Abstract
Protein translation is at the heart of cellular metabolism and its in-depth characterization is key for many lines of research. Recently, ribosome profiling became the state-of-the-art method to quantitatively characterize translation dynamics at a transcriptome-wide level. However, the strategy of library generation affects its outcomes. Here, we present a modified ribosome-profiling protocol starting from yeast, human cells and vertebrate brain tissue. We use a DNA linker carrying four randomized positions at its 5′ end and a reverse-transcription (RT) primer with three randomized positions to reduce artifacts during library preparation. The use of seven randomized nucleotides allows to efficiently detect library-generation artifacts. We find that the effect of polymerase chain reaction (PCR) artifacts is relatively small for global analyses when sufficient input material is used. However, when input material is limiting, our strategy improves the sensitivity of gene-specific analyses. Furthermore, randomized nucleotides alleviate the skewed frequency of specific sequences at the 3′ end of ribosome-protected fragments (RPFs) likely resulting from ligase specificity. Finally, strategies that rely on dual ligation show a high degree of gene-coverage variation. Taken together, our approach helps to remedy two of the main problems associated with ribosome-profiling data. This will facilitate the analysis of translational dynamics and increase our understanding of the influence of RNA modifications on translation.
Keywords: Ribosome profiling, RNA modification, Translation, Sequencing bias, Codon-translation speed, Translational control
1. Introduction
Protein translation is the most energy-consuming cellular process and lies at the heart of metabolism. The tight regulation of translation and preserving its fidelity is therefore absolutely essential. Interestingly, all major classes of RNA molecules critical for translation contain chemically modified nucleosides that influence translation dynamics [1]. In particular, transfer RNAs (tRNAs), the adaptor molecules that link the genetic information of the codons to their respective amino acids carry a plethora of modifications affecting tRNA folding, aminoacylation or codon recognition thereby influencing protein synthesis. Modifications of the anticodon loop have been linked to translation and shown to be critical to maintain proteome integrity [2], [3], [4]. Thus, methods to analyze how modified nucleosides influence global and local translational dynamics are becoming increasingly important. Recently, ribosome profiling has dramatically improved our ability to characterize translation in vivo [5]. The method allows to study transcriptome-wide ribosome dynamics and has been used for the characterization of several complex biological processes [6]. Ribosome profiling generally relies on the use of translation inhibitors to arrest ribosomes in defined conformations [5], [7], [8]. Subsequently, the parts of mRNA that are not actively read at the moment of lysis and hence not protected by ribosomes are digested by the addition of nucleases. This digest leads to a strong enrichment of ribosome-protected fragments (RPFs) that can be analyzed by deep sequencing [5], [9]. Importantly, ribosome profiling more information than just levels of gene translation. Due to the precise mapping of the A- and P-sites, ribosome profiling can accurately provide positional information of translation at sub-codon resolution. This feature has been used to analyze the function of modified tRNA nucleosides from yeast to mice [3], [4], [10], [11], [12].
Even though its development has sparked huge excitement in fields related to translational control, ribosome profiling is still an emerging technology, which warrants an improvement in its accuracy and sensitivity. The protocol described by Ingolia and coworkers is complex and requires time and experience [5], [7]. This has led to various attempts to shorten it or to reduce hands on time by using commercial kits for library preparation (e.g. [13], [14]). Furthermore, recent improvements found sophisticated methods to use less input material, a strategy that needs to be balanced with the necessity to use suboptimal concentrations of material in the enzymatic reactions and to amplify libraries by PCR [15]. However, not all of these updates were systematically compared to the original protocol and may cause undesirable biases. Here we present a ribosome-profiling protocol that relies on randomized linker and reverse-transcription (RT) primer sequences fully compatible with standard protocols [7] that reduces biases and allows to identify artifacts during downstream bioinformatics analysis. Furthermore, we provide examples of how to funnel different types of input materials, such as yeast, cultured cells or vertebrate tissues into the library-preparation process (see Supplementary protocol for additional information). Originally, the protocol was optimized for codon-speed analyses as needed in the characterization of tRNA modifications. However, our protocol will be useful for all applications of ribosome profiling with limited amounts of input material.
2. Protocol
2.1. Library preparation
2.1.1. Lysis
Isolation of intact ribosome-mRNA complexes that accurately represent in vivo translation is a critical step during sample preparation. Prolonged treatment with translation inhibitors can distort coverage or codon-level profiles [16]. While, more recent isolation procedures recommend cryogenic grinding, if done excessively, it too can distort polysome integrity. Thus, pre-treatment with translation inhibitors and the subsequent lysis method need to be optimized depending on sample type. As a starting point for optimization, we recommend the following methods for lysate preparation:
-
1.
100–250 ml yeast cultures are treated for 1 min with 100 μg/ml CHX to stabilize elongating ribosomes and harvested by rapid vacuum filtration (∼45 s) and flash freezing. Cells are cryogenically pulverized in a SPEX 6750 Freezer-Mill (SPEX 6750 SamplePrep) at 5 cps in yeast lysis buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100 with freshly added 0.5 mM DTT and 100 μg/ml CHX).
-
2.
Cultured mammalian cells are treated with medium containing 100 μg/ml CHX for 1 min, washed with ice cold PBS containing 100 μg/ml CHX and harvested by scraping in vertebrate lysis buffer (10 mM Tris-HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100 with freshly added 0.5 mM DTT, 0.5% deoxycholate (w/v) and 100 μg/ml CHX).
-
3.
Flash-frozen brain tissue is thawed in vertebrate lysis buffer and lysed by mechanical force with a pestle followed by titurating at least 10 times through a 20-G needle to ensure complete lysis.
Immediately after the lysis, cell debris is removed by centrifugation at 4 °C, 3000 g for 3 min followed by clarifying the supernatant by additional centrifugation at 4 °C, 10,000g for 5 min. Lysates are flash-frozen in liquid nitrogen until digestion.
2.1.2. Digestion of polysomes
Nuclease digestion of polysomes to obtain RPFs requires a compromise between efficient degradation of unprotected mRNA and rRNA contamination resulting from over digestion. Therefore, we recommend performing polysome profiling with undigested sample and titrating the digestion strength to find the best conditions: a clearly visible monosome peak without polysomes, ultimately resulting in libraries with coverage in coding exons and little rRNA contamination. Great care needs to be taken at this step, since incomplete digest will result in spurious reads e.g. in untranslated regions (UTR) that do not stem from translating ribosomes.
Thawed lysates (from 2.1.1) are treated with RNase I (Ambion) at a sample type-dependent concentration (11.25 U and 30 U of RNase I/A260 unit of extract for yeast and vertebrate samples, respectively) and incubated for 1 h (yeast) or 10 min (vertebrates) at 22 °C with continuous agitation. RNase I digestion is inhibited by adding 150 U SUPERase In (Ambion). Deoxycholate concentration in vertebrate samples is increased to 1% (w/v) to relieve ribosome aggregates and samples are loaded onto linear 10–50% (w/v) sucrose gradients (50 mM Tris-HCl pH = 7.5, 50 mM NH4Cl, 12 mM MgCl2, 0.5 mM DTT, 100 μg/ml CHX). Subsequently, samples are treated in an identical fashion irrespective of their origin.
2.1.3. Polysome profiling and footprint isolation
Sucrose gradients are centrifuged for 3 h at 35,000 rpm, 4 °C in a TH-641/SW-41 rotor (Thermo Scientific/Beckman Coulter). Gradients are fractionated using a density gradient fractionator (Isco) and a SYR-101 syringe pump (Brandel) with 60% sucrose (flow rate = 0.75 ml/min) and continuous monitoring of A254. SDS (final concentration 1%) is immediately added to fractions corresponding to the monosome peak, which are pooled, flash-frozen and stored at −80 °C. Total RNA is recovered from monosome samples using the hot-acid-phenol method which involves incubating samples for 5 min at 65 °C with intermittent vortexing, followed by three rounds of acid-phenol (pH 4.3) chloroform (5:1 ratio) extraction. The aqueous phase is extracted once with chloroform and total RNA is recovered by ethanol precipitation. 10 μg of total RNA from monosome fractions are recommended as input for RPF isolation, but we have made libraries from as little as 1 μg. RPFs are size-selected by separating RNA on 15% polyacrylamide-TBE-Urea gels (8 M urea, 1 × TBE) followed by staining with SYBR Gold (Life Technologies) and excision of 28–30 nt bands.
For mammalian samples rich in rRNA fragments the Ribo-Zero Gold rRNA removal kit (Illumina) can be used at this stage according to the manufacturer’s instructions, but using only half of the recommended amounts of reagents.
2.1.4. Isolation and fragmentation of samples for poly(A) RNA sequencing
Yeast total RNA is extracted from the fractions of cultures that are used for ribosome profiling. Cells are harvested by rapid vacuum filtration, resuspended in ice-cold 50 mM NaOAc (pH 5.5), 10 mM EDTA, followed by addition of SDS (final concentration 1%), and homogenized in a Percellys 24 bead beater (Bertin Technologies) at power setting 6.5 for two cycles of 20 s. Lysates are clarified by centrifugation at 4 °C, 10,000g for 5 min.
For cultured mammalian cells and brain tissue, SDS (final concentration 1%) is added to clarified aliquots from pre-digest lysates.
Subsequently, samples are treated in exactly the same manner irrespective of their origin. Samples are incubated with 100 μg/ml Proteinase K for 10 min at 60 °C. Total RNA is extracted using the hot-acid-phenol method (see above) and 100 μg of it is treated with TURBO DNase (Ambion) followed by poly(A) RNA purification by two rounds of selection with the Poly(A)Purist MAG kit (Ambion) according to the manufacturer’s instructions. Poly(A) RNA is subjected to random fragmentation via alkaline hydrolysis in 50 mM sodium bicarbonate (pH 9.2) and 1 mM EDTA for 20 min at 95 °C. RNA is purified by ethanol precipitation and fragments of 50–80 nt are extracted from a 15% polyacrylamide TBE-urea gel (8 M urea, 1 × TBE) stained with SYBR Gold (Life Technologies).
2.1.5. Library preparation
Libraries from ribosome footprints and fragmented poly(A) RNA are essentially prepared as described by Ingolia and coworkers [7]. Briefly, RNA fragments are dephosphorylated at their 3′ end using T4 PNK (NEB) for 1 h at 37 °C, extracted with acid-phenol:chloroform (5:1) and precipitated in ethanol for 1 h at 80 °C. Dephosphorylated RNA is ligated to 0.25–0.75 μg of adapters depending on the amounts of input RNA, with total RNA and yeast footprint samples using approximately 0.75 μg and vertebrate samples with limited footprint inputs using 0.25 μg. Adapter and target RNAs are denatured for 90 s at 80 °C followed by ligation using T4 RNA ligase 2, truncated (NEB) for 4 h at 22 °C. Samples are precipitated at −80 °C with ethanol and ligation fragments of ∼50 nt (RPFs) are excised from 15% polyacrylamide-TBE-Urea gels (8 M urea, 1 × TBE). Adaptor ligated RNAs are denatured for 90 s at 80 °C with 1 μl of 10 μM randomized or standard RT primer in a 12 μl reaction and reverse transcribed using SuperScript III (ThermoFisher) for 40 min at 55 °C. Reverse transcribed libraries are extracted from 10% polyacrylamide-TBE-Urea gels (8 M urea, 1 × TBE) at ∼135 nt (RPFs).
Depending on the degree of rRNA contamination, biotinylated oligonucleotides complementary to reverse transcribed rRNA can be employed at this stage. The quantity and type of contaminating rRNA species depends on the type of starting material and digestion strength. In our hands rRNA contamination of yeast libraries was negligible and did not require subtraction. Samples are denatured in 2× SSC with 200 μM biotinylated oligonucleotides at 90 °C and cooled to 37 °C at 0.05 °C/s. Oligonucleotides with bound reverse transcribed cDNA are removed by incubation with Dynabeads® MyOne™ Streptavidin C1 (Thermo Fisher) and samples precipitated with ethanol. ssDNA libraries are circularized with CircLigase I or II (Epicentre) for 3 h at 60 °C followed by enzyme inactivation for 10 min at 80 °C. The optimum PCR cycle number required to generate the final library is experimentally determined by performing a test PCR (typically stopping the reaction at 10, 12 and 14 cycles). The optimum cycle number is determined by running the test-PCR reactions on a gel (see below), selecting conditions with a clearly visible library and no detectable background smear. PCR is performed using Phusion polymerase (NEB), the forward primer described by Ingolia and coworkers [7] and a standard Illumina TrueSeq index adapter. Libraries of ∼175 nt size (RPFs) are isolated from 8% polyacrylamide-TBE gels and precipitated in ethanol at −80 °C. Resuspended libraries are quantified using the Qubit dsDNA HS Assay (Thermo Fisher) and sequenced on a suitable Illumina sequencing platform.
2.2. Bioinformatics pipeline
Read counting and mapping for ribosome-profiling data is comparable to RNA-seq data analysis. We, therefore, use similar methods for mapping ribosome-profiling and RNAseq datasets. However, libraries made using randomized linkers require additional steps of filtering for PCR duplicates and removal of randomized linkers compared to traditional libraries.
2.2.1. Linker clipping
We use the tool fastx_clipper (http://hannonlab.cshl.edu/fastx_toolkit/) for initial clipping of fixed linker sequences and to discard the reads lacking the linker or having unidentified nucleotides (represented by an N). Processed reads longer than 30 nt for mRNA and 25 nt for RPFs are kept for further analyses.
2.2.2. Amplification duplicate removal
To differentiate amplification duplicates from actual biological duplicates we use a customized python script (available upon request). Briefly, the script accepts fastq files containing sequences with clipped randomized linkers, filters amplification duplicates, trims the randomized nucleotides in the reads and returns a text file containing information about detected duplicates and a fastq file containing processed reads. Additionally, the script can generate graphs showing the ligation events in the samples and tables that can be used to perform an in-depth analysis of the ligation process. Furthermore, the script can be used to remove identical reads by setting the randomized linker lengths to 0.
2.2.3. Removal of rRNA sequences
Despite its depletion during library preparation, rRNA remains an abundant contaminant in ribosome-profiling datasets. Contaminating reads are removed by aligning against a reference containing all the rRNA of the organism of interest using a short read aligner such as Bowtie [17]. During this process mismatches can be accepted and multi-mapping is allowed, since rRNA reads might map to more than one genomic copy of rRNA genes. rRNA read removal can also be performed by local alignment before amplification duplicate and randomized linker removal.
2.2.4. Genome alignment
The remaining reads are aligned to a genome or transcriptome using a splice-aware aligner (for example TopHat2 [18]) with stringent parameters not allowing mismatches and multi-mapping. We suggest performing optional codon-composition analyses only in cases where the performance of the randomized linkers needs to be assessed or if the quality of the library indicates that a significant fraction of the reads might originate from artifacts.
2.2.5. Codon occupancy
To determine transcriptome-wide A-site-codon occupancy we use a published strategy [3], [19]. Briefly, we analyze 28–30 nt long RPFs without mismatch to annotated coding sequences, which show the highest level of codon periodicity in these samples. The offset used to determine the A-site position of a read is based on a characteristic inflection of ribosome occupancy at 12–13 nt upstream of annotated start codons. To pinpoint A-site positions we therefore choose positions 15–16 nt from the 5′ end of reads. We select reads mapped to canonical ORFs in the 0 or −1 frame, using offsets of 15 and 16 nt respectively. Occupancy of A-site codons is normalized by the frequency of the same codon in the non-decoded +1, +2 and +3 triplets relative to the A-site. In contrast to the −1, −2 and −3 triplets, these codons have not been read by the ribosome. Thus, their frequency is not influenced by ribosome dynamics and reflects codon usage without biases. A script to generate plots of transcriptome-wide A-site codon occupancy (e.g. Fig. 1E) is available upon request.
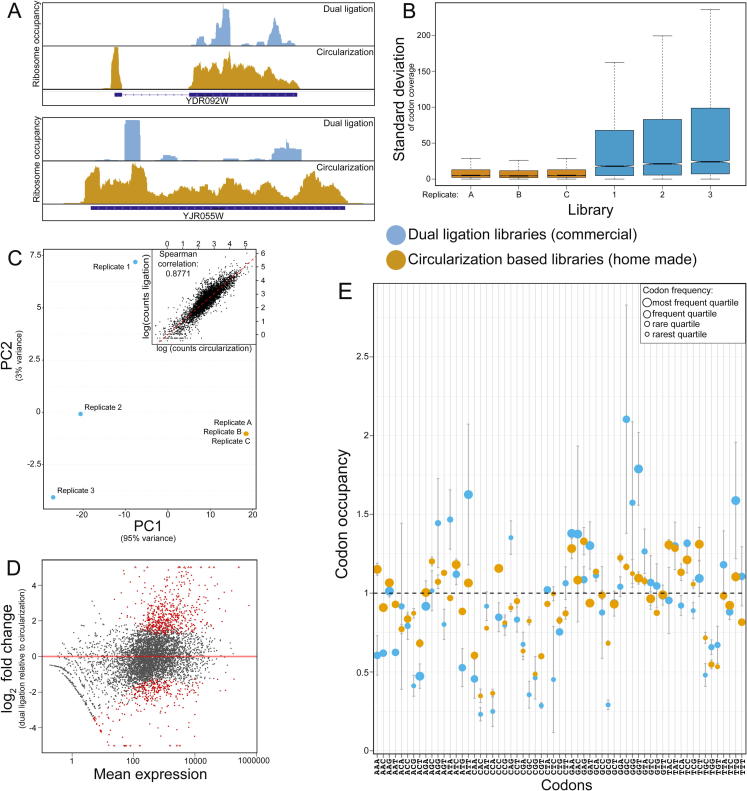
Fig. 1.
Comparison of library preparation methods. (A) RPF coverage of representative yeast ORFs made with a commercial dual ligation kit or according to a circularization based approach. Coverage of circularization-based libraries is given in orange, dual ligation libraries in light blue (colors match in A, B, C and E). The annotated ORF is indicated in dark blue. (B) Boxplot showing standard deviations of codon coverage of individual transcripts in three replicates of the ribosome-profiling libraries shown in A. Sequencing density was corrected for library size and the first 15 codons in each gene were omitted from analysis. Higher values and wider boxplots point to uneven coverage of transcripts. (C) Principal Component Analysis (PCA) comparing transcript read counts from both library types using three replicates each. Top right: Regression analysis of transcript read counts from both libraries, regression line in red. Spearman correlation: 0.8771. (D) Differential expression of two-linker ligation libraries relative to circularization libraries (n = 3) using DESeq2 with a log2 fold change threshold of 0.8 and an adjusted p-value of 0.05. Differentially expressed genes are indicated in red. (E) Codon-specific A-site ribosome occupancy relative to downstream sites (mean ± SD, n = 3). Symbol size indicates the relative frequency of codons in the A-site.
3. Strong biases in dual ligation libraries
We compared yeast ribosome-profiling libraries generated from isolated RPFs that were further processed by a commercial kit using dual ligation to libraries generated according to the circularization protocol [3] using the same conditions for harvesting, lysis, digestion and isolation of protected fragments but not from the same extract. We noticed uneven coverage of ORFs in the dual-ligation libraries (Fig. 1A). This is in agreement with previous reports showing an inherent bias of RNA ligases [20], [21]. To assess the extent of this phenomenon, we plotted the standard deviation of codon coverage per transcript after correcting the values according to library size by dividing each value by the sample median of the mean-coverage values per gene, similar to the size-correction method used in the DEseq2 package [24]. We observed a higher coverage variation in dual-ligation libraries even after excluding the coverage peak in the first 15 codons of each gene (Fig. 1B). To test whether the less even coverage of ribosome-profiling libraries is inherent to the dual-ligation method, we analyzed published ribosome-profiling datasets from yeast [22], [23]. Indeed, we found that libraries generated by dual-ligation were overall more variable (Fig. S1). This effect leads to significant differences in the observed translation levels in our libraries. Both library types are clearly separated by their principal components and replicates of dual-ligation libraries were less consistent, while differences between circularization libraries were negligible (Fig. 1C). Strikingly, the correlation of average ribosome occupancy between the libraries is <0.9, a poor value for ribosome profiling. We used DESeq2 [24] to compare gene expression between these libraries made from the same yeast strain and lysed according to the same protocol. 881 genes are called as differentially translated arguably based on the library generation protocol (Fig. 1D). To test whether codon-occupancy measurements are similarly affected, we determined transcriptome-wide A-site codon occupancy in both libraries (Fig. 1E) [3]. Similar to translation level, in the library constructed by the dual-ligation strategy several codons deviate clearly and exhibit a higher degree of sample-to-sample variation, an effect that is independent of overall codon frequency. This strategy would thus compromise the use of ribosome profiling to study the impact of tRNA modifications on the translation speed of specific codons. Therefore, we do not recommend the use of dual-ligation libraries for codon analysis. Importantly, the original protocol uses cDNA circularization to avoid biases during 5′ linker ligation [5], [7]. This strategy improves the libraries, but potential biases resulting from 3′ linker ligation remain. Finally, the PCR amplification of cDNA may selectively amplify certain sequences thus distorting the relative abundance of reads [25], [26].
4. A dual randomization strategy for bias prevention and removal
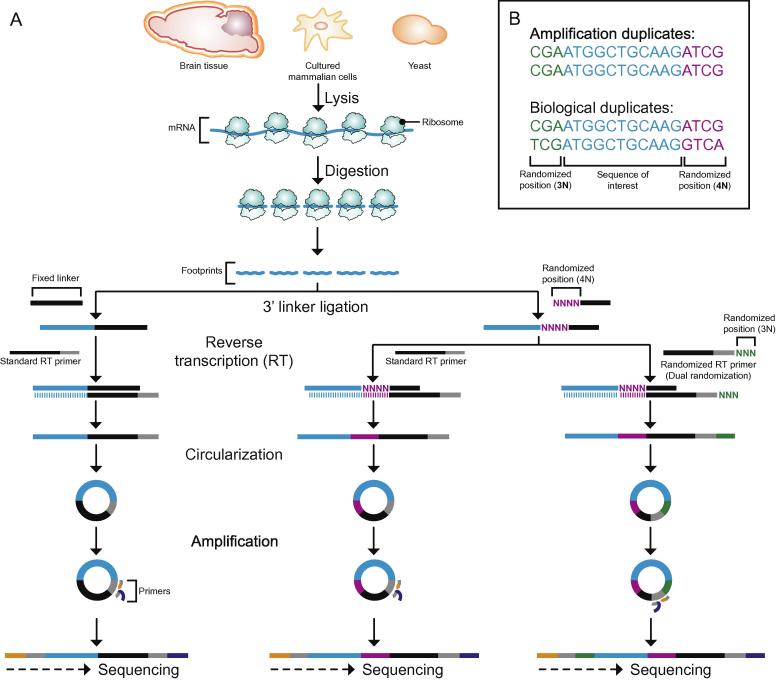
To be able to detect subtle differences in translation levels and codon occupancy, we sought to reduce library artifacts. In miRNA sequencing, the 5′ end of the DNA linker was shown to affect ligation efficiency, thereby compromising miRNA quantification [27]. Randomization of the 5′ end of the linker circumvents this effect by providing an optimal linker sequence for each target sequence [27]. A similar strategy is used in CLIP and CRAC protocols [28], [29]. Introducing this approach to the generation of ribosome-profiling libraries is more likely to reflect abundance of RPF. Furthermore, randomized nucleotides can be used as barcodes to remove reads that are a result of PCR over-amplification. However, if a specific sequence preferentially ligates to a specific randomized linker sequence, this strategy would mistake the abundance of this RPF as an amplification artifact. To distinguish such cases from PCR over-amplification, we further randomized 3 nucleotides at the 3′ end of the RT primer. Overall, our strategy introduces seven randomized nucleotides to reduce biases of 3′ linker ligation and circularization besides identifying and removing PCR duplicates.
To analyze the effects of randomization on library preparation, we generated RPF and mRNA libraries from two independent yeast extracts. From each replicate, we generated three libraries using different combinations of DNA-linker and RT-primer (Fig. 2): First, a commercial non-randomized DNA-linker and the standard RT-primer (fixed linker) [7], second, a DNA-linker randomized at its four most 5′ positions and the standard RT-primer (randomized linker) and third, a DNA-linker randomized at its four most 5′ positions and a RT-primer randomized at the last three nucleotides (dual randomization). We did not include dual-ligation in this experiment as this strategy had performed poorly in our comparison.
Fig. 2.
Schematic overview of ribosome-profiling library preparation. (A) Schematic overview of ribosome-profiling library preparation from footprint sequences (blue), using fixed or randomized linkers in combination with a standard or randomized RT primer. Illumina sequencing of the resulting libraries includes randomized sequences allowing for downstream analysis and artifact removal. (B) Example of a dual randomized sequencing read showing three randomized positions (3 N) derived from the RT-primer (green), the footprint (blue) and the four randomized positions (4 N) of the linker (purple). This allows to distinguish amplification duplicates (top) with identical randomized positions from biological duplicates with different barcodes (bottom).
5. Randomization allows duplicate identification
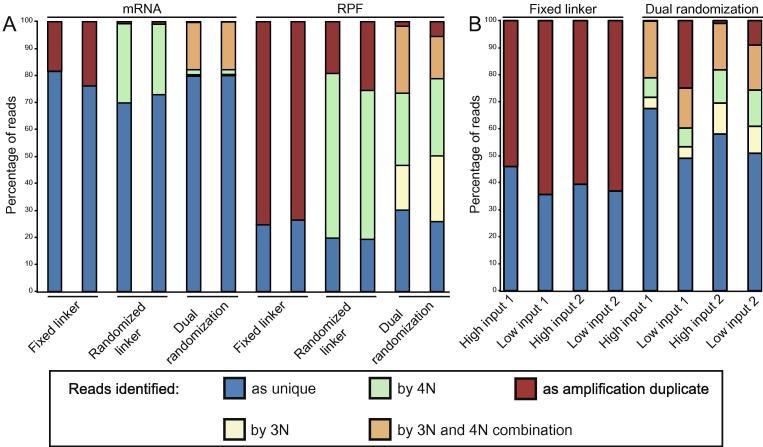
Duplicate reads can stem from individual ribosomes – and thus reflect independent translation events (biological duplicates) – or they can stem from artifacts of library preparation (library duplicates). In the mRNA libraries, we found that roughly 80% of the reads were unique using a fixed linker, while 20% scored as duplicates based on multiple reads having identical sequences. It is impossible to distinguish the artifacts among these duplicates from biologically relevant sequences without randomization. The use of randomized linker or dual randomization drastically reduced the occurrence of duplicates (1% and 0.3%), since the reads could be classified with high confidence using the randomized sequences as barcodes.
However, mRNA samples are approximately 50 nt long, providing ample sequence space to distinguish individual reads. In contrast, RPF reads are only 28–30 nt long, complicating their distinction. Indeed, only 25% of all reads were unique using the fixed linker, while 75% were classified as duplicates. Randomized linker and dual randomization reduced the number of reads identified as amplification duplicates to 22% and 5%, respectively. This shows that 7 randomized positions are generally sufficient to distinguish seemingly identical reads and to increase confidence in the data even for short reads.
6. Duplicates arise when input material is limiting
Importantly, because yeast is translationally very active, we started from sufficient input material allowing for optimal reaction conditions during the different steps of the protocol. To assess whether randomization will be more powerful in cases with limited input material e.g. postmitotic vertebrate tissues, we tested the impact of two potential sources of artifacts. First, we performed 10 additional PCR cycles on the cDNA libraries that we had analyzed before, sequenced those and compared the results. This, however, did not change the number of amplification duplicates (data not shown). Second, we tested whether starting from less input material affects the outcome by using 3–6 times less input and an additional 20× dilution before library amplification. We found that this increased the number of duplicates in the libraries by up to 57%, based on the replicate and type of linker used (Fig. 3B). However, the barcoding through randomized positions allows to identify and to remove the amplification duplicates, which constitute 18–48% of the potential duplicate reads. This emphasizes the importance of barcoding to remove the significant number of duplicate reads in many low input ribosome-profiling libraries.
Fig. 3.
Identification of duplicates in ribosome-profiling reads. (A) Compositions of mRNA and RPF libraries prepared with fixed linkers or two different randomization strategies. Unique reads (blue) are sequences only present once in the library regardless of randomized sequences. Amplification duplicates (red) are reads that cannot be identified as unique based on randomization. The remaining sequences appear multiple times, but can be distinguished by randomization of the DNA linker (green), the RT primer (yellow) or a unique combination of both (orange). (B) Composition of two replicates of RPF libraries (as in A) with fixed linkers or dual randomization using a high input (samples as in A) or a 2–6× lower input of the same monosomal RNA for ribosome profiling. Library pairs (high:low) were randomly downsampled to an equal number of reads to accurately reflect differences in library composition.
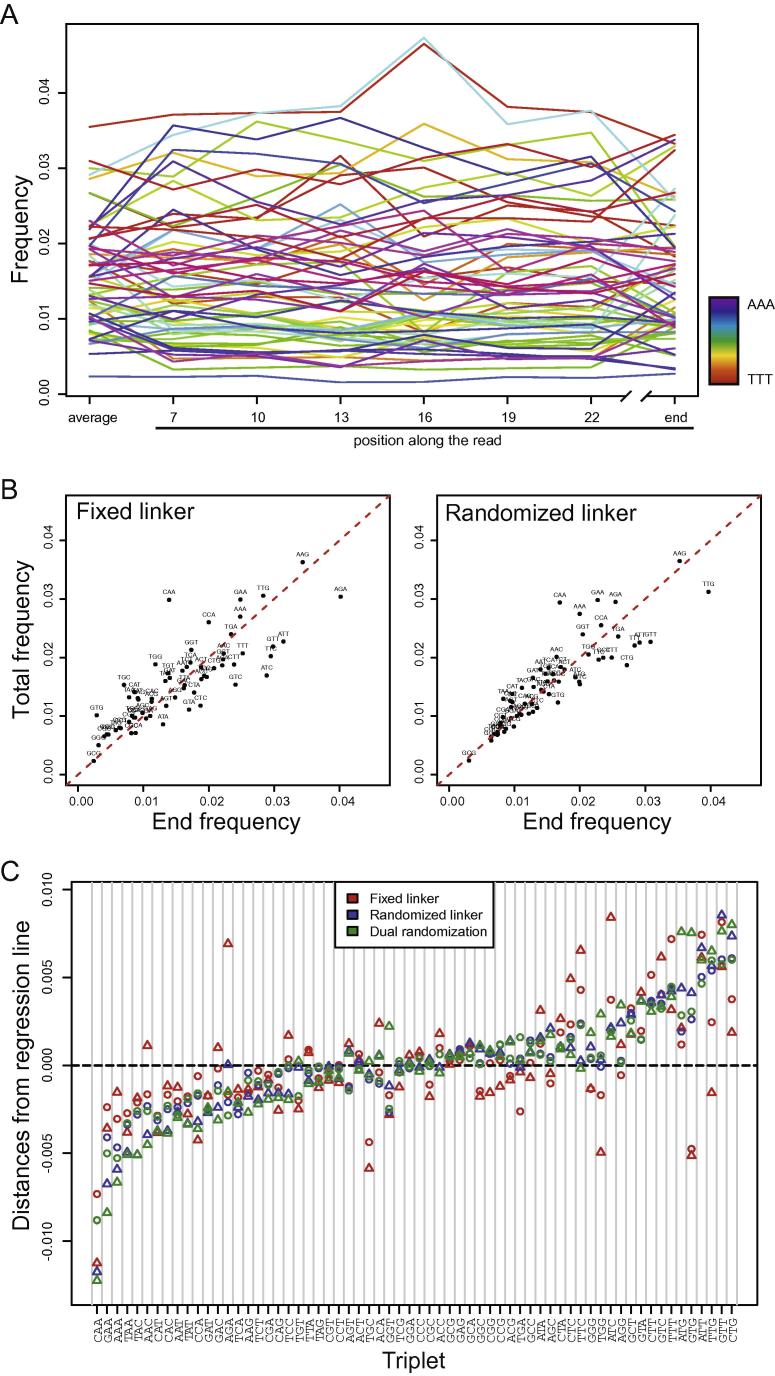
7. Randomized linkers alleviate end biases by enhanced fragment ligation
Next, we analyzed whether certain triplets are more likely to occur in the different high-input, high sequencing-depth libraries and found that the global frequency of triplets correlated well between all datasets (data not shown). Thus, all three strategies perform well in generating a global snapshot of the transcriptome and translatome. To study the influence of randomization on linker-ligation biases, we quantified whether specific triplets are enriched or depleted at the 3′ end of the reads relative to the rest of the read. This can be critical, since this region of the read is used for normalization of codon occupancy [3]. We calculated the frequency of triplets in all positions of the reads and found that the frequency is relatively constant throughout the mRNA reads (Supplementary Fig. 2A), while RPF reads (Fig. 4A) show a more diverse composition. This more diverse composition of RPF reads is likely due to their shorter length and the large influence of codons in particular in the A-site of the ribosome on read-composition. However, the frequency at the last position of the reads is most distinct from the word-composition of the overall read and deviates between library types (Fig. 4B). We then calculated the distance of each triplet from the ideal correlation and plotted the result for all RPF libraries (Fig. 4C) and for all mRNA libraries (Supplementary Fig. 2B). It is apparent that the libraries with randomized linkers perform very similarly. In contrast, in libraries generated with fixed linkers certain words clearly deviate in their end frequency. Among the most striking changes is the underrepresentation of TGC and GTG triplets or rather the larger GTGC, CTGC, TTGC and GGTG 4-mers (Supplementary Fig. 3), which might reflect a ligation bias. When plotting the frequency of each triplet at the end relative to the occurrence of each triplet in the libraries, only very few triplets deviate. Thus, all three circularization strategies lead to valid outcomes if analyzing codon-occupancy on a global level. However, this may differ when analyzing translation of individual genes on a codon-by-codon basis.
Fig. 4.
Sequence bias in ribosome-profiling libraries. (A) Frequencies of triplets along RPF reads. (B) End frequency vs. total frequency of triplets in fixed-linker (left) and dual-randomized (right) libraries. The midline (red) represents a perfect match of frequencies. (C) Distance of data points in (B) to the line midline. Triplets are ordered based on differences in randomized-linker libraries. Similar shapes represent samples derived from the same biological replicate.
8. Concluding remarks
Ribosome profiling is a powerful technique to quantitatively analyze the cellular translation landscape in a high-throughput manner [6]. The methodological improvement that we describe here allows for a more reliable recovery of individual codons. Our protocol will help to further the characterization of translational dynamics by providing a strategy for the generation of high-quality ribosome-profiling libraries from different starting material. In particular, this method proves powerful when input material is limiting, a common situation when working with cell-culture or tissue samples.
We introduce a total of seven randomized positions to the DNA linker and to the RT primer. The use of randomized nucleotides allows to decrease the overall noise in ribosome-profiling and small RNA datasets via reliable detection and removal of amplification duplicates [27], [30]. When performing general gene-translation analysis starting from sufficient material, the use of randomized linkers does not appear to be critical. In these cases the use of the standard circularization protocol achieves sufficient information. Importantly, library artifacts occur when preparing ribosome-profiling libraries under conditions that are realistic for mammalian cells, where input material is limiting. Our strategy allows to correct for this and to further reduce the required input material taking ribosome profiling one step closer to analysis on the single-cell level.
Another critical step for codon-specific sequence analysis appears to be the use of the previously established circularization protocol [5], [7]. Dual-ligation protocols using fixed linker proved to severely perturb gene-translation analysis at the level of global gene expression, codon-specific translation and coverage of individual ORFs. We therefore, do not recommend its use for the generation of ribosome-profiling datasets despite the relative ease of library preparation.
In summary, the implementation of our adapted protocol will help to facilitate ribosome profiling as a technique to address open questions not only in the field of RNA modifications.
Conflict of interest statement
The authors have no conflicts to declare.
Acknowledgements
We thank C. Gräf and F. Rabert for technical support during sequencing, all members of the Leidel group for helpful discussions and Sandra Kienast for help with editing. This work received support by the Max Planck Society, the North Rhine-Westphalian Ministry for Innovation, Science and Research (314-40001009), and the European Research Council (ERC-2012-StG 310489-tRNAmodi) to S.A.L.; D.D.N. received an EMBO Long-Term Fellowship (ALTF1291-2010). J.S., B.S.N. and P.S. are supported by the International Max Planck Research School-Molecular Biomedicine and the Cells in Motion Graduate School. J.S. received a Cells-in-Motion bridging Fellowship. A.L., J.M.V. and S.A.L. are members of the Muenster Graduate School for Evolution.
Accession Numbers Sequencing data are available at the Gene Expression Omnibus. Wild-type libraries using the circularization protocol used in Fig. 1 are from GSE67387 (GSM1646015, GSM1646016 & GSM1646017). All other sequencing data is part of GSE84746.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ymeth.2016.07.011.
Appendix A. Supplementary data
Boxplot showing standard deviations of codon coverage of individual transcripts in published ribosome-profiling datasets from yeast. Data is taken from the circularization based meiosis timecourse of [22] (yellow) and the two-linker ligation based wild-type libraries generated from [23] (blue).
Sequence bias in mRNA libraries. (A) Frequencies of triplets along mRNA reads. (B) Distance of end frequency of triplets vs. overall frequency to a model with equal frequency of the end position. Triplets are ordered based on differences in randomized-linker libraries. Similar shapes represent samples derived from the same biological replicate.
Ligation of abundant RPF fragment ends to randomized linker positions. (A) Ligation frequency of GGTG-ending reads with the 40 most abundant and 20 least abundant 4N sequences. (B) As in (A), but showing CTGC-ends of RPF fragments. (C) As in (A), but showing TTGC-ends of RPF fragments. (D) As in (A), but showing GTGC-ends of RPF fragments.
References
- 1.Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H., editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009. pp. 1–18. [Google Scholar]
- 2.Agris P.F., Vendeix F.A.P., Graham W.D. TRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Nedialkova D.D., Leidel S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiaville P.C., Legendre R., Rojas-Benitez D., Baudin-Baillieu A., Hatin I., Chalancon G. Global translational impacts of the loss of the tRNA modification t(6)A in yeast. Microb. Cell. 2016;3:29–45. doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science (New York, NY) 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingolia N.T. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lareau L.F., Hite D.H., Hogan G.J., Brown P.O. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife; 3; 2014. p. e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steitz J.A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969;224:957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- 10.Zinshteyn B., Gilbert W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laguesse S., Creppe C., Nedialkova D.D., Prévot P.-P., Borgs L., Huysseune S. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell. 2015;35:553–567. doi: 10.1016/j.devcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Tuorto F., Herbst F., Alerasool N., Bender S., Popp O., Federico G. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015 doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aeschimann F., Xiong J., Arnold A., Dieterich C., Grosshans H. Transcriptome-wide measurement of ribosomal occupancy by ribosome profiling. Methods. 2015;85:75–89. doi: 10.1016/j.ymeth.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Reid D.W., Shenolikar S., Nicchitta C.V. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods. 2015;91:69–74. doi: 10.1016/j.ymeth.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyer E.E., Ozadam H., Ricci E.P., Cenik C., Moore M.J. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res. 2015;43:e2. doi: 10.1093/nar/gku1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussmann J.A., Patchett S., Johnson A., Sawyer S., Press W.H. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadler M., Fire A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA. 2011 doi: 10.1261/rna.02890211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romaniuk E., McLaughlin L.W., Neilson T., Romaniuk P.J. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur. J. Biochem. 1982;125:639–643. doi: 10.1111/j.1432-1033.1982.tb06730.x. [DOI] [PubMed] [Google Scholar]
- 21.Hafner M., Renwick N., Brown M., Mihailović A., Holoch D., Lin C. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science (New York, NY) 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiaville P.C., Legendre R., Rojas-Benítez D. Global translational impacts of the loss of the tRNA modification t 6 A in yeast. Microbial. 2015 doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozarewa I., Ning Z., Quail M.A., Sanders M.J., Berriman M., Turner D.J. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aird D., Ross M.G., Chen W.-S., Danielsson M., Fennell T., Russ C. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayaprakash A.D., Jabado O., Brown B.D., Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huppertz I., Attig J., D’Ambrogio A., Easton L.E., Sibley C.R., Sugimoto Y. ICLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohnsack M.T., Tollervey D., Granneman S. Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods Enzymol. 2012;511:275–288. doi: 10.1016/B978-0-12-396546-2.00013-9. [DOI] [PubMed] [Google Scholar]
- 30.Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016;14:1787–1799. doi: 10.1016/j.celrep.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxplot showing standard deviations of codon coverage of individual transcripts in published ribosome-profiling datasets from yeast. Data is taken from the circularization based meiosis timecourse of [22] (yellow) and the two-linker ligation based wild-type libraries generated from [23] (blue).
Sequence bias in mRNA libraries. (A) Frequencies of triplets along mRNA reads. (B) Distance of end frequency of triplets vs. overall frequency to a model with equal frequency of the end position. Triplets are ordered based on differences in randomized-linker libraries. Similar shapes represent samples derived from the same biological replicate.
Ligation of abundant RPF fragment ends to randomized linker positions. (A) Ligation frequency of GGTG-ending reads with the 40 most abundant and 20 least abundant 4N sequences. (B) As in (A), but showing CTGC-ends of RPF fragments. (C) As in (A), but showing TTGC-ends of RPF fragments. (D) As in (A), but showing GTGC-ends of RPF fragments.