Abstract
Tissue engineering and regenerative medicine technologies offer promising therapies for both medicine and dentistry. Our long-term goal is to create functional biomimetic tooth buds for eventual tooth replacement in humans. Here, our objective was to create a biomimetic 3D tooth bud model consisting of dental epithelial (DE) – dental mesenchymal (DM) cell sheets (CSs) combined with biomimetic enamel organ and pulp organ layers created using GelMA hydrogels. Pig DE or DM cells seeded on temperature-responsive plates at various cell densities (0.02, 0.114 and 0.228 cells 106/cm2) and cultured for 7, 14 and 21 days were used to generate DE and DM cell sheets, respectively. Dental CSs were combined with GelMA encapsulated DE and DM cell layers to form bioengineered 3D tooth buds. Biomimetic 3D tooth bud constructs were cultured in vitro, or implanted in vivo for 3 weeks. Analyses were performed using micro-CT, H&E staining, polarized light (Pol) microscopy, immunofluorescent (IF) and immunohistochemical (IHC) analyses. H&E, IHC and IF analyses showed that in vitro cultured multilayered DE-DM CSs expressed appropriate tooth marker expression patterns including SHH, BMP2, RUNX2, tenascin and syndecan, which normally direct DE-DM interactions, DM cell condensation, and dental cell differentiation. In vivo implanted 3D tooth bud constructs exhibited mineralized tissue formation of specified size and shape, and SHH, BMP2 and RUNX2and dental cell differentiation marker expression. We propose our biomimetic 3D tooth buds as models to study optimized DE-DM cell interactions leading to functional biomimetic replacement tooth formation.
Keywords: Tooth development, dental stem cells, tissue engineering, regenerative medicine, biomaterials
Introduction
Tooth loss due to genetic disorders or microbial diseases, iatrogenic, traumatic, or therapeutic insults, patient negligence and poor oral hygiene continues to affect most adults at some point during their lives [3, 4]. It is estimated that approximately 150 million adults currently suffer from tooth loss, and that over 10 Million new cases of edentulism will arise during this decade [3]. Dental clinical procedures such as root canal treatment and dental implants are commonly used as tooth repair/replacement therapies [5, 6]. However, root canal therapy results in loss of tooth sensitivity and vitality, hence, the tooth cannot respond immunologically to subsequent infections, and also become brittle over time [5]. Dental implants, such as titanium implants, are not equivalent to natural teeth, either in function nor aesthetics, because they lack periodontal and cementum tissues which function to cushion and modulate the mechanical stress of mastication, while at the same time promoting healthy alveolar bone turnover [7, 8]. These disadvantages have prompted an ongoing search for alternative methods that would avoid the need for root canal and for dental implants. Examples of proposed approaches to engineer biological teeth include: tissue engineering scaffolds; stimulation of third dentition formation; cell-tissue recombination; chimeric tooth tissue engineering; and gene-manipulated tooth regeneration [8–11].
To date, our group has been using tissue engineering approaches to identify optimal scaffold materials and designs that promote dental epithelial (DE) and mesenchymal (DM) cell interactions leading to replacement tooth formation [4, 12, 13]. For the study presented here, we have used DE and DM progenitor cells isolated from un-erupted molar tooth buds extracted from 5–6 month old porcine jaws, which consisted of enamel organ, dental papilla, and dental follicle tissues and cells. The enamel organ is derived from the ectoderm, while the dental papilla and dental follicle are derived from the neural crest (also called ectomesenchyme) [14]. Briefly, harvested tooth bud tissues were used to create single cell suspensions, which were then cultured in vitro as previously described [4, 15]. Differentiated DE and DM cells die during this process, resulting in highly enriched populations of undifferentiated DE and DM cells. The rationale for using pig teeth at these developmental stages was to obtain sufficient numbers of dental progenitor cell populations, also known as dental stem cells (DSCs), that can be used to generate teeth and supporting tissues [16]. The progenitor DE cells generated in this manner will differentiate into enamel forming ameloblasts, while the DM cells will give rise to odontoblasts, dental pulp stem cells (DPSCs), periodontal ligament stem cells (PDLSCs), stem cells from apical papilla (SCAP) and dental follicle precursor cells (DFPCs) [17]. In many instances, our prior published reports have shown that instead of creating bioengineered teeth that adopted the size and shape of the scaffold, we observed that many small tooth crowns formed throughout the implant, suggesting that these models lacked the proper ECM molecule gradients present in naturally formed teeth, which provide essential cues for proper tooth development, and for periodontal tissue and surrounding alveolar bone formation [4, 12, 13, 15].
To address this, we recently established a 3D biomimetic tooth bud model using photopolymerizable gelatin methacrylamide (GelMA) hydrogel formulas, designed to facilitate DE and DM cell interactions leading to ameloblast and odontoblast differentiation, respectively, and the formation of bioengineered teeth of predictable size and shape [18, 19]. GelMA hydrogels exhibit many properties that make it an attractive material for tissue engineering applications including [20, 21]: i) it is largely composed of denatured collagen and is relatively inexpensive; ii) it retains collagen’s natural RGD adhesive domains and MMP sensitive sites which are known to enhance cell binding and cell-mediated matrix degradation, respectively; iii) the physical properties of GelMA hydrogels can be tuned by varying GelMA and/or photoinitiator (PI) concentrations; and iv) GelMA is suitable for cell encapsulation at 37°C, and promotes cell viability and proliferation. In these reports we identified GelMA formulas that exhibited elastic moduli similar to those of natural tooth bud derived enamel organ and pulp organ tissue, and in addition to DE and DM cells we incorporated Human Umbilical Vein Endothelial Cells (HUVECs) to promote neo-vasculature formation to facilitate in vivo engraftment with host tissues [18, 19]. Our published 3D tooth bud model consisted of a biomimetic enamel organ layer (DE-HUVEC encapsulated in 3% GelMA) and biomimetic pulp organ (DM-HUVEC encapsulated in 5% GelMA). In vitro culture and in vivo implantation studies showed that the 3D GelMA biomimetic tooth bud constructs supported DE and DM cell attachment, spreading, metabolic activity, neo-vasculature formation, and mineralized tissue formation of specified size and shape in vivo [19]. However, limitations to this model included cell mixing between GelMA layers, and lack of distinct enamel or dentin layers.
Here, we aimed to improve our 3D tooth bud model, and overcome such limitations by using successive photocrosslinking of individual dental cell-seeded GelMA layers, and to increase DE-DM cell interactions by introducing DE-DM cell sheet (CSs) layers between the biomimetic enamel and pulp organs of our 3D tooth bud model (Figure 1). Our results demonstrate the successful creation of multilayered DE-DM cell sheet containing GelMA (CSG) 3D biomimetic tooth bud constructs. We also show that in vivo CSG implanted constructs exhibited distinct biomimetic enamel and pulp layers, and that DE and DM cells express dental cell differentiation marker expression (DSPP, OC, and AM). We propose this novel 3D bioengineered tooth bud model as a means to study DE and DM cell interactions leading to biomimetic replacement tooth formation.
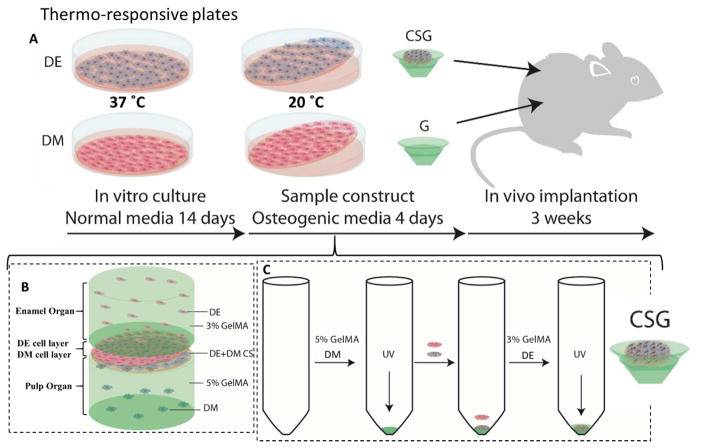
Figure 1. Experimental design and culture of 3D GelMA-CS tooth buds.
A. DE and DM cells were seeded on thermo-responsive plates and cultured in normal DE and DM media, respectively, for 14 days. DE and DM CSs were detached by temperature reduction (20ºC) and layered over GelMA constructs to create experimental 3D tooth bud constructs (CSG = DE and DM CSs layered over dental cells encapsulated in GelMA; G = GelMA alone). For in vivo analyses, replicate constructs were cultured in osteogenic media for 4 days and implanted subcutaneously onto the backs of the rats. B. Bioengineered 3D CS - GelMA tooth bud model. The bottom layer mimics the pulp organ (5% GelMA encapsulating DM cells) and the top layer mimics the enamel organ (3% GelMA encapsulating DE cells). The DE and DM CS layers mimic polarized DE-DM cell layers normally observed in developing teeth. C. Steps used to prepare the constructs. DM cells (3×107 cells/ml) were re-suspended in 100 μL of 5% GelMA and photo-crosslinked. DM and DE cell sheets were layered over the polymerized DM 5% GelMA. DE cells (3×107 cells/ml) re-suspended in 100 μL 3% GelMA and 100 μL, layered over construct and photo-crosslinked.
Materials and methods
Primary dental cell isolation, in vitro culture and expansion
Porcine DE and DM progenitor cells were obtained and cultured as previously published [4, 15]. Briefly, DE and DM progenitor cells were isolated from un-erupted tooth buds extracted from 5 month old porcine jaws. Single cell suspensions of DE and DM tooth bud cells were prepared, seeded in T175 cm2 flasks (Corning Inc., Corning, NY, USA), and expanded using epithelial medium [LHC-8 (GIBCO), 10 % FBS, 1 % PSA, 0.5 g/mL Epinephrine] or mesenchymal medium [Advanced DMEMF12 (GIBCO), 10 % FBS, 25 μg/mL Ascorbic Acid, 1 % PSA, 1 % Glutamax], respectively, in 5% CO2 at 37°C. Expanded cells were cryopreserved in 10% DMSO in appropriate culture media until use.
Dental cell sheet fabrication
DE and DM tooth bud cells were recovered from cryopreservation and expanded in T175 cm2 flasks. DE and DM cells were trypsinized and seeded on UpCell thermo-responsive plates (CellSeed, Tokyo, Japan) at densities of 0.02, 0.114 and 0.228 × 106 cells/cm2 for 7, 14 and 21 days, in 5% CO2 at 37°C. Cell sheets (CSs) were detached from the thermo-responsive plates by decreasing the temperature to 20ºC and harvested by gentle flushing with media. The formation of layered CS was made by careful stacking of the CSs [22].
Creation of 3D tooth bud constructs
Biomimetic 3D tooth bud constructs were fabricated as depicted in Figure 1. DE and DM cell sheets were harvested after 14 days at plating cell densities of 0.228 and 0.114 × 106 cells/cm2, respectively. Lyophilized GelMA was fully dissolved in DMEM/F12 media (w/v), and photo-initiator (Irgacure 2959, Sigma, St. Louis, MO) was added to create 3 and 5 percent GelMA formulations, denoted as 3% GelMA and 5% GelMA. Dental cell containing GelMA bilayers were created by sequential photo-crosslinking via exposure to 9.16 W/cm2 UV light for 20 seconds using an Omnicure S2000 (Lumen Dynamics Group Inc., Mississauga (ON) Canada). For in vitro studies, harvested cell sheets were layered over dental cell encapsulated and acellular GelMA constructs and cultured in osteogenic media (DMEM/F12:LCH8 basal media supplemented with, 1% PSA, 10% FBS, 100 nM Dexamethasone, 10mM beta Glycerol Phosphate, 0.05 mM Ascorbic Acid) for 24 hours, and for 4, 7 and 12 days, in 5% CO2 at 37°C (Figure 1).
In vivo implantation of biomimetic tooth bud constructs
For in vivo analyses, bioengineered 3D CS-GelMA constructs were cultured in osteogenic media for 4 days in 5% CO2 at 37°C, and randomly implanted subcutaneously onto the backs of immunocompromised 5 month old female Rowett Nude rats (Charles River Laboratories, Willmington, MA). All animal surgeries were performed using Tufts University approved Institutional Animal Care and Use Committee (IACUC) protocols and Mandatory Animal Care and Use (MACU) regulations. Four replicate bioengineered 3D tooth buds and acellular GelMA constructs were implanted and harvested after 3 weeks (Figure 1).
Dental Cell sheet 3D tooth bud analyses
Harvested in vitro and in vivo constructs were fixed in 10% formalin overnight and washed in Dulbecco’s Phosphate-Buffered Saline (DPBS). In vivo mineralization was quantified via micro-CT (SkyScan 1076, Bruker), and bone mineral density was measured using CT-Analyser software (Version 1.14.4.1, Bruker). Data were statistically analyzed using GraphPad Prism software (version 6.01). The parametric t-test with Welch correction was applied. P values lower than 0.05 were considered statistically significant in the analysis of the results. Following the micro-CT analyses, the mineralized constructs were immersed in fresh decalcification solution (22.5% formic acid + 10% sodium citrate) every 3 days until fully decalcified. Decalcification was defined by lack of ammonium oxalate-calcium precipitate formation after 20 minutes. For paraffin sectioning, the constructs were dehydrated through a graded ethanol and xylene series, submerged in molten paraffin for 18 hours, and then embedded into paraffin blocks and serially sectioned (6 μm thick) as previously described [23]. Hematoxylin & Eosin, and Picrosirius Red (Polysciences, Warrington, PA) stains were used to analyze selected sections.
For Immunofluorescent (IF) analyses, paraffin sections were blocked for 20 min in 5% BSA and incubated for 1 hour with primary antibodies: rabbit anti-Cytokeratin 18 (CK18, SC-28264, 1:25, Santa Cruz Biotechnology), rabbit anti-Ecadherin (ECAD, ABIN1858334, 1:20, Antibodies Online, Atlanta, GA), rabbit anti-Runt-Related Transcription Factor 2 (RUNX2, SC-10758, 1:25, Santa Cruz Biotechnology), rabbit anti-Sonic Hedgehog (SHH, SC-9024, 1:25, Santa Cruz Biotechnology), rabbit anti- Bone Morphogenetic Protein 2 (BMP2, AB14933, 1:25, Abcam), rabbit anti-CD31 (CD31, LS-B5577, 1:25, LifeSpan BioSciences Inc), and mouse anti-vimentin (VM, BS-0756R, 1:25, Bioss, Woburn, MA). Sections were incubated for an additional hour with fluorescent conjugated secondary antibodies donkey anti-mouse IgG-Alexa Fluor 488 (515-545-003, Jackson ImmunoResearch Laboratories, West Grove, PA) and donkey anti-rabbit IgG-Alexa Fluor 568 (A11011, 1:50, Invitrogen, Carlsbad, CA). Sections were cover slipped with DAPI mounting medium (H-1500, 1: 100, Vector Laboratories, Burlingame, CA).
For immunohistochemical (IHC) analyses, sections were incubated for 1 hour with primary antibody: mouse anti-Osteocalcin (OC, ab13418, 1:400, Abcam); rabbit anti-Dentin Sailophosphoprotein (DSPP, GTX60194, 1:50, Genetex, Irvine, CA); rabbit anti-focal adhesion kinase (FAK, SC-557, 1:400, Santa Cruz Biotechnology); goat anti-syndecan-4 (SYN4, SC-33912, 1:100, Santa Cruz Biotechnology); rabbit anti-tenascin-C (TEN, SC-20932, 1:100, Santa Cruz Biotechnology), goat anti-Amelobastin (AMBN, SC-33102, 1:100, Santa Cruz Biotechnology); rabbit anti-Amelogenin (AM, ABT260, 1:500, Millipore). Sections were incubated for 45 min with Biotin-SP conjugated secondary antibody donkey anti-rabbit IgG (711-065-152, 1:500, Jackson ImmunoResearch), donkey anti-goat IgG (705-065-147, 1:500, Jackson ImmunoResearch), donkey anti-mouse IgG (715-065-1501:500, Jackson ImmunoResearch), 45 min in ABC reagent (PK-4000, Vector Laboratories), and 5 min in DAB (D4293, Sigma, St. Louis, MO). All slides were counterstained with 0.1% fast green (F7252, Sigma).
Sections were analyzed using a Zeiss Axiophot Imager microscope and digital camera (Zeiss, Germany), and images acquired and processed with AxioVision software (Zeiss, Germany, V.4.9.1). Collagen deposition was performed using polarized light (Pol) microscopy (Zeiss, Germany). Figures were created using Adobe Photoshop.
Results
Optimization of dental cell sheet fabrication
To date, cell sheets have been successfully used to regenerate cornea, heart, skin, bone, cartilage, bio-root, dental pulp/dentin complex and periodontal tissues [22, 24–29]. Cell sheets fabricated from dental cells such as stem cells from root apical papilla (SCAPs), periodontal ligament (PDL), pulp and dental follicle (DFCs) have also been studied [25–28, 30]. A variety of techniques can be used to detach cell sheets from cell culture plates, including the use of temperature responsive polymer coatings, cell scrapers, magnetic force, ionic solution, electrochemical polarization, reduced pH, and light [26, 31, 32]. In this study, temperature responsive polymer coated Up-Cell plates were used to create multilayered DE and DM cell sheets. To accomplish this, DE and DM cells were cultured to confluence on thermos-responsive polymer (PIPAAm) immobilized substrate that is hydrophobic at 37ºC, and contiguous cell sheets were harvested at 20ºC, at which temperature the substrate is hydrophilic. This technique also allows for the preservation of the extracellular matrix formed by the cell sheets, including adhesive proteins such as fibronectin. In addition to the obvious advantages of using cell sheet techniques to achieve a highly efficient cell delivery system over the traditional cell suspension systems, the layered dental cell sheets are anticipated to promote the in vivo organized deposition of matrix formation observed in natural tooth development [26].
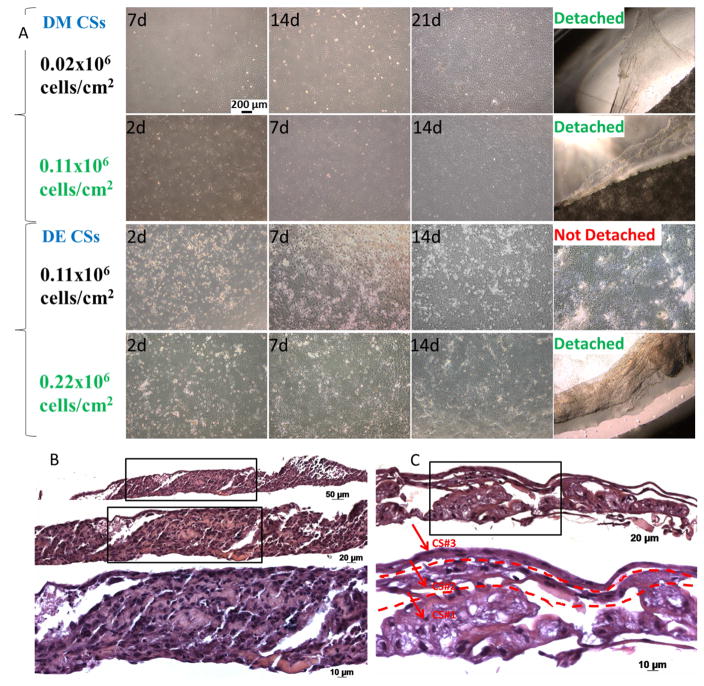
To define optimized methods to create DE and DM cell sheets, DE and DM cells were seeded onto thermos-responsive plates at various cell seeding densities (Figure 2A). DE cells exhibited a typical cobble-stone like morphology, and DM cells showed a typical spindle shaped fibroblast morphology [23]. As anticipated and as previously published, DM cells proliferated faster than the DE cells when cultured at the same cell seeding density, 0.11×106 cells/cm2, after 7 days. Both types of cells retained their characteristic morphologies after achieving one hundred percent confluence. We could observe that as the in vitro culture time increased, the cells spontaneously condensed into an aggregate indicating deposition of ECM. The abundant ECM secreted during the cell sheet formation is anticipated to mimic the microenvironment created by the embryonic basement membrane, including inductive signals responsible for directing dentinogenesis and tooth morphogenesis [27]. DM cells seeded on thermo-responsive plates at cell seeding densities of 0.02×106 and 0.11×106 cells/cm2 both spontaneously detached after 21 and 14 days in vitro culture. DE cells seeded at 0.11×106 cells/cm2 did not detach from the plates after 14 days. However, increasing the cell density to 0.22×106 cells/cm2 resulted in the formation of DE CSs that spontaneously detached after 14 days (Figure 2).
Figure 2. DM and DE cell sheet formation at indicated cell-seeding densities and in vitro culture times, and formation of multilayered dental cell sheets constructs.
A. Optimized cell-seeding densities for CS formation was 0.11 × 106 cells/cm2 for DM cells and 0.22 × 106 cells/cm2 for DE cells (scale bar 200μm). B. Two layered DM CSs. C. Three layered DE CSs.
Dental cell sheet layers
After detachment of the dental CS from the thermo-responsive plates by decreasing the temperature from 37ºC to 20ºC, layered DE-DM CSs were made by carefully stacking the CSs [22]. H&E analyses of paraffin embedded and sectioned specimens reveled organized two and three multilayered CS formation (Figure 2, B and C). The abundant natural ECM secreted by the CSs serves as an endogenous scaffold that not only provides a physical support for the cells in adhesion and cell migration, but can also store and release endogenous growth factors. However, cell sheets have poor mechanical properties and are therefore difficult to maneuver [27]. Therefore, 3D pelleted cell sheets can be created to produce dense cellular constructs that exhibit even cell distribution, satisfactory size and improved handling properties [27].
3D tooth bud constructs in vitro
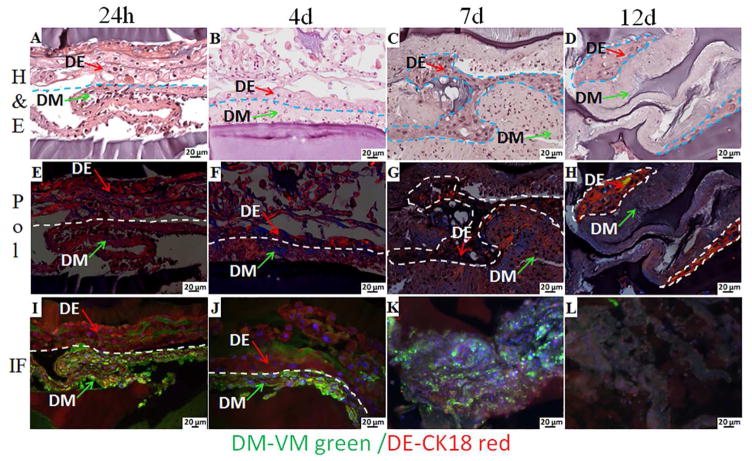
Figure 3 shows H&E, Pol and IF of multilayered 3D dental CS GelMA tooth bud constructs cultured in osteogenic media for 24 hours, and for 4, 7 and 12 days. H&E-staining revealed the morphology of the extracellular matrix formed by the DE and DM cells, between the bottom and the top layers of bilayered GelMA construct (Figure 3A, B, D, C, arrows). Based on the fabrication process, we could easily identify DM CSs in contact with bottom 5% GelMA layer, and DE CSs in contact with the top 3% GelMA layer. The bottom 5% GelMA layer also appeared relatively thicker than the 3% GelMA layer, due to the cone shaped falcon tube used in the fabrication process. The ECM of DE CS appeared loose, with open spaces apparent between the DE cells, while that of DM CS appeared more compact. We also observed close contact between DE and DM CSs, consistent with DE-DM cell layer interactions in natural tooth development. IF analyses of VM expressing (green) DM cells and CK18 expressing (red) DE cells revealed organization of the bilayered GelMA CS constructs, (Figure 3I, J). Polarized light imaging was also used to interrogate collagen deposition and organization in CS derived ECM. After culturing for 12 days in osteogenic media, the nuclei of the cells within the CSs were no longer visible by H&E staining, indicative of cell death (Figure 3D, H, L). Therefore, we chose to limit our in vitro culture of the constructs in osteogenic media for 4 days prior to in vivo implantation.
Figure 3. 3D GelMA-CS constructs cultured in osteogenic media for 24 h, 4, 7 and 12 days.
H&E images (A, B, C, D) revealed extracellular matrix formation and the morphology of the DE and DM cell sheets within the bilayer GelMA constructs. The arrows indicate the DE and the DM CSs. Pol images (E, F, G, H) show the organized collagen in the extracellular matrix. IF imaging (I, J, K, L) showed the expression of VM (green) by DM cells and CK18 (red) by the DE cells.
DE and DM cells sheet interactions
It is well known that reciprocal tissue interactions regulate morphogenesis and cell differentiation during embryonic development [33, 34]. Tooth development is an excellent example of an organ that starts as an epithelial bud surrounded by condensed mesenchyme, and subsequently undergoes a complex morphogenetic process that is tightly regulated by reciprocal dental epithelial-dental mesenchymal cell interactions. During these interactions, cells send and respond to well characterized inductive signals, and differentiate according to their developmental history and position [33]. One of the goals of this study was to demonstrate that our biomimetic 3D tooth bud model can be used to study DE and DM cell interactions both in vitro and in vivo.
We therefore used immunohistochemical analyses to study interactions between in vitro cultured and in vivo implanted DE and DM CSs. Figure 4 shows higher magnification H&E images and IHC analyses of FAK, TEN and SYN4 expression in multilayered DE DM CSs GelMA constructs cultured in osteogenic media for 24 h and 4 days. FAK is a crucial signaling component that is activated by numerous stimuli and functions as a biosensor or integrator to control cell motility, adhesion and shape [35, 36]. It is a highly phosphorylated tyrosine containing protein that is localized at integrin-enriched cell adhesion sites known as focal contacts. Focal contacts are formed at ECM–integrin junctions that bring together cytoskeletal and signaling proteins during the processes of cell adhesion, spreading and migration [35]. Our IHC analyses showed that FAK is expressed when DE and DM CSs are in contact and cultured in osteogenic media for 24 hours and 4 days (Figure 4B and F), demonstrating DE and DM CSs focal contact formation in our bioengineered 3D tooth bud constructs. FAK expression was also observed at sites where DM CSs were attached to the DM-cell encapsulated GelMA surface.
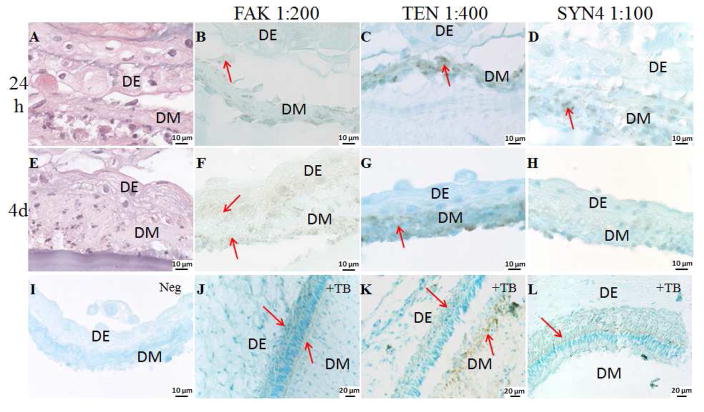
Figure 4. Dental CSs interaction in in vitro cultured GelMA constructs.
High magnification H&E images and IHC analyses of multilayered DE DM CSs GelMA constructs cultured in osteogenic media for 24 h and 4 days, stained with FAK, TEN and SYN4. Arrows indicate expression. Cell sheets are identified as epithelial (DE) and mesenchymal (DM). A. H&E stained DE DM CSs GelMA constructs cultured in osteogenic media for 24 h. FAK, TEN and SYN4 staining (B, C and D) were detected in the DM CSs cultured in osteogenic media for 24 h. E. H&E image of DE DM CSs GelMA constructs cultured in osteogenic media for 4 days. F. FAK staining was detected in DE and DM CSs cultured in osteogenic media for 4 days. G. TEN was detected in the DM CSs cultured in osteogenic media for 4 days. H. Faint SYN4 staining was detected in DM CSs cultured in osteogenic media for 4 days. I. No staining was detected in the negative controls. Specific staining was detected on the natural tooth bud (J. FAK, K. TEN and L. SYN4).
Two of the first ECM molecules found to be highly expressed in the condensed dental mesenchyme at bud stage are the glycoprotein tenascin (TEN) and the cell membrane proteoglycan syndecan (SYN) [33, 34]. TEN functions as a substrate adhesion molecule and is involved in regulating numerous developmental processes, such as morphogenetic cell migration and organogenesis. IHC analyses of developing in vitro cultured 3D GelMA-CS tooth bud constructs showed that TEN is highly expressed by the DM CSs after 24 hours and at 4 days in vitro culture, while no TEN expression was observed in DE CSs (Figure 4, C and G). SYNs are type I integral membrane proteoglycans that contain both chondroitin sulfate and heparan sulfate groups. They are involved in cell-extracellular matrix adhesion, growth factor binding, and are expressed in areas of high morphogenetic activity such as epithelial-mesenchymal interfaces and mesenchymal condensations [33, 34]. SYN4 interacts with a variety of ligands, including vascular endothelial growth factors (VEGFs), platelet-derived growth factors (PDGFs) and fibroblast growth factors (FGFs), and stabilizes the interactions between ligands and receptors by forming a ternary complex [37]. Our IHC analyses showed that SYN4 is expressed by in vitro cultured DM CSs after 24 hours, and exhibited decreased expression after 4 days in culture, but was not expressed in DE CSs (Figure 4, D and H). The co-localization of TEN and SYN4 in the DM CS may suggest roles in regulating changes in cell morphology, growth factor binding, cell adhesion and proliferation to promote DE and DM cell interactions and induce dental cell differentiation, as observed in natural tooth development.
Activation of signaling pathways in dental CSs in vitro
The process of tooth development is under strict genetic control [14]. Dental epithelial-mesenchymal cell interactions are mediated by conserved growth factor signaling pathways and downstream transcriptional partners [14, 38, 39]. The major signaling pathways associated with tooth development include fibroblast growth factor (FGF), bone morphogenetic protein (BMP), transforming growth factor-β (TGF-β), Notch, nuclear transcription factor kappa-B (NF-κB), runt-related transcription factor 2 (RUNX2), and sonic hedgehog (SHH) [14, 38–41]. In this study, we investigated the activation of several signaling pathways associated with tooth development including SHH, BMP2, and RUNX2. SHH and BMP are expressed in both dental epithelium and mesenchyme during several stages of tooth development [38, 42]. In this study, IF analyses of in vitro cultured biomimetic tooth buds showed that SHH (Figure 5, Panels B, F) and BMP2 (Figure 5, Panels D, H) were expressed in multilayered DE-DM cell sheet - GelMA constructs cultured in osteogenic media for 24 h and 4 days. We also investigated the expression of RUNX2, a transcription factor that is essential for tooth development, and is intimately involved in the development of calcified tooth tissue [40]. In vitro IF analyses of in vitro cultured biomimetic tooth bud constructs revealed RUNX2 expression in DE and DM CSs after 24h and 4 days (Figure 5C, G).
Figure 5. Activation of signaling pathways in dental CSs in vitro.
H&E images and IF analyses of multilayered DE DM CSs GelMA constructs cultured in osteogenic media for 24 h and 4 days, stained with SHH, RUNX2 and BMP2 in green, and VM positive DM cells in red. The red staining identifies the DM CSs, while, the absence of red staining identifies the DE cells. Arrows indicate expression. H&E stained DE DM CSs GelMA constructs cultured in osteogenic media for 24 h (A) and 4 days (E). SHH staining was detected in DE and DM CSs after 24 h (B) and 4 days (F). RUNX2 staining was faintly detected at the interface of DE and DM CSs (C), but strongly detected at the second layer of DE CSs after 24 h (inset in the image C), and detected in DE and DM CSs after 4 days (G). BMP2 staining was detected in DE and DM CSs after 24 h (D) and 4 days (H). No staining was detected in the negative controls (I, J and K).
In vivo implantation of 3D tooth bud constructs
We next characterized in vivo implanted biomimetic 3D tooth bud CSG constructs as described in Materials and Methods. After in vitro culture in osteogenic media for 4 days, the constructs were subcutaneously implanted on the backs of rats and harvested after 3 weeks. Harvested in vivo implanted constructs (Figure 6A), and bright field images of the explanted constructs after 3 weeks revealed distinct features (Figure 6B, C).
Figure 6. In vivo implanted bioengineered 3D CSG tooth bud constructs.
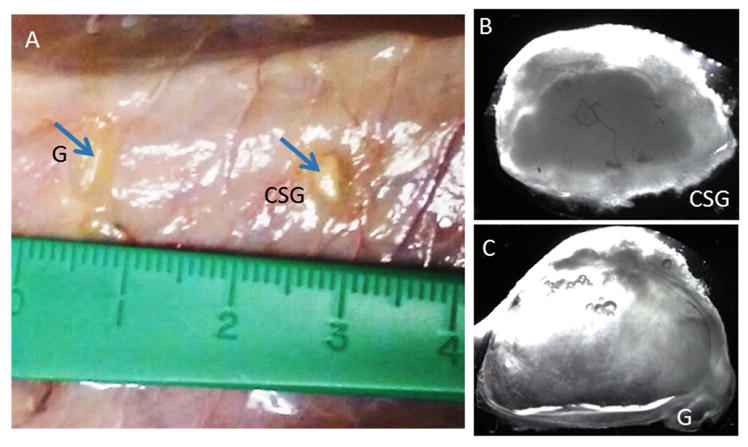
A. In vivo implanted 3 week constructs at harvest (G is acellular GelMA, CSG is biomimetic 3D CSs GelMA construct). B. Bright field images of an in vivo CSG construct. C. Bright field image of an in vivo acellular GelMA constructs.
Micro-CT analyses of in vivo grown 3D CS GelMA (CSG) tooth bud constructs
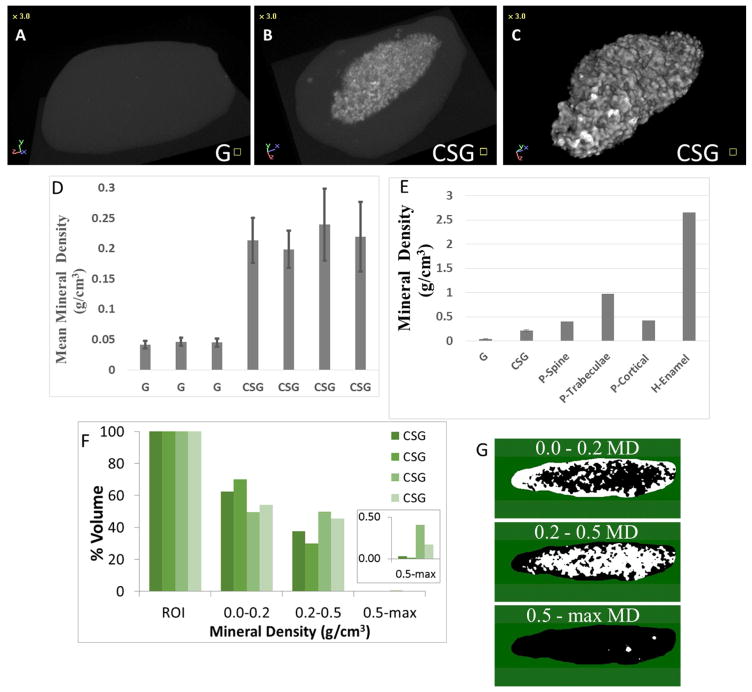
Harvested in vivo grown constructs were fixed in formalin and analyzed using micro-CT for mineralized tissue formation. Hard tissue formation was observed in 4/4 (100%) of the CSG constructs, while all acellular GelMA (G) constructs appeared negative for hard tissue formation (Figure 7A versus 7B). 3D modeling of mineralized tissue formation within the in vivo implanted constructs is shown in the Figure 7C. The volume and density of mineralized tissue formed in each construct was then determined using CT-Analyser software (Figure 7D). CSG constructs displayed significantly higher mineral density than the acellular GelMA control constructs (p<0.05). We also compared the mineral density obtained from in vivo implanted CSG constructs (mean 0.22±0.02 g/cm3) with those reported for naturally formed mineralized tissues (Figure 7E). Mineralized tissue formed by CSG constructs implanted in vivo for 3 weeks appeared less dense than pig spine, trabecular bone, cortical bone and human enamel. However, small areas within the CSG constructs appeared brighter by micro-CT (maximum mineral density of 0.9 g/cm3), but were not reflected in the calculated mean mineral densities. We therefore calculated the percent volume of mineralized tissue in three ranges of mineral density: 0.0–0.2; 0.2–0.5; and 0.5-max g/cm3 (Figure 7F). We also calculated the location of mineralized tissues of various densities within each construct, where the color white color represents the location of mineralized tissue of each density range (Figure 7G). These results showed that the least dense mineralized tissue (0.0–0.2 g/cm3) was located around the periphery of the constructs, while more highly mineralized tissues with densities in the range of 0.2–0.5 g/cm3 were localized toward the center of the constructs. These results suggested that 3D construct mineralization may have begun within the center of the constructs, and subsequently expanded to the outer edges. The brightest areas of the micro-CT images, representing mineralized tissues in the range of 0.5-max g/cm3, (Figure 7F), also appeared to localize to the center of the constructs, again consistent with the fact that more mature mineralized tissues were located in the center of the constructs, and that longer in vivo implantation times may result in increased mineral density of the entire construct over time.
Figure 7. MicroCT analyses of in vivo bioengineered CSG constructs.
A. No mineralized tissue formation was observed in the acellular GelMA constructs (G). B. Mineralized tissue formation was observed in the CSG constructs. C. 3D model of the mineralized tissue. D. Quantification of mineral density (g/cm3) of the CSG constructs. E. Comparison of mineral densities from engineered and natural mineralized tissues (pig spine, trabecular bone, cortical bone and human enamel) [1, 2]. F. Percent volume of mineralized tissue within ranges of mineral density (ROI – region of interest corresponds to the whole mineralized tissue). G. Representation of areas of mineralized tissue within the ranges of mineral densities (white color represents areas within the range). Abbreviations: MD, mineral density.
Histological and Immunohistochemical analyses of 3D tooth bud constructs
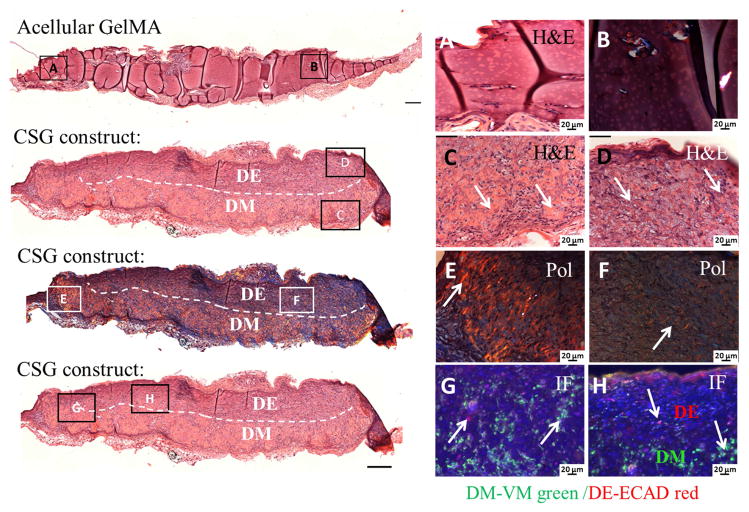
We next used histological approaches to characterize the cellular nature of the in vivo implanted 3D CSG tooth bud constructs. The mineralized constructs were decalcified, embedded in paraffin and sectioned. H&E staining and polarized light (Pol) imaging confirmed that no tissue formation was observed in acellular GelMA constructs after 3 weeks in vivo implantation (Figure 8, A and B). In contrast, in vivo grown 3D CSG tooth bud constructs exhibited high cellularity, extensive extracellular matrix formation, and mineralized osteodentin-like tissue formation, with dashed lines indicating the biomimetic enamel organ (on the top) and pulp organ (on the bottom) layers (Figure 8, C and D). These analyses showed that the GelMA scaffolds in dental cell-encapsulated GelMA constructs were largely biodegraded by the cells over time, only detectable at the edges of the DM cell layer and more so in the DE cell encapsulated biomimetic enamel organ layer. We did not detect distinct layered DE-DM cell sheets within the CSG constructs.
Figure 8. In vivo implanted bioengineered 3D CSG tooth bud constructs exhibited elaborate extracellular matrix formation after 3 weeks.
No tissue formation was observed in the acellular GelMA constructs, H&E (A) and Pol (B) images. H&E stained embedded paraffin and sectioned constructs exhibited high cellularity (C, D), extensive extracellular matrix and dentin/bone-like tissue formation at the DM GelMA layer. The dashed line separates the biomimetic pulp organ (DM in the bottom layer) from the biomimetic enamel organ (DE in the top layer). Pol images (E, F) revealed organized collagen formation within the CSG constructs. IF images (G, H) show the expression of VM (green) by DM cells in the biomimetic pulp organ layer, and ECAD (red) by the DE cells in the biomimetic enamel organ of the CSG constructs.
Polarized light microscopy, a useful technique to study the molecular organization of collagens [43], revealed significant organized collagen present within the CSG constructs (Figure 8E and F, Pol). The collagen produced in the biomimetic DM-GelMA pulp organ GelMA layer appeared more organized than that produced by the DE-GelMA biomimetic enamel organ layer. Collagen fiber alignment within the GelMA constructs appeared to be oriented perpendicular to the encapsulating host tissue, distinguishing collagen secretion by the encapsulated dental cell seeded GelMA constructs as compared to that of the surrounding rat host tissue, consistent with our previously published results [19].
We next used immunofluorescence (IF) to characterize DM and DE cells within the in vivo implanted CSG constructs (Figure 8G and H). VM positive DM cells were identified in the biomimetic pulp organ layer of the CSG constructs (Figure 8H), and E-cad positive DE cells were identified in the biomimetic enamel organ layer. At the interface of the two layers, both DE and DM cell populations were identified, consistent with the location of the DE and DM cell sheets (Figure 8G, arrows).
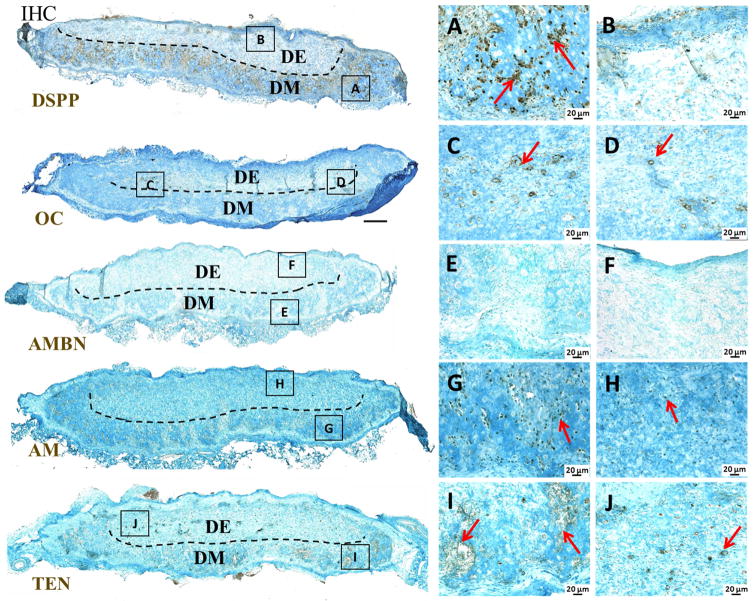
IHC was used to examine dental cell differentiation marker expression within harvested in vivo CSG constructs (Figure 9). Distinct and robust DSPP expression was detected in the biomimetic pulp organ layer of in vivo implanted CSG constructs, indicative of DM cell derived odontoblast cell differentiation (Figure 9A). DSPP was also faintly expressed in DE cells of the biomimetic enamel organ layer of in vivo implanted CSG constructs (Figure 9B). Strong OC expression was localized toward the center of in vivo implanted CSG constructs, and at the interface of the DE and DM cell layers, the location of the dental CSs, indicative of DM cell derived odontoblast and osteoblast cell differentiation (Figure 9C and D). AM was expressed in both biomimetic enamel and pulp organ layers (Figure 9G, H), indicating DE cell differentiation toward ameloblast cell fates, and possible mixing of DE and DM containing GelMA layers prior to photo-polymerization. However, no AMBN expression was observed in in vivo implanted CSG constructs (Figure 9E, F). TEN was detected in both biomimetic enamel and pulp organ layers, appearing more highly expressed in the pulp organ layer (Figure 9I, J). No SYN4 expression was observed in in vivo implanted CSG constructs (data not shown).
Figure 9. Dental cell differentiation within in vivo grown CSG tooth bud constructs after 3 weeks.
IHC analyses of dentin, enamel and bone specific markers. The odontoblast differentiation marker DSPP was highly expressed throughout the biomimetic pulp organ layer (A, B). Odontoblast/Osteoblast differentiation marker OC was expressed in the centers of the CSG constructs (C, D). Ameloblast differentiation marker AM was expressed throughout the CSG constructs in both biomimetic pulp and enamel organ layer (G, H), while AMBN was not detected in the CSG constructs (E, F). TEN was detected in the both biomimetic pulp and enamel organ layers of the CSG constructs (I, J).
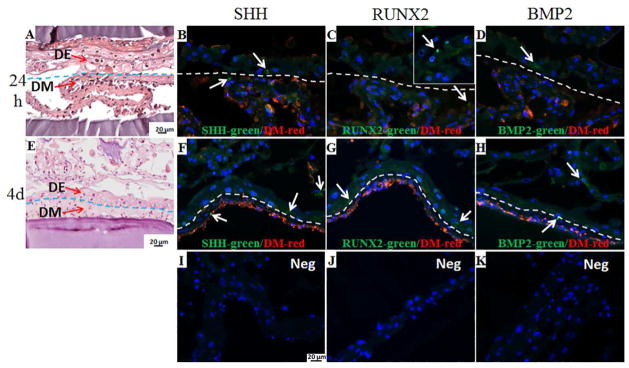
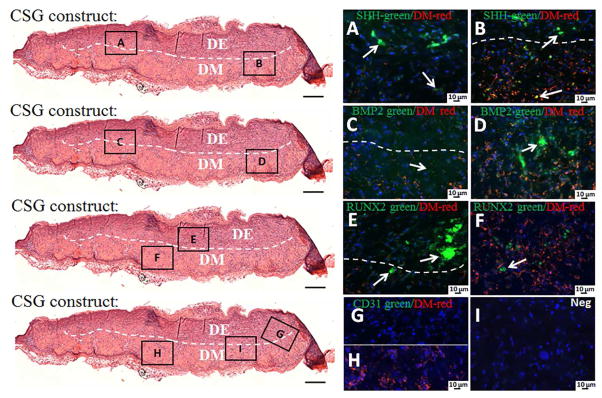
IF was used to investigate tooth differentiation signaling pathways, and to detect host blood vessel formation within 3 week in vivo grown CSG tooth bud constructs (Figure 10). The red staining indicates the VM expressing DM cell biomimetic pulp organ layer, and the absence of red staining indicates the biomimetic DE containing enamel organ layer. We found that SHH was expressed throughout the CSG constructs in both biomimetic enamel and pulp organ layer (Figure 10A, B), however, stronger SHH expression was observed in the biomimetic enamel organ layer (Figure 10A, arrows). BMP2 was also expressed throughout the CSG constructs, however, stronger BMP2 expression was observed at the biomimetic pulp organ layers (Figure 10C, D). RUNX2 was expressed in both biomimetic pulp and enamel organ layers (Figure 10E, F), but, stronger RUNX2 expression was observed in the biomimetic epithelial organ layer (Figure 10F, arrows). These results are consistent with our analyses of natural porcine teeth, and published literature (See Supplemental Figure 1).
Figure 10. Activation of signaling pathways and host blood vessel detection within in vivo grown CSG tooth bud constructs after 3 weeks.
IF analyses of SHH, RUNX2, BMP2 and CD31 in green and VM positive DM cells in red. The red staining identifies the biomimetic pulp organ layer where the DM cells are localized, while, the absence of red staining identifies the biomimetic enamel organ layer where the DE cells are localized. SHH was expressed throughout the CSG constructs in both biomimetic enamel and pulp organ layer (A, B). BMP2 was also expressed throughout the CSG constructs, but stronger BMP2 expression was observed in biomimetic pulp organ layers (C, D). RUNX2 was expressed in both biomimetic enamel and pulp organ layer (E, F), however, stronger RUNX2 expression was observed in the biomimetic epithelial organ layer (E, arrows). No CD31 positive staining was detected in the constructs (G, H). No positive staining was detected in the negative controls (I).
H&E staining revealed blood vessels at locations adjacent to, but not within, the in vivo implanted and grown biomimetic constructs. To better identify host derived endothelial cells and neo-vasculature in the in vivo construct, we performed IF analysis using a CD31 antibody. No CD31 positive expression was detected within the in vivo implanted constructs (Figure 10G, H). Indeed, these results were not unexpected, since in this specific study we did not include endothelial cells in the biomimetic constructs. Our previously published reports have shown that neo-vasculature formation within GelMA constructs was observed when human umbilical vein endothelial cells (HUVECs) were included [19].
Discussion
Bioengineered tooth buds that can develop and remodel in a manner similar to that of natural teeth, have the potential to serve as superior living, functional permanent replacement teeth as compared to the currently used standard prosthetic teeth, dental implants [8]. Previously, we described the design and characterization of dental and endothelial cell encapsulated GelMA constructs as 3D biomimetic tooth bud models, consisting of biomimetic enamel and pulp organ bilayers [18]. We used in vitro analyses to identify GelMA formulas that supported DE and DM cell attachment, spreading, metabolic activity, and neo-vasculature formation by co-seeded endothelial cells (HUVECs) [19]. We also demonstrated that selected GelMA formulas supported dental cell differentiation, vascularization, and in vivo formation of mineralized tissues of specified size and shape. We also identified some limitations of our original model, including the fact that extensive DE-DM cell mixing and no distinct enamel or dentin layers formation was observed. To address these limitations, here we performed successive photo-crosslinking of individual cell-seeded GelMA layers, to generate distinct bilayered constructs. Moreover, to create a biomimetic tooth bud, and to increase the DM-DM, DE-DE, and DE-DM cell interactions observed in natural tooth bud development, DE-DM dental cell sheets were sandwiched between biomimetic 3D GelMA enamel and pulp organs. We optimized the cell seeding density and in vitro culture time to create DE and DM cell sheets grown on thermo-responsive UpCell plates. DM and DE CSs were obtained at cell densities of 0.11×106 cells/cm2 and 0.22×106 cells/cm2, respectively, after 14 days (Figure 2A), and organized multilayered CSs were created by stacking layers of DM and DE CSs (Figure 2B, C).
The design of our fabricated 3D biomimetic tooth bud constructs was intended to facilitate cross-talk between the DE-DM cell sheets, and also with the adjacent biomimetic enamel and pulp organ layers. First, we studied the effect of the osteogenic media on multilayered 3D cell sheets GelMA tooth bud construct differentiation in in vitro culture for 24 hours, 4, 7 and 12 days (Figure 3). The cell sheets survived in vitro culture at day 7, but distinct cell nuclei were not observed after 12 days in culture, suggesting lack of long-term cell survival in in vitro culture. A possible explanation for lack of long-term survival is due to oxygen and nutrient diffusion limitations through the 3D GelMA constructs. Our previous results showed that optimized GelMA formulations supported cell attachment, morphology, and metabolic and MMP activities of encapsulated cells after 4 weeks in vitro culture [18, 19].
Our in vitro results also demonstrated that our CSG 3D tooth bud model can be used to study DE and DM interactions leading to bioengineered tooth development (Figure 4). High resolution H&E and IHC images were used to show that DE and DM CSs were in close contact with each other, and expressed focal adhesion molecules including FAK, TEN and SYN4, indicative of cellular cross-talk. These results are consistent with DE and DM CS interact and focal contacts, and expression of proteins normally expressed during natural tooth development [33, 34, 37]. We also demonstrated expression of many of the signaling pathways that control tooth development [41]. IF analyses of in vitro cultured biomimetic tooth buds revealed that 3D DE and DM CSG constructs expressed SHH, BMP2 and RUNX2 (Figure 5), consistent with and providing evidence for DE and DM cell interactions, formation, maintenance, homeostasis, and differentiation.
Our in vivo biomimetic tooth bud constructs formed robust mineralized tissues that adopted the size and shape of the original 3D constructs. Micro-CT analysis showed that more dense mineralized tissue was located toward the center of the constructs, while less dense mineralized tissues were located at the periphery. Our interpretation of these results is that mineralized tissue formation may have initiated in the center of the CSG constructs, perhaps at the location of the DE-DM dental CS. These results are in contrast to published report that bioengineered mineral tissue formation normally progresses from the defect borders toward the center, when using an injectable bone substitute in a rabbit distal femoral condyle model [44].
The density of the biomimetic CSG mineralized tissue was less than that of a variety of natural formed mineralized tissues (Figure 7E). These results indicate that additional modifications, such as increasing in vivo implantation time, may result in increased mineral density of the bioengineered tooth constructs [1, 2]. In addition, we compared the mineralized tissue density of 3D biomimetic tooth bud constructs grown with cell sheets (CSG) and without cell sheets (unpublished data), and found no statistically significant differences. However, the 3D biomimetic CSG tooth bud constructs exhibited higher cellularity and more extensive extracellular matrix formation as compared with the biomimetic tooth bud constructs without DE-DM cell sheets.
DE-DM cell interactions regulate all stages of tooth development [34]. The expression of several molecules, including TEN and SYNs, are regulated by the epithelium [34, 37]. Our results showed that TEN was expressed in both the biomimetic enamel and pulp organs of in vivo grown CSG constructs (Figure 9), and that SYN4 expression was not detected in biomimetic tooth buds, but was detectable in ameloblasts and odontoblasts of the natural pig tooth buds. In contrast, we found that TEN was highly expressed in in vitro cultured DM CSs and not in the DE CSs after 24 hours and 4 days in osteogenic media, and that SYN4 was more highly expressed in the DM CSs after 24 hours (Figure 4). A possible explanation for the absence of SYN4 expression in in vitro cultured DE cell sheets could be that DE cells had not yet differentiated into ameloblasts after 24 hours. It has been demonstrated that SYN4 is mainly expressed in the oral epithelium, the dental epithelial cells of enamel organs in the molars, and in the cervical loops in the incisors during the late bell stage of mouse development [37]. When the inner enamel epithelial cells gave rise to ameloblasts, the loss of SYN4 expression was evident. SYN4 was also expressed in stratum intermedium cells in the incisors and in DM cells adjacent to the cervical loops in molars and postnatal incisors [37]. Based on these studies, we hypothesize that morphogenetic cell migration and organogenesis might be occurring in our in vivo grown biomimetic enamel organ, which would explain the expression of TEN in both biomimetic enamel and pulp organ. This result is consistent with the expression of TEN in natural tooth buds, in that we observed TEN expression at the ameloblast – odontoblast interface, and a gradient of TEN expression in the pulp. It has been reported that the expression of TEN is induced in the dental mesenchyme in the same area as SYN1, and that TEN and SYN1 expression increased with the duration of epithelial-mesenchymal cell contact, and that TEN and SYN1 expression is stage dependent [33, 34]. For example, in natural bud stage teeth, the expression of TEN and SYN1 showed closely correlated expression in the condensed dental mesenchyme, and that cell division was localized to the same area. In contrast, SYN1 expression persists until cap stage in natural tooth buds, then is downregulated as tooth development proceeds. TEN expression is downregulated in the DM in natural cap stage teeth, upregulated during bell stage, and thereafter persists in the dental pulp, even in mature teeth [33, 34].
Dental pulp cells can contribute to the formation of dentin and bone-like tissue in response to external stimuli [45]. However, the molecular mechanisms regulating their induction and elaboration of mineralized dentin tissue remain largely unknown. Our results shows that GelMA encapsulated DM cells produce dense mineralized tissue in the biomimetic pulp organ layer of the CSG constructs, where DSPP was highly expressed. In contrast, DSPP was only faintly detected in the biomimetic enamel organ layer of the 3D CSG tooth bud constructs. Indeed, DSPP is transiently expressed in the early stage of secretory ameloblasts in natural tooth development [46], where the secretion of ameloblast-derived DSPP is short-lived, and correlates to the establishment of the dentin-enamel junction (DEJ). These observations are consistent with roles for DSPP in creating the specialized first-formed enamel adjacent to the DEJ. Once DSPP is secreted, it is subjected to proteolytic cleavage that results in the formation of two distinct proteins, referred to as dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) [46]. It has also been reported that DSPP mRNA and protein are detected in odontoblasts and pre-ameloblasts during tooth development [47]. OC expression is reduced as compared to DSPP, indicative of DM cell derived odontoblast cell differentiation and dentin-like matrix formation, rather than osteoblast derived bone matrix formation. Interestingly, in our bioengineered 3D tooth bud constructs, we detected OC expression at the interface between the biomimetic enamel and pulp organs, where the DE and DM cell sheets were originally located, but which were not distinctly observed as discrete structures in either the in vitro cultured or in vivo grown tooth bud constructs. Possible explanations for the lack of discrete cell sheet structures in harvested constructs could be that the cell sheet layers reorganized and remodeled over time [48]. One of the major functions of the ECM is to provide structural support to cells and tissues, and cells respond differently to ECM proteins when they are cultured in 2D as compared to 3D cell culture, and also depending on the rigidity of the matrix [48]. Several studies using dynamic imaging approaches with fluorescently tagged ECM molecules have demonstrated that ECM networks are highly dynamic structures that are subjected to constant stretching and contracting as well as reorganization mediated by cell and tissue movements [48]. It was reported that enamel matrix derivative (EMD) containing AM had a stimulatory effect on mesenchymal cells and tissues in the context of alveolar bone regeneration [49]. EMD had a positive effect on mineralization, inducing increased alkaline phosphatase activity, and osteocalcin and collagen type I expression [49]. Therefore, it is important to understand the mechanisms of assembly and reorganization of ECM proteins from a 3D perspective in cell and organ systems. For example, our results show that collagen fiber alignment within the biomimetic constructs appeared to be oriented perpendicular to that of the encapsulating host tissue (Figure 8E and F). During tooth development, cell differentiation and ECM secretion occur initially as DM cells differentiate into odontoblasts producing dentin, and secondly as DE cells differentiate into ameloblasts producing enamel. DE cells interact with extracellular matrix components including the basement membrane, which plays a crucial role in maintaining ameloblast differentiation, expression of enamel matrix proteins AMBN, AM and enamelin, and subsequent enamel formation [50, 51]. In natural early tooth development, AMBN is expressed before AM, both proteins are expressed in later staged of tooth development, while even later AMBN is downregulated while AM expression is maintained [52]. AM is detectable in both dental epithelial and dental mesenchymal cells and tissues at the epithelial-mesenchymal cell interface at early stages of tooth development [53]. Our results show that AM was expressed in both biomimetic enamel and pulp organ layers (Figure 9), while AMBN was not detected. We also demonstrated that mineralized dentin-like tissue formed initially at the interface of the biomimetic pulp and enamel organs, at the location of the DE and DM CSs, while enamel-like tissue formation was not observed in the biomimetic enamel organ after 3 weeks in vivo growth. We hypothesize that the encapsulated dental cells may be recapitulating their natural developmental process by upregulating AM and downregulating AMBN, forming dentin-like tissue, and perhaps if grown for longer periods of time ultimately would form enamel-like tissue.
During the past years, researchers have extensively studied signaling networks regulating tooth development [38–42, 54]. These studies have shown exquisitely regulated temporal expression of growth factors and their downstream transcription factors, in dental epithelial and mesenchymal tissues throughout tooth development [38]. In many instances, reiterative growth factor expression is observed during successive stages of tooth morphogenesis, to promote signaling in both directions between DE and DM tissues. Our results show that SHH, BMP2 and RUNX2 were expressed in in vivo cultured biomimetic tooth buds in both enamel organ and pulp organ layers (Figure 10), and that in general, expression in in vivo cultured constructs appeared stronger than in in vitro cultured constructs (Figure 5). SHH and RUNX2 were highly expressed in biomimetic epithelial organ layers (Figure 10A and F), while BMP2 was highly expressed at the interface of the biomimetic epithelial organ and pulp organ layers (Figure 10D). In natural tooth development, SHH is involved in the proliferation and differentiation of both DE and DM stem cells, while SHH expression is limited to the dental epithelium, suggesting that SHH functions as an autocrine signal in dental epithelium and as a paracrine signal in dental mesenchyme [41, 42]. Thus, SHH exerts crucial functions in tooth germ growth, morphogenesis, and tissue-tissue interactions. In natural teeth, BMP2 is expressed during embryonic and post-natal tooth development during odontoblast and ameloblast differentiation [41]. The differential fate of dental epithelial stem cells is controlled by BMP/SHH signaling network, which partially accounts for the different postnatal growth potential of teeth [54]. Therefore, the crosstalk between the two signaling pathways, BMP and SHH, is very important in regulating the fate of epithelial stem cells during organogenesis. RUNX2 is also essential for the later stages of tooth formation during development of calcified tooth tissue (enamel organ and dentin organ formation) [40]. Moreover, RUNX2 regulates the alveolar remodeling process essential for tooth eruption and may play a role in the maintenance of the periodontal ligament [40]. Although enamel-like tissue formation was not observed in our biomimetic enamel organ tissues after 3 weeks in vivo growth, the dental epithelial cells were actively expressing SHH, BMP2 and RUNX2 similar to expression patterns observed in naturally developing teeth. Future studies that include increased in vivo implantation times are needed to better understand the behavior and fate of the DE cells in the enamel organ of our biomimetic 3D tooth bud constructs.
Future improvements to our model include methods to fabricate biomimetic tooth roots that can maintain viability and support tooth functions, as an important contribution to the field of regenerative dentistry [25]. We previously demonstrated that our bioengineered 3D biomimetic tooth bud constructs containing HUVECs exhibited functional vascularization resembling natural neovasculature organization, which is very important to maintain tooth functions [19]. Future implantation of bioengineered 3D biomimetic GelMA-CS-HUVEC tooth buds in a jaw bone socket may facilitate periodontal tissue formation [26]. Together, these approaches suggest the potential to create biomimetic living, functional bioengineered tooth replacements.
Conclusions
Natural tooth formation is a highly complex process where the tightly controlled spatiotemporal expression and interactions of growth factors, cytokines, and transcription factors direct macro-morphological and micro-morphological aspects of tooth development. Proper control of these interactions is essential in order to generate bioengineered teeth of specified size and shape. In this study we demonstrate, for the first time, the successful creation of multilayered DE-DM cell sheets that express proteins involved in the DE-DM interactions, condensation and differentiation. The combined use of DE-DM cell sheets with dental cell encapsulated 3D GelMA constructs significantly increased dental ECM formation in both of the biomimetic enamel and pulp organ layers, and upregulated dental cell differentiation marker expression. Future studies will focus on increasing in vivo implantation duration in order to better understand the behavior and fate of DE and DM cells in the respective biomimetic enamel organ and pulp organs, for eventual enamel and dentin formation. Moreover, 3D in vitro culture conditions that include mechanical stimulation could be used study how these forces contribute to natural tooth development, and influence dental cell differentiation. We conclude that dental cell sheets provide a new and useful tool to study DE and DM cell interactions leading to ameloblast and odontoblast cell differentiation, respectively, and that biomimetic 3D GelMA-CS tooth bud constructs provide a novel and promising model for the development of functional, bioengineered replacement teeth of specified size and shape.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIDCR R01DE016132 (PCY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nelson Monteiro, Email: Nelson.Monteiro@tufts.edu.
Elizabeth E. Smith, Email: Elizabeth.Smith@tufts.edu.
Shantel Angstadt, Email: Shantel.Angstadt@tufts.edu.
Weibo Zhang, Email: Weibo.Zhang@tufts.edu.
Ali Khademhosseini, Email: alik@bwh.harvard.edu.
Pamela C. Yelick, Email: Pamela.Yelick@tufts.edu.
References
- 1.Huang TT, Jones AS, He LH, Darendeliler MA, Swain MV. Characterisation of enamel white spot lesions using X-ray micro-tomography. Journal of Dentistry. 2007;35(9):737–43. doi: 10.1016/j.jdent.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Inui A, Itamoto K, Takuma T, Tsutsumi H, Tanigawa M, Hayasaki M, et al. Age-related changes of bone mineral density and microarchitecture in miniature pigs. Journal of Veterinary Medical Science. 2004;66(6):599–609. doi: 10.1292/jvms.66.599. [DOI] [PubMed] [Google Scholar]
- 3.Cooper LF. The current and future treatment of edentulism. Journal of Prosthodontics. 2009;18(2):116–22. doi: 10.1111/j.1532-849X.2009.00441.x. [DOI] [PubMed] [Google Scholar]
- 4.Young CS, Terada S, Vacanti JP, Honda M, Bartlett JD, Yelick PC. Tissue engineering of complex tooth structures on biodegradable polymer scaffolds. Journal of Dental Research. 2002;81(10):695–700. doi: 10.1177/154405910208101008. [DOI] [PubMed] [Google Scholar]
- 5.Ravindran S, George A. Biomimetic extracellular matrix mediated somatic stem cell differentiation: applications in dental pulp tissue regeneration. Front Physiol. 2015;6:118. doi: 10.3389/fphys.2015.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monteiro N, Yelick P. Alveolar complex regeneration. In: Tolstunov L, editor. Horizontal Alveolar Ridge Augmentation in Implant Dentistry: A Surgical Manual. Wiley-Blackwell; 2015. p. 360. [Google Scholar]
- 7.Yen AH, Yelick PC. Dental tissue regeneration - a mini-review. Gerontology. 2011;57(1):85–94. doi: 10.1159/000314530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteiro N, Yelick P. Advances and Perspectives in Tooth Tissue Engineering. Journal of Tissue Engineering and Regenerative Medicine. 2016 doi: 10.1002/term.2134. Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai WF, Lee JM, Jung HS. Molecular and engineering approaches to regenerate and repair teeth in mammals. Cellular and Molecular Life Sciences. 2014;71(9):1691–701. doi: 10.1007/s00018-013-1518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirayama M, Oshima M, Tsuji T. Development and Prospects of Organ Replacement Regenerative Therapy. Cornea. 2013;32:S13–S21. doi: 10.1097/ICO.0b013e3182a18e6c. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Kiso H, Saito K, Togo Y, Tsukamoto H, Huang B, et al. Feasibility of Gene Therapy for Tooth Regeneration by Stimulation of a Third Dentition. 2013 [Google Scholar]
- 12.Duailibi MT, Duailibi SE, Young CS, Bartlett JD, Vacanti JP, Yelick PC. Bioengineered teeth from cultured rat tooth bud cells. Journal of Dental Research. 2004;83(7):523–8. doi: 10.1177/154405910408300703. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Ahluwalia IP, Literman R, Kaplan DL, Yelick PC. Human dental pulp progenitor cell behavior on aqueous and hexafluoroisopropanol based silk scaffolds. Journal of Biomedical Materials Research Part A. 2011;97(4):414–22. doi: 10.1002/jbm.a.33062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nanci A, Cate ART. Ten Cate’s oral histology: development, structure, and function. Mosby; 2003. [Google Scholar]
- 15.Young CS, Abukawa H, Asrican R, Ravens M, Troulis MJ, Kaban LB, et al. Tissue-engineered hybrid tooth and bone. Tissue Engineering. 2005;11(9–10):1599–610. doi: 10.1089/ten.2005.11.1599. [DOI] [PubMed] [Google Scholar]
- 16.Yelick PC, Vacanti JP. Bioengineered teeth from tooth bud cells. Dental Clinics of North America. 2006;50(2):191–203. viii. doi: 10.1016/j.cden.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Martens W, Bronckaers A, Politis C, Jacobs R, Lambrichts I. Dental stem cells and their promising role in neural regeneration: an update. Clinical Oral Investigations. 2013;17(9):1969–83. doi: 10.1007/s00784-013-1030-3. [DOI] [PubMed] [Google Scholar]
- 18.Smith EE, Yelick PC, Khademhosseini A, editors. Optimization of a biomimetic model for tooth regeneration. Bioengineering Conference (NEBEC), 2014 40th Annual Northeast; 2014 25–27 April. [Google Scholar]
- 19.Smith E, Zhang W, Schiele NR, Khademhosseini A, Kuo CK, Yelick PC. Developing a biomimetic 3D model for tooth regeneration. Journal of Tissue Engineering and Regenerative Medicine. 2016 doi: 10.1002/term.2246. Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichol JW, Koshy S, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31(21):5536–44. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosseini V, Ahadian S, Ostrovidov S, Camci-Unal G, Chen S, Kaji H, et al. Engineered contractile skeletal muscle tissue on a microgrooved methacrylated gelatin substrate. Tissue Engineering Part A. 2012;18(23–24):2453–65. doi: 10.1089/ten.TEA.2012.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pirraco RP, Iwata T, Yoshida T, Marques AP, Yamato M, Reis RL, et al. Endothelial cells enhance the in vivo bone-forming ability of osteogenic cell sheets. Laboratory Investigation. 2014;94(6):663–73. doi: 10.1038/labinvest.2014.55. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Ahluwalia IP, Yelick PC. Three dimensional dental epithelial-mesenchymal constructs of predetermined size and shape for tooth regeneration. Biomaterials. 2010;31(31):7995–8003. doi: 10.1016/j.biomaterials.2010.07.020. http://dx.doi.org/10.1016/j.biomaterials.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okano T. Current Progress of Cell Sheet Tissue Engineering and Future Perspective. Tissue Engineering Part A. 2014;20(9–10):1353–4. doi: 10.1089/ten.tea.2013.0783. [DOI] [Google Scholar]
- 25.Wei F, Song T, Ding G, Xu J, Liu Y, Liu D, et al. Functional tooth restoration by allogeneic mesenchymal stem cell-based bio-root regeneration in swine. Stem Cells and Development. 2013;22(12):1752–62. doi: 10.1089/scd.2012.0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Li Y, Mao L, Peng H. Periodontal healing by periodontal ligament cell sheets in a teeth replantation model. Archives of Oral Biology. 2012;57(2):169–76. doi: 10.1016/j.archoralbio.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Na S, Zhang H, Huang F, Wang W, Ding Y, Li D, et al. Regeneration of dental pulp/dentine complex with a three-dimensional and scaffold-free stem-cell sheet-derived pellet. Journal of Tissue Engineering and Regenerative Medicine. 2013 doi: 10.1002/term.1686. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 28.Honjo K, Yamamoto T, Adachi T, Amemiya T, Mazda O, Kanamura N, et al. Evaluation of a dental pulp-derived cell sheet cultured on amniotic membrane substrate. Bio-Medical Materials and Engineering. 2015;25(2):203–12. doi: 10.3233/bme-151270. [DOI] [PubMed] [Google Scholar]
- 29.Gomes JA, Geraldes Monteiro B, Melo GB, Smith RL, Cavenaghi Pereira da Silva M, Lizier NF, et al. Corneal reconstruction with tissue-engineered cell sheets composed of human immature dental pulp stem cells. Investigative Ophthalmology and Visual Science. 2010;51(3):1408–14. doi: 10.1167/iovs.09-4029. [DOI] [PubMed] [Google Scholar]
- 30.Yang B, Chen G, Li J, Zou Q, Xie D, Chen Y, et al. Tooth root regeneration using dental follicle cell sheets in combination with a dentin matrix - based scaffold. Biomaterials. 2012;33(8):2449–61. doi: 10.1016/j.biomaterials.2011.11.074. [DOI] [PubMed] [Google Scholar]
- 31.Owaki T, Shimizu T, Yamato M, Okano T. Cell sheet engineering for regenerative medicine: Current challenges and strategies. Biotechnology Journal. 2014;9(7):904–14. doi: 10.1002/biot.201300432. [DOI] [PubMed] [Google Scholar]
- 32.Ito A, Hibino E, Kobayashi C, Terasaki H, Kagami H, Ueda M, et al. Construction and delivery of tissue-engineered human retinal pigment epithelial cell sheets, using magnetite nanoparticles and magnetic force. Tissue Engineering. 2005;11(3–4):489–96. doi: 10.1089/ten.2005.11.489. [DOI] [PubMed] [Google Scholar]
- 33.Vainio S, Jalkanen M, Thesleff I. Syndecan and tenascin expression is induced by epithelial-mesenchymal interactions in embryonic tooth mesenchyme. Journal of Cell Biology. 1989;108(5):1945–53. doi: 10.1083/jcb.108.5.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thesleff I, Vaahtokari A, Vainio S, Jowett A. Molecular mechanisms of cell and tissue interactions during early tooth development. Anatomical Record. 1996;245(2):151–61. doi: 10.1002/(SICI)1097-0185(199606)245:2<151::AID-AR4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 35.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nature Reviews Molecular Cell Biology. 2005;6(1):56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 36.Premaraj S, Souza I, Premaraj T. Focal adhesion kinase mediates beta-catenin signaling in periodontal ligament cells. Biochemical and Biophysical Research Communications. 2013;439(4):487–92. doi: 10.1016/j.bbrc.2013.08.097. [DOI] [PubMed] [Google Scholar]
- 37.Yan Z, Chen G, Yang Y, Sun L, Jiang Z, Feng L, et al. Expression and roles of syndecan-4 in dental epithelial cell differentiation. International Journal of Molecular Medicine. 2014;34(5):1301–8. doi: 10.3892/ijmm.2014.1910. [DOI] [PubMed] [Google Scholar]
- 38.Thesleff I, Sharpe P. Signalling networks regulating dental development. Mechanisms of Development. 1997;67(2):111–23. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- 39.Jussila M, Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harbor Perspectives in Biology. 2012;4(4):a008425. doi: 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camilleri S, McDonald F. Runx2 and dental development. European Journal of Oral Sciences. 2006;114(5):361–73. doi: 10.1111/j.1600-0722.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu G, Ma S, Zhou Y, Lu Y, Jin L, Wang Z, et al. Signaling Pathways in Dental Stem Cells During Their Maintenance and Differentiation. In: Şahin F, Doğan A, Demirci S, editors. Dental Stem Cells. Cham: Springer International Publishing; 2016. pp. 69–92. [Google Scholar]
- 42.Wu C, Shimo T, Liu M, Pacifici M, Koyama E. Sonic hedgehog functions as a mitogen during bell stage of odontogenesis. Connective Tissue Research. 2003;44(Suppl 1):92–6. [PubMed] [Google Scholar]
- 43.Komatsu K, Mosekilde L, Viidik A, Chiba M. Polarized light microscopic analyses of collagen fibers in the rat incisor periodontal ligament in relation to areas, regions, and ages. The Anatomical Record. 2002;268(4):381–7. doi: 10.1002/ar.10179. [DOI] [PubMed] [Google Scholar]
- 44.Barbeck M, Christiane H, Sader R, Fabian P, Wolf-Dietrich H, Kirkpatrick CJ, et al. Injectable bone substitute based on beta-TCP combined with a hyaluronan-containing hydrogel contributes to regeneration of a critical bone size defect. Journal of Oral Implantology. 2015 doi: 10.1563/aaid-joi-D-14-00203. [DOI] [PubMed] [Google Scholar]
- 45.Hosoya A, Yukita A, Yoshiba K, Yoshiba N, Takahashi M, Nakamura H. Two distinct processes of bone-like tissue formation by dental pulp cells after tooth transplantation. Journal of Histochemistry and Cytochemistry. 2012;60(11):861–73. doi: 10.1369/0022155412459741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White SN, Paine ML, Ngan AY, Miklus VG, Luo W, Wang H, et al. Ectopic expression of dentin sialoprotein during amelogenesis hardens bulk enamel. Journal of Biological Chemistry. 2007;282(8):5340–5. doi: 10.1074/jbc.M604814200. [DOI] [PubMed] [Google Scholar]
- 47.Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. European Journal of Oral Sciences. 1998;106(5):963–70. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 48.Dallas SL, Chen Q, Sivakumar P. Dynamics of assembly and reorganization of extracellular matrix proteins. Current Topics in Developmental Biology. 2006;75:1–24. doi: 10.1016/s0070-2153(06)75001-3. [DOI] [PubMed] [Google Scholar]
- 49.Reseland JE, Reppe S, Larsen AM, Berner HS, Reinholt FP, Gautvik KM, et al. The effect of enamel matrix derivative on gene expression in osteoblasts. European Journal of Oral Sciences. 2006;114(Suppl 1):205–11. doi: 10.1111/j.1600-0722.2006.00333.x. discussion 54–6, 381–2. [DOI] [PubMed] [Google Scholar]
- 50.Fukumoto S, Yamada A, Nonaka K, Yamada Y. Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation. Cells, Tissues, Organs. 2005;181(3–4):189–95. doi: 10.1159/000091380. [DOI] [PubMed] [Google Scholar]
- 51.Ravindranath RM, Devarajan A, Uchida T. Spatiotemporal expression of ameloblastin isoforms during murine tooth development. Journal of Biological Chemistry. 2007;282(50):36370–6. doi: 10.1074/jbc.M704731200. [DOI] [PubMed] [Google Scholar]
- 52.Torres-Quintana MA, Gaete M, Hernandez M, Farias M, Lobos N. Ameloblastin and amelogenin expression in posnatal developing mouse molars. Journal of Oral Science. 2005;47(1):27–34. doi: 10.2334/josnusd.47.27. [DOI] [PubMed] [Google Scholar]
- 53.Bronckers AL, D’Souza RN, Butler WT, Lyaruu DM, van Dijk S, Gay S, et al. Dentin sialoprotein: biosynthesis and developmental appearance in rat tooth germs in comparison with amelogenins, osteocalcin and collagen type-I. Cell and Tissue Research. 1993;272(2):237–47. doi: 10.1007/BF00302729. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, et al. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Developmental Cell. 2015;33(2):125–35. doi: 10.1016/j.devcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.