Abstract
RAF kinase inhibitor protein (RKIP) is a seminal regulator of intracellular signaling and exhibits both antimetastatic and antitumorigenic properties. Decreased expression of RKIP has been described in several human malignancies, including acute myelogenous leukemia (AML). As the mechanisms leading to RKIP loss in AML are still unclear, we aimed to analyze the potential involvement of miRNAs within this study. miRNA microarray and qPCR data of more than 400 AML patient specimens revealed correlation between decreased expression of RKIP and increased expression of miR-23a, a member of the miR-23a/27a/24-2 cluster. In functional experiments, overexpression of miR-23a decreased RKIP mRNA and protein expression, whereas miR-23a inhibition caused the opposite effect. By using an RKIP 3′-untranslated region luciferase reporter construct with and without mutation or deletion of the putative miR-23a–binding site, we could show that RKIP modulation by miR-23a is mediated via direct binding to this region. Importantly, miR-23a overexpression induced a significant increase of proliferation in hematopoietic cells. Simultaneous transfection of an RKIP expression construct lacking the miR-23a–binding sites reversed this phenotype, indicating that this effect is truly mediated via downregulation of RKIP. Finally, by analyzing more than 4,300 primary patient specimens via database retrieval from The Cancer Genome Atlas, we could highlight the importance of the miR-23a/RKIP axis in a broad range of human cancer entities. In conclusion, we have identified miR-23a as a negative regulator of RKIP expression in AML and have provided data that suggest the importance of our observation beyond this tumor entity.
Introduction
RAF kinase inhibitor protein (RKIP) has been described as a negative regulator of various intracellular signaling cascades, most notably the RAS-mitogen–activated protein kinase/extracellular signal regulated kinase (MAPK/ERK) pathway and the nuclear factor-kB–Snail circuitry (1–3). The seminal role of RKIP as a gatekeeper in these signaling processes is further highlighted by the fact that complete or partial loss of this protein has been observed in a variety of human malignancies, including carcinomas of the prostate, colon, and breast (1, 4–8). Noteworthy, decreased expression of RKIP correlated with a higher level of metastasis formation on the one hand, and with impaired sensitivity to chemo- and radiotherapy on the other hand (1, 4). Indeed, the functional involvement of RKIP within these processes could be corroborated in a broad range of in vitro and in vivo models (1, 9, 10). Recently, we described loss of RKIP in up to 20% of primary specimens of acute myelogenous leukemia (AML), an aggressive malignancy of the hematopoietic system (11, 12). Most importantly, functional in vitro studies demonstrated that RKIP inhibits the proliferation and oncogenic transformation of hematopoietic cells (11, 12), which suggests an additional tumor-suppressor function within the hematopoietic system.
Although much progress has been made in evaluating the consequences of RKIP loss, relatively little is known about the reasons behind its downregulation. Clarification of the causative mechanisms will provide further insight into the processes of oncogenesis and may form the basis for the development of specific therapeutic approaches, aiming to restore its expression. This is of particular relevance for AML, as overall survival (OS) with current treatment strategies is still dismal and new therapeutic options are desperately needed (13, 14). However, we and others have previously screened for mutations and deletions within RKIP but could not identify any abnormalities (1, 11, 12). Even though methylation of CpG-rich islands within the RKIP promoter (15) as well as the overexpression of specific transcriptional repressors (16, 17) have been linked to RKIP downregulation in specific tumor entities, we failed to observe these mechanisms in AML.
miRNAs are small non-coding RNAs with a length of approximately 21 to 23 bp. They typically inhibit the translation and/or stability of messenger RNAs (mRNAs), thereby fulfilling a central role in the regulation of gene expression (18, 19). Indeed, aberrant miRNA expression levels can be detected in most human malignancies and their functional relevance for carcinogenesis could be proven in numerous in vitro and in vivo studies (18–21). Recently, aberrant miRNA expression profiles have been linked to the modulation of RKIP, as increased expression of miR-224 and miR-27a caused the downregulation of RKIP in breast and gastric cancer cell lines (22–24). Here, we aimed to evaluate whether miRNAs are involved in the development of RKIP loss in AML.
Materials and Methods
Patient samples and cell lines
AML patient samples for miRNA qPCR as well as for RKIP qPCR and immunoblot analyses were collected at the Division of Hematology, Medical University of Graz, Graz, Austria, and processed as previously described (25–29). For RKIP and miRNA array analysis in the Dutch-Belgian AML cohort II, a total of 214 AML patients were collected at the Erasmus University of Rotterdam, the Netherlands, as previously reported (11, 30–32). Classification of AML was performed according to French-American-British (FAB) and World Health Organization (WHO) guidelines (13, 33). THP-1, U937, and NB4 cell lines were obtained in 2007 from the German National Resource Center for Biological Material (DSMZ, Braunschweig, Germany), where they were characterized by variable number of tandem repeat DNA profiling (VNTR). Low passage stocks were frozen and cells were always passaged for fewer than 6 months after resuscitation. In addition, stocks were regularly re-authenticated in our laboratory by VNTR as previously described (4, 11, 25). HEK-293 cells were obtained from the Center for Medical Research at the Medical University of Graz in 2015, where they were tested by VNTR and passaged for fewer than 6 months after resuscitation in our laboratory. The study was approved by the institutional review board of the Medical University of Graz (24-036 ex 11/12) and informed consent was obtained from all individuals.
Cell culture, lentiviral transduction, and transfection
Cell lines were maintained at 37°C/5% CO2 in RPMI-1640 (for U937, NB4, and THP-1) and DMEM (for HEK-293 and 293T packaging cells), respectively, supplemented with 10% heat-inactivated FCS and 1X Antibiotic-Antimycotic (Thermo Fisher Scientific; comprising 100 U/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL amphotericin B). U937 with stable expression of pMSCV-FLAG-hRKIP or empty vector (as previously described in ref. 11) were additionally maintained with 2.5 μg/mL puromycin. For RKIP knockdown, U937 were lentivirally transduced with either RKIP shRNA or empty control (both psi-LVRU6GP, obtained from Genecopeia) as previously described (34) and stable selection was performed using 2.5 μg/mL puromycin. miR-23a-3p mimics, hairpin inhibitors, and scrambled controls (all obtained from Dharmacon and Qiagen) were transfected at a concentration of 20 nmol/L using DharmaFECT2 (Dharmacon; for U937 and THP-1) or Lipofectamine RNAiMAX (Thermo Fisher Scientific; for HEK-293) in 6-well plates according to the manufacturer's instructions. Cells were harvested after 24 to 48 hours for further analyses. For luciferase reporter assays, HEK-293 were transfected with 0.25 ng/μl pCS2-red fluorescent protein (RFP), together with 0.125 ng/μL RKIP-3′-untranslated region (UTR)-pMirTarget (Origene) as well as with miR-23a mimics or scrambled control (both at a concentration of 20 nmol/L). The RFP-construct was included to enable the compensation of differences in transfection efficiency. Mutation and deletion of binding site 1, respectively, were performed as previously described (35). DNA and Mimics were mixed with 4 μL jetPrime transfection reagent (Polyplus, Illkirch-Graffenstaden, France) in jetPrime buffer (Polyplus) to reach a final volume of 200 μL transfection mix per well. After briefly vortexing and incubating for 10 minutes at room temperature, the mix was added dropwise to each well. Transfection was carried out in complete DMEM medium.
qPCR expression analysis
RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA with TaqMan Reverse Transcription (RT) Reagents (Applied Biosystems) for the mRNA and miScript II RT Kit (Qiagen) for miRNA, respectively. Random hexamers were used for RT of mRNA whereas stem-loop RT primers were used for RT of miRNA. Real-time quantitative PCR (qPCR) for miRNA expression analysis was performed on a LightCycler 480 Instrument II (Roche Life Sciences) using the miScript SYBR Green PCR Kit (Qiagen). qPCR for mRNA expression analysis was performed on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems) using the SYBR Green method (Invitrogen). Expression levels were evaluated using the ΔΔCt method as previously described (11, 36). B2M and GUSB served as control genes for analyses performed with mRNA, RNU6, and SNORD44 for those performed with miRNA. Primer sequences are displayed in Supplementary Table S1. NB4 AML cells (for miRNA qPCR of the Austrian AML cohort I) as well as cells transfected with the empty vector/scrambled control (for all other experiments) served as calibrators.
Immunoblot analysis
Cells were lysed in 100 μL ice-cold lysis RIPA-Buffer (Sigma-Aldrich), supplemented with Protease and Phosphatase inhibitor cocktails (Sigma-Aldrich and Thermo Fisher Scientific). Immunoblots were then performed as previously described (11, 12, 35) using Mini-PROTEAN TGX gels (Bio-Rad) for electrophoresis and the Bio-Rad Trans Blot TurboBlotting System for transfer. Polyvinylidene difluoride membranes (Bio-Rad) were incubated with anti-RKIP (#07-137, Merck Millipore), anti-IκB (#9242S, Cell Signaling Technology), anti–β-actin (#A5441; Sigma-Aldrich) and anti-Vinculin (#Ab129002, Abcam) as previously reported (11, 12, 35). Band intensities were compared using ImageJ (37).
Luciferase reporter assays
Twenty-four hours after transfection, cells were harvested and lysis was performed using 1X cell culture lysis reagent (CCLR, Promega) according to the manufacturer's instructions. Subsequently, luciferase activity and RFP fluorescence values were determined using a Plate chameleon reader (Hidex). The reader injected automatically 100 μL of luciferase Assay System Reagent (Promega) into each well of a 96-well plate containing 20 μL of cell lysate, CCLR was used as background control. Luminescence was measured for 10 seconds after a delay of 2 seconds. Fluorescence was detected with a 544/616 nm filter set.
Analysis of cell growth and proliferation
For growth curves, U937 were seeded at a density of 1 × 104 cells/mL in reduced serum conditions (5%). The amount of viable cells was measured 5 days later using a Bio-Rad TC20 automated cell counter (Bio-Rad) via trypan blue exclusion assay. Proliferation was additionally assessed via bromodeoxyuridine (BrdUrd)/7-AAD staining using the APC BrdUrd Flow Kit (BD Pharmingen) according to the manufacturer's instructions and as previously described (11). Fifty μmol/L BrdUrd was added for 30 minutes and cells were stained with APC-anti–BrdUrd (1:50) for 20 minutes. Ten-thousand stained cells were measured on a BD-LSRII Flow Cytometer (BD Biosciences) and analyzed with Kaluza Flow Cytometry Analysis Software v1.2 (Beckman Coulter).
Database retrieval and statistical analyses
Expression data for RKIP (as obtained by RNA Sequencing V2 RSEM as well as by Affymetrix U133 and Agilent microarrays) and miRNAs (as obtained by miRNA arrays and miRNA Sequencing) were downloaded and analyzed from The Cancer Genome Atlas (TCGA, www.cancergenome.nih.gov; Supplementary Table S2). In case of availability, data were downloaded and analyzed using the cBioPortal for Cancer Genomics (www.cbioportal.org/public-portal/index.do; refs. 38, 39). In addition, RKIP mRNA and miRNA expression values were analyzed in previously published array and qPCR datasets (data available at http://www.ncbi.nlm.nih.gov/geo under accession numbers GSE49665, GSE1159 and GSE6891; refs. 30–32, 40). A comparison of miRNA expression levels between primary patient samples with and without RKIP loss was performed using the Mann–Whitney–Wilcoxon test. The same test was applied for comparison of RKIP and miRNA expression, respectively, as well as between AML patients with and without monocytic phenotypes (AML M4/M5 vs. other AML subgroups). To test for a correlation between RKIP and miRNA expression levels, Spearman-Rho correlation coefficients were calculated. miR-23a expression was related to overall and relapse-free survival in AML cohort II and to OS in AML cohort III by Cox Regression analysis. For analysis of in vitro experiments, Student t test was calculated from at least three independent experiments. SPSS 22.0 (SPSS Inc.) and R 3.2.2 (www.r-project.org) were used for analysis. All tests were performed two-sided and a P value of <0.050 was considered statistically significant.
Results
Increased expression of miR-23a in AML correlates with RKIP loss
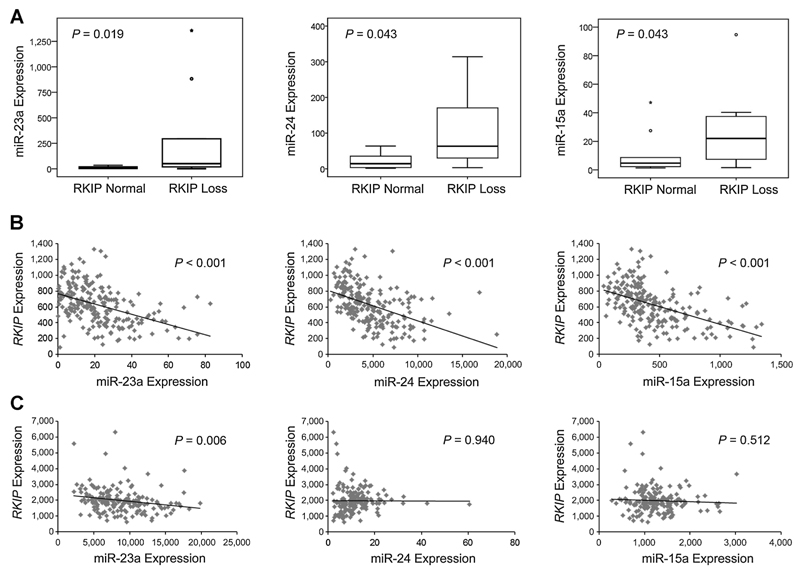
In a first step, we aimed to identify miRNAs, which are associated with decreased expression of RKIP in AML. Therefore, we analyzed 33 AML specimens, which had been evaluated for RKIP protein expression by immunoblot, as well as for the expression of more than 600 miRNAs by microarray analyses previously (11, 40). By comparing miRNA expression profiles between patients defined as harboring either normal (n = 27) or decreased (n = 6) RKIP expression (for definition of RKIP loss see Supplementary Fig. S1 and ref. 11), we identified a set of seven miRNAs with statistically significant differential expression between these groups (miR-23a, miR-23b, miR-24, miR-15a, miR-320a, miR-518b and miR-519d, data not shown). We then sought to validate these results by the means of qPCR in an additional cohort of 20 AML patient specimens (cohort I). Out of these, 10 demonstrated normal RKIP expression, both at mRNA and protein level, whereas the other 10 exhibited loss of RKIP. Importantly, a set of three miRNAs could be validated and demonstrated increased expression in samples with RKIP loss (miR-23a, miR-24, and miR-15a; Fig. 1A). To further corroborate these findings, we analyzed RKIP mRNA as well as miR-23a, miR-24, and miR-15a expression in two additional and independent datasets comprising almost 400 AML patient specimens. Cohort II consisted of 214 Dutch-Belgian AML patients and was previously analyzed by mRNA arrays and miRNA qPCRs (30–32), whereas cohort III consisted of 173 specimens and was acquired via TCGA database retrieval. Although the techniques of expression analysis differed between these cohorts, correlation of decreased RKIP expression with increased levels of miR-23a could be validated in all of them (Fig. 1B and C). Although cohort II revealed an additional correlation of decreased RKIP expression with increased levels of miR-24 and miR-15a (Fig. 1B), no additional correlations with statistical significance could be observed in cohort III (Fig. 1C). Of note, miR-224 and miR-27a, which have been linked to the modulation of RKIP in solid cancer cell lines previously (22–24), failed to correlate with RKIP expression (data not shown).
Figure 1.
Increased expression of miR-23a correlates with decreased expression of RKIP in AML. A, box plots showing a significant increase of miR-23a, miR-24, and miR-15a expression in AML patient specimens defined as RKIP loss in AML cohort I (n = 20). miRNA expression values were assessed by qPCR and are displayed as x-fold expression of the calibrator (NB4 cells). RKIP expression was evaluated by qPCR and immunoblot. RKIP loss was defined as described in Supplementary Fig. S1 and as previously reported (11). B, scatter plots showing a significant and inverse correlation between the expression of RKIP mRNA (displayed as microarray expression values at the y-axis) and the miRNAs miR-23a, miR-24, and miR-15a (displayed as miRNA array expression values at the x-axis) in 214 patient specimens of the Dutch-Belgian AML cohort II (30–32). C, scatter plots showing a significant and inverse correlation between the expression of RKIP mRNA (displayed as RNA Sequencing V2 RSEM expression values at the y-axis) and miR-23a (displayed as miRNA-sequencing expression values at the x-axis) in the TCGA AML cohort III (n = 173).
RKIP loss and increased expression of miR-23a correlate to myelomonocytic and monocytic AML phenotypes
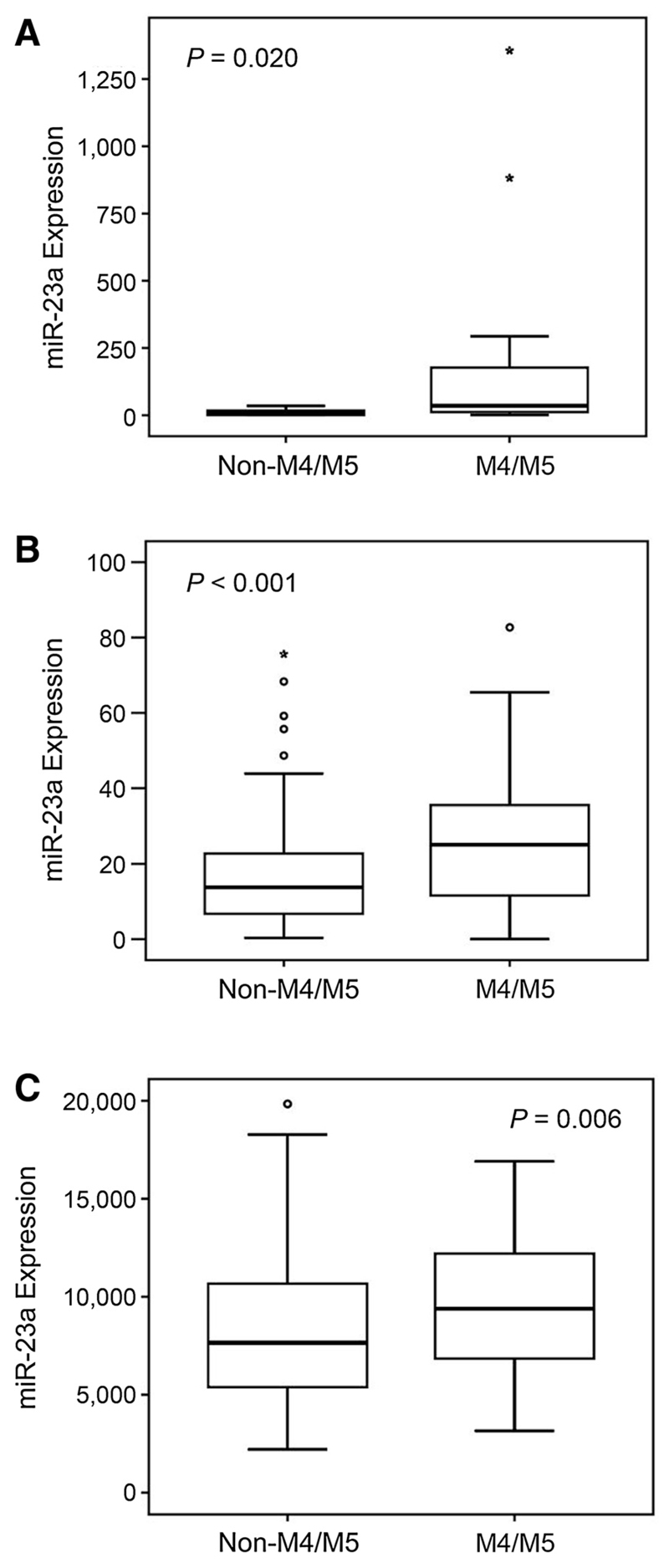
We previously have shown that decreased expression of RKIP correlates with AML subgroups with myelomonocytic and monocytic differentiation (FAB subgroups M4/M5; ref. 11). Therefore, we were interested, whether this applies for increased expression of miR-23a as well. When looking at the 20 AML specimens of cohort I, AML M4/M5 phenotypes were significantly enriched in the 10 samples defined as RKIP loss (Supplementary Fig. S2). In agreement with our hypothesis, miR-23a expression was significantly increased in AML specimens characterized as AML M4/M5 (Fig. 2A). We then tried to corroborate these findings in the two additional AML cohorts comprising almost 400 patients. In line with our previous studies, RKIP expression levels were significantly decreased in AML M4/M5 as compared with all other subtypes in both cohorts (Supplementary Fig. S3 and ref. 11). Importantly, the expression of miR-23a was indeed increased in AML M4/M5 phenotypes (Fig. 2B and C). Of note, miR-23a expression failed to correlate with both overall and relapse-free survival in these AML cohorts (data not shown).
Figure 2.
miR-23a demonstrates increased expression in AML with myelomonocytic and monocytic differentiation. A, box plots showing a significant increase of miR-23a expression (measured by qPCR and displayed as x-fold expression of a calibrator; NB4 cells) in AML phenotypes with myelomonocyitc and monocytic phenotypes (M4/M5) in the Austrian AML cohort I (n = 20). B and C, these results could be corroborated in the Dutch-Belgian AML cohort II (B; n = 214; miR-23a expression displayed as miRNA array expression values; refs. 30–32) and in the TCGA cohort III (C; n = 173; miR-23a expression displayed as miRNA-sequencing expression values).
RKIP expression is regulated by miR-23a
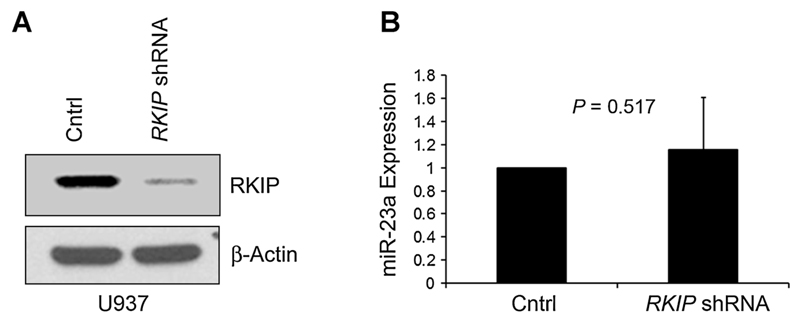
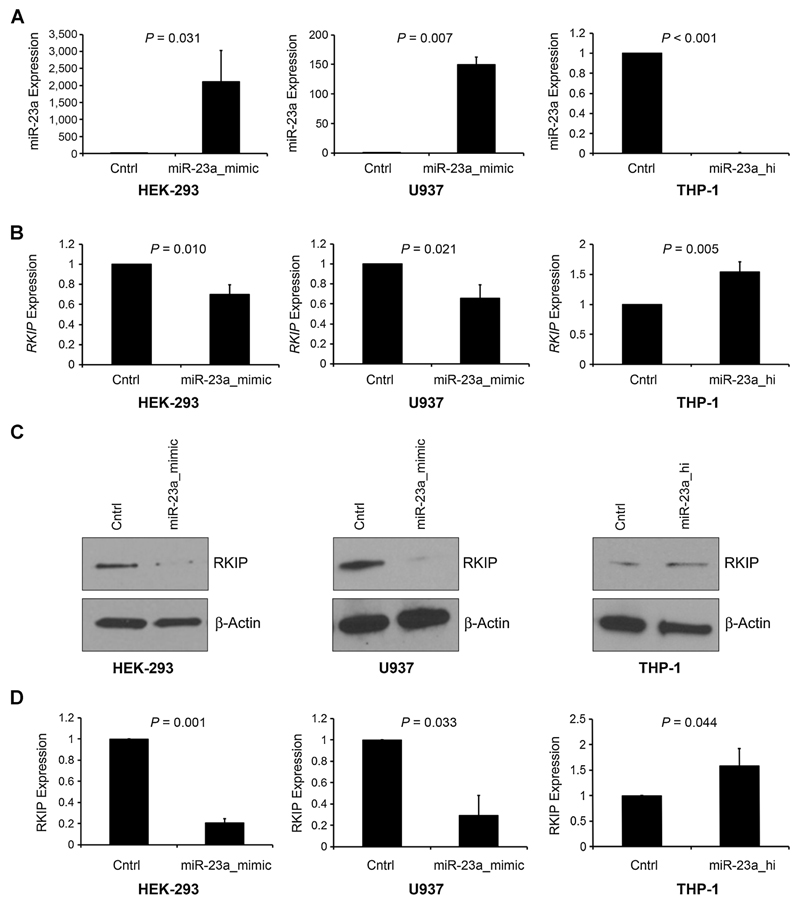
As outlined above, we observed inverse correlation between RKIP and miR-23a expression in AML. To further evaluate a potential functional involvement of miR-23a in the downregulation of RKIP in AML, we used a series of AML and nonhematopoietic cell lines, including U937, THP1, and HEK293. Initially, we aimed to test the possibility that RKIP loss is the primary event, which in turn causes the miRNA upregulation, as shown for miR-98 and miR-200 (41, 42). Therefore, we performed a lentiviral shRNA-based knockdown of RKIP in U937 AML cells and subsequently monitored miR-23a expression levels. No significant differences could be observed between cells with and without RKIP knockdown (Fig. 3 and Supplementary Fig. S4), indicating that increased expression of miR-23a in AML is no effect of RKIP loss. In a next step, we aimed to clarify the role of miR-23a in the downregulation of RKIP. HEK-293 cells were transiently transfected with miR-23a mimics and RKIP expression was monitored by qPCR and immunoblot. Importantly, miR-23a overexpression caused a statistically significant downregulation of RKIP expression, both at the mRNA and protein level (Fig. 4A–C, left). To corroborate these findings in the context of AML, we repeated these experiments in U937, a cell line exhibiting strong expression of endogenous RKIP (11). Again, miR-23a transfection induced a statistically significant downregulation of RKIP (Fig. 4A–C, middle). To further prove the mechanistic involvement of miR-23a in RKIP regulation, we transfected an miR-23a hairpin inhibitor in THP-1 AML cells, which has been shown to express RKIP at a low level previously (11). Indeed, miR-23 knockdown caused the statistically significant upregulation of RKIP (Fig. 4A–C, right). These results were confirmed with different miR-23a mimics and hairpin inhibitors that were obtained from different companies (for details see Materials and Methods; data not shown). Taken together, these data indicate that miR-23a suppresses the expression of RKIP in AML.
Figure 3.
Increased expression of miR-23a has no effect on RKIP loss. A, U937 cells were lentivirally transduced with either RKIP-shRNA or an empty vector control (Cntrl). Successful knockdown was confirmed by immunoblot. β-actin was used as a loading control. B, qPCR expression analysis demonstrated that miR-23a expression is not affected by the knockdown of RKIP. Graphs represent the mean of three independent experiments ± SD; expression values are given as x-fold expression of control (Cntrl).
Figure 4.
miR-23a modulates the expression of RKIP. A, HEK-293 and U937 (both exhibiting strong RKIP expression; ref. 11) cells were transfected with either miR-23a mimic or unspecific control (Cntrl). THP-1 (exhibiting decreased expression of RKIP; ref. 11) was transfected with either miR-23a hairpin inhibitor (miR-23a_hi) or control. miR-23a expression was assessed by qPCR and is displayed as x-fold expression of the control-transfected cells. B, RKIP mRNA expression analysis by qPCR of the conditions mentioned above demonstrated significant downregulation of RKIP after transfection with miR-23a mimic and significant upregulation after transfection with miR-23a_hi. RKIP expression is displayed as x-fold expression of the control-transfected cells. C, immunoblot analyses demonstrating decreased expression of RKIP protein in cells transfected with miR-23a mimic and increased expression in conditions where miR-23a_hi had been used. β-Actin was used as a loading control. D, densitometric quantification of the immunoblot results, displayed as x-fold expression of the control-transfected cells. Graphs represent the mean of three independent experiments ± SD; expression values are given as x-fold expression of control.
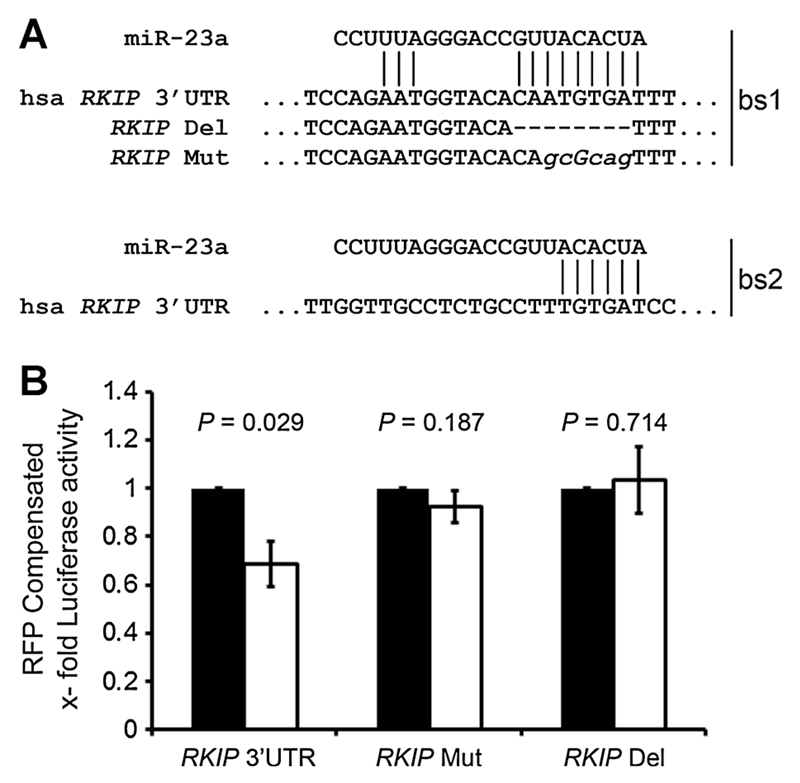
miR-23a modulates RKIP expression by direct binding to the RKIP 3′UTR
Having proven that miR-23a regulates RKIP expression, we next sought to identify, whether this regulation is mediated via direct binding of miR-23a within the RKIP 3′UTR. Therefore, we initially performed an in silico analysis using 10 miRNA target prediction algorithms and identified RKIP as a putative direct target of miR-23a in 5 of 10 (Supplementary Fig. S5). In more detail, we identified two potential binding sites for miR-23a within the 3′UTR region with binding site 1 displaying the strongest match within the seed region (Fig. 5A). In a next step, we continued to work with an expression construct, where the 3′UTR of RKIP was attached to the coding region of luciferase. Importantly, transfection of this construct together with miR-23a mimics resulted in a significant downregulation of luciferase activity, whereas scrambled miR-controls failed to do so (Fig. 5B). To further prove that regulation of RKIP by miR-23a is caused by direct interaction, we altered the 3′UTR of RKIP either by mutation (RKIP Mut) or by deletion (RKIP Del) of the putative binding site 1 (Fig. 5A). Indeed, both approaches inhibited the miR-23a–mediated downregulation of luciferase activity (Fig. 5B), indicating that miR-23a regulates the expression of RKIP by direct interaction with binding site 1 within the 3′UTR.
Figure 5.
miR-23a modulates the expression of RKIP via direct interaction with the 3′UTR. A, sequence of the human RKIP 3′UTR (hsa RKIP) showing the two miR-23a–binding sites (bs1-2). Matching regions are highlighted by lines. Alteration of bs1 by either deletion (RKIP Del) or mutation (RKIP Mut) is displayed. B, HEK-293 cells were transfected with either miR-23a mimic (white bars) or unspecific control (Cntrl, black bars) together with luciferase reporter clones, containing (i) wild-type RKIP 3′UTR (RKIP 3′UTR), (ii) RKIP Del, or (iii) RKIP Mut. pCS2-RFP was included in all transfections to compensate for different transfection efficiencies. Graphs represent the mean RFP-compensated luciferase activity of three independent experiments ± SD; values are given as x-fold expression of the respective control-transfected setting.
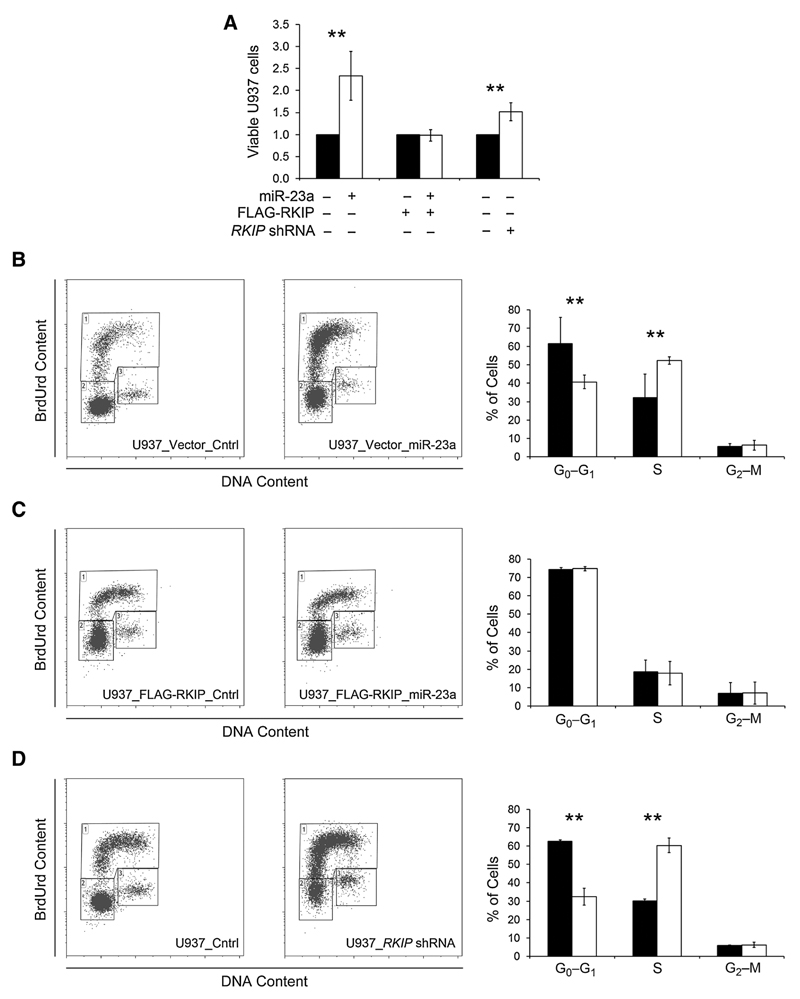
miR-23a increases proliferation of hematopoietic cells via downregulation of RKIP
We previously demonstrated that stable overexpression of FLAG-RKIP in AML cell lines caused a significant decrease of cellular growth. This was mediated via decreased proliferation as assessed in BrdUrd/PI cell-cycle assays (11). To delineate the role of miR-23a within these processes, we now performed additional transfection of miR-23a mimics within these cells. Interestingly, when vector transfected controls without RKIP over-expression were studied, miR-23a overexpression caused a significant increase in cellular growth (Fig. 6A), corresponding to an increased rate of proliferating cells in BrdUrd/7-AAD assays (Fig. 6B). To further prove that these biologic effects are indeed mediated via downregulation of RKIP, we repeated these experiments in U937 with stable overexpression of FLAG-RKIP. Without a 3′UTR, this construct is resistant to miR-23a–induced decay and should therefore rescue the biologic effects caused by miR-23a. Indeed, transfection of miR-23a failed to induce a phenotype in this cellular system (Fig. 6A and C). In agreement with these data, lentiviral shRNA knockdown of RKIP in parental U937 caused increased cellular growth (Fig. 6A) as well as increased proliferation in BrdUrd/7-AAD assays (Fig. 6D).
Figure 6.
miR-23a induces the proliferation of hematopoietic cells via downregulation of RKIP. A, U937 with stable expression of FLAG-RKIP and empty vector, respectively, were transfected with either miR-23a mimic or unspecific control (Cntrl) as indicated. In addition, parental U937 cells were lentivirally transduced with either RKIP shRNA or control. Cells were seeded and maintained as described in Materials and Methods and viable cells were counted after 5 days. For comparison of the different conditions, respective Cntrl situations were set at a value of 1 and the relative increase of viable cells in the miR-23a/RKIP shRNA conditions was calculated using the ratio viable cells miR-23a/RKIP shRNA to viable cells control. B–D, BrdUrd/7-AAD cell-cycle/proliferation assays were performed in all the settings described above to evaluate the percentage of cells in S-phase (top gate), G0–G1-phase (left bottom gate), and G2–M-phase (right bottom gate), respectively. Black bars, control; white bars, miR-23a/RKIP shRNA. The graphs summarize the results of at least three independent experiments. Data are expressed as mean values ± SD; *, P < 0.05; **, P < 0.01.
The miR-23a/27a/24-2/RKIP axis is of general relevance for other cancer entities as well
Up to this point, we could show that increased expression of miR-23a causes downregulation of RKIP within AML. Of note, miR-23a is part of the miR-23a/27a/24-2 cluster, and the members within this network have been shown to share many common targets (43). Indeed, miR-27a has been identified as RKIP modulator in lung cancer previously (24). We thus were interested whether an miR-23a/27a/24-2–mediated RKIP downregulation is of relevance for other malignancies as well, and to possibly identify the responsible cluster member. Therefore, we performed a data-base retrieval via TCGA and analyzed tumor entities, where (i) decreased expression of RKIP has been previously reported, and (ii) data for RKIP and miR-23a/27a/24-2 expression levels were available. All together, we were able to analyze 14 additional cancer entities comprising 4,342 primary patient specimens. A statistically significant inverse correlation between the expression of RKIP and at least one of the cluster miRNAs could be shown in 10 of 14 cancers. miR-23a proved to be the predominant cluster member with significant correlations in nine of these entities (Table 1).
Table 1.
Correlation between RKIP and miRNA expression in human cancer
| Cancer entity (n = 14) | No. of patient samples (total n = 4,342) | miR-23a |
miR-24-2 |
miR-27a |
|||
|---|---|---|---|---|---|---|---|
| Coefficient | P | Coefficient | P | Coefficient | P | ||
| Lung squamous cell carcinoma (LUSC) | 104 | −0.131 | 0.186 | 0.176 | 0.074 | −0.291 | 0.003b |
| Lung adenocarcinoma (LUAD) | 447 | −0.201 | <0.001b | −0.021 | 0.661 | 0.172 | <0.001b |
| Prostate adenocarcinoma (PRAD) | 493 | −0.190 | <0.001b | −0.174 | <0.001b | −0.095 | 0.060 |
| Kidney renal clear cell carcinoma (KIRC) | 162 | −0.280 | <0.001b | −0.224 | 0.004b | −0.272 | <0.001b |
| Bladder urothelial carcinoma (BLCA) | 405 | −0.306 | <0.001b | −0.187 | <0.001b | −0.354 | <0.001b |
| Invasive breast carcinoma (BRCA) | 299 | −0.188 | 0.001b | −0.141 | 0.015b | −0.158 | 0.006b |
| Ovarian serous cystadenocarcinoma (OV) | 513 | −0.137 | 0.002b | −0.084 | 0.058 | −0.051 | 0.253 |
| Uterine corpus endometrial carcinoma (UCEC) | 174 | −0.089 | 0.242 | 0.025 | 0.747 | −0.098 | 0.196 |
| Pancreatic adenocarcinoma (PAAD) | 179 | −0.492 | <0.001b | −0.368 | <0.001b | −0.297 | <0.001b |
| Liver hepatocellular carcinoma (LIHC) | 369 | −0.425 | <0.001b | −0.277 | <0.001b | −0.407 | <0.001b |
| Colon and rectal adenocarcinoma (COAD) | 78 | 0.1 | 0.382 | 0.001 | 0.997 | 0.073 | 0.526 |
| Head and neck squamous cell carcinoma (HNSC) | 476 | −0.104 | 0.023a | −0.126 | 0.006b | −0.07 | 0.087 |
| Skin cutaneous melanoma (SKCM) | 447 | 0.044 | 0.354 | 0.039 | 0.412 | 0.069 | 0.145 |
| Glioblastoma multiforme (GBM) | 196 | 0.048 | 0.506 | 0.002 | 0.975 | 0.044 | 0.542 |
NOTE: Expression values of RKIP mRNA (as obtained by RNA Sequencing V2 RSEM, Affymetrix U133, or Agilent microarrays) and miRNAs (as obtained by miRNA arrays) were correlated using Spearman-Rho. Negative coefficients equal to inverse correlation.
P < 0.05.
P < 0.01.
Discussion
Loss of RKIP has been described in a significant subset of patients with AML and has shown to be functionally relevant for the pathogenesis of this aggressive malignancy (11, 12). In this study, we aimed at delineating the underlying mechanisms behind the downregulation of RKIP in this disease, and therefore studied the role of miRNA involvement. By analyzing more than 400 primary AML patient specimens, we identified a strong correlation of RKIP loss with increased expression of miR-23a. In subsequent in vitro studies, we could show that enforced expression of miR-23a indeed causes a downregulation of RKIP, and that this downregulation is mediated via direct binding to the 3′UTR of RKIP. In agreement with our data, increased expression of miR-23a has been reported in acute leukemias previously (43, 44). Moreover, the proleukemogenic effects of increased miR-23a expression were recently demonstrated in a murine in vivo model (45). In agreement with this publication, we now demonstrate that overexpression of miR-23a—just like knockdown of RKIP— induces increased proliferation of hematopoietic cells. This phenotype could be rescued by additional overexpression of an RKIP expression construct without the miR-23a–binding region, indicating that at least some of the leukemogenic effects of miR-23a are truly mediated via downregulation of RKIP. Noteworthy, increased expression of miR-23a failed to prove as a prognostic marker within this study. This is unexpected, as we previously demonstrated a link between decreased expression of RKIP and favorable prognosis in AML (11). Although the reasons behind this phenomenon remain to be elucidated, one might speculate that miR-23a additionally regulates other, RKIP-independent targets as well. Irrespective of its effects on RKIP, miR-23a could thereby influence other properties of leukemic cells with prognostic relevance, such as therapeutic resistance. Hence, analysis of the genetic associations of RKIP loss and increased miR-23a expression within larger, preferably prospective and randomized patient cohorts with well-documented clinical and molecular data will be of great interest.
Previous work further suggested that increased expression of miR-23a is specific for acute leukemias with myeloid differentiation, as evidenced by the fact that its expression levels were significantly lower in specimens of acute lymphatic leukemia (43, 44). Our results, showing a significantly increased miR-23a expression and a decrease of RKIP in AMLs with monocytic and myelomonocytic phenotypes are in line with these observations and suggest a possible relation of the miR-23a/RKIP axis to the myelomonocytic differentiation of acute leukemias. In particular, increased expression of miR-23a might serve as a biomarker with diagnostic potential in this setting, which is based on the fact that the small size of miRNAs offers protection during tissue processing, thereby guaranteeing stability and reproducibility of the expression analyses (46). Indeed, miRNA expression profiling has been shown to be feasible from formalin-fixed paraffin-embedded tissues, including decalcified bone marrow biopsies (46). Whether the miR-23a/RKIP axis is indeed functionally involved in the myeloid lineage commitment of hematopoietic stem and progenitor cells, thereby facilitating a myeloid and/or monocytic differentiation of the leukemic clone, is currently unclear.
miR-23a is part of the miR-23a/27a/24-2 cluster, a network of closely related miRNAs showing a variety of similar functions and targets. Deregulation and overexpression of the whole cluster or its single miRNAs has been shown in a variety of human cancers and proved to be functionally involved in malignant transformation (43). Of particular interest, this cluster has been related to RKIP modulation previously. Li and colleagues (24) demonstrated that enforced expression of miR-27a causes downregulation of RKIP, thereby conferring cisplatin resistance in lung cancer cell lines. Although the members of this cluster share many common targets, these miRNAs itself might underlie a completely different regulation and processing, which might vary between different disease states. Indeed, only miR-23a correlated to the expression of RKIP in AML, whereas the other cluster members failed to do so. Therefore, although our data further corroborate the functional involvement of the miR-23a/27a/24-2 cluster in the pathogenesis of RKIP loss, they suggest that miR-23a is the causative miRNA in AML. The recent description of proleukemogenic effects of miR-23a overexpression in a murine in vivo model further strengthens this conclusion (45).
In consideration of the relevance of the miR-23a/RKIP axis in AML and the importance of the miR-23a/27a/24-2 cluster in RKIP regulation, we were interested, whether such an association exists in other cancers with decreased RKIP expression as well. Therefore, we performed a database retrieval via the TCGA and analyzed expression levels of RKIP and the miR-23a/27a/24-2 cluster members. Altogether, we were able to extract 14 additional cancer entities, comprising more than 4,300 primary patient specimens. Importantly, an inverse correlation between RKIP and one or more members of the miR-23a/27a/24-2 cluster could be observed in ten out of fourteen cancer entities, suggesting that the miR-23a/27a/24-2/RKIP cascade is of general relevance for human carcinogenesis. With the exception of squamous cell lung carcinoma, where statistical significance could only be reached for miR-27a, increased expression of miR-23a correlated to decreased RKIP levels in all of these cancers making it the predominant cluster member. The relevance of this finding is further highlighted by the fact that aberrant miR-23a expression levels have been described in most of these cancer entities previously. Moreover, its functional relevance for the pathogenesis of these diseases as well as a role in the development of chemoresistance have been suggested (43, 47–50).
In summary, we have identified that increased expression of miR-23a, a member of the miR-23a/27a/24-2 cluster, is correlated with a loss of RKIP in AML and that both events are linked to myelomonocytic and monocytic AML phenotypes. We could further show that miR-23a overexpression is indeed causative for RKIP decrease in AML by directly binding to its 3′UTR. Finally, we could highlight the importance of the miR-23a/27a//24-2/RKIP axis in a broad range of human cancer entities and thereby provide a mechanism for RKIP downregulation with general relevance for the field of cancer research. Therefore, our results might provide the basis to develop specific therapies with the potential to restore the expression of RKIP.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Acknowledgments
The authors would like to acknowledge the TCGA Research Network (http://cancergenome.nih.gov/) for generating datasets analyzed within this article. In this respect, the authors are also thankful to the contribution of the appropriate specimen donors and research groups.
Grant Support
This work was supported by the Austrian Science Fund under Grant No. P 26619-B19 (A. Zebisch). Work in the laboratory of A. Zebisch, H. Sill, and A. Wölfler is further supported by Leukaemiehilfe Steiermark. Work of M. Pichler is supported by the Oesterreichische Nationalbank (Anniversary Fund, project no. 14869). Work by M. Scheideler is supported by the Austrian Science Fund (P257-29-B26).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: S. Hatzl, O. Geiger, V. Caraffini, M. Pichler, M. Scheideler, J. Troppmair, H. Sill, A. Zebisch
Development of methodology: S. Hatzl, O. Geiger, M.K. Kuepper, V. Caraffini, T. Seime, E. Nussbaumer, M. Pichler, A. Zebisch
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Hatzl, O. Geiger, M.K. Kuepper, V. Caraffini, T. Furlan, E. Nussbaumer, R. Wieser, K. Nowek, M. Jongen-Lavrencic, J. Troppmair, A. Zebisch
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Hatzl, O. Geiger, M.K. Kuepper, V. Caraffini, T. Seime, M. Pichler, M. Scheideler, M. Jongen-Lavrencic, F. Quehenberger, A. Wölfler, J. Troppmair, H. Sill, A. Zebisch
Writing, review, and/or revision of the manuscript: S. Hatzl, O. Geiger, V. Caraffini, R. Wieser, M. Pichler, M. Scheideler, K. Nowek, M. Jongen-Lavrencic, A. Wölfler, H. Sill, A. Zebisch
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): T. Seime, K. Nowek, H. Sill
Study supervision: A. Zebisch
References
- 1.Al-Mulla F, Bitar MS, Taqi Z, Yeung KC. RKIP: much more than raf kinase inhibitory protein. J Cell Physiol. 2013;228:1688–702. doi: 10.1002/jcp.24335. [DOI] [PubMed] [Google Scholar]
- 2.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, et al. Suppression of raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–7. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 3.Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, et al. Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol Cell Biol. 2001;21:7207–17. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamiman K, Keller JM, Mizokami A, Zhang J, Keller ET. Survey of raf kinase inhibitor protein (RKIP) in multiple cancer types. Crit Rev Oncog. 2014;19:455–68. doi: 10.1615/critrevoncog.2014011987. [DOI] [PubMed] [Google Scholar]
- 5.Granovsky AE, Rosner MR. Raf kinase inhibitory protein: a signal transduction modulator and metastasis suppressor. Cell Res. 2008;18:452–7. doi: 10.1038/cr.2008.43. [DOI] [PubMed] [Google Scholar]
- 6.Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, et al. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol. 2006;24:5672–9. doi: 10.1200/JCO.2006.07.5499. [DOI] [PubMed] [Google Scholar]
- 7.Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R, Rhodes D, et al. Metastasis suppressor gene raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–56. doi: 10.1002/pros.20319. [DOI] [PubMed] [Google Scholar]
- 8.Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–89. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 9.Escara-Wilke J, Keller JM, Ignatoski KM, Dai J, Shelley G, Mizokami A, et al. Raf kinase inhibitor protein (RKIP) deficiency decreases latency of tumorigenesis and increases metastasis in a murine genetic model of prostate cancer. Prostate. 2015;75:292–302. doi: 10.1002/pros.22915. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, et al. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J Biol Chem. 2004;279:17515–23. doi: 10.1074/jbc.M313816200. [DOI] [PubMed] [Google Scholar]
- 11.Zebisch A, Wolfler A, Fried I, Wolf O, Lind K, Bodner C, et al. Frequent loss of RAF kinase inhibitor protein expression in acute myeloid leukemia. Leukemia. 2012;26:1842–9. doi: 10.1038/leu.2012.61. [DOI] [PubMed] [Google Scholar]
- 12.Zebisch A, Haller M, Hiden K, Goebel T, Hoefler G, Troppmair J, et al. Loss of RAF kinase inhibitor protein is a somatic event in the pathogenesis of therapy-related acute myeloid leukemias with C-RAF germline mutations. Leukemia. 2009;23:1049–53. doi: 10.1038/leu.2009.68. [DOI] [PubMed] [Google Scholar]
- 13.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the european LeukemiaNet. Blood. 2010;115:453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 14.Sill H, Olipitz W, Zebisch A, Schulz E, Wolfler A. Therapy-related myeloid neoplasms: pathobiology and clinical characteristics. Br J Pharmacol. 2011;162:792–805. doi: 10.1111/j.1476-5381.2010.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li DX, Cai HY, Wang X, Feng YL, Cai SW. Promoter methylation of raf kinase inhibitory protein: a significant prognostic indicator for patients with gastric adenocarcinoma. Exp Ther Med. 2014;8:844–50. doi: 10.3892/etm.2014.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72:3091–104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 17.Beach S, Tang H, Park S, Dhillon AS, Keller ET, Kolch W, et al. Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene. 2008;27:2243–8. doi: 10.1038/sj.onc.1210860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbri M, Croce CM, Calin GA. Micrornas. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 20.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L, Dai T, Lin X, Zhao X, Chen X, Wang C, et al. MicroRNA-224 targets RKIP to control cell invasion and expression of metastasis genes in human breast cancer cells. Biochem Biophys Res Commun. 2012;425:127–33. doi: 10.1016/j.bbrc.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Li P, Li B, Sun P, Zhang J, Wang B, et al. RKIP suppresses gastric cancer cell proliferation and invasion and enhances apoptosis regulated by microRNA-224. Tumour Biol. 2014;35:10095–103. doi: 10.1007/s13277-014-2303-4. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Wang Y, Song Y, Fu Z, Yu W. miR-27a regulates cisplatin resistance and metastasis by targeting RKIP in human lung adenocarcinoma cells. Mol Cancer. 2014;13:193. doi: 10.1186/1476-4598-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz E, Klampfl P, Holzapfel S, Janecke AR, Ulz P, Renner W, et al. Germline variants in the SEMA4A gene predispose to familial colorectal cancer type X. Nat Commun. 2014;5:5191. doi: 10.1038/ncomms6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fried I, Wolfler A, Quehenberger F, Hoefler G, Sill H, Zebisch A. Mutations inDNMT3A and loss of RKIP are independent events in acute monocytic leukemia. Haematologica. 2012;97:1936–7. doi: 10.3324/haematol.2012.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz E, Valentin A, Ulz P, Beham-Schmid C, Lind K, Rupp V, et al. Germline mutations in the DNA damage response genes BRCA1, BRCA2, BARD1 and TP53 in patients with therapy related myeloid neoplasms. J Med Genet. 2012;49:422–8. doi: 10.1136/jmedgenet-2011-100674. [DOI] [PubMed] [Google Scholar]
- 28.Geiger O, Hatzl S, Kashofer K, Hoefler G, Wolfler A, Sill H, et al. Deletion of SPRY4 is a frequent event in secondary acute myeloid leukemia. Ann Hematol. 2015;94:1923–4. doi: 10.1007/s00277-015-2445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zebisch A, Sill H. Are mouthwashes a reliable source of constitutional DNA in patients with leukemia? Leuk Res. 2008;32:1164–5. doi: 10.1016/j.leukres.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–28. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 31.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. Micro-RNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–85. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 32.Nowek K, Sun SM, Dijkstra MK, Bullinger L, Dohner H, Erkeland SJ, et al. Expression of a passenger miR-9* predicts favorable outcome in adults with acute myeloid leukemia less than 60 years of age. Leukemia. 2016;30:303–9. doi: 10.1038/leu.2015.282. [DOI] [PubMed] [Google Scholar]
- 33.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the WHO classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 34.Deutsch AJ, Rinner B, Wenzl K, Pichler M, Troppan K, Steinbauer E, et al. NR4A1-mediated apoptosis suppresses lymphomagenesis and is associated with a favorable cancer-specific survival in patients with aggressive B-cell lymphomas. Blood. 2014;123:2367–77. doi: 10.1182/blood-2013-08-518878. [DOI] [PubMed] [Google Scholar]
- 35.Zebisch A, Staber PB, Delavar A, Bodner C, Hiden K, Fischereder K, et al. Two transforming C-RAF germ-line mutations identified in patients with therapy-related acute myeloid leukemia. Cancer Res. 2006;66:3401–8. doi: 10.1158/0008-5472.CAN-05-0115. [DOI] [PubMed] [Google Scholar]
- 36.Milewska M, Romano D, Herrero A, Guerriero ML, Birtwistle M, Quehenberger F, et al. Mitogen-inducible gene-6 mediates feedback inhibition from mutated BRAF towards the epidermal growth factor receptor and thereby limits malignant transformation. PLoS One. 2015;10:e0129859. doi: 10.1371/journal.pone.0129859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rommer A, Steinleitner K, Hackl H, Schneckenleithner C, Engelmann M, Scheideler M, et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer. 2013;13:364. doi: 10.1186/1471-2407-13-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun M, Gomes S, Chen P, Frankenberger CA, Sankarasharma D, Chung CH, et al. RKIP and HMGA2 regulate breast tumor survival and metastasis through lysyl oxidase and syndecan-2. Oncogene. 2014;33:3528–37. doi: 10.1038/onc.2013.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z, Cheng Q, Ma Z, Xi H, Peng R, Jiang B. Overexpression of RKIP inhibits cell invasion in glioma cell lines through upregulation of miR-98. Biomed Res Int. 2013;2013:695179. doi: 10.1155/2013/695179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–6. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang YC, Ye H, Zeng Z, Chin YE, Huang YN, Fu GH. The NF-kappaB p65/miR-23a-27a-24 cluster is a target for leukemia treatment. Oncotarget. 2015;6:33554–67. doi: 10.18632/oncotarget.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon JE, Wong JJ, Rasko JE. MicroRNAs in myeloid malignancies. Br J Haematol. 2013;162:162–76. doi: 10.1111/bjh.12364. [DOI] [PubMed] [Google Scholar]
- 47.Jin AH, Wei ZL. Molecular mechanism of increased sensitivity of cisplatin to ovarian cancer by inhibition of microRNA-23a expression. Int J Clin Exp Med. 2015;8:13329–34. [PMC free article] [PubMed] [Google Scholar]
- 48.Qu WQ, Liu L, Yu Z. Clinical value of microRNA-23a upregulation in non–small cell lung cancer. Int J Clin Exp Med. 2015;8:13598–603. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang XW, Liu N, Chen S, Wang YE, Sun KL, Xu ZM, et al. Upregulation of microRNA-23a regulates proliferation and apoptosis by targeting in laryngeal carcinoma. Oncol Lett. 2015;10:410–6. doi: 10.3892/ol.2015.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao L, Zhao J, Dai X, Wang Y, Ma R, Su Y, et al. Correlation between miR-23a and onset of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2014;38:318–30. doi: 10.1016/j.clinre.2013.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.