cancer Res 9: 645–648. [PubMed] [Google Scholar]
- Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, Zamboni N, Schulze A. 2012. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov 2: 328–343. [DOI] [PubMed] [Google Scholar]
- Rosales KR, Reid MA, Yang Y, Tran TQ, Wang WI, Lowman X, Pan M, Kong M. 2015. TIPRL inhibits protein phosphatase 4 activity and promotes H2AX phosphorylation in the DNA damage response. PLoS One 10: e0145938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara R, Kato M, Okamura N, Nakagawa T, Komada Y, Tominaga N, Shimojo M, Fukasawa M. 1997. Characterization of a human placental fructose-6-phosphate, 2-kinase/fructose-2,6-bisphosphatase. J Biochem 122: 122–128. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israel A, Rajewsky K, Pasparakis M. 2003. Mature T cells depend on signaling through the IKK complex. Immunity 19: 377–389. [DOI] [PubMed] [Google Scholar]
- Schoonbroodt S, Ferreira V, Best-Belpomme M, Boelaert JR, Legrand-Poels S, Korner M, Piette J. 2000. Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of IκBα in NF-κB activation by an oxidative stress. J Immunol 164: 4292–4300. [DOI] [PubMed] [Google Scholar]
- Schoors S, De Bock K, Cantelmo AR, Georgiadou M, Ghesquiere B, Cauwenberghs S, Kuchnio A, Wong BW, Quaegebeur A, Goveia J, et al. 2014. Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab 19: 37–48. [DOI] [PubMed] [Google Scholar]
- Storz P, Toker A. 2003. Protein kinase D mediates a stress-induced NF-κB activation and survival pathway. EMBO J 22: 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P, Doppler H, Toker A. 2004. Protein kinase Cδ selectively regulates protein kinase D-dependent activation of NF-κB in oxidative stress signaling. Mol Cell Biol 24: 2614–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MP, Nelson JA, Feldman S, Van Nguyen B. 1988. Pharmacokinetic and phase I study of intravenous DON (6-diazo-5-oxo-L-norleucine) in children. Cancer Chemother Pharmacol 21: 78–84. [DOI] [PubMed] [Google Scholar]
- Takada Y, Mukhopadhyay A, Kundu GC, Mahabeleshwar GH, Singh S, Aggarwal BB. 2003. Hydrogen peroxide activates NF-κB through tyrosine phosphorylation of IκBα and serine phosphorylation of p65: evidence for the involvement of IκBα kinase and Syk protein–tyrosine kinase. J Biol Chem 278: 24233–24241. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Nussinov R, Keskin O. 2011. Predicting protein–protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 6: 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaftingen E, Hue L, Hers HG. 1980. Fructose 2,6-bisphosphate, the probably structure of the glucose- and glucagon-sensitive stimulator of phosphofructokinase. Biochem J 192: 897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaftingen E, Jett MF, Hue L, Hers HG. 1981. Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proc Natl Acad Sci 78: 3483–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. 1999. The SCFβ–TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev 13: 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. 2010. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 35: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MS, Raab RM, Rigoutsos I, Stephanopoulos GN, Kelleher JK. 2004. Metabolic and transcriptional patterns accompanying glutamine depletion and repletion in mouse hepatoma cells: a model for physiological regulatory networks. Physiol Genomics 16: 247–255. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Stine ZE, Xia J, Lu Y, O'Connor RS, Altman BJ, Hsieh AL, Gouw AM, Thomas AG, Gao P, et al. 2015. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Invest 125: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A, Clem BF, Imbert-Fernandez Y, Ozcan SC, Peker S, O'Neal J, Klarer AC, Clem AL, Telang S, Chesney J. 2014. 6-Phosphofructo-2-kinase (PFKFB3) promotes cell cycle progression and suppresses apoptosis via Cdk1-mediated phosphorylation of p27. Cell Death Dis 5: e1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Takano N, Ishiwata K, Ohmura M, Nagahata Y, Matsuura T, Kamata A, Sakamoto K, Nakanishi T, Kubo A, et al. 2014. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat Commun 5: 3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Xiang J, Lin Y, Ma J, Zhang J, Zhang H, Sun J, Danial NN, Liu J, Lin A. 2013. Inactivation of BAD by IKK inhibits TNFα-induced apoptosis independently of NF-κB activation. Cell 152: 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.