Dear Editor,
Recently, deletions of SPRY4, a negative regulator of the intracellular RAS-mitogen activated protein kinase/extracellular signal regulated kinase (MAPK/ERK) signaling cascade, have been described in de-novo and therapy-related acute myeloid leukemia (AML) [1]. Importantly, employing both in-vitro and in-vivo approaches, the authors could prove the functionality of these events in myeloid leukemogenesis. To further extend these findings, we screened for SPRY4 deletions in 45 patients with secondary AML, developing from an antecedent myelodysplastic syndrome (MDS) or myeloproliferative neoplasm (MPN).
SPRY4 copy number was evaluated by real-time quantitative PCR (qPCR) using the comparative ddCT method as previously described [2–5]. Briefly, DNA gene dosage of the target gene SPRY4 (5q31.3) from a sample of interest was first normalized to two control genes F8 (Xq28) and ABL1 (9q34.12), and then related to a healthy calibrator of the same sex, which was always included on the same qPCR plate. While this approach resulted in a SPRY4 value of 1 for the calibrator, deletion of SPRY4 in the sample of interest was defined by a value below 0.65, which is in accordance with previously published recommendations [6]. When analyzing DNA obtained at the diagnosis of sAML, we detected a heterozygous deletion of SPRY4 in eight out of 45 sAML specimens (18%; median copy number 0.51, range 0.2 - 0.64; Supplementary Table 1). Values for patients defined as carrying both SPRY4 alleles were consistently higher than 0.83, confirming a clear discrimination between the groups. For a more detailed characterization of SPRY4 deleted patients, we initially reviewed the cytogenetic information available. Most interestingly, complex karyotypes were significantly enriched in the SPRY4 deleted group as compared to patients carrying both alleles (100% vs 7%; P<0.001). In a next step, we focused on the molecular profile and therefore performed next generation sequencing (NGS) of sAML material in patients with SPRY4 deletion using an amplicon panel covering 18 genes, which are recurrently mutated in AML [7] (Supplementary Table 2). In agreement with Zhao and coworkers [1], we identified an overrepresentation of TP53 mutations and an underrepresentation of gain-of-function RAS pathway mutations (KRAS, NRAS, FLT3 and/or KIT), thereby suggesting that the pathogenetic mechanisms of SPRY4 deletion in sAML do not differ from those developing de-novo.
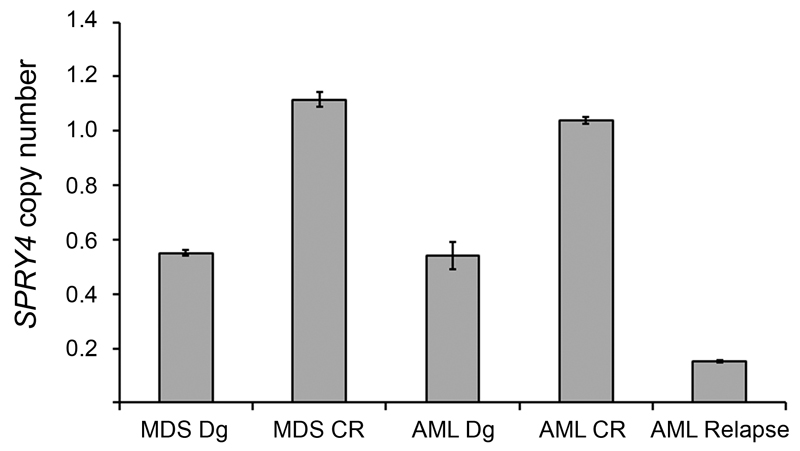
Finally, we studied diagnostic MDS/MPN specimens of patients harboring a SPRY4 copy number loss. Although DNA was available for three out of eight patients only, deletion of SPRY4 could be unambiguously detected in all early phase specimens suggesting that it describes an early event in myeloid leukemogenesis. Interestingly, we were also able to study the course of the disease in one patient (Fig.1). SPRY4 deletion was consistently observed at stages of disease progression or recurrence, indicating that it is a stable event during the clonal evolution of AML.
Fig. 1.
Detection of SPRY4 deletion in AML evolution. SPRY4 gene dosage was measured in sequential samples of case UPN 7324, obtained at MDS diagnosis (MDS Dg), MDS in complete cytogenetic remission after azacitidine therapy (MDS CR), AML diagnosis (AML Dg), AML in complete remission (AML CR) and AML relapse. Deletion of SPRY4 was clearly detectable at the diagnosis of MDS and at stages of disease progression or recurrence. Graphs represent the averaged results of both reference genes ± standard deviation (SD)
Taken together, we describe deletion of SPRY4 as a frequent event in sAML. It is associated to complex sAML karyotypes and to mutations in TP53. Ultimately, our data suggest that deletion of SPRY4 describes an early and stable event during myeloid leukemogenesis.
Supplementary Material
Funding
This work was supported by the Austrian Science Fund under Grant No. P 26619-B19 (to AZ). Work in the laboratories of AW, HS and AZ is further supported by Leukämiehilfe Steiermark.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval and informed consent: This study was approved by the institutional review board of the Medical University of Graz, Austria, and was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all individual participants included in this study.
Authors’ contributions: OG and SH carried out the qPCR experiments, analyzed the data and drafted the manuscript. KK and GH carried out NGS experiments, analyzed the data and drafted the manuscript. AW and AZ reviewed the clinical data, performed statistical analyses and drafted the manuscript. HS and AZ designed the study, analyzed the data and drafted the manuscript. All authors read and approved the final typescript.
References
- 1.Zhao Z, Chen CC, Rillahan CD, et al. Cooperative loss of RAS feedback regulation drives myeloid leukemogenesis. Nat Genet. 2015;47:539–543. doi: 10.1038/ng.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zebisch A, Sill H. Are mouthwashes a reliable source of constitutional DNA in patients with leukemia? Leuk Res. 2008;32:1164–1165. doi: 10.1016/j.leukres.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Zebisch A, Staber PB, Delavar A, et al. Two transforming C-RAF germ-line mutations identified in patients with therapy-related acute myeloid leukemia. Cancer Res. 2006;66:3401–3408. doi: 10.1158/0008-5472.CAN-05-0115. [DOI] [PubMed] [Google Scholar]
- 4.Zebisch A, Haller M, Hiden K, et al. Loss of RAF kinase inhibitor protein is a somatic event in the pathogenesis of therapy-related acute myeloid leukemias with C-RAF germline mutations. Leukemia. 2009;23:1049–1053. doi: 10.1038/leu.2009.68. [DOI] [PubMed] [Google Scholar]
- 5.Zebisch A, Wolfler A, Fried I, et al. Frequent loss of RAF kinase inhibitor protein expression in acute myeloid leukemia. Leukemia. 2012;26:1842–1849. doi: 10.1038/leu.2012.61. [DOI] [PubMed] [Google Scholar]
- 6.Wilke K, Duman B, Horst J. Diagnosis of haploidy and triploidy based on measurement of gene copy number by real-time PCR. Hum Mutat. 2000;16:431–436. doi: 10.1002/1098-1004(200011)16:5<431::AID-HUMU8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.