Abstract
A recent study reported that Acinetobacter baumannii could induce autophagy, but the recognition and clearance mechanism of intracytosolic A. baumannii in the autophagic process and the molecular mechanism of autophagy induced by the pathogen remains unknown. In this study, we first demonstrated that invading A. baumannii induced a complete, ubiquitin-mediated autophagic response that is dependent upon septins SEPT2 and SEPT9 in mammalian cells. We also demonstrated that autophagy induced by A. baumannii was Beclin-1 dependent via the AMPK/ERK/mammalian target of rapamycin pathway. Of interest, we found that the isochorismatase mutant strain had significantly decreased siderophore-mediated ferric iron acquisition ability and had a reduced the ability to induce autophagy. We verified that isochorismatase was required for the recognition of intracytosolic A. baumannii mediated by septin cages, ubiquitinated proteins, and ubiquitin-binding adaptor proteins p62 and NDP52 in autophagic response. We also confirmed that isochorismatase was required for the clearance of invading A. baumannii by autophagy in vitro and in the mouse model of infection. Together, these findings provide insight into the distinctive recognition and clearance of intracytosolic A. baumannii by autophagy in host cells, and that isochorismatase plays a critical role in the A. baumannii–induced autophagic process.—Wang, Y., Zhang, K., Shi, X., Wang, C., Wang, F., Fan, J., Shen, F., Xu, J., Bao, W., Liu, M., Yu, L. Critical role of bacterial isochorismatase in the autophagic process induced by Acinetobacter baumannii in mammalian cells.
Keywords: bacteria clearance, septin, pathway
Acinetobacter baumannii is a gram-negative, opportunistic pathogen. A. baumannii infection could lead to a wide range of infectious diseases with high mortality rates. Currently, A. baumannii has become increasingly important because of its strong survival ability in the medical environment and the increased spread of multiple antibiotic-resistant strains (1). A. baumannii was previously regarded as an extracellular bacteria, but several recent studies have shown that A. baumannii could attach to and invade several mammalian cell lines, and that it could also survive in infected host cells (2, 3). Thus, an intriguing topic of study is whether the host innate immunity adopted the same defense mechanism against intracellular A. baumannii as with other traditional intracellular pathogenic bacteria.
Autophagy, a cellular degradative pathway, plays a key role during starvation conditions and in protection of the cytosol from bacterial colonization (4, 5). Both the classic pathways of autophagy, Atg7–Atg4–Atg8 (Atg8 is also known as LC3 in mammals) and Atg12–Atg7–Atg5, are Atg6 dependent (Atg6 is also known as Beclin-1 in mammals) (6). The Akt/mammalian target of rapamycin (mTOR)/p70S6K pathway regulates the autophagy of ATP and amino acids, whereas the Beclin-1/Atg7/Atg8 and MEK/ERK pathways have been demonstrated to regulate autophagy induced by some pathogen infections (7, 8). Recent reports also demonstrate that AMPK and mTOR coordinate mammalian autophagy initiation (9). Selective autophagy is a lysosome-dependent pathway by which large cytosolic components are selectively sequestered and degraded, and substrate selectivity is conferred by ubiquitination and recruitment of ubiquitin-binding receptors (e.g., NDP52 and p62) (10–13). The different adaptor proteins are matched to various intracytosolic bacteria and could lead to diverse autophagy pathways and outcomes (14). Previous studies have demonstrated that bacterial virulence factor, such as α-hemolysin, is required for the activation of the autophagic pathway in host cells (15). A recent study reported that out membrane protein (Omp) 33–36 of A. baumannii could induce autophagy (16); however, it is still unclear how host cells recognize and clear intracellular A. baumannii through autophagy and what the possible mechanism of autophagy may be.
Active Fe3+ uptake from the environment is an important process for bacterial growth, and proliferation and is achieved by siderophore-mediated ferric iron acquisition (17, 18). It was demonstrated that isochorismatase-like hydrolases were necessary for bacterial siderophore-mediated ferric iron acquisition, as the siderophore is hydrolyzed from isochorismate with isochorismatase (19). A. baumannii could encode an isochorismatase superfamily hydrolase (20). A recent report has demonstrated that iron starvation could induce autophagy in mammal cells (21), but the role of isochorismatase in the innate immune response to bacterial infection was unclear.
In this experiment, we show that A. baumannii isochorismatase is critical for siderophore-mediated ferric iron acquisition from the environment, and isochorismatase is required for activation of autophagic response, for recognition of intracytosolic A. baumannii mediated by septin cages and adaptor proteins, and for clearance of invading A. baumannii by autophagy in vitro and in vivo, and we clarified the molecular mechanism of autophagy caused by A. baumannii.
MATERIALS AND METHODS
Antibodies, chemicals, plasmids, and strains
Unless stated otherwise, all reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). See Table 1 for a list of plasmids and strains used in our study. All strains were grown in tryptic soy broth medium. Antibodies used in our study are described in the Supplemental Data.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| A. baumannii | ||
| ATCC 19606T | Clinical isolate-type strain | ATCC (Manassas, VA, USA) |
| 19606T-GFP | ATCC 19606T derivative producing GFP encoded by a gene on pMU125; Ampr | This study |
| 98-37-09 | Clinical isolate | Ref. 19 |
| ACJ6 | EZ-Tn5::A1S_3278 derivative of 98-37-09 | Ref. 19 |
| ΔACJ6 | Complementation strain | This study |
| Plasmids | ||
| pWH1266 | A. baumannii-E. coli shuttle vector | Ref. 50 |
| pMU125 | pWH1266 harboring GFP; Ampr | Ref. 50 |
| pMUiso | pWH1266 harboring isochorismatase; Ampr | This study |
| pmRFP-LC3 | Mammalian expression of rat LC3 fused to mRFP | Ref. 51; Addgene (Cambridge, MA, USA) |
| ptfLC3 | Mammalian expression of rat LC3 fused to mRFP and EGFP | Ref. 51; Addgene |
| pEGFP-LC3 | Mammalian expression of rat LC3 fused to EGFP | Ref. 52; Addgene |
| shBeclin-1 | pGPH1/Neo; target: 5′-GGAGTCTCTGACAGACAAATC-3′; Neor | GenePharma (Shanghai, China) |
| shAtg7 | pGPH1/Neo; target: 5′-GCTTCCAGAAATGGCATTTAG-3′; Neor | GenePharma |
| shSEPT2 | pGPH1/Neo; target: 5′-AAGGTGAATATTGTGCCTGTC-3′; Neor | GenePharma |
| shSEPT9 | pGPH1/Neo; target: 5′-GGCAGCCCATCATGAAGTTCA-3′; Neor | GenePharma |
| shRNA negative control | pGPH1/Neo; target: 5′-GTTCTCCGAACGTGTCACGT-3′; Neor | GenePharma |
ATCC, American Type Culture Collection; EGFP, enhanced green fluorescent protein; mRFP, monomeric red fluorescent protein.
Construction of the isochorismatase complementation strain ΔACJ6
The assay was performed as previously reported (22). The isochorismatase parental allele was PCR amplified by using total strain 98-37-09 DNA as a template, Pfu DNA polymerase, and primers (forward: 5′-CGGGATCCATGAAACAAGCACTATTAGTTATC-3′; and reverse: 5′-CGCCTAGGTTAAGATTTTGCTAGAAAGTCAG-3′), both of which included BamHI restriction sites. Amplicon was ligated into the BamHI site of A. baumannii-Escherichia coli shuttle vector pWH1266 and transformed into E. coli cells. Plasmid DNA from an ampicillin-resistant, tetracycline-sensitive transformant, named pMUiso, was conjugated to the isochorismatase mutant ACJ6 cells by using triparental mating with E. coli DH5α that harbored pRK2073 as a helper. A. baumannii transconjugants that harbored complementing plasmid pMUiso were recovered by plating onto Simmons citrate agar that contained kanamycin (40 μg/ml) and ampicillin (500 μg/ml). The presence and stability of pMUiso in the complemented strain ΔACJ6 were confirmed by restriction analysis and DNA sequencing of plasmid DNA isolated from cells cultured in LB broth that contained 200 μg/ml ampicillin.
Quantitative real-time PCR
Total RNA was isolated from bacteria by using GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) and reverse transcribed by using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany). RNA concentration was determined with Nanodrop (Nanodrop Technologies, Wilmington, DE, USA). Quantitative real-time PCR was performed as previously reported (16). The following primers were used: Omp33–36 expression gene mapA: forward: 5′-CAAGATGCTGTAACTGCTCGTACT-3′; reverse: 5′-CAATAGCCATGTTAGTGCCATC-3′; isochorismatase gene: forward: 5′-ACTCGTGATCACAGGCATGA-3′; reverse: 5′-ACTCGTGATCACAGGCATGA-3′; and 16sRNA, which was used to normalize other gene expression: forward: 5′-CAGCTCGTGTCGTGAGATGT-3′; reverse: 5′-CGTAAGGGCCATGATGACTT-3′.
Detection of the iron metabolism indexes
Siderophore production in strains was analyzed as previously described (23). The siderophore chelate ring of the strain 98-37-09, ACJ6 strains, and the isochorismatase mutant reverant ΔACJ6 were observed and the radius of each chelate ring was detected. Presence of catechol in supernatants of bacterial cultures grown for 48 h was detected as previously described (24). Phenolic compounds were measured in the solution by using the Arnow assay (25). The number of siderophores in solution was also determined and calculated by measuring the optical density (OD) at 630 nm with a microtiter ELISA reader (Molecular Devices, Foster City, CA, USA) (23).
Cells and cell cultures
HeLa, THP-1, and MH-S cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and maintained in RPMI 1640 medium that was supplemented with 10% fetal bovine serum. All cell culture reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Cells were incubated at 37°C with 5% CO2. THP-1 cells were differentiated into macrophage-like cells with 10 ng/ml phorbol myristate acetate for 24 h.
Mice
Male BALB/c mice (age 5–8 wk) were maintained under specific pathogen-free conditions in individual ventilated cages. All experiments were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China and approved by the Animal Welfare and Research Ethics Committee at Jilin University (IZ-2009-008). Protocols were reviewed and approved by the committee.
Infection experiments in vitro
Bacteria were grown overnight in tryptic soy broth medium at 37°C with vigorous shaking. The next day, bacteria were pelleted by centrifugation at 8000 g and resuspended in RPMI 1640 medium. Thereafter, OD600 was measured, and the density was adjusted to an OD of 0.6 (1 × 108 cells/ml). Cells were cultured in serum-free and antibiotic-free RPMI 1640 medium for 12 h before infection (10).
In vivo A. baumannii infections
A. baumannii infection mouse model and detection of the autophagic response were as previously described (26, 27). Three groups of 3 mice each were i.p. infected with 0.5 ml of a bacterial suspension that contained 3.5 × 104 colony-forming units (CFU) of A. baumannii. After 24 h, mice were i.p. injected with rapamycin at 1 mg/kg/d or 3-methyladenine (3-MA) at 24 mg/kg/d for 4 d. Stock solutions were diluted in PBS. A control group of 3 mice and rapamycin positive control group of 3 mice were also used. All mice were euthanized on d 5. Peritoneal macrophages were isolated and spleens were collected, then homogenized in saline. Proteins were extracted to detect LC3 expression, and CFUs were determined by plating serial dilutions of cells on trypticase soy agar plates. The strain used in this experiment includes the wild strain 98-37-09, the isochorismatase mutant ACJ6, and the complementation strain ΔACJ6.
Transmission electron microscopy
Cell cultures were washed twice with PBS and pelleted at the bottom of 1.5-ml Eppendorf tubes by centrifugation at 1000 rpm for 5 min. Samples were performed as previously described (10). Finally, autophagosome-like vesicles were examined under a transmission electron microscope (H-7650; Hitachi, Tokyo, Japan).
Western blotting
Western blots were performed as previously described (10). Protein bands were detected by using the ECL Plus kit (P0018; Beyotime Biotechnology, Jiangsu, China), and images were obtained by using a CanoScan LiDE 100 scanner (Canon, Tokyo, Japan). Protein blots were analyzed by using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Confocal microscopy and immunofluorescence staining
We performed immunofluorescence staining as previously described (14). Images were captured by using an Olympus FV1000 confocal laser scanning microscope (Olympus, Tokyo, Japan) with a ×60 objective lens. Image analyses and export were performed by using a FluoView (v1.7.3.0; Olympus) (12).
Transfection and short hairpin RNA–mediated knockdown of specific protein expression in HeLa cells
Transfection and knockdown was performed as previously described (10). In brief, HeLa cells were grown to 60% confluence in glass-bottomed cell culture dishes and 6-well plates and were transfected with plasmids by using the X-tremeGene HP DNA transfection regent (Roche, Indianapolis, IN, USA) according manufacturer instructions. Gene knockdown efficiency was evaluated by Western blotting and observed by a confocal laser-scanning microscope. A nontargeting vector was used as negative control.
Analysis of bacterial growth within cells
The internalization and phagocytosis of bacteria within cells and the ability of macrophages to remove bacteria were determined by using the gentamicin protection assay. Number of intracellular bacteria was also determined by CFU quantification by using a previously described assay (28–31).
Statistical analysis
Results are expressed as means ± sd. Data were analyzed using a 2-tailed Student’s t test. Differences were considered significant when P < 0.05. All experiments were performed at least three times.
RESULTS
Isochorismatase is critical for siderophore-mediated ferric iron acquisition in A. baumannii
In this study, we first investigated the relationship between isochorismatase and siderophore-mediated ferric iron acquisition ability in the A. baumannii strain. Some indexes related to iron metabolism were detected by comparing differences in the iron metabolism among the isochorismatase mutant A. baumannii strain ACJ6, its wild-type strain 98-37-09, and the complementation strain ΔACJ6. Chrome azurol S assays showed that both strain 98-37-09 and ΔACJ6 could form the obvious siderophore chelate ring, whereas the radius of the chelate ring of the ACJ6 strain was significantly smaller than that of strain 98-37-09 and ΔACJ6 (Supplemental Fig. S1A, B; P < 0.01). Similarly, the presence of catechol in supernatants of bacterial cultures and arnow colorimetric assay showed that the production of catechol in the supernatants of the ACJ6 cultures was significantly less than that of strain 98-37-09 and ΔACJ6 (Supplemental Fig. S1C, D). These results indicated that the ACJ6 isochorismatase mutant strain produced much fewer siderophores than its parent strain (P < 0.05).
Isochorismatase is required for invading A. baumannii to induce autophagic response in vitro and in vivo
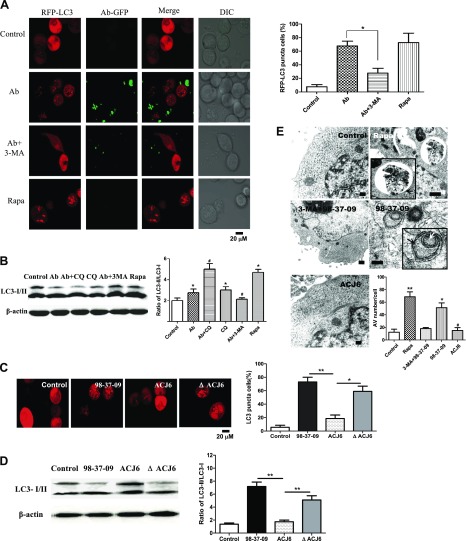
Next, we verified that A. baumannii infection induced an autophagic response in vitro. The pmRFP-LC3 plasmid was used to transfect HeLa cells, and confocal microscopy images were semiquantitatively analyzed to determine the portion of significant LC3 puncta in the cells. It was observed that the LC3 puncta in the cells were significantly increased in the A. baumannii–infected and rapamycin (an autophagy activator)-treated HeLa cells (Fig. 1A), whereas the number of puncta in the 3-MA (an autophagy inhibitor)-pretreated, A. baumannii–infected cells was lower than that of infected cells. Results demonstrated that A. baumannii could induce autophagy in HeLa cells, which was inhibited by pretreatment with 3-MA. To further prove the A. baumannii infection-induced autophagic response, we analyzed the changes in LC3 expression level in HeLa cells after A. baumannii ATCC 19606T infection. Results showed that the ratio of LC3-II/LC3-I in the infected group was much higher than that in the control group (P < 0.05; Fig. 1B), and the lysosomotropic agent chloroquine-treated cells showed increased expression of lipidated LC3-II after A. baumannii ATCC 19606T infection (P < 0.05); the accumulation of LC3-II in the A. baumannii–infected host cells did not result from inhibiting degradation. We also found that there were distinct levels of LC3 expression among the autophagic responses induced by different A. baumannii clinical strains, including ATCC 19606T, S1, J1, M2, and 98-37-09, and strain 98-37-09 exhibited the most significantly increased ratio of LC3-II/I (P < 0.05; Supplemental Fig. S2A). Our data also proved that the A. baumannii–induced autophagy occurred in a dose- and time-dependent manner by detecting the LC3 expression level with Western blotting; the ratio of LC3-II/I increased significantly at 2 h postinfection, the optimal infection time is 3 h, and the optimal multiplicity of infection is 10:1 (Supplemental Fig. S2B, C).
Figure 1.
Isochorismatase might play an important role in autophagy induced by A. baumannii. A) HeLa cells were transfected with the pmRFP-LC3 plasmid for 24 h and then infected with A. baumannii strain ATCC19606T-GFP for 2 h [multiplicity of infection (MOI) = 10:1]. Before infection, cells were pretreated with rapamycin (Rapa; 5 μM, 12 h) and 3-MA (3 μM, 3 h). The puncta in each cell were counted, and cells with >10 puncta were considered to be LC3-RFP puncta cells. Values are from 100 cells/sample. B) Western blots of LC3 in ATCC19606T-infected HeLa cells. HeLa cells were infected with ATCC19606T for 2 h (MOI = 10:1). Before infection, HeLa cells were pretreated with rapamycin, chloroquine (CQ; 50 μM, 3 h), or 3-MA. *P < 0.05 compared with control group; #P < 0.05 compared with the Ab group. C) HeLa cells were transfected with the pmRFP-LC3 plasmid for 24 h. Then, cells were infected with the A. baumannii strain 98-37-09, ACJ6, or ΔACJ6 for 2 h and observed by confocal microscopy. The percentage of LC3-RFP puncta cells was calculated. *P < 0.05; **P < 0.01. D) HeLa cells were infected with strain 98-37-09, ACJ6, or ΔACJ6 for 3 h, and Western blotting for LC3 was performed. **P < 0.01. E) Autophagosome formation after infection with A. baumannii in HeLa cells was examined by transmission electron microscopy. HeLa cells were infected with strain 98-37-09 or ACJ6 (MOI = 10:1) for 2 h. Before infection, cells were treated with rapamycin or 3-MA. Black arrows indicate the autophagosomes with double membranes, white arrow indicates the bacteria inside the autophagic vesicles (AVs). Scale bars, 1 μm. The number of AVs in each cell from 20 cells in each sample was determined. *P < 0.05 and **P < 0.01 compared with control group; #P < 0.05 compared with wild-type strain 98-37-09. Data are representative of 3 experiments with similar results. DIC, differential interference contrast.
Because isochorismatase was identified to be involved in iron metabolism, and iron starvation could induce the autophagic response (32), we compared autophagy levels in HeLa cells infected with the A. baumannii isochorismatase mutant strain ACJ6, its wild-type strain 98-37-09, and the complementation strain ΔACJ6. We observed that strain ACJ6 did not induce a detectable autophagic response, whereas strain 98-37-09 and ΔACJ6 induced a significant autophagic response, as determined by the number of LC3 puncta (Fig. 1C) or LC3 expression analysis (Fig. 1D). Together, these data suggested that isochorismatase might be necessary for the autophagy induced by A. baumannii in vitro.
To further investigate the autophagic response caused by A. baumannii in vivo and the related effect of isochorismatase, we detected LC3 expression in A. baumannii–infected mouse peritoneal macrophages or in the spleen of a mouse model of infection. The ratio of LC3-II/I was significantly increased in A. baumannii–infected mouse peritoneal macrophages in vitro compared with control group (P < 0.05) and was higher than that of the 3-MA–treated group (P < 0.05; Supplemental Fig. S2D). Similar results were found in the mouse model of infection. The ratio of LC3-II/I in spleens of the A. baumannii–infected group was much higher than in the control group (Supplemental Fig. S2E). These data showed that isochorismatase was also necessary for autophagy induced by A. baumannii in primary macrophages.
To investigate whether Omp33–36 of A. baumannii could induce autophagic response, we detected the expression level of Omp33–36 and isochorismatase gene in strains 98-37-09, ACJ6, and ΔACJ6. Results showed that no significant difference of Omp33–36 expression was observed among these strains (Supplemental Fig. S1E), whereas expression levels of the isochorismatase gene in strains 98-37-09 and ΔACJ6 were much higher than in the mutant ACJ6 (Supplemental Fig. S1F). It indicated that the difference in autophagic response between 98-37-09 and ACJ6 was only related to isochorismatase.
We also detected the existence of newly formed autophagosomes in HeLa cells. The number of autophagic vesicles with double membranes was significantly increased in the 98-37-09–infected HeLa group compared with control group, and the bacteria was observed inside the vesicles clearly (Fig. 1E; P < 0.05). This phenomenon was similar to that found in cells treated with positive autophagy inducer rapamycin, whereas 3-MA decreased the formation of autophagosomes in A. baumannii–infected cells. Moreover, the isochorismatase mutant strain ACJ6 induced fewer autophagosomes and autophagic vesicles (P < 0.05).
Together, the results of the transmission electron microscopy organelle analysis reconfirmed that isochorismatase is required for autophagosome formation caused by A. baumannii within host cells.
A. baumannii induces a complete autophagic response
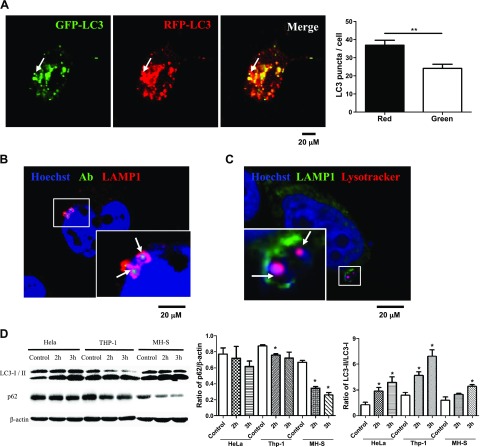
Autophagy can clear some pathogens via autolysosome degradation (10) and can also help some pathogens survive in the infected cells when incomplete autophagy occurs (33). To discover whether A. baumannii induced a complete autophagy, we used a mRFP-GFP tandem fluorescently tagged LC3 plasmid (ptfLC3), which contains both red and green fluorescent proteins tagged to the LC3 protein. When the autophagosomes form in the cells, red and green puncta could be observed at the same point (10); however, when autolysosomes form, the GFP signal is lost, but RFP-LC3 continues to be expressed and represents the LC3 puncta. After infection with strain 98-37-09, we found increased numbers of red and green LC3 puncta, and there were more red puncta than green puncta in the HeLa cells (Fig. 2A). To further corroborate whether intracytosolic A. baumannii induced the fusion of autophagosomes with lysosomes, we incubated FITC-labeled, A. baumannii–infected HeLa cells with Hoechst 33342 and the antibody to lysosomal-associated membrane protein-1 (LAMP-1), a marker of late endosomes/lysosomes that to colocalizes with bacteria during autolysosome maturation. Results showed that FITC-labeled A. baumannii was surrounded by LAMP-1 (Fig. 2B). To determine whether A. baumannii resides in an acidic vesicle, we stained HeLa cells with antibodies to LAMP-1 and LysoTracker DND 99 (Fig. 2C), a weak lysosomotrophic base that accumulates and fluoresces within acidic vesicles. Results suggested that at 2 h postinfection, a majority of A. baumannii are inside LAMP-1–surrounded acidic vesicles. Our data indicated that the autophagosomes efficiently fuse with the lysosomes in A. baumannii–infected cells (34, 35). Previous studies have demonstrated that decreased p62 expression was also an important index for determining autophagic degradation (36); therefore, we simultaneously detected the LC3 and p62 expression levels in 3 cell lines, HeLa, MH-S, and THP-1 cells (Fig. 2D). The level of p62 expression was decreased in all 3 cell lines (P < 0.05), whereas that of LC3-II was increased (P < 0.05). This demonstrated that the A. baumannii–induced autophagy was a common response in mammalian cells. Together, these data confirmed that intracellular A. baumannii could induce a complete autophagic response and promote autophagic degradation in mammalian cells.
Figure 2.
A. baumannii infection up-regulated autophagic degradation. A) HeLa cells were transfected with ptfLC3 plasmid for 24 h, infected with strain 98-37-09 for 2 h [multiplicity of infection (MOI) = 10:1], and observed by using confocal microscopy. Arrows indicate the LC3 puncta, which could only be detected in the RFP channel. Red and green LC3 puncta were counted in 50 LC3 puncta cells respectively. B) HeLa cells were infected with FITC-labeled strain 98-37-09 for 2 h (MOI = 10:1) and were then immunostained for LAMP-1 and analyzed by confocal microscopy. Arrows indicate the bacteria inside the autophagic vesicles (AVs) marked with LAMP-1. C) HeLa cells were infected with strain 98-37-09 for 2 h (MOI = 10:1), incubated with 50 nM of LysoTracker for 1 h at 37°C, and then immunostained for LAMP-1 and analyzed by confocal microscopy. Arrows indicate the acidic LysoTracker and bacteria inside the AV marked with LAMP-1. D) HeLa, THP-1, and MH-S cells were infected with strain 98-37-09 for 2 and 3 h. Then, Western blotting for LC3 and p62 was performed and ratios of LC3-II/I and p62/β-actin were calculated. Data are representative of 3 experiments with similar results. *P < 0.05 compared with control group from the same cell line; **P < 0.01.
Isochorismatase is required for the recognition of intracytosolic A. baumannii mediated by septin cages, ubiquitinated proteins, and ubiquitin-binding adaptor proteins in autophagic response
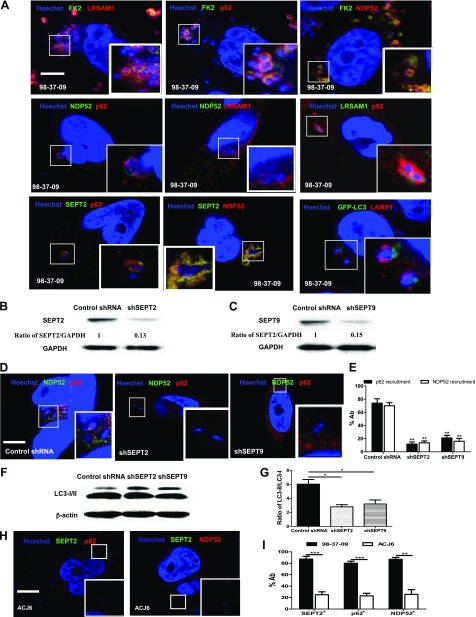
To investigate whether the E3 ubiquitin ligase, leucine rich repeat and sterile alpha motif containing 1 (LRSAM1), and the ubiquitin-binding adaptor proteins, p62 and NDP52, are associated with the recognition of intracytosolic A. baumannii during autophagy, we used immunofluorescence staining in the A. baumannii–infected HeLa cells. First, colocalization between the internalized bacteria and the endogenous LRSAM1, NDP52, p62, or ubiquitin proteins in HeLa cells was analyzed. The kinetics showed that peak LRSAM1 localization to the bacteria (60 min) occurred before peak bacterial ubiquitin, P62, and NDP52 colocalization (90 min; Supplemental Fig. S3A). Then, we observed colocalization between different adaptor proteins, the LC3 puncta, and the intracellular bacteria (Fig. 3A). Further analysis showed that at 75 min postinfection, 84.00 ± 6.08% of ubiquitinated proteins (FK2 mAb to ubiqutinated proteins) in the bacteria colocalized with LRSAM1, and 81.67 ± 7.54% of LRSAM1+ bacteria were colocalized with ubiquitinated proteins (Supplemental Fig. S3B). We also found that 80.00 ± 6.43% of ubiqutinated proteins in the bacteria colocalized with p62, and 85.33 ± 4.33% of the p62+ bacteria colocalized with ubiquitinated proteins. In addition, we observed that 83.67 ± 6.98% of ubiquitinated proteins in the bacteria colocalized with NDP52, and 89.33 ± 4.81% of NDP52+ bacteria colocalized with ubiquitinated proteins (Supplemental Fig. S3B). These results indicated that most of the intracellular ubiqutinated proteins in the A. baumannii strains colocalized with LRSAM1, p62, or NDP52. This suggests that LRSAM1, p62, and NDP52 were involved in the early event of autophagy and may be recruited to bind to the ubiquitinated proteins that surround the intracellular A. baumannii. Pairwise combination staining was performed to further investigate the relationship among p62 and NDP52 adaptor proteins and the ubiquitin ligase LRSAM1 in the autophagic response induced by intracytosolic A. baumannii. We found that there was cooperation between each pair of p62, NDP52, and LRSAM1 proteins during A. baumannii infection (Supplemental Fig. S3B). Our results showed that at 75 min postinfection, 71.33 ± 7.51% of the NDP52+ bacteria colocalized with p62, whereas 78.33 ± 6.06% of p62+ bacteria colocalized with NDP52, and 74.33 ± 8.09% of NDP52+ bacteria colocalized with LRSAM1. Thus, p62 and NDP52 are recruited to the same bacteria, with similar kinetics. This is consistent with previous reports in Salmonella that show that LRSAM1 may be localized to the intracellular bacteria and generate the bacteria-associated ubiquitin signal. Then, it would be bound by the ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52, which results in the recruitment of the autophagic machinery (12). Of interest, when we inspected the subcellular distribution of ubiquitin-binding adaptor proteins p62 and NDP52, we found that the puncta labeled with one adaptor protein did not colocalize with the other during intracellular A. baumannii infection, similar to results reported in Salmonella (37). We suspect that p62 and NDP52 can target nonoverlapping microdomains around bacteria. To investigate the course of the autophagic response, LAMP-1 and the LC3 puncta were detected at 75 min postinfection (Supplemental Fig. S3B), and colocalization level of LAMP-1 with the LC3+ bacteria was 80.33 ± 5.18%.
Figure 3.
Septin cages and adaptor proteins targeted the invading A. baumannii to induce isochorismatase-dependent autophagy. A) Adaptor proteins bound to the ubiquitinated proteins surrounding A. baumannii. HeLa cells were infected with strain 98-37-09 for 75 min [multiplicity of infection (MOI) = 10:1]. Cells were then coimmunostained with different antibodies [ubiquitinated protein (FK2), LRSAM1, NDP52, p62 and SEPT2] and Hoechst 33342 and analyzed for colocalization of LAMP-1, LC3 puncta, and intracellular bacteria. B, C) SEPT2 (B) and SEPT9 (C) were knocked down in HeLa cells and identified by Western blotting. D) HeLa cells were infected with strain 98-37-09 for 75 min (MOI = 10:1) after being transfected with shSEPT2, shSEPT9 plasmids, or a negative control shRNA plasmid for 24 h and then coimmunostained with an NDP52 antibody, a p62 antibody, and Hoechst 33342. E) The number of colocalized bacteria in 50 infected cells per group was measured by confocal fluorescence microscopy. F, G) LC3 expression level (F) of the SEPT2 or SEPT9 knockout HeLa cells infected with strain 98-37-09 for 2 h (MOI = 10:1) was analyzed by Western blotting (G). H) HeLa cells were infected with ACJ6 for 75 min (MOI = 10:1) and then coimmunostained with NDP52, p62, and SEPT2 antibodies and Hoechst 33342. I) The number of colocalized bacteria in 50 infected cells per group were measured by confocal fluorescence microscopy. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Scale bars, 20 μm. *P < 0.05; **P < 0.01; ***P < 0.001.
SEPT2 and SEPT9 are 2 cytoskeletal septins that have been reported to be involved in the initial steps of autophagy (13, 14). Some studies have also indicated that ubiquitin-binding adaptor proteins, p62 and NDP52, target some intracytosolic bacteria, such as Shigella, to a septin-dependent autophagy pathway (13). Our results showed that at 75 min postinfection, 89.67 ± 3.48% of the A. baumannii–septin cages recruited NDP52, whereas 90.67 ± 3.93% of A. baumannii–septin cages recruited p62. Most p62 and NDP52 were recruited to the same A. baumannii entrapped by septin cages, which suggests that the recruitment of p62 and NDP52 is dependent upon septins (Fig. 3A and Supplemental Fig. S5B). To further corroborate whether the recruitment of NDP52 or P62 is dependent upon septins, we used short hairpin RNA (shRNA) to deplete SEPT2 or SEPT9 in HeLa cells (Fig. 3B, C) and evaluated NDP52 or P62 recruitment to A. baumannii 98-37-09 in these septin-depleted cells. In the SEPT2- or SEPT9-depleted cells, both NDP52 and p62 recruitment were significantly reduced (P < 0.01; Fig. 3D, E). These results indicated that p62/SQSTM1 and NDP52 target intracytosolic A. baumannii to septin-dependent autophagy pathways. We also detected LC3 expression level, and the results showed that SEPT2 or SEPT9 knockout significantly decreased the autophagic response induced by A. baumannii (Fig. 3F, G).
In particular, our results showed that the ACJ6 isochorismatase mutant strain rarely induced ubiquitinated proteins or p62, NDP52, and SEPT2 recruitment compared with the wild-type strain (Fig. 3H, I), which demonstrated that A. baumannii isochorismatase is required in the recognition step of the autophagic response. Collectively, these data indicated that the ubiquitinated protein, the E3 ligase LRSAM1, the adaptor proteins p62 and NDP52, and septins were recruited and cooperated to target the intracellular A. baumannii and induce the autophagic response.
Isochorismatase is required for the clearance of invading A. baumannii by autophagy in vitro and in vivo
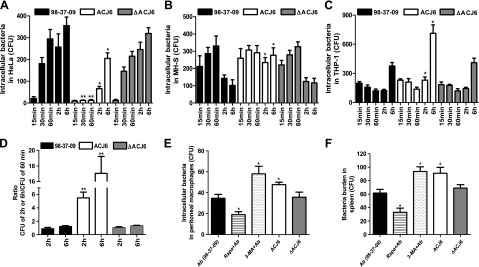
To examine the role of isochorismatase in the clearance of invading A. baumannii from host cells in vitro, we used strain 98-37-09, its mutant ACJ6, and the complementation strain ΔACJ6 to infect HeLa, MH-S, and THP-1 cells. Within 60 min, the CFU of intracellular bacteria were regarded as the internalization of HeLa cells, or the phagocytosis of macrophages. After 120 min postinfection, the CFU presented the results of the clearance effect by autophagy. Results of strain ΔACJ6 were similar to 98-37-09 in all 3 cell lines. No difference was found between the number of the intracellular strain 98-37-09 and that of ACJ6 in MH-S cells and THP-1 cells within 60 min postinfection (Fig. 4B, C). The number of intracellular strain 98-37-09 was found to be significantly less than that of ACJ6 in MH-S and THP-1 cells at 2 and 6 h postinfection (Fig. 4B, C). This implied that isochorismatase had no effect on phagocytosis of A. baumannii in macrophages, whereas isochorismatase was essential for the clearance of invading A. baumannii by autophagy in macrophages. However, the number of intracellular strain 98-37-09 was found to be much greater than that of ACJ6 at 30 min, 60 min, 2 h, and 6 h postinfection in HeLa cells (Fig. 4A). We also calculated the ratio of the number of intracellular bacteria at 2 and 6 h to that at 60 min postinfection In HeLa cells (Fig. 4D). It was found that the ratio of the number of the mutant strain was much higher than that of the wild-type strain, whether 2 h to 60 min postinfection or 6 h to 60 min postinfection. These data showed that isochorismatase of A. baumannii might play a different and complex role in HeLa cells.
Figure 4.
Isochorismatase is required for the clearance of A. baumannii in vitro and in vivo. A−C) HeLa (A), MH-S (B), and THP-1 cells (C) were infected with strain 98-37-09, ACJ6, or ΔACJ6 for different time [multiplicity of infection (MOI) = 10:1]. Then, infected cells were cultured in medium containing 100 μg gentamicin ml−1 for 1 h to remove extracellular bacteria. Intracellular bacteria were counted and expressed as CFUs. D) The ratio of the intracellular bacteria at 2 or 6 h postinfection to the intracellular bacteria at 60 min postinfection. E) Mice peritoneal macrophages were pretreated with rapamycin (Rapa) or 3-MA and were then infected with A. baumannii strain 98-37-09, ACJ6, or ΔACJ6 for 2 h (MOI = 10:1), and the number of intracellular bacteria in each group was counted and expressed as CFUs. F) Autophagy was induced by A. baumannii in a mouse infection model. Bacteria burden in spleen were counted and showed as CFU. Data are representative of 3 experiments with similar results. *P < 0.05 compared with 98-37-09 group; **P < 0.01 compared with 98-37-09 group.
To further investigate the effect of isochorismatase on the autophagic response caused by A. baumannii in vivo, we detected the bacterial burden in A. baumannii–infected mouse peritoneal macrophages or in the spleen of a mouse model of infection. The number of intracellular strain 98-37-09 and ΔACJ6 was much lower than that of its mutant ACJ6 (P < 0.05) in infected peritoneal macrophages (Fig. 4E). Similar results were found in the bacterial burden in the spleen. The bacterial burden in the spleens of strain 98-37-09 and ΔACJ6 infected group was significantly lower than that of ACJ6 (Fig. 4F). Results showed that isochorismatase also plays an important role in clearance of A. baumannii in vivo. Collectively, our data indicate that isochorismatase is required for clearance in host cells defense against invading A. baumannii by autophagy in vitro and in vivo.
A. baumannii induces a Beclin-1–dependent autophagy via the AMPK/ERK/mTOR pathway
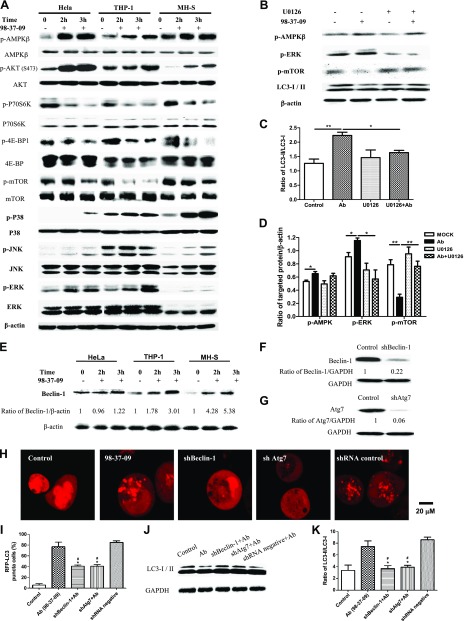
To assay the possible pathways that are involved in autophagic response induced by A. baumannii (38), we detected the expression of some key kinases of the AMPK/ERK/mTOR pathway. Our results showed that the phosphorylations of mTOR, p70S6K, and 4E-BP1 were significantly decreased in HeLa, THP-1, and MH-S cells at 2 h postinfection with strain 98-37-09 compared with uninfected cells, and reached a minimum at 3 h, whereas phosphorylations of Akt and AMPK were increased (Fig. 5A). Phosphorylations of ERK, JNK, and p38 were increased after 2 h of infection with strain 98-37-09 in HeLa, THP-1, and MH-S cells, and phosphorylation levels reached a maximum at 3 h (Fig. 5A). Pretreatment with MEK inhibitor U0126 (10 μM) inhibited ERK phosphorylation and increased mTOR phosphorylation, whereas it had no obvious effect on AMPK phosphorylation (Fig. 5D). U0126 could decrease the level of LC3-II expression in cells infected with strain 98-37-09 (Fig. 5B, C). These data suggest that A. baumannii induces autophagy via the AMPK/ERK/mTOR pathway, in which AMPK was upstream of ERK, whereas mTOR was downstream of ERK.
Figure 5.
A. baumannii induced Beclin-1–dependent autophagy via the AMPK/ERK/mTOR pathway. A) HeLa, THP-1, and MH-S cells were infected with strain 98-37-09 (multiplicity of infection = 10:1). Western blotting for AMPK, p-AMPK, Akt, p-Akt, mTOR, p-mTOR, p70S6K, p-70S6K, 4E-BP1, p-4E-BP1, ERK, pERK, p-JNK, JNK, p-P38, P38, and LC3 was performed by using proteins from the 98-37-09–infected HeLa, THP-1, and MH-S cells. B) HeLa cells were infected with strain 98-37-09 for 2 h and pretreated with U0126 (10 μM) for 1 h. Western blotting for p-AMPKβ, p-ERK, and p-mTOR was performed. C) The ratio of LC3II/I was calculated. D) The ratio of p-AMPKβ, p-ERK, and p-mTOR to β-actin was calculated. E) Western blotting of Beclin-1 in 98-37-09–infected HeLa, THP-1, and MH-S cells. F, G) Western blotting of Beclin-1 (F) and Atg7 (G) in HeLa cells. H) HeLa cells were transfected with pmRFP-LC3 plasmid for 24 h and then transfected with the shRNA negative control, shBeclin-1, or shAtg7 plasmids for 48 h before cells were infected with strain 98-37-09 for 2 h. Confocal images show the induction of the LC3 puncta. I) The number of puncta in each cell was determined, and cells with >10 puncta were considered LC3-RFP puncta cells. Data are representative of 100 cells. J, K) Western blotting for LC3 was performed. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Data are representative of 3 experiments with similar results. *P < 0.05 compared with 98-37-09–infected cells without shRNA plasmid; **P < 0.01; #P < 0.05 compared with 98-37-09–infected cells with negative control shRNA plasmid.
In addition, we detected the expression level of Beclin-1 in A. baumannii–infected HeLa, THP-1, and MH-S cells. We found that A. baumannii infection increased Beclin-1 expression in all 3 infected cell lines (Fig. 5E), which implied that the A. baumannii–induced autophagic response may be mediated by Beclin-1. To further corroborate the correlation between Beclin-1 and A. baumannii–induced autophagy, we used specific shBeclin-1 plasmids to knock down the expression of Beclin-1 (Fig. 5F). It was found that the number RPF-LC3 puncta in 98-37-09–infected HeLa cells transfected with the shBeclin-1 plasmid was significantly decreased compared with cells transfected with the negative shRNA plasmid and the control group (P < 0.05; Fig. 5H, I). To further examine this pathway, we analyzed an autophagy-related major downstream protein, Atg7. Atg7-specific knockdown (Fig. 5G) significantly decreased the number of RPF-LC3 puncta in 98-37-09–infected HeLa cells compared with the control group (P < 0.05; Fig. 5H, I). We also found that shBeclin-1 and shAtg7 significantly decreased the level of LC3 expression in the 98-37-09–infected HeLa cells compared with cells transfected with the negative shRNA plasmid and the control group (Fig. 5J, K). Together, these results indicated that Beclin-1 plays a central role in the mechanisms of autophagy induced by A. baumannii.
DISCUSSION
Recently, many publications have indicated that autophagy is very important in pathogen infections. Some microorganisms induce autophagy to help host cells clear these organisms, whereas other microorganisms may have the opposite effect (10, 29, 30). In this study, the puncta labeling or Western blot assays showed that A. baumannii induced an increase of LC3-II in HeLa, THP-1, and MH-S cells, whereas the AJ6 isochorismatase mutant strain rarely induced this autophagic response, and the complementation strain ΔACJ6 could induce obvious autophagy. In addition, strain ACJ6 induced fewer autophagosomes and autophagic vesicles. These findings were confirmed by transmission electron microscopy, which is regarded as the only solid proof of autophagy induction. Results of organelle analysis reconfirmed that A. baumannii infection can induce autophagy and that isochorismatase may play a role in autophagic induction.
It has been reported that ubiquitinated proteins are involved in bacterial autophagy (39), and subsequent studies have revealed that p62 and NDP52 act as adaptors to ubiquitinated proteins that surround Salmonella typhimurium, Shigella flexneri, Listeria monocytogenes, invasive E. coli, and Burkholderia cenocepacia in bacteria-induced autophagy (11, 37, 40, 41). Recently, LRSAM1, an E3 ubiquitin ligase, has been shown to aid in the recognition of intracellular bacteria (12). Our kinetics results showed that LRSAM1 localization to A. baumannii (40 min) peaked before peak bacterial ubiquitin, P62, and NDP52 colocalization (60 min) and verified that LRSAM1 was responsible for autophagy-associated ubiquitination of A. baumannii. We found that most of the intracellular ubiqutinated proteins in the A. baumannii strains colocalized with LRSAM1, p62, or NDP52 at 45 min postinfection, and a population of LC3+ A. baumannii colocalized with p62, NDP52, or LRSAM1. This suggested that ubiquitin, LRSAM1, NDP52, and p62 participated in initiating autophagy in A. baumannii–infected HeLa cells, and the pairwise combination staining results showed that there was cooperation between each pair of p62, NDP52, and LRSAM1 proteins during A. baumannii infection. This indicated that LRSAM1 may localize to intracellular A. baumannii to generate a ubiquitin signal and then be bound by the ubiquitin adaptor proteins p62/SQSTM1 and NDP52, which results in recruitment of the autophagic machinery. Furthermore, we found that p62 and NDP52 are recruited to the same bacteria with similar kinetics, which was similar to the results in Salmonella (12); however, although LRSAM1 and NDP52 each colocalized with intracellular bacteria, they never colocalized with each other at the same point. It indicated that they seemed to localize to separate subdomains around bacteria spatially. The same situation was also found in the relationship of p62 and NDP52. These conclusions were similar to previous reports (12, 40). Recent reports have also demonstrated that septins, a cytoskeletal component, were found to be recruited along with autophagy proteins to cage S. flexneri in the cytosol of infected cells and restrict bacterial dissemination (13, 14). We found that both SEPT2 and SEPT9 depletion decreased the recruitment of ubiquitin, p62, and NDP52 to wild-type A. baumannii. Thus, accumulation of ubiquitinated proteins and targeting of A. baumannii for autophagy by p62 and NDP52 are dependent on septins. We found that most intracellular A. baumannii could recruit ubiquitinated proteins, NDP52, p62, LRSAM1, and SEPT2, but only some of them induced LC3 puncta formation at the earlier time point. This may imply specific kinetics for the recognition of A. baumannii by ubiquitinated proteins and ubiquitin adaptors and the subsequent induction of the autophagic response.
Isochorismatase-like hydrolases were required for siderophore-mediated ferric iron acquisition (27). Isochorismate was hydrolyzed by isochorismatase to form siderophore, which could combine with iron ions. Siderophores are regarded as key virulence factors in many pathogens, and iron starvation caused by some bacterial infections could induce autophagy in host cells (21, 32). A. baumannii encodes an isochorismatase superfamily hydrolase (20), but its roles in A. baumannii virulence and immunoreactions have not been studied. Our study proved that the ACJ6 isochorismatase mutant strain produced much fewer siderophores than its parent strain 98-37-09 and the complementation strain ΔACJ6. And we found the isochorismatase mutant strain could hardly induce autophagy.
On the basis of our results that the isochorismatase mutant strain could not induce autophagy, further studies were performed to identify the role of isochorismatase in the autophagy process. Our immunofluorescence staining results showed that strain ACJ6 could not induce ubiquitinated proteins p62, NDP52, and SEPT2 recruitment compared with the wild-type strain. This demonstrated that A. baumannii isochorismatase is required in the recognition step of the autophagic response. We suggested that A. baumannii isochorismatase affected the level of bacteria-induced autophagy in host cells by enhancing recruitment of adaptor proteins. We also compared the internalization difference between strain 98-37-09 and its mutant ACJ6 in HeLa cells, the phagocytosis difference in MH-S and THP-1 cells, and the autophagic clearance difference in these 3 cell lines. In previous pathogen infection-related studies, the optimal time of internalization in epithelial cells and the best time of phagocytosis in macrophages was between 30 min and 2 h, whereas most studies used 2 h postinfection as the time of bacteria clearance to detect the effect of autophagy (28–30). A reported study about Acinetobacter strains proved the phagocytosis of Acinetobacter strains in macrophages (J774A.1 cells) started after 5 min postinfection, and 1 h was the time of maximal uptake (31). According the previous reports, we compared the internalization of 98-37-09 and its mutant in HeLa cells and phagocytosis in MH-S cells and THP-1 cells at 15, 30, and 60 min postinfection and the clearance of bacteria at 2 and 6 h postinfection. Our data showed that isochorismatase was necessary in the clearance of intracellular bacteria by autophagy in macrophages, but in HeLa cells, isochorismatase might play a different and complex role. Internalization and clearance of intracellular bacteria by autophagy were influenced by isochorismatase of A. baumannii in HeLa cells.
Autophagy, a complex process, involves a series of steps, such as initiation, nucleation, maturation, merging with lysosomes, etc. In the initiation phase, inducers or nutrition status could regulate some signal molecules, such as AMPK, MEK, ERK, mTOR, and Akt to initiate autophagy (42). In this study, we demonstrated that A. baumannii induced autophagy via the AMPK/ERK/mTOR pathway. A recent study has indicated that AA starvation can cause the dispersion of mTOR complex 1 from late endosomes/lysosomes. Therefore, we suggested that A. baumannii–induced mTOR suppression is a result of host amino acid starvation, which is similar to results from Shigella and Salmonella infection (7). Recent findings demonstrated that the ERK pathway is crucial to autophagy activation and provides a mechanistic link between the innate immune receptors and induction of autophagy against cytoplasm-invading microbes, such as L. monocytogenes (8). AMPK is an energy sensor in all eukaryotes and is activated by ATP depletion or glucose starvation (43, 44). AMPK was believed to activate autophagy via inhibition of mTOR complex 1 (45). Ouchi et al. (46) demonstrated that AMPK activation increased Akt signaling. In this study, we noticed that Akt phosphorylation was increased in the 3 cell lines after A. baumannii infection. Some previous reports have indicated that Akt phosphorylation mirrored mTOR phosphorylation (47). Similar to our findings, recent studies indicated that Akt phosphorylation was correlated with Pseudomonas aeruginosa invasion, and that the PI3K signaling pathway was necessary and sufficient for P. aeruginosa entry (10, 48). Thus, we speculated that Akt activation may be related to the invasion and internalization of A. baumannii. By using MEK inhibitor U0126, we demonstrated that A. baumannii infection may cause glucose starvation, which activates AMPK, then activates ERK and down-regulates mTOR, followed by the activation of autophagy (49). Our results also demonstrated Beclin-1 might take part in the second step of autophagy, a nucleation process. Recently, the Beclin-1-Atg7-Atg5 pathway was suggested to be involved in autophagy induced by P. aeruginosa (10). In our study, we found that autophagy induced by A. baumannii was Beclin-1–dependent through knocking out Beclin-1 and Atg7.
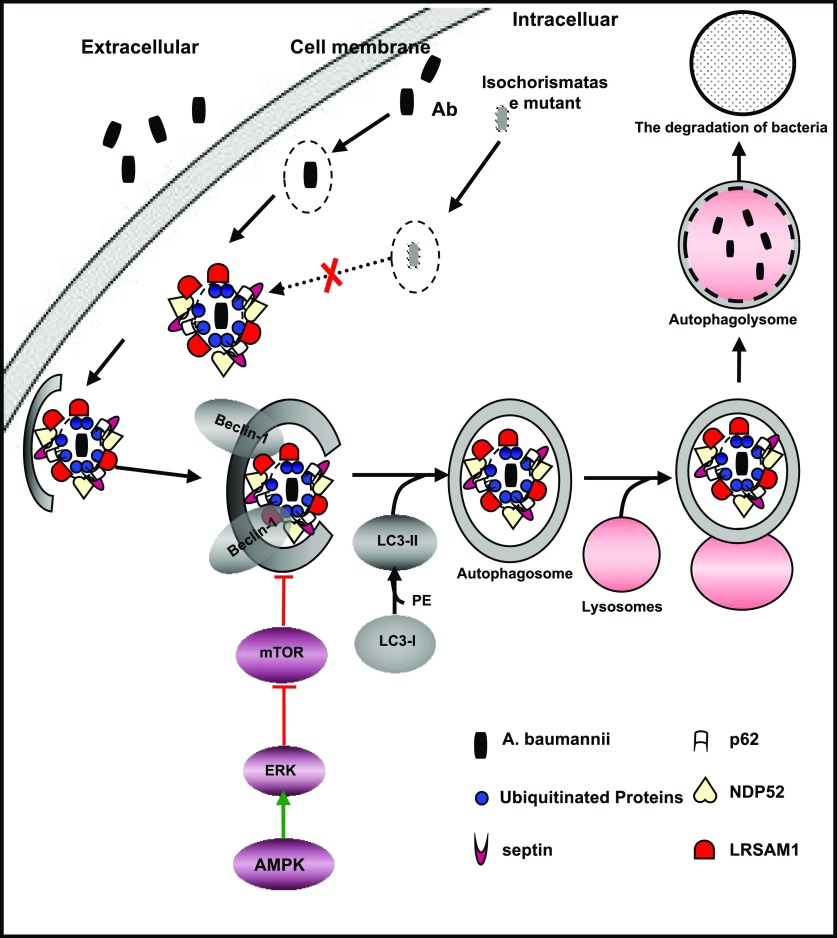
On the basis of the results of this study, autophagy induced by A. baumannii infection was complicated. We have summarized the bacterial recognition and the pathways involved in A. baumannii–induced autophagy in macrophages in the schematic shown in Fig. 6. Of most importance, our data also confirmed that isochorismatase was required for the recognition of A. baumannii and the activation of the autophagic pathway both in vitro and in vivo.
Figure 6.
The pathways involved in A. baumannii–induced autophagy in macrophages. →, direct stimulatory modification; ⊥, direct inhibitory modification.
In summary, this study demonstrated for the first time, to our knowledge, that intracellular A. baumannii can induce complete autophagy in host cells, and it illustrated the mechanism of recognition and pathogen clearance associated with autophagy, which depended on A. baumannii isochorismatase. These findings provide basic data for pertinent treatment options on the basis of the innate immune response against A. baumannii infection.
ACKNOWLEDGMENTS
This work was supported by funds form the National Nature Science Foundation of China (31172364 and 31271951), fund for Science and Technology Development of Jilin Province (20150101108JC), the Important National Science and Technology Specific Projects (2012ZX10003002), the Program for New Century Excellent Talents in University (NCET-09-0434; NCET-13-0245), China Postdoctoral Science Foundation (2013M530142), Fundamental Research Program of Shenzhen (JCYJ20130401172016183 and ZDSY20120616141302982), and Shenzhen Science and Technology Research and Development funds (JCYJ20130401173155808). The authors thank Dr. Luis A. Actis (Miami University, Miami, FL, USA) for providing the pMU125 plasmid; Dr. Yoshimori Tamotsu (Research Institute for Microbial Diseases, Osaka University, Japan) for providing plasmids pmRFP-LC3, ptfLC3, and pEGFP-LC3; Dr. Philip N. Rather (Research Service Veterans Affairs Medical Center, Decatur, GA, USA) for providing strains M2 and abaI::Km M2′; and Dr. Paul M. Dunman (University of Rochester School of Medicine and Dentistry, Rochester, NY, USA) for providing strain 98-37-09, ACJ6.
Glossary
- 3-MA
3-methyladenine
- CFU
colony-forming unit
- EGFP
enhanced green fluorescent protein
- LAMP-1
lysosomal-associated membrane protein-1
- LRSAM1
leucine rich repeat and sterile alpha motif containing 1
- mRFP
monomeric red fluorescent protein
- mTOR
mammalian target of rapamycin
- OD
optical density
- Omp
out membrane protein
- shRNA
short hairpin RNA
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
L. Yu designed research; Y. Wang, J. Fan, and J. Xu analyzed data; Y. Wang, K. Zhang, X. Shi, C. Wang, and F. Shen performed research; Y. Wang wrote the paper; M. Liu contributed analytic tools; and F. Wang and W. Bao developed software necessary to perform and record experiments.
REFERENCES
- 1.Peleg A. Y., Seifert H., Paterson D. L. (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaddy J. A., Arivett B. A., McConnell M. J., López-Rojas R., Pachón J., Actis L. A. (2012) Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu H., KuoLee R., Harris G., Van Rooijen N., Patel G. B., Chen W. (2012) Role of macrophages in early host resistance to respiratory Acinetobacter baumannii infection. PLoS One 7, e40019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. (2004) Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 5.Harris J. (2011) Autophagy and cytokines. Cytokine 56, 140–144 [DOI] [PubMed] [Google Scholar]
- 6.Ohsumi Y., Mizushima N. (2004) Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 15, 231–236 [DOI] [PubMed] [Google Scholar]
- 7.Tattoli I., Sorbara M. T., Philpott D. J., Girardin S. E. (2012) Bacterial autophagy: the trigger, the target and the timing. Autophagy 8, 1848–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anand P. K., Tait S. W., Lamkanfi M., Amer A. O., Nunez G., Pagès G., Pouysségur J., McGargill M. A., Green D. R., Kanneganti T. D. (2011) TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J. Biol. Chem. 286, 42981–42991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang L., Wang X. (2011) AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy 7, 924–926 [DOI] [PubMed] [Google Scholar]
- 10.Yuan K., Huang C., Fox J., Laturnus D., Carlson E., Zhang B., Yin Q., Gao H., Wu M. (2012) Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J. Cell Sci. 125, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., Wandel M. P., Li F., Liu Z., He C., Wu J., Shi Y., Randow F. (2013) Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Sci. Signal. 6, ra9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huett A., Heath R. J., Begun J., Sassi S. O., Baxt L. A., Vyas J. M., Goldberg M. B., Xavier R. J. (2012) The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe 12, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mostowy S., Bonazzi M., Hamon M. A., Tham T. N., Mallet A., Lelek M., Gouin E., Demangel C., Brosch R., Zimmer C., Sartori A., Kinoshita M., Lecuit M., Cossart P. (2010) Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 8, 433–444 [DOI] [PubMed] [Google Scholar]
- 14.Mostowy S., Sancho-Shimizu V., Hamon M. A., Simeone R., Brosch R., Johansen T., Cossart P. (2011) p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem. 286, 26987–26995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mestre M. B., Fader C. M., Sola C., Colombo M. I. (2010) Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy 6, 110–125 [DOI] [PubMed] [Google Scholar]
- 16.Rumbo C., Tomás M., Fernández Moreira E., Soares N. C., Carvajal M., Santillana E., Beceiro A., Romero A., Bou G. (2014) The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect. Immun. 82, 4666–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neilands J. B. (1991) A brief history of iron metabolism. Biol. Met. 4, 1–6 [DOI] [PubMed] [Google Scholar]
- 18.Goral A. M., Tkaczuk K. L., Chruszcz M., Kagan O., Savchenko A., Minor W. (2012) Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica. J. Struct. Funct. Genomics 13, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caruthers J., Zucker F., Worthey E., Myler P. J., Buckner F., Van Voorhuis W., Mehlin C., Boni E., Feist T., Luft J., Gulde S., Lauricella A., Kaluzhniy O., Anderson L., Le Trong I., Holmes M. A., Earnest T., Soltis M., Hodgson K. O., Hol W. G., Merritt E. A. (2005) Crystal structures and proposed structural/functional classification of three protozoan proteins from the isochorismatase superfamily. Protein Sci. 14, 2887–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs A. C., Hood I., Boyd K. L., Olson P. D., Morrison J. M., Carson S., Sayood K., Iwen P. C., Skaar E. P., Dunman P. M. (2010) Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78, 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy T. A., Moreland S. M., Andrews-Polymenis H., Detweiler C. S. (2013) The ferric enterobactin transporter Fep is required for persistent Salmonella enterica serovar typhimurium infection. Infect. Immun. 81, 4063–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiester S. E., Nwugo C. C., Penwell W. F., Neary J. M., Beckett A. C., Arivett B. A., Schmidt R. E., Geiger S. C., Connerly P. L., Menke S. M., Tomaras A. P., Actis L. A. (2015) Role of the carboxy terminus of SecA in iron acquisition, protein translocation, and virulence of the bacterial pathogen Acinetobacter baumannii. Infect. Immun. 83, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwyn B., Neilands J. B. (1987) Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56 [DOI] [PubMed] [Google Scholar]
- 24.Rioux C., Jordan D. C., Rattray J. B. (1983) Colorimetric determination of catechol siderophores in microbial cultures. Anal. Biochem. 133, 163–169 [DOI] [PubMed] [Google Scholar]
- 25.Arnow L. (1937) Colorimetric determination of the components of 3, 4-dihydroxyphenylalanine-tyrosine mixtures. J. Biol. Chem. 118, 531–537 [Google Scholar]
- 26.Zhang X., Goncalves R., Mosser D. M. (2008) The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 14, Unit 14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dauphinee S. M., Richer E., Eva M. M., McIntosh F., Paquet M., Dangoor D., Burkart C., Zhang D. E., Gruenheid S., Gros P., Behr M., Malo D. (2014) Contribution of increased ISG15, ISGylation and deregulated type I IFN signaling in Usp18 mutant mice during the course of bacterial infections. Genes Immun. 15, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaPenta D., Rubens C., Chi E., Cleary P. P. (1994) Group A streptococci efficiently invade human respiratory epithelial cells. Proc. Natl. Acad. Sci. U S A 91, 12115–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Q., Ahn S. H., Fowler V. G. Jr (2015) Macrophage phagocytosis assay of Staphylococcus aureus by flow cytometry. Bio. Protoc. 5, e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiedemann A., Rosselin M., Mijouin L., Bottreau E., Velge P. (2012) Involvement of c-Src tyrosine kinase upstream of class I phosphatidylinositol (PI) 3-kinases in Salmonella enteritidis Rck protein-mediated invasion. J. Biol. Chem. 287, 31148—31154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tayabali A. F., Nguyen K. C., Shwed P. S., Crosthwait J., Coleman G., Seligy V. L. (2012) Comparison of the virulence potential of Acinetobacter strains from clinical and environmental sources. PLoS One 7, e37024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirienko N. V., Ausubel F. M., Ruvkun G. (2015) Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 112, 1821–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Checroun C., Wehrly T. D., Fischer E. R., Hayes S. F., Celli J. (2006) Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA 103, 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arroyo D. S., Soria J. A., Gaviglio E. A., Garcia-Keller C., Cancela L. M., Rodriguez-Galan M. C., Wang J. M., Iribarren P. (2013) Toll-like receptor 2 ligands promote microglial cell death by inducing autophagy. FASEB J. 27, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y., Yu S., Ding N., Meng C., Meng S., Zhang S., Zhan Y., Qiu X., Tan L., Chen H., Song C., Ding C. (2014) Autophagy benefits the replication of Newcastle disease virus in chicken cells and tissues. J. Virol. 88, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A. Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cemma M., Kim P. K., Brumell J. H. (2011) The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy 7, 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu B., Zhou Y., Xu F., Shuai J., Li X., Fang W. (2012) Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J. Virol. 86, 12003–12012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perrin A. J., Jiang X., Birmingham C. L., So N. S., Brumell J. H. (2004) Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr. Biol. 14, 806–811 [DOI] [PubMed] [Google Scholar]
- 40.Thurston T. L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 41.Zheng Y. T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J. H. (2009) The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183, 5909–5916 [DOI] [PubMed] [Google Scholar]
- 42.Ozpolat B., Benbrook D. M. (2015) Targeting autophagy in cancer management - strategies and developments. Cancer Manag. Res. 7, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mihaylova M. M., Shaw R. J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardie D. G. (2011) AMPK and autophagy get connected. EMBO J. 30, 634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouchi N., Kobayashi H., Kihara S., Kumada M., Sato K., Inoue T., Funahashi T., Walsh K. (2004) Adiponectin stimulates angiogenesis by promoting cross-talk between AMP-activated protein kinase and Akt signaling in endothelial cells. J. Biol. Chem. 279, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu S., Shen G., Khor T. O., Kim J. H., Kong A. N. (2008) Curcumin inhibits Akt/mammalian target of rapamycin signaling through protein phosphatase-dependent mechanism. Mol. Cancer Ther. 7, 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kierbel A., Gassama-Diagne A., Mostov K., Engel J. N. (2005) The phosphoinositol-3-kinase-protein kinase B/Akt pathway is critical for Pseudomonas aeruginosa strain PAK internalization. Mol. Biol. Cell 16, 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakahira K., Choi A. M. (2013) Autophagy: a potential therapeutic target in lung diseases. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L93–L107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomaras A. P., Dorsey C. W., Edelmann R. E., Actis L. A. (2003) Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484 [DOI] [PubMed] [Google Scholar]
- 51.Kimura S., Noda T., Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 52.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]