Abstract
The role of tumor heterogeneity in regulating disease progression is poorly understood. We hypothesized that interactions between subpopulations of cancer cells can affect the progression of tumors selecting for a more aggressive phenotype. We developed an in vivo assay based on the immortalized nontumorigenic breast cell line MCF10A and its Ras-transformed derivatives AT1 (mildly tumorigenic) and CA1d (highly tumorigenic). CA1d cells outcompeted MCF10A, forming invasive tumors. AT1 grafts were approximately 1% the size of CA1d tumors when initiated using identical cell numbers. In contrast, CA1d/AT1 mixed tumors were larger than tumors composed of AT1 alone (100-fold) or CA1d (3-fold), suggesting cooperation in tumor growth. One of the mechanisms whereby CA1d and AT1 were found to cooperate was by modulation of TGF-α and TGF-β signaling. Both of these molecules were sufficient to induce changes in AT1 proliferative potential in vitro. Reisolation of AT1 tumor-derived (AT1-TD) cells from these mixed tumors revealed that AT1-TD cells grew in vivo, forming tumors as large as tumorigenic CA1d cells. Cooperation between subpopulations of cancer epithelium is an understudied mechanism of tumor growth and invasion that may have implications on tumor resistance to current therapies.—Franco, O. E., Tyson, D. R., Konvinse, K. C., Udyavar, A. R., Estrada, L., Quaranta, V., Crawford, S. E., Hayward, S. W. Altered TGF-α/β signaling drives cooperation between breast cancer cell populations.
Keywords: epithelial paracrine interactions, intratumor cooperation, tumor heterogeneity
Intratumor cellular heterogeneity is increasingly recognized as a critical factor in disease progression. Tumor stroma is a diverse environment including fibroblasts, endothelium, nerves, and immune/inflammatory infiltrate (1–5). Stromal–epithelial interactions are known drivers of tumor progression. Interactions between distinct fibroblast subpopulations within the stroma have been shown to promote tumor growth and invasion (5–7). Inflammatory infiltrate, comprising several cell types, can also directly affect progression (8, 9).
Although the epithelial compartment of tumors is itself heterogeneous, interactions between and among distinct subpopulations of cancer epithelial cells are not well understood (10–12). The possibility that such interactions may contribute to tumorigenesis is suggested by theoretical studies indicating that interepithelial cell communication may facilitate tumor progression by supporting survival of at least some subpopulations under harsh tumor microenvironmental conditions (13). Such views are supported by recent genetic studies of tumor cell diversity and by evolutionary theory (14, 15). While the struggle between “winner” and “loser” cancer cells may determine the outcome among competing populations in tumors, preservation of tumor heterogeneity can also be beneficial to poorly invasive cancer cells (16–20).
Our expectation at the inception of this work was that the aggressive cells would outcompete slower growing populations (competition) and would come to dominate tumors started with mixtures of epithelial cells. However, a contrary suggestion is the idea that tumor epithelial cells might work together to their mutual benefit to promote tumor progression (cooperation). Experiments using a zebrafish-melanoma xenograft model have shown that poorly invasive cells benefit from exposure to highly tumorigenic clones, increasing the deposition extracellular matrix components, notably fibronectin, and inducing the simultaneous invasion of participant clones, a phenomenon termed cooperative invasion (18). Moreover, evidence of cooperative interactions among cancer cells was also shown in a transgenic model of breast cancer containing different levels of Wnt1 in basal and luminal cells (19). In this model, the presence of biclonal tumors was a prerequisite for the maintenance and restoration (upon Wnt withdrawal) of interclonal cooperation during tumor progression. In our xenograft model using human cancer cell lines, we observed cooperative invasion and growth, but also the acquisition during this process of a more aggressive phenotype by the previously poorly tumorigenic clones. We used 3 related cell lines: MCF10A, AT1, and CA1d, representing nonmalignant; partly transformed but noninvasive (poorly tumorigenic); and aggressive, invasive mammary epithelium, respectively (21). MCF10A cells are a commonly used model of nontumorigenic mammary epithelial cells; AT1 and CA1d cells have each been derived from MCF10A by the introduction of oncogenic H-Ras (Harvey rat sarcoma virus oncogene) and passaging in immunocompromised mice as xenografts (AT1 from a single passage and CA1d after 4 rounds of xenografting). This panel of cell lines provides a model system with which to examine how epithelial cells (originating from the same progenitor) demonstrating varying degrees of malignancy interact and contribute to tumor growth and progression.
Here we show that in addition to the view of clonal selection and survival of the fittest, epithelial subpopulations can cooperate to form more aggressive tumors than those achieved by either cell line alone. The result from this interaction is the permanent acquisition of increased tumorigenicity of poorly invasive cells even in the absence of a highly tumorigenic environment. Furthermore, we provide evidence that the underlying mechanism could rely on cooperation driven by a complex interplay of secreted growth factors, including TGF-α and TGF-β. These results provide a framework for the use of agents that target intraepithelial tumor cell signaling as a component of cancer therapy.
MATERIALS AND METHODS
Culture conditions for cell lines and isolation of TD cells
MCF10A cells were a gift from J. Brugge (Harvard Medical School, Boston, MA, USA), and AT1 and CA1d were a gift from F. Miller (Karmanos Cancer Institute, Detroit, MI, USA). MCF10A cells were derived from a patient with benign fibrocystic disease and are characterized as nontumorigenic (22). AT1 and CA1d are derivative cell lines with increasingly malignant phenotypes (23). These cells and other MCF10A derivatives can be obtained from P. Robert (Barbara Ann Karmanos Cancer Institute, Detroit, MI, USA, under Material TransferAgreement) (23, 24). Authentication of cell lines was validated by short tandem repeat (STR) DNA profiling analysis using the Cell Authentication Testing Service at American Type Culture Collection (ATCC; Manassas, VA, USA). All cells matched the profile of the MCF10A cell line in the ATCC STR database (CRL-10317). Cells were cultured in DMEM/F-12 with 5% horse serum, 20 ng/ml epidermal growth factor (Upstate Biotechnology, Lake Placid, NY, USA), 10 μg/ml insulin (Sigma-Aldrich, St. Louis, MO, USA), 100 ng/ml cholera enterotoxin (Calbiochem, San Diego, CA, USA), and 0.5 μg/ml hydrocortisone (Sigma-Aldrich). For experiments in which cell lines were mixed, cells were transduced with the pBabe–green fluorescent protein (GFP; Addgene, Cambridge, MA, USA) and red fluorescent protein (RFP; Clontech Laboratories, Mountain View, CA, USA) using a retroviral and lentiviral system followed by fluorescence-activated cell sorting (FACS) isolation of transduced cells, as previously described (7).
Tumor tissues derived from xenografts were finely chopped with sterile scalpels, and the pieces were digested in 10 ml DMEM/F-12 medium containing 10% horse serum and 150 units/ml collagenase (Sigma-Aldrich), and incubated in a shaking water bath for 90 min. Then partly digested tissue was washed (2×) with fresh medium, cells counted and plated in a T25 flask at a density of 5 × 105 cells per flask, and expanded for about 1 wk before undergoing FACS separation into individual clones [tumor-derived (TD) derivatives].
Xenograft studies
Animal care and experiments were performed with the approval of and in accordance with Vanderbilt University Institutional Animal Care and Use Committee guidelines. For the in vivo studies, a total of 5 × 105 cells were pelleted and resuspended in 50 μl neutralized type I rat tail collagen, as described previously (7). After incubation at 37°C overnight, the tissue recombinant was grafted under the renal capsule of adult (8 wk old) intact female CB17Icr/Hsd-SCID mice (Harlan Industries, Indianapolis, IN, USA). Hosts were humanely killed 12 wk after grafting. The kidneys were removed and imaged before processing for histology. Graft dimensions were measured, and the resultant tumor volume was calculated using the following formula: volume = width × length × depth × (π/6). This formula underestimates the volume of large invasive tumors as a result of irregularity in the tumor burden.
Assessment of cell numbers
Cells were seeded at a density of 3 × 103 cells per well in 96-well plates in regular medium. For experiments in which 2 different types of cells were used, 1.5 × 103 cells for each group were combined in a 1:1 ratio, maintaining the total density of 3 × 103 cells per well. Cells were allowed to adhere and were incubated overnight at 37°C. The following day, complete medium was removed, cells were washed with PBS (3×), replaced with serum-free DMEM/F-12, and incubated for another 24 h before being exposed to the different experimental conditions. Conditioned medium (CM) was obtained by plating 1 × 106 CA1d cells in 10 cm dishes with complete medium, replacing medium 24 h later with serum-free DMEM/F-12 medium, and conditioning for 72 h before collecting, centrifuging, and filtering through a 0.45 µm pore size membrane. After serum starvation, cells were treated with 10 ng/ml TGF-α (R&D Systems, Minneapolis, MN, USA) and/or 5 ng/ml TGF-β1. CM was incubated with 10 μg/ml of the pan-TGF-β blocking antibody 2G7 (which can block all 3 TGF-β isoforms) and/or 2 μM epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor erlotinib (Erl; LC Laboratories, Woburn, MA, USA) to block TGF-α ligand activity (25). Each experimental condition was performed in triplicate. At d 6, DAPI (Sigma-Aldrich) to a final concentration of 0.1 μg/ml was added to cells in culture, followed by imaging at ×20 and 32 magnification. Addition of DAPI enabled us to identify and quantify GFP- and/or RFP-expressing cells. Quantification of cell numbers was performed using ImageJ software (Image Processing and Analysis in Java; National Institutes of Health, Bethesda, MD, USA) (26).
RNA isolation and microarray analysis
Total RNA was isolated from 5 × 106 cells using an RNAeasy Mini kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocol. For cDNA synthesis, 1 μg total RNA was added to a reaction mix using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). For real-time semiquantitative PCR, 1 μl cDNA template was added to IQ RealTime SYBR Green PCR Supermix (Bio-Rad). Relative quantitation was calculated by the ΔΔCt method normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primers were purchased from RealTime Primers (Elkins Park, PA, USA).
Immunohistochemistry and immunofluorescence
Sections (5 μm) of paraffin-embedded tissue samples were prepared as previously described (7). Samples were then incubated with primary antibodies against human GFP (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), RFP (1:500; Abcam, Cambridge, MA, USA), Ki-67 (1:250; Abcam), P-Smad2 (1:100; Cell Signaling Technology, Danvers, MA, USA), P-Akt (1:25; Cell Signaling Technology), and p-Erk (1:25; Cell Signaling Technology). After washing in PBS, the slides were incubated in Alexa Fluor 488–conjugated anti-mouse or 546 anti-rabbit secondary antibodies (Sigma-Aldrich) or biotinylated anti-rabbit secondary antibody (Dako, Glostrup, Denmark) for 1 h. After extensive washing, the slides were mounted with Vectashield mounting medium with propidium iodide (Vector Laboratories, Burlingame, CA, USA) or DAPI. For immunohistochemical staining, the slides were incubated in ABC solution (Vector Laboratories) to amplify the signals before visualizing with 3,3′-diaminobenzidine. Slides were counterstained with hematoxylin before imaging.
Western blot analysis
AT1 cells were seeded in 6-well plates at density of 1 × 105 cells per well. The following day, the medium was replaced with serum-free DMEM/F-12. Twenty-four hours later, cells were either treated with growth factors (10 ng/ml TGF-α and/or 5 ng/ml TGF-β1) or CM from CA1d cells exposed to blocking antibodies (10 μg/ml 2G7 and/or 2 μM Erl). Treatment groups were performed in triplicate. After 3 d, cells were lysed with RIPA buffer. Proteins (about 20 μg per well) were loaded and electrophoresed through 10% NuPage Bis-Tris Gel (Thermo Fisher Scientific, Waltham, MA, USA) and electrophoretically transferred to nitrocellulose membranes. Membranes were incubated with primary antibody overnight at 4°C. The next day, membranes were washed and incubated with horseradish peroxidase–conjugated secondary antibody (1:1000; Amersham–GE Healthcare, Pittsburgh, PA, USA). Amersham ECL plus detection reagent (Amersham–GE Healthcare) was used to visualize protein bands. Antibodies (1:1000; Cell Signaling Technology) against P-Smad2, Smad2, Smad3, P-Akt (1:500), P-p44/42, P-p38, P–stress-activated protein kinase (SAPK)/JNK, P-MKK7, and P-MKK4 were used. The monoclonal β-actin antibody (1:5000; Sigma-Aldrich) was used for loading control. Densitometric quantitation was performed using the Band Analysis tools of ImageLab 5.2.1 software (Bio-Rad) to select and determine the background-subtracted density of the bands in all the gels and blots according to the manufacturer’s specification. The phosphorylated protein density of each band was normalized to its corresponding total protein control.
Wound healing assay
To study motility, cells were cultured in CM at an initial density of 5 × 104 cells/ml to a confluent monolayer in 12-well tissue culture plates for 24 h. A sterile 200 μl pipette tip was used to scratch the cell monolayer to form a crosslike wound area approximately 10 × 1 mm wide. Then the medium was replaced with serum-free medium supplemented with 10 ng/ml TGF-α and/or 5 ng/ml TGF-β or CM treated with 2G7 (10 μg/ml) and/or Erl (2 μM). Treatment groups were performed in triplicate. In order to monitor migration in the wound area, serial pictures were taken at 0, 3, 6, 12, and 24 h after exposure to the different conditions. The extent of wound closure was quantified by measuring the area of the wound before and 24 h after migration, using ImageJ software and expressed as percentage change.
Array-based comparative genomic hybridization analysis
Genomic DNA from MCF10A, AT1, CA1d, and TD cell lines (AT1-TD1, AT1-TD2, and CA1d-TD1) were extracted with a DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA was fragmented and random-prime labeled as described elsewhere (27, 28) and hybridized to human oligonucleotide microarrays. The oligonucleotide array contains unique 236,000 elements designed for array-based comparative genomic hybridization (array-CGH) profiling (Human Genome CGH 244A; Agilent Technologies, Santa Clara, CA, USA). The median interval between mapped elements was 8.9 kb. Fluorescence ratios of scanned images of the arrays were processed to identify statistically significant transitions in copy number by using a circular binary segmentation algorithm (28). In this study, significant copy number changes were determined on the basis of segmented profiles only. To define ploidy level, array-CGH assay was supplemented with G-band karyotyping. At least 40 DAPI-stained metaphase spreads were evaluated per sample.
Statistical analysis
Data are presented as means ± sd representing 3 independent experiments performed in triplicate. One-way ANOVA and the Dunnett’s multiple comparison test were used to determine the significance of difference between groups. Differences were considered statistically significant when P < 0.05.
RESULTS
Enhancement of tumor growth and invasion by mixing related but distinct cell populations
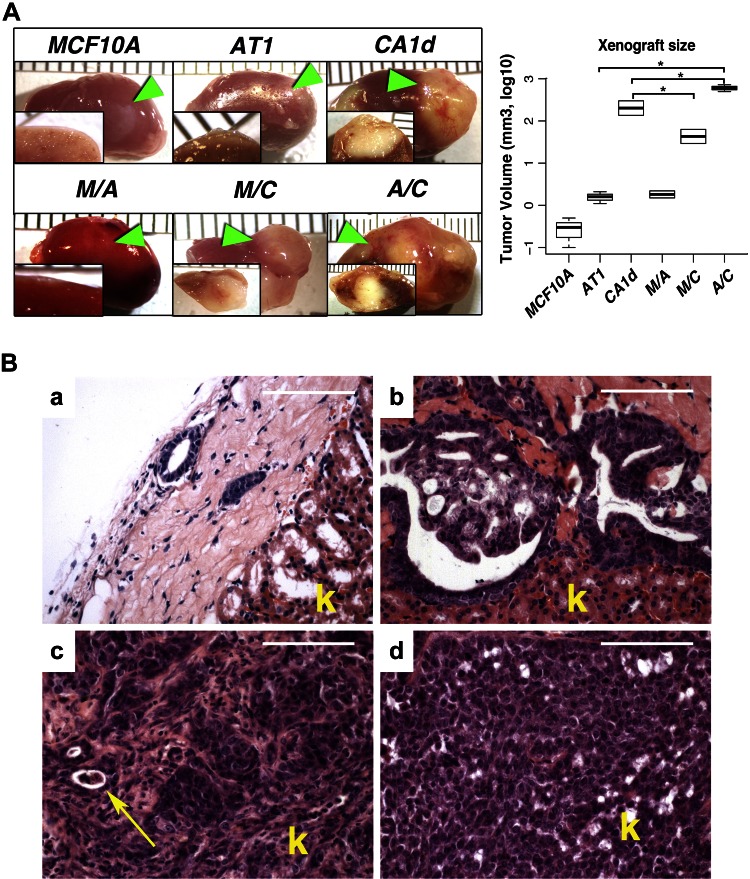
To mimic interactions between cell populations in patients, we chose MCF10A and its derivatives, AT1 and CA1d. We examined the tumorigenic properties of MCF10A, AT1, and CA1d in vivo, grafted under the renal capsule of SCID mice. Most in vivo models use the subcutaneous space; however, the subrenal approach offers an excellent scenario to study the initial steps of tumor progression by measuring local invasion of cancer cells in the surrounding tissues. The behavior of these cell lines in our model was consistent with reports showing benign, premalignant, and tumorigenic characteristics (Fig. 1A, top row) (29).
Figure 1.
In vivo growth of MCF10A, AT1, and CA1d breast cell lines. A) MCF10A and AT1 grafts demonstrated minimal growth, grossly appearing small and white, while CA1d grafts were significantly larger (>700-fold larger than MCF10A and >100-fold than AT1) with signs of invasion. Mixed tumors were very aggressive, destroying >50% of renal parenchyma. Arrowheads show position of tumors in kidney. Graph depicts tumor volume (mean ± sd) on log10 scale. *P < 0.05, 1-way ANOVA with Dunnett’s multiple comparison test. B) a) Histologically, MCF10A cells formed limited numbers of round glandular benign structures with small lumens and without any sign of malignancy. b) Premalignant AT1 cell line produced lesions consisting of simple ducts with some areas of benign hyperplasia and others resembling carcinoma-in-situ without any evident invasion into kidney. c) CA1d grafts were composed of invasive adenocarcinomas with large areas of undifferentiated cells. Arrow indicates glomerulus remnant surrounded by cancer cells. d) Mixed AT1/CA1d tumors showed more solid pattern compared to CA1d tumors. These tumors, although less obviously invasive at histologic level, were highly aggressive. Scale bars, 200 μm. k, kidney.
To understand the consequences of heterogeneous epithelial populations in tumors, we mixed pairs of cell lines in an equal ratio [MCF10A/AT1 (M/A), MCF10A/CA1d(M/C), and AT1/CA1d (A/C)] and grafted them into SCID mice. Compared to parental CA1d cells, MCF10A significantly decreased the size of M/C tumors and had no significant effect on the size of M/A tumors compared to those formed by each of the parental lines (Fig. 1A). However, A/C produced highly invasive tumors that were ∼3-fold larger than CA1d tumors and more than 100-fold larger than AT1 xenografts (Fig. 1A). Gross anatomic inspection of the A/C tumors showed that cancer cells destroyed a large proportion of the kidney with unclear margins. In contrast, in M/C and CA1d tumors, invasion was more delineated, with a clear separation between the invasive front and the kidney parenchyma. Histologic examination revealed that the A/C tumors demonstrated a heterogeneous phenotype containing poorly and moderately differentiated adenocarcinoma, with no sign of benign components (Fig. 1B). In contrast, M/C tumors consisted of poorly differentiated cells but had areas of well-differentiated adenocarcinoma. The fraction of Ki-67-positive cells was significantly higher in A/C tumors (≈300 positive nuclei per field) compared to those composed of only CA1d (≈150 positive nuclei per field) or AT1 (≈20 positive nuclei per field) cells (Supplemental Fig. 1A). A/C tumors also exhibited desmoplastic changes in the stroma, as detected by Masson trichrome staining (Supplemental Fig. 1B). In summary, the mixed AT1/CA1d tumors grew much larger than CA1d alone and exhibited increased proliferation and stromal changes, but because AT1 and CA1d cells in these tumors were not distinguishable, the proliferation rate of individual cell types and their relative contribution was not discernible.
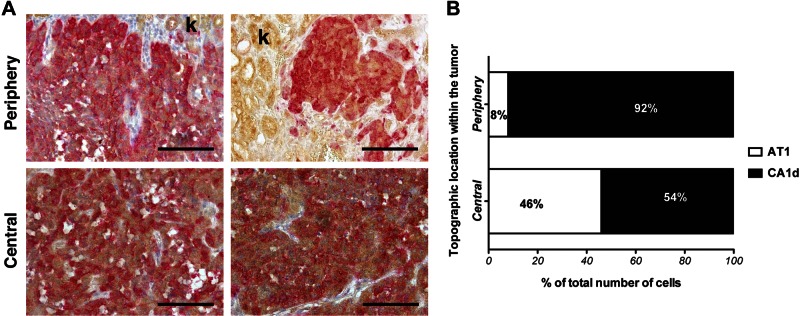
To address this limitation, we uniquely labeled each cell type using viral transduction of GFP or RFP tags and repeated the experiment to examine the respective contribution of AT1 and CA1d cells to the tumors (Fig. 2). The center of the tumor was composed of an even mixture of AT1 and CA1d cells (46 and 54%, respectively). There was a close interaction between both cell types with areas containing pockets of either AT1 or CA1d cells. In contrast, the periphery (or invasive front) was composed mainly of CA1d cells (90%) with a relatively low percentage of AT1 cells. These data, along with the proliferation studies described above, demonstrate that both cell types increase their proliferation in the combination model, with the largest change being the increase in AT1 cell proliferation necessary. To further examine the relationship of CA1d cells to the invasive front, we performed in vitro experiments in which cells were mixed within collagen plugs on culture dishes and allowed to grow in culture for several days. Again, CA1d cells were enriched at the invasive edge (data not shown). In addition, in a wound-closure assay of cell migration, the mixture of AT1/CA1d exhibited faster wound-closing kinetics than either cell line, alone suggesting that enhanced motility may contribute to the more aggressive behavior of the mixed tumor cell population. Taken together, these data suggest that mildly tumorigenic cells can contribute to the aggressive phenotype of tumors when mixed with more aggressive cells by accelerating proliferation and increasing the invasiveness of the resultant tumor. Our results demonstrate that at least in this model, more aggressive cells (CA1d) tend to lead the invasive front and may benefit from the presence of AT1 cells in the interior of the tumor. The close proximity of these 2 cell types suggests that direct cell–cell contact, or juxtacrine or paracrine signaling may contribute to these effects.
Figure 2.
Topographical localization of AT1 and CA1d cells in mixed tumors. A) Localization of AT1 and CA1d in mixed tumors revealed distinct pattern of distribution. Double immunohistochemical staining was performed of GFP-labeled AT1 (dark brown) and RFP-labeled CA1d cells (red) in 2 different tumors. Invasive front was composed mainly of CA1d cells and fewer AT1 cells. Kidney parenchyma shows light brown nonspecific background staining (k). B) While both cell types occupied center of tumor in similar ratio, CA1d cells dominated margins of tumor. Scale bars, 200 μm. k, kidney.
Accelerated growth and transformation of AT1 cells exposed to CA1d cells in vivo
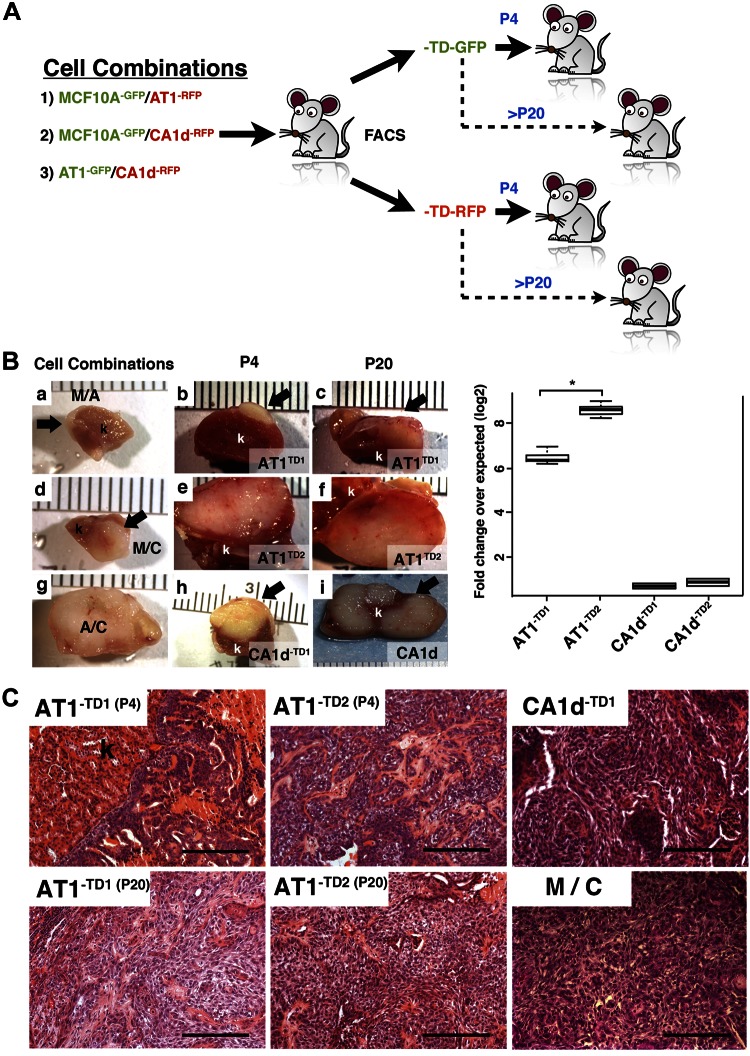
We have previously shown that carcinoma-associated fibroblasts impart permanent phenotypic and genetic changes on proximal epithelia (30, 31). However, it is not clear whether juxtaposing AT1 and CA1d in vivo resulted in new phenotypes that could be maintained after reisolation and culture in the absence of the other cell type. To test this, we repeated the in vivo mixing experiments, except that after 12 wk, a sample of each tumor was collected, dissociated, and briefly cultured (for 1 wk) to reisolate AT1 and CA1d cells by flow sorting (Fig. 3A). Tumor size and morphology essentially replicated the observations shown in Fig. 1, indicating that the fluorescent labels (Fig. 3Ba, d, g) did not alter the behavior of the cell lines or mixtures in vivo. Notably, no MCF10A cell derivatives could be isolated, suggesting either that these cells were lost or died during the in vivo growth, thus hinting at competition with the more aggressive CA1d cells. We were able to isolate CA1d-derived cells from the mixtures with MCF10A cells, indicating that only viable tumorigenic CA1d cells were present within the tumors. Two AT1-derived clone lines were isolated from 2 combined CA1d/AT1 tumors and designated as AT1-TD1 and AT1-TD2. A similar approach was performed for the CA1d cells and the 2 CA1d-TD1 and CA1d-TD2 cell lines we isolated (Fig. 3A). These cells were maintained in culture for 4 passages. Each TD cell line and its parental controls (separately) were then grafted for 3 mo to test their tumor growth properties (Fig. 3B). The resultant CA1d-TD tumors (Fig. 3Bh) were similar in size to parental CA1d (Fig. 3Bi), and all showed frank invasion, indicating that the aggressiveness of CA1d was maintained in CA1d-TD. In contrast, AT1-TD1 and AT1-TD2 formed tumors that were equal to or larger than CA1d and were likewise invasive (Fig.3Bb, c, e, f). The AT1 parental line showed minimal growth in vivo (as previously observed), with no sign of invasion. Similar results were obtained after passaging the cells 10 or 20 times in culture (Fig. 3C), indicating that the enhanced aggressiveness of the AT1-TD cell lines is a stable trait. This is consistent with previous mathematical predictions that the tumor microenvironment can induce subpopulations with greater tumorigenic potential (32).
Figure 3.
Characterization of TD AT1 and CA1d cells. A) Isolation of TD cells. Cells were combined in 1:1 ratio and grafted in female SCID mice. Cells were allowed to grow in vivo for 80 d and reisolated by FACS. Sorted cells were expanded separately and then regrafted alone at passage 4 and passage 20. B) Quantitation of tumor volume. Left: gross appearance of renal xenograft. Compared to MCF10A/AT1 (a) or MCF10A/CA1d (d), AT1/CA1d (g) tumors (arrows) increased in size. Reisolated AT1 cells from mixed A/C formed larger tumors compared to parental cells. At both passage 4 (P4) and passage 20 (P20), AT1-TD2 (e, f) cells formed larger tumors than AT1-TD1 (b, c). Cells grafted after 20 passages revealed that early changes (P4) were permanent. CA1d-TD1, and CA1d-TD2 (h) grew slightly better than parental CA1d (i). Right: AT1-derivative tumors (AT1-TD1 and AT1-TD2) were significantly larger compared to parental AT1 cell. AT1-TD2 clone tumor size was significantly bigger than AT1-TD1. Tumors derived from tumorigenic CA1d cells (CA1d-TD1 and CA1d-TD2) were slightly larger than tumors from their expected control line. k, kidney. Graph depicts tumor volume (means ± sd) on log2 scale. *P < 0.05, 1-way ANOVA with Dunnett’s multiple comparison test. C) Histologic analysis of grafts. Tumors were composed of highly invasive cancer cells with strong stromal reaction, consistent with malignant phenotype of these tumors. AT1-TD1 and AT1-TD2 grafted at P4 showed cancer cells with invasive phenotype. However, both AT1-TD cell lines at P20 were more aggressive and had mesenchymal type similar to CA1d-TD1 and CA1d-TD2 cells. Scale bars, 200 μm.
Tumorigenic AT1 derivative lines have altered gene expression profiles but no major genomic lesions
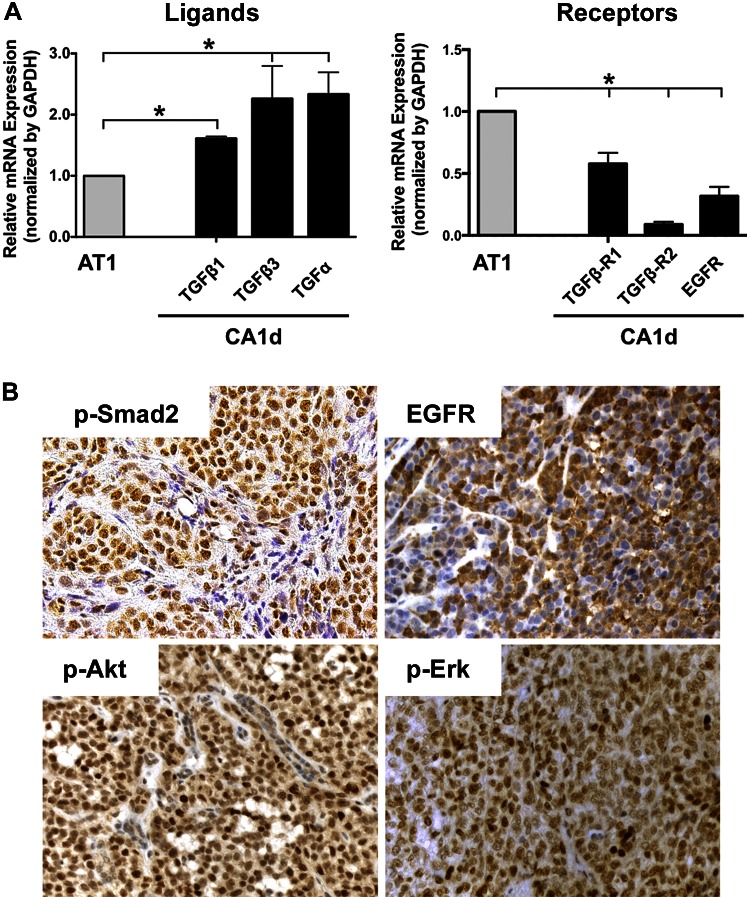
Array CGH confirmed the identity and common origin of MCF10A, AT1, and CA1d cells. No major genomic amplifications or deletions were observed by array CGH when the TD derivatives were compared to their respective parental lines (Supplemental Fig. 2A), suggesting that genomic amplifications or deletions do not explain the newly acquired aggressive behavior of AT1-TD. We performed microarray-based gene expression analyses and found that 306 genes were differentially expressed between parental AT1 and AT1-TD1. Functional enrichment using Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated dysregulation of several pathways in AT1-TD1 (Supplemental Fig. 2B), including TGF-β, EGFR, Wnt, IL-1, nuclear receptors, metabolism, and focal adhesions. Of particular interest was the down-regulation of some inhibitors of the TGF-β pathway, including Follistatin, Smad1, Smad7, and Decorin. In addition, expression of several EGFR signaling effectors, including TGF-α, were altered in AT1-TD1, AT1-TD2, and CA1d compared to parental AT1. To corroborate these observations, we examined expression of EGF and TGF-β receptors and ligands by quantitative real-time PCR and found significantly increased TGF-α and TGF-β 1 and 3 and decreased TGF-β receptor 1 (TGF-βR) and -2 in CA1d compared to AT1 (Fig. 4A). Immunohistochemical and quantitative analysis of these receptors and downstream effectors in mixed A/C tumors were significantly up-regulated (Fig. 4B and Supplemental Fig. 3). These results suggest the possibility that paracrine interactions between AT1 and CA1d involving TGF-α and TGF-β may contribute to the phenotypic changes of AT1-TD. The mechanism by which these changes fixed is not clear but could be an epigenetic phenomenon.
Figure 4.
Expression of TGF-α and TGF-β ligands and their receptors in parental and tumor-derived cell lines. A) TGF-α and TGF-β family members and their corresponding receptor mRNA levels were quantitated in AT1 and CA1d cells. Data were normalized to GAPDH, and expression of target genes was compared to AT1 levels. Left: CA1d cells expressed higher levels of ligands compared to AT1 cells. Right: expression of corresponding receptors was significantly increased in AT1 cells, suggesting that these cells have ability to respond to presence of paracrine-secreted factors. *P < 0.05, 1-way ANOVA with Dunnett’s multiple comparison test. B) Immunohistochemical staining of mixed A/C tumors. To corroborate presence of TGF-β and EGFR activation in our in vivo tumors, immunohistochemistry of P-Smad2, EGFR, p-Erk, and P-Akt was performed. While pSmad2 and EGFR showed relatively uniform staining, pattern of P-Akt and p-Erk depicted high-expressing intermingled with low-expressing cells.
TGF-α and TGF-β enhance proliferation and motility in AT1 cells
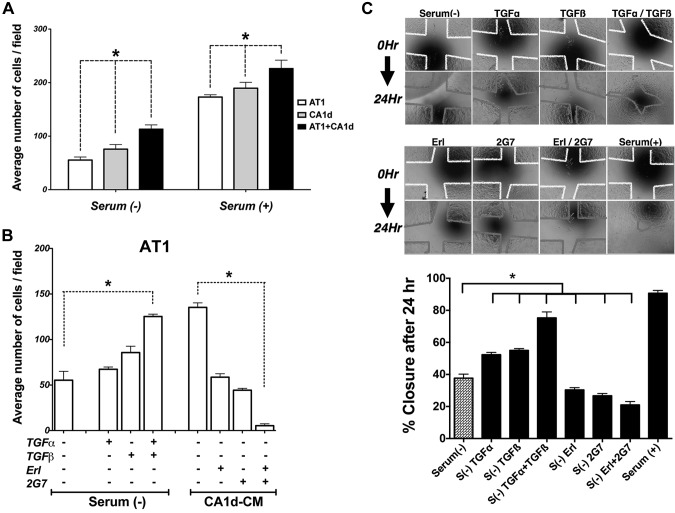
To assess whether secreted factors diffusing between the 2 epithelial cell populations can directly affect cellular behavior, we compared the proliferation of cells cultured alone to cells grown within a heterogenous mixture in vitro. Whether in the absence or presence of serum, the A/C mixed cultures proliferated more than either line alone (Fig. 5A). Interestingly, medium conditioned by tumorigenic CA1d cells produced a similar increase in proliferation of AT1 cells (Fig. 5B), indicating that factors secreted by CA1d cells were able to enhance AT1 proliferation. This ability to induce AT1 proliferation was partially inhibited by the pan-TGF-β blocking antibody 2G7 or by the EGFR tyrosine kinase inhibitor Erl. The combination of 2G7 and Erl reduced proliferation further than either agent alone (Fig. 5B), indicating that both EGFR and TGF-βR signaling contribute to CA1d-induced paracrine proliferation of AT1 cells in vitro. To assess the direct effect or effects of TGF-α or TGF-β alone or in combination, proliferation of AT1 cells was quantitated (Fig. 5B). Both showed increased effects; however, together, these 2 growth factors had an additive effect and induced significantly more proliferation of AT1 cells compared to either factor alone. In contrast, addition of TGF-α and/or TGF-β ligands had minimal effects on CA1d proliferation (data not shown). Interestingly blocking these pathways with Erl and/or 2G7 produced significant growth inhibition, suppressing the autocrine loop in CA1d cells (Supplemental Fig. 4B). These results support the notion that paracrine mechanisms, in these experiments TGF-α and TGF-β, amplified the proliferative response in AT1 cells and that this mechanism could also have implications in the emergence of the AT1-TD proliferative phenotype observed in vivo.
Figure 5.
Effect of TGF-α and TGF-β on cell proliferation and motility. A) Cells were cultured under different conditions and their proliferation quantitated. Increased cell number was observed in presence of serum. Proliferation of mixed A/C cells significantly increased compared to AT1 or CA1d cells alone. *P < 0.05, 1-way ANOVA with Dunnett’s multiple comparison test. B) Addition of TGF-β alone or TGF-α + TGF-β increased proliferation of AT1 cells. When AT1 cells were exposed to CM from CA1d cells, the pan-TGF-β and EGFR inhibitors (2GT7 and Erl, respectively), alone or in combination, were able to significantly reduced growth of AT1 cells. *P < 0.05, 1-way ANOVA and multiple comparisons. C) Wound-healing assay was undertaken to assess motility of AT1 mixed with CA1d cells under different conditions. Photomicrograph of cells at beginning of experiment (top) and 24 h later (bottom). Colored lines indicate edge of scratch at beginning (0 h, yellow) and at end of experiment (24 h, red). Percentage of wound closure was calculated. Both inhibitors Erl and 2G7 had negative effect alone or in combination. Presence of serum had highest effect and almost completely closed gap between cells. *P < 0.05, 1-way ANOVA with Dunnett’s multiple comparison test.
In addition to their contributions to proliferation, TGF-α or TGF-β enhanced wound closure of AT1 cells in a scratch assay (Fig. 5C). TGF-α and TGF-β in combination were more effective than either factor alone, suggesting a functional interaction. In A/C mixtures, Erl or 2G7 significantly blocked the wound closure effect, and the combination of inhibitors was even more effective than either alone, suggesting an additive effect. This indicates that both EGFR signaling and TGF-β signaling contribute to cell migration, likely enhancing the invasive phenotype. Consistent with the in vivo results, the leading edge of wound closure was occupied mostly by CA1d cells (Fig. 5D).
Activation of TGF-α and TGF-β pathways in AT1 cells
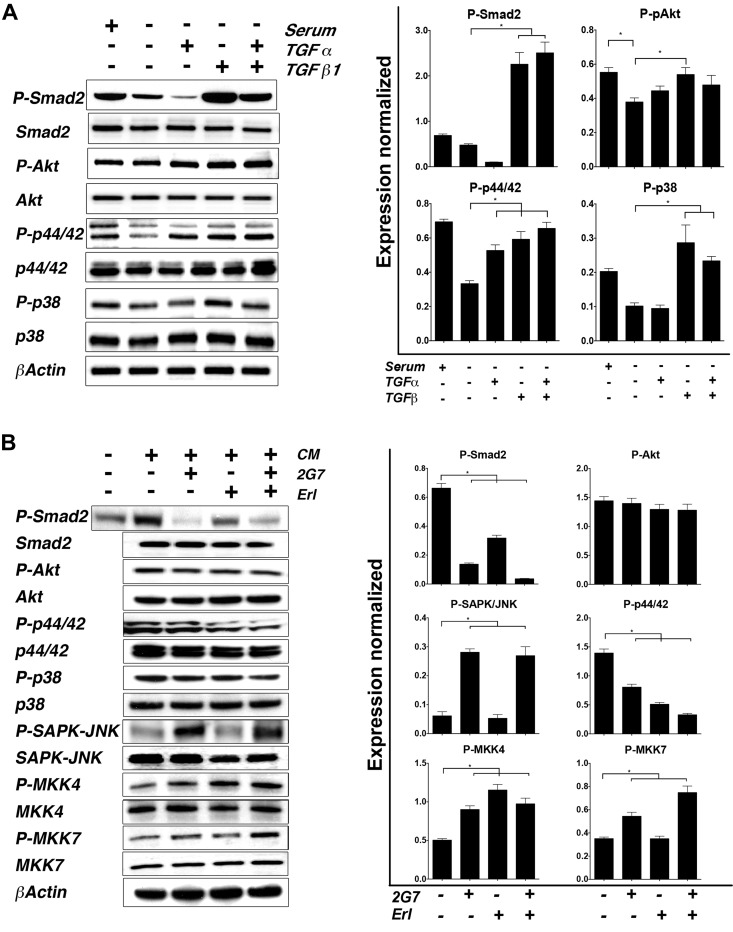
To investigate molecular events downstream of TGF-α and TGF-β in AT1 cells, we examined activation of relevant nonreceptor kinases and signaling proteins 48 h after stimulation with TGF-α and TGF-β alone or in combination. TGF-β activated canonical and noncanonical downstream signaling. Individually, TGF-α and TGF-β increased phosphorylation of p44/42 MAPK; however, the combined effect of the 2 ligands was greater (>2-fold compared to the control) (Fig. 6A). No activation of the Akt pathway was observed. Only TGF-β was able to increase phosphorylation of p38. The signaling response of AT1 to TGF-α and TGF-β provides a molecular correlate for the effects on proliferation and migration. To confirm this possibility, we tested whether the same inhibitors used in functional assays also affected signaling events induced by CA1d CM (Fig. 6B).
Figure 6.
Downstream effects of TGF-α and TGF-β activation in AT1 cells. A) Response of AT1 cells to TGF-α and/or TGF-β stimulation was evaluated. Increased activation of TGF-β pathway was observed in presence of serum, TGF-β, and combination of ligands compared to basal (serum −). Phosphorylation of p44/42 increased in presence of ligands compared to basal levels. Interestingly P-p38 MAPK had dramatic positive response only in presence of TGF-β. Right: average relative density of each band from 3 different experiments was quantitated and normalized against their corresponding control (total protein). Left: representative blot. *P < 0.05, 1-way ANOVA with Dunnett’s multiple comparison test. B) To address influence of secreted factors, AT1 cells were cultured in presence of CA1d CM and treated with TGF-β and/or TGF-α inhibitors 2G7 and Erl. TGF-β pathway was impaired with both inhibitors. While Akt pathway remained unchanged, P-p44/42 and P-p38 decreased with addition of inhibitors. Blocking TGF-β pathway significantly increased phosphorylation of SAPK/JNK and its MAPK2 activator MKK7, while MKK4 phosphorylation was increased in presence of both 2G7 and Erl. Right: quantitation of bands followed similar approach used in (A). Left: representative blot.
Phosphorylation of Smad2, a key indicator of activation of the TGF-β canonical pathway, was inhibited by either the TGF-β ligand-neutralizing antibody 2G7 (>50%), or Erl (>75%). Remarkably, their combination resulted in undetectable levels of pSmad2. Phosphorylation of p44/42 MAPK showed a similar trend, while P-Akt and p38 were not significantly affected. Interestingly, in the presence of the TGF-β inhibitor 2G7, we detected activation of SAPK/JNK. To further identify potential upstream TGF-β-mediators of SAPK/JNK, we evaluated the phosphorylation status of the MAPKKs MKK4 (also known as SEK1) and MKK7, which are known activators of JNKs. While P-MKK4 was higher in the presence of inhibitors, phosphorylation of MKK7 specifically correlated with the presence of 2G7 similar to P-SAPK/JNK (1.8-fold higher with 2G7 only and 2.1-fold higher in the 2G7/Erl group), suggesting that MKK7 activation may be responsible of JNK phosphorylation when the TGF-β pathway is blocked. Exposure of CA1d cells to AT1-CM did not alter TGF-β signaling, and the degree of response (activation of downstream signaling pathways) of CA1d when exposed to TGF-β/TGF-α ligands was lower but not negligible when compared to AT1 cells (data not shown). Taken together, these data highlight the complex molecular downstream effects of paracrine activators in heterogeneous tumors.
DISCUSSION
The data presented here imply that cooperation between disparate phenotypes can contribute to tumor growth and invasion; that paracrine mechanisms involving secreted growth factors (in this model, TGF-β and TGF-α) can mediate such cooperation; and that less aggressive cells may undergo a stable conversion to an aggressive phenotype under the influence of aggressive cancer cells.
The contribution of stromal–epithelial interactions to tumor progression has been well established (33, 34). More recently, stromal heterogeneity has been observed in vivo and has been experimentally demonstrated to contribute to the cancer fibroblast phenotype (5–7). Here we show another level of interaction that parallels these previous observations but occurs between epithelial cell types instead.
It has been suggested that during tumor progression, only the fittest populations survive (20). However, less malignant cells can be found in different regions of both primary and metastatic tumors (35). Although competition may be a common factor (13, 36), the cooperation of cancer cells in which cells at various stages of initiation/transformation interact to promote tumor progression has been more challenging to study experimentally and has not been so thoroughly investigated.
The lines interrogated here represent a useful model of breast cancer progression from a benign state (MCF10A) to a premalignant phenotype (AT1) and a fully transformed aggressive phenotype (CA1d) (23). The changes from AT1 to CA1d were a result of environmental pressure, in which the most aggressive cells were selected from common progenitors. Heterogeneous tumors composed of premalignant AT1 cells and malignant CA1d cells grew much larger than expected (based on analysis of the individual cell lines) and were composed approximately equally of both cell types (Fig. 1). We expected that these experiments would show competition, with the most aggressive cell type winning out, a phenomenon observed in the MCF10A/CA1d mixtures and consistent with similar observations in some other model systems (37). However, the results obtained suggested that each cell line benefits from the other. Such cooperation has been postulated to play a role in tumor evolution by maintaining the survival and proliferation of various tumor cell populations, to the benefit of the entire tumor (16). A mouse model of small-cell lung cancer showed that cell populations with different phenotypic characteristics can dictate tumor behavior (38). Here we provide experimental evidence (using human cells) consistent with this phenomenon. A noninitiated parental MCF10A cell population was outcompeted by the other cell lines and did not contribute to tumor progression, suggesting that initiation may be a prerequisite for cooperative interactions during tumor progression. Although the model has limitations—for example, understating the number of populations present in a given tumor—we provide evidence that such interactions can occur and suggest one mechanism by which epithelial subpopulations could communicate to promote cancer progression.
Another important observation was the maintenance of the aggressive phenotype acquired by cooperative interactions in the absence of the cooperating, more aggressive tumor cell types. Two distinct clones, AT1-TD1 and AT1-TD2, derived from the mixed tumors and passaged in culture for multiple generations, maintained a substantially more aggressive phenotype compared to the parental AT1 cells. These 2 clones could not be distinguished from the AT1 parental cells by coarse genetic profiling. They showed minimal changes not obviously related to aggressiveness. It has been hypothesized that mutations accumulate over time after transformation but remain undetectable until a major environmental driver exerts a breakthrough effect conferring a greater advantage on growth, invasion, or drug resistance (39). We have previously shown in prostate cancer models that tumor microenvironment can elicit similar initiated-to-malignant phenotypic conversion. In that model system, we observed and described genomic rearrangements associated with malignant progression (30, 31). It should be noted that in the prostate model, we used a cell line that expressed SV40 T antigen, making it highly susceptible to genomic instability. No such modifier was present in this study. Thus, the major changes seen in the present study were in gene expression rather than genomic alterations. The 2 independently isolated sublines exhibited similar phenotypic changes, suggesting the possibility of environmentally regulated epigenetic changes as a driver of increased aggressiveness.
To understand our biologic observations at a molecular level, we focused our screening on the expression of potential paracrine mediators. We identified TGF-α and TGF-β as candidate signaling molecules to regulate the malignant phenotype of the combination of AT1 and CA1d cells on the basis of differential mRNA expression between the parental AT1 cells and the TD sublines. High circulating levels of these 2 ligands have been linked to tumor progression (40, 41). Our in vitro experiments showing that Erl and the pan-TGF-β antibody 2G7 abolish the paracrine effects exerted by CA1d cells on AT1 support their role in modulating tumor cell aggressiveness. In addition, although we have not explored them, it is possible that when both EGFR- and TGF-βR-mediated pathways are activated or inhibited, novel signaling responses are induced that could not be elicited by either pathway alone.
Although TGF-α had no an obvious effect on the SAPK/JNK pathway, blocking the TGF-β pathway in CA1d-CM-treated AT1 cells increased the phosphorylation of JNK and its upstream kinase MKK7, known regulators of apoptosis and senescence (42). We hypothesize that TGF-β may act as a prosurvival factor in AT1 cells by providing a stress buffer. Inhibiting this pathway would result in activation of the MKK7-SAPK/JNK complex that could trigger apoptosis. We suggest that in a heterogeneous tumor, high levels of TGF-β secreted by cancer cells and/or the surrounding stroma could allow AT1 cells to avoid apoptosis and instead transform toward a malignant phenotype.
The importance of epithelial heterogeneity within a single neoplasm is increasingly accepted (32). Activation of multiple mechanisms/pathways are likely to be present in these tumors, which can make single-target therapy less effective. However, the impact of multiple populations on cancer progression is not well understood. Human tumors have long been known to be heterogeneous with respect to their genotype (43, 44). It is clear that in a given tumor, several cancer cell clones with different malignant potential coexist during the progression to the invasive and metastatic phenotype. Several models have been proposed to explain the complex effects of epithelial cell heterogeneity on progression (45).
In summary, we have shown that cooperation between cancer cells can play a role in determining tumor growth. Competition can restrict overall growth, at least in the short term; however, epithelial cells within a tumor can also cooperate with each other, promoting growth, invasion, and ultimately transformation of the premalignant component of the lesion. This observation is in line with previously stated theoretical predictions (33). This model opens new exploratory avenues and may provide a new framework for designing therapies.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank L. Chin (University of Texas System at Austin, Austin, TX, USA), A. Protopopov (University of Texas M. D. Anderson Cancer Center), and Y. Xiao (Lion Bioscience, Heidelberg, Germany) for help with the array-CGH experiments and analysis. The authors also thank J. R. Nevins and M. West (Duke University Durham, NC, USA) for performing the cDNA microarray experiments and for their guidance on the analysis. The authors are grateful to J. Jourquin (Susan G. Komen, Nashville, TN, USA) for his help in the analysis of microarray expression data and pathway enrichment analysis. This work was supported by the U.S. National Institutes of Health/National Cancer Institute Integrative Cancer Biology Program (5U54 CA113007 and 5U01 CA151924).
Glossary
- A/C
AT1/CA1d
- array CGH
array-based comparative genomic hybridization
- CM
conditioned medium
- EGFR
epithelial growth factor receptor
- Erl
erlotinib
- FACS
fluorescence-activated cell sorting
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- H-Ras
Harvey rat sarcoma virus oncogene
- M/A
MCF10A/AT1
- M/C
MCF10A/CA1d
- MKK
mitogen-activated protein kinase
- RFP
red fluorescent protein
- SAPK
stress-activated protein kinase
- STR
short tandem repeat
- TD
tumor derived
- TGF-βR
TGF-β receptor
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
S. W. Hayward, V. Quaranta, D. R. Tyson, and L. Estrada were involved in the research design; O. E. Franco, K. C. Konvinse, and A. Udyavar performed research; O. E. Franco, D. R. Tyson, and S. W. Hayward analyzed data; S. E. Crawford performed the histopathologic evaluation of mouse xenografts, O. E. Franco and D. R. Tyson wrote the paper; and S. W. Hayward and V. Quaranta reviewed and approved the final version of the manuscript.
REFERENCES
- 1.Franses J. W., Baker A. B., Chitalia V. C., Edelman E. R. (2011) Stromal endothelial cells directly influence cancer progression. Sci. Transl. Med. 3, 66ra5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franco O. E., Shaw A. K., Strand D. W., Hayward S. W. (2010) Cancer associated fibroblasts in cancer pathogenesis. Semin. Cell Dev. Biol. 21, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancino M., Ametller E., Gascón P., Almendro V. (2011) The neuronal influence on tumor progression. Biochim. Biophys. Acta 1816, 105–118 [DOI] [PubMed] [Google Scholar]
- 4.Ayala G., Tuxhorn J. A., Wheeler T. M., Frolov A., Scardino P. T., Ohori M., Wheeler M., Spitler J., Rowley D. R. (2003) Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin. Cancer Res. 9, 4792–4801 [PubMed] [Google Scholar]
- 5.Sugimoto H., Mundel T. M., Kieran M. W., Kalluri R. (2006) Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 5, 1640–1646 [DOI] [PubMed] [Google Scholar]
- 6.Kiskowski M. A., Jackson R. S. II, Banerjee J., Li X., Kang M., Iturregui J. M., Franco O. E., Hayward S. W., Bhowmick N. A. (2011) Role for stromal heterogeneity in prostate tumorigenesis. Cancer Res. 71, 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco O. E., Jiang M., Strand D. W., Peacock J., Fernandez S., Jackson R. S. II, Revelo M. P., Bhowmick N. A., Hayward S. W. (2011) Altered TGF-β signaling in a subpopulation of human stromal cells promotes prostatic carcinogenesis. Cancer Res. 71, 1272–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D., Coussens L. M. (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 [DOI] [PubMed] [Google Scholar]
- 9.Coussens L. M., Zitvogel L., Palucka A. K. (2013) Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 339, 286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polyak K. (2011) Heterogeneity in breast cancer. J. Clin. Invest. 121, 3786–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almendro V., Marusyk A., Polyak K. (2013) Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 8, 277–302 [DOI] [PubMed] [Google Scholar]
- 12.Heppner G. H. (1993) Cancer cell societies and tumor progression. Stem Cells 11, 199–203 [DOI] [PubMed] [Google Scholar]
- 13.Anderson A. R., Hassanein M., Branch K. M., Lu J., Lobdell N. A., Maier J., Basanta D., Weidow B., Narasanna A., Arteaga C. L., Reynolds A. B., Quaranta V., Estrada L., Weaver A. M. (2009) Microenvironmental independence associated with tumor progression. Cancer Res. 69, 8797–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser J. (2013) The downside of diversity. Science 339, 1543–1545 [DOI] [PubMed] [Google Scholar]
- 15.Swanton C. (2012) Intratumor heterogeneity: evolution through space and time. Cancer Res. 72, 4875–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axelrod R. (2012) Launching “the evolution of cooperation.” J. Theor. Biol. 299, 21–24 [DOI] [PubMed] [Google Scholar]
- 17.Miller B. E., Miller F. R., Leith J., Heppner G. H. (1980) Growth interaction in vivo between tumor subpopulations derived from a single mouse mammary tumor. Cancer Res. 40, 3977–3981 [PubMed] [Google Scholar]
- 18.Chapman A., Fernandez del Ama L., Ferguson J., Kamarashev J., Wellbrock C., Hurlstone A. (2014) Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 8, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleary A. S., Leonard T. L., Gestl S. A., Gunther E. J. (2014) Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamori Y., Deng W. M. (2011) Cell competition and its implications for development and cancer. J. Genet. Genomics 38, 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller F. R. (2000) Xenograft models of premalignant breast disease. J. Mammary Gland Biol. Neoplasia 5, 379–391 [DOI] [PubMed] [Google Scholar]
- 22.Soule H. D., Maloney T. M., Wolman S. R., Peterson W. D. Jr., Brenz R., McGrath C. M., Russo J., Pauley R. J., Jones R. F., Brooks S. C. (1990) Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 [PubMed] [Google Scholar]
- 23.Santner S. J., Dawson P. J., Tait L., Soule H. D., Eliason J., Mohamed A. N., Wolman S. R., Heppner G. H., Miller F. R. (2001) Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65, 101–110 [DOI] [PubMed] [Google Scholar]
- 24.Bessette D. C., Tilch E., Seidens T., Quinn M. C., Wiegmans A. P., Shi W., Cocciardi S., McCart-Reed A., Saunus J. M., Simpson P. T., Grimmond S. M., Lakhani S. R., Khanna K. K., Waddell N., Al-Ejeh F., Chenevix-Trench G. (2015) Using the MCF10A/MCF10CA1a breast cancer progression cell line model to investigate the effect of active, mutant forms of EGFR in breast cancer development and treatment using gefitinib. PLoS One 10, e0125232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arteaga C. L., Hurd S. D., Winnier A. R., Johnson M. D., Fendly B. M., Forbes J. T. (1993) Anti–transforming growth factor (TGF)-beta antibodies inhibit breast cancer cell tumorigenicity and increase mouse spleen natural killer cell activity. Implications for a possible role of tumor cell/host TGF-beta interactions in human breast cancer progression. J. Clin. Invest. 92, 2569–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguirre A. J., Brennan C., Bailey G., Sinha R., Feng B., Leo C., Zhang Y., Zhang J., Gans J. D., Bardeesy N., Cauwels C., Cordon-Cardo C., Redston M. S., DePinho R. A., Chin L. (2004) High-resolution characterization of the pancreatic adenocarcinoma genome. Proc. Natl. Acad. Sci. USA 101, 9067–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Protopopov A., Feng B., Chin L. (2008) Full complexity genomic hybridization on 60-mer oligonucleotide microarrays for array comparative genomic hybridization (aCGH). Methods Mol. Biol. 439, 87–100 [DOI] [PubMed] [Google Scholar]
- 29.Strickland L. B., Dawson P. J., Santner S. J., Miller F. R. (2000) Progression of premalignant MCF10AT generates heterogeneous malignant variants with characteristic histologic types and immunohistochemical markers. Breast Cancer Res. Treat. 64, 235–240 [DOI] [PubMed] [Google Scholar]
- 30.Hayward S. W., Wang Y., Cao M., Hom Y. K., Zhang B., Grossfeld G. D., Sudilovsky D., Cunha G. R. (2001) Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res. 61, 8135–8142 [PubMed] [Google Scholar]
- 31.Phillips J. L., Hayward S. W., Wang Y., Vasselli J., Pavlovich C., Padilla-Nash H., Pezullo J. R., Ghadimi B. M., Grossfeld G. D., Rivera A., Linehan W. M., Cunha G. R., Ried T. (2001) The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 61, 8143–8149 [PubMed] [Google Scholar]
- 32.Li J., Wang K., Jensen T. D., Li S., Bolund L., Wiuf C. (2010) Tumor heterogeneity in neoplasms of breast, colon, and skin. BMC Res. Notes 3, 321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olumi A. F., Grossfeld G. D., Hayward S. W., Carroll P. R., Tlsty T. D., Cunha G. R. (1999) Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 59, 5002–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhowmick N. A., Chytil A., Plieth D., Gorska A. E., Dumont N., Shappell S., Washington M. K., Neilson E. G., Moses H. L. (2004) TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303, 848–851 [DOI] [PubMed] [Google Scholar]
- 35.Gerlinger M., Rowan A. J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N. Q., Butler A., Jones D., Raine K., Latimer C., Santos C. R., Nohadani M., Eklund A. C., Spencer-Dene B., Clark G., Pickering L., Stamp G., Gore M., Szallasi Z., Downward J., Futreal P. A., Swanton C. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson A. R., Weaver A. M., Cummings P. T., Quaranta V. (2006) Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127, 905–915 [DOI] [PubMed] [Google Scholar]
- 37.Kajita M., Fujita Y. (2015) EDAC: epithelial defence against cancer-cell competition between normal and transformed epithelial cells in mammals. J. Biochem. 158, 15–23 [DOI] [PubMed] [Google Scholar]
- 38.Calbo J., van Montfort E., Proost N., van Drunen E., Beverloo H. B., Meuwissen R., Berns A. (2011) A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19, 244–256 [DOI] [PubMed] [Google Scholar]
- 39.Shibata D. (2012) Cancer. Heterogeneity and tumor history. Science 336, 304–305 [DOI] [PubMed] [Google Scholar]
- 40.Rhee J., Han S. W., Cha Y., Ham H. S., Kim H. P., Oh D. Y., Im S. A., Park J. W., Ro J., Lee K. S., Park I. H., Im Y. H., Bang Y. J., Kim T. Y. (2011) High serum TGF-α predicts poor response to lapatinib and capecitabine in HER2-positive breast cancer. Breast Cancer Res. Treat. 125, 107–114 [DOI] [PubMed] [Google Scholar]
- 41.Ivanović V., Todorović-Raković N., Demajo M., Nesković-Konstantinović Z., Subota V., Ivanisević-Milovanović O., Nikolić-Vukosavljević D. (2003) Elevated plasma levels of transforming growth factor-beta 1 (TGF-beta 1) in patients with advanced breast cancer: association with disease progression. Eur. J. Cancer 39, 454–461 [DOI] [PubMed] [Google Scholar]
- 42.Haeusgen W., Herdegen T., Waetzig V. (2011) The bottleneck of JNK signaling: molecular and functional characteristics of MKK4 and MKK7. Eur. J. Cell Biol. 90, 536–544 [DOI] [PubMed] [Google Scholar]
- 43.Magee J. A., Piskounova E., Morrison S. J. (2012) Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ene C. I., Fine H. A. (2011) Many tumors in one: a daunting therapeutic prospect. Cancer Cell 20, 695–697 [DOI] [PubMed] [Google Scholar]
- 45.Shackleton M., Quintana E., Fearon E. R., Morrison S. J. (2009) Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 138, 822–829 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.