Abstract
Peroxisomes are single membrane-bound organelles, whose basic enzymatic constituents are catalase and H2O2-producing flavin oxidases. Previous reports showed that peroxisome is involved in numerous processes including primary and secondary metabolism, plant development and abiotic stress responses. However, knowledge on the function of different peroxisome genes from rice and its regulatory roles in salt and other abiotic stresses is limited. Here, a novel prey protein, OsPEX11 (Os03g0302000), was screened and identified by yeast two-hybrid and GST pull-down assays. Phenotypic analysis of OsPEX11 overexpression seedlings demonstrated that they had better tolerance to salt stress than wild type (WT) and OsPEX11-RNAi seedlings. Compared with WT and OsPEX11-RNAi seedlings, overexpression of OsPEX11 had lower level of lipid peroxidation, Na+/K+ ratio, higher activities of antioxidant enzymes (SOD, POD, and CAT) and proline accumulation. Furthermore, qPCR data suggested that OsPEX11 acted as a positive regulator of salt tolerance by reinforcing the expression of several well-known rice transporters (OsHKT2;1, OsHKT1;5, OsLti6a, OsLti6b, OsSOS1, OsNHX1, and OsAKT1) involved in Na+/K+ homeostasis in transgenic plants under salinity. Ultrastructural observations of OsPEX11-RNAi seedlings showed that they were less sensitive to salt stress than WT and overexpression lines. These results provide experimental evidence that OsPEX11 is an important gene implicated in Na+ and K+ regulation, and plays a critical role in salt stress tolerance by modulating the expression of cation transporters and antioxidant defense. Thus, OsPEX11 could be considered in transgenic breeding for improvement of salt stress tolerance in rice crop.
Keywords: Oryza sativa, protein interaction, OsPEX11, transgene, salt tolerance
Introduction
The gradual salinization of arable land is a serious constraint in the sustainable development of crop production. Among the cereals, rice is considered as one of the most salt sensitive crops (Zhu, 2001; Munns and Tester, 2008). With the development of biotechnology and genetic engineering, cloning and transferring of salt tolerant genes, could not only increase the utilization ratio of saline soils, but also supply a number of novel germplasm for breeding and sustainable food production (Geng et al., 2013; Dinneny, 2015). Thus, it is important to search and identify salt tolerant genes for successful development of high yielding cultivars for rice.
In the past decade, high-throughput transcriptomics and proteomics studies have provided immense data on gene function of plant salt tolerance (Zhuang et al., 2014). A few of important genes and/or proteins for osmolyte synthesis, ion channels and ROS scavenging enzymes have been founded from the previous studies (Parker et al., 2006; Jiang et al., 2007; Witzel et al., 2009; Kishor and Sreenivasulu, 2014), which have revealed the fundamental functions of the genes/proteins in crops’ response and adaptation to salinity. These bioinformatics data offer an integral view of salt-responsive genes in different plants. However, because of post-translational modifications and complication in salt response regulated networks, it is of utmost importance to elucidate exact biological function of these candidate genes/proteins under saline stress conditions.
Cyclophilins (CYPs), a class of highly conserved molecular chaperone, possessed peptidyl-prolyl cis-trans isomerase (PPIase) activity. These ubiquitous proteins are involved in a wide variety of biological processes such as protein assembly and transporting, RISC assembly and miRNA activity (Smith et al., 2009; Iki et al., 2012; Campos et al., 2013). In our previous report, we identified and characterized a rice cyclophilin (OsCYP2; Os02g0121300) that confers salt tolerance when overexpressed (Ruan et al., 2011). Until now, many cyclophilins genes have been discovered in Glycine max, Arabidopsis thaliana, and O. sativa, and they can be induced by a serial of abiotic stress, and heterologous expression confers toward multiple abiotic stresses (Mainali et al., 2014; Vasudevan et al., 2014; Lee et al., 2015).
Yeast two-hybrid is considered as a powerful tool to allow the identification of specific protein–protein interactions, which are more directly related to signal transduction processes under salt stress. However, the full list of peroxisomal proteins is not yet completely known (Dixit et al., 2010), which indicates the functions of peroxisome are still obscure. In this study, a peroxisomal biogenesis factor 11 family protein (OsPEX11; Os03g0302000) which interacted with OsCYP2 was screened by yeast two-hybrid assay in cDNA library of rice. Peroxisomal proteins are ubiquitous components of eukaryotic cells which are encoded by nuclear genes, synthesized on free cytosolic ribosomes and imported post-translationally (Gould and Valle, 2000; Nito et al., 2007; Kumar et al., 2014). They have been implicated in numerous metabolic processes, ranging from hydrogen peroxide metabolism to biosynthesis of lipids (Dixit et al., 2010; Pieuchot and Jedd, 2012; Odendall et al., 2014). Mutations of peroxisome biogenesis proteins in various eukaryotes result in serious developmental deficiencies and stress sensitivities (Aung and Hu, 2011; Burkhart et al., 2013; Cassin-Ross and Hu, 2014). They had been identified for involving in lipid catabolism, photorespiration and hormone biosynthesis in Arabidopsis (Nito et al., 2007). Our work is the first exploration on the biological functions of OsPEX11. The phenotype, physiological and expression level of candidate interacted protein (OsPEX11) were analyzed in overexpression and RNAi transgenic lines under salt stress. Therefore, these results will provide peroxisomal biogenesis factor mediated molecular and physiological responses of crop salt tolerance.
Materials and Methods
Construction of cDNA Library
Total RNA which was extracted from mix sample (leave, shoots, and roots) of 10-day-old wild type (WT) seedlings (O. sativa L. cv. Aichi-ashahi) was used to synthesize a cDNA library. The mRNA was purified by Dynabeads mRNA Purification Kit (Thermo Scientific, 61006). First and second strand synthesis and size fractionation were conducted according to the method of cDNA Library Construction Kit (Clontech, 634901) with minor modification. Then, cDNA library was directly cloned into the pGADT7AD vector encoding the GAL4 activation domain with EcoRI and XhoI. The size of inserted fragment was detected by using specific primers (Supplementary Table S1).
Yeast Two-Hybrid Assay
Yeast two-hybrid analysis was performed in accordance with the Matchmaker Gold Yeast Two-Hybrid Kit (Clontech, 630489). The coding region of OsCYP2 (519 bp) was amplified from rice leaves (O. sativa L. cv. Aichi-ashahi) by high-fidelity PCR, restricted and fused in-frame with GAL4 DNA binding domain into pGBKT7 for constructing bait vector. Then it was transformed into yeast strain Y2H through the lithium acetate method. After 3 days, auto-activation and toxicity assays were confirmed by SD/-Trp, SD/-Trp/X-α-gal, and SD/-Trp/X-α-gal/AbA selected plates. After that, the yeast two-hybrid screening between OsCYP2 and previous cDNA library was done according to the co-transformation protocol of Y2H strain. The candidate clones (blue) were selected by SD/-Trp/-Leu/X-α-gal/AbA plate. We patched out all the blue colonies that grew on SD/-Trp/-Leu/X-α-gal/AbA plate onto higher stringency SD/-Trp/-Leu/-His/-Ade/X-α-gal/AbA plate using yellow pipette tip. To increase the chance of rescuing the positive prey plasmid, we streaked 2–3 times for each selected single blue clone on SD/-Trp/-Leu/X-α-gal (no Aureobasidin A) plate. Then the candidate prey plasmid (blue clone) was rescued by using the Easy Yeast Plasmid Isolation Kit (Clontech, 630467) and sequenced with T7 primer. Co-transform BD or BD-OsCYP2 with rescued AD-prey plasmids into Y2H strains by small scale yeast transformation on selective media plates to distinguish positive interaction from false positive interaction.
SDS-PAGE and GST Pull-down Assays
The genuine positive was further confirmed by GST pull-down assays. The OsCYP2 and OsPEX11 were cloned into pGEX-4T-1 and pET-28a vectors, respectively, for expressing fusion protein with glutathione-S-transferase (GST) and histidine (His), in Escherichia coli strain BL21. The MagneHis Protein Purification System (Promega, V8500) and MagneGST Pull Down System (Promega, V8870) were used for fused protein purification and GST pull-down, respectively. The purified GST, GST-OsCYP2 and His-OsPEX11 proteins were analyzed with 12% SDS-PAGE and stained by coomassie brilliant blue R-250. Western blotting signals were detected by Horseradish Peroxidase (HRP) DAB (3, 3-diaminobenzidine) staining with either the His tag antibody (Genescript, A00612) or GST tag antibody (Genescript, A00130).
Plasmid Constructs and Plant Transformation
The full-length and partial cds of OsPEX11 gene were cloned into pCAMBIA1300-Ubi and pCAMBIA1300-35S-RNAi vectors, respectively. The primers which contained different restriction enzyme sites were listed in Supplementary Table S1. Then, both of two vectors were transformed into Agrobacterium tumefaciens strain EHA105. Plant transformation was conducted into O. sativa L. cv. Aichi-ashahias previously described with minor modification (Hiei et al., 1994).
Plant Growth and Quantitative RT-PCR
The seeds of WT, OsPEX11 overexpression and RNAi were germinated and hydroponically grown in a greenhouse. The culture was maintained at 32°C/28°C, with the photoperiod of 16 h/8 h (light/dark). After 10 days, seedlings of each genotype were treated by 200 mM NaCl for 24 h and their phenotypes were identified. Six plants per treatment were sampled for the measurement of plant height, root length and leaves angle. Average values of these six plants were considered as one replicate. To examine the mRNA expression pattern of OsPEX11 and seven crucial genes which code Na+ and K+ transport proteins, fresh leaves from each genotype were sampled for RNA isolation. Total RNA was extracted using Trizol (Thermo Scientific, 15596-026) according to the manufacturer’s instructions. Total RNA was used to synthesize the first strand cDNA with RevertAidTM First-Strand cDNA Synthesis kit (Thermo Scientific, K1622). Quantitative real time PCR reactions were performed using three biological replicates for each different genotype (three technical replicates as one biological replicate) on Bio-Rad CFX 96. Primers used in this experiment were listed in Supplementary Table S1. The expression level of OsPEX11 and other genes related to salt was calculated following Livak and Schmittgen (2001).
Determination of Lipid Peroxidation and Antioxidant Enzyme
Lipid peroxidation was determined by malondialdehyde (MDA) contents according to the method of Hodges et al. (1999). Fresh leaves (0.5 g) of 10-day-old seedlings were homogenized in 10 ml of precooled potassium phosphate buffer (pH 7.0) by grinding with a mortar and pestle in an ice bath. The mixture was centrifuged at 4°C for 20 min at 12000 rpm. The supernatants were immediately used for the determination of various antioxidant enzymes. SOD, POD, and CAT activity were performed according to Dhindsa and Matowe (1981), Aebi (1984), and Rao et al. (1996), respectively. Moreover, proline content was measured according to Bates et al. (1973). For the determination of sodium and potassium ions, leaves were dried and ground. About 0.1 g of the ground leaf was digested with H2SO4 and H2O2, and then sodium and potassium contents were analyzed through atomic absorption spectrophotometry (Munns et al., 2010).
Transmission Electron Microscopy
For electron-microscopic study, leaf fragments without veins (about 1 mm2) were fixed in 2.5% (v/v) glutaraldehyde in 0.1 M sodium phosphate buffer (PBS, pH 7.4) overnight and then washed three times with PBS. The samples were post fixed in 1% (m/v) OsO4 for 1 h and washed again three times with PBS. After that, the samples were dehydrated in a graded series of ethanol (50, 60, 70, 80, 90, 95, and 100%, v/v) for 15–20 min each and then in absolute acetone for 20 min. After dehydration, the samples were embedded in Spurr’s resin overnight. The specimens were heating at 70°C for 9 h, the ultra-thin sections (80 nm) were cut and mounted on copper grids for observation in the transmission electron microscope (TEM 1230EX, JEOL, Japan) at 60.0 kV.
Statistical Analysis
The results presented here are the means of three replicates. Treatment means were compared by the analysis of variance (ANOVA) and using Tukey’s multiple range tests at the 5% level of significance.
Results
cDNA Library Quality and Yeast Two-Hybrid Screening
Total RNA and mRNA were extracted and purified from mix sample (shoot and root), respectively (Supplementary Figure 1). EcoRI and XhoI was introduced into the double strand cDNA which was synthesized by reverse transcription. Then, the synthesized cDNA was size fractionated to removing short fragments (Supplementary Figure 1). Then, double strand cDNA library was directional cloned into linearized pGADT7 AD vector with EcoRI and XhoI digested. The titer of cDNA library was more than 1.7 × 106 cfu, and the size of inserted fragments were 1 kb approximately (Supplementary Figure 2).
There was an identical size of Y2H strain on SD/-Trp plate containing bait vector (pGBKT7-OsCYP2) or not, meanwhile, pale blue and no clone on SD/-Trp/X-α-gal and SD/-Trp/X-α-gal/AbA plate, respectively (Supplementary Figure 3). That means the bait vector containing OsCYP2 without auto-activation and toxicity, was suitable for yeast two-hybrid cDNA library screening. Then, 5 μg of bait and 10 μg of prey were used for yeast two-hybrid library co-transformation. The blue colonies that grew on DDO (SD/-Trp/-Leu/X-α-gal/AbA) plate were patched out onto higher stringency QDO (SD/-Trp/-Leu/-Ade/-His/X-α-gal/AbA) plate to eliminate the false interaction (Supplementary Figure 4). The result of sequencing and blast showed that rescued positive prey (No. 2) was a peroxisomal biogenesis factor 11 family protein (Os03g0302000). This prey protein was also identified by co-transforming into Y2H with pGBKT7 or pGBKT7-OsCYP2 on the selective media (Figure 1).
FIGURE 1.
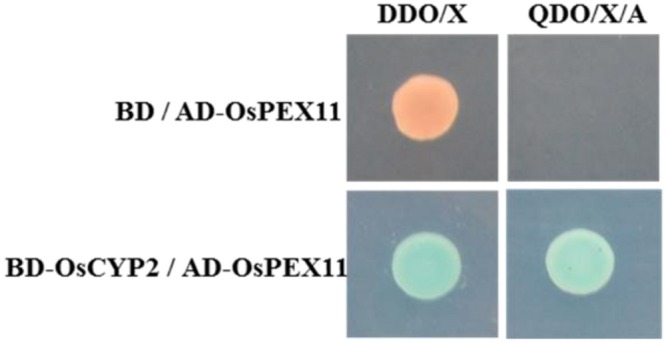
OsCYP2 interacts with OsPEX11 protein by in vivo assay. Yeast clones were grown on DDO/Xor QDO/X/A plates that contained X-α-gal. The blue color in the colony co-transformed with BD-OsCYP2 and AD-OsPEX11 indicates interaction between the two proteins. The empty pAD vector was used as negative control. DDO/X medium: SD/-Trp-Leu/X-α-gal; QDO/X/A medium: SD/-Trp-Leu-Ade-His/X-α-gal/AbA; AD-OsPEX11: pGADT7+OsPEX11; BD: pGBKT7 vector; BD-OsCYP2: pGBKT7+OsCYP2.
OsCYP2 Directly Interacts with OsPEX11
The GST and GST-OsCYP2 protein were extracted from E. coli strain BL21 which contained pGEX4T-1 and pGEX4T-1 plus OsCYP2, respectively. Twenty-six kDa (GST) and 45 kDa (GST-OsCYP2) were showed by SDS-PAGE and coomassie brilliant blue staining (Figure 2A). We conducted the western blot to detect the specific signal by using HRP conjugated GST antibody and DAB staining (Figure 2B). In in vitro GST pull-down assay, OsCYP2 could directly interact with OsPEX11 (Figure 3).
FIGURE 2.
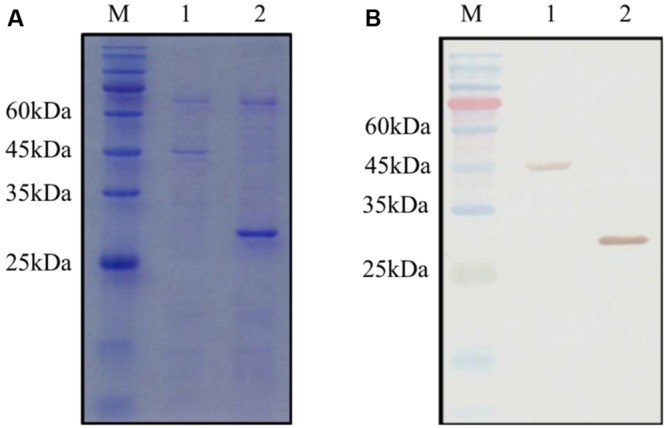
Detection of GST (glutathione-S-transferase) and recombinant GST-OsCYP2. The GST and GST-OsCYP2 proteins were extracted and purified from E. coli strain BL21 using MagneGST Pull Down System (Promega). The proteins were analyzed with 12% SDS-PAGE. (A) SDS-PAGE gel was stained by coomassie brilliant blue R-250. (B) Horseradish Peroxidase DAB staining was used in western blotting. 1: GST-OsCYP2. 2: GST. M: Pre-Stained Protein Marker.
FIGURE 3.

Confirmation of interaction between OsCYP2 and OsPEX11 protein using in vitro assay. Recombinant glutathione-S-transferase (GST) and GST-OsCYP2 was used as bait and incubated with 1 μg of prey protein (His-OsPEX11), respectively. After incubation, GST and GST-OsCYP2 were retrieved with glutathione beads, and the pulled-down proteins were detected on Western blots with antibodies to Histidine. GST: pGEX-4T-1; GST-OsCYP2: pGEX-4T-1+OsCYP2; His-PEX11: pET28a+OsPEX11.
Phenotypic Characterization of OsPEX11 Transgenic Plants
To evaluate the stress response of transgenic plants, 10-day-old OsPEX11 overexpression and RNAi seedlings were treated by 200 mM NaCl. After 24 h, leaves of WT and RNAi lines showed wilting, especially RNAi lines exhibited even more chlorosis, whereas the overexpressing plants remained normal growth condition (Figure 4). Moreover, the plant height was significantly decreased under 200 mM NaCl treatment in each genotype, however, root length and leave angles were significantly higher in OsPEX11 overexpression seedlings compared with WT and OsPEX11 RNAi (Supplementary Table S2). On the other hand, the transcript level of OsPEX11 gene also showed significantly higher and lower expression in overexpression and RNAi lines, respectively, compared to that of WT. During salt stress treatment, OsPEX11 was significantly induced in WT, and its expression level was up-regulated in overexpression lines (Figure 5).
FIGURE 4.

The morphological changes of 10-day-old wild type (WT), OsPEX11 over-expression (OE1) and RNAi (RNAi1) seedlings under control and salt stress (200 mM NaCl). The seedlings were treated with 200 mM NaCl for 24 h. (A) WT, (B) WT + NaCl, (C) 35S-OsPEX11, (D) 35S-OsPEX11 + NaCl, (E) RNAi-OsPEX11, and (F) RNAi-OsPEX11 + NaCl.
FIGURE 5.
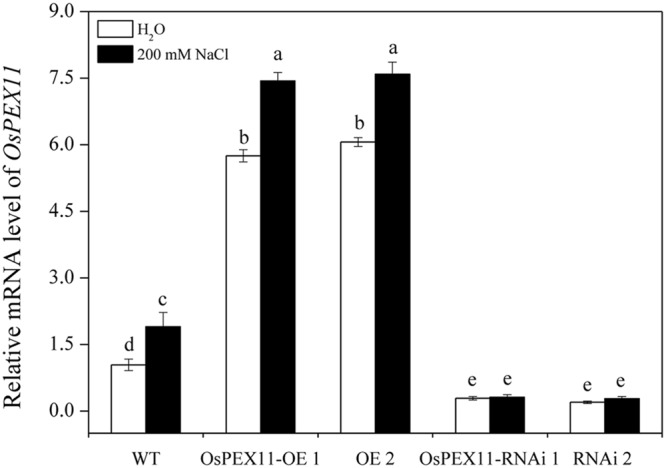
Relative transcript level of OsPEX11 responses to H2O and 200 mM NaCl stress in 10-day-old WT, overexpression (OE1 and OE2), and RNA interference (RNAi1 and RNAi2) lines. The seedlings were treated with 200 mM NaCl for 24 h. Relative expression levels were measured by qRT-PCR analysis using actin as an internal standard. The relative fold expression of CK was considered as 1. Values are means of three biological replicates and significant differences between means, as determined by Turkey test (P < 0.05), are indicated by different letters.
Sodium/Potassium Accumulation
Plant salt tolerance is mainly associated with the low maintenance of cytosolic Na+/K+ ratio. Thus, the sodium and potassium contents in the leaves of WT and transgenic plants were determined by atomic absorption spectrophotometry. There were no significant differences among three genotypes under control conditions, but in the presence of salt (200 mM NaCl), the Na+/K+ ratio was significantly elevated in OsPEX11-RNAi seedlings (Figure 6A). Compared with WT, the highest increase in Na+ concentration (2.5 fold) was observed in OsPEX11-RNAi plants. Similarly, K+ content maintained relatively higher in OsPEX11-OE seedlings (1.25 fold) (Supplementary Table S3). These results suggested that overexpression of OsPEX11 can enhance salt tolerance and maintain a low Na+/K+ ratio through selective uptake of K+ over Na+.
FIGURE 6.
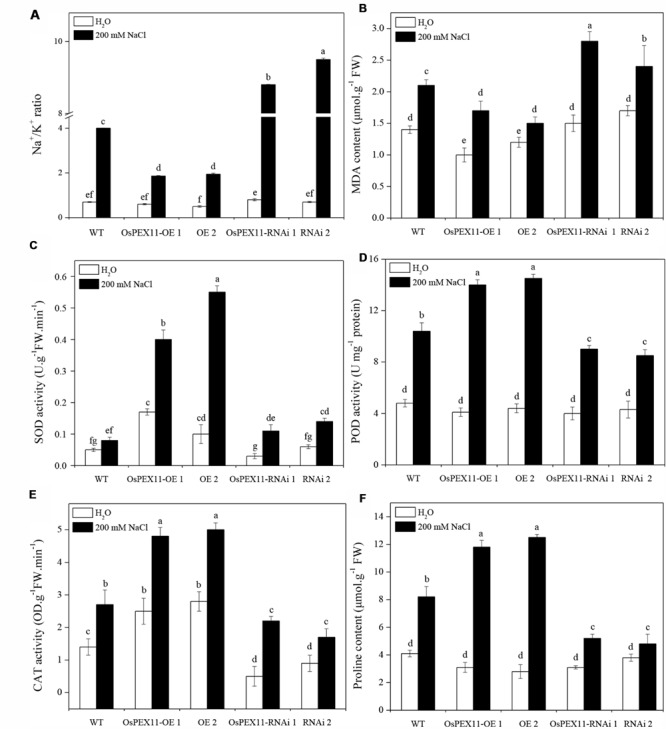
Comparison of lipid peroxidation and ROS scavenging in leaves of 10-day-old WT and OsCYP2-transgenic (OE1 and OE2, RNAi1 and RNAi2) seedlings under control and salt stress. The rice seedlings were treated with 200 mM of NaCl for 24 h. The Na+/K+ ratio (A) and activities of antioxidant enzymes MDA (B), SOD (C), POD (D), CAT (E), and proline content (F) were assayed. Values are means of three biological replicates followed by the same letter did not significantly differ at P ≤ 0.05 according to Turkey’s multiple range test.
Physiological Responses to Saline Stress of Transformants
To further investigate the role of gene OsPEX11 in plant tolerance to high salinity, we measured the effect of salt induced oxidative stress on MDA and ROS-scavenging anti-oxidative enzymes. As shown in Figures 6B–F, saline stress differentially modulated the accumulation of MDA and activities of SOD, POD, CAT enzymes and proline content in each type of seedlings. The MDA level was significantly lower in OsPEX11 overexpression seedlings OE1 (15.35%) and OE2 (29.14%), while the SOD, POD, CAT enzyme activities were significantly increased (OE1: 5.00, 1.35, 1.78 fold; OE2: 6.88, 1.39, and 1.85 fold, respectively) as compared to WT plants. Accumulation of proline was accompanied by changes in ROS scavenging enzyme activities. Here, we found that the proline content was significantly elevated in OsPEX11-OE1 and OE2 seedlings (3.81 and 4.46 fold, respectively) under saline stress treatment. These results suggested that gene OsPEX11 may have a better protection against salt induced ROS by dynamic modulation of antioxidant enzymes (SOD, POD, and CAT) and proline accumulation, which result in reduced lipid peroxidation under salt stress condition.
Expression Pattern of Na+/K+ Transporter Proteins
To better understand the mechanisms of underlying Na+/K+ accumulation variation in OsPEX11 transgenic seedlings, we utilized qRT-PCR to detect the expression level of genes encoding Na+/K+ transport proteins. The data showed that except the other five genes, OsHKT2;1 and OsHKT1;5 (Na+ transporters) were significantly up/down-regulated in OsPEX11 overexpression/RNAi lines under control condition. Saline stress treatment (200 mM NaCl) repressed the expression of the OsHKT2;1 and OsHKT1;5 (Na+ transporters) in each genotype (Figures 7A,B), however, OsLti6a, OsLti6b (two homologous PMP3 genes involved in Na+ excess entry in plant cells), OsSOS1 (Na+/H+ antiporter salt overly sensitive1), OsNHX1 (vacuolar Na+, K+, and H+ antiporter) and OsAKT1 (K+ transporter) performed an opposite effect (Figures 7C–G). In addition, OsNHX1 was the most strongly induced (>20-fold) under salt stress overexpression seedlings compared with RNAi and WT (Figure 7F).
FIGURE 7.
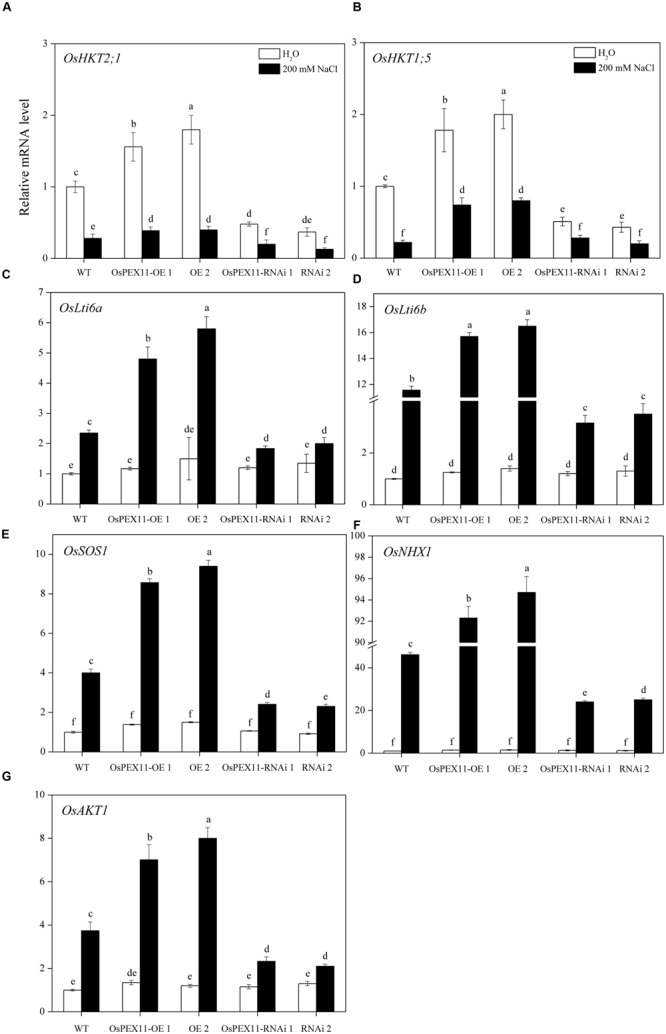
Relative fold expression of the genes encoding Na+ and K+ transporter proteins. (A) OsHKT2;1; (B) OsHKT1;5; (C) OsLti6a; (D) OsLti6b; (E) OsSOS1; (F) OsNHX1; (G) OsAKT1 in the leaves of 10-day-old rice seedlings treated with 200 mM of NaCl. The relative fold expression of CK was considered as 1. An actin was used as internal standard. Values are the means of three biological replications ±SD. Variants possessing the same letter are not statistically significant at P < 0.05.
Salinity Induces Ultrastructural Changes
Figure 8 showed the representative TEM images of the chloroplast ultrastructure of WT and transgenic rice plants with/without saline stress conditions. In WT and OsPEX11 overexpression plants, chloroplast was elliptical, the granum and stroma thylakoids were in an orderly arrangement, the lamellar structure was relatively tight, and the chloroplast envelope was intact with much larger size of starch grains under control condition (Figures 8A,B), while the structure of the chloroplast in PEX11 RNAi plants was integrated, and the shape was round with swollen mitochondria (Figure 8C). Under saline stress condition, the shape of chloroplast in WT plants deformed with fewer thylakoid stacks of grana and lamella being observed. Additionally, the grana stacks were irregular and distributed unevenly with larger size of plastoglobuli, suggesting that the chloroplast structure was degraded (Figure 8D). The thylakoids of OsPEX11-OE1 plants were loose, the stroma thylakoids were in an orderly arrangement and chloroplast maintained relatively normal shape (Figure 8E). The chloroplast shape of OsPEX11-RNAi plants was round and dilated grana with fewer and abnormal shapes of mitochondria were also observed (Figure 8F).
FIGURE 8.
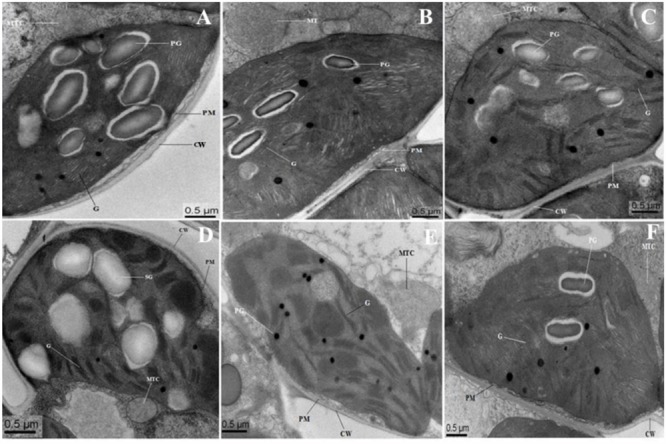
Electron micrographs of leaf mesophyll cells of 10-day-old hydroponically grown seedlings of O. sativa under control condition and 200 mM NaCl treatment. Under control condition, grana (G), mitochondria (MTC), and plastoglobuli (PG) well developed. TEM micrographs of leaf mesophyll cells of WT showed that ultrastructure of chloroplast is disrupted with enlarged size PG under 200 mM NaCl treatment (D). Chloroplasts of OsPEX11-OE1 plants maintained normal shape under salt stress condition with dilated thylakoids arrangement (E), whereas, chloroplasts of OsPEX11-RNAi1 plants were round in shape with fewer numbers of PG and swollen mitochondria (MTC) as compared to its respective control (F). (A–C) control: WT (Left); OsPEX11-OE1 (Middle); OsPEX11-RNAi1 (Right); (D–F) 200 mM NaCl: WT (Left); OsPEX11-OE1 (Middle); OsPEX11-RNAi1 (Right).
Discussion
Peroxisomes play key role in the regulation of metabolic process by modulating cellular redox homeostasis in a cell (Yun et al., 2012). It has been showed that peroxisomes can rapidly modify their metabolism and dynamics according to the subsequent ROS level in the cell. Mutations of peroxisome biogenesis proteins in various eukaryotes result in serious developmental deficiencies and stress sensitivities (Aung and Hu, 2011; Burkhart et al., 2013; Cassin-Ross and Hu, 2014). Under stress conditions, coordinated functioning of peroxisomal proteins provides highly dynamic responses of peroxisomal metabolism to adjust its redox metabolism at steady-state. In the present study, the overexpression of OsPEX11 enhanced the growth and survival of transgenic plants under higher saline stress conditions, which further clarify it important role in reducing Na+ induced toxicity that protect the plants against saline stress.
Previous studies reported that enhanced activities of antioxidant enzymes (SOD, POD, and CAT) act as a coping strategy to scavenge ROS (Munns and Gilliham, 2015). Overexpression of genes involved ROS detoxification resulted in lower cellular damage, and the maintenance of photosynthetic energy capture under saline conditions (Roy et al., 2014). Similarly, OsPEX11 gene substantially increased the antioxidant enzymes activities. It is likely that the peroxisome protein can suppress glycolate oxidase (Sousa et al., 2015) or harbor enzymes that can breakdown ROS (Antonenkov et al., 2010), and therefore led to decreased MDA contents as well as less damage to organelles membranes (Guan et al., 2015; Camejo et al., 2016). Proline is not only compatible osmolyte and osmoprotectant but also acts as a signaling molecule to modulate ion balance, protect enzyme activity and trigger the expression of specific genes, which are essential for plant recovery from stresses (Liu et al., 2015). In the present study, we found enhanced accumulation of proline in overexpression plants compared to RNAi plants (Figure 6). The increased content of proline may help to decrease Na+ toxicity in overexpression plant as compared to the RNAi plants (Islam et al., 2015, 2016). Therefore, OsPEX11 overexpression could alleviate ion toxicity and oxidant damage in transgenic plants by enhancing proline accumulation and antioxidant defense.
The accumulation of K+ and Na+ in plants is important parameter to understand salt tolerance mechanisms (Noreen et al., 2010). In the present investigation, overexpression plants accumulated high K+ and maintained lower Na+/K+ ratio and Na+ accumulation under stress conditions as compared to RNAi plants. The higher accumulation of Na+ in RNAi plant might be due to enhanced accumulation of Na+ in roots, passive diffusion of sodium ions from damaged membranes and reduced efficient mechanism of sodium diffusion (Islam et al., 2015). Thus, OsPEX11 overexpression could improve the growth of rice seedlings by reducing Na+ uptake under saline stress condition as compared to the RNAi plants. Furthermore, the ability of the plants to restrict the transportation and accumulation of Na+ is considered as a critical aspect of plant salt tolerance/adaptation (Apel and Hirt, 2004; Foyer and Noctor, 2005; Munns and Tester, 2008). To understand the salt-tolerant mechanism in OsPEX11 overexpression plants, we compared the transcript levels of marker genes related to Na+ and K+ regulation under control and stress conditions (Figure 7). The OsPEX11 overexpression plants exhibit lower leaf Na+ concentration showed better adaptation to the salinity. This controlled entry of Na+ to the leaf is often associated with the lowered Na+/K+ ratio (Figure 6A), which is vital for the normal functioning of a cell under saline stress conditions (Su et al., 2015). Na+ exclusion/restriction is usually under the control of different Na+ transport proteins. Such as, high affinity potassium transporter (HKT) genes encode Na+ selective transporter proteins which demonstrated essential salinity tolerance mechanism in different crop plants including rice (Hamamoto et al., 2015). To understand underlying mechanism of restricted Na+ transport in the leaves of transgenic and WT plants, we analyzed the expression of OsHKT1;5 both under control and salt stress conditions. OsPEX11 overexpression plants showed considerable induction in transcript level of OsHKT1;5 as compared to the RNAi plants, which may suggest that downregulation of OsHKT1;5 expression in RNAi plants may be the reason of higher accumulation of Na+ in leaves as compared to the overexpressed plants, that restricted the unregulated Na+ transport to the leaves and protect the plants from growth impairment due to Na+ toxicity (Figure 7). Previously, decreased expression of HKT1;5 through RNAi in bread wheat was linked with increased Na+ accumulation in the leaves, which suggest a critical role of HKT1;5 transporter in the restriction of Na+ transport from root to leaves in saline stress plants (Byrt et al., 2014).
Plasma membranes proteins 3 (PMP3) are conserved hydrophobic proteins induced in wide range of abiotic stress conditions, suggesting their significant role in membranes stability under stress conditions (Chidambaram et al., 2015). The overexpression of PMP3 homolog proteins in Arabidopsis and Avicennia marina plants show lowered shoot Na+ levels and enhanced plant performance under stress conditions (Mitsuya et al., 2006; Chidambaram et al., 2015). In the present study, the expression level of two rice homologous PMP3 genes, OsLti6a and OsLti6b was downregulated under saline stress condition in RNAi plants as compared to the overexpression and WT plants, which clearly indicate the involvement of these genes in Na+ entry in the RNAi plants (Figure 7). Such as, a loss of function of PMP3 in yeast mutants resulted in Na+ accumulation, salt sensitivity and membrane hyperpolarization (Nylander et al., 2001). Additionally, Mekawy et al. (2015) suggested that reduction in the expression levels of PMP3 genes may facilitate Na+ entry in sensitive rice cultivar as compared to the resistant one. The enhanced activation of PMP3 proteins in OsPEX11 overexpression plants may be due to the induction of other Na+ transporter proteins, because PMP3 proteins contribute indirectly to cation homeostasis within cells by interaction with other ion transporters (Fu et al., 2012). Plasma membrane Na+/H+ exchanger (SOS 1) and tonoplast Na+/H+ exchanger (NHX) are considered as main transporters mediating the efflux and compartmentalization of Na+ under saline stress conditions (de la Garma et al., 2015).
Both antiporters are ubiquitous membrane proteins that catalyze the electroneutral exchange of Na+ or for H+ across the membrane, thereby playing important roles in cellular Na+/K+ homeostasis. SOS1 is not only involved in Na+ exclusion from cytoplasm but also maintain optimum level of Na+ by xylem loading under low salt stress, while Na+ removal from xylem under saline stress conditions. In the present investigation, upregulation of OsSOS1 in overexpression plants as compared to the RNAi and WT plants probably facilitated exclusion of toxic Na+ into root apoplast and thus resulted in higher K+/Na+ ratio of leaves (Figure 6A). Additionally, Na+/ K+/H+ antiporter NHX1 is involved in the intracellular Na+/K+ sequestration in vacuoles, depending on the salt concentration in the cell. We observed that NHX1 gene expression levels were more upregulated in overexpression plants which may be the reason of higher accumulation of K+ uptake into the vacuole and lower Na+ concentration in cytoplasm, which relieves the toxic effect on cytosolic enzymes, maintaining turgor pressure and cell expansion under saline stress conditions (Cao et al., 2016).
Moreover, ultrastructural observations of leaf mesophyll cells were also conducted to confirm the salt tolerance of OsPEX11 gene. Because, chloroplast and mitochondria are sensitive to the salinity. Salinity may change the functionality and integrity of chloroplast, that can affect energy metabolism of the mesophyll cells. The ultrastructural alternations in the chloroplast and mitochondrial structures in our study are consistent with the previous reports (Tripathy et al., 2015), suggesting that the accumulation of excess Na+ in RNAi and WT plants damaged the chloroplasts ultimately causing decreased photosynthesis efficiency at the whole plant level and thereby reduced growth and biomass production. Because, productivity and maintenance of the structural integrity of chloroplast is directly linked with the conversion of light energy during photosynthesis. Taken together, the overexpression of OsPEX11 gene enhanced the growth and survival of plants under salinity but further research is needed to explore the relationship between OsPEX11 and salt stress tolerance using advanced physiological and molecular technologies.
Conclusion
In our present study, a novel prey protein (OsPEX11) was screened and identified using in vivo (yeast two-hybrid) and in vitro (GST pull-down) assays. Under saline stress, leaves of OsPEX11-RNAi lines showed wilting and exhibited even more chlorosis compared to OsPEX11 overexpressing plants. OsPEX11 gene may have a better protection against salt induced ROS by dynamic modulation of antioxidant enzymes (SOD, POD, and CAT) and proline accumulation, which can reduce lipid peroxidation under salt stress condition. We also demonstrated that OsPEX11 overexpression can better protect plants from saline stress by restricting the entry of excess Na+ through dynamic regulation of Na+/K+ transporter proteins and its subsequent sequestration through NHX1 in vacuoles. Moreover, ultrastructural observations of OsPEX11 overexpression seedlings demonstrated that they had better tolerance to salt stress than RNAi and WT plants.
Author Contributions
Conceived and designed the experiments: HL and WZ. Performed the experiments: PC, FI, LL, and MF. Analyzed the data: HL, WZ, LL, SR, and PC. Contributed reagents/materials/analysis tools: SR, WZ, HL, and MF. Wrote and revised the paper: PC, HL, FI, and WZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (201303022), National Natural Science Foundation of China (31301272, 31570434), the Fund from Zhejiang A & F University (2013FR022), and Zhejiang Provincial Top Key Discipline of Biology and its Open Foundation (2015D19).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.0135
References
- Aebi H. (1984). Catalase in vitro. Methods Enzymol. 105 121–126. 10.1016/S0076-6879(84)05016-3 [DOI] [PubMed] [Google Scholar]
- Antonenkov V. D., Grunau S., Ohlmeier S., Hiltunen J. K. (2010). Peroxisomes are oxidative organelles. Antioxid. Redox Signal. 13 525–537. 10.1089/ars.2009.2996 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Aung K., Hu J. (2011). The Arabidopsis tail-anchored protein peroxisomal and mitochondrial division factor1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23 4446–4461. 10.1105/tpc.111.090142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L., Waldren R., Teare I. (1973). Rapid determination of free proline for water stress studies. Plant Soil 39 205–207. 10.1007/BF00018060 [DOI] [Google Scholar]
- Burkhart S. E., Lingard M. J., Bartel B. (2013). Genetic dissection of peroxisome associated matrix protein degradation in Arabidopsis thaliana. Genetics 193 125–141. 10.1534/genetics.112.146100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt C. S., Xu B., Krishnan M., Lightfoot D. J., Athman A., Jacobs A. K., et al. (2014). The Na+ transporter, TaHKT1; 5-D, limits shoot Na+ accumulation in bread wheat. Plant J. 80 516–526. 10.1111/tpj.12651 [DOI] [PubMed] [Google Scholar]
- Camejo D., Guzmán-Cedeño Á., Moreno A. (2016). Reactive Oxygen Species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 103 10–23. 10.1016/j.plaphy.2016.02.035 [DOI] [PubMed] [Google Scholar]
- Campos B. M., Sforça M., Ambrosio A. L. B., Domingues M. N., de Souza T. D. C. B., Barbosa J. A. R. G., et al. (2013). A redox 2-Cys mechanism regulates the catalytic activity of divergent cyclophilins. Plant Physiol. 162 1311–1323. 10.1104/pp.113.218339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Long D., Zhang M., Liu C., Xiang Z., Zhao A. (2016). Molecular characterization and expression analysis of the mulberry Na+/H+ exchanger gene family. Plant Physiol. Biochem. 99 49–58. 10.1016/j.plaphy.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Cassin-Ross G., Hu J. (2014). Systematic phenotypic screen of Arabidopsis peroxisomal mutants identifies proteins involved in β-Oxidation. Plant Physiol. 166 1546–1559. 10.1104/pp.114.250183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidambaram R., Venkataraman G., Parida A. (2015). Analysis of transcriptional regulation and tissue-specific expression of Avicennia marina Plasma Membrane Protein 3 suggests it contributes to Na+ transport and homoeostasis in A. marina. Plant Sci. 236 89–102. 10.1016/j.plantsci.2015.03.013 [DOI] [PubMed] [Google Scholar]
- de la Garma J. G., Fernandez-Garcia N., Bardisi E., Pallol B., Rubio-Asensio J. S., Bru R., et al. (2015). New insights into plant salt acclimation: the roles of vesicle trafficking and reactive oxygen species signaling in mitochondria and the endomembrane system. New Phytol. 205 216–239. 10.1111/nph.12997 [DOI] [PubMed] [Google Scholar]
- Dhindsa S. R., Matowe W. (1981). Drought tolerance in two mosses: correlated with enzymatic defense against lipid peroxidation. J. Exp. Bot. 32 79–91. 10.1093/jxb/32.1.79 [DOI] [Google Scholar]
- Dinneny J. R. (2015). Traversing organizational scales in plant salt-stress responses. Curr. Opin. Plant Biol. 23 70–75. 10.1016/j.pbi.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Dixit E., Boulant S., Zhang Y. J., Lee A. S. Y., Odendall C., Shum B., et al. (2010). Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141 668–681. 10.1016/j.cell.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2005). Oxidant and antioxidant signaling in plants: are-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 28 1056–1071. 10.1111/j.1365-3040.2005.01327.x [DOI] [Google Scholar]
- Fu J., Zhang D. F., Liu Y. H., Ying S., Shi Y. S., Song Y. C., et al. (2012). Isolation and characterization of maize PMP3 genes involved in salt stress tolerance. PLoS ONE 7:e31101 10.1371/journal.pone.0031101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Wu R., Wee C. W., Xie F., Wei X., Chan P. M. Y., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25 2132–2154. 10.1105/tpc.113.112896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Valle D. (2000). Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 16 340–345. 10.1016/S0168-9525(00)02056-4 [DOI] [PubMed] [Google Scholar]
- Guan Q., Wang Z., Wang X., Takano T., Liu S. (2015). A peroxisomal APX from Puccinelliatenuiflora improves the abiotic stress tolerance of transgenic Arabidopsis thaliana through decreasing of H2O2 accumulation. J. Plant Physiol. 175 183–191. 10.1016/j.jplph.2014.10.020 [DOI] [PubMed] [Google Scholar]
- Hamamoto S., Horie T., Hauser F., Deinlein U., Schroeder J. I., Uozumi N. (2015). HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr. Opin. Biotechnol. 32 113–120. 10.1016/j.copbio.2014.11.025 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. 10.1046/j.1365-313X.1994.6020271.x [DOI] [PubMed] [Google Scholar]
- Hodges D. M., DeLong J. M., Forney C. F., Prange R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207 604–611. 10.1007/s004250050524 [DOI] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M. (2012). Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31 267–278. 10.1038/emboj.2011.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam F., Ali B., Wang J., Farooq M. A., Gill R. A., Ali S., et al. (2016). Combined herbicide and saline stress differentially modulates hormonal regulation and antioxidant defense system in Oryza sativa cultivars. Plant Physiol. Biochem. 107 82–95. 10.1016/j.plaphy.2016.05.027 [DOI] [PubMed] [Google Scholar]
- Islam F., Yasmeen T., Ali S., Ali B., Farooq M. A., Gill R. A. (2015). Priming-induced antioxidative responses in two wheat cultivars under saline stress. Acta Physiol. Plant. 37 1–12. 10.1007/s11738-015-1897-5 [DOI] [Google Scholar]
- Jiang Y., Yang B., Harris N. S., Deyholos M. K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58 3591–3607. 10.1093/jxb/erm207 [DOI] [PubMed] [Google Scholar]
- Kishor P. B. K., Sreenivasulu N. (2014). Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue. Plant Cell Environ. 37 300–311. 10.1111/pce.12157 [DOI] [PubMed] [Google Scholar]
- Kumar S., Kawalek A., van der Klei I. J. (2014). Peroxisomal quality control mechanisms. Curr. Opin. Microbiol. 22 30–37. 10.1016/j.mib.2014.09.009 [DOI] [PubMed] [Google Scholar]
- Lee S. S., Park H. J., Jung W. Y., Lee A., Yoon D. H., You Y. N., et al. (2015). OsCYP21-4, a novel Golgi-resident cyclophilin, increases oxidative stress tolerance in rice. Front. Plant Sci. 6:797 10.3389/fpls.2015.00797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Yu Y., Li R., Duan X., Zhu D., Sun X., et al. (2015). A novel hybrid proline-rich type gene GsEARLI17 from Glycine soja participated in leaf cuticle synthesis and plant tolerance to salt and alkali stresses. PCTOC 121 633–646. 10.1007/s11240-015-0734-2 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mainali H. R., Chapman P., Dhaubhadel S. (2014). Genome-wide analysis of cyclophilin gene family in soybean (Glycine max). BMC Plant Biol. 14:282 10.1186/s12870-014-0282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekawy A. M. M., Assaha D. V., Yahagi H., Tada Y., Ueda A., Saneoka H. (2015). Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol. Biochem. 87 17–25. 10.1016/j.plaphy.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Mitsuya S., Taniguchi M., Miyake H., Takabe T. (2006). Over-expression of RCI2A decreases Na+ uptake and mitigates salinity-induced damages in Arabidopsis thaliana plants. Physiol. Plant. 128 95–102. 10.1111/j.1399-3054.2006.00714 [DOI] [Google Scholar]
- Munns R., Gilliham M. (2015). Salinity tolerance of crops-what is the cost? New Phytol. 208 668–673. 10.1111/nph.13519 [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Munns R., Wallace P. A., Teakle N. L., Colmer T. D. (2010). Measuring soluble ion concentrations (Na+, K+, Cl-) in salt-treated plants. Methods Mol. Biol. 639 371–382. 10.1007/978-1-60761-702-0_23 [DOI] [PubMed] [Google Scholar]
- Nito K., Kamigaki A., Kondo M., Hayashi M., Nishimura M. (2007). Functional calssfication of Arabidopsis peroxisome biogenesis factors proposed analyses of knockdown mutants. Plant Cell Physiol. 48 763–774. 10.1016/S0168-9525(00)02056-4 [DOI] [PubMed] [Google Scholar]
- Noreen Z., Ashraf M., Akram N. (2010). Salt-induced regulation of some key antioxidant enzymes and physio-biochemical phenomena in five diverse cultivars of turnip (Brassica rapa L.). J. Agro. Crop. Sci. 196 273–285. [Google Scholar]
- Nylander M., Heino P., Helenius E., Palva E. T., Ronne H., Welin B. V. (2001). The low-temperature- and salt-induced RCI2A gene of Arabidopsis complements the sodium sensitivity caused by a deletion of the homologous yeast gene SNA1. Plant Mol. Biol. 45 341–352. 10.1023/A:1006451914231 [DOI] [PubMed] [Google Scholar]
- Odendall C., Dixit E., Stavru F., Bierne H., Franz K. M., Durbin A. F., et al. (2014). Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15 717–726. 10.1038/ni.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Flowers T. J., Moore A. L., Harpham N. V. (2006). An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J. Exp. Bot. 57 1109–1118. 10.1093/jxb/erj134 [DOI] [PubMed] [Google Scholar]
- Pieuchot L., Jedd G. (2012). Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu. Rev. Microbiol. 66 237–263. 10.1146/annurev-micro-092611-150126 [DOI] [PubMed] [Google Scholar]
- Rao L., Perez D., White E. (1996). Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 135 1441–1455. 10.1083/jcb.135.6.1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. J., Negrao S., Tester M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26 115–124. 10.1016/j.copbio.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Ruan S. L., Ma H. S., Wang S. H., Fu Y. P., Xin Y., Liu W. Z., et al. (2011). Proteomic identification of OsCYP2, a rice cyclophilin that confers salt tolerance in rice (Oryza sativa L.) seedlings when overexpressed. BMC Plant Biol. 11:34 10.1186/1471-2229-11-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Willmann M. R., Wu G., Berardini T. Z., Moller B., Weijers D. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. PNAS 106 5424–5429. 10.1073/pnas.0812729106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R. H. V., Carvalho F. E. L., Ribeiro C., Passaia G., Cunha J. R., Lima-Melo Y., et al. (2015). Peroxisomal APX knockdown triggers antioxidant mechanisms favorable for coping with high photorespiratory H2O2 induced by CAT deficiency in rice. Plant Cell Environ. 383 499–513. 10.1111/pce.12409 [DOI] [PubMed] [Google Scholar]
- Su Y., Luo W., Lin W., Ma L., Kabir M. H. (2015). Model of cation transportation mediated by high-affinity potassium transporters (HKTs) in higher plants. Biol. Proced. Online 17 484–488. 10.1186/s12575-014-0013-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy M. K., Tiwari B. S., Reddy M. K., Deswal R., Sopory S. K. (2015). Ectopic expression of PgRab7 in rice plants (Oryza sativa L.) results in differential tolerance at the vegetative and seed setting stage during salinity and drought stress. Protoplasma 252 1–16. 10.1007/s00709-015-0914-2 [DOI] [PubMed] [Google Scholar]
- Vasudevan D., Fu A. G., Luan S., Swaminathan K. (2014). Crystal structure of Arabidopsis cyclophilin38 reveals a previously uncharacterized immunophilin fold and a possible autoinhibitory mechanism. Plant Cell 24 2666–2674. 10.1105/tpc.111.093781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzel K., Weidner A., Surabhi G. K., Borner A., Mock H. P. (2009). Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J. Exp. Bot. 60 3545–3557. 10.1093/jxb/erp198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B.-W., Spoel S. H., Loake G. J. (2012). Synthesis of and signaling by small, redox active molecules in the plant immune response. Biochim. Biophys. Acta 1820 770–776. 10.1016/j.bbagen.2011.06.015 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6 66–71. 10.1016/S1360-1385(00)01838-0 [DOI] [PubMed] [Google Scholar]
- Zhuang J., Zhang J., Hou X. L., Wang F., Xiong A. S. (2014). Transcriptomic, proteomic, metabolomic and functional genomic approaches for the study of abiotic stress in vegetable crops. Crit. Rev. Plant Sci. 33 225–237. 10.1080/07352689.2014.870420 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


