Abstract
Nematodes lack a heme biosynthetic pathway and must acquire heme from exogenous sources. Given the indispensable role of heme, this auxotrophy may be exploited to develop drugs that interfere with heme uptake in parasites. Although multiple heme-responsive genes (HRGs) have been characterized within the free-living nematode Caenorhabditis elegans, we have undertaken the first study of heme transport in Brugia malayi, a causative agent of lymphatic filariasis. Through functional assays in yeast, as well as heme analog, RNAi, and transcriptomic experiments, we have shown that the heme transporter B. malayi HRG-1 (BmHRG-1) is indeed functional in B. malayi. In addition, BmHRG-1 localizes both to the endocytic compartments and cell membrane when expressed in yeast cells. Transcriptomic sequencing revealed that BmHRG-1, BmHRG-2, and BmMRP-5 (all orthologs of HRGs in C. elegans) are down-regulated in heme-treated B. malayi, as compared to non–heme-treated control worms. Likely because of short gene lengths, multiple exons, other HRGs in B. malayi (BmHRG-3–6) remain unidentified. Although the precise mechanisms of heme homeostasis in a nematode with the ability to acquire heme remains unknown, this study clearly demonstrates that the filarial nematode B. malayi is capable of transporting exogenous heme.—Luck, A. N., Yuan, X., Voronin, D., Slatko, B. E., Hamza, I., Foster, J. M. Heme acquisition in the parasitic filarial nematode Brugia malayi.
Keywords: iron metabolism, infectious disease, parasite metabolism
Human filarial nematode infections responsible for lymphatic filariasis (caused by Wuchereria bancrofti, Brugia malayi, and Brugia timori) and onchocerciasis (caused by Onchocerca volvulus) affect nearly 150 million people worldwide (1). Medical treatment relies on sustained mass drug administrations of microfilaricidal therapeutics (e.g., albendazole, DEC, ivermectin) to disrupt transmission of the disease (2) but is contraindicated in regions where another filarial nematode, Loa loa, is endemic. Furthermore, as growing evidence of drug resistance in filarial nematodes emerges (3, 4), the development of safer macrofilaricidal treatment options has become an urgent need.
Although most cases of lymphatic filariasis are caused by W. bancrofti, B. malayi is frequently the subject of investigation because of its ability to maintain its life cycle in a laboratory setting. As with other filarial nematodes, transmission of B. malayi requires an arthropod vector (blood-feeding female mosquitoes) and a mammalian host (normally humans, although other mammals, e.g., Mongolian jirds, are used in the laboratory). Within an infected mammalian host, B. malayi adult males and females reside in the lymphatic vessels, where they reproduce and release microfilariae (mf). The mf migrate to the capillaries from which they can be ingested by a mosquito during a blood meal. Within the insect vector, mf penetrate the midgut, enter the thoracic muscle cells, and remain intracellular for 2 molts before migrating via the hemolymph to the mouthparts of the mosquito.
Tetrapyrroles, such as heme, are used in every kingdom of life and have become indispensable to many biologic processes by serving as a cofactor for numerous proteins. Most organisms are readily able to synthesize heme (5); however, all nematodes (either free-living or parasitic) studied to date lack a complete and functional heme biosynthetic pathway (6). As heme auxotrophs, helminths must acquire heme from an exogenous source. Given the essential role of heme, this auxotrophy in nematodes may be exploited to develop drugs that interfere with heme uptake and utilization. Although B. malayi contains a functional ferrochelatase gene (the final step in the heme biosynthetic pathway and a likely product of lateral gene transfer from a Rhizobales-related species) (7), like other nematodes, B. malayi is incapable of synthesizing heme (6). However, unlike most nematodes, B. malayi (and most other filarial nematodes) contain Wolbachia, an obligate α-proteobacterial endosymbiont present within the lateral cords of male and female worms and in the developing oocytes and embryos in females that are required for worm fertility and development. Genomic sequencing of Wolbachia from B. malayi (wBm) revealed a complete and likely functional heme pathway (8). Certain trypanosomatids are also incapable of synthesizing heme, but contain a β-proteobacterial endosymbiont capable of synthesizing and supplying the vital cofactor (9, 10). Precisely how heme homeostasis is maintained in B. malayi, which may acquire heme from its environment, as well as perhaps from its Wolbachia endosymbiont, has remained an unanswered question.
Multiple heme responsive genes (HRGs) have been identified and assigned various functions within Caenorhabditis elegans (11–13). Paralogs C. elegans HRG-4 and -1 (CeHRG-4 and -1) (11), are both high affinity regulated heme transporters involved in uptake and trafficking of heme within the intestine of C. elegans. Two other CeHRG-4 paralogs, CeHRG-5 and -6, are also involved in heme uptake within the intestine (13). Although heme uptake into the intestine is redundant (CeHRG-4–6 are all involved at various heme concentrations), heme export from the intestine is accomplished via the ABC-transporter C. elegans multidrug resistance protein 5 (CeMRP-5) (14). In addition, CeHRG-2 (a type I transmembrane protein involved in heme uptake and utilization within the hypodermis) (15) and CeHRG-3 (likely a heme chaperone involved in delivering maternal heme to developing oocytes) (12) have both been well characterized in C. elegans.
The B. malayi orthologs of CeHRG-1 [B. malayi HRG-1 (BmHRG-1), Bm5182, WormBase ID: WBGene00225443], CeHRG-2 (BmHRG-2, Bm2383, WormBase ID: WBGene00222644), and CeMRP-5 [B. malayi multidrug resistance protein 5 (BmMRP-5, Bm3373, WormBase ID: WBGene00223634] have all been identified, on the basis of protein sequence homology. However, probably because of short gene lengths that are split into multiple exons and large gaps in the genome assembly, BmHRG-3–6 have not been identified. Despite this, and to further understand the biology of heme metabolism in filarial worms, a potential target for filariasis control, we have undertaken this first study of heme homeostasis in B. malayi.
MATERIALS AND METHODS
Yeast strains and growth medium
The Saccharomyces cerevisiae strains used in this study were derived from the W303 and YPH499 backgrounds. The hem1Δ(6D) and OPY102 strains were constructed as described elsewhere (16, 17). To construct hem1Δ fre1Δ fre2Δ MET3-FRE1, plasmid pRS404-MET3-FRE1 was linearized with NdeI and integrated into the TRP1 locus of OPY102. Cells were maintained in yeast peptone dextrose (YPD) or appropriate synthetic complete (SC) medium supplemented with 250 μM δ-aminolevulinic acid (ALA) (Frontier Scientific, Inc., Logan, UT, USA) (18).
A codon optimized BmHRG-1 ORF (±HA tag) for yeast expression was synthesized by IDT (Integrated DNA Technology, Coralville, IA, USA), amplified by PCR using gene-specific primers containing the BamHI and XbaI sites, digested, and ligated to pYES-DEST52 vector (Thermo Fisher Scientific Life Sciences, Carlsbad, CA, USA) digested with the same enzymes.
Spot growth assay
The hem1Δ S. cerevisiae yeast strain lacks the first enzyme in the heme biosynthetic pathway, ALA synthase (ALAS). Because of the lack of ALAS, ALA (the product of ALAS) or excess hemin must be supplied exogenously in the growth medium, for the hem1Δ strain to grow. Plasmids for BmHRG-1 expression were transformed into strain hem1Δ(6D) using the lithium acetate method (19). Transformants were selected on 2% w/v glucose SC-Ura plates supplemented with 250 μM ALA. Five or 6 transformed colonies were picked and streaked on 2% w/v raffinose SC-Ura plates supplemented with 250 μM ALA for 48 h to deplete glucose. Before spotting, the cells were cultivated in 2% w/v raffinose SC-Ura medium for 18 h to deplete hemin. Cells were then suspended in water to an OD600 of 0.2. Ten-fold serial dilutions of each transformant were spotted (10 μl/spot) onto 2% w/v raffinose SC-Ura plates supplemented with 0.4% w/v glucose and 250 μM ALA (positive control) or 0.4% w/v galactose and different concentrations of hemin and then incubated at 30°C for 3 d before imaging.
Ferrireductase assay
The strain hem1Δ fre1Δ fre2Δ MET3-FRE1 was used for the ferrireductase assay. The iron- and copper-regulated endogenous genes for FRE1 and FRE2 (20, 21) have been deleted in this strain, which instead contains only 1 ferric reductase (FRE1) under control of the inducible MET3 promoter, thus making it possible to directly assay any changes in intracellular heme via ferric reductase activity caused by the expression of HRG-1 (22). Yeast transformation and selection were performed as described above using respective SC auxotrophic medium supplemented with 250 μM ALA. After being depleted of hemin in 2% w/v raffinose SC-Ura, -Trp, -Met medium for 12 h, cells were suspended in 2% w/v raffinose SC-Ura, -Trp medium supplemented with 0.4% w/v galactose, 0.1 mM Na2S, and various concentrations of hemin to an OD600 of 0.3. These were cultivated in 96-well plates at 30°C, with shaking at 225 rpm for 16 h and assayed for ferrireductase activity (20). The cells were washed with washing buffer (2% bovine serum albumin, 0.1% Tween-20 in 2× PBS) 3 to 4 times to remove residual hemin in the medium, washed twice with reaction buffer [(5% glucose and 0.05 M sodium citrate buffer (pH 6.5)], suspended in reaction buffer and the OD600 determined using a plate reader. Equal volume of assay buffer (2 mM bathophenanthroline disulfonate, 2 mM FeCl3 in reaction buffer) was added to the cells (t = 0 min) and incubated at 30°C in the dark until red color developed. OD535 and OD610 were determined, and ferrireductase activity (nmol/106cells/min) was calculated as:
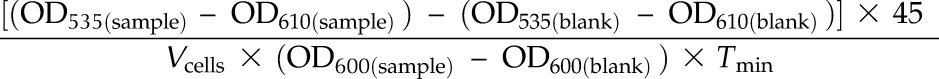 |
β-Galactosidase reporter assay
The plasmids for BmHRG-1 expression were cotransformed into strain hem1Δ(6D) with pCYC1-LacZ. Selection of transformants was performed as described above using appropriate SC auxotrophic medium supplemented with 250 μM ALA. Cells were depleted of hemin in 2% w/v raffinose SC-Ura, -Trp medium for 12 h, and then were suspended in 10 ml 2% w/v raffinose SC-Ura, -Trp medium supplemented with 0.4% w/v galactose, and different concentrations of hemin to an OD600 of 0.1. Cells were cultivated at 30°C, with shaking at 225 rpm for 12 h and assayed for β-galactosidase activity, as described elsewhere (23). β-Galactosidase activities were normalized to total protein concentration.
Immunoblot analysis
For Western blot analysis experiments, yeast transformants were resuspended in lysis buffer [1% SDS, 8 M urea, 10 mM Tris-HCl (pH 8.0), and 10 mM EDTA] with protease inhibitors (1 mM PMSF, 4 mM benzamidine, 2 μg/ml leupeptin, and 1 μg/ml pepstatin) and 0.5 mm diameter acid-washed glass beads. Cells were heated at 65°C for 10 min and disrupted in FastPrep-24 (MP Biomedicals, Santa Ana, CA, USA) 3 times for 30 s each at the 6.5 m/s setting. Cell lysates were collected, and the total protein concentration was quantified with the Bradford reagent (Bio-Rad, Hercules, CA, USA). Protein samples were resolved on 12% SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Bio-Rad). For immunoblot analysis, the membranes were incubated with rabbit anti-HA (Sigma-Aldrich, St. Louis, MO, USA) as primary antibody at a 1:5,000 dilution for 16 h at 4°C, followed by HRP-conjugated goat anti-rabbit antibody at a 1:10,000 dilution for 1 h at room temperature. Signal was detected using SuperSignal chemiluminescence reagents (Thermo Fisher Scientific Life Sciences) in a gel documentation system (Bio-Rad).
Immunofluorescence
Yeast transformants were cultivated in 2% w/v raffinose SC-Ura medium supplemented with 0.4% w/v galactose and 250 μM ALA to midlog phase and then fixed with 4% formaldehyde for 1 h at room temperature. Immunofluorescence microscopy was performed as described elsewhere (23). Images were taken using a DM IRE2 epifluorescence microscope (Leica, Wetzlar, Germany) connected to a Retiga 1300 cooled Mono 12-bit camera (Retiga, QImaging, Surry, BC, Canada).
B. malayi in vitro culture
Unless otherwise noted, B. malayi mf and adult worms (TRS Labs, Athens, GA, USA) were incubated in RPMI 1640 medium (containing 25 mM HEPES, 5 mM glutamine, 200 μg/ml penicillin, and 200 μg/ml streptomycin) at 37°C, 5% CO2. All hemin and heme analog solutions were prepared in 300 mM ammonium hydroxide and pH adjusted to pH 8.0 with 6 M HCl before filter sterilization.
Production of rabbit polyclonal antibodies to BmHRG-1
Anti-BmHRG-1 serum was raised against a peptide of the 18 C-terminal residues of BmHRG-1 conjugated to KLH via an N-terminal cysteine using m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS; Thermo Fisher Scientific Life Sciences) (24). Sera were raised in rabbits by Covance Immunology Services (Princeton, NJ, USA). Antibodies were purified according to a published procedure (25).
B. malayi protein extraction and immunoblot analysis
Live B. malayi mf and adult male and female worms were incubated for 24 h in RPMI-1640 containing 0 (control), 5, 20, or 100 μM hemin chloride (Frontier Scientific, Inc.) before being flash frozen at −80°C. For extraction of total protein, frozen worm samples were thawed on ice before being washed 3 times with 200 μl of 1× PBS (pH 7.4). Samples were resuspended in 200 μl of tissue extraction reagent I (Thermo Fisher Scientific Life Sciences) containing protease cocktail inhibitor (Sigma-Aldrich). Worm samples were then homogenized with ceramic beads in CK14 tubes (3 × 30-s 5000 rpm pulses, with 1 min ice in between) using a Minilys homogenizer (Precellys-Bertin Technologies, Rockville, MD, USA). Protein concentrations were determined with Bradford reagent (Bio-Rad) (26). Protein extracts were resolved by SDS-PAGE on 10–20% Tris-glycine gels (Thermo Fisher Scientific Life Sciences). Proteins were transferred to nitrocellulose (200 mA, constant for 2 h). Following overnight blocking in 5% milk/ Tris-buffered saline/0.1% Tween-20 (TBST), the immunoblot was analyzed using the purified anti-BmHRG-1 rabbit sera described above (1:5000 dilution in 2% milk/TBST) as the primary antibody. A horseradish peroxidase-linked anti-rabbit secondary antibody (Cell Signaling Technology, Danvers, MA, USA) and the ECL Western Blot analysis Detection Kit (GE Healthcare, Pittsburgh, PA, USA) were used to detect bound antibody.
Measurement of intracellular B. malayi heme concentration
The heme concentrations within B. malayi adults and mf were determined by using a published pyridine hemochrome method (27, 28). In brief, adult worms (18 females and 27 males) were washed once in PBS. For mf studies, to normalize each sample by number of mf, a 1:100 dilution of each mf sample (1 ml) was counted before centrifugation and a subsequent wash in PBS. All samples were then stored at −80°C until further use. For heme extraction and quantification, the samples were resuspended in 840 μl of 1 mM Tris-HCl (pH 8.0) and homogenized with ceramic beads in CK14 tubes using a Minilys homogenizer (Precellys-Bertin Corp.) (30-s 5000 rpm pulses 3 times, with 1 min ice in between). Another 840 μl of 1 mM Tris-HCl (pH 8.0) was added, and mixed (total volume 1680 μl). After 840 μl was transferred into each of two 13 × 100 mm glass tubes (duplicates), 100 μl of 1 N NaOH was added, and the tube vortexed. After 2 min, 200 μl of pyridine solution (Sigma-Aldrich) was added and mixed by vortexing. Samples were transferred to a 1 ml cuvette, and the baseline absorbance (at 541 and 557 nm) was read. Sodium hydrosulfite (Sigma-Aldrich) crystals (2–3 mg) were added, the sample was mixed by gentle pipetting and the reduced absorbance at 541 and 557 nm acquired. Heme concentrations were calculated based on millimolar absorption coefficient of 20.7 for the difference in absorption between the spectrum peak at 557 nm and the valley at 541 nm. B. malayi mf heme concentrations were normalized based on mf counts in each sample.
Ex vivo motility of heme-treated B. malayi
B. malayi adults were cultured (1 male or female/well, in 24-well plates) in 1 ml RPMI-1640 medium (control), supplemented with 5, 20, or 100 μM hemin chloride at 37°C. Medium was changed every 2 d. Motility was scored daily, based on a previously described motility scoring system (7), where 0 is nonmotile and 4 is highly active.
Gallium protoporphyrin IX toxicity assays
B. malayi adults were cultured (6 adult male or female worms/group) in RPMI-1640 medium supplemented with either gallium protoporphyrin IX (GaPPIX; Frontier Scientific, Inc.) or gallium chloride (GaCl3; Sigma-Aldrich). The effect of heme on GaPPIX cytotoxicity was examined by incubating B. malayi adults with 2, 5, or 20 μM GaPPIX and increasing hemin concentrations (10, 20, or 50 μM heme). Motility was assayed as described above. Viability of GaCl3-treated worms was determined by the previously described MTT assay (29, 30).
Zinc mesoporphyrin pulse-chase analysis
B. malayi adults were cultured (1 male or female/well, in 24-well plates) in RPMI-1640 medium containing 40 μM zinc mesoporphyrin (ZnMP; Frontier Scientific, Inc.) for 18 h at 37°C (previously determined conditions, data not presented). Fluorescently labeled worms were then transferred into fresh RPMI-1640 medium (1 adult male or female/well, in 24-well plates) containing unlabeled hemin chloride. Control worms were transferred into fresh RPMI-1640 medium containing no heme analog. At timed intervals (0, 2, 4, 8, and 24 h), aliquots of worms (8 adult male and female worms/timepoint) were fixed in 80% ethanol, mounted on a slide, and immediately analyzed using an epifluorescence microscope [differential interference contrast (DIC) and rhodamine channels; Axiovert 200M; Zeiss, Oberkochen, Germany].
BmHRG-1 hairpin small interfering RNA preparation
The preparation of BmHRG-1 hairpin small interfering (hsi)RNA was as previously described by Landmann et al. (31). Total RNA (700 ng from B. malayi females) was used as a template for the production of cDNA, with random primers and the ProtoScript AMV First Strand cDNA Synthesis Kit (New England BioLabs, Inc.). DNA templates for in vitro transcription were generated by PCR using Crimson Taq DNA Polymerase (New England BioLabs, Inc.) and BmHRG-1 gene–specific PCR primers (designed to yield a PCR product corresponding to ∼400 bp and also containing a T7 promoter sequence followed by 2 guanine bases at the 5′ end for transcription by T7 RNA polymerase; left primer: 5′-TAATACGACTCACTATAGGGGGCTTTGACATGCAAGATGA-3′, right primer: 5′-TAATACGACTCACTATAGGGATACCACGCCGAAAGCATAG-3′) (Integrated DNA Technology). BmHRG-1 specific double-stranded (ds)RNA was prepared using the T7 Quick High Yield RNA Synthesis Kit (New England BioLabs, Inc.) and purified by isopropanol precipitation. The dsRNA was processed into hsiRNA using ShortCut RNase III (New England BioLabs, Inc.), purified by ethanol precipitation and resuspended in distilled H2O. Agarose gel electrophoresis alongside the siRNA Marker (New England BioLabs, Inc.) was used to examine the size and purity of the BmHRG-1 hsiRNA. A Nanodrop spectrophotometer (Thermo Fisher Scientific Life Sciences) was used to quantify BmHRG-1 hsiRNA.
In vitro hsiRNA soaking and analysis of BmHRG-1 knockdown
In vitro RNA interference was accomplished by adding various amounts of BmHRG-1 hsiRNA (0–5 μM) to a 12-well plate and adding warm RPMI-1640 medium (1 ml final volume). Adult female worms (1 worm/well, 6 worms/treatment group) were incubated at 37°C (5% CO2), and the hsiRNA/medium was changed every 12 h. Microfilarial output from female worms was assessed after every hsiRNA treatment by counting the number of mf in 10 μl of medium from each well.
BmHRG-1 knockdown was assessed by using various experimental methods: 1) Following 48 h (4 total treatments) of hsiRNA (0, 1 or 5 μM), worms were soaked in RPMI-1640 medium containing 40 μM ZnMP for 18 h before the fluorescence was visualized, as described above. 2) After 24 h of soaking in 0.5 μM BmHRG-1 hsiRNA, worms were soaked in RPMI-1640 containing 20 μM heme (with or without RNAi) for 7 d (medium was changed every 24 h before heme was extracted, and total intracellular heme content determined. 3) After 24 h of soaking in BmHRG-1 hsiRNA (0.5 or 1 μM), worms were soaked in RPMI-1640 containing GaPPIX (0, 1, 2, 5, or 20 μM, with or without RNAi). Medium was changed every 24 h, and motility was scored daily as described above. Control worms were treated in a similar manner in the absence of any hsiRNA.
Transcriptome sequencing and bioinformatic analysis
B. malayi mf and adult worms (1 male or female/well, in 24-well plates) were incubated in RPMI 1640 medium (control) supplemented with 20 or 100 μM heme (hemin chloride) at 37°C . After 24 h, the worms were harvested, flash frozen in liquid nitrogen and stored at −80°C until further use. For RNA preparation, samples were homogenized with ceramic beads in CK14 tubes using a Minilys homogenizer (as above) and total RNA was extracted by organic extraction with Trizol (Thermo Fisher Scientific Life Sciences). Samples were treated with DNase I (Thermo Fisher Scientific Life Sciences) before further Trizol extraction and final purification. The RNA integrity, purity and concentration of all samples were assessed with a Bioanalyzer 2100 (Agilent Technologies, Lexington, MA, USA). Samples were prepared for sequencing by using the NEBNext mRNA Library Prep Master Mix Set for Illumina (New England BioLabs, Inc.). Library quality was assessed before transcriptomic sequencing of 50 bp single-end reads on an Illumina Genome Analyzer IIx sequencer.
All data was analyzed with a local version of Galaxy (32–34). Sequence reads from each tissue sample were first assessed for quality using FastQC (v., 1.0.0; Babraham Institute, Babraham, United Kingdom; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (35), and further analyzed by using the Tuxedo protocol (36). RNA-Seq reads from each sample were aligned to the B. malayi genome (Wormbase, v. WS236) using TopHat, (v. 1.4.1; Johns Hopkins University, Baltimore, MD, USA; http://ccb.jhu.edu/software/tophat/index.shtml (37). Default parameters were used, except that the maximum number of alignments allowed was set to 40. Reads aligned using TopHat were assembled into transcripts by using Cufflinks (v. 1.3.0; Cole Laboratory, University of Washington, Seattle, WA, USA; http://cole-trapnell-lab.github.io/cufflinks/). Default parameters were used. Cufflinks assemblies from all samples were merged by Cuffmerge (v.1.0.0) and used for differential expression testing by Cuffdiff (v.1.3.0), with the false-discovery rate set to 0.01.
In situ hybridization
B. malayi cDNA was synthesized from 1 μg of adult female total RNA using the Protoscript II First Strand cDNA Synthesis Kit (New England BioLabs, Inc.). BmHRG-1 primers were designed with Primer 3 and synthesized by Integrated DNA Technology. BmHRG-1 was amplified by PCR using Taq DNA polymerase (New England BioLabs, Inc.) from the B. malayi 1st strand cDNA. Amplified BmHRG-1 fragments were ligated into the pCRII vector (Thermo Fisher Scientific Life Sciences). Ligated plasmids were transformed into INVαF′ competent cells (Thermo Fisher Scientific Life Sciences). Transformed cells were grown on X-Gal/IPTG plates, the recombinants selected and grown overnight before plasmid isolations, using the Monarch Plasmid Miniprep Kit (New England BioLabs, Inc.). BmHRG-1 inserts were confirmed by sequencing and used as templates for RNA probe labeling.
Sense (negative control) and antisense (experimental) RNA probes were prepared by in vitro transcription from linearized plasmids (1.25 μg) containing BmHRG-1 inserts with flanking Sp6 and T7 RNA polymerase transcription start sites. In brief, reactions (60 μl) contained 1× RNA polymerase buffer (New England BioLabs, Inc.); 20 nmol each of dCTP, dGTP, dATP, and fluorescein-12-dUTP (FITC-dUTP; Thermo Fisher Scientific Life Sciences); 40 units RNase Inhibitor (New England BioLabs, Inc.); and 80 units of RNA polymerase (Sp6 or T7; New England BioLabs, Inc.) for 4 h at 40°C. Synthesized RNA probes were DNAse I (Thermo Fisher Scientific, Inc.) treated (37°C for 30 min), purified by ethanol precipitation, resuspended in 1× TE buffer, and stored at −80°C.
For the visualization of the RNA probe, B. malayi adult females were fixed and permeabilized with 4% formaldehyde in PBS (Sigma-Aldrich) with 0.1% Triton-X100 for 20 min. During fixation, worms were cut several times to improve the penetration of the reagents. Samples were then washed 3 times in PBS. Additional permeabilization of samples was performed by incubating with Proteinase K [20 µg/ml in 50 mM Tris-HCl, (pH 7.4); New England BioLabs, Inc.] 15 min at 37°C. The samples were washed with PBS and prehybridized for 2 h at 58°C in hybridization buffer [45% deionized formamide, 4× saline sodium citrate (SSC) buffer, 10 mM DTT, 100 µg/ml yeast transfer RNA, and 40 µg/ml denatured and sheared salmon sperm DNA). The buffer was replaced with fresh hybridization buffer containing 10 ng of the probe, and the samples were incubated at 58°C overnight. After the hybridization, samples were washed 2 times (15 min each) with 2× SSC buffer at 37°C, 2 times (15 min each) with 1× SSC buffer at 37°C, and 2 times (15 min each) with 0.5× SSC buffer at room temperature. To digest unbound single-stranded RNA probe, samples were incubated with RNase A [20 µg/ml, in 500 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA (pH 8.0)] at 37°C for 1 h. The samples were washed in PBS and mounted in glycerol/PBS (1:1) solution on a slide. The samples on the slides were viewed using a 510 Meta confocal microscope (Zeiss, Oberkochen, Germany). The same procedure was performed for both sense- and antisense-oriented RNA probes.
RESULTS
BmHRGs
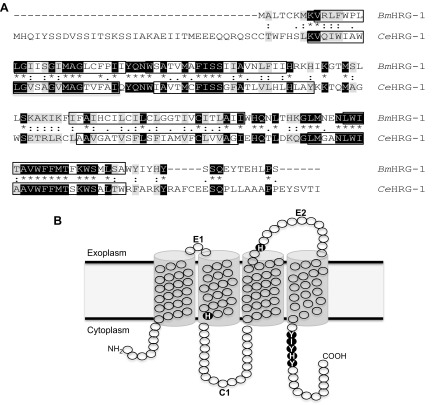
To date, only the B. malayi orthologs of CeHRG-1 (BmHRG-1), CeHRG-2 (BmHRG-2), and CeMRP-5 (BmMRP-5) have been identified based on sequence homology. BmHRG-1, although slightly smaller in size (148 vs. 194 aa), was identified because of its sequence homology to CeHRG-1 (∼39% identical) (Fig. 1A). The BmHRG-1 (also known as Bm5182, WormBase ID: WBGene00225443) locus (Bmal_v3_scaffold110:17681-19284) is composed of 4 exons. As with CeHRG-1, BmHRG-1 is predicted to contain 4 transmembrane helices connected by 2 exoplasmic loops, whereas the N and C termini are predicted to be cytoplasmic (Fig. 1B; TMHMM, v. 2.0; Prediction of Transmembrane Helices in Proteins; Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark; http://www.cbs.dtu.dk/services/TMHMM/). Two histidine residues, one in extracellular loop 2 and another in the second transmembrane domain (His90/His135 in CeHRG-1) identified as being critical to heme transport, are conserved in BmHRG-1 (His55/His100) (38). Moreover, a C-terminal cluster of aromatic and basic amino acids (FARKY in CeHRG-1) (38), which may be involved in translocation of heme into the cytoplasm, is conserved in BmHRG-1 (YIYHY).
Figure 1.
A) ClustalW amino acid alignment shows similarities between BmHRG-1 and CeHRG-1. Identical residues are shown in black, while conserved residues are in gray. B) The putative membrane topology of BmHRG-1 includes 4 transmembrane domains (boxed regions in A, as predicted by TMHMM 2.0) as well as a conserved histidine residue (His100) and C-terminal motif (YIYHY, dark circles) important for heme uptake (38).
HRG-2, through experiments in C. elegans, has been shown to be a slightly different member of the HRG family. A single-pass type I transmembrane protein, CeHRG-2 localizes to the endoplasmic reticulum and apical plasma membrane of hypodermal cells (15). As a single-pass membrane protein, CeHRG-2 is unlikely to function as a heme transporter, but contains a thioredoxin-like fold and glutathione S-transferase domain in the C terminus that may allow it to function as an oxidoreductase (13). Although the 2 are fairly similar (∼31% identical/51% similar), BmHRG-2 is slightly larger in size (290 vs. 279 aa) than its C. elegans homolog and does not contain any predicted transmembrane domains. However, BmHRG-2 does maintain the thioredoxin-like fold and glutathione-S transferase domain found in CeHRG-2 and may also therefore serve as an oxidoreductase, although its intracellular location remains unclear.
In C. elegans, an ABC transporter, CeMRP-5, localizes to the basolateral membrane of the intestine and appears to be the only mechanism for heme export from the intestine of C. elegans (14). The B. malayi homolog, BmMRP-5, is fairly similar (48% identical/66% similar) to CeMRP-5 (1473 vs. 1400 aa) and is likely to function in much the same manner.
BmHRG-1 overexpression, localization, and in vivo functionality
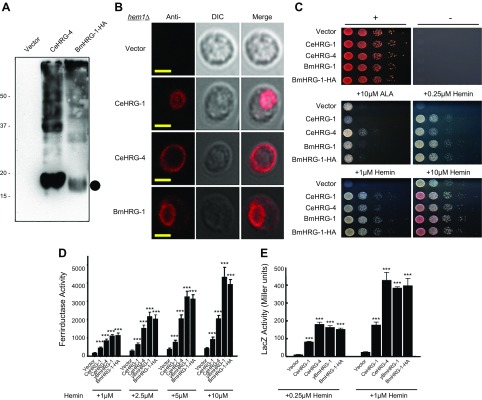
Immunoblot analysis of lysates from the yeast hem1Δ strain expressing a codon-optimized C-terminal HA-tagged BmHRG-1 revealed that BmHRG-1 migrates at the predicted molecular mass (∼17 kDa; Fig. 2A). However, expression of BmHRG-1 was much lower than the CeHRG-1 paralog CeHRG-4. As with overexpression of other HRGs in yeast, multiple oligomeric forms of BmHRG-1 are detected. Although CeHRG-1-HA appears to localize to endosomal compartments (yeast vacuole), and CeHRG-4-HA localizes to the plasma membrane, BmHRG-1-HA appears to localize to both membranes, as indicated by indirect immunofluorescence of the transformed hem1Δ strain (Fig. 2B).
Figure 2.
BmHRG-1 overexpression, localization, and functionality in S. cerevisiae. A) Immunoblot analysis of yeast transformants overexpressing C-terminal HA-tagged versions of CeHRG-4 (5 μg total extract) or BmHRG-1 (30 μg total extract) in the hem1Δ strain. The predicted molecular mass (∼17 kDa) of BmHRG-1 is indicated by the filled circle. B) Indirect immunofluorescence images of the hem1Δ strain transformed with vector alone, CeHRG-1-HA, CeHRG-4-HA, or BmHRG-1-HA. Scale bars, 5 μm. C) Spot-growth assay. Yeast (hem1Δ) transformed with empty vector, CeHRG-4, CeHRG-1, BmHRG-1, or BmHRG-1-HA spotted in serial dilutions on plates supplemented with 10 μM ALA or the indicated hemin concentration and incubated at 30°C for 3 d before imaging. Positive control (top left): +0.4% glucose, +250 μM ALA; negative control (top right): −ALA, −hemin. D) The hem1Δfre1Δfre2ΔMET3-FRE1 strain was transformed with empty vector, CeHRG-1, CeHRG-4, BmHRG-1, or BmHRG-1-HA and ferric reductase activity (nanomoles/106cells/min) was measured in the presence of hemin (as indicated). E) The hem1Δ strain stably expressing either the pYes-DEST52 vector alone, CeHRG-1, CeHRG-4, or BmHRG-1 (untagged as well as HA tagged) were transformed with pCYC1-LacZ. Following growth in the indicated concentrations of hemin after 12 h, β-galactosidase activity of the cell lysates was determined. The results are means ± sem of 3 independent experiments. ***P < 0.001, Student’s t test.
To verify the functionality of BmHRG-1, spot growth assays were performed using the hem1Δ S. cerevisiae yeast strain, growth of which necessitates that either ALA or hemin be exogenously supplied (Fig. 2C). As with CeHRG-1 and -4, we found that yeast transformed with BmHRG-1 (with or without the HA-tag) rescued growth of the hem1Δ strain in relatively low concentrations of hemin (0.25 vs. 10 μM for the vector control).
Using the transformed hem1Δ yeast strain, we assayed changes in the intracellular heme pool as a result of expressing BmHRG-1. First, we measured the ferrireductase activity (a heme-dependent enzyme) in the hem1Δfre1Δfre2ΔMET3-FRE1 strain transformed with CeHRG-1, CeHRG-4, BmHRG-1, or BmHRG-1-HA. Heme imported by BmHRG-1 was indeed incorporated into intracellular hemoproteins, as evidenced by a significant increase in ferrireductase activity (Fig. 2D). Next we measured intracellular heme as a function of Hap1-5 regulated β-galactosidase activity from a CYC1::lacZ promoter–reporter fusion. In this experiment, β-galactosidase activity is a direct measure of intracellular heme content, as lacZ expression is dependent on Hap1-5, a heme-binding transcription factor (38). As with CeHRG-1 or -4, the hem1Δ yeast strain expressing the CYC1::lacZ promoter-reporter fusion construct showed a significant increase in β-galactosidase activity when expressing BmHRG-1 (Fig. 2E). Based on both the ferrireductase and β-galactosidase activity assays, BmHRG-1 is at least as effective as CeHRG-1 or -4 at transporting heme.
BmHRGs promote heme uptake and increase the heme content of B. malayi
Immunoblot analysis of protein lysates (30 μg total protein extract) from B. malayi mf, adult females and males treated with increasing concentrations of heme revealed that endogenous protein levels of BmHRG-1 were much higher in mf than adult females and almost undetectable in adult males (data not shown), suggesting that heme requirements may be more significant in adult females and the microfilarial stage. As with overexpression of HRGs in yeast, multiple oligomeric forms of BmHRG-1 are detected within B. malayi. Moreover, the monomeric form detected by immunoblot analysis is slightly larger than the predicted monomer molecular mass (∼17 kDa), possibly suggesting the existence of in vivo posttranslational modifications.
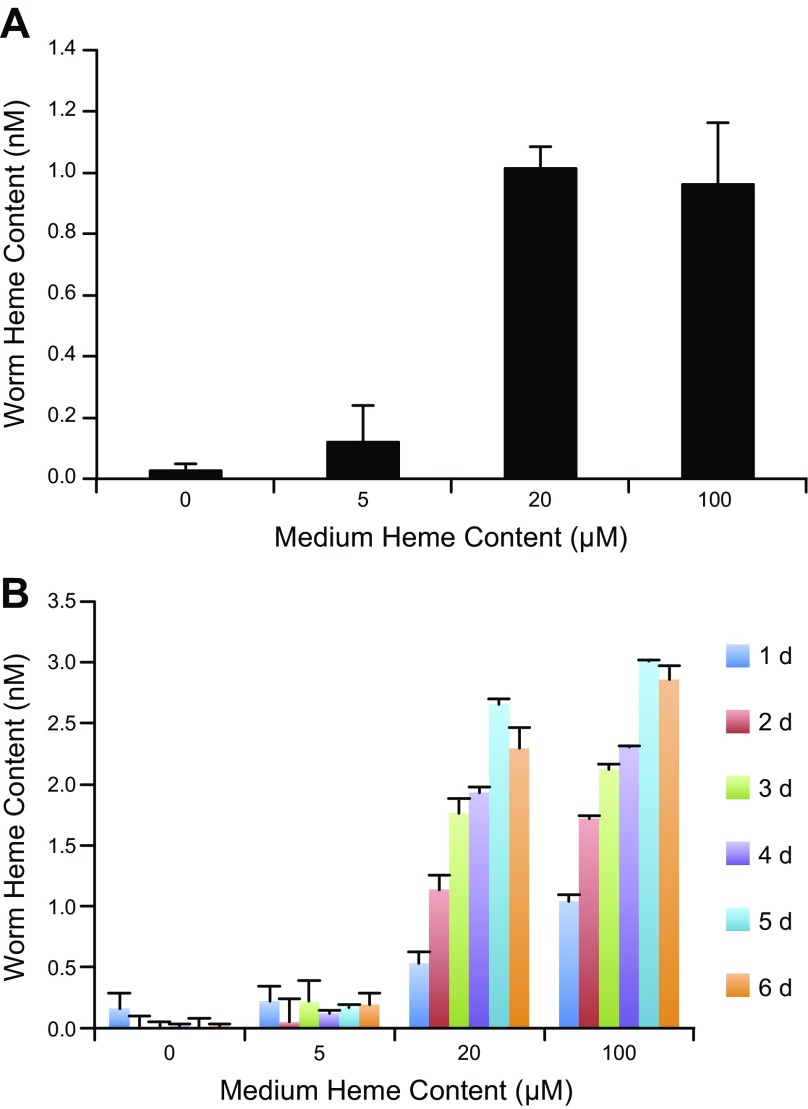
To evaluate the functionality of BmHRGs in nematodes, the effect of exogenous heme on the motility of adult B. malayi males and females was examined. Most likely because of the cytotoxic effects of heme, worm motility was reduced by exposure to heme as compared to the untreated controls in both adult B. malayi females and males (Supplemental Fig. S1A, B). In addition, elevated concentrations of total heme were observed in B. malayi adult females and mf after being cultured in the presence of increasing concentrations of heme (Fig. 3). However, no significant differences in total worm heme content were observed above 20 μM heme exposure, suggesting total heme content is regulated in vivo. Heme concentrations were undetectable from a pool of 27 adult male worms, likely either because of a lack of biomass or limited heme uptake in this life cycle stage.
Figure 3.
BmHRG-1 and its effects on heme content in B. malayi. A) Heme content of 18 adult female worms after 7 d in culture with increasing concentrations of heme. B) Heme content of mf incubated in increasing concentrations of heme over time (6 d). Heme content of mf was normalized by the number of mf in each sample.
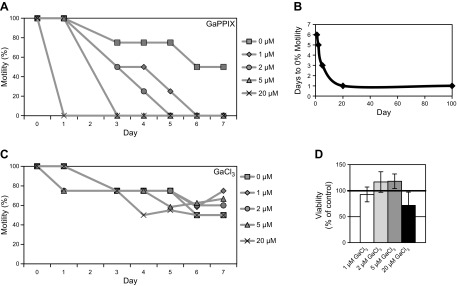
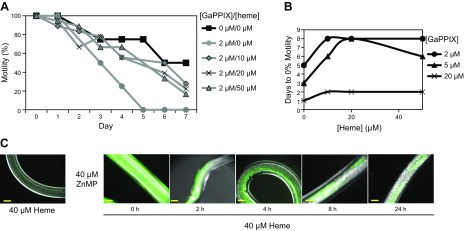
Highly cytotoxic noniron metalloporphyrins commonly exploit heme transport machinery and serve as potent antimicrobials (39). Studies have shown that CeHRG-1 is capable of mediating the transport of 6 different noniron metalloporphyrins (6, 11). The functionality of BmHRG-1 was further demonstrated by using the structurally similar heme analogs, GaPPIX and ZnMP. Gallium effectively mimics iron because of its similar atomic radius, electronic configuration, and +3 oxidation state (40). However, unlike Fe3+, Ga3+ cannot be reduced to Ga2+ under normal physiologic conditions (40), making it highly cytotoxic. Therefore, as expected, GaPPIX has been determined to be more than 800-fold more cytotoxic to C. elegans than is iron-containing heme (6). To determine whether GaPPIX may be taken up via the same mechanisms as heme in B. malayi and have similar cytotoxic effects, we exposed B. malayi adult females, ex vivo, to increasing concentrations of GaPPIX and monitored effects on motility. Exposure to GaPPIX had a much more pronounced negative effect on adult female B. malayi motility than heme (Fig. 4A). Exposure to only a 5 μM concentration of GaPPIX caused adult worms (both male and female) to become immotile within 3 d in culture, whereas adult female B. malayi cultured in the same concentration of heme (5 μM) displayed ∼90% of the motility of control worms on d 3 (Supplemental Fig. S1A). Moreover, concentrations of GaPPIX as low as 1 μM eliminated worm motility after 6 d in culture (Fig. 4B). Neither motility nor viability (as measured by the MTT assay) of B. malayi adult females was significantly affected by exposure to the gallium salt, GaCl3 (Fig. 4C, D), suggesting the cytotoxic effect observed with GaPPIX was not because of spontaneous release of free gallium.
Figure 4.
Effects of the heme analog GaPPIX on B. malayi motility. A) Motility of B. malayi females cultured with 0, 1, 2, 5, or 20 μM GaPPIX. B) Female B. malayi immotility as a function of GaPPIX concentration. C) Motility of B. malayi females cultured with 0, 1, 2, 5, or 20 μM GaCl3. Motility was assessed on a scale from 0 (nonmotile) to 4 (highly motile). Each data point represents motility from a single experiment where worms were scored as a group (6 adult worms/treatment group) (A, C). D) Viability of B. malayi females cultured in the presence of GaCl3 for 7 d. Viability was measured by the previously described MTT assay and the value of the control group (0 μM GaCl3) was set to 100 for comparison.
To further investigate heme uptake in B. malayi, live adult worms were soaked in ZnMP, a fluorescent heme analog. Fluorescence microscopy revealed concentrations as low as 5 μM ZnMP for 18 h resulted in detectable fluorescence (data not shown). Given the diffuse fluorescent signal of ZnMP, it is unclear what tissues or organs within the worm accumulate the heme analog. Concentrations of 40 μM ZnMP were used for further experiments, because this was the lowest concentration tested that produced the most consistent fluorescent accumulation.
To correlate the effects observed with the various heme analogs with heme transport, adult B. malayi females were exposed to 2 μM GaPPIX in the presence of increasing concentrations of heme (0, 10, 20, or 50 μM). The presence of heme in the medium provided protection from the GaPPIX-induced cytotoxicity, as indicated by the fact that worms incubated with GaPPIX + heme maintained ∼20% motility even after 7 d, whereas worms treated with only 2 μM GaPPIX were reduced to 0% motility after only 5 d in culture (Fig. 5A). Furthermore, this heme-induced protection was evident even at higher concentrations of the toxic GaPPIX (5 and 20 μM) (Fig. 5B). To correlate ZnMP fluorescence with heme transport, the competitive effect of heme on ZnMP fluorescence was determined in a pulse–chase analysis. Worms were first fluorescently labeled by incubation in 40 μM ZnMP for 18 h before being washed to remove nonspecifically bound ZnMP and then incubated in 40 μM heme. The ZnMP fluorescence accumulated during the pulse slowly diminished over time in worms incubated in heme (Fig. 5C). Experiments performed in the absence of any unlabeled heme in the chase periods (to test for nonspecific fluorescence loss) showed no significant depreciation in ZnMP fluorescence in the 24-h time period (data not shown). Likewise, negligible fluorescence was detected in worms incubated only in 40 μM heme (Fig. 5C).
Figure 5.
Characterization of heme uptake in B. malayi. A) B. malayi females were cultured in RPMI-1640 medium containing 2 μM GaPPIX and increasing hemin (0, 10, 20, or 50 μM). Motility was assessed as described in Materials and Methods on a scale from 0 (nonmotile) to 4 (highly motile). Each data point represents motility from a single experiment where worms were scored as a group (6 adult worms/treatment group) (A, B). The control groups (negative control: 0 μM GaPPIX/0 μM heme; positive control: 2 μM GaPPIX/0 μM heme) are taken from Fig. 4A as a reference. B) GaPPIX-induced immotility in female B. malayi is attenuated in the presence of heme. B. malayi females (6 worms/treatment group) were cultured in RPMI-1640 medium containing 2, 5, or 20 μM GaPPIX and increasing hemin (0, 10, 20, or 50 μM). C) Fluorescent labeling of female B. malayi worms incubated with 40 μM ZnMP for 18 h followed by a chase with 40 μM heme (hemin chloride). Worms were analyzed by epifluorescence microscopy at the indicated time points throughout the chase period (6 worms/time point) Scale bars, 100 μm. Representative images are shown.
BmHRG-1 in vitro knockdown via hsiRNA
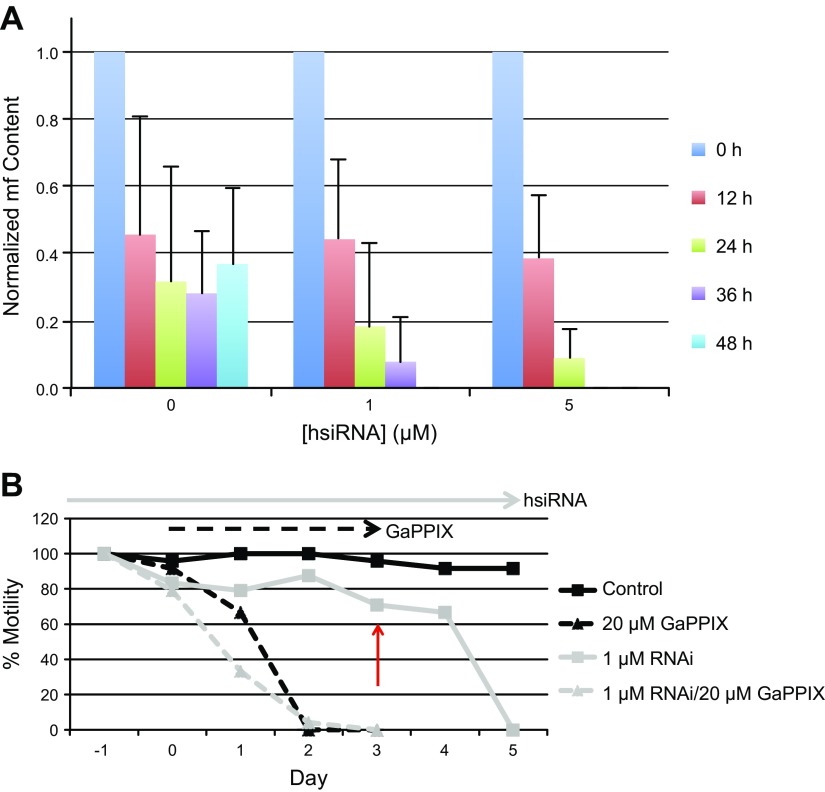
To investigate the role of BmHRG-1 in vitro, adult B. malayi females were soaked in hsiRNA designed to target the BmHRG-1 transcript to knockdown endogenous expression. Although motility of the worms was slightly affected (data not shown), mf production was severely affected by BmHRG-1 knockdown (Fig. 6A). Females treated with 1 and 5 μM BmHRG-1 RNAi ceased mf production within 48 and 36 h, respectively. This is in stark contrast to control worms treated with no hsiRNA that were still readily producing mf after 2 d in culture. BmHRG-1 knockdown was assessed using various experimental methods. Presoaking worms in 0.5 μM BmHRG-1 hsiRNA followed by exposure to GaPPIX (0, 1, 2, 5, or 20 μM ± RNAi) provided a slight improvement in worm motility within the first 2 d (Supplemental Fig. S2). However, the GaPPIX-induced decline in worm motility was unaffected by the presence of BmHRG-1 RNAi after d 2. Increasing the concentration of hsiRNA to 1 μM did not produce this same protective effect as the lower RNAi concentration and actually exacerbated the GaPPIX-induced cytotoxicity (Fig. 6B). Moreover, pretreatment with BmHRG-1 hsiRNA for 4 d before exposure to GaPPIX did not provide any substantial protection from GaPPIX toxicity. In addition, presoaking adult female B. malayi with BmHRG-1 hsiRNA had a concentration-dependent effect on ZnMP fluorescence (Supplemental Fig. S3A), whereas heme accumulation was seemingly unaffected (Supplemental Fig. S3B). Taken together, these results suggest that, although knockdown of BmHRG-1 has an effect on mf production in adult females in vivo, other compensatory mechanisms (potentially other as of yet unidentified BmHRGs) transport heme and heme analogs.
Figure 6.
Knockdown of BmHRG-1 by hsiRNA soaking. A) B. malayi females were cultured in RPMI-1640 medium containing increasing concentrations of BmHRG-1 hsiRNA (0, 1, or 5 μM). Medium was changed every 12 h, at which point mf output for each treatment group was determined. Each hsiRNA treatment group mf count is normalized based on the 0-h timepoint. B) B. malayi females (6 worms/treatment group) were pretreated with or without 1 μM BmHRG-1 hsiRNA for 24 h (d −1 to 0) before the addition of 20 μM GaPPIX (d 0). Motility was assessed as described in Materials and Methods on a scale from 0 (nonmotile) to 4 (highly motile). Each data point represents motility from a single experiment where worms were scored as a group (6 adult worms/treatment group). At d 3, 20 μM GaPPIX was added to the RNAi-only treatment group (red arrow) to determine if longer pretreatment with BmHRG-1 hsiRNA would provide significant protection from GaPPIX cytotoxicity.
Heme responsiveness of HRGs
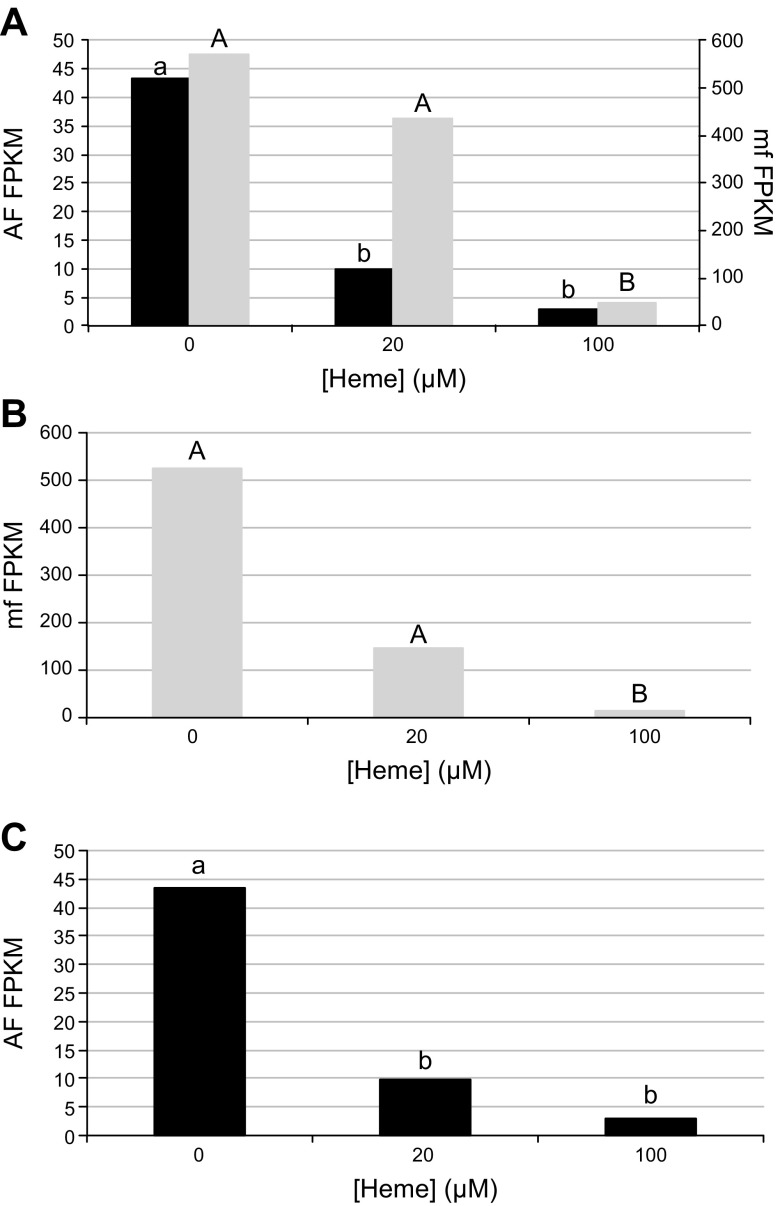
To assess whether the currently annotated B. malayi HRGs were indeed heme responsive, we performed RNA-sequencing on B. malayi that were exposed to various concentrations of exogenous heme (0, 20, or 100 μM) for 24 h. BmHRG-1 expression is up-regulated in the mf stages, as compared to the adult stages (Fig. 7A). Based on fragments per kilobase of transcript per million mapped reads (FPKM) values, BmHRG-1 is down-regulated in a dose-dependent manner in both adult females (at heme concentrations as low 20 μM, black bars) and mf (at heme concentrations greater than 20 μM, gray bars). BmHRG-1 was expressed in adult males at all heme concentrations tested; however, no significant differences were observed (data not shown).
Figure 7.
HRG expression in B. malayi adult females (AF) and mf in response to heme. A) FPKM values show expression of BmHRG-1 is significantly down-regulated (q ≤ 0.01) in the presence of heme in B. malayi adult females (black bars) and mf (gray bars). At every heme concentration tested, BmHRG-1 expression is significantly higher (q ≤ 0.01) in mf (gray bars, uppercase letters) than female worms (black bars, lowercase letters) (note difference in y-axis scales). B) BmHRG-2 expression is significantly down-regulated (q ≤ 0.01) in mf at high (100 μM) heme concentrations. C) BmMRP-5 expression is significantly down-regulated (q ≤ 0.01) in adult female B. malayi at 20 μM heme and above.
Similar to BmHRG-1 expression, BmHRG-2 was significantly up-regulated in the mf compared to adults at every heme concentration examined (data not shown). BmHRG-2 was significantly down-regulated in mf at high heme concentrations (100 μM; Fig. 7B), however, no significant differences were seen at any heme concentration for BmHRG-2 in adult males or females.
Although up-regulated by low heme in C. elegans (13), the only significant difference in BmMRP-5 was observed in adult females, in which BmMRP-5 was significantly down-regulated at heme concentrations of 20 μM or above (Fig. 7C). No significant differences in BmMRP-5 expression were observed between the different life cycle stages, except at 100 μM heme, where BmMRP-5 was significantly up-regulated in mf in comparison to adult B. malayi (data not shown).
BmHRG-1 expression in adult females
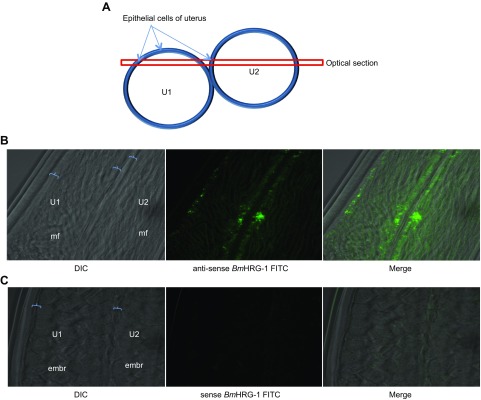
Transcriptomic studies in both B. malayi and D. immitis indicate that heme biosynthesis genes are up-regulated in the microfilarial stage as well as the female body wall, intestine, and uterus (ref. 41 and unpublished results). Whole-mount in situ hybridization of adult B. malayi females revealed that BmHRG-1 is indeed expressed in intrauterine mf, with the greatest expression observed in the epithelial cells of the uterine wall (Fig. 8).
Figure 8.
BmHRG-1 expression by whole-mount in situ hybridization. A) Cross-section of B. malayi adult female showing 2 uterine branches (U1 and U2) containing intrauterine stages [mf or embryos (embr)]. B) Expression of BmHRG-1 by whole-mount in situ hybridization with anti-sense FITC-labeled probe 120 d after infection. C) BmHRG-1 sense probe image indicates negligible background staining.
DISCUSSION
Originally identified because of its similarity to CeHRG-1 (∼39%), BmHRG-1 (Bm5182, WormBase ID: WBGene00225443) is slightly smaller than its C. elegans homolog, but maintains all the proven functional residues and motifs to make it an efficient transporter of heme. Although BmHRG-1 displays sequence similarity to other C. elegans HRGs [CeHRG-4 (29%), CeHRG-5 (27%), and CeHRG-6 (27%)], these other HRGs have not yet been identified in B. malayi. Presumably because of the lack of the C-terminal sorting motif that is present in CeHRG-1, BmHRG-1 expressed in yeast appears to localize, not only to the endocytic compartments, as CeHRG-1 does, but also to the plasma membrane, as does CeHRG-4. As with other HRGs, immunoblot analysis indicates BmHRG-1 (expressed in yeast and from worm extracts) migrates as a monomer, as well as larger oligomeric forms. The monomeric form of endogenous BmHRG-1 (from B. malayi extracts) is slightly larger than the expected 17 kDa, suggesting that it may be posttranslationally modified in vivo. A potential N-linked glycosylation site is present at position 34 of BmHRG-1 (Asn34 in the E1 loop) and may account for the discrepancy in electrophoretic mobility between the yeast- and worm-derived extracts. Based on the RNAi-induced knockdown experiments, other heme transporters (BmHRG-4, -5, or -6) are likely encoded within the B. malayi genome, yet remain unidentified.
It is evident that various stages of B. malayi (mf and adult males and females) can indeed acquire heme and heme analogs via specific heme transporters. Furthermore, both heme uptake assays in live worms and immunoblot analysis of whole worm extracts suggest protein levels and functional heme transport via BmHRG-1 may be most critical during the mf stage. In agreement with our findings, transcriptomic studies of the B. malayi life cycle found that mature mf displayed higher expression levels of BmHRG-1 than adults (42, 43). Previously, the highest levels of transcription for BmHRG-1 were observed in the larval stage 3 (L3) (which we did not test in this study); however, the exact level of transcription at this stage was relatively uncertain, given the large variation in RPKM mapped read values (42, 43). In addition, the recent transcriptomic study of the life cycle of the related filarial nematode, Dirofilaria immitis, found that the 2 potential DiHRGs (nDi.2.2.2.g03420 and nDi.2.2.2.g07804) are both most highly expressed in the mf stage compared to L3 and L4, as well as adult males and females (44). Furthermore, gene ontology (GO) term analysis revealed that tetrapyrrole/heme binding functions were overrepresented in mf-associated transcripts compared to the other life cycle stages (44). The suggestion that heme transporters are highly expressed during the mf stage is particularly interesting, given that B. malayi mf typically survive for long periods in the blood of the mammalian host, where heme is plentiful. In contrast, adults are generally found feeding on the lymphatic fluid which typically contains only 32% as much heme (globulin content) as blood (45). Why heme transporters would be prevalent when the nematode is essentially bathed in heme remains unanswered. However, it may be related to heme availability, or lack thereof, in the insect vector stages of B. malayi development. Plasmodium spp. readily synthesizes heme despite the ability to acquire heme from hemoglobin during blood stages of infection (46). However, the same investigation found that the capability of synthesizing heme is critical for malarial parasite development in the liver and mosquito stages, suggesting that heme availability may be limiting within the mosquito. An analogous situation may exist for B. malayi development: mf may be accumulating stores of heme from the blood before mosquito-induced heme deprivation where the nematode may be forced to rely more on heme biosynthesis from Wolbachia.
CeHRG-1 and -4 are expressed primarily in the intestine of the worm (6, 11). Like CeHRG-1 and CeMRP-5, BmHRG-1 has a potential heme-responsive element (CGACATGTGATGAACTAATAATC) located 163 bp upstream of the transcriptional start site; however, in B. malayi it lacks critical elements required for intestinal expression (47). In addition, the intestine of B. malayi is relatively poorly developed or completely lacking (in the mf stage, L2, and L3) (48) and is thought to play little if any role in nutrient absorption (49). It is clear that many other nutrients (including leucine, adenosine and d-glucose) are selectively transported across the cuticle in adult and infective L3 B. malayi (50). Therefore, given the lack of a distinctly formed intestinal tract in B. malayi mf and that other nutrients are clearly obtained via transcuticular absorption, it seems plausible that heme uptake via HRGs may occur through the body wall and not the intestine.
Another question that arises is the availability of free unbound heme in any of the microenvironments experienced by B. malayi throughout its life cycle (human lymph: adult/L4; human blood: mf/infective L3; mosquito thoracic muscle cells and hemolymph: L1–L3). Estimates suggest the human body contains ∼3.5 g of iron, approximately 70% of which is contained as heme (51, 52), making it a vast iron resource. However, most heme in the blood is contained within red blood cells (essentially membranous sacs full of hemoglobin). Extracellular free heme, rarely found in the body because of its ability to induce the formation of radical oxygen species, is normally bound to hemopexin. However, serum hemoglobin levels (presumably bound to haptoglobin in a 2:1 hemoglobin:haptoglobin complex) are estimated to be anywhere between 80–800 nM (53). Various mechanisms exist in pathogenic species to capture and liberate intact heme and iron from host proteins (54); however, exactly how heme is liberated (possibly from free serum hemoglobin-haptoglobin complexes) before transport by BmHRG-1 in vivo remains unclear. Although receptors specific for hemoglobin-haptoglobin and heme-hemopexin complexes are present in humans (CD163 and CD91) (55), such receptors involved in heme uptake (if present) could be potential filarial drug target candidates, but as of yet have not been identified in these nematodes.
Although many pathogenic bacterial species encode homologs of mammalian heme oxygenases that cleave the porphyrin ring of heme to liberate the iron (54), no heme oxygenase, which would serve to detoxify the peroxidase activity of excessive heme, as well as liberate iron for other uses (formation of Fe-S clusters), has been identified in nematodes. Studies in C. elegans have found that iron-deprived worms were unable to grow in the presence of normally adequate heme concentrations and were rescued only by increasing heme concentrations in the growth medium (6), suggesting that although heme is taken up and incorporated into hemoproteins, very little heme is broken down and utilized as a free iron source. Recent discoveries of not only a heme transporter, but a ferrous iron transporter and ferric iron reductase in Leishmania (56), reinforce the possibility that iron, as well as heme, may be transported and utilized within other parasites, such as B. malayi.
The acquisition of exogenous heme is critical for the survival of most nematodes, including C. elegans, which lack the ability to synthesize the essential cofactor. Filarial nematodes, such as B. malayi that contain the Wolbachia endosymbiont, may not be exclusive heme auxotrophs, but may also procure heme synthesized by their symbiont. Although little is known about iron metabolism in wBm, insect Wolbachia have been shown to be sensitive and responsive to host iron content (57).
The complementarity of the heme uptake–heme synthesis relationship of B. malayi and Wolbachia remains unclear. Genomic sequencing has revealed that Wolbachia contain the full repertoire of heme biosynthesis genes (8), whereas the host nematode does not (58). Biochemical studies suggest the Wolbachia-encoded heme biosynthetic pathway is essential for worm development and survival (59). Further, Wolbachia expression studies suggest heme regulates the encoded heme biosynthesis genes (unpublished results), yet B. malayi has the functional genes for heme uptake and distribution, as well as the laterally transferred functional gene for the last biochemical step (ferrochelatase). Transcriptomic studies in both B. malayi and D. immitis indicate that Wolbachia heme biosynthesis genes are up-regulated in male and female body wall, intestine, uterus, and testis, along with Wolbachia secretion systems (Sec and type IV secretion system components) (unpublished results). Taken together, the data lead us to speculate that Wolbachia helps supply heme to B. malayi for 2 potential purposes: 1) for fertility (oogenesis) and other major heme requirements and/or 2) for worm survival in the mosquito host component of the life cycle (mf stage through infective L3), where environmental heme is likely to be negligible or difficult to obtain. Heme biosynthesis remains a viable antifilarial target, but, as such, is likely not the only biochemical process involved in the mutualistic symbiosis.
Further investigations are needed, to better understand the complicated interactions between nematode and symbiont. What about other filarial nematodes that do not contain Wolbachia? Do they use only the one mechanism, that of heme uptake? It will be of interest to see how B. malayi worms cured of their Wolbachia endosymbionts (through tetracycline treatments) or filarial worms lacking Wolbachia (such as Acanthocheilonema viteae) react to exogenous heme.
ACKNOWLEDGMENTS
The authors thank Drs. Donald Comb, William Jack, Clotilde Carlow, and James Ellard (New England BioLabs, Inc.) for their continued scientific support. This work was supported by New England BioLabs, Inc., New York Blood Center intramural funding, and U.S. National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grants R01DK85035 and R01DK074797 (to I.H.). The authors declare no conflicts of interest.
Glossary
- ALA
δ-aminolevulinic acid
- ALAS
δ-aminolevulinic acid synthase
- BmHRG-1–6
Brugia malayi heme-responsive gene 1–6
- BmMRP-5
Brugia malayi multidrug resistance protein 5
- CeHRG-1–6
Caenorhabditis elegans heme-responsive gene 1–6
- CeMRP-5
Caenorhabditis elegans multidrug resistance protein 5
- DIC
differential interference contrast
- FPKM
fragments per kilobase of transcript per million mapped reads
- GaPPIX
gallium protoporphyrin IX
- HRG
heme-responsive gene
- hsi
hairpin small interfering
- L1–4
larval stage 1–4
- mf
microfilariae
- RNAi
RNA interference
- RPKM
reads per kilobase of transcript per million mapped reads
- SC
synthetic complete
- SSC
saline sodium citrate
- wBm
Wolbachia from Brugia malayi
- ZnMP
zinc mesoporphyrin
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
A. N. Luck, X. Yuan, B. E. Slatko, I. Hamza, and J. M. Foster conceived and designed the experiments; A. N. Luck, D. Voronin, and X. Yuan performed the experiments and analyzed the data; A. N. Luck and X. Yuan drafted the manuscript; and all authors read, edited, and approved the final manuscript.
REFERENCES
- 1.Taylor M. J., Hoerauf A., Bockarie M. (2010) Lymphatic filariasis and onchocerciasis. Lancet 376, 1175–1185 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. (2010) GPELF Progress Report 2000-2009 and Strategic Plan 2010-2020. WHO/HTM/NTD/PCT/2010.6. Available at: http://www.searo.who.int/entity/vector_borne_tropical_diseases/topics/lymphatic_filariasis/LFREP.pdf
- 3.Taylor M. J., Awadzi K., Basáñez M. G., Biritwum N., Boakye D., Boatin B., Bockarie M., Churcher T. S., Debrah A., Edwards G., Hoerauf A., Mand S., Matthews G., Osei-Atweneboana M., Prichard R. K., Wanji S., Adjei O. (2009) Onchocerciasis control: vision for the future from a Ghanian perspective. Parasit. Vectors 2, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osei-Atweneboana M. Y., Awadzi K., Attah S. K., Boakye D. A., Gyapong J. O., Prichard R. K. (2011) Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl. Trop. Dis. 5, e998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korený L., Lukes J., Oborník M. (2010) Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? Int. J. Parasitol. 40, 149–156 [DOI] [PubMed] [Google Scholar]
- 6.Rao A. U., Carta L. K., Lesuisse E., Hamza I. (2005) Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. USA 102, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B., Novelli J., Jiang D., Dailey H. A., Landmann F., Ford L., Taylor M. J., Carlow C. K., Kumar S., Foster J. M., Slatko B. E. (2013) Interdomain lateral gene transfer of an essential ferrochelatase gene in human parasitic nematodes. Proc. Natl. Acad. Sci. USA 110, 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster J., Ganatra M., Kamal I., Ware J., Makarova K., Ivanova N., Bhattacharyya A., Kapatral V., Kumar S., Posfai J., Vincze T., Ingram J., Moran L., Lapidus A., Omelchenko M., Kyrpides N., Ghedin E., Wang S., Goltsman E., Joukov V., Ostrovskaya O., Tsukerman K., Mazur M., Comb D., Koonin E., Slatko B. (2005) The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3, e121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alves J. M., Voegtly L., Matveyev A. V., Lara A. M., da Silva F. M., Serrano M. G., Buck G. A., Teixeira M. M., Camargo E. P. (2011) Identification and phylogenetic analysis of heme synthesis genes in trypanosomatids and their bacterial endosymbionts. PLoS One 6, e23518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kořený L., Oborník M., Lukeš J. (2013) Make it, take it, or leave it: heme metabolism of parasites. PLoS Pathog. 9, e1003088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopal A., Rao A. U., Amigo J., Tian M., Upadhyay S. K., Hall C., Uhm S., Mathew M. K., Fleming M. D., Paw B. H., Krause M., Hamza I. (2008) Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453, 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C., Samuel T. K., Sinclair J., Dailey H. A., Hamza I. (2011) An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell 145, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair J., Hamza I. (2015) Lessons from bloodless worms: heme homeostasis in C. elegans. Biometals 28, 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korolnek T., Zhang J., Beardsley S., Scheffer G. L., Hamza I. (2014) Control of metazoan heme homeostasis by a conserved multidrug resistance protein. Cell Metab. 19, 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C., Samuel T. K., Krause M., Dailey H. A., Hamza I. (2012) Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by heme-responsive gene-2. J. Biol. Chem. 287, 9601–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crisp R. J., Pollington A., Galea C., Jaron S., Yamaguchi-Iwai Y., Kaplan J. (2003) Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278, 45499–45506 [DOI] [PubMed] [Google Scholar]
- 17.Protchenko O., Shakoury-Elizeh M., Keane P., Storey J., Androphy R., Philpott C. C. (2008) Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot. Cell 7, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 19.Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. (1990) Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgatsou E., Alexandraki D. (1994) Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shatwell K. P., Dancis A., Cross A. R., Klausner R. D., Segal A. W. (1996) The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J. Biol. Chem. 271, 14240–14244 [DOI] [PubMed] [Google Scholar]
- 23.Adams A., Gottschling D. E., Kaiser C. A., Sterns T. (2000) Methods in Yeast Genetics, Cold Spring Harbor Press, Cold Spring Harbor, NY, USA [Google Scholar]
- 24.Kitagawa T., Aikawa T. (1976) Enzyme coupled immunoassay of insulin using a novel coupling reagent. J. Biochem. 79, 233–236 [DOI] [PubMed] [Google Scholar]
- 25.Sun L., Ghosh I., Xu M. Q. (2003) Generation of an affinity column for antibody purification by intein-mediated protein ligation. J. Immunol. Methods 282, 45–52 [DOI] [PubMed] [Google Scholar]
- 26.Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27.Huynh C., Yuan X., Miguel D. C., Renberg R. L., Protchenko O., Philpott C. C., Hamza I., Andrews N. W. (2012) Heme uptake by Leishmania amazonensis is mediated by the transmembrane protein LHR1. PLoS Pathog. 8, e1002795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul K. G., Throrell H., Akeson A. (1953) The molar light absorption of pyridine ferroprotoporphyrin (pyridine haemochromogen). Acta Chem. Scand. 7, 1284–1287 [Google Scholar]
- 29.Comley J. C., Rees M. J., Turner C. H., Jenkins D. C. (1989) Colorimetric quantitation of filarial viability. Int. J. Parasitol. 19, 77–83 [DOI] [PubMed] [Google Scholar]
- 30.Townson S., Tagboto S., McGarry H. F., Egerton G. L., Taylor M. J. (2006) Onchocerca parasites and Wolbachia endosymbionts: evaluation of a spectrum of antibiotic types for activity against Onchocerca gutturosa in vitro. Filaria J. 5, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landmann F., Foster J. M., Slatko B. E., Sullivan W. (2012) Efficient in vitro RNA interference and immunofluorescence-based phenotype analysis in a human parasitic nematode, Brugia malayi. Parasit. Vectors 5, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Miller W., Kent W. J., Nekrutenko A. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blankenberg, D., Von Kuster, G., Coraor, N., Ananda, G., Lazarus, R., Mangan, M., Nekrutenko, A., and Taylor, J. (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19, Unit 19 10 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goecks J., Nekrutenko A., Taylor J.; Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed June 20, 2016
- 36.Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan X., Protchenko O., Philpott C. C., Hamza I. (2012) Topologically conserved residues direct heme transport in HRG-1-related proteins. J. Biol. Chem. 287, 4914–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stojiljkovic I., Kumar V., Srinivasan N. (1999) Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31, 429–442 [DOI] [PubMed] [Google Scholar]
- 40.Kelson A. B., Carnevali M., Truong-Le V. (2013) Gallium-based anti-infectives: targeting microbial iron-uptake mechanisms. Curr. Opin. Pharmacol. 13, 707–716 [DOI] [PubMed] [Google Scholar]
- 41.Luck A. N., Anderson K. G., McClung C. M., VerBerkmoes N. C., Foster J. M., Michalski M. L., Slatko B. E. (2015) Tissue-specific transcriptomics and proteomics of a filarial nematode and its Wolbachia endosymbiont. BMC Genomics 16, 920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi Y. J., Ghedin E., Berriman M., McQuillan J., Holroyd N., Mayhew G. F., Christensen B. M., Michalski M. L. (2011) A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl. Trop. Dis. 5, e1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WormBase, website. www.wormbase.org (2014)
- 44.Luck A. N., Evans C. C., Riggs M. D., Foster J. M., Moorhead A. R., Slatko B. E., Michalski M. L. (2014) Concurrent transcriptional profiling of Dirofilaria immitis and its Wolbachia endosymbiont throughout the nematode life cycle reveals coordinated gene expression. BMC Genomics 15, 1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutili G., Arfors K. E. (1977) Protein concentration in interstitial and lymphatic fluids from the subcutaneous tissue. Acta Physiol. Scand. 99, 1–8 [DOI] [PubMed] [Google Scholar]
- 46.Nagaraj V. A., Sundaram B., Varadarajan N. M., Subramani P. A., Kalappa D. M., Ghosh S. K., Padmanaban G. (2013) Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PLoS Pathog. 9, e1003522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinclair J., Hamza I. (2010) A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans. J. Biol. Chem. 285, 39536–39543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott A. L. (2000) Lymphatic-dwelling filariae. In Lymphatic Filariasis Nutman T. B., ed. Vol. 1 p. 5–39, Imperial College Press, London: [Google Scholar]
- 49.Munn E. A., Munn P. D. (2002) Feeding and digestion. In The Biology of Nematodes Lee D. L., ed, p. 211–232, CRC Press, Boca Raton: [Google Scholar]
- 50.Chen S. N., Howells R. E. (1979) The uptake in vitro of dyes, monosaccharides and amino acids by the filarial worm Brugia pahangi. Parasitology 78, 343–354 [DOI] [PubMed] [Google Scholar]
- 51.Andrews N. C., Schmidt P. J. (2007) Iron homeostasis. Annu. Rev. Physiol. 69, 69–85 [DOI] [PubMed] [Google Scholar]
- 52.Theil E. C., Goss D. J. (2009) Living with iron (and oxygen): questions and answers about iron homeostasis. Chem. Rev. 109, 4568–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schryvers A. B., Stojiljkovic I. (1999) Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32, 1117–1123 [DOI] [PubMed] [Google Scholar]
- 54.Runyen-Janecky L. J. (2013) Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol. 3, 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schultz I. J., Chen C., Paw B. H., Hamza I. (2010) Iron and porphyrin trafficking in heme biogenesis. J. Biol. Chem. 285, 26753–26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flannery A. R., Renberg R. L., Andrews N. W. (2013) Pathways of iron acquisition and utilization in Leishmania. Curr. Opin. Microbiol. 16, 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kremer N., Voronin D., Charif D., Mavingui P., Mollereau B., Vavre F. (2009) Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog. 5, e1000630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J. E., Delcher A. L., Guiliano D. B., Miranda-Saavedra D., Angiuoli S. V., Creasy T., Amedeo P., Haas B., El-Sayed N. M., Wortman J. R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S. L., Schobel S., Pertea M., Pop M., White O., Barton G. J., Carlow C. K., Crawford M. J., Daub J., Dimmic M. W., Estes C. F., Foster J. M., Ganatra M., Gregory W. F., Johnson N. M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B. W., Li W., Lindblom T. H., Lustigman S., Ma D., Maina C. V., Martin D. M., McCarter J. P., McReynolds L., Mitreva M., Nutman T. B., Parkinson J., Peregrín-Alvarez J. M., Poole C., Ren Q., Saunders L., Sluder A. E., Smith K., Stanke M., Unnasch T. R., Ware J., Wei A. D., Weil G., Williams D. J., Zhang Y., Williams S. A., Fraser-Liggett C., Slatko B., Blaxter M. L., Scott A. L. (2007) Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu B., Novelli J., Foster J., Vaisvila R., Conway L., Ingram J., Ganatra M., Rao A. U., Hamza I., Slatko B. (2009) The heme biosynthetic pathway of the obligate Wolbachia endosymbiont of Brugia malayi as a potential anti-filarial drug target. PLoS Negl. Trop. Dis. 3, e475 [DOI] [PMC free article] [PubMed] [Google Scholar]