Abstract
DYX1C1, DCDC2, and KIAA0319 are three of the most replicated dyslexia candidate genes (DCGs). Recently, these DCGs were implicated in functions at the cilium. Here, we investigate the regulation of these DCGs by Regulatory Factor X transcription factors (RFX TFs), a gene family known for transcriptionally regulating ciliary genes. We identify conserved X-box motifs in the promoter regions of DYX1C1, DCDC2, and KIAA0319 and demonstrate their functionality, as well as the ability to recruit RFX TFs using reporter gene and electrophoretic mobility shift assays. Furthermore, we uncover a complex regulation pattern between RFX1, RFX2, and RFX3 and their significant effect on modifying the endogenous expression of DYX1C1 and DCDC2 in a human retinal pigmented epithelial cell line immortalized with hTERT (hTERT-RPE1). In addition, induction of ciliogenesis increases the expression of RFX TFs and DCGs. At the protein level, we show that endogenous DYX1C1 localizes to the base of the cilium, whereas DCDC2 localizes along the entire axoneme of the cilium, thereby validating earlier localization studies using overexpression models. Our results corroborate the emerging role of DCGs in ciliary function and characterize functional noncoding elements, X-box promoter motifs, in DCG promoter regions, which thus can be targeted for mutation screening in dyslexia and ciliopathies associated with these genes.—Tammimies, K., Bieder, A., Lauter, G., Sugiaman-Trapman, D., Torchet, R., Hokkanen, M.-E., Burghoorn, J., Castrén, E., Kere, J., Tapia-Páez, I., Swoboda, P. Ciliary dyslexia candidate genes DYX1C1 and DCDC2 are regulated by Regulatory Factor (RF) X transcription factors through X-box promoter motifs.
Keywords: reading disorder, ciliopathies, gene regulation, hTERT-RPE1 cell line, ciliary proteins
Developmental dyslexia (DD) is a common neurodevelopmental learning disorder that affects 5–10% of school-age children worldwide (1, 2); a highly heritable trait with a demonstrated biological basis (3). Human genetic studies have implicated multiple loci to be involved in the etiology of DD. Thus far, only a handful of dyslexia candidate genes (DCGs) have been identified from these loci and subsequently characterized. To date, DYX1C1 (dyslexia 1 candidate 1 gene), DCDC2 (doublecortin domain-containing 2 gene), and KIAA0319 are the most replicated and best-studied DCGs (3–7). Functionality of the DD-associated single nucleotide polymorphisms (SNPs) has only been shown in a few instances. For example, we demonstrated that SNPs in the promoter region of DYX1C1 affect the binding of transcription factor complexes that depend both on the specific allele and on exposure to estrogen (8, 9). Likewise, associated variants in the promoter of KIAA0319 can affect its expression (10).
By use of cell and rodent models, the 3 DCGs, DYX1C1, DCDC2, and KIAA0319, have been linked strongly to cytoskeletal organization and to neuronal migration in the brain (4, 5, 11–13). Recently, studies have shown an entirely novel connection between DCGs and cilia. Cilia are nearly ubiquitous, hair-like eukaryotic subcellular organelles that project off polarized cell surfaces. In mammals, cilia are present on many different cell types, including most types of neurons (14, 15). Primary cilia house signal reception and transduction components that are critical for development, for example, the Hedgehog and Wnt pathways. Defects in the function of cilia and cilia-associated genes can lead to a class of human genetic disorders broadly called ciliopathies (16, 17). Furthermore, cilia have a demonstrated role in brain development, and dysfunctions in neuronal cilia can lead to brain abnormalities (18, 19).
After our initial discovery that increased expression of DCDC2 controls the length and signaling of primary cilia (41), more evidence has emerged that links DCGs to cilia. Recent genetic findings support a critical role for these genes in ciliary biology. Knockout mouse and zebrafish morpholino models have further shown that lack of Dyx1c1 leads to outer dynein arm and inner dynein arm defects, which likely cause impaired ciliary motility (20, 21). Mutations in DYX1C1 have been found in patients with primary ciliary dyskinesia (PCD) (21, 22) and mutations in DCDC2 in patients with nephronophthisis-related ciliopathy (23) and heritable deafness (24).
Cilia are composed of hundreds of different protein components. Studies primarily in Caenorhabditis elegans, Drosophila, and Danio rerio have shown that Regulatory Factor X transcription factors (RFX TFs) directly regulate many ciliary genes, including components of the intraflagellar transport complex, axonemal dyneins, and Bardet-Biedl syndrome proteins (25–30), a mechanism that is likely common across many different species (31). In humans, the RFX TF gene family consists of 8 members (RFX1–8), which can be divided into groups on the basis of protein domain homologies (26). The RFX TF family of proteins is defined by a highly conserved DNA binding domain that recognizes a sequence motif known as the X-box, typically located in gene promoters (26). RFX TFs are important in brain development and many of the members, namely RFX1–5 and RFX7, are highly expressed in the brain (32, 33), with RFX1-3 being the most studied.
On the basis of the recently uncovered links between DCGs and cilia, we hypothesized that ciliogenic RFX TFs are involved in the transcriptional regulation of DCGs. This would further strengthen the emerging concept that DCGs play important roles at the cilium and impact the view on the still unknown cell biology that underlies a human brain condition such as DD.
MATERIALS AND METHODS
Identification of X-box promoter motifs
To identify putative X-box motifs in the promoter regions of DCGs, a bioinformatics approach was taken as described by Henriksson et al. (34). In short, genomic sequences 3 kb upstream from the start of exon 1 for DYX1C1, DCDC2, and KIAA0319 on the basis of Ensembl protein-coding transcripts (release 75, February 2014) were retrieved by using the UCSC Genome Browser (GRCh37/hg19; University of California, Santa Cruz, Santa Cruz, CA, USA). Thereafter, the nucleic acid pattern search tool EMBOSS fuzznuc (http://emboss.bioinformatics.nl/cgi-bin/emboss/fuzznuc) was used to identify putative X-box motif sequences within these promoter regions. The search was based on 7 different X-box motif consensus sequences (Supplemental Table S1) and conducted on both the coding and noncoding strands. Further ranking and prioritization of the identified X-box motifs was done by using the mean PhyloP score on the basis of 100 vertebrates (retrieved from the UCSC Genome Browser) (35). To visualize the conservation of motifs, we aligned the most highly ranked of the identified X-box sequences against 4 other mammalian species—mouse, dog, cow, and cat—and made a consensus sequence logo by using WebLogo (University of California, Berkeley, Berkeley, CA, USA).
Cell culture
The human retinal pigmented epithelial cell line immortalized with hTERT (hTERT-RPE1; CRL-4000; American Type Culture Collection, Manassas, VA, USA) was cultured in DMEM/F12, 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 0.01 mg/ml hygromycin B at 5% CO2. To induce ciliogenesis, cells were starved by using Opti-MEM reduced serum medium for 24 h unless indicated otherwise. Human neuroblastoma cell line, subline of the neuroblastoma cell line SK-N-SH (SH-SY5Y) was cultured as described previously (13). In brief, cells were cultured in MEM with Earle’s salts and GlutaMax-I that was supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Luciferase reporter assays
Up to 2 kb of promoter sequence was amplified from human genomic DNA for each of the 3 DCGs, DYX1C1, DCDC2, and KIAA0319. Fragments were then cloned upstream of the luciferase reporter gene in promoterless pGL3basic vector (Promega, Madison, WI, USA; Supplemental Table S2). Genomic regions that were cloned were upstream of DCG exon 1 and contained the identified X-box promoter motifs. To disrupt the X-box motif, we changed up to 8 nucleotides following previously published information and principles (36, 37) using site-directed mutagenesis (Supplemental Table S2). The resulting clones were sequence verified before use. Luciferase assays were performed as described previously (5). In brief, hTERT-RPE1 or SH-SY5Y cells were cotransfected at 80–90% confluence with either the wild-type (WT) or the corresponding X-box mutated pGL3basic construct, together with 10 ng of the pRLTK vector, which constitutively expresses Renilla luciferase and functions as an internal normalization control. For hTERT-RPE1 cells that underwent ciliogenesis, standard cultivation medium was replaced by serum-reduced Opti-MEM medium. After 24 h, we lysed and measured luciferase activity by using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer protocol and a Tecan Luminometer Infinite 200 (Tecan AG, Männedorf, Switzerland). Relative luciferase expression was normalized and results were presented as relative response ratios compared with the indicated WT promoter construct. Experiments were performed in triplicate and were repeated independently 3 times. Statistical significance was analyzed by using 2-sided Student’s t test.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed by using biotin end-labeled probes that contained sequences surrounding the conserved X-boxes of the DCGs DYX1C1, DCDC2, and KIAA0319 (Fig. 1A). Probes consisted of double-stranded WT sequences, 29 or 30 bp in length. Sequences are as follows: wt-DYX: forward, 5′-CCCAGCTTCCCTAGCAACCAAGCAGGC-3′; wt-DYX: reverse, 5′-GCCTGCTTGGTTGCTAGGGAAGCTGGGGTT-3′; mut-DYX: forward, 5′-AACCCCACATTGACTCTCAAACAAGCAGGC-3′; mut-DYX: reverse, 5′-GCCTGCTTGTTTGAGAGTCAATGTGGGGTT-3′; wt1-DCDC2: forward, 5′-GAGGCTGCGGTTGCTATGGAAACAGCGGGA-3′; wt1-DCDC2: reverse, 5′-TCCCGCTGTTTCCATAGCAACCGC AGCCTC-3′; mut1-DCDC2: forward, 5′-AGGCTGCGACTGGTATGCAAATAGCGGGA-3′; mut1-DCDC2: reverse, 5′-TCCCGCTATTTGCATACCAGTCGCAGCCT-3′; wt-KIAA0319: forward, 5′-CGCGCGCGTCGCCGTGGTAACCGCGGCGGC-3′; wt-KIAA0319: reverse, 5′-GCCGCCGCGGTTACCACGGCGACGCGCGCG-3′; mut-KIAA0319: forward, 5′-CGCGCGCGTCGTCTAGATAACCGCGGCGGC-3′; mut-KIAA0319: reverse, 5′-GCCGCCGCGGTTATCTAGACGACGCGCGCG-3′. Nuclear extracts from serum-starved hTERT-RPE1 cells were obtained by using the NE-PER nuclear and cytoplasmic extraction reagents, according to manufacturer instructions (Thermo Fisher Scientific, Waltham, MA, USA). Each probe was incubated with 10 μg of nuclear protein extracts for 15 min at room temperature. The remaining steps followed the LightShift Chemiluminescent EMSA Kit protocol (Thermo Fisher Scientific). Samples were electrophoresed on 6% Novex TBE gels (Thermo Fisher Scientific) in 0.5 × TBE (0.09 M Tris borate, 2 mM EDTA) at 100 V. Gel was blotted on to a nylon membrane (Thermo Fisher Scientific) and the detection was performed according to the manufacturer protocol. For super shift assays, antibodies against RFX1 (H-11, sc-166468), RFX2 (A-18, sc-10659), and RFX3 (T-17, sc-10662; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used. RFX2 and RFX3 antibodies were used before in similar experiments (38–40) and their specificity was tested by Western blot (data not shown).
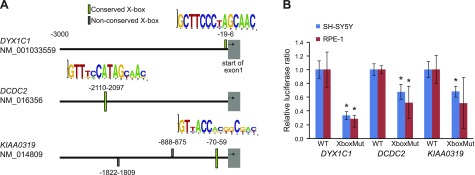
Figure 1.
The promoters of DYX1C1, DCDC2, and KIAA0319 contain functional X-box promoter motifs. A) Location of putative X-box promoter motifs upstream of DYX1C1, DCDC2, and KIAA0319 identified in the bioinformatics screen. X-box sequence logos on the basis of human, mouse, dog, cow, and cat sequence alignments are shown above 3 well-conserved motifs. B) Luciferase expression assays: pGL3 basic vector containing promoter sequences of DYX1C1, DCDC2, and KIAA0319 (containing WT or mutated X-box motifs) were transfected into starved hTERT-RPE1 and SH-SY5Y cells under normal growth conditions. Luciferase expression was measured and data are shown as relative ratio to the WT construct. Data are displayed as means ± sem. *P < 0.05 of 2-sided Student’s t test.
Small interfering RNA knockdowns
hTERT-RPE1 and SH-SY5Y cells were seeded in 6-well plates and grown to 80% confluence. Cells were transfected with 25 µM of each siGENOME SMARTpool targeting RFX TF genes (RFX1: M-010147-01; RFX2: M-011129-00; RFX3: M-011764-00) at a final concentration of 75 µM for any combination of the 3 genes or adjustment of the concentration with a control pool (nontargeting #2: D-001206-14-20; all Thermo Fisher Scientific) using DharmaFECT transfection reagent 1 (T-2001-03; Thermo Fisher Scientific). After 48 h, hTERT-RPE1 cells were serum starved for 24 h and harvested. For SH-SY5Y cells, culture media was changed after 48 h and cells were then harvested after 24 h. RNA from both cell lines was prepared by using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and RNase-free DNase Set (Qiagen) according to manufacturer instructions. All experiments were performed in triplicate and were repeated independently 3 times.
Quantitative real-time PCR
cDNA was synthesized with SuperScript III First-Strand Synthesis SuperMix kit (11752-250; Thermo Fisher Scientific) by using 500 ng RNA. Quantitative real-time PCR (qRT-PCR) was analyzed with cDNA diluted 1:5 using TaqMan expression assays and TaqMan fast Universal PCR Master Mix [4352042; Thermo Fisher Scientific; DYX1C1: Hs00370049_m1; DCDC2: Hs00393203_m1; KIAA0319: Hs00207788_m1; HPRT1 (hypoxanthine phosphoribosyltransferase 1): Hs02800695_m1; CDK1 (cyclin-dependent kinase 1): Hs00938777_m1; TUBA1A (tubulin α1a): Hs00362387_m1; RFX2: 01100925_m1] or using SYBR green primers for RFX1, RFX3, and HPRT1 (10 µM) and the Fast SYBR Green Master Mix (4385612; Applied Biosystems, Foster City, CA, USA). The following SYBR Green primers were used: RFX1: forward, 5′-CAGACGAGCGTGCAGGCCAA-3′; RFX1: reverse, 5′-TGGCCACCTTTGCTGCCTGG-3′; RFX3: forward, 5′-TGGCGATTGAGACGCTGCAAA-3′; RFX3: reverse, 5′-TGGGAAGGCTCACTCCTTCTGCT-3′; HPRT: forward, 5′-TCAGGCAGTATAATCCAAAGATGGT -3′; HPRT: reverse, 5′-AGTCTGGCTTATATCCAACACTTCG-3′. HPRT1 was used as an internal control to normalize expression levels. Reactions were amplified by using the 7500 Fast Real-Time PCR system (Applied Biosystems). The ΔΔCt method was used to calculate relative expression levels and shown as fold change (2−ΔΔCt). For samples in which the gene was down-regulated, −2ΔΔCt was calculated. The statistical significance between different conditions of each analyzed gene was analyzed by using ANOVA followed by Tukey’s post hoc test.
Immunofluorescence
hTERT-RPE1 cells were seeded on coverslips, grown to 80% confluence, and serum starved for 24 h. Cells were incubated on ice for 45 min, then fixed in a 50:50 solution of 4% formaldehyde (Sigma-Aldrich, St. Louis, MO, USA) and MeOH or in pure MeOH for 15 min at −20°C. Fixed cells were blocked and permeabilized in 5% horse serum (Thermo Fisher Scientific) and 0.05% PBS-Tween for 1 h at room temperature and incubated overnight at 4°C with a primary antibody (rabbit anti-DYX1C1, 1:500; Sigma-Aldrich; rabbit anti-DCDC2, 1:200; Abcam, Cambridge, MA, USA; mouse anti-acetylated tubulin, 1:5000; Sigma-Aldrich; mouse anti–γ-tubulin, 1:500; Abcam). The specificities of anti-DYX1C1 and anti-DCDC2 antibodies have been assessed previously (21, 23). Cells were incubated for 1 h at room temperature with secondary antibodies (donkey anti-rabbit 568; Thermo Fisher Scientific; donkey anti-mouse 568; Thermo Fisher Scientific). Nuclei were stained with DRAQ5 [1,5-bis{[2-(di-methylamino)ethyl]amino}s-4,8-dihydroxyanthracene-9,10-dione] (Cell Signaling Technology, Cambridge, United Kingdom) at 1:1000 for 10 min at room temperature and coverslips were mounted in Prolong Gold antifade reagent (Thermo Fisher Scientific). Images were captured on an LSM 510 confocal microscope (Zeiss, Jena, Germany) using Plan-Neofluar ×40/NA 1.3 oil and Plan-Apochromatic ×63/NA 1.4 oil objectives and the Zeiss Laser Scanning Microscope LSM 510 software (version 4.2 SP1; Zeiss). Images were captured in multitrack scanning mode in 8-bit format using Z-stack and subsequent maximum intensity projection. Brightness and contrast were adjusted by using LSM 510 software and the far-red channel was pseudo colored in blue. Cilia and nuclei were quantified manually by using ImageJ (National Institutes of Health, Bethesda, MD, USA).
RESULTS
Conserved X-box motifs in the promoters of DCGs
We performed a bioinformatics search to identify candidate X-box motifs within promoter regions (3 kb upstream from exon 1) of the DCGs DYX1C1, DCDC2, and KIAA0319. By using the EMBOSS fuzznuc tool with 7 known X-box motif (consensus) sequences as query patterns (Supplemental Table S1), we identified a total of 7 candidate X-box motifs in the promoter sequences (Fig. 1A, Supplemental Table S3). Conservation estimation using the PhyloP 100 vertebrates mean score revealed that 5 of the candidate X-box motifs (at 3 independent genomic sites) were well conserved across species (PhyloP score >1.0), 1 in each DCG promoter (Fig. 1A, Supplemental Table S3). We considered the 3 well-conserved DCG promoter sites with their X-box motifs as good candidates for being involved in the regulation of DCG gene expression.
X-box motifs in the promoters of DYX1C1, DCDC2, and KIAA0319 are functional
We next addressed whether the identified, conserved X-box motifs are functional and required for gene expression. We cloned the X-box–containing promoter regions of DYX1C1, DCDC2, and KIAA0319 into a vector with the luciferase reporter gene and produced mutated versions of the X-box motifs by changing multiple nucleotides at essential positions of the motif sequences to abolish any possible RFX TF binding (Supplemental Table S2). We then transfected WT or mutant constructs into 2 different human cell lines, hTERT-RPE1 (ciliated upon starvation) and SH-SY5Y (nonciliated), that both express high levels of endogenous RFX1-3, RFX5, and RFX7 (data not shown). All 3 cloned regions comprised functional promoters that were sufficient to initiate luciferase gene expression well above background. We observed a significant decrease in luciferase activity for X-box mutant constructs for DYX1C1 and DCDC2 promoters compared with promoter activity of the corresponding WT motif (Fig. 1B). A similar trend was observed for the KIAA0319 X-box mutant construct, but a significant change was only observed in SH-SY5Y cells. These results suggest that the identified X-box motifs are functional and required for proper expression of the corresponding downstream gene.
RFX1 and RFX2 bind to conserved X-box motif sequences in the promoters of DYX1C1 and DCDC2
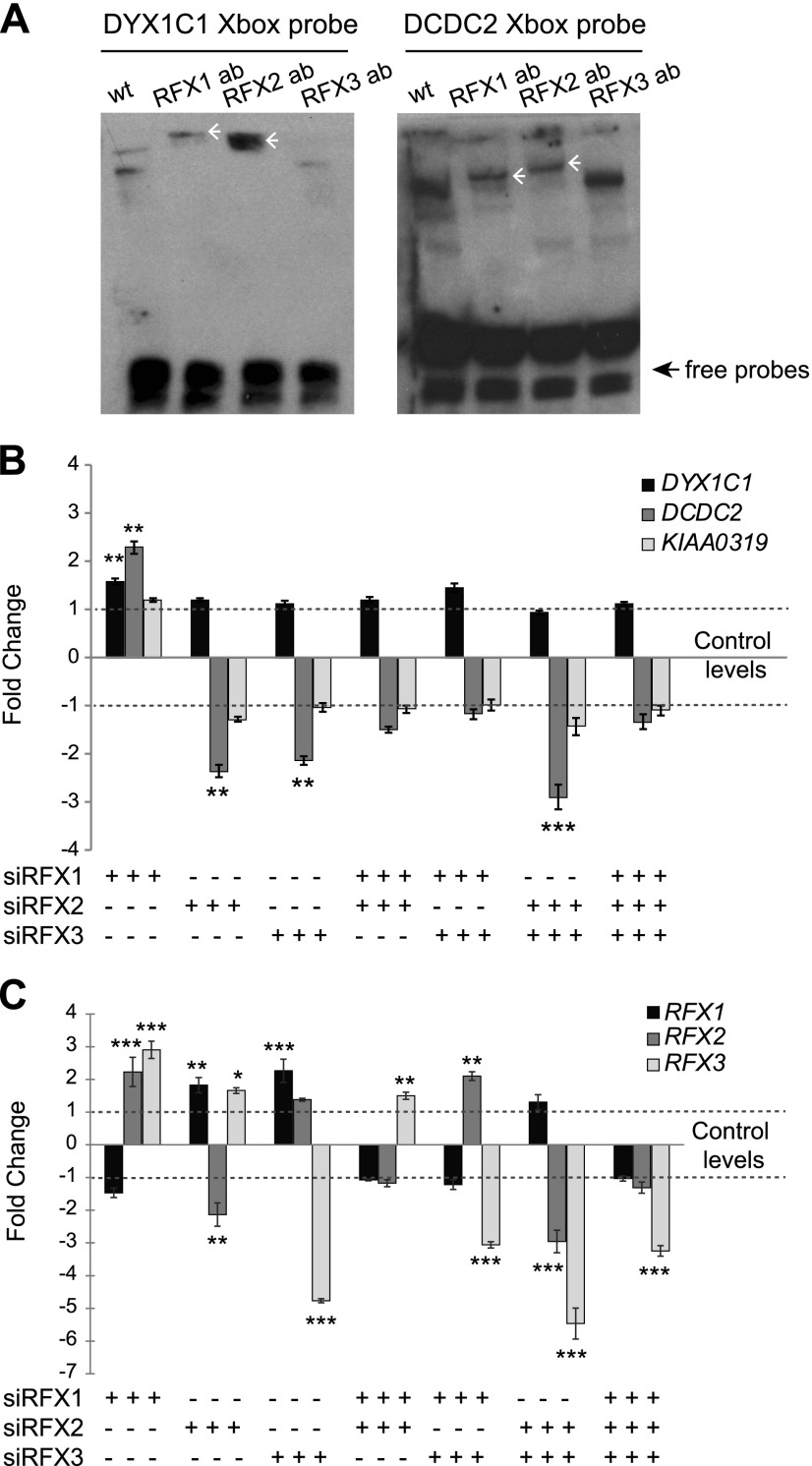
We then performed EMSA experiments with nuclear extracts from ciliated hTERT-RPE1 cells to directly demonstrate in vitro the binding of RFX TFs to X-box motif sequences. We observed 2 binding complexes when incubating hTERT-RPE1 nuclear extracts with duplex oligonucleotide probes spanning the X-box promoter motifs from DYX1C1 or DCDC2 (Fig. 2A); however, no binding was observed for the KIAA0319 X-box promoter motif (data not shown). In addition, RFX-to-X-box binding was abolished upon incubation with an excess of unlabeled, biotin-free probes (data not shown). We further demonstrated direct binding of RFX TFs by using EMSA supershift assays by adding anti-RFX1, anti-RFX2, or anti-RFX3 antibodies to the binding reactions (38). We observed only 1 supershifted complex after addition of anti-RFX1 or anti-RFX2 antibodies, which indicated that both RFX1 and RFX2 bind to the X-box motifs in the promoter regions of DYX1C1 and DCDC2 (Fig. 2A). Experiments that employed the anti-RFX3 antibody were in parts inconclusive (Fig. 2A and data not shown). All experiments were repeated at least 3 times with similar results. In conclusion, these in vitro experiments showing direct binding of RFX TFs to DCG X-boxes underscore that at least the DCGs DYX1C1 and DCDC2 are direct RFX target genes.
Figure 2.
Knockdown of RFX1, RFX2, and RFX3 affect the expression of DYX1C1 and DCDC2 but not KIAA0319 in hTERT-RPE1 cells. A) Binding of RFX1 and RFX2 to the X-box motifs present in DYX1C1 and DCDC2 promoters. Biotinylated probes spanning the X-box motifs were incubated with nuclear extracts from serum-starved hTERT-RPE1 cells with or without antibodies (ab) against RFX1, RFX2, and RFX3. Supershifts for both probes were detected for RFX1 and RFX2 antibodies (white arrowheads). One representative experiment of 3 is shown. B) Fold-change differences in the expression of DCGs upon knockdown of RFX TFs. By using siRNA against RFX1, RFX2, and RFX3, alone or in different combinations, genes were silenced in hTERT-RPE1 cells. Cells were thereafter starved for 24 h to induce ciliogenesis, and expression levels of DYX1C1, DCDC2, and KIAA0319 were measured by using qRT-PCR. C) Fold-change difference in expression levels of RFX1, RFX2, and RFX3 upon siRNA silencing. qRT-PCR data are summarized by using the ΔΔCt method as displayed as mean fold-change (2−ΔΔCt or −2ΔΔCt) ± sem. Significance of the post hoc tests after ANOVA is presented as ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05 after multiple correction of simultaneous comparisons.
Knockdown of RFX TFs affects expression of DYX1C1 and DCDC2
Next, we asked whether endogenous RFX TFs drive expression of DYX1C1, DCDC2, and KIAA0319. To address this issue, we performed knockdown of RFX1, RFX2, and RFX3 in ciliated hTERT-RPE1 cells and measured expression levels of the DCGs. In addition, we tested for potential combined effects among the RFX TFs by performing double and triple knockdowns of these factors. The level of DYX1C1 expression was changed significantly only upon knockdown of RFX1 (Fig. 2B). Expression of DCDC2 was up-regulated significantly upon silencing of RFX1 (Fig. 2B). The opposite effect was observed upon reducing the levels of RFX2 alone or together with RFX3. The combination of small interfering RNAs (siRNAs) against RFX2 and RFX3 had the most significant effect on DCDC2 levels, which resulted in a 3-fold down-regulation (Fig. 2B). Expression of KIAA0319 was not changed in any of the conditions. Of interest, when we performed similar experiments in the nonciliated neuroblastoma cell line SH-SY5Y, no significant changes were observed for the expression levels of any of the 3 DCGs (Supplemental Fig. S1). These results suggest that in ciliated cells RFX1 acts as a repressor for DYX1C1 and DCDC2 and that RFX2 and RFX3 act as an activator complex for DCDC2.
It is noteworthy that we observed that in our double or triple siRNA experiments, the effect on the target gene varied on the basis of the combination of silenced genes. To test for the possibility of regulation between these factors, we measured the expression levels of RFX1–3 in all siRNA conditions. Indeed, we observed that knockdown of RFX1 led to significant increase in the levels of RFX2 and RFX3, which further supported its role as a transcriptional repressor (Fig. 2C). Knockdown of RFX2 alone and RFX3 alone increased the expression of RFX1 (Fig. 2C); however, combined knockdowns of both RFX2 and RFX3 did not affect RFX1 expression. Finally, expression levels of RFX3 were significantly changed in all conditions. Taken together, these data suggest a complex mutual regulatory mechanism among different RFX TFs, whereby—depending on their relative expression levels in a given cell type—they can up- or down-regulate expression levels of targets, such as DYX1C1 and DCDC2.
Expression of DCGs increases upon induction of ciliogenesis
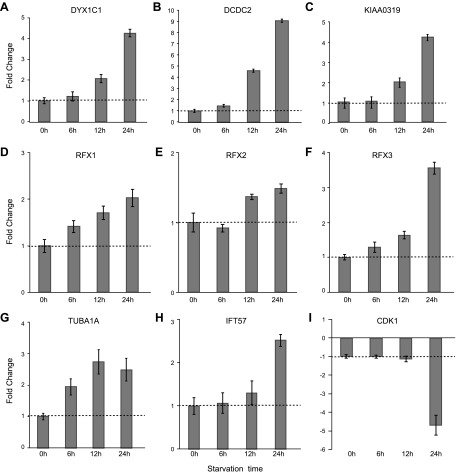
Given the well-documented role of RFX TFs in ciliary gene regulation (25, 26) and the fact that DCGs DYX1C1 and DCDC2 are direct RFX TF targets, we studied the expression of DCGs during ciliogenesis. We performed qRT-PCR at different time points during cilia induction. For this purpose, hTERT-RPE1 cell extracts were isolated at 0, 6, 12, and 24 h after serum starvation. We observed that expression of DYX1C1, DCDC2, and KIAA0319 was up-regulated during ciliogenesis, with the highest expression at 24 h (Fig. 3). In parallel, we measured the expression of RFX1, RFX2, and RFX3. Like the 3 DCGs, RFX TF genes also showed a similar increase of expression, but to a lesser extent (Fig. 3). As a control for the growth of cilia, we measured the expression of known cilia-specific genes, IFT57 (intraflagellar transport 57) and TUBA1A (tubulin α1a). As expected, their expression was up-regulated during ciliogenesis (Fig. 3G, H). At the same time, expression of CDK1 (cyclin-dependent kinase 1), a cell-cycle gene, was down-regulated as cells enter G0 and undergo ciliogenesis (Fig. 3I). In parallel and independently, using immunocytochemistry against the ciliary marker acetylated tubulin, we analyzed the process of ciliogenesis at different time points after serum starvation (Supplemental Fig. S2). We observed that after 12 h, >85% of cells had grown cilia. Finally, the same set of experiments was carried out in hTERT-RPE1 cells without serum starvation, and no differential expression of DCGs or of control genes was observed (Supplemental Fig. S3). Taken together, these time course experiments indicate that induction of RFX TF and DCG expression correlate well with the process of ciliogenesis.
Figure 3.
Induction of ciliogenesis in hTERT-RPE1 cells affects the expression levels of DCGs, RFX TF genes, and ciliogenesis/cell-cycle marker genes. hTERT-RPE1 cells were starved for 6, 12, and 24 h to induce ciliogenesis. Fold-change of difference for each time point compared with 0 h is shown for DYX1C1 (A), DCDC2 (B), KIAA0319 (C), RFX1 (D), RFX2 (E), RFX3 (F), TUBA1A (tubulin α1a) (G), IFT57 (intraflagellar transport 57) (H), and CDK1 (cyclin-dependent kinase 1) (I). qRT-PCR data are summarized by using the ΔΔCt method as displayed as mean fold-change (2−ΔΔCt or −2ΔΔCt) ± sem.
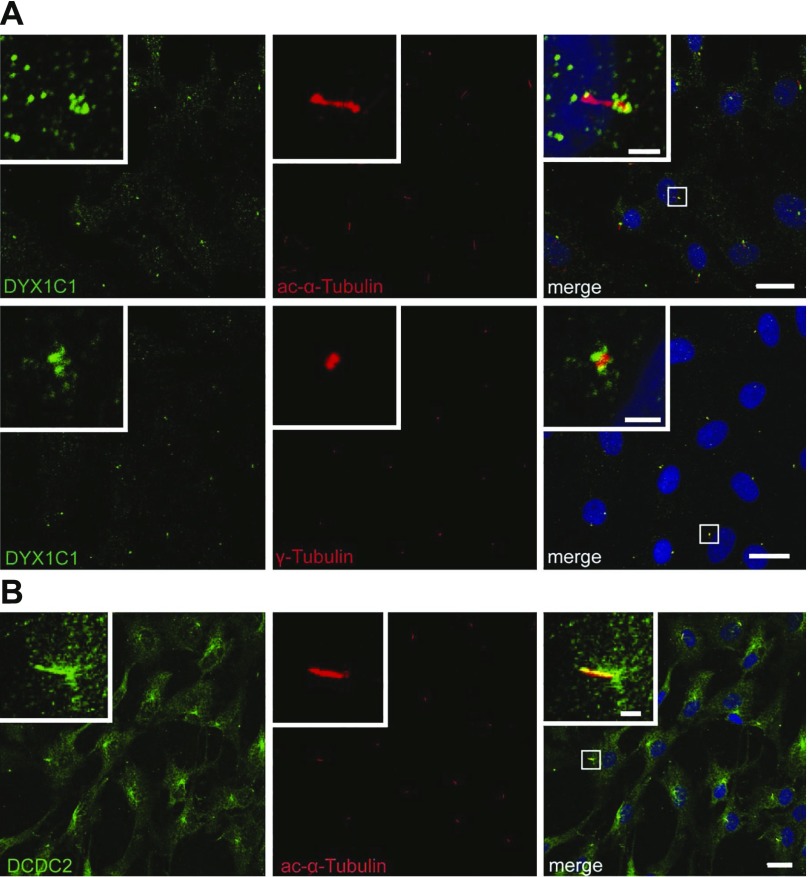
Endogenous DYX1C1 and DCDC2 proteins localize to ciliary structures in hTERT-RPE1 cells
On the basis of our and others’ previous results that show exogenous, overexpressed DCDC2 localizes to primary cilia in rat hippocampal neurons and in NIH/3T3 cells (41), and on the basis of results obtained in human ciliated tissues and hTERT-RPE1 cells (23), we tested whether the endogenous proteins DYX1C1 and DCDC2 localize to the primary cilium using immunocytochemistry in serum-starved hTERT-RPE1 cells. When costaining for DYX1C1 together with the cilia-specific marker acetylated α-tubulin or the centrosome-specific marker γ-tubulin, we observed DYX1C1 accumulation around the basal body of the cilium (Fig. 4A). When we costained for DCDC2 together with the cilia-specific marker acetylated α-tubulin, specific accumulation of DCDC2 along the entire ciliary axoneme was observed (Fig. 4B). These results—colocalization experiments of endogenous proteins—strongly support a role for DYX1C1 and DCDC2 at the basal body and the cilium, respectively.
Figure 4.
Subcellular localization of endogenous DYX1C1 (A) and DCDC2 (B) in hTERT-RPE1 cells. hTERT-RPE1 cells were grown to 80% confluence, serum starved for 24 h to induce ciliogenesis, then fixed and stained for DYX1C1, DCDC2, acetylated α-tubulin (cilia marker), or γ-tubulin (centrosome marker). Nuclei are stained with DRAQ5. Scale bars, 20 µm, 2 µm (insets).
DISCUSSION
Here, we describe for the first time, to our knowledge, that RFX TFs directly regulate 2 of the most replicated and best-characterized DCGs: DYX1C1 and DCDC2. This regulation of DCG expression by RFX TFs occurs via evolutionarily well-conserved X-box promoter motifs, which we demonstrate by using a combination of bioinformatics and in vitro approaches. In addition, we demonstrate a novel complex regulatory pattern between the RFX TFs, which was previously only suggested by protein-binding assays (28, 43–45). RFX TFs are well known as key regulators of genes involved in cilia structure and function (26). We provide strong evidence that DYX1C1 and DCDC2 play a role in human primary cilia and are regulated by RFX TFs in a human cell model.
Our results are in agreement with an emerging body of work that points toward a role for DCGs in ciliary biology. DYX1C1, DCDC2, and KIAA0319—the 3 most replicated DCGs—have been shown to be up-regulated in ciliated tissues (42, 46). Dyx1c1 and Dcdc2, as well as Rfx2 and Rfx3, have been identified in ciliated ependymal cells of the adult mouse brain by using a single-cell RNA-sequencing approach (47). The strongest functional evidence for DYX1C1 being needed for the proper assembly of cilia was obtained by Chandrasekar et al. (20) by morpholino knockdowns of the zebrafish ortholog dyx1c1, which resulted in full ciliopathy phenotypes, including situs inversus, hydrocephalus, defects in body curvature, shortening of cilia in the Kupffer vesicle, and lack of inner dynein arm and outer dynein arm in motile cilia. Shortly thereafter, rare homozygous or compound heterozygous deleterious mutations were identified in DYX1C1 from patients with PCD (21, 22). Similarly, Dyx1c1-knockout mice also displayed a PCD phenotype (21). Despite its reported role in cilia, localization of DYX1C1 to cilia and centrosomes has so far only been achieved in overexpression cell lines (11, 13, 42). Here, we advance by clearly demonstrating that endogenous DYX1C1 localizes specifically to the base of cilia in human hTERT-RPE1 cells (Fig. 4). For DCDC2, further evidence for being a ciliary gene emerged when it was identified as a causative gene for nephronophthisis-related ciliopathy in 2 individuals (23) and for heritable hearing loss (24). These results are consistent with a reported localization to the cilium of overexpressed DCDC2 (41) and in human ciliated tissues (23). Again, here we advance by clearly demonstrating that endogenous DCDC2 localizes along the entire axoneme of cilia in human hTERT-RPE1 cells (Fig. 4). In future, experiments should also be conducted to delineate the ciliary role of DYX1C1 and DCDC2 in neuronal cells. For KIAA0319, there is some evidence from the literature that it could be involved in ciliary processes (42, 46); however, our work offers only indirect clues for such a possible ciliary role for KIAA0319 as we show that an evolutionarily well-conserved X-box promoter motif is located in its proximal promoter and its expression is up-regulated during the process of ciliogenesis.
Whereas DCGs DYX1C1, DCDC2, and KIAA0319 share demonstrated functions in neuronal migration, the role of cilia in neuronal migration and brain development is currently a matter of debate; however, intriguing evidence has emerged. For instance, a number of ciliary genes are needed for proper brain development (18, 19) and many ciliopathies lead to deficits in cognitive functions, which indicates disrupted brain development under these conditions (16). Another possibility for links between cilia and neuronal development, brain patterning, and morphogenesis is offered by the Shh and Wnt signaling pathways, where crucial pathway components with ciliary localization are involved in key signaling steps of primary cilia (48). Furthermore, both motile and nonmotile cilia are needed to setup the left/right (LR) asymmetry during embryonic development, which could have implications for brain structure and connectivity. Of interest, early studies by Galaburda et al. (49, 50) reported reduced planum temporale asymmetry in 8 postmortem brain specimens from individuals with DD. Recently, PCSK6 (proprotein convertase subtilisin/kexin type 6), a gene that is involved in LR patterning, was associated with LR relative hand skills in individuals with DD, which further indicated that the cilia-regulated process of determining LR asymmetry and DD might share the same biologic pathways (51). Rfx2 and Rfx3 have also been implicated in the regulation of LR asymmetry in the mouse (52, 53).
It is conceivable that RFX TFs exert their impact on neuronal and brain development and possibly on brain-related conditions, such as DD, directly via their target genes. Proven targets from the ciliogenic gene battery include genes TMEM138 (transmembrane protein 138) and TMEM216 (transmembrane protein 216), which are expressed in human embryonic brain and cause Joubert Syndrome when mutated (54), and the gene ALMS1 (Alström Syndrome 1), which is important for neurosensory development and causes Alström Syndrome upon mutation (55). Our study tentatively adds DD to this body of work by assigning the ciliary DCGs DYX1C1 and DCDC2 as direct targets of RFX TFs. Our work is further supported by independent findings from genome-wide approaches. By using chromatin immunoprecipitation sequencing (ChIP-seq), RFX3 was found to bind the DCGs DYX1C1, DCDC2, KIAA0319, and KIAA0319L, ciliary genes, such as Arl13b (ADP-ribosylation factor-like protein 13B), as well as other RFX genes (44). In addition, a combination of rfx2 ChIP-seq and RNA sequencing of rfx2 morpholino knockdown in Xenopus laevis identified dcdc2 in the group of differentially regulated genes (28). In future, it will be important to study how other RFX TFs not analyzed here affect the expression of DCGs and whether they occupy X-box motifs in different cell systems. Furthermore, alternative possibilities include nonciliogenic RFX TF target gene batteries that impact neuronal and brain development and thus brain-related conditions, such as DD. RFX TF–mediated regulation of target genes and cellular processes other than ciliogenesis and the adaptive immune response (56, 57) are still not well understood and, as such, are a highly speculative topic. Nonciliogenic examples of RFX TF regulation would be transforming growth factor β2, which is important for neuronal cell proliferation (58), or RFX TF targets, which are important for neuronal synapse structure and function as originally discovered for the C. elegans RFX TF DAF-19 (dauer formation gene 19) (59).
We have successfully focused on a noncoding element of the genome, the X-box promoter motif, and have used it as a tool to uncover the transcriptional regulation of DCGs and how these genes are linked to cilia. The identified X-box in the promoter of DYX1C1 is located at a transcriptional hub, which contains the DD-associated SNP-3G/A (rs3743205), and where many different TFs bind depending on cell type, state, and environmental stimuli (8, 9). In recent years, increased interest has focused on understanding the functional elements in the noncoding parts of the genome, as exemplified by the Encyclopedia of DNA Elements (60) and Functional Annotation of the Human Genome (61, 62) projects. With advances in whole-genome sequencing, this characterization will become even more important as rare mutations and variants are often detected outside protein coding regions. For example, Bae et al. (63) identified a functional X-box in the promoter region of the gene GPR56 (GPCR 56) by analyzing homozygous small deletions in individuals with brain gyrification defects, including the Broca’s area. The authors further demonstrated the binding of RFX1 and RFX3 to the X-box, thereby linking these TFs to the regulation of genes that are expressed in language-related areas of the brain. Thus, mutation screening of the noncoding genomic regions characterized here could lead to the identification of causal mutations for DD or for ciliopathies associated with these DCGs. Another approach, looking for X-boxes in already described loci for DD, could facilitate the identification of the respective candidate genes.
In summary, our work with human cells clearly demonstrates that DCGs are regulated by RFX TFs via X-box promoter motifs and are linked to the development, structure, or function of cilia. Our work is in agreement with work in other systems that describes the role of DCGs in ciliary biology. Combined, these results lend strong support to an emerging theme that assigns cilia a potentially crucial role in the etiology of DD. Whereas the link between DCGs and cilia is strongly supported by data, the next step—the link between DD and cilia—is much more tenuous and hypothetical. As brain imaging studies suggest overlapping regions of functional importance and involvement of white matter for the 3 DCGs studied here, these regions could serve as starting points for further research to connect the pathways (64). To acquire this more comprehensive understanding, future studies need to clarify which cells of the brain are crucial for the etiology of DD, what the function of cilia is for these cells, and how such results could explain the observed genetic effect on white matter.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Swedish Brain Foundation (Hjärnfonden), the Swedish Research Council, the Torsten Söderberg, Åhlén, Lars Hiertas Minne, and Sigrid Jusélius Foundations, the Swedish Foundation for Strategic Research, the Karolinska Institutet (KI) Strategic Neuroscience Program, and the Academy of Finland. This study was performed, in part, at the Live Cell Imaging Unit/Nikon Center of Excellence at the KI Department of Biosciences and Nutrition, which is supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the KI Center for Innovative Medicine, and the Jonasson donation to the School of Technology and Health, Royal Institute of Technology.
Glossary
- ChIP-seq
chromatin immunoprecipitation sequencing
- DCDC2
doublecortin domain-containing 2 gene
- DCG
dyslexia candidate gene
- DD
developmental dyslexia
- DYX1C1
dyslexia 1 candidate 1 gene
- hTERT-RPE1
human retinal pigmented epithelial cell line immortalized with hTERT
- LR
left/right
- PCD
primary ciliary dyskinesia
- RFX TF
Regulatory Factor X transcription factor
- SH-SY5Y
human neuroblastoma cell line, subline of the neuroblastoma cell line SK-N-SH
- siRNA
small interfering RNA
- SNP
single nucleotide polymorphism
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
K. Tammimies, A. Bieder, G. Lauter, D. Sugiaman-Trapman, R. Torchet, M.-E. Hokkanen, J. Burghoorn, and I. Tapia-Páez performed experiments; K. Tammimies, E. Castrén, J. Kere, I. Tapia-Páez, and P. Swoboda conceived and designed the study; K. Tammimies, A. Bieder, and P. Swoboda wrote the manuscript; and all authors critically revised the manuscript.
REFERENCES
- 1.Katusic S. K., Colligan R. C., Barbaresi W. J., Schaid D. J., Jacobsen S. J. (2001) Incidence of reading disability in a population-based birth cohort, 1976-1982, Rochester, Minn. Mayo Clin. Proc. 76, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 2.Shaywitz S. E., Shaywitz B. A., Fletcher J. M., Escobar M. D. (1990) Prevalence of reading disability in boys and girls. Results of the Connecticut Longitudinal Study. JAMA 264, 998–1002 [PubMed] [Google Scholar]
- 3.Kere J. (2014) The molecular genetics and neurobiology of developmental dyslexia as model of a complex phenotype. Biochem. Biophys. Res. Commun. 452, 236–243 [DOI] [PubMed] [Google Scholar]
- 4.Meng H., Smith S. D., Hager K., Held M., Liu J., Olson R. K., Pennington B. F., DeFries J. C., Gelernter J., O’Reilly-Pol T., Somlo S., Skudlarski P., Shaywitz S. E., Shaywitz B. A., Marchione K., Wang Y., Paramasivam M., LoTurco J. J., Page G. P., Gruen J. R. (2005) DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc. Natl. Acad. Sci. USA 102, 17053–17058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paracchini S., Thomas A., Castro S., Lai C., Paramasivam M., Wang Y., Keating B. J., Taylor J. M., Hacking D. F., Scerri T., Francks C., Richardson A. J., Wade-Martins R., Stein J. F., Knight J. C., Copp A. J., Loturco J., Monaco A. P. (2006) The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum. Mol. Genet. 15, 1659–1666 [DOI] [PubMed] [Google Scholar]
- 6.Taipale M., Kaminen N., Nopola-Hemmi J., Haltia T., Myllyluoma B., Lyytinen H., Muller K., Kaaranen M., Lindsberg P. J., Hannula-Jouppi K., Kere J. (2003) A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc. Natl. Acad. Sci. USA 100, 11553–11558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher J., Anthoni H., Dahdouh F., König I. R., Hillmer A. M., Kluck N., Manthey M., Plume E., Warnke A., Remschmidt H., Hülsmann J., Cichon S., Lindgren C. M., Propping P., Zucchelli M., Ziegler A., Peyrard-Janvid M., Schulte-Körne G., Nöthen M. M., Kere J. (2006) Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am. J. Hum. Genet. 78, 52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tammimies K., Tapia-Páez I., Rüegg J., Rosin G., Kere J., Gustafsson J. A., Nalvarte I. (2012) The rs3743205 SNP is important for the regulation of the dyslexia candidate gene DYX1C1 by estrogen receptor β and DNA methylation. Mol. Endocrinol. 26, 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tapia-Páez I., Tammimies K., Massinen S., Roy A. L., Kere J. (2008) The complex of TFII-I, PARP1, and SFPQ proteins regulates the DYX1C1 gene implicated in neuronal migration and dyslexia. FASEB J. 22, 3001–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis M. Y., Paracchini S., Scerri T. S., Prokunina-Olsson L., Knight J. C., Wade-Martins R., Coggill P., Beck S., Green E. D., Monaco A. P. (2009) A common variant associated with dyslexia reduces expression of the KIAA0319 gene. PLoS Genet. 5, e1000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Paramasivam M., Thomas A., Bai J., Kaminen-Ahola N., Kere J., Voskuil J., Rosen G. D., Galaburda A. M., Loturco J. J. (2006) DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience 143, 515–522 [DOI] [PubMed] [Google Scholar]
- 12.Rosen G. D., Bai J., Wang Y., Fiondella C. G., Threlkeld S. W., LoTurco J. J., Galaburda A. M. (2007) Disruption of neuronal migration by RNAi of Dyx1c1 results in neocortical and hippocampal malformations. Cereb. Cortex 17, 2562–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tammimies K., Vitezic M., Matsson H., Le Guyader S., Bürglin T. R., Ohman T., Strömblad S., Daub C. O., Nyman T. A., Kere J., Tapia-Páez I. (2013) Molecular networks of DYX1C1 gene show connection to neuronal migration genes and cytoskeletal proteins. Biol. Psychiatry 73, 583–590 [DOI] [PubMed] [Google Scholar]
- 14.Gerdes J. M., Davis E. E., Katsanis N. (2009) The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J. H., Gleeson J. G. (2010) The role of primary cilia in neuronal function. Neurobiol. Dis. 38, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valente E. M., Rosti R. O., Gibbs E., Gleeson J. G. (2014) Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 10, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildebrandt F., Benzing T., Katsanis N. (2011) Ciliopathies. N. Engl. J. Med. 364, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J., Higginbotham H., Li J., Nichols J., Hirt J., Ghukasyan V., Anton E. S. (2015) Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 6, 7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkisian M. R., Guadiana S. M. (2015) Influences of primary cilia on cortical morphogenesis and neuronal subtype maturation. Neuroscientist 21, 136–151 [DOI] [PubMed] [Google Scholar]
- 20.Chandrasekar G., Vesterlund L., Hultenby K., Tapia-Páez I., Kere J. (2013) The zebrafish orthologue of the dyslexia candidate gene DYX1C1 is essential for cilia growth and function. PLoS One 8, e63123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarkar A., Loges N. T., Slagle C. E., Francis R., Dougherty G. W., Tamayo J. V., Shook B., Cantino M., Schwartz D., Jahnke C., Olbrich H., Werner C., Raidt J., Pennekamp P., Abouhamed M., Hjeij R., Köhler G., Griese M., Li Y., Lemke K., Klena N., Liu X., Gabriel G., Tobita K., Jaspers M., Morgan L. C., Shapiro A. J., Letteboer S. J., Mans D. A., Carson J. L., Leigh M. W., Wolf W. E., Chen S., Lucas J. S., Onoufriadis A., Plagnol V., Schmidts M., Boldt K., Roepman R., Zariwala M. A., Lo C. W., Mitchison H. M., Knowles M. R., Burdine R. D., Loturco J. J., Omran H.; UK10K (2013) DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 45, 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casey J. P., McGettigan P. A., Healy F., Hogg C., Reynolds A., Kennedy B. N., Ennis S., Slattery D., Lynch S. A. (2015) Unexpected genetic heterogeneity for primary ciliary dyskinesia in the Irish Traveller population. Eur. J. Hum. Genet. 23, 210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schueler M., Braun D. A., Chandrasekar G., Gee H. Y., Klasson T. D., Halbritter J., Bieder A., Porath J. D., Airik R., Zhou W., LoTurco J. J., Che A., Otto E. A., Böckenhauer D., Sebire N. J., Honzik T., Harris P. C., Koon S. J., Gunay-Aygun M., Saunier S., Zerres K., Bruechle N. O., Drenth J. P., Pelletier L., Tapia-Páez I., Lifton R. P., Giles R. H., Kere J., Hildebrandt F. (2015) DCDC2 mutations cause a renal-hepatic ciliopathy by disrupting Wnt signaling. Am. J. Hum. Genet. 96, 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grati M., Chakchouk I., Ma Q., Bensaid M., Desmidt A., Turki N., Yan D., Baanannou A., Mittal R., Driss N., Blanton S., Farooq A., Lu Z., Liu X. Z., Masmoudi S. (2015) A missense mutation in DCDC2 causes human recessive deafness DFNB66, likely by interfering with sensory hair cell and supporting cell cilia length regulation. Hum. Mol. Genet. 24, 2482–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas J., Morle L., Soulavie F., Laurencon A., Sagnol S., Durand B. (2010) Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol. Cell 102, 499–513 [DOI] [PubMed] [Google Scholar]
- 26.Choksi S. P., Lauter G., Swoboda P., Roy S. (2014) Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141, 1427–1441 [DOI] [PubMed] [Google Scholar]
- 27.Swoboda P., Adler H. T., Thomas J. H. (2000) The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5, 411–421 [DOI] [PubMed] [Google Scholar]
- 28.Chung M. I., Kwon T., Tu F., Brooks E. R., Gupta R., Meyer M., Baker J. C., Marcotte E. M., Wallingford J. B. (2014) Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife 3, e01439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efimenko E., Bubb K., Mak H. Y., Holzman T., Leroux M. R., Ruvkun G., Thomas J. H., Swoboda P. (2005) Analysis of xbx genes in C. elegans. Development 132, 1923–1934 [DOI] [PubMed] [Google Scholar]
- 30.Laurençon A., Dubruille R., Efimenko E., Grenier G., Bissett R., Cortier E., Rolland V., Swoboda P., Durand B. (2007) Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 8, R195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piasecki B. P., Burghoorn J., Swoboda P. (2010) Regulatory factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc. Natl. Acad. Sci. USA 107, 12969–12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aftab S., Semenec L., Chu J. S., Chen N. (2008) Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol. Biol. 8, 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashique A. M., Choe Y., Karlen M., May S. R., Phamluong K., Solloway M. J., Ericson J., Peterson A. S. (2009) The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Sci. Signal. 2, ra70 [DOI] [PubMed] [Google Scholar]
- 34.Henriksson J., Piasecki B. P., Lend K., Bürglin T. R., Swoboda P. (2013) Finding ciliary genes: a computational approach. Methods Enzymol. 525, 327–350 [DOI] [PubMed] [Google Scholar]
- 35.Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L. W., Richards S., Weinstock G. M., Wilson R. K., Gibbs R. A., Kent W. J., Miller W., Haussler D. (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burghoorn J., Piasecki B. P., Crona F., Phirke P., Jeppsson K. E., Swoboda P. (2012) The in vivo dissection of direct RFX-target gene promoters in C. elegans reveals a novel cis-regulatory element, the C-box. Dev. Biol. 368, 415–426 [DOI] [PubMed] [Google Scholar]
- 37.Gajiwala K. S., Chen H., Cornille F., Roques B. P., Reith W., Mach B., Burley S. K. (2000) Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403, 916–921 [DOI] [PubMed] [Google Scholar]
- 38.Hsu Y. C., Kao C. Y., Chung Y. F., Chen M. S., Chiu I. M. (2012) Ciliogenic RFX transcription factors regulate FGF1 gene promoter. J. Cell. Biochem. 113, 2511–2522 [DOI] [PubMed] [Google Scholar]
- 39.Wolfe S. A., van Wert J., Grimes S. R. (2006) Transcription factor RFX2 is abundant in rat testis and enriched in nuclei of primary spermatocytes where it appears to be required for transcription of the testis-specific histone H1t gene. J. Cell. Biochem. 99, 735–746 [DOI] [PubMed] [Google Scholar]
- 40.Wolfe S. A., Vanwert J. M., Grimes S. R. (2008) Transcription factor RFX4 binding to the testis-specific histone H1t promoter in spermatocytes may be important for regulation of H1t gene transcription during spermatogenesis. J. Cell. Biochem. 105, 61–69 [DOI] [PubMed] [Google Scholar]
- 41.Massinen S., Hokkanen M. E., Matsson H., Tammimies K., Tapia-Páez I., Dahlström-Heuser V., Kuja-Panula J., Burghoorn J., Jeppsson K. E., Swoboda P., Peyrard-Janvid M., Toftgård R., Castrén E., Kere J. (2011) Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PLoS One 6, e20580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoh R. A., Stowe T. R., Turk E., Stearns T. (2012) Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One 7, e52166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung M. I., Peyrot S. M., LeBoeuf S., Park T. J., McGary K. L., Marcotte E. M., Wallingford J. B. (2012) RFX2 is broadly required for ciliogenesis during vertebrate development. Dev. Biol. 363, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jolma A., Kivioja T., Toivonen J., Cheng L., Wei G., Enge M., Taipale M., Vaquerizas J. M., Yan J., Sillanpää M. J., Bonke M., Palin K., Talukder S., Hughes T. R., Luscombe N. M., Ukkonen E., Taipale J. (2010) Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20, 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lubelsky Y., Reuven N., Shaul Y. (2005) Autorepression of rfx1 gene expression: functional conservation from yeast to humans in response to DNA replication arrest. Mol. Cell. Biol. 25, 10665–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivliev A. E., ’t Hoen P. A., van Roon-Mom W. M., Peters D. J., Sergeeva M. G. (2012) Exploring the transcriptome of ciliated cells using in silico dissection of human tissues. PLoS One 7, e35618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeisel A., Muñoz-Manchado A. B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., Rolny C., Castelo-Branco G., Hjerling-Leffler J., Linnarsson S. (2015) Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 [DOI] [PubMed] [Google Scholar]
- 48.Abdelhamed Z. A., Wheway G., Szymanska K., Natarajan S., Toomes C., Inglehearn C., Johnson C. A. (2013) Variable expressivity of ciliopathy neurological phenotypes that encompass Meckel-Gruber syndrome and Joubert syndrome is caused by complex de-regulated ciliogenesis, Shh and Wnt signalling defects. Hum. Mol. Genet. 22, 1358–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galaburda A. M., Kemper T. L. (1979) Cytoarchitectonic abnormalities in developmental dyslexia: a case study. Ann. Neurol. 6, 94–100 [DOI] [PubMed] [Google Scholar]
- 50.Galaburda A. M., Sherman G. F., Rosen G. D., Aboitiz F., Geschwind N. (1985) Developmental dyslexia: four consecutive patients with cortical anomalies. Ann. Neurol. 18, 222–233 [DOI] [PubMed] [Google Scholar]
- 51.Brandler W. M., Paracchini S. (2014) The genetic relationship between handedness and neurodevelopmental disorders. Trends Mol. Med. 20, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnafe E., Touka M., AitLounis A., Baas D., Barras E., Ucla C., Moreau A., Flamant F., Dubruille R., Couble P., Collignon J., Durand B., Reith W. (2004) The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol. Cell. Biol. 24, 4417–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bisgrove B. W., Makova S., Yost H. J., Brueckner M. (2012) RFX2 is essential in the ciliated organ of asymmetry and an RFX2 transgene identifies a population of ciliated cells sufficient for fluid flow. Dev. Biol. 363, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J. H., Silhavy J. L., Lee J. E., Al-Gazali L., Thomas S., Davis E. E., Bielas S. L., Hill K. J., Iannicelli M., Brancati F., Gabriel S. B., Russ C., Logan C. V., Sharif S. M., Bennett C. P., Abe M., Hildebrandt F., Diplas B. H., Attié-Bitach T., Katsanis N., Rajab A., Koul R., Sztriha L., Waters E. R., Ferro-Novick S., Woods C. G., Johnson C. A., Valente E. M., Zaki M. S., Gleeson J. G. (2012) Evolutionarily assembled cis-regulatory module at a human ciliopathy locus. Science 335, 966–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purvis T. L., Hearn T., Spalluto C., Knorz V. J., Hanley K. P., Sanchez-Elsner T., Hanley N. A., Wilson D. I. (2010) Transcriptional regulation of the Alström syndrome gene ALMS1 by members of the RFX family and Sp1. Gene 460, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devaiah B. N., Singer D. S. (2013) CIITA and its dual roles in MHC gene transcription. Front. Immunol. 4, 476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reith W., Mach B. (2001) The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19, 331–373 [DOI] [PubMed] [Google Scholar]
- 58.Feng C., Zuo Z. (2012) Regulatory factor X1-induced down-regulation of transforming growth factor β2 transcription in human neuroblastoma cells. J. Biol. Chem. 287, 22730–22739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Senti G., Swoboda P. (2008) Distinct isoforms of the RFX transcription factor DAF-19 regulate ciliogenesis and maintenance of synaptic activity. Mol. Biol. Cell 19, 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kellis M., Wold B., Snyder M. P., Bernstein B. E., Kundaje A., Marinov G. K., Ward L. D., Birney E., Crawford G. E., Dekker J., Dunham I., Elnitski L. L., Farnham P. J., Feingold E. A., Gerstein M., Giddings M. C., Gilbert D. M., Gingeras T. R., Green E. D., Guigo R., Hubbard T., Kent J., Lieb J. D., Myers R. M., Pazin M. J., Ren B., Stamatoyannopoulos J. A., Weng Z., White K. P., Hardison R. C. (2014) Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA 111, 6131–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., Ntini E., Arner E., Valen E., Li K., Schwarzfischer L., Glatz D., Raithel J., Lilje B., Rapin N., Bagger F. O., Jørgensen M., Andersen P. R., Bertin N., Rackham O., Burroughs A. M., Baillie J. K., Ishizu Y., Shimizu Y., Furuhata E., Maeda S., Negishi Y., Mungall C. J., Meehan T. F., Lassmann T., Itoh M., Kawaji H., Kondo N., Kawai J., Lennartsson A., Daub C. O., Heutink P., Hume D. A., Jensen T. H., Suzuki H., Hayashizaki Y., Müller F., Forrest A. R., Carninci P., Rehli M., Sandelin A.; FANTOM Consortium (2014) An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.FANTOM Consortium and the RIKEN PMI and CLST (DGT) (2014) A promoter-level mammalian expression atlas. Nature 507, 462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bae B. I., Tietjen I., Atabay K. D., Evrony G. D., Johnson M. B., Asare E., Wang P. P., Murayama A. Y., Im K., Lisgo S. N., Overman L., Šestan N., Chang B. S., Barkovich A. J., Grant P. E., Topçu M., Politsky J., Okano H., Piao X., Walsh C. A. (2014) Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 343, 764–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darki F., Peyrard-Janvid M., Matsson H., Kere J., Klingberg T. (2012) Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo-parietal white matter structure. Biol. Psychiatry 72, 671–676 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.