Abstract
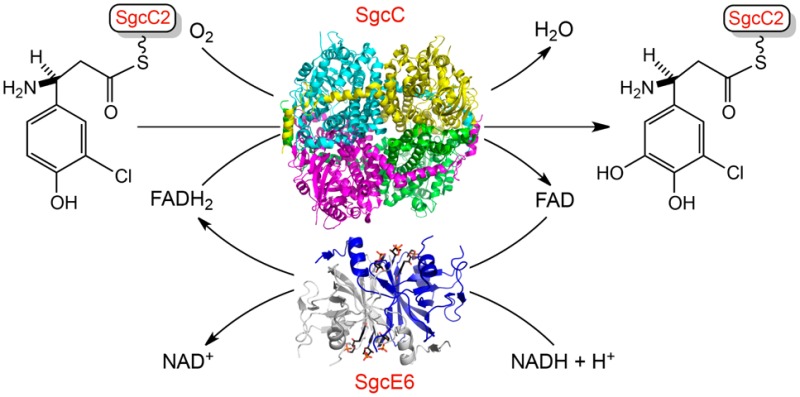
C-1027 is a chromoprotein enediyne antitumor antibiotic produced by Streptomyces globisporus. In the last step of biosynthesis of the (S)-3-chloro-5-hydroxy-β-tyrosine moiety of the C-1027 enediyne chromophore, SgcE6 and SgcC compose a two-component monooxygenase that hydroxylates the C-5 position of (S)-3-chloro-β-tyrosine. This two-component monooxygenase is remarkable for two reasons. (i) SgcE6 specifically reacts with FAD and NADH, and (ii) SgcC is active with only the peptidyl carrier protein (PCP)-tethered substrate. To address the molecular details of substrate specificity, we determined the crystal structures of SgcE6 and SgcC at 1.66 and 2.63 Å resolution, respectively. SgcE6 shares a similar β-barrel fold with the class I HpaC-like flavin reductases. A flexible loop near the active site of SgcE6 plays a role in FAD binding, likely by providing sufficient space to accommodate the AMP moiety of FAD, when compared to that of FMN-utilizing homologues. SgcC shows structural similarity to a few other known FADH2-dependent monooxygenases and sheds light on some biochemically but not structurally characterized homologues. The crystal structures reported here provide insights into substrate specificity, and comparison with homologues provides a catalytic mechanism of the two-component, FADH2-dependent monooxygenase (SgcE6 and SgcC) that catalyzes the hydroxylation of a PCP-tethered substrate.
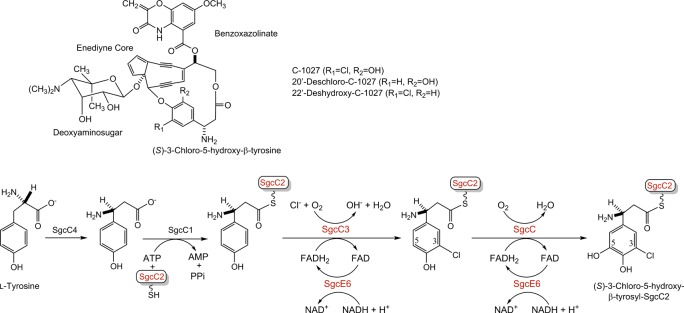
C-1027 is a chromoprotein enediyne antitumor antibiotic isolated from Streptomyces globisporus, consisting of an apoprotein and the C-1027 chromophore.1 The structure of the C-1027 chromophore is comprised of a nine-membered enediyne core with three appended moieties, including a deoxyaminosugar, benzoxazolinate, and (S)-3-chloro-5-hydroxy-β-tyrosine (Figure 1A).1 The enediynes share a common mechanism of action. The enediyne core undergoes a Bergman cycloaromatization to generate a transient benzenoid diradical capable of abstracting hydrogen atoms from DNA, leading to double-strand breaks.2 C-1027 is unique in that it can also generate interstrand cross-links under anaerobic conditions.3,4 The β-tyrosine moiety of C-1027 plays an important role in determining the ratio of double-strand breaks to interstrand cross-links, such that removal of the C-20′ chloride or the C-22′ hydroxyl functionalities almost abolishes interstrand cross-links and decreases the number of double-strand breaks by 2–20-fold.4,5
Figure 1.
Structures of the C-1027 chromophore and engineered analogues. (A) The C-1027 chromophore consists of four components: a nine-membered enediyne core, a benzoxazolinate, a deoxyaminosugar, and (S)-3-chloro-5-hydroxy-β-tyrosine. (B) Biosynthesis of the (S)-3-chloro-5-hydroxy-β-tyrosyl moiety of the C-1027 chromophore from l-tyrosine, featuring SgcC3-catalyzed chlorination and SgcC-catalyzed hydroxylation, both of which prefer a SgcC2-tethered substrate.
Deletion of the halogenase gene sgcC3 and monooxygenase gene sgcC from S. globisporus afforded mutant strains that produced 20′-deschloro-C-1027 and 22′-deshydroxy-C-1027, respectively (Figure 1A).1,6 Characterization of the (S)-3-chloro-5-hydroxy-β-tyrosyl moiety biosynthetic pathway (Figure 1B) demonstrated that (i) SgcC4 is an aminomutase that converts l-tyrosine to (S)-β-tyrosine,7 (ii) SgcC1 is a free-standing nonribosomal peptide synthetase (NRPS) adenylation protein that loads (S)-β-tyrosine onto the free-standing peptidyl carrier protein (PCP), SgcC2, leading to (S)-β-tyrosyl-S-SgcC2,6,8 (iii) SgcC3 is a FAD-dependent halogenase that catalyzes regioselective chlorination of (S)-β-tyrosyl-S-SgcC2 to form (S)-3-chloro-β-tyrosyl-S-SgcC2,9 and finally (iv) SgcC is a FADH2-dependent monooxygenase that catalyzes the hydroxylation of (S)-3-chloro-β-tyrosyl-S-SgcC2 to form (S)-3-chloro-5-hydroxy-β-tyrosyl-S-SgcC2.10 The SgcE6 flavin reductase supplies FADH2 to both SgcC3 and SgcC.11
Three families of enzymes catalyze most hydroxylation reactions in natural product biosynthesis, cytochrome P450 monooxygenases, α-ketoglutarate-dependent non-heme iron monooxygenases, and flavin-dependent monooxygenases.12 Flavin-dependent monooxygenases are further divided into two different classes, one-component and two-component monooxygenases, which are not related by similarities in sequence or structure.13−15 The one-component systems use an FMN or FAD prosthetic group and can directly reduce it via NAD(P)H, while the two-component systems consist of a flavin reductase component and a flavin-dependent monooxygenase component.16−21 The flavin reductase and monooxygenase components have been further divided into subfamilies, classes, or groups. The flavin reductase components can be divided into two groups on the basis of whether flavin is a substrate (class I) or a prosthetic group (class II). Class I flavin reductases can be further divided into at least three subfamilies based on sequence and structure as exemplified by (i) Fre from Escherichia coli, (ii) FRaseI from Vibrio fischeri, and (iii) HpaC from E. coli W (EcHpaC).20 The monooxygenase components have been divided into groups, A–H, based on sequence/structure and function, with groups C–F having a flavin reductase component.14,22
SgcE6 and SgcC form a two-component monooxygenase system. On the basis of previous bioinformatics analysis, SgcE6 belongs to the HpaC-like flavin reductase subfamily possessing conserved (S/T/C)XXPP and GDH motifs.11,20 This subfamily contains a dual-substrate binding pocket to accommodate FAD/FMN and NAD(P)H. The HpaC-like family catalyzes a two-electron transfer from NAD(P)H to generate reduced flavin.23 SgcE6 uses FAD and NADH but not NADPH or FMN as substrates and provides FADH2 to SgcC and SgcC3 through free diffusion.11 On the basis of previous bioinformatics analysis, SgcC belongs to a group of monooxygenases that act on p-hydroxyphenylacetate (4HPA), such as HpaB from E. coli strain W (EcHpaB)24 and HpaA from Pseudomonas aeruginosa (PaHpaA).25 Thus, SgcC falls into “group D”, whereas most monooxygenases in natural product biosynthesis are in “group A”.22 The “group D” monooxygenases share structural homology with acyl-CoA dehydrogenases (ACAD) and are split into two types represented by HpaB from E. coli W (EcHpaB) and HpaH from Acinetobacter baumanii (AbHpaH), which are 520 and 422 residues in length, respectively.22 Structures of HpaB from Thermus thermophilis HB8 (TtHpaB),17 which is ∼25% identical to EcHpaB and similar in length, reveals mechanistic insights into the catalysis of EcHpaB and SgcC homologues.
In vitro, SgcC efficiently catalyzes the regioselective hydroxylation of 3-substituted β-tyrosyl-S-SgcC2 analogues, including F-, Cl-, Br-, I-, and methyl-substituted analogues; however, a 3-OH-substituted analogue was not hydroxylated.10 Two-component monooxygenases typically catalyze hydroxylations of small molecule substrates,17,19,26−29 whereas SgcC is active on only carrier protein-tethered substrates. The other known carrier protein-dependent oxidase is BtrO that participates in the biosynthesis of butirosin.30 However, BtrO is an FMN-dependent, two-component monooxygenase, which catalyzes hydroxylation at the α carbon of the acyl-S-ACP substrate and falls into “group C”. Other putative homologues of SgcC that might act on carrier protein-tethered substrate can be found in the nine-membered enediyne biosynthetic pathways of kedarcidin (KedY),31 maduropeptin (MdpC),32 and sporolides A and B (SpoT3).33 There have been few studies investigating two-component monooxygenases in which the carrier protein plays an essential role; thus, a structure of SgcC could provide insight into its family.
Enediynes are promising candidates for anticancer therapies, and alteration of the appended moieties can have important impacts on biological function. Homologues of sgcC and sgcE6 are widespread in nine-membered enediyne biosynthetic gene clusters, with sgcE6 being conserved. Here, we present the crystal structures of the PCP-dependent two-component monooxygenase, SgcC, and the flavin reductase, SgcE6. The structure of SgcC reveals insight into the “group D” class of flavin-dependent monooxygenases that act on carrier protein-tethered substrates. The molecular details responsible for the substrate specificity of SgcC could now be explored and exploited for protein engineering, potentially leading to new enediyne analogues.
Materials and Methods
Gene Cloning and Production and Purification of SgcE6
The sgcE6 gene from S. globisporus was amplified from genomic DNA by polymerase chain reaction (PCR) using two primers, SgcE6-F and SgcE6-R (Table S1), and subcloned into expression vector pMCSG73,34 yielding APC109096 (pBS1159). This construct produced a fusion protein containing an N-terminal NusA, followed by a His6 tag and a TEV protease cleavage site with the target protein, which leaves an N-terminal Ser-Asn-Ala sequence after TEV cleavage.
To overproduce the selenomethionyl (SeMet)-SgcE6 protein, the APC109096 construct was transformed into E. coli BL21(DE3)-Gold (Stratagene), and the bacterial culture was then grown at 37 °C and 190 rpm in 1 L of enriched M9 medium35 until it reached an OD600 of 1.0. After the sample had been cooled in air at 4 °C for 60 min, methionine biosynthesis inhibitory amino acids (l-valine, l-isoleucine, l-leucine, l-lysine, l-threonine, and l-phenylalanine, each at 25 mg/L) and 90 mg/L selenomethionine were added. Protein overproduction was induced by 0.5 mM isopropyl β-d-thiogalactoside (IPTG). The cells were incubated overnight at 18 °C and subsequently harvested and resuspended in lysis buffer [500 mM NaCl, 5% (v/v) glycerol, 50 mM HEPES (pH 8.0), 20 mM imidazole, and 10 mM β-mercaptoethanol]. The cells were disrupted by sonication. The insoluble cellular material was removed by centrifugation. SeMet-SgcE6 was purified using Ni-NTA affinity chromatography and the ÄKTAxpress system (GE Healthcare Life Sciences). The N-terminal tag was cleaved from pure protein using recombinant His6-tagged TEV protease (Sigma), and an additional step of Ni-NTA affinity chromatography was performed to remove the protease, uncut protein, and affinity tag. Pure SeMet-SgcE6 was concentrated using Amicon Ultra-15 concentrators (Millipore) in 20 mM HEPES buffer (pH 8.0), 250 mM NaCl, and 2 mM dithiothreitol (DTT). Protein concentrations were determined from the absorbance at 280 nm using a calculated molar absorption coefficient (ε280 = 12615 M–1 cm–1).36 The concentration of protein samples used for crystallization was 30.2 mg/mL.
Crystallization of SgcE6
SgcE6 crystallization screens were prepared with a Mosquito liquid dispenser (TTP Labtech) using the sitting-drop vapor-diffusion technique in 96-well CrystalQuick plates (Greiner Bio-one). For each condition, 0.4 μL of protein and 0.4 μL of crystallization formulation were mixed; the mixture was equilibrated against 140 μL of the reservoir in the well. The protein–ligand complex was prepared by mixing protein with 27.7 mM FAD and 27.7 mM NADH at 4 °C for several hours before setting up crystallizations. The following commercially available crystallization screens were used: MCSG-1-3 (Microlytic Inc.) at 24 °C for the ligand-free protein and MCSG-1-4 (Microlytic Inc.) at 24 °C for the protein–ligand complexes. The crystals for the ligand-free protein were obtained under 25% PEG 3350, 0.1 M HEPES (pH 7.5), and 0.2 M ammonium sulfate. The best crystal of the protein–ligand complex of SgcE6 was produced under 20% PEG 8000, 0.1 M MES (pH 6.0), and 0.2 M calcium acetate.
Data Collection and Structure Determination of SgcE6
The diffraction data of ligand-free SeMet-SgcE6 (apo-SgcE6) were collected at Argonne National Laboratory on the APS (19-ID) beamline using a wavelength of 0.97912 Å with the ADSC QUANTUM 315r CCD detector. The data sets were collected to a resolution of 1.90 Å. For the complex structure of SeMet-SgcE6 (SgcE6-FAD), the diffraction data were collected on the same beamline using a wavelength of 0.97935 Å with the same detector. The data sets were collected to a resolution of 1.66 Å. The diffraction data were indexed, integrated, and scaled using HKL-3000.37 Both apo-SgcE6 and SgcE6-FAD structures were determined by the single-wavelength anomalous diffraction (SAD) method, and the initial models were built using HKL-3000.37 The structures were completed with alternating rounds of manual model building with COOT38 and refinement with phenix.refine.39 Waters were added and updated during refinement. The final structures were refined to the same resolution limit as in data collection with favorable Rcryst and Rfree values (Table 1).
Table 1. Data Collection, Phasing, and Refinement Statistics for Structures.
| apo-SgcE6 | SgcE6-FAD | SgcC | |
|---|---|---|---|
| Data Collection | |||
| Protein Data Bank entry | 4HX6 | 4R82 | 4OO2 |
| space group | P21 | P21 | P212121 |
| cell dimensions | |||
| a, b, c (Å) | 56.37, 213.31, 56.72 | 42.05, 62.83, 70.09 | 99.16, 173.73, 113.86 |
| α, β, γ (deg) | 90.00, 88.83, 90.00 | 90.00, 92.06, 90.00 | 90.00, 90.00, 90.00 |
| wavelength (Å) | 0.97912 | 0.97935 | 0.91165 |
| resolutiona (Å) | 30.10–1.90 (1.93–1.90) | 34.93–1.66 (1.69–1.66) | 47.67–2.63 (2.72–2.63) |
| Rsymb or Rmergec (%) | 9.5 (45.2) | 9.0 (52.1) | 29.6 (231.4) |
| CC1/2 (%) | 99.2 (84.1) | 99.4 (69.5) | 98.0 (17.3) |
| I/σ | 19.5 (2.4) | 22.6 (2.0) | 5.3 (0.62) |
| completeness (%) | 98.3 (80.9) | 99.5 (97.3) | 96.7 (97.9) |
| redundancy | 3.3 (2.8) | 3.7 (2.9) | 5.2 (4.3) |
| Refinement | |||
| resolution (Å) | 30.10–1.90 | 34.93–1.66 | 47.67–2.63 |
| no. of reflections | 104829 | 43097 | 296449 |
| Rwork/Rfree | 0.169/0.212 | 0.164/0.188 | 0.216/0.246 |
| Ramachandran plot (%)d | |||
| favored | 99 | 98 | 95 |
| outliers | 0 | 2 | 0 |
| no. of atoms | |||
| protein | 10513 | 2515 | 15938 |
| ligand/ion | 50 | 176 | 8 |
| water | 774 | 240 | 217 |
| B-factor (Å2) | |||
| protein | 29.0 | 21.1 | 57.1 |
| ligand/ion | 53.8 | 22.4 | 61.9 |
| water | 37.0 | 30.9 | 47.8 |
| root-mean-square deviation | |||
| bond lengths (Å) | 0.007 | 0.006 | 0.002 |
| bond angles (deg) | 1.083 | 1.158 | 0.67 |
Numbers in parentheses are values for the highest-resolution bin.
Rmerge = ∑hkl∑i|Ii(hkl) – I̅(hkl)|/∑hkl∑iIi(hkl), where Ii(hkl) is the ith observation of reflection hkl and I̅(hkl) is the weighted average intensity for all observations i of reflection hkl.
Rmeas = ∑hkl[N/(N – 1)1/2]∑i|Ii(hkl) – I̅(hkl)|/∑hkl∑iIi(hkl).
As defined by MolProbity.
Gene Cloning and Production and Purification of SgcC
The sgcC gene from S. globisporus was amplified from the genomic DNA by PCR using two primers, SgcC-F and SgcC-R (Table S1), and subsequently subcloned into expression vector pBS3080,40 yielding APC109081 (pBS1160), which produced SgcC with an N-terminal His6 tag and a TEV protease cleavage site, which leaves an N-terminal Ser-Pro sequence after TEV cleavage. For SeMet-SgcC production, the APC109081 construct was transformed into E. coli methionine auxotroph strain B834 (DE3) pLysS (Invitrogen). SeMet-SgcC overproduction was accomplished using autoinduction medium at 25 °C according to the standard protocol.41 The cells were disrupted by sonication. The insoluble cellular material was removed by centrifugation. Ni-NTA affinity chromatography was used to initially purify the His6-tagged protein with a supplement of 0.5 mM FAD during purification. The His6 tag was then removed by TEV protease (Sigma) cleavage at 4 °C overnight in 50 mM Tris (pH 8.0) containing 200 mM NaCl, 1 mM DTT, 0.5 mM EDTA, and 0.5 mM FAD. The sample after TEV protease cleavage was run through Ni-NTA resin again under loading buffer conditions, and the flow-through was collected, which contained the purified untagged SeMet-SgcC. The untagged protein was subsequently run through a Supderdex 200 size-exclusion column using the ÄKTAxpress system (GE Healthcare Life Sciences) in 20 mM Tris buffer (pH 7.5) containing 100 mM NaCl. SeMet-SgcC was concentrated to 15 mg/mL by Amicon Ultra-15 concentrators (Millipore) for protein crystallization. Protein concentrations were determined from the absorbance at 280 nm using a calculated molar absorption coefficient (ε280 = 94435 M–1 cm–1).36
Crystallization of SgcC
SeMet-SgcC was screened for crystallization conditions as described for SgcE6. SeMet-SgcC was crystallized by the hanging drop vapor diffusion method at 20 °C by mixing 1 μL of 15 mg/mL SeMet-SgcC with 1 μL of reservoir solution [100 mM MES buffer (pH 6.5), 60 mM mixture of MgCl2 and CaCl2, 100 mM imidazole, and 30% mixture of PEG 550 MME and PEG 20000].
Data Collection and Structure Determination of SgcC
The diffraction data of SgcC were collected at Argonne National Laboratory on the GM/CA (23-ID-B) beamline using a wavelength of 0.911650 Å with the MAR 300 CCD detector. The data sets were collected to a resolution of 2.63 Å. The data were indexed, integrated, and scaled using XDS.42 The SgcC structure was determined by molecular replacement using Phaser-MR43 and Autobuild programs44 of the PHENIX suite39 with monomer polypeptide coordinates without ligands or water using the oxygenase component of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus (TtHpaB) as the search model [Protein Data Bank (PDB) entry 2YYK]. The structural refinement of SgcC was conducted with the same procedures that were used for SgcE6. The final data collection and refinement statistics can be found in Table 1.
Size-Exclusion Chromatography
Size-exclusion chromatography was performed using a Superdex 200 16/600 column (GE Healthcare Life Sciences) with an ÄKTAxpress system (GE Healthcare Life Sciences) at 4 °C and 500 μL of sample loaded per run. The column was calibrated with a size-exclusion calibration kit (GE Healthcare Life Sciences) and developed with the elusion buffer [200 mM NaCl and 100 mM Tris (pH 8.0)] at a flow rate of 0.5 mL/min with UV detection at 280 nm.
Protein–Protein Docking of SgcC and SgcC2
Two methods of SgcC–SgcC2 docking were performed, both using a model of SgcC2 generated by I-TASSER45 (Figure S1, trimmed conserved core sequence). The ClusPro server46 was used to compute a global blind docking, and the Rosie server47 was used for local flexible docking (allowing local rotation and tilting), which generated similar results.
Results and Discussion
In Nine-Membered Enediyne Biosynthetic Gene Clusters, Homologues of SgcE6 Are Conserved and Those of SgcC Are Widespread
Our previous studies showed that SgcE6 supplies FADH2 to both SgcC and SgcC3 in biosynthesis of the β-tyrosyl moiety of the C-1027 chromophore.9,10 Bioinformatics analysis of the C-1027 biosynthetic gene cluster revealed the existence of other genes encoding putative flavin-dependent enzymes, such as sgcE9, sgcD2, and sgcL, the functions of which are still unknown.1 A genome neighborhood network of enediyne biosynthetic gene clusters from the NCBI and JGI databases48 revealed that sgcE6 homologues are conserved in enediyne biosynthetic gene clusters, especially those encoding nine-membered enediynes, suggesting a role in enediyne core biosynthesis (Figure S2). On the other hand, sgcC homologues are conserved in the aromatic amino acid-containing enediyne biosynthetic gene clusters, kedarcidin,31 maduropeptin,32 and sporolides A and B,33 indicating this group of flavin-dependent monooxygenases specifically catalyzes hydroxylation onto aromatic rings. Homologues of sgcC are also present in the other 14 putative nine-membered enediyne biosynthetic gene clusters (Figure S3) with high levels of amino acid sequence identity among them (56.6–99.8%). Hydroxylation of an aromatic amino acid moiety is therefore thought to be involved in their biosynthetic pathways. We previously determined that SgcE6 is kinetically capable of providing more reduced flavin than is necessary for both SgcC and SgcC3, further supporting a broader role for SgcE6 in the biosynthesis of enediynes.11
Structure Solution, Refinement, and Quality of SgcE6 Crystal Structures
The crystal structures of the ligand-free form of SgcE6 (apo-SgcE6) was obtained in monoclinic space group P21 and was determined by the single-wavelength anomalous diffraction (SAD) method; the summary of crystallographic data is given in Table 1. Apo-SgcE6 was refined to a resolution of 1.90 Å with an Rwork factor of 16.9% and an Rfree factor of 21.2%. The structure of apo-SgcE6 contains eight polypeptide chains, five acetates, and six sulfates per asymmetric unit. The eight polypeptide chains had varying density for the N- and C-termini, with the start of the chain varying from residue −2 to 8 and ending between residues 175 and 180. Many monomers also lack interpretable density for the region between residues 97 and 105, which is part of the SgcE6 “flexible loop” and will be discussed further below (Figure S4).
Crystals of the FAD complex form of SgcE6 (SgcE6-FAD) were prepared by mixing protein with high concentrations of FAD and NADH via cocrystallization under various conditions. SgcE6-FAD crystallized in the P21 space group, but was not isomorphous with apo-SgcE6, as it had different cell dimensions. SgcE6-FAD was refined to a high resolution of 1.66 Å with an Rwork of 16.4% and an Rfree of 18.8%. The asymmetric unit of SgcE6-FAD contains two polypeptide chains, each with associated electron density that is modeled unambiguously as FMN and two adenosine 5′-monophosphates (AMP) in the substrate/cofactor binding cavity (Figure 2C). On average, the atoms from the FMN and AMP molecules had B factors similar to those of the surrounding protein, indicating a lack of disorder. The FMN moiety is from FAD, and the two AMPs are from either FAD or NADH due to the large excess used in crystallization and apparent degradation. Each monomer could be modeled from residue 9 to 99 and from residue 104 to 177 and again was lacking density in the “flexible loop”. Each monomer is associated with one Mg2+, two Ca2+, two Cl–, and two acetates.
Figure 2.
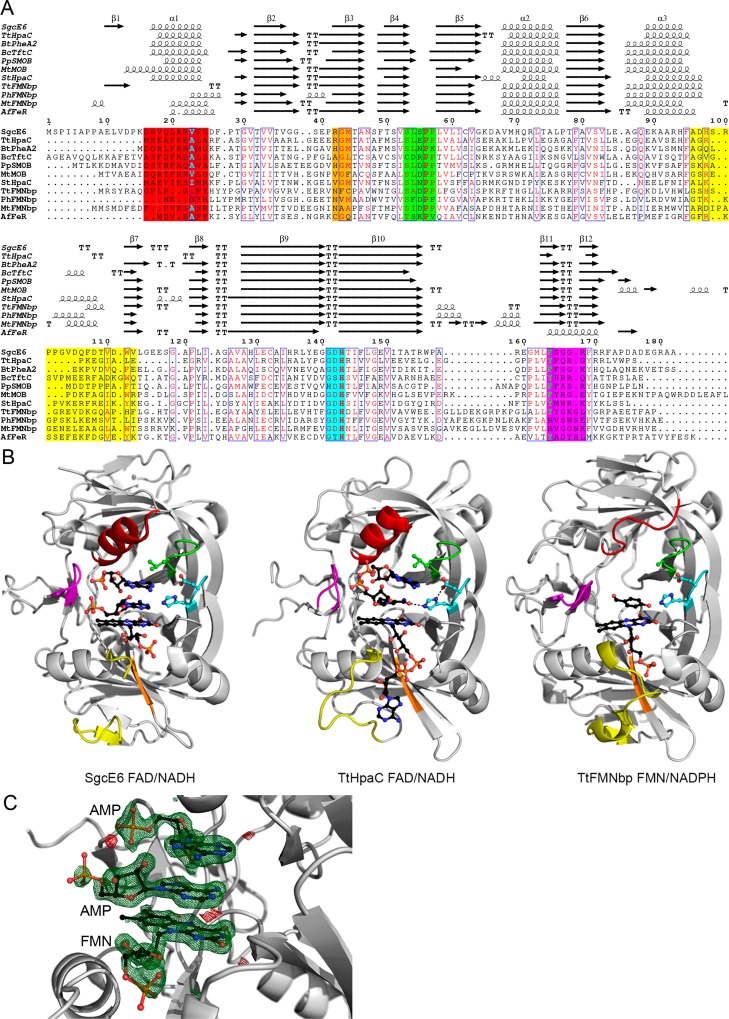
Structure-based sequence alignment of SgcE6 and homologues. (A) The sequence alignment is colored according to the loops lining the active and flavin binding sites. The sequence/structure highlighted in red or magenta correlates with either NADH or NADPH preference, respectively; the regions highlighted in orange or yellow correlate with FMN or FAD preference, respectively, and the regions highlighted in green and cyan highlight the (S/T/C)XXPP and GDH motifs, respectively. The yellow region corresponds to the “flexible AMP binding loop”. The alignment was created with RaptorX,59 edited by hand, and rendered with ESPript 3.0.60 (B) Structural comparison of SgcE6 and other HpaC-like flavin reductases. The labels indicate the preferred substrates. The bound ligands are displayed as black balls and sticks, with key hydrogen bonds shown as black dashes. The structures of SgcE6, TtHpaC, and TtFMNbp are from PDB entries 4R82, 2ED4, and 3ZOE, respectively. Further comparisons are shown in Figure S3. (C) σ-A-weighted difference (mFo – DFc) omit map contoured at 3σ with the positive and negative features shown as green and red mesh, respectively. The modeled FMN and AMP moieties are accurately modeled.
Overall Structure Establishing SgcE6 as a Member of the HpaC-like Subfamily of Flavin Reductases
As previously mentioned, bioinformatics analyses revealed that SgcE6 is a member of the HpaC-like flavin reductase subfamily as determined by conserved (S/T/C)XXPP and GDH sequence motifs. Although the sequence identities between SgcE6 and the other HpaC-like flavin reductases are low and vary from 16 to 34%, they all share similar three-dimensional structures with root-mean-square deviations (rmsds) of 0.52–1.24 Å for Cα atom superposition. These SgcE6 homologues include HpaC from T. thermophilus HB8 (TtHpaC),16 PheA2 from Geobacillus thermoglucosidasius (GtPheA2),18 NTA-MoB from Mycobacterium thermoresistible (MtMoB),49 TftC from Burkholderia cepacia AC1100 (BcTftC),19 SMOB from Pseudomonas putida S12 (PpSMOB),50 HpaC from Sulfolobus tokodaii strain 7 (StHpaC),51 FMN binding protein form Pyrococcus horikoshii (PhFMNbp),52T. thermophilus (TtFMNbp),52 and Methanothermobacter thermautotrophicus (MtFMNbp),53 and FeR from Archaeoglobus fulgidus (AfFeR)54 (Figure 2 and Figure S5). Because of the low degrees of similarity between many of these members, we generated a structure-based alignment that captures nonconserved structural elements and regions where residues overlap in space rather than based on sequence alone (Figure 2 and Figure S5). Important regions will be discussed in detail below in reference to substrate binding.
All of the HpaC-like flavin reductases are homodimers, with monomers related by 2-fold rotational axes. Not surprisingly, in the crystal structures of apo-SgcE6 and SgcE6-FAD, each asymmetric unit contains dimers, which were confirmed by size-exclusion chromatography (Figure S6A). PISA analysis55 of SgcE6 reveals that the dimerization interface is stabilized by many hydrogen bonds and electrostatic interactions and buries ∼2770 Å2 of accessible surface area (ASA). In addition, β-sheets 4 and 5 and a loop containing the (S/T/C)XXPP motif are swapped between dimers and make up portions of the active site, making the HpaC-like flavin reductases obligate homodimers.
Flavin Binding and Preference in SgcE6 and Homologues
Previous characterization of SgcE6 revealed a preference for FAD over FMN and revealed that there were no interactions necessary for FADH2 transfer to SgcC or SgcC3. The presence of FMN in the active site of SgcE6-FAD reveals how the flavin ring and ribose are bound. However, because of degradation of FAD during crystallization, how SgcE6 differentiates between FAD and FMN remains unknown. Nevertheless, insight can be gained through comparison of SgcE6-FAD with homologues and apo-SgcE6.
Superposition of the SgcE6 monomers reveals significant conformational differences in the “flexible loop” near the flavin binding site, which is flanked by α-helix 3 and β-sheet 7 (Figure 2 and Figures S4 and S5). The differences in conformation depend greatly on crystal contacts with the symmetry mates. Comparison of SgcE6 with the other HpaC-like flavin reductases reveals that for members preferring FAD [TtHpaC, BtPheA2, BcTftC, and PpSMOB (Table S2)], the flavin rings and ribose are bound in identical conformations; however, the AMP portion of FAD is bound in various conformations (Figure S5). We did not try to model the AMP moiety due to the conformational heterogeneity of the nonconserved “flexible loop” and because the conformation of AMP differs among homologues. SgcE6 homologues that prefer FMN [StHpaC, TtFMNbp, PhFMNbp, MtFMNbp, and AfFeR (Table S2)] also bind the flavin rings and ribose in an identical location; however, the “flexible loop” and more specifically α-helix 3 are extended into places where the AMP moieties would bind. Thus, it has been suggested that longer “flexible loops” correlate with FMN preference; however, SgcE6 has an extended “flexible loop” yet strictly binds FAD. Rather, the length of α-helix 3 and residues emanating from β-sheet 3 correlate with flavin preference (Figure 2).
Reductant Preference of SgcE6 and Homologues
The previous characterization of SgcE6 revealed it had a preference for NADH as the reductant. Four regions interact with the NAD(P)H reductant, α-helix 1, β-sheets 11 and 12, and the (S/T/C)XXPP and GDH sequence motifs (Figure 2 and Figure S5). In most structures with NAD+ bound (TtHpaC, BtPheA2, BcTftC, and StHpaC), it is found in a “U” conformation with the nicotinamide ring sandwiched between the isoalloxazine rings of the flavin and the adenine ring (Figure 2C and Figure S5). The binding of NADPH remains a mystery for most of the members preferring it, but a single-residue insertion in β-sheet 11 and shortened α-helix 1 distinguishes TtFMNbp, PhFMNbp, and MtFMNbp from NADH-utilizing members. As an outlier, AfFeR has a bound NADP+ moiety, where the adenine ring is bound by an α-helix in place of β-sheets 11 and 12. Nevertheless, binding of NADPH to TtFMNbp, PhFMNbp, and MtFMNbp may be similar to that of AfFeR. The two AMP moieties bound in the active site of SgcE6, and comparison with TtHpaC/BtPheA2/BcTftC/StHpaC structures reveals that the “U” conformation binding mode of NADH is identical.
Structure Solution and Refinement of SgcC
SgcC crystallized in orthorhombic space group P212121. The anomalous signal from the SeMets was weak; thus, the structure was determined by molecular replacement. The structure of TtHpaB (PDB entry 2YYK) was used as the search model as it was the most similar in the PDB (identity and similarity scores of 34 and 52%, respectively) (Figure 3). Four copies of the polypeptide chain were found and built in the asymmetric unit. The chains could be reliably traced between residues 18–167 and 178–526, with the region between residues 168 and 177 lacking significant density for three of the monomers. Two Ca2+ ions were built into the model based on Fo – Fc regular and anomalous electron density maps, even though a mixture of Mg2+ and Ca2+ was included in the crystallization condition. The Ca2+ ions fit the following criteria better than Mg2+ ions based on the refinement of occupancy, B factor, coordination number, and distance. The asymmetric unit also contained one molecule of glycerol. The crystal structure of SgcC was refined to a resolution of 2.63 Å with an R factor of 21.6% and an Rfree factor of 24.6%. Data processing and refinement statistics are summarized in Table 1.
Figure 3.
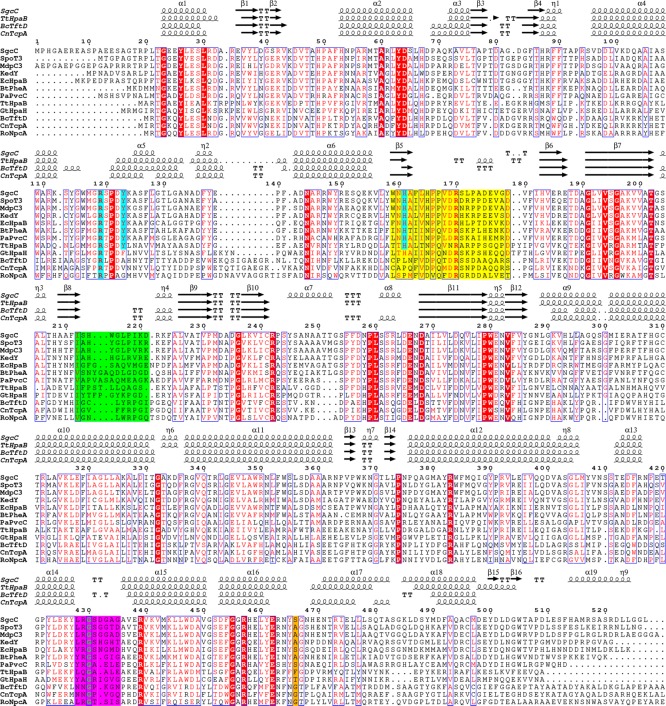
Structure-based sequence alignment of SgcC and homologues. According to the crystal structures of TtHpaB, the regions colored yellow (flavin binding loop), green (substrate binding loop), and magenta (AMP responsive loop) change conformation upon FAD binding, and the region colored green further changes conformation upon substrate binding. The residues highlighted in cyan are involved in binding the phenolic hydroxyl group of their substrates. The residue highlighted in orange differs from TtHpaB to the others, which likely opens the active site.
Overall Structure Establishing SgcC as a Member of the ACAD-Fold Flavin-Dependent Monooxygenases
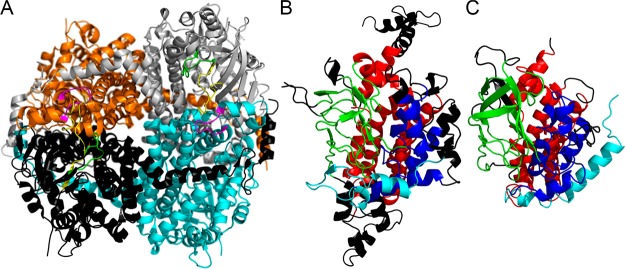
The crystal structure of SgcC was found to be packed as a tetramer in the asymmetric unit (Figure 4A), which was confirmed in solution by size-exclusion chromatography (Figure S6B). PISA analysis revealed that the tetrameric interface is formed by several tight hydrogen bonds and salt bridges and buries ∼8395 Å2 of ASA. Dali alignments56 reveal that the SgcC monomer has a core structure with an ACAD fold as judged by comparison with mammalian mitochondrial medium chain ACAD with a rmsd of 3.2 Å for 312 Cα atoms (Figure 4B,C). ACADs are also tetrameric. SgcC, EcHpaB, and homologues are much longer than ACADs by at least 100 residues. The extra residues are found in many loops around the structure; however, the tetramerization interfaces are relatively conserved. ACAD and SgcC are comprised of three domains, an N-terminal α-helical domain, a core domain containing a β-barrel-like structure, and a C-terminal domain containing a group of α-helices. The N-terminal α-helix 1 and β-sheets 1 and 2 of SgcC are found in a conformation completely different from that of ACADs, and the C-terminal α-helices 18 and 19 and β-sheets 15 and 16 of SgcC are lacking from ACADs. As mentioned previously, flavin-dependent monooxygenases with ACAD folds fall into two types, those similar to EcHpaB/TtHpaB or AbHpaH. AbHpaH is very similar to ACADs with very little deviation in structure, while SgcC is more similar to EcHpaB and TtHpaB.
Figure 4.
Overall structure of SgcC and comparison with acyl-CoA dehydrogenase (PDB entry 3MDD). (A) The tetramer of SgcC is colored as follows: monomer A in gray, monomer B in cyan, monomer C in black, and monomer D in orange. The active site loops “flavin binding loop”, “AMP responsive loop”, and “substrate binding loop” are colored as in Figure 3. (B) A monomer from SgcC is colored by homology to acyl-CoA dehydrogenase as seen in panel C. The N-terminal α-helices and loops are in completely different orientations and colored cyan. The rest of the N-terminal α-helical bundle is colored blue, the β-barrel green, and the C-terminal α-helical bundle red. The black regions are extended regions not found in acyl-CoA dehydrogenases when compared to SgcC and regions that are not homologous in structure and deviate by ≥5 Å.
Other structurally characterized ACAD-fold enzymes that are less similar to SgcC include AnaB, a prolyl-ACP dehydrogenase that acts on a carrier protein-tethered substrate; FkbI, a dehydrogenase proposed to act on an acyl carrier protein (ACP)-tethered aldehyde to generate hydroxylmalonyl-S-ACP; and KijD3 and EvdC, which are amino sugar oxidizing enzymes. Interestingly, all these enzymes function in natural product biosynthetic pathways.
Structural and Sequence Comparisons of SgcC with EcHpaB/TtHpaB-Type Monooxygenases Reveal the Binding Site for Substrates
Only three structures in the PDB are highly similar to that of SgcC. These include the structures of TtHpaB,17 2,4,5-trichlorophenol 4-monooxygenase from B. cepacia AC1100 (BcTftD),19 and 2,4,6-trichlorophenol 4-monooxygenase from Cupriacidus necator JMP134 (CnTcpA).26 Despite the low levels of sequence identity among them (20–29%), they all share nearly identical structures (Figure 3). SgcC shares high to low levels of sequence similarity with various TtHpaB-type monooxygenases that act on phenolic substrates and afford ortho hydroxylated products and a lower level of sequence similarity with monooxygenases that act on highly halogenated or nonphenolic aromatic substrates (Table S3, Figure 3, and Figures S7 and S8). Structural comparisons of apo-SgcC and FAD/4HPA-bound TtHpaB reveal the substrate binding residues Arg119, Tyr123, His161, and Thr202 of SgcC are conserved (numbering throughout will be based on the SgcC sequence and alignment in Figure 3), along with the tertiary structure of most of the residues surrounding the active site.
The Structure of SgcC Is Most Similar to That of Apo-TtHpaB, Revealing That Conformational Changes Are Necessary for FAD and Substrate Binding
TtHpaB was crystallized in three states, (i) in the absence of substrates as an apo form, (ii) with FAD, and (iii) with FAD and 4HPA.17 Three loops in the active site undergo changes in conformation from apo to substrate-bound and consist of a “flavin binding loop”, “AMP responsive loop”, and “substrate binding loop” (Figures 4A, 5, and S9). The loop between residues 159 and 179, “flavin binding loop”, carries His161 and Phe163, and the backbone of these residues moves to form hydrogen bonds with the flavin ring (Figure S9). In addition, an “AMP responsive loop” emanating from an adjacent monomer comprised of residues 428–435 also undergoes a change in conformation to interact with the “flavin binding loop”, which orders the conserved Arg170 to interact with the FAD diphosphate moiety. A “substrate binding loop” that interacts with 4HPA consists of residues 212–221, the sequence of which is highly dependent on substrate, undergoes an order to disorder transition upon FAD binding, and again becomes ordered upon 4HPA binding. The “flavin binding loop” and “substrate binding loop” of SgcC have the largest deviations between the monomers, and the “flavin binding loop” contains disordered residues (Figure 5A). A comparison of SgcC to each of the TtHpaB structures reveals that the structure of SgcC is most similar to that of apo-TtHpaB; thus, conformational changes are necessary for FAD and substrate binding from the current SgcC structure (Figure S9). As conformational changes are necessary, we avoided modeling to further understand FAD binding.
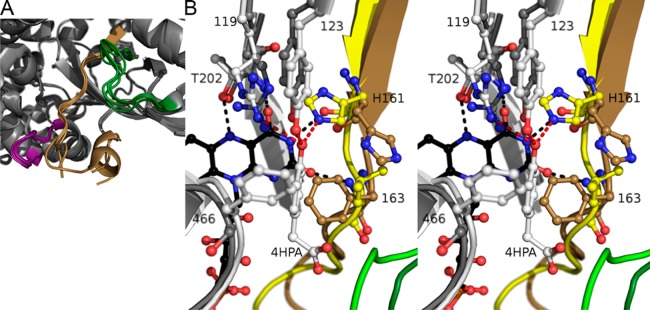
Figure 5.
Comparison of SgcC monomers and SgcC with TtHpaB. (A) Comparison of the SgcC monomers, with the “flavin binding loop” colored tan, the “AMP responsive loop” purple, and the “substrate binding loop” dark green. This highlights the comparative disorder found in the active site loops compared to the surrounding regions. (B) Stereoimage of an overlay of the active site of SgcC shown in gray and dark colors as in panel A, and TtHpaB in white and light colors with FAD in black and the 4HPA substrate bound. Hydrogen bonds and interactions with 4HPA are colored red and those with FAD black. This reveals the conformational changes required for SgcC to bind FAD and substrate; namely, the loop containing His161 and Phe163 needs to rearrange, and the side chains need to rotate. In addition, the catalytic Arg119 side chain would need to be repositioned. This figure also reveals the stacking of the TtHpaB Phe at position 466 over the 4HPA substrate, which would preclude a modification at position C-3; however, in SgcC, this residue is a serine, which makes plenty of space for a modification.
Putative Binding Mode of the 3-Chloro-β-tyrosyl Moiety of the SgcC Substrate
To gain some insight into FAD and substrate binding, we compared the structure of SgcC with that of TtHpaB substrate complex (Figure 5B). The major differences in substrate binding between the 3-chloro-β-tyrosyl moiety and 4HPA are the presence of the 3-chloro group and an extended chain in place of the acetic acid portion of 4HPA; however, both substrates contain a p-phenol. In the structure of TtHpaB, the 4HPA phenol is bound by Arg119, Tyr123, His161, and hydrophobic contacts are made with a phenylalanine and leucine at positions 466 and 163, respectively. For the FAD and substrate to bind, the backbone and side chain of His161 and Phe163 and the side chain of the conserved Arg119 rearrange. Specificity for a phenol and accommodation of the 3-chloro and pantetheine arm will be discussed below.
For SgcC homologues acting on phenol-bearing substrates, Tyr123 and His161 are conserved. However, these residues are replaced by residues incapable of hydrogen bonding in GtHpaH, which acts on anthranilate as a substrate, as well as the nitrophenol- and chlorophenol-degrading homologues.57 Furthermore, comparison of the active sites of AbHpaH with TtHpaH reveals that FAD is bound almost identically, but Tyr123 is replaced with a histidine and His161 with a serine (Figure S10). The histidine and serine bind the common 4HPA substrate phenol such that position 3 is within 5 Å of the C-4a atom of the flavin, which upon formation of the C-4a peroxyflavin intermediate would be close enough for reaction with the peroxide. This suggests that p-hydroxy-bearing substrates are bound similarly within both AbHpaH and EcHpaB/TtHpaB families.
If a 3-chloro-β-tyrosyl moiety is superimposed on the ring of 4HPA, the chloro group would clash with the phenylalanine in position 466 of TtHpaB; however, this residue is a serine in SgcC and either a glycine, alanine, or serine in homologues (Figure 3). In the structure of SgcC, there does not appear to be residues that would fill this space, allowing for the halogenated substrate to bind. This suggests that the 3-chloro-β-tyrosyl moiety of the SgcC substrate is bound in a manner identical to that of the 4HPA substrate of TtHpaB, with the halide facing away from the flavin. The acetic acid moiety of 4HPA interacts specifically with the “substrate binding loop”, and this loop is not conserved between TtHpaB and SgcC. However, this loop is conserved between the SgcC homologues proposed to act on PCP-tethered substrates (Figure S3), suggesting that there is a common mode to accommodate the pantetheine and thioester moieties of their substrates.
Implications for the Proposed Substrate Binding Model
SgcC efficiently catalyzes the regioselective hydroxylation of (S)-3-substituted β-tyrosyl-S-SgcC2 analogues, including F-, Cl-, Br-, I-, and Me-substituted derivatives. The reactivity order of these analogues is as follows: I > Br > Cl > Me > F > H.10 The reactivity trend indicates, in general, that higher electronegativity decreases the hydroxylation activity, with the exception of methyl and hydrogen. Highly electronegative functional groups such as fluorine limit the π-electrons from attacking the hydroxy group of the hydroperoxyflavin intermediate (FADHOOH). The electronegativity of the 3-substitutent therefore likely plays a role in catalysis larger than that of specific interactions with active site residues. The apparent lack of activity for the hydrogen-bearing substrate is perplexing in light of the activity of homologues for 4HPA. This suggests that changing Ser466 and other active site residues could increase the activity of SgcC for the non-3-substituted substrate to increase yields of 20′-deschloro-C-1027, should SgcC activity become limiting in engineering efforts.
Putative SgcC2–SgcC Binding Interface
SgcC specifically acts on its SgcC2-tethered substrate, as the free substrate is not recognized, implying an interaction between SgcC and SgcC2. Comparison of SgcC to the ACADs is not useful for gaining insight into the binding of the shared phosphopantetheine moieties, as the structures are not homologous in the regions around the “substrate binding loop”. Similarly, comparisons with the structural homologues AnaB and FkbI did not provide insight. Therefore, SgcC–SgcC2 docking models were used to predict the interaction interface (Figure S11). The majority of the top-ranked models reveals that the phosphopantetheine attachment point, Ser31, of SgcC2 points toward the active site of SgcC (Figure S11A). According to this SgcC–SgcC2 docking model, SgcC2 is bound at the center of the SgcC tetramer, contacting each of the subunits, with an area of 985 Å (Figure S11B), and detailed interactions are shown in Figure S11C. While this docking experiment seems reasonable, it was, however, performed using an SgcC tetramer as the protein acceptor and an SgcC2 monomer as the protein donor and may not reflect the true binding mode of the SgcC tetramer, which most likely interacts with multiple molecules of SgcC2.
Modifications catalyzed by tailoring enzymes acting on PCP-tethered substrates are known, including cyclization, halogenation, methylation, oxidation, and reduction. In the biosynthesis of C-1027, SgcC3 and SgcC catalyze chlorination and hydroxylation of SgcC2-tethered (S)-β-tyrosine, respectively. By covalently tethering, PCPs sequester the substrates from endogenous metabolite pools, thereby increasing their effective concentrations at the active sites for catalysis.58 In addition, PCP-tethered substrates also prevent promiscuous activity from occurring on free substrates.
Proposed Overall Reaction Scheme of SgcE6, SgcC, and SgcC2
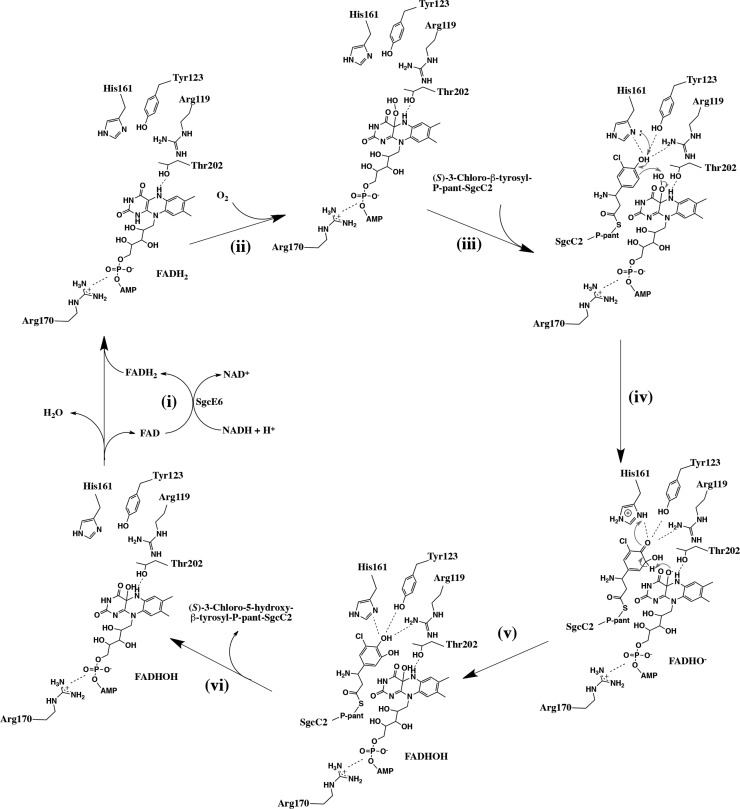
The E. coli flavin reductase (Fre) could be substituted for SgcE6 without impacting the activity of SgcC, implying that SgcC produces FADH2 from SgcE6 by diffusion rather than delivery via a specific protein–protein interaction.11 The catalytic mechanisms of flavin-dependent monooxygenases that share a mechanism common to SgcC are well studied.17,27 According to the crystal structures of SgcE6 and SgcC and the docking models, a general mechanism could now be proposed as shown in Figure 6.
Figure 6.
Proposed catalytic mechanism of SgcC. (i) SgcE6 is responsible for the production of FADH2 at the expense of NADH. (ii) FADH2 diffuses to SgcC, where Arg119 activates an O2 molecule to form a C-4a-hydroperoxyflavin intermediate. (iii) SgcC2 forms protein–protein interactions with SgcC and guides the (S)-3-chloro-β-tyrosyl moiety into the active site. (iv) The (S)-3-chloro-β-tyrosyl moiety is activated by a conserved general base His161, which plays a key role in deprotonation of the 4-hydroxy group. The electrons delocalize into the aromatic ring system and undergo electrophilic attack of the hydroxy group of the C-4a-hydroperoxyflavin intermediate, introducing a hydroxy group at the C-5 position of the (S)-3-chloro-β-tyrosyl moiety. (v) The hydroxyflavin (FADHO–) abstracts a proton from the C-5 atom of the intermediate, resulting in the formation of the re-aromatizated (S)-3-chloro-5-hydroxy-β-tyrosyl-S-SgcC2, which then dissociates from SgcC. (vi) Finally, the hydroxyflavin (FADHOH) undergoes dehydration to form FAD, which is released and recycled by SgcE6.
Conclusion
In the last step of biosynthesis of the (S)-3-chloro-5-hydroxy-β-tyrosyl moiety of the C-1027 chromophore, ScgE6 and SgcC comprise a two-component monooxygenase that catalyzes the hydroxylation of (S)-3-chloro-β-tyrosine into (S)-3-chloro-5-hydroxy-β-tyrosine only upon tethering of the substrate to the PCP of SgcC2. In this study, we determined the crystal structures of SgcE6 and SgcC, to investigate the substrate selectivity and the enzyme catalytic mechanism. The crystal structures show that SgcE6 possesses a flexible loop, which provides sufficient space to accommodate FAD. This structural feature is consistent with our previous studies that showed SgcE6 specifically reacts with FAD but not FMN. The crystal structure of SgcC is highly homologous to that of TtHpaB, providing insight into substrate binding and catalysis. Finally, the crystal structures of SgcE6 and SgcC give new insight into the intriguing group of carrier protein-dependent two-component monooxygenases in a three-protein system and could be explored and exploited to engineer C-1027 diversity by structure-based enzyme and biosynthetic pathway engineering.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00713.
Primers and vectors used in this study (Table S1), homologues of SgcE6 discussed in this study (Table S2), homologues of SgcC discussed in this study (Table S3), the top 10 threading templates used by I-TASSER for SgcC2 modeling (Figure S1), sequence alignment of SgcE6 with other nine-membered enediyne gene cluster homologues (Figure S2), sequence alignment of SgcC with other nine-membered enediyne gene cluster homologues (Figure S3), stereoimage of the SgcE6 monomer alignment (Figure S4), structural comparison of SgcE6 with other flavin reductases (Figure S5), molecular weight estimation of SgcE6 and SgcC by size-exclusion chromatography (Figure S6), reactions performed by SgcC homologous enzymes (Figure S7), structure-based sequence alignment identity matrix (Figure S8), comparison of SgcC and TtHpaB in stereograms (Figure S9), active site of AbHpaH (Figure S10), and the SgcC–SgcC2 docking model (Figure S11) (PDF)
Author Contributions
C.-Y.C. and J.R.L. contributed equally to this work.
This work is supported in part by National Institutes of Health Grants GM098248 (G.N.P.), GM109456 (G.N.P.), GM094585 (A.J.), and CA078747 (B.S.). The use of Structural Biology Center beamlines at the Advanced Photon Source was supported in part by the U.S. Department of Energy, Office of Biological and Environmental Research, Grant DE-AC02-06CH11357 (M.E.C. and A.J.). C.-Y.C. and J.D.R. are supported in part by postdoctoral fellowships from Academia Sinica-The Scripps Research Institute Postdoctoral Talent Development Program and the Arnold and Mabel Beckman Foundation, respectively. We also thank Shonda Clancy for help with gene cloning.
The authors declare no competing financial interest.
Supplementary Material
References
- Liu W.; Christenson S. D.; Standage S.; Shen B. (2002) Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297, 1170–1173. 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- Galm U.; Hager M. H.; Van Lanen S. G.; Ju J.; Thorson J. S.; Shen B. (2005) Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem. Rev. 105, 739–758. 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- Kennedy D. R.; Ju J.; Shen B.; Beerman T. A. (2007) Designer enediynes generate DNA breaks, interstrand cross-links, or both, with concomitant changes in the regulation of DNA damage responses. Proc. Natl. Acad. Sci. U. S. A. 104, 17632–17637. 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman T. A.; Gawron L. S.; Shen B.; Kennedy D. R. (2014) The radiomimetic enediyne, 20′-deschloro-C-1027 induces inter-strand DNA crosslinks in hypoxic cells and overcomes cytotoxic radioresistance. DNA Repair 21, 165–170. 10.1016/j.dnarep.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D. R.; Gawron L. S.; Ju J.; Liu W.; Shen B.; Beerman T. A. (2007) Single chemical modifications of the C-1027 enediyne core, a radiomimetic antitumor drug, affect both drug potency and the role of ataxia-telangiectasia mutated in cellular responses to DNA double-strand breaks. Cancer Res. 67, 773–781. 10.1158/0008-5472.CAN-06-2893. [DOI] [PubMed] [Google Scholar]
- Van Lanen S. G.; Lin S.; Dorrestein P. C.; Kelleher N. L.; Shen B. (2006) Substrate specificity of the adenylation enzyme SgcC1 involved in the biosynthesis of the enediyne antitumor antibiotic C-1027. J. Biol. Chem. 281, 29633–29640. 10.1074/jbc.M605887200. [DOI] [PubMed] [Google Scholar]
- Christianson C. V.; Montavon T. J.; Van Lanen S. G.; Shen B.; Bruner S. D. (2007) The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry 46, 7205–7214. 10.1021/bi7003685. [DOI] [PubMed] [Google Scholar]
- Van Lanen S. G.; Dorrestein P. C.; Christenson S. D.; Liu W.; Ju J.; Kelleher N. L.; Shen B. (2005) Biosynthesis of the beta-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring beta-amino acyl-S-carrier protein intermediates. J. Am. Chem. Soc. 127, 11594–11595. 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- Lin S.; Van Lanen S. G.; Shen B. (2007) Regiospecific chlorination of (S)-beta-tyrosyl-S-carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027. J. Am. Chem. Soc. 129, 12432–12438. 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Van Lanen S. G.; Shen B. (2008) Characterization of the two-component, FAD-dependent monooxygenase SgcC that requires carrier protein-tethered substrates for the biosynthesis of the enediyne antitumor antibiotic C-1027. J. Am. Chem. Soc. 130, 6616–6623. 10.1021/ja710601d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen S. G.; Lin S.; Horsman G. P.; Shen B. (2009) Characterization of SgcE6, the flavin reductase component supporting FAD-dependent halogenation and hydroxylation in the biosynthesis of the enediyne antitumor antibiotic C-1027. FEMS Microbiol. Lett. 300, 237–241. 10.1111/j.1574-6968.2009.01802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich R.; Hofrichter M. (2007) Enzymatic hydroxylation of aromatic compounds. Cell. Mol. Life Sci. 64, 271–293. 10.1007/s00018-007-6362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou D. P.; Entsch B.; Cole L. J. (2005) Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem. Biophys. Res. Commun. 338, 590–598. 10.1016/j.bbrc.2005.09.081. [DOI] [PubMed] [Google Scholar]
- van Berkel W. J.; Kamerbeek N. M.; Fraaije M. W. (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689. 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Ziegler D. M. (2002) An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev. 34, 503–511. 10.1081/DMR-120005650. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Hisano T.; Iwasaki W.; Ebihara A.; Miki K. (2008) Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity. Proteins: Struct., Funct., Genet. 70, 718–730. 10.1002/prot.21534. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Hisano T.; Takeda K.; Iwasaki W.; Ebihara A.; Miki K. (2007) Crystal structure of the oxygenase component (HpaB) of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8. J. Biol. Chem. 282, 33107–33117. 10.1074/jbc.M703440200. [DOI] [PubMed] [Google Scholar]
- van den Heuvel R. H.; Westphal A. H.; Heck A. J.; Walsh M. A.; Rovida S.; van Berkel W. J.; Mattevi A. (2004) Structural studies on flavin reductase PheA2 reveal binding of NAD in an unusual folded conformation and support novel mechanism of action. J. Biol. Chem. 279, 12860–12867. 10.1074/jbc.M313765200. [DOI] [PubMed] [Google Scholar]
- Webb B. N.; Ballinger J. W.; Kim E.; Belchik S. M.; Lam K. S.; Youn B.; Nissen M. S.; Xun L.; Kang C. (2010) Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Biol. Chem. 285, 2014–2027. 10.1074/jbc.M109.056135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan B.; Diaz E.; Prieto M. A.; Garcia J. L. (2000) Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new Flavin:NAD(P)H reductase subfamily. J. Bacteriol. 182, 627–636. 10.1128/JB.182.3.627-636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge D. R.; Corbett M. D. (1984) Primary arylamine oxidation by a flavinhydroperoxide. A study of the basis for the substrate specificity of the flavoprotein monooxygenase. Biochem. Pharmacol. 33, 3615–3619. 10.1016/0006-2952(84)90146-1. [DOI] [PubMed] [Google Scholar]
- Huijbers M. M.; Montersino S.; Westphal A. H.; Tischler D.; van Berkel W. J. (2014) Flavin dependent monooxygenases. Arch. Biochem. Biophys. 544, 2–17. 10.1016/j.abb.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Sucharitakul J.; Tinikul R.; Chaiyen P. (2014) Mechanisms of reduced flavin transfer in the two-component flavin-dependent monooxygenases. Arch. Biochem. Biophys. 555–556, 33–46. 10.1016/j.abb.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Xun L.; Sandvik E. R. (2000) Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 66, 481–486. 10.1128/AEM.66.2.481-486.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S.; Ortiz-Maldonado M.; Entsch B.; Ballou D. P. (2010) Studies on the mechanism of p-hydroxyphenylacetate 3-hydroxylase from Pseudomonas aeruginosa: a system composed of a small flavin reductase and a large flavin-dependent oxygenase. Biochemistry 49, 372–385. 10.1021/bi901454u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes R. P.; Webb B. N.; Subramanian A. K.; Nissen M.; Popchock A.; Xun L.; Kang C. (2012) Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134. Int. J. Mol. Sci. 13, 9769–9784. 10.3390/ijms13089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri A.; Fersini F.; Ruangchan N.; Prongjit M.; Chaiyen P.; Mattevi A. (2007) Structure of the monooxygenase component of a two-component flavoprotein monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 104, 1177–1182. 10.1073/pnas.0608381104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. L.; Palfey B. A.; Lah M. S.; Entsch B.; Massey V.; Ballou D. P.; Ludwig M. L. (1994) The mobile flavin of 4-OH benzoate hydroxylase. Science 266, 110–114. 10.1126/science.7939628. [DOI] [PubMed] [Google Scholar]
- Schreuder H. A.; Mattevi A.; Obmolova G.; Kalk K. H.; Hol W. G.; van der Bolt F. J.; van Berkel W. J. (1994) Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring. Biochemistry 33, 10161–10170. 10.1021/bi00199a044. [DOI] [PubMed] [Google Scholar]
- Li Y.; Llewellyn N. M.; Giri R.; Huang F.; Spencer J. B. (2005) Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem. Biol. 12, 665–675. 10.1016/j.chembiol.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Lohman J. R.; Huang S. X.; Horsman G. P.; Dilfer P. E.; Huang T.; Chen Y.; Wendt-Pienkowski E.; Shen B. (2013) Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics. Mol. BioSyst. 9, 478–491. 10.1039/c3mb25523a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen S. G.; Oh T. J.; Liu W.; Wendt-Pienkowski E.; Shen B. (2007) Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J. Am. Chem. Soc. 129, 13082–13094. 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey R. P.; Nett M.; Moore B. S. (2008) Unraveling the biosynthesis of the sporolide cyclohexenone building block. J. Am. Chem. Soc. 130, 2406–2407. 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- Eschenfeldt W. H.; Lucy S.; Millard C. S.; Joachimiak A.; Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115. 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. I.; Zhou M.; Millard C. S.; Clancy S.; Stols L.; Eschenfeldt W. H.; Collart F. R.; Joachimiak A. (2006) An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expression Purif. 47, 446–454. 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C.; von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326. 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Minor W.; Cymborowski M.; Otwinowski Z.; Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 62, 859–866. 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman J. R.; Bingman C. A.; Phillips G. N. Jr.; Shen B. (2013) Structure of the bifunctional acyltransferase/decarboxylase LnmK from the leinamycin biosynthetic pathway revealing novel activity for a double-hot-dog fold. Biochemistry 52, 902–911. 10.1021/bi301652y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenath H. K.; Bingman C. A.; Buchan B. W.; Seder K. D.; Burns B. T.; Geetha H. V.; Jeon W. B.; Vojtik F. C.; Aceti D. J.; Frederick R. O.; Phillips G. N. Jr.; Fox B. G. (2005) Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expression Purif. 40, 256–267. 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010) XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C.; Grosse-Kunstleve R. W.; Afonine P. V.; Moriarty N. W.; Zwart P. H.; Hung L. W.; Read R. J.; Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr., Sect. D: Biol. Crystallogr. 64, 61–69. 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinf. 9, 40. 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau S. R.; Gatchell D. W.; Vajda S.; Camacho C. J. (2004) ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50. 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- Lyskov S.; Gray J. J. (2008) The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 36, W233–238. 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J. D.; Yan X.; Shen B. (2016) Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J. Ind. Microbiol. Biotechnol. 43, 261–276. 10.1007/s10295-015-1671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Edwards T. E.; Begley D. W.; Abramov A.; Thompkins K. B.; Ferrell M.; Guo W. J.; Phan I.; Olsen C.; Napuli A.; Sankaran B.; Stacy R.; Van Voorhis W. C.; Stewart L. J.; Myler P. J. (2011) Structure of nitrilotriacetate monooxygenase component B from Mycobacterium thermoresistibile. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 67, 1100–1105. 10.1107/S1744309111012541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E.; Kantz A.; Gassner G. T.; Sazinsky M. H. (2013) Structure and mechanism of styrene monooxygenase reductase: new insight into the FAD-transfer reaction. Biochemistry 52, 6063–6075. 10.1021/bi400763h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okai M.; Kudo N.; Lee W. C.; Kamo M.; Nagata K.; Tanokura M. (2006) Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(−)free, NAD(+)(−)bound, and NADP(+)(−)bound. Biochemistry 45, 5103–5110. 10.1021/bi052313i. [DOI] [PubMed] [Google Scholar]
- Steinkellner G.; Gruber C. C.; Pavkov-Keller T.; Binter A.; Steiner K.; Winkler C.; Lyskowski A.; Schwamberger O.; Oberer M.; Schwab H.; Faber K.; Macheroux P.; Gruber K. (2014) Identification of promiscuous ene-reductase activity by mining structural databases using active site constellations. Nat. Commun. 5, 4150. 10.1038/ncomms5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christendat D.; Yee A.; Dharamsi A.; Kluger Y.; Savchenko A.; Cort J. R.; Booth V.; Mackereth C. D.; Saridakis V.; Ekiel I.; Kozlov G.; Maxwell K. L.; Wu N.; McIntosh L. P.; Gehring K.; Kennedy M. A.; Davidson A. R.; Pai E. F.; Gerstein M.; Edwards A. M.; Arrowsmith C. H. (2000) Structural proteomics of an archaeon. Nat. Struct. Biol. 7, 903–909. 10.1038/82823. [DOI] [PubMed] [Google Scholar]
- Chiu H. J.; Johnson E.; Schroder I.; Rees D. C. (2001) Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+. Structure 9, 311–319. 10.1016/S0969-2126(01)00589-5. [DOI] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Holm L.; Rosenstrom P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549. 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Dong Y.; Li X.; Ren Y.; Li Y.; Wang W.; Wang L.; Feng L. (2010) Characterization of the anthranilate degradation pathway in Geobacillus thermodenitrificans NG80–2. Microbiology 156, 589–595. 10.1099/mic.0.031880-0. [DOI] [PubMed] [Google Scholar]
- Lin S.; Huang T.; Shen B. (2012) Tailoring enzymes acting on carrier protein-tethered substrates in natural product biosynthesis. Methods Enzymol. 516, 321–343. 10.1016/B978-0-12-394291-3.00008-3. [DOI] [PubMed] [Google Scholar]
- Wang S.; Peng J.; Xu J. (2011) Alignment of distantly related protein structures: algorithm, bound and implications to homology modeling. Bioinformatics 27, 2537–2545. 10.1093/bioinformatics/btr432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P.; Courcelle E.; Stuart D. I.; Metoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308. 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.