Abstract
The strict anaerobe Clostridium difficile is the most common cause of nosocomial diarrhea, and the oxygen-resistant spores that it forms have a central role in the infectious cycle. The late stages of sporulation require the mother cell regulatory protein σK. In Bacillus subtilis, the onset of σK activity requires both excision of a prophage-like element (skinBs) inserted in the sigK gene and proteolytical removal of an inhibitory pro-sequence. Importantly, the rearrangement is restricted to the mother cell because the skinBs recombinase is produced specifically in this cell. In C. difficile, σK lacks a pro-sequence but a skinCd element is present. The product of the skinCd gene CD1231 shares similarity with large serine recombinases. We show that CD1231 is necessary for sporulation and skinCd excision. However, contrary to B. subtilis, expression of CD1231 is observed in vegetative cells and in both sporangial compartments. Nevertheless, we show that skinCd excision is under the control of mother cell regulatory proteins σE and SpoIIID. We then demonstrate that σE and SpoIIID control the expression of the skinCd gene CD1234, and that this gene is required for sporulation and skinCd excision. CD1231 and CD1234 appear to interact and both proteins are required for skinCd excision while only CD1231 is necessary for skinCd integration. Thus, CD1234 is a recombination directionality factor that delays and restricts skinCd excision to the terminal mother cell. Finally, while the skinCd element is not essential for sporulation, deletion of skinCd results in premature activity of σK and in spores with altered surface layers. Thus, skinCd excision is a key element controlling the onset of σK activity and the fidelity of spore development.
Author Summary
Clostridium difficile, a major cause of antibiotic-associated diarrhea, produces resistant spores that facilitate its persistence in the environment including hospitals. C. difficile transmission is mediated by contamination of gut by spores. Understanding how this complex developmental process is regulated is fundamental to decipher the C. difficile transmission and pathogenesis. A less tight connection between the forespore and mother cell lines of gene expression is observed in C. difficile compared to Bacillus subtilis especially at the level of the late sigma factor, σK. In C. difficile, the sigK gene is interrupted in most of the strains by a prophage-like intervening sequence, skinCd, which is excised during sporulation. Contrary to B. subtilis, CD1231 encoding the large serine recombinase required for skinCd excision, is constitutively expressed and a recombination directionality factor, whose synthesis is detected only in the mother cell, restricts skinCd excision to this terminal cell. These two proteins are necessary and sufficient to trigger skinCd excision promoting the timely appearance of σK, which in turn switches-on late sporulation events. While several strains of C. difficile lack a skin element, we show that deletion of skinCd results in premature σK activity and in spores with altered surface layers, a property that might be important for host colonization.
Introduction
Endosporulation is an ancient bacterial cell differentiation process allowing the conversion of a vegetative cell into a mature spore through a series of morphological steps [1, 2]. Many bacilli, clostridia and related organisms form bacterial spores. The spores have the ability to withstand extreme physical and chemical conditions and their resistance properties allow them to survive for long periods in a variety of environments. Spores serve as the infectious vehicle for several pathogens such as Bacillus anthracis, Bacillus cereus and Clostridium difficile [3, 4]. C. difficile is the main cause of antibiotic-associated diarrhea. Disruption of the intestinal flora caused by antibiotherapy increases the risk to develop a C. difficile infection. After ingestion, C. difficile spores germinate in the intestine in the presence of specific bile salts [5]. Then, vegetative forms multiply and produce two toxins, TcdA and TcdB, which are the main virulence factors [6]. These toxins cause enterocyte lysis and inflammation leading to diarrhea, colitis, pseudomembranous colitis or more severe symptoms including bowel perforation, sepsis and death. During the infection process, C. difficile also forms spores in the gut that are essential for transmission of this strict anaerobe and contribute to the establishment of reservoirs in the environment including the host and hospital settings [7, 8].
Despite the importance of spores in the infectious cycle, our knowledge of the molecular mechanisms underlying spore development in C. difficile is still scarce. Sporulation has been extensively studied in the model organism Bacillus subtilis [9, 10]. At the onset of sporulation, an asymmetric division forms a forespore and a mother cell. A key developmental transition is when the mother cell finishes engulfing the forespore, which becomes fully surrounded by the mother cell. The mother cell maintains metabolic potential in the forespore and contributes to assembly of the spore protective structures and to the release of mature spores. The developmental program of sporulation is mainly governed by the sequential appearance of four cell type-specific sigma factors: σF in the forespore and σE in the mother cell control early stages of development, prior to engulfment completion, and are replaced by σG and σK following engulfment completion. The main morphological stages of sporulation are conserved among spore-formers, which also share a core of sporulation genes [11, 12]. Nevertheless, recent work has highlighted important differences in the genetic control of sporulation between the aerobic bacilli and the anaerobic clostridia [13–15].
In C. difficile, the main functions and periods of activity of the sporulation σ factors are largely conserved relative to B. subtilis [16–18]. In B. subtilis, several mechanisms including signaling pathways between the two compartments and the architecture of the mother cell- and forespore-specific lines of gene expression, formed by interlocked feed-forward loops (FFLs), converge for the timely activation of the σ factors at specific developmental stages [9, 19]. However, in C. difficile, the communication between the forespore and the mother cell appears less effective, contributing for a weaker connection between morphogenesis and gene expression [16–18]. Indeed, the activation of the σE regulon in the mother cell just after asymmetric division, is rigorously dependent on σF in B. subtilis, but is partially independent of σF in C. difficile. Likewise, the synthesis of the forespore signaling protein SpoIIR, essential for pro-σE processing, is strictly dependent on σF in B. subtilis but partially independent of σF in C. difficile [18]. Furthermore, in B. subtilis, the onset of σG activity coincides with engulfment completion and requires the activity of σE, while in C. difficile σG activity is detected in pre-engulfment sporangia and this early activity is independent of σE [20, 21]. Finally, several levels of regulation ensure that the activity of σK in B. subtilis is restricted to the mother cell following engulfment completion. Firstly, the sigK gene is interrupted by an intervening prophage-like element, skinBs. Secondly, expression of sigK and of spoIVCA encoding a member of the large serine recombinases (LSRs) superfamily [22] responsible for skinBs excision is under the control of σE and requires the transcriptional regulator SpoIIID [19, 23]. Expression of spoIIID is also controlled by σE, but since SpoIIID is auto-regulated [24], a coherent FFL delays expression of the spoIVCA and sigK genes towards the end of engulfment [19, 25]. Moreover, σK activity depends on the cleavage of an inhibitory pro-sequence, a step controlled by σG. Finally, σK directs expression of an anti-sigma factor, CsfB that inhibits σE, thereby promoting transition from σE- to σK-controlled stages in the mother cell [26]. σK is required for assembly of the spore cortex and the more external coat, the main spore surface structures, as well as for mother cell lysis. The segregation of σK activity to post-engulfment sporangia in B. subtilis is explained by multi-level regulation of σK synthesis and activation. Redundancy ensures fail-safe solutions and increases robustness of the developmental process.
In C. difficile, σK is dispensable for cortex biogenesis but is required for the assembly of the spore coat and of the exosporium and for mother cell lysis [17]. sigK is interrupted by a skinCd element, which is excised during sporulation [27]. The skin elements of B. subtilis and C. difficile have different sizes and gene content and are inserted at different sites and in opposite orientation indicating that integration into sigK has occurred independently during evolution [27]. Previous studies have also shown that some transcription of sigK takes place during engulfment [17]. However, σK of C. difficile lacks a pro-sequence [27] and accordingly, σG is dispensable for σK activation [16–18]. In the absence of this cleavage at the end of engulfment, skinCd excision is likely a crucial element in the regulation of σK activity in C. difficile. Importantly, given the role of σK in the assembly of the spore surface layers together with the observation that some strains of C. difficile lack skinCd [27, 28], it is imperative to better understand excision of the element in this organism.
The CD1231 gene, located within skinCd, codes for a protein similar to the SpoIVCA recombinase of skinBs [27, 29]. Surprisingly, σE does not control CD1231 expression in C. difficile [18, 30], which, as we now show, is expressed constitutively. In this work, we studied the role of CD1231 in sporulation and in skinCd excision. We demonstrated that another factor present in the skinCd element, CD1234 encoded by a gene under σE and SpoIIID control is required for skinCd excision but not integration. Thus, CD1234 is a recombination directionality factor that restricts skinCd excision to the mother cell. Importantly, we showed that skinCd is a key element in controlling the onset of σK activity, which in turn is important for proper spore morphogenesis and function.
Results
Role of CD1231 in sporulation
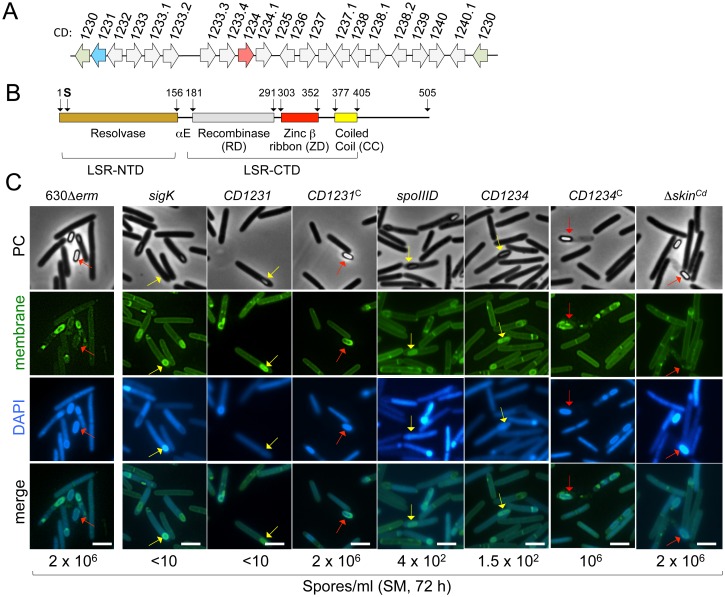
The skinCd element of strain 630Δerm inserted into the sigK gene contains 19 genes (Fig 1A). CD1231, located immediately upstream of the 3´-moiety of sigK (CD1230), is the unique gene within the skinCd element coding for a protein with similarity to large serine recombinases (LSRs) superfamily [22]. The first 300 amino acid residues of CD1231 share 24% identity with SpoIVCA, the recombinase encoded by skinBs [31], and 30% identity over its entire length to the SprA protein, responsible for the excision of the B. subtilis SPβ prophage [32]. CD1231 is also similar to other C. difficile recombinases associated with conjugative transposons. A domain analysis of CD1231 (Fig 1B) identifies the three main structural domains of LSRs and the motifs that connect them: the N-terminal resolvase domain (LSR-NTD) bearing the conserved catalytic nucleophile (Ser at position 10) and additional catalytic residues, followed by a recombinase domain (RD), a zinc β-ribbon domain (ZD) and a coiled coil (CC) motif (Figs 1B and S1). The RD, the ZD and the CC form the C-terminal domain (LSR-CTD); the NTD and CTD are linked by a long α-helix (αE) while a short linker connects the RD to the ZD [22]. In CD1231 as in other LSRs, the CTD is followed by an extension of variable length, which is mostly α-helical (S1 Fig) [22].
Fig 1. The skinCd gene CD1231, coding for a serine recombinase, is required for sporulation.
A: schematic representation of the sigK-skinCd region of the C. difficile 630Δerm chromosome. The two halves of the sigK gene (5‘ and 3’ part of CD1230) are shown in green, and the CD1231 gene, coding for a protein of the large serine recombinase family is shown in blue. The CD1234 gene is shown in pink. B: domain organization of the CD1231 serine recombinase. The horizontal black line is a linear representation of the amino acid sequence. The three conserved domains identified are color-coded: brown, resolvase domain (PF00239, which forms the N-terminal domain (NTD); grey, recombinase domain (RD; PF07508); red, a zinc ribbon domain (ZD; PF13408); yellow, a coiled-coil (CC) motif. The recombinase domain, the zinc finger and the coiled-coil form the C-terminal domain (CTD). A long α-helix linking the NTD and CTD domains is indicated as αE. The catalytic serine, close to the N-terminal end of the protein, is represented. C: cells of the WT strain 630Δerm, the ΔskinCd, spoIIID, sigK, CD1231 and CD1234 mutants and the complemented strains, CDIP533 and CDIP397, carrying multicopy alleles of CD1231 (CD1231C) or CD1234 (CD1234C) expressed under the control of their native promoters were collected after 24 h of growth in liquid SM, stained with the DNA stain DAPI and the membrane dye MTG and examined by phase contrast and fluorescence microscopy. The red arrows point to phase bright spores and the yellow arrows to phase grey spores. Scale bar, 1μm. The titer of heat resistant spores measured for each strain after 72 h of growth in SM is indicated below the panels. The titer of heat resistant spores at 48 h was 0.75 x 106 for strain 630Δerm and for the ΔskinCd mutant.
In B. subtilis, the excision of the skinBs element occurs in the mother cell, and creates an intact sigK gene, essential for sporulation. Since σK is required for sporulation in C. difficile [15], inactivation of CD1231, which is probably involved in skinCd excision, would cause a block in the process. We constructed a CD1231 mutant (CDIP526) using the Clostron system (S2 Fig) as well as a complemented strain (CDIP533) carrying CD1231 under the control of its promoter (pMTL84121-CD1231; see below). We then examined the morphology of the strains by phase contrast and fluorescence microscopy after 24 h of growth in sporulation medium (SM), and we tested the efficiency of heat-resistant spore formation at 72 h. The 630Δerm strain produced 2 x106 heat-resistant CFU/ml and phase bright spores, either free or still inside the mother cell, were seen (Fig 1C). In contrast, less than 10 heat resistant CFU/ ml were detected for the CD1231 mutant. While some phase gray spores were seen in cultures of the mutant at 24 h, free spores were not detected (Fig 1C). Complementation of the CD1231 mutation restored the wild-type phenotype (Fig 1C). Therefore, inactivation of the CD1231 gene severely impaired sporulation. The phenotype caused by the CD1231 mutation phenocopied that imposed by a sigK mutation in that phase gray, heat-sensitive spores were formed that often were seen in a angle relative to the long axis of the cell (Fig 1C) [16, 17]. Moreover, as found for a sigK mutant, formation of the phase gray spores was not accompanied by loss of viability of the mother cell, as is the case for the wild-type strain [17]. These observations strongly suggest that σK is inactive in this mutant.
Constitutive expression of the CD1231 gene
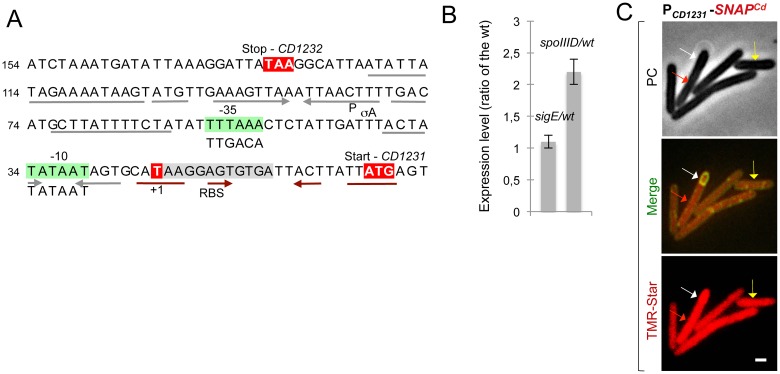
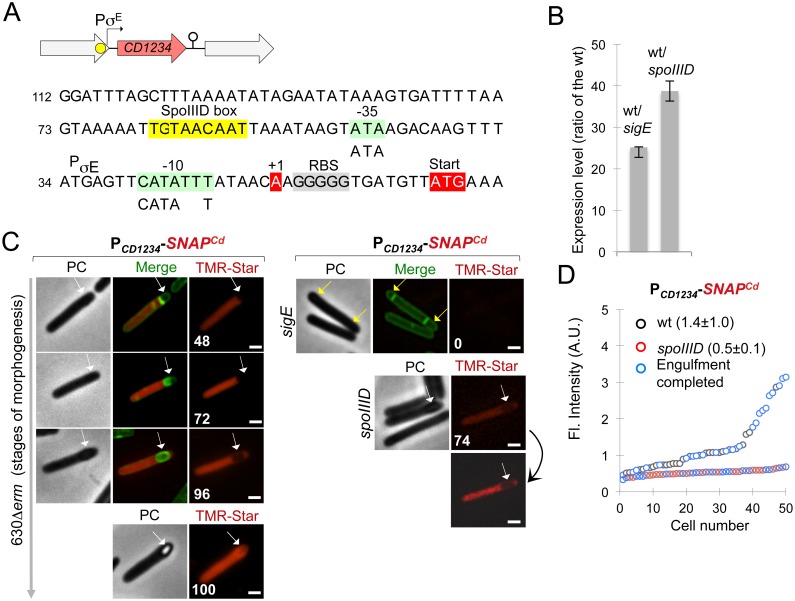
Expression of the spoIVCA gene in B. subtilis is under the dual control of the mother cell proteins σE and SpoIIID leading to the restriction of skinBs excision to this compartment [19, 33]. However, previous transcriptome studies suggested that σE or SpoIIID does not control CD1231 expression in C. difficile [18, 30]. Moreover, qRT-PCR using RNA extracted from SM cultures of the strain 630Δerm, a sigE mutant and a spoIIID mutant did not show variations in the level of CD1231 expression in the sigE mutant and only a slight increase in the spoIIID mutant relative to the wild-type strain (Fig 2B). In our genome-wide mapping of promoters in strain 630Δerm [34], a transcriptional start site (TSS) was found 21 bp upstream of the CD1231 start codon and -35 (TTTAAA) and -10 (TATAAT) sequences for σA-dependent promoters are present upstream of this TSS while no consensus for σE is found (Fig 2A). This suggests that expression of CD1231 is under the control of σA and therefore probably not confined to the mother cell. To test this possibility, we constructed a PCD1231-SNAPCd fusion and this fusion was transferred to the 630Δerm strain. Samples of cultures expressing PCD1231- SNAPCd were collected at 24 h of growth in SM, and the cells doubly labeled with the SNAP substrate TMR-Star and the membrane dye MTG. Expression of PCD1231- SNAPCd was detected in 93% of the vegetative cells scored, consistent with the presence of a σA-type promoter. However, expression of PCD1231-SNAPCd was also detected in sporulating cells in both the forespore and the mother cell (78% of the sporangia) (Fig 2C). Thus, in agreement with the absence of a requirement for σE and SpoIIID for its expression as determined by qRT-PCR, CD1231 is not a mother cell-specific gene.
Fig 2. Constitutive expression of the skinCd gene CD1231.
A: promoter region of the CD1231 gene. The mapped transcriptional start sites (+1, red) [34] and the -10 and -35 promoter elements (green boxes) that match the consensus for σA recognition are indicated. Also represented are the stop codon of CD1232 and the start codon of CD1231 (red). Inverted repeats upstream (grey arrows) and downstream (brown) of the +1 position are indicated as well as a possible RBS overlapping the left arm of these repeats. B: qRT-PCR analysis of CD1231 transcription in strain 630Δerm, and in sigE or spoIIID mutant. RNA was extracted from cells collected 14 h (sigE mutant) or 15 h (spoIIID mutant) after inoculation in liquid SM. Expression is represented as the fold ratio between the indicated mutants and the wild-type (WT). Values are the average ± SD of two independent experiments. C: cells of the C. difficile 630Δerm strain carrying a PCD1231-SNAPCd transcriptional fusion were collected after 24 h of growth in liquid SM, stained with TMR-Star and the membrane dye MTG, and examined by phase contrast (PC) and fluorescence microscopy. The merged images show the overlap between the TMR-Star (red) and MTG (green) channels. The yellow arrow shows a vegetative cell expressing PCD1231-SNAPCd, the white arrow shows expression in the forespore and the red arrow expression in the mother cell. Scale bar, 1 μm.
Control of sigK transcription and σK activity by SpoIIID
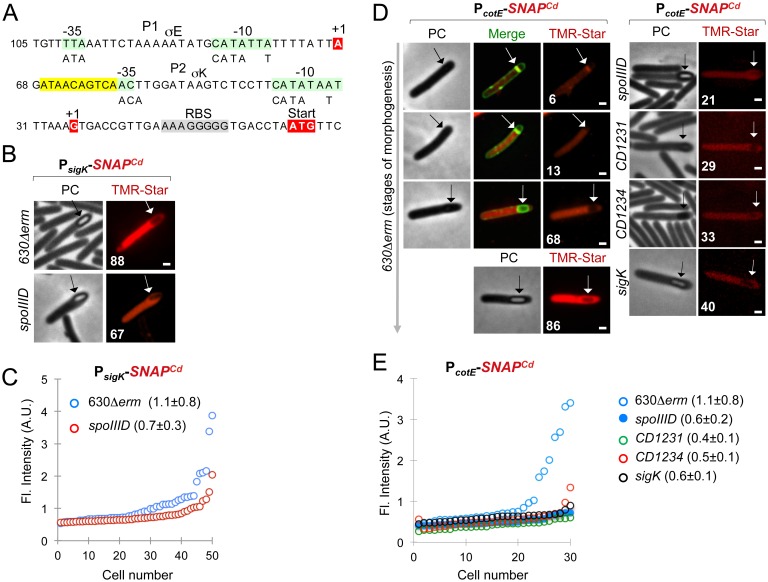
Previous work indicated that the skinCd element is excised from the chromosome only during sporulation [27]. This suggests that a factor is required in addition to CD1231 to trigger excision during sporulation. In a first step to search for this factor, we wanted to establish the requirements for sigK transcription and σK activity. Previous work has shown that SpoIIID is required for sporulation and for the transcription of the sigK gene in C. difficile [16, 18, 30]. Importantly, expression of a skinCd-less version of the sigK gene from a SpoIIID-independent promoter largely bypasses the requirement for SpoIIID for sporulation [30]. While showing that a critical function of SpoIIID in sporulation is to ensure efficient sigK expression, this result does not discard a possible role of SpoIIID in skinCd excision. Here, we examined the effect of a spoIIID mutation on sigK transcription using a PsigK-SNAPCd fusion and on the activity of σK using a fusion of the σK-controlled PcotE to SNAPCd [17]. Two TSSs have been mapped in the sigK promoter region (Fig 3A) [18]. The upstream promoter (P1) matches the consensus for σE recognition, whereas the downstream promoter (P2) matches the consensus for σK recognition [18]. Using the consensus of SpoIIID of B. subtilis [19], a possible SpoIIID binding site is found upstream of P2 and downstream of P1 (Fig 3A). Transcription of sigK was detected in the mother cell of the wild-type strain soon after septation and during engulfment [17], but increased following engulfment completion, when it was detected in 88% of the sporangia (Fig 3B). In contrast, transcription of sigK was detected in only 67% of the spoIIID mutant sporangia in which engulfment was completed (Fig 3B). Moreover, quantification of the fluorescence signal from PsigK-SNAPCd in those cells revealed a reduction of the signal per sporangia, from 1.1±0.8 arbitrary units (A.U.) in the wild-type strain, to 0.7±0.3 A.U. in the spoIIID mutant (Fig 3C). The arrangement of the sigK promoter region suggests that the low level of transcription during engulfment may arise from P1 whereas the main period of sigK transcription could involve utilization of P2 possibly by σE first, then by σK, with the assistance of SpoIIID. P2 may also be involved in the late, positive auto-regulation of σK in cells carrying phase bright spores [17, 18] (see also the Discussion). Thus, following engulfment completion, transcription of sigK is reduced, but not abolished, in the absence of SpoIIID both in terms of the number of cells in which transcription is activated and, although a less pronounced effect, in the level of expression per cell. The reduction in sigK transcription in the spoIIID mutant is in line with earlier results [18, 30].
Fig 3. Control of sigK expression and of σK activity.
A: promoter region of the sigK gene. The transcriptional start sites (+1, red) as previously mapped [34], the -10 and -35 promoter elements (green) that match the consensus for σE or σK recognition (represented below the sequence), the SpoIIID box (yellow), and the start codon of the sigK gene, are indicated. B: microscopy analysis of C. difficile cells carrying a PsigK-SNAPCd fusion in strain 630Δerm and in the spoIIID mutant. The cells collected after 24 h of growth in SM were stained with TMR-Star and examined by phase contrast (PC) and fluorescence microscopy. The morphological stage at which sigK transcription reaches its maximum, concomitant with the appearance with phase gray spores, is illustrated. The position of the forespore is clearly seen in the PC images. The numbers represent the percentage of cells at the indicated stage that show expression of the reporter fusion. C: quantitative analysis of the fluorescence intensity (Fl., in arbitrary units, A.U.) in sporulating cells (as in B) expressing PsigK- SNAPCd. The numbers in the legend represent the average fluorescence intensity ± SD (a minimum of 50 cells were scored). D: microscopy analysis of cells carrying fusions of the σK-dependent cotE promoter to SNAPCd in strain 630Δerm and in the spoIIID, CD1231, CD1234 and sigK mutants. Cells were collected and processed for imaging as indicated in panel B. However, MTG staining was used to visualize the forespore membranes and the stage of sporulation during engulfment; the merged images show the overlap between TMR-star (red) and MTG (green). Note that merged images (MTG/TMR-Star) are not shown whenever the position of the forespore is seen in the PC images. The panels are representative of the expression patterns observed for different sporulation stages, ordered from early to late. For the mutant strains, the morphological stage characteristic of each mutant is shown. The numbers refer to the percentage of cells at the represented stage showing SNAPCd fluorescence. E: quantitative analysis of the fluorescence (Fl.) intensity in the various strains expressing PcotE-SNAPCd. The numbers in the panels are the average fluorescence intensity ± SD (30 cells were analyzed). In B and D, the arrows show the position of developing spores. Scale bar, 1 μm.
As a measure of σK activity, PcotE-driven production of SNAPCd was detected in the mother cell in 6–13% of the wild-type sporangia during engulfment but increased to 68% just after engulfment completion and was seen in 86% of the sporangia when phase bright spores became visible (Fig 3D). Expression of PcotE-SNAPCd was detected in only 40% of the sigK mutant sporangia that reached late stages of morphogenesis to form partially refractile spores, and the average fluorescence signal per sporangia decreased to 0.6±0.1 A.U. (Fig 3E). Importantly, disruption of spoIIID or CD1231 reduced expression of PcotE-SNAPCd to only 21% and 29% of the sporangia that reached late stages of morphogenesis. Moreover, the average fluorescence intensity per cell was of 0.6±0.2 A.U. and 0.4±0.1 A.U. in the spoIIID or CD1231 mutant, respectively as compared to 1.1±0.8 A.U. for the wild-type strain (Fig 3D and 3E). Thus, disruption of sigK, spoIIID, or CD1231 reduced expression of the PcotE-SNAPCd fusion approximately to the same extent. In any event, the increase in σK activity following engulfment completion is not seen in CD1231 and spoIIID mutants, compatible with a role for CD1231 and SpoIIID in the control of σK activity. This strongly suggests that SpoIIID may play a role in skinCd excision.
The cell type-specific excision of skinCd requires SpoIIID
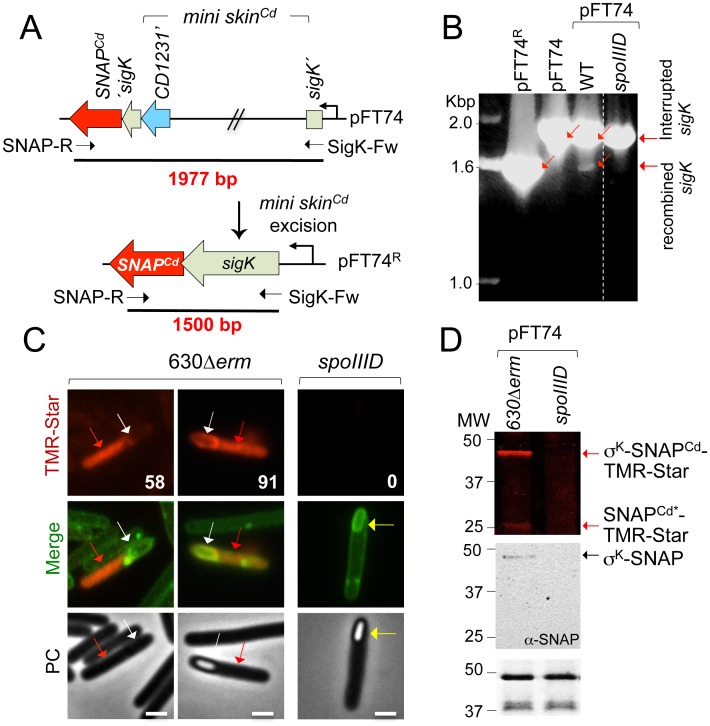
To examine the time and requirements of skinCd excision, we devised an assay to monitor reconstitution of a functional sigK gene in C. difficile. We previously described a plasmid, pFT38, carrying a sigK gene disrupted by a mini-skinCd element bearing a deletion of all the skinCd genes except CD1231 [17]. We modified this plasmid in order to create a translational fusion between the C-terminal moiety of σK and SNAPCd and to remove the 5´-end of CD1231 (i.e., only the chromosomal CD1231 is functional) (Fig 4A). This plasmid, pFT74, was introduced in strain 630Δerm and in the spoIIID mutant. Excision through recombination involving sequences at the ends of the mini-skinCd element reconstitutes the sigK gene (Fig 4A). The recombined sigK in pFT74 named pFT74R was first detected by PCR using an oligonucleotide hybridizing to the 5’ moiety of the sigK gene and a second in the SNAPCd gene: a fragment of 1500 bp is expected upon mini-skinCd excision instead of 1977 bp for pFT74 (Fig 4A). The 1500 bp PCR fragment was detected in strain 630Δerm but not in the spoIIID mutant (Fig 4B) indicating that SpoIIID is necessary for mini-skinCd excision. This is also the case for the chromosomal copy of the skinCd element as described below.
Fig 4. Time of excision of a mini-skinCd element as detected by production of a σK-SNAPCd fusion protein.
A: schematic representation of plasmid pFT74, containing the SNAPCd reporter fused in frame to the 3´-end of sigK. The sigK gene is interrupted by a mini-skinCd element carrying the 3´end of the CD1231 gene (blue). When skinCd excision occurs to form pFT47R, σK-SNAPCd is produced. The size of the inserts before and after recombination is indicated. The primers used for PCR analysis are indicated. B: analysis of skinCd excision by PCR using the primer pair indicated in panel A and DNA extracted from cultures of the strain 630Δerm, and the spoIIID mutant after 24 h of growth in SM. The red arrows point to the recombined sigK and the interrupted gene. pFT74 and pFT74R were used as controls for non-recombined and recombined sigK, respectively. C: fluorescence microscopy analysis of sporulating cells producing σK-SNAPCd in the strain 630Δerm and in the spoIIID mutant. Cells grown in SM were collected at 24 h and stained with the membrane dye MTG and TMR-Star. The red arrows point to the mother cell, and the white arrow to the developing spore. The yellow arrow shows the position of a spore in a spoIIID sporangium. The numbers refer to the percentage of cells at the represented stage showing production of the fusion. Scale bar, 1 μm. D: accumulation of σK-SNAPCd in extracts produced from sporulating cells of the WT and spoIIID mutant. The cells were collected from SM cultures 24 h after inoculation, labeled with the TMR-Star substrate. Proteins in whole cell lysates were resolved by SDS-PAGE, visualized by fluoroimaging or subject to immunoblot analysis with anti-SNAP antibodies. A section of the corresponding Coomassie-stained gel is shown as a loading control. The arrows indicate the position of the TMR-Star-labeled σK-SNAPCd fusion (expected size 47 kDa). The asterisk points to a possible degradation product of σK-SNAPCd (SNAPCd*) of about 25 kDa. The position of molecular mass markers (in kDa) is shown on the left side of the panels.
To gain further insight into the time of excision relative to the course of spore morphogenesis, we monitored production of the σK-SNAPCd translational fusion formed after mini-skinCd excision at the single cell level by fluorescence microscopy (Fig 4C). Production of σK-SNAPCd was detected only in the mother cell in 58% of the wild-type sporangia in which spores were not yet discernible in the mother cell by phase contrast microscopy but that were close to or just after engulfment completion as judged from the MTG staining pattern (membranes almost fused or fused) (Fig 4C). However, σK-SNAPCd was detected in 91% of the sporangia in which partially phase bright or phase bright spores were visible by phase contrast microscopy (Fig 4C). This parallels the pattern of sigK transcription and σK activity [17] and suggests that σK is active as soon as it is produced after skinCd excision. In contrast, no accumulation of σK-SNAPCd was detected in the spoIIID mutant (Fig 4C) even if the sigK gene remains expressed in this mutant (Fig 3A). σK-SNAPCd (47 kDa) was detected by Western blotting using anti-SNAP antibodies and by fluorimaging in crude extracts of strain 630Δerm but not in extracts prepared from the spoIIID mutant (Fig 4D). These results strongly suggest that SpoIIID is required for skinCd excision in C. difficile as also observed for B. subtilis. Moreover, since the main period of sigK transcription, σK accumulation and σK activity appear to coincide during the course of morphogenesis, skinCd excision and sigK transcription seem to concur to delay the onset of σK activity.
CD1234 is a mother cell-specific skinCd gene
Given that skinCd excision did not take place in vegetative cells or in the forespore in spite of CD1231 expression in these cells and the suspected role of SpoIIID in controlling σK activity via skinCd excision, we inferred that a factor probably encoded within skinCd and produced under the joint control of σE and SpoIIID could modulate CD1231 synthesis and activity. Among the 19 skinCd genes, only CD1234 (Fig 5A) was down-regulated in a sigE and in a spoIIID mutant in transcriptome analyses [18, 30]. CD1234 codes for a small protein of 72 amino acids, with a predicted pI of 5.5 and no significant similarity to proteins found in databases. We confirmed by qRT-PCR using RNA extracted from SM cultures that CD1234 expression decreased 25-fold and 40-fold in a sigE and in a spoIIID mutant, respectively as compared to the wild-type strain (Fig 5B). We mapped a TSS 14 bp upstream of the start codon of CD1234, and a consensus sequence for σE recognition was detected upstream of this TSS [18]. Using the SpoIIID consensus sequence of B. subtilis [19], we also identified a putative SpoIIID binding motif (TGTAACAAT) centered 46 bp upstream of the CD1234 TSS (Fig 5A) in agreement with the positive control of CD1234 expression by SpoIIID.
Fig 5. CD1234 is a mother cell-specific gene.
A: the panel represents the region of the CD1234 gene within the skinCd element (top) and the sequence of its promoter region (bottom). The transcriptional start site (+1, red), the -10 and -35 promoter elements (green) that match the consensus for the σE binding (represented below the sequence), and a putative SpoIIID binding site (yellow) are represented. B: qRT-PCR analysis of CD1234 transcription in strain 630Δerm (WT), and in sigE or spoIIID mutant. RNA was extracted from cells collected 14 h (sigE mutant) and 15 h (spoIIID mutant) after inoculation in liquid SM. Expression is represented as the fold ratio between the WT strain and the indicated mutants. Values are the average ± SD of two independent experiments. C: microscopy analysis of cells of the 630Δerm strain and of the sigE and spoIIID mutants carrying a PCD1234-SNAPCd transcriptional fusion. The cells were collected 24 h after inoculation in liquid SM, stained with TMR-Star and the membrane dye MTG, and examined by phase contrast (PC) and fluorescence microscopy. Merged images (MTG/TMR-Star) are not shown whenever the position of the forespore is clearly seen in the phase contrast images. The panels are representative of the expression patterns observed for different stages of sporulation, ordered from early to late. For the mutant strains, the morphological stage characteristic of each mutant is shown. The numbers in the panels refer to the percentage of cells at the represented stage showing SNAPCd fluorescence. The white arrows show the position of the developing spore in WT or spoIIID sporangia, and the yellow arrows the two-forespore compartments characteristic of a sigE mutant (disporic phenotype). Note that the TMR-Star panel for the spoIIID mutant is shown repeated with enhanced contrast (the two panels linked by a curved arrow) so that the lack of signal in the forespore is clearly seen. Scale bar, 1 μm. D: quantitative analysis of the fluorescence intensity in sporulating cells of the strains shown in C. The numbers in the legend represent the average ± SD of fluorescence intensity (50 cells were scored in each case) in the WT or spoIIID mutant. No difference was observed for sporangia before (black symbol in the curve for the WT and red symbols in the curve for the spoIIID mutant) or after engulfment completion (blue symbols in both curves).
To examine the compartment and time of CD1234 expression during sporulation, we constructed a PCD1234-SNAPCd transcriptional fusion. This fusion was transferred by conjugation into the 630Δerm strain and the sigE or spoIIID mutant. In the wild-type strain, SNAP production confined to the mother cell, was detected in 48% and 72% of the cells just after asymmetric division and during engulfment, respectively and persisted until late stages in development, when phase bright spores were seen (Fig 5C). SNAPCd production was eliminated by mutation of sigE (Fig 5C) while the average intensity of the SNAPCd-TMR signal decreased from 1.4±1 in the 630Δerm strain to 0.5±0.1 in a spoIIID mutant (Fig 5D). Together, these results indicate that CD1234 is a mother cell-specific gene expressed under the joint control of σE and SpoIIID.
The CD1234 gene is required for sporulation
To investigate the role of CD1234 in sporulation, we constructed a CD1234 mutant, CDIP396, using the Clostron system (S2 Fig). A complemented strain, CDIP397, carrying the CD1234 gene expressed under the control of its native promoter was also constructed. We examined the morphology of the CD1234 mutant using phase contrast and fluorescence microscopy. Some phase bright or partially phase bright spores were present in cultures of the CD1234 mutant but free spores were not detected. The mutant formed only 1.5 x 102 heat-resistant spores/ml of culture at 72 h of growth in SM (i.e. 104 less than the wild-type) (Fig 1C). Importantly, the wild-type phenotype was restored in the complemented strain (Fig 1C). Thus, inactivation of the CD1234 gene strongly impaired sporulation. Nevertheless, the CD1234 mutant like the spoIIID mutant was not as severely affected in sporulation as the sigE and sigK mutants [16, 17] or CD1231 mutant (Fig 1C).
CD1231 and CD1234 are required for σK activity
Since both CD1231 and CD1234 are likely required for skinCd excision, a prerequisite for σK activation, we tested the impact of CD1231 and CD1234 inactivation on the expression of σK or σE targets using qRT-PCR (Table 1). We extracted RNA from strain 630Δerm, the CD1231 and CD1234 mutants and the complemented strains after 24 h of growth in SM, a time where σK target genes are highly expressed [18]. As expected, expression of CD1231 and CD1234 was strongly reduced in the CD1231 and CD1234 mutants, respectively (Table 1). The expression of three σE targets (spoIIIAA, spoIVA, spoIIID) was not significantly altered in the CD1231 and CD1234 mutants as compared to the wild-type strain. In sharp contrast, expression of six σK target genes strongly decreased in the CD1231 or CD1234 mutant compared to the wild-type strain (Table 1), as observed previously for a sigK mutant [18]. CD1231 and CD1234 were about 10-fold more expressed in the CD1231 (pMTL84121-CD1231) and CD1234 (pMTL84121-CD1234) strains than in 630Δerm (Table 1), and expression of the σK target genes was fully or partially restored in these strains. Accordingly, expression of a PcotE-SNAPCd fusion decreased in a CD1231 mutant compared to strain 630Δerm as described above and was reduced in the CD1234 mutant. Only 33% of the sporangia that reached late stages of development expressed the fusion when CD1234 is inactivated compared to 86% for the wild-type strain (Fig 3D). Moreover, the average intensity of the fluorescence signal decreased from 1.1±0.8 A.U. (WT) to 0.5±0.1 (CD1234), similar to the intensity seen for the CD1231 mutant (Fig 3E). In conclusion, expression of σK-dependent but not of σE-dependent genes requires CD1231 and CD1234 as expected for proteins involved in sigK reconstruction through skinCd excision.
Table 1. Effect of CD1231 or CD1234 inactivation on the expression of σK or σE targets.
| Fold change | ||||
|---|---|---|---|---|
| Gene | 630Δerm/ CD1231 | 630Δerm/ CD1231C | 630Δerm/ CD1234 | 630Δerm/ CD1234 C |
| CD1231 | 11.5 +/-1.5 | 0.1 +/-0.01 | 1.24 +/-0.02 | 1.3 +/-0.03 |
| σE targets | ||||
| spoIIIAA | 1.3 +/-0.3 | ND | 0.85 +/- 0.25 | ND |
| spoIVA | 2.9 +/-1 | ND | 1.7 +/- 0.1 | ND |
| spoIIID | 2.45 +/-0.5 | ND | 1.3 +/- 0.7 | ND |
| sigK | 167 +/-12 | 2.55 +/- 0.1 | 206 +/-32 | 1.9 +/- 0.1 |
| CD1234 | 1.85 +/- 0.05 | 2 +/- 0.1 | 545 +/-140 | 0.11 +/-0.01 |
| σK targets | ||||
| sleC | 95 +/- 19 | 4.75 +/-1.25 | 86 +/- 20 | 3.75 +/- 0.25 |
| cotBC | 2045 +/- 49 | 9 +/- 1 | 1650 +/-430 | 1 +/- 0.4 |
| bclA1 | 88 +/- 4.5 | 2.6 +/- 1.3 | 53 +/- 6 | 2.8 +/- 1.1 |
| cotE | 149 +/-5 | 8.5 +/- 3 | 89 +/- 8 | 6 +/-0.2 |
| CD1133 | 31 +/- 2 | 2.7 +/- 0.8 | 30 +/-1 | 2.2 +/-0.8 |
| CD3580 | 70 +/-15 | 2.9 +/- 0.9 | 50 +/-5 | 3 +/- 0.5 |
Total RNAs were extracted from C. difficile 630Δerm strain, the CD1231 and CD1234 mutants, and the complementation strains CD1231C (CD1231 mutant with pMTL84121-CD1231) and CD1234C (CD1234 mutant with pMTL84121-CD1234) after 24 h of growth in SM medium. After reverse transcription, specific cDNAs were quantified by qRT-PCR using the DNApolIII gene for normalization. The results presented correspond to the mean of at least two independent experiments.
skinCd excision during growth and sporulation requires both CD1231 and CD1234
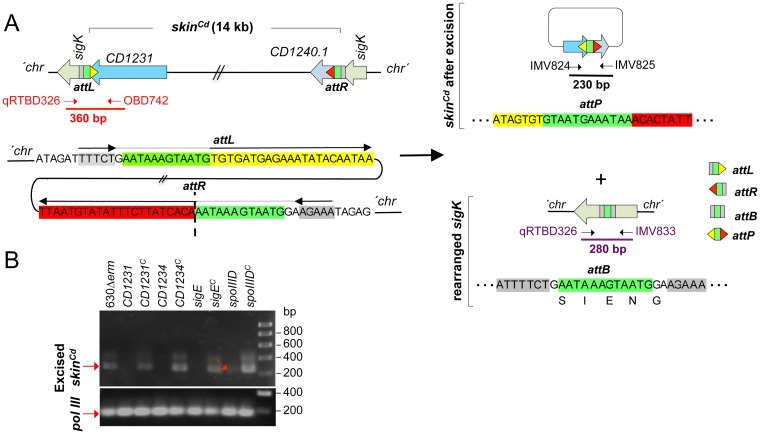
The temporal control of skinCd excision and its confinement to the terminal mother cell may thus require the σE- and SpoIIID-controlled gene, CD1234. We tested excision of the chromosomal skinCd in strain 630Δerm and in the CD1231, CD1234, sigE and spoIIID mutants (Fig 6). In B. subtilis, skinBs excision occurs within two 5 bp inverted repeats that flank an imperfect 21 bp repeat [35]. In C. difficile, excision is expected to occur by means of a recombination event involving attL and attR (by analogy with the sequences involved in phage excision) at the left and right ends of skinCd (Fig 6A) [27]. As in B. subtilis, attL and attR consist of two half-sites formed by a 5 bp inverted repeat external to a longer 22 bp imperfect inverted repeat (Fig 6A) and of two conserved 12 bp motifs, one in attL and one in attR, within which recombination take place (in green in Fig 6A). The intervening DNA is excised as a circular molecule carrying attP (by analogy with sequences responsible for phage integration), leaving behind a chromosomal attB site (analogous to prophage insertion sequences in the bacterial chromosome) (Fig 6A).
Fig 6. Requirements for skinCd excision in C. difficile.
A: schematic representation of skinCd excision from the 630Δerm chromosome and of the recombination products derived from the process. The attL (the half-sites are represented by the green box and the yellow triangle) and attR (the green box and the red triangle) in the chromosome, attP in the excised skinCd (half-sites correspond to yellow and red triangles) and attB in the chromosome (green and grey boxes) are represented. The horizontal arrows represent inverted repeats. The vertical dashed line represents the point of junction of the sequenced following recombination. The oligonucleotides used to amplify the 5’ junction of skinCd (OBD742-qRTBD326), the reconstructed sigK gene (qRTBD326-IMV833) and the circularized skinCd (IMV824-IMV825) are indicated, as well as the expected size of the respective products. B: detection of skinCd excision in different strains by PCR using primers IMV824 and IMV825 and DNA extracted from the indicated strains grown in liquid SM medium for 24 h. CD1231C denotes the CD1231 mutant complemented with pMTL84121-CD1231 and CD1234C the CD1234 mutant complemented with pMTL84121-CD1234. sigEC and spoIIIDC correspond to the sigE or spoIIID mutant complemented with pMTL84121-sigE or pMTL84121-spoIIID, respectively.
The excised circular element obtained after excision of skinCd can be monitored by PCR using primers annealing upstream and downstream of attP (Fig 6A). During sporulation, skinCd excision was detected in strain 630Δerm (Fig 6B, lane 1), but not in the CD1231 (lane 2), CD1234 (lane 4) and sigE (lane 6) mutants. A very faint band corresponding to the excised skinCd was detected in the spoIIID mutant (lane 8, red arrow and red dot). Plasmids bearing the disrupted chromosomal genes restored skinCd excision to all mutants (Fig 6B, lane 3, 5, 7, 9). Thus, skinCd excision during sporulation requires both CD1231 and CD1234. Moreover, the results confirm the key role of SpoIIID and σE in skin excision likely through their control of the cell type-specific production of CD1234.
To test whether expression of CD1234 could result in skinCd excision during vegetative growth, we constructed a plasmid carrying CD1234 under the control of an ATc-inducible promoter (pDIA6103-PtetCD1234). This plasmid or the empty vector pDIA6103 were introduced into strain 630Δerm and the sigE, spoIIID, CD1231 and CD1234 mutants. The resulting strains were grown for 4 h in TY medium. Following induction of CD1234 expression, the cells were either plated onto BHI or harvested for DNA extraction. qPCR was then performed with 2 primer pairs, one corresponding to DNApolIII as a control and the second to sigK on both sides of attB for the detection of skinCd excision. The ΔCt (CtsigK-CtpolIII) was determined for each strain carrying either pDIA6103 or pDIA6103-PtetCD1234. The ΔCt was >10 for all strains containing pDIA6103. Interestingly, the ΔCt was reduced to < 1 for all strains containing pDIA6103-Ptet-CD1234 with the exception of the CD1231 mutant where a ΔCt of 18 was observed, as was also the case for the strain CD1231 (pDIA6103) (Table 2). In parallel, chromosomal DNA was extracted from 8 independent clones obtained after seeding BHI plates with samples from the cultures of the different strains carrying pDIA6103-PtetCD1234. For each clone, we tested the presence of skinCd in the chromosome by PCR amplification of the 5’ junction of the skin (attL) using one oligonucleotide located in the 3’ part of CD1231 and the second in sigK (Fig 6A). While the skinCd/chromosome junction was detected in only 1 out of 8 clones tested for strains 630Δerm, sigE, spoIIID or CD1234, this junction was amplified for the 8 clones of the CD1231 mutant (Table 2). This confirmed that skinCd was excised after induction of CD1234 expression during growth of the wild-type strain and of the sigE, spoIIID and CD1234 mutants. In contrast, skinCd remained integrated in the CD1231 mutant under similar conditions.
Table 2. Excision of skinCd during growth in strains producing CD1234 under Ptet control.
| ΔCt = CtsigK-CtpolIII | Detection of skinCd junction by PCR | ||
|---|---|---|---|
| Strain | pDIA6103 | pDIA6103-Ptet-CD1234 | |
| 630Δerm | 10.9+/-0.5 | 0.8+/-0.4 | 0/8 |
| sigE::erm | 17.9+/-0.5 | 0.6+/-0.5 | 1/8 |
| spoIIID::erm | 16.3+/-0.15 | 0.85+/-0.35 | 1/8 |
| CD1231::erm | 18.3+/-0.25 | 18+/-1 | 8/8 |
| CD1234::erm | 17.9+/-0.25 | 0.65+/-0.35 | 1/8 |
Strains 630Δerm, sigE::erm, spoIIID::erm, CD1234::erm and CD1231::erm containing either pDIA6103 or pDIA6103-Ptet-CD1234 were grown in TY medium for 4 h at which time ATc (100 ng/ml) was added. After 2 h of induction, cells were serially diluted and plated on BHI or collected and DNA extracted. qPCR was performed on with 2 primer pairs: one corresponding to DNApolIII as a control and the second to sigK on both sides of the skin insertion site (qRTBD325-qRTBD326). Chromosomal DNA extracted from 8 independent clones obtained after plating the cultures on BHI was used to amplify the 5’ junction of the skin using one primer in sigK (qRTBD326) and one in CD1231 (OBD742) (See Fig 6A). The number of positive clones among the eight tested is indicated.
In conclusion, these results indicate that during C. difficile growth: i) excision occurs if and only if CD1234 is produced; ii) under these conditions skinCd excision is independent of σE and SpoIIID; and iii) excision is absolutely dependent on the skinCd CD1231 recombinase, even if CD1234 is induced. Together, these results indicate that both CD1231 and CD1234 are necessary for skinCd excision in C. difficile.
Evidence that CD1231 and CD1234 directly interact
Since CD1231 was not specifically transcribed during sporulation and its expression was not altered in a CD1234 background (Table 1), we reasoned that CD1234 could post-transcriptionally control the synthesis or activity of CD1231. The integration reaction catalyzed by the LSRs is unidirectional, in that excision often requires an additional recombination directionality factor (RDF) that modulates the LSR activity by direct protein-protein interactions [22, 36, 37]. We therefore tested whether CD1231 and CD1234 could interact using pull-down assays. Whole cell extracts prepared from E. coli BL21(DE3) strains producing separately CD1234-His6 and CD1231-Strep or co-producing the two proteins under the control of PT7lac were prepared. None of the proteins was detected by Coomassie staining, but they were detected by immunoblotting with antibodies to their C-terminal tags (S4A and S4B Fig). The extracts were then incubated with Ni2+-NTA agarose beads, and following washing and elution, the bound proteins were identified by immunoblotting. While a protein of about 30 kDa recognized by the Strep-tag antibody seems to bind non-specifically to the beads, the full length CD1231-Strep as well as two probable degradation products, of about 37 and 40 kDa, were only detected in the presence of CD1234-His6 (S4B Fig). The two likely degradation fragments of CD1231-Strep may contain the CTD domain followed by the C-terminal extension (residues 405–505) consistent with the existence of a protease-sensitive site just downstream of the NTD in the recombinases from phages C31 and Bxb1 and from transposon TnpX [22, 38] (S4D Fig). These fragments may be retained by the Ni2+ column because they bind to the full-length CD1231-Strep recombinase or to CD1234-His6 (S4D Fig). In a different set of experiments, full-length CD1231-Strep was retained by the beads when these were pre-incubated with extracts prepared from BL21(DE3) cells producing CD1234-His6 and not Tgl-His6, an unrelated spore-associated protein from B. subtilis [39, 40], which accumulated to much higher levels than CD1234-His6 (S4C Fig). Together, these results indicate that CD1234 and CD1231 were part of a complex that formed in E. coli and suggest that CD1234 might control the activity of CD1231 by direct interaction.
CD1231 and CD1234 are necessary and sufficient for skinCd excision in E. coli
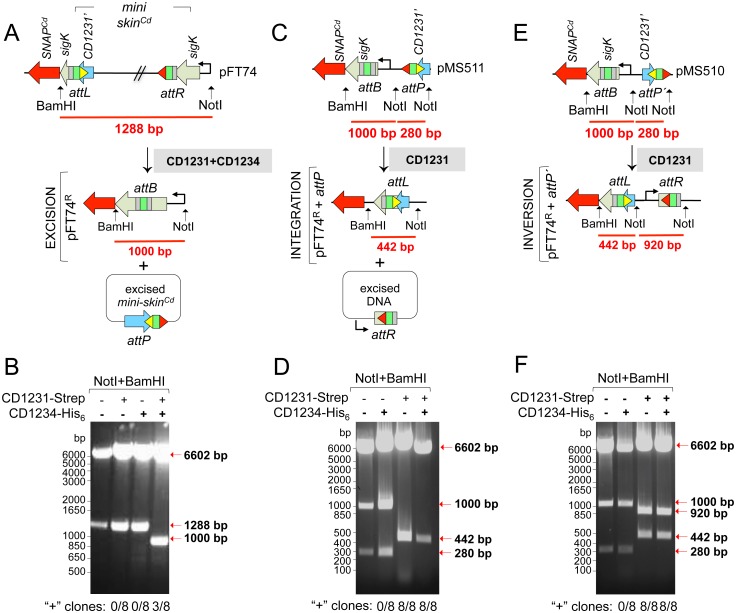
To test whether CD1234 and CD1231 were sufficient for skinCd excision, we used a heterologous host E. coli. The plasmid pFT74 carrying a mini skinCd element integrated into sigK was introduced in E. coli carrying plasmids for expression of CD1231, CD1234 or the co-expression of both genes. The cells were first induced to produce CD1231, CD1234 or both. The plasmid pFT74 was then purified and examined for the recombination reaction by digestion with BamHI and NotI (Fig 7A). Digestion of the resulting recombined plasmid obtained after mini skinCd excision, termed pFT74R, with BamHI and NotI should produce a fragment of 1000 bp as compared to a fragment of 1288 bp for the parental plasmid carrying the mini-skin (pFT74). A fragment of 1000 bp was not isolated from cells producing neither CD1234 alone nor CD1231 alone or a mutant allele of CD1231 in which the putative catalytic serine in the NTD was changed to an alanine (S10A) (Figs 1B, 7B and S5). By contrast, pFT74R was detected in cells in which both proteins were produced (Fig 7B). Together, these results show that CD1231 requires CD1234 as an auxiliary factor for the recombination reaction between attL and attR that results in skinCd excision.
Fig 7. Detection of recombinase activity in E. coli.
E. coli cells were transformed with plasmids pFT74 (A) or pFT74R-attP carrying a skinCd-less sigK gene and attP in both possible orientations (C and E). The resulting strains were then transformed with plasmids containing the CD1231-Strep, CD1234-His6 or both genes (“+” signs) under the control of an IPTG inducible promoter. A, C and E: schematic representation of the recombination event (skinCd excision in A, skinCd integration in C and DNA inversion in E), the expected recombination products and the size of the intact and recombined inserts between the BamHI and NotI sites. attP´denotes an inverted attP site. B, D and F: analysis of the recombinant events following pFT74 or pFT74R-attP recovery from E. coli strains producing CD1231-Strep, CD1234-His6 or both genes (“+” signs) and digestion with BamHI and NotI. pFT74 and pFT74R+attP from strains that did not produce CD1231-Strep or CD1234-His6 were used as controls for non-recombined and recombined sigK, respectively. The size of molecular marker (in bp) is indicated on the left side of the panels. The “+” sign at the bottom of the panels indicate clones where the expected recombination event has occurred (8 clones were analyzed).
CD1231 is sufficient for skinCd integration in E. coli
While both CD1231 and CD1234 are required for skinCd excision in E. coli, we also wanted to test the involvement of these proteins in integration reactions. With that purpose, the attP site of the prophage-like element was inserted into pFT74R, which already contains the integration sequence attB as the result of the excision reaction (Fig 7A). Two plasmids were constructed, one with attP and attB in the same orientation (pMS511; Fig 7C) and one with attB and attP in opposite orientation (attP´ in pMS510; Fig 7E). These plasmids were introduced in E. coli cells carrying the plasmids for expression of CD1231, CD1234 or both. Cells were induced to produce CD1231, CD1234 or both and then the plasmids were purified and examined for recombination events between attP and attB by digestion with BamHI and NotI. Two types of recombination events serve as readouts for the ability of CD1231 to catalyze DNA integration. A recombination event between attP and attB should result in the removal of the intervening DNA for pMS511 (Fig 7C) or in the inversion of the intervening DNA in the case of pMS510 (Fig 7E). We showed that CD1231 is necessary and sufficient for both types of recombination events involving attP and attB (Fig 7D and 7F). No recombined products of pMS510 or pMS511 were retrieved when the catalytically inactive CD1231 bearing the S10A substitution was produced, alone or together with CD1234 (S5 Fig).
These results showed that CD1231 is sufficient for the integration event that results from the recombination between attP and attB, but requires CD1234 for the excision event that results from the recombination reaction involving attL and attR. Thus, CD1234 is a recombination directionality factor (RDF) assisting CD1231 in skinCd excision.
Deletion of the skinCd element causes premature activity of σK
While the regulated excision of skinCd has been suggested as a critical mechanism for efficient sporulation in C. difficile [17, 27], a more recent study suggests that skinCd is not essential for the formation of heat-resistant spores in this organism [30]. However, skinCd excision controls the onset of σK activity. The deletion of the skin in a pro-less sigK strain in B. subtilis imposes changes in the mother cell line of gene expression leading to altered spore structure and functional properties, while the final titer of spores formed is reduced compared to the wild-type [41, 42]. To analyze more precisely the involvement of skinCd in sporulation, we took advantage of the 630Δerm, which expressed CD1234 under Ptet control, to obtain a congenic derivative of strain 630Δerm lacking skinCd (summarized in S6A Fig). Addition of ATc during growth led to skinCd excision and after plating of the cells, DNA was extracted from isolated colonies. We identified several clones that carried a reconstructed sigK gene (S6B Fig, lane 1) but lacked the 5’ junction of skinCd in the chromosome (S6B Fig, lane 2) and the excised form of skinCd that was lost after cellular division (S6B Fig, lane 3). In a second step, a clone carrying an intact sigK gene was cured of the pDIA6103-PtetCD1234 plasmid by successive cycles of growth and dilution in TY medium. After plating, Tm-sensitive clones that had lost pDIA6103-PtetCD1234 were isolated. One clone was named 630Δerm ΔskinCd. Phase contrast microscopy experiments revealed the presence of free spores in both the 630Δerm and 630Δerm ΔskinCd strains (Fig 1C) and the titer of heat resistant spores measured 48 h and 72 h after inoculation in SM medium was almost identical for both strains (Fig 1C). Moreover, the percentage of sporulation measured for the two strains at 12 h (0.4% for 630Δerm and 0.3% for 630Δerm Δskin), 18 h (1.6% and 1.1%) and 24 h (6.4% and 5.4%) following inoculation into SM also did not differ significantly. Thus, in agreement with the previous results using a skinCd-less sigK gene expressed from a SpoIIID-independent promoter [30], deletion of skinCd did not appear to affect the final titer of spores and the kinetics of sporulation.
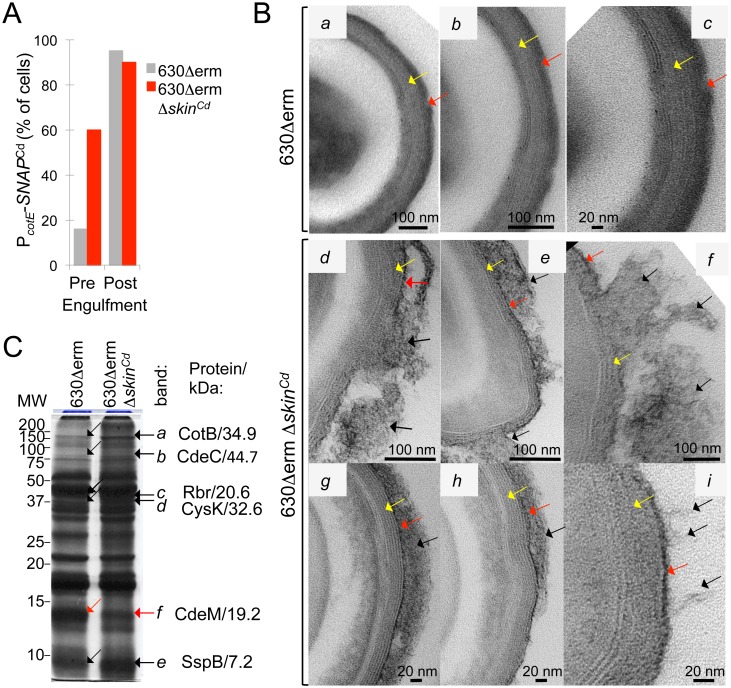
We then analyzed transcription of sigK and of σK target genes by qRT-PCR in the 630Δerm and 630Δerm ΔskinCd strains. We first harvested the cultures between 10 h and 24 h of growth in SM. After RNA extraction, we tested the expression of sigK using oligonucleotides located on both sides of the skinCd insertion into sigK and of σK target genes. The results showed that the expression of sigK and of several sigK targets (cotE, cotBC, sleC, cdeC, bclA1 and bclA3) was higher in the Δskin strain than in the 630Δerm strain (S2 Table). Lastly, we monitored the activity of σK at the single cell level using a PcotE-SNAPCd transcriptional fusion in the wild-type and ΔskinCd background. Fluorescence microscopy revealed that the signal intensity from the accumulation of TMR-Star-labeled SNAPCd did not differ significantly between wild-type and ΔskinCd sporangia, before or after engulfment completion. Strikingly, however, the ΔskinCd mutation increased the fraction of cells that showed PcotE-SNAPCd expression and hence σK activity, prior to engulfment completion, from 20% (WT) to 60% (ΔskinCd) (Fig 8A). Thus, expression of a skinCd-less sigK gene from its native promoter results in premature σK activity.
Fig 8. skinCd controls the time of σK activation and the fidelity of morphogenesis.
A: quantitative analysis of the cells with fluorescence before and after the completion of engulfment in C. difficile cells carrying the PcotE-SNAPCd fusion (σK-responsive) in strain 630Δerm and 630Δerm ΔskinCd. SNAPCd production was monitored as described in the legend for fig 3. B: purified spores were processed for imaging by transmission electron microscopy. Sections showing details of the spore surface layers for representative specimens are shown (the entire spores are shown in S7 Fig). The yellow arrow indicates the lamellar, inner structure of the coat, whereas the red arrow points to the more electrondense surface layer. The black arrows indicate material peeling off the surface of the spores. C: spores of the 630Δerm and 630Δerm ΔskinCd strains were purified by density gradient centrifugation, the spore surface proteins extracted and resolved by 15% SDS-PAGE. Bands a to d and e show increased representation in spores of the ΔskinCd strain, whereas band f shows reduced representation in ΔskinCd spores as compared to the 630Δerm. The proteins identified in these bands by mass spectrometry are indicated, together with their predicted molecular weight.
Deletion of the skinCd element in strain 630Δerm affects the assembly of the spore surface layers
Activation of σK in B. subtilis is tightly linked to engulfment completion, and mutations that cause its premature activation result in alterations in the properties of the resulting spores [41, 42]. σK plays an important role in the assembly of the spore coat [16, 17] and because σK was active prior to engulfment completion in a larger fraction of the 630Δerm ΔskinCd sporangia as compared to the 630Δerm strain, we examined ultrastructure and the polypeptide composition of the spore surface layers in the two strains. Spores were density gradient purified from cultures of the 630Δerm and 630Δerm ΔskinCd strains, and processed for electron microscopy. Under our conditions, spores of the 630Δerm strain showed a more internal lamellar coat (Fig 8B, yellow arrows in panels a to c), covered by a more external electrondense layer (red arrows); this layer had a uniform and compact appearance around the entire spore (Figs 8B and S7; see also [17]). It may correspond to an exosporium-like layer, which in C. difficile seems to be closely apposed to the underlying coat ([3]; see also below). In contrast, the electrondense outer layer was less compacted in spores of the ΔskinCd strain, and absent in some sections (Fig 8B, black arrows in panels d to i). The disorganization of the outer layer presumably allowed the visualization of a thin electrondense layer forming the edge of the lamellar coat and in close apposition to it (Fig 8B, red arrows in panels d to i). However, this thin layer was missing in some sections and material from the inner lamellar layer appeared to peel off the spore in those sections (Fig 8B, black arrows in panels d and f). Overall, the lamellar coat layer also appeared to be less dense, making its structural organization more apparent (Fig 8B, d to i and S7 Fig). Spore surface proteins from both the coat and exosporium layers were extracted from spores of the 630Δerm and ΔskinCd strains and resolved by SDS-PAGE. Several proteins showed increased extractability from spores of the ΔskinCd strain relative to the wild-type strain (Fig 8C, black arrows, bands a through d and e), whereas a protein of about 12 kDa (red arrow, band f) showed reduced extractability. Mass spectrometry analysis indicates that band a (size close to 200 kDa) contains CotB, whereas band b (size around 80 kDa) contains CdeC. Because the predicted sizes of CotB and CdeC are 34.9 kDa and 44.7 kDa, respectively, these two species may represent cross-linked products of these proteins. CotB and CdeC are likely critical determinants for the assembly of the spore exosporium [3]. Rubrerythrin (Rbr) with a predicted size of 20.6 kDa and CysK, with a predicted size of 32.6 kDa, are detected in bands c and d, at about 37 kDa (Fig 8C). At least Rbr, found in a band of about twice its predicted molecular weight, may form cross-linked homodimers, or be cross-linked to a protein of about 20 kDa. In contrast, a likely proteolytic fragment of the 19.2 kDa-exosporium protein CdeM, shows decreased representation or extractability in the mutant (Fig 8C, band f). The alterations in the assembly of CotB, CdeC and CdeM may explain at least in part the morphology of the outer spore layers in the ΔskinCd strain (Fig 8B); possibly, the Δskin deletion affects mainly the assembly of the exosporium-like layer that in C. difficile seems to be juxtaposed to the coat, while the morphological alterations seem at the level of the coat may be in part a consequence of a misassembled exosporium-like layer. The increased representation of SspB (a forespore-specific protein) [18] may indicate that the spores of the mutant are more permeable or more sensitive to the extraction procedure (Fig 8C, band e). In toto, we conclude that deletion of skinCd affects the assembly of the C. difficile spore surface layers. The alterations in the assembly of the spore coat and of a more external possible exosporium-like layer are most likely due to the premature activity of σK in 630Δerm ΔskinCd sporangia.
Discussion
The mother cell-specific excision of the skin element, in either B. subtilis or C. difficile, is essential for the production of a functional sigK gene. In B. subtilis, expression of the gene coding for SpoIVCA, the LSR responsible for skinBs excision is under the joint control of σE and SpoIIID [33]. Hence, skinBs excision is restricted to the terminal mother cell. Excision of the skinCd element is absolutely dependent on CD1231 encoding a SpoIVCA homologue, but expression of this gene is not restricted to the mother cell. Rather, CD1231 is transcribed from a σA-dependent promoter and is expressed in vegetative cells and during sporulation in both compartments. An important finding of the present investigation is that excision of skinCd requires, in addition to CD1231, the product of a gene, CD1234, whose expression is under the control of both σE and SpoIIID (Fig 9). It is the requirement for CD1234 that restricts skinCd excision to the mother cell. However, the inactivation of CD1234 or SpoIIID does not reduce sporulation to the level observed for the CD1231 or sigK gene disruption. CD1234 and SpoIIID thus appear less crucial for sporulation than σK or the catalytic function of the LSR, CD1231. It seems possible that CD1231 occasionally performs excision of skinCd without CD1234. Nevertheless, the normal requirement of CD1234 for skinCd excision in C. difficile is paralleled by its requirement for the CD1231-dependent excision of a mini-skinCd in a heterologous host, E. coli but conversely, CD1234 is not required for the CD1231-dependent skinCd integration. Thus, our recombination assays show that CD1234 is a RDF [22, 32, 37]. RDFs likely stimulate formation of the synapse complex between attL and attR by competing for inhibitory interactions involving the CC motifs of LSRs or inhibit formation or otherwise destabilize the synaptic complex formed between attB and attP possibly by stabilizing the auto-inhibitory activity of the CC motifs in this complex [22, 37]. Accordingly, the RDFs interact with the recombinase in solution, in line with our observation that CD1231 and CD1234 are part of a complex that formed in E. coli. Both structural studies and the isolation of mutations close or within the CC motif of the ϕC31 LSR that allows it to recombine attL and attR in the absence of the cognate RDF, suggest binding of the CC motifs by the RDF [22, 37]. More studies are needed to unravel the mechanism by which CD1234 cooperates with the CD1231 LSR to control skinCd excision. RDFs have been identified for several LSRs. However, they do not share significant sequence similarity and most of them appear to be small, basic proteins. In contrast, CD1234 is acidic like SprB, the RDF of phage SPβ [32]. SprB is required for the SprA recombinase-mediated excision of phage SPβ. A RDF assisting SpoIVCA in the excision of skinBs has not been identified, but the function of the skinBs-encoded genes has not been inspected individually.
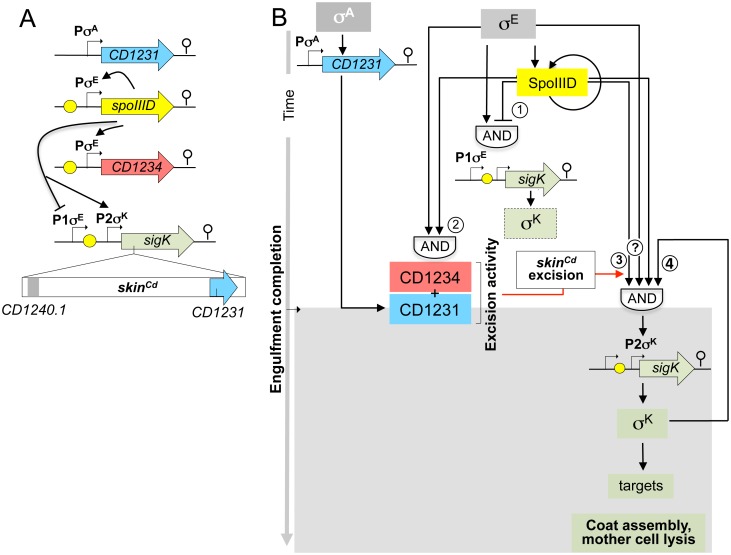
Fig 9. The sporulation network leading to skinCd excision and σK activation in C. difficile.
A: schematic representation of the genes involved in skinCd excision and σK activation in C. difficile. Bent arrows show the promoters and yellow dots represent putative SpoIIID binding sites. The lines represent the control exerted by SpoIIID over the indicated genes. B: model of the organization of the mother cell transcriptional network. σE drives production of SpoIIID, which then activates transcription of both CD1234 and sigK P2. CD1234 and probably also sigK P2 (the later indicated by a question mark in A and B) are under the control of a coherent FFL with an AND gate logic (2 and 3 in the figure) established by σE and SpoIIID thought to delay skinCd excision (which also requires the CD1231 recombinase) and the main period of sigK transcription to post-engulfment sporangia. Then, a positive auto-regulatory loop combined with a positive control by SpoIIID leads to an increase in sigK transcription at P2 (4 in the figure) and to σK activity. SpoIIID may also repress transcription from the σE-dependent sigK P1; the two regulatory proteins could thus form an incoherent FFL with an AND gate logic (1 in the figure), which could result in a low level of sigK transcription during engulfment. Transcription from sigK P1 would be shut-off coincidently with activation of P2. Possibly this would lead to a pulse in the production of σK in some cells in which skinCd is excised during engulfment (indicated as a dotted box in figure). As shown by correlating transcription of σK-dependent genes with stages of morphogenesis using single cell analysis, the onset of the main period of σK activity coincides with the transition from phase grey to phase bright spores, presumably when synthesis of the spore cortex is finalized and the final stages in the assembly of the spore coat and exosporium begins.
Recent studies have examined in detail the sporulation program of C. difficile and provided evidence that the temporal compartmentalization of sigma factor activity is less tightly regulated in C. difficile as compared to B. subtilis [16–18]. A difference between the sporulation programs in the two organisms may be the existence of a higher degree of redundancy in the control of gene expression at key stages in morphogenesis in B. subtilis. The control over spoIVCA and sigK transcription, skinBs excision, and proteolytical removal of the inhibitory pro-sequence ensures that σK is only active following engulfment completion in B. subtilis [41–43]. Since σK in C. difficile lacks a pro-sequence, the main period of σK activity may be mainly dictated by the time of CD1234 production and skinCd excision (Fig 9). In B. subtilis, both the sigK and spoIVCA genes are controlled by a coherent FFL involving σE and SpoIIID [19]. Thus, both the SpoIVCA-mediated skinBs excision and the transcription of sigK are delayed towards the end of the engulfment sequence, when the forespore signal that leads to pro-σK processing results in “just-in-time” activation of σK. In C. difficile, transcription of CD1234 and sigK decreased in a spoIIID mutant. We detected a common binding motif in the promoter region of these genes suggesting a direct effect of SpoIIID on their transcription. This motif is also present upstream of spoIIID itself allowing us to propose an auto-regulation for SpoIIID and to define a first consensus for the SpoIIID binding site of C. difficile (KRTAACARK) (S3A and S3B Fig) sharing partial similarity with the SpoIIID consensus of B. subtilis (S3C Fig) [19].
Our results show that expression of CD1234 in C. difficile relies on a coherent FFL involving σE and SpoIIID (Fig 9B) [19]. Although transcription of CD1234 is detected in the mother cell soon after asymmetric division and does not increase during or following engulfment completion, it is possible that the CD1234 protein only reaches a threshold level following engulfment completion. The effect of a spoIIID deletion on the transcription of sigK as assessed here using SNAPCd labeling and scoring of cells in which engulfment had been completed is less pronounced than previously reported on the basis of a more sensitive qRT-PCR analysis [18]. It seems possible that binding of SpoIIID to the sigK regulatory region represses the σE-dependent P1 promoter towards the end of the engulfment process, while allowing activation of transcription from P2 (Fig 3A). If so, deletion of spoIIID would allow prolonged utilization of P1 by σE, possibly explaining the reduced effect of the mutation following engulfment completion as seen here (Fig 3B). We do not presently know whether P2 is initially utilized by σE with the help of SpoIIID, and later by σK. Further work is needed to test these possibilities. In any event, positive auto-regulation of sigK transcription at P2 may then increase production of active σK locking the cells in a late-mode of gene expression [16–18] (Fig 9B). This genetic architecture may allow delaying sigK transcription and skinCd excision to ensure that the main period of σK activity follows engulfment completion [17]. Thus, although the pro-sequence is absent from σK, some level of redundancy also appears to be embedded in the activation of σK in C. difficile, with delayed transcription and rearrangement contributing to its timely activation. As we show here, the rearrangement is important, since expression of a skinCd-less sigK gene increases σK activity during engulfment. In this regard, we comment on the possible function of sigK P1. If indeed this promoter is repressed through binding of SpoIIID to the downstream box, P1 would be subject to a type I incoherent FFL, which could result in a pulse of sigK transcription (Fig 9B) [44]. Provided skinCd excision also occurs, a pulse in sigK transcription may result in σK activation during engulfment in some cells in a population (Fig 9B). Given that a skinCd-less sigK allele leads to alteration in the spore surface structure, we speculate that this regulatory scheme could produce spores with different structure and functional properties in a fraction of the population.
The presence of a skin element remains an exception in clostridia. Although the still more familiar designation of C. difficile was used herein, the organism differs significantly from typical Clostridial species, and was recently placed in the Peptostreptococcaceae family and renamed Peptoclostridium difficile [45]. Nevertheless, a prophage integrated into sigK is found in Clostridium tetani [46], in some strains of C. botulinum and in a strain of C. perfringens [47]. Absence of a skin element may be related to the involvement of σK in functions in addition to its role as a late mother cell-specific transcription factor. For instance, in C. acetobutylicum and C. botulinum, σK is required for entry into sporulation and for solventogenesis in the former organism and for cold and salt stress in the latter, while in C. perfringens σK is also involved in enterotoxin production [48]. It thus seems likely that skinCd together with the compartmentalized expression of CD1234 helps preventing ectopic and heterochronic activity of σK. This suggestion is in agreement with the observation that bypassing both the recombination and pro-protein processing levels of control leads to some activity of σK during stationary phase under conditions that do not support efficient sporulation by B. subtilis [42]. This inappropriate activity of σK is most likely the result of auto-regulation, and shows that even in B. subtilis, transcriptional control alone is not sufficient to restrict the activity of σK to the mother cell during late stages of spore development but also for proper sporulation as a pro-less skin-less strain of B. subtilis shows reduced sporulation [41, 42]. In contrast, the early activity of σK in the ΔskinCd strain of C. difficile, while causing alterations in coat assembly, does not affect the final titer of heat resistant spores. It is possible that the sporulation program in C. difficile is more permissive to changes in the proper timing of morphogenetic events [16–18]. It is also possible, however, that an additional as yet unidentified mechanism, controls the activity of σK in C. difficile.
Why the expression of CD1231 is not restricted to the mother cell is intriguing and might suggest the possible existence of other functions than skin excision for this recombinase. CD1231 may also function occasionally in the forespore or in vegetative cells, in the absence of CD1234, resulting in permanent elimination of skinCd, as it is known that some strains of C. difficile including epidemic strains lack this element [27, 28, 49]. This possibility also raises the question of whether in those strains σK is recruited to additional functions, by analogy with C. perfringens, C. botulinum and C. acetobutylicum. Importantly, as we show that the premature activity of σK during sporulation in C. difficile imposes alterations to the assembly of the spore surface layers, we speculate that spores of the skinCd-less epidemic strains may have alterations in structural and/or functional properties important for spore persistence and infection.
Reminiscent of the situation with skin, whose deletion alters assembly and function of the spore surface layers, it is noteworthy that the B. subtilis SPβ prophage is inserted into the spsM gene, required for the glycosylation of proteins at the spore surface, and that SPβ excision during sporulation depends on the mother cell-specific expression of the SPβ gene coding for the SprB RDF. Remarkably, SPβexcision during sporulation does not result in the assembly of phage particles, most likely because the prophage genes lack mother cell-specific promoters [32]. In different spore-formers, several other sporulation genes such as spoVFB encoding the dipicolinate synthase and spoVR involved in cortex formation are interrupted by phage-like elements that are able to excise during sporulation [32, 50]. Moreover, developmentally-controlled, recombinase-dependent excision of phage-like intervening elements also takes place in cyanobacteria where these elements, interrupting genes required for nitrogen fixation, are excised during heterocyst differentiation [51]. Insertion of phages or phage like elements in genes that are specifically expressed in terminal cell lines ensures their vertical transmission and minimizes cost for the host. However, regulated prophage excision may also occur in non-terminally differentiated cells. The Listeria monocytogenes comK gene, coding for the competence master regulatory protein, is interrupted by a prophage [52]. The DNA-uptake complex formed by the Com proteins is required for L. monocytogenes to escape from macrophage phagosomes and remarkably, excision of the comK intervening sequence is specifically induced during intracellular growth within phagosomes [52]. Also remarkably, prophage excision from comK as observed for SPβ excision in B. subtilis sporangia does not result in the production of phage particles [52]. Clearly, whether or not in terminally differentiated cells, host “domestication” of prophage excision, may lead to additional levels of genetic control over important cellular functions [22, 53].
Materials and Methods
Bacterial strains, growth and sporulation conditions
C. difficile strains and plasmids used in this study are presented in Table 3. C. difficile strains were grown anaerobically (5% H2, 5% CO2, and 90% N2) in Brain Heart Infusion (BHI, Difco), which was used for selection of conjugants, in TY (Bacto tryptone 30 g.L-1, yeast extract 20 g.L-1, pH 7.4) or in sporulation medium (SM) [54], which was used for sporulation assays. SM medium contained per liter: 90 g Bacto tryptone, 5 g Bacto peptone, 1 g (NH4)2SO4, 1.5 g Tris Base. When necessary, cefoxitin (Cfx; 25 μg/ml), thiamphenicol (Tm; 15 μg.ml-1) or erythromycin (Erm; 2.5 μg.ml-1) was added to C. difficile cultures. E. coli strains were grown in LB broth. When indicated, ampicillin (100 μg.ml-1) or chloramphenicol (15 μg.ml-1) was added to the culture medium. The antibiotic analog anhydrotetracycline (ATc; 100 ng.ml-1) was used for induction of the Ptet promoter present in derivatives of the C. difficile pDIA6103 vector [34].
Table 3. Bacterial strains used in this study.
| Strain | Relevant Genotype/phenotype | Origin/Reference |
|---|---|---|
| E. coli | ||
| Top10 | F- mcrA Δ (mrr-hsdRMS-mcrBC) f80lacZΔM15 ΔlacX74 deoR, recA1 araD139 Δ (ara-leu)7697 galK rpsL(StrR) endA1 nupG | Invitrogen |
| HB101 (RP4) | supE44 aa14 galK2 lacY1 Δ(gpt-proA) 62 rpsL20 (StrR)xyl-5 mtl-1 recA13 Δ(mcrC-mrr) hsdSB(rB-mB) RP4 (Tra+ IncP ApR KmR TcR) | Laboratory stock |
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB- mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Novagen |
| C. difficile | ||
| 630Δerm | wild-type | Laboratory stock |
| AHCD532 | 630Δerm sigE::ermB | [17] |
| AHCD535 | 630Δerm sigK::ermB | [17] |
| AHCD601 | 630Δerm pFT47- PsigK -SNAPCd | [17] |
| AHCD667 | 630Δerm pFT47-PCD1231-SNAPCd | This work |
| AHCD690 | 630Δerm spoIIID::ermB pFT47- PsigK -SNAPCd | This work |
| AHCD695 | 630Δerm pFT47- PcotE- SNAPCd | [17] |
| AHCD699 | 630Δerm sigK::ermB pFT47- PcotE- SNAPCd | [17] |
| AHCD700 | 630Δerm spoIIID::ermB pFT47- PcotE- SNAPCd | This work |
| AHCD758 | 630Δerm CD1234::ermB pFT47- PcotE- SNAPCd | This work |
| AHCD763 | 630Δerm CD1231::ermB pFT47- PcotE- SNAPCd | This work |
| AHCD787 | 630Δerm pFT74 (SigK-SNAP, skin+) | This work |
| AHCD800 | 630Δerm spoIIID::ermB pFT74 (SigK-SNAP, skin+) | This work |
| AHCD851 | 630Δerm ΔskinCd pFT47- PcotE-SNAPCd | This work |
| CDIP224 | 630Δerm spoIIID::ermB | [18] |
| CDIP345 | 630Δerm sigE::ermB pFT47-PCD1234-SNAPCd | This work |
| CDIP389 | 630Δerm pFT47-PCD1234-SNAPCd | This work |
| CDIP396 | 630Δerm CD1234::ermB | This work |
| CDIP397 | 630Δerm CD1234::ermB pMTL84121-CD1234 | This work |
| CDIP399 | 630Δerm spoIIID::ermB pFT47-PCD1234-SNAPCd | This work |
| CDIP526 | 630Δerm CD1231::ermB | This work |
| CDIP533 | 630Δerm CD1231::ermB pMTL84121-CD1231 | This work |
| CDIP560 | 630Δerm pDIA6103 | This work |
| CDIP561 | 630Δerm pDIA6103-CD1234 | This work |
| CDIP562 | 630Δerm spoIIID::ermB pDIA6103 | This work |
| CDIP563 | 630Δerm spoIIID::ermB pDIA6103-CD1234 | This work |
| CDIP564 | 630Δerm sigE::ermB pDIA6103 | This work |
| CDIP565 | 630Δerm sigE::ermB pDIA6103-CD1234 | This work |
| CDIP566 | 630Δerm CD1231::ermB pDIA6103 | This work |
| CDIP567 | 630Δerm CD1231::ermB pDIA6103-CD1234 | This work |
| CDIP568 | 630Δerm CD1234::ermB pDIA6103 | This work |
| CDIP569 | 630Δerm CD1234::ermB pDIA6103-CD1234 | This work |
| CDIP583 | 630Δerm ΔskinCd | This work |
| Plasmids | ||
| pMTL007 | ClosTron plasmid; catP; intron containing ermB::RAM (CmR/TmR)1 | [55] |
| pMTL84121 | Clostridia modular plasmid; catP (CmR/TmR) | [56] |
| pDIA6103 | pRPF185 ΔgusA | [34] |
| pFT38 | pMTL84121-sigKskin+ (CmR/TmR) | [17] |
| pFT47 | pMTL84121-SNAPCd (CmR/TmR) | [17] |
| pFT51 | pFT47 PsigK-SNAPCd | [17] |
| pFT69 | pFT47 PcotE-SNAPCd | [17] |
| pFT74 | pMTL84121 P-sigK5’-skin+-SigK-SNAPCd | This work |
| pMS469 | pFT47 PCD1231-SNAPCd | This work |
| pMS484 | pET33b-CD1234-His6 | This work |
| pMS486 | pET16b-CD1231-Strep tag | This work |
| pMS494 | pETDuet-1- CD1234-His6 | This work |
| pMS499 | pETDuet-1- CD1231-Strep tag | This work |
| pMS500 | pETDuet-1- CD1234-His6- CD1231-Strep tag | This work |
| pMS510 | pMTL84121-sigK-SNAPCd-attP’ | This work |
| pMS511 | pMTL84121-sigK-SNAPCd-attP | This work |
| pMS512 | pETDuet-1- CD1231S10A-Strep tag | This work |
| pMS513 | pETDuet-1- CD1234-His6- CD1231S10A-Strep tag | This work |
| pDIA6314 | pMTL007::Cdi-CD1234-89a | This work |
| pDIA6346 | pMTL007::Cdi-CD1231-206a | This work |
| pDIA6192 | pFT47-PCD1234-SNAPCd | This work |
| pDIA6307 | pMTL84121-CD1234 | This work |
| pDIA6348 | pMTL84121-CD1231 | This work |
| pDIA6353 | pDIA6103-CD1234 | This work |
| pDIA6382 | pGEMT-attP | This work |
| pLOM4 | pET30a(+)-tgl-His6 | [40] |
Sporulation assays were performed as follows. After 12 h, 18 h, 24 h, 48 h or 72 h of growth in SM medium, 1 ml of culture was divided into two samples. To determine the total number of cells, the first sample was serially diluted and plated on BHI with 0.1% taurocholate (Sigma-Aldrich) to ensure efficient spore germination [5]. To determine the number of spores, the vegetative bacteria of the second sample were heat killed by incubation for 30 min at 65°C prior to plating on BHI with 0.1% taurocholate. The rate of sporulation was determined as the ratio between the number of spores/ml and the total number of bacteria/ml (x100).
Construction of C. difficile strains
The ClosTron gene knockout system [55] was used to inactivate the CD1231 and CD1234 genes in the strain 630Δerm, to produce strains CDIP526 (CD1231::erm) and CDIP396 (CD1234::erm) (Table 3). Primers to retarget the group II intron of pMTL007 to these genes (S1 Table) were designed using the Targetron design software (http://www.sigmaaldrich.com). The PCR primer sets were used with the EBS universal primer and intron template DNA to generate, by overlap extension PCR, a 353-bp product that would facilitate intron retargeting to CD1231 or CD1234. The PCR products were cloned between the HindIII and BsrGI sites of pMTL007 to yield pDIA6346 (pMTL007::Cdi-CD1231-206a) and pDIA6314 (pMTL007::Cdi-CD1234-89a). pDIA6346 and pDIA6314 were introduced into E. coli HB101 (RP4) and the resulting strains subsequently mated with C. difficile 630Δerm. C. difficile transconjugants were selected on BHI agar containing Tm and Cfx and then plated on BHI agar containing Erm. Chromosomal DNA of transconjugants was purified using the Instagene kit (Biorad). The Erm resistance phenotype was due to the splicing of the group I intron from the group II intron following integration, as shown by PCR using the ErmRAM primers (ErmF and ErmR). To verify integration of the Ll.LtrB intron into the right gene targets, we used PCR with two primers flanking the insertion site in CD1231 (IMV736/IMV737) or CD1234 (IMV695/IMV696), the intron primer EBSu and the CD1231- (IMV736) or CD1234-specific primers (IMV696) (S2 Fig). The Southern blot probe was generated by PCR using pMTL007 as a template and the primer pair OBD522 and OBD523 (S1 Table), yielding a 374 bp PCR product that hybridized with the group II intron. Southern blot analyses were performed as previously described [17, 18].
For complementation studies, the CD1231 gene with its promoter (positions -192 to +1569 from the translational start site) and the CD1234 gene with its promoter (-168 to +343 from the translational start site) were amplified by PCR using primers IMV728 and IMV729 or IMV695 and IMV696, respectively (S1 Table). The PCR fragments were cloned between the XhoI and BamHI sites of pMTL84121 [56] to produce pDIA6307 (CD1234) and pDIA6348 (CD1231). These plasmids were introduced into E. coli HB101 (RP4) and then transferred by conjugation into CDIP526 (CD1231::erm) to produce CDIP533, and into CDIP396 (CD1234::erm) to produce CDIP397 (Table 3).
Transcriptional SNAPCd fusions
To construct transcriptional SNAPCd fusions to the CD1231 and CD1234 promoters, 485 and 139 bp DNA fragments containing the promoter region of each gene were PCR-amplified using genomic DNA from strain 630Δerm and primer pairs CDsigK3’ Fw/CDsigK3’_EcoRI Rev or IMV677/IMV678, respectively. These fragments were cloned into pFT47 [17] to create pMS469 and pDIA6192 (Table 3). Plasmid pDIA6192 was transferred to 630Δerm, 630Δerm spoIIID::erm or 630Δerm sigE::erm and pMS469 was transferred to 630Δerm by conjugation from derivatives of E. coli HB101 (RP4) (Table 3).
Construction of a translational reporter for skinCd excision
Primers PCDsigK Fw and CDsigK5’Rev (S1 Table) were used to amplify a fragment from the sigK gene using pFT38 [17] as the template. The resulting 819 bp fragment encompasses 415 bp of the sigK regulatory region and the 5´-end of the interrupted coding region, 404 bp downstream of the sigK translational start site [18]. This fragment was cleaved with NotI and EcoRI. In a second PCR, the region containing the 3´-end of the coding sequence of sigK, and 180 bp upstream of this position, was PCR amplified using primers RecFP-Fw and SigK-SNAP Rev, yielding a 445 bp fragment. This fragment was cleaved with EcoRI and BamHI. The two fragments were inserted between the NotI and BamHI sites of pFT58 to produce pFT74 (Fig 4A). pFT74 was transferred by conjugation into C. difficile 630Δerm and 630Δerm spoIIID::erm (Table 3).
Construction of CD1234 inducible strains
To express the CD1234 gene under the control of a Ptet inducible promoter, the CD1234 gene with its ribosome-binding site (positions -19 to + 343 from the translational start codon) was amplified using primers IMV720 and IMV696 (S1 Table). The resulting PCR product was digested with StuI and BamHI and cloned into pDIA6103, a derivative of pRPF185 lacking the gusA gene [34]. Using the E. coli HB101 (RP4) strain containing either pDIA6103 or pDIA6353 (pDIA6103-CD1234), these plasmids were transferred by conjugation into C. difficile 630Δerm, 630Δerm spoIIID::erm, 630Δerm sigE::erm, 630Δerm CD1234::erm or 630Δerm CD1231::erm.
Detection of skinCd excision by PCR or qPCR
The various strains of C. difficile containing either pDIA6103 or pDIA6103-CD1234 (Table 3) were grown overnight in TY containing 0.025% of taurocholate. The pre-cultures were diluted 100-fold in TY medium and the resulting cultures incubated at 37°C for 4 h. To induce expression of CD1234, ATc (100 ng/ml) was then added to the medium. After 2 h of incubation, the cells were harvested by centrifugation and the chromosomal DNA was extracted from each strain using the Puregene Yeast/Bact kit (QIAGEN). To quantify the reconstruction of sigK associated to skinCd excision, quantitative PCR (qPCR) was performed using primers upstream (QRTBD325) and downstream (QRTBD326) of the skinCd insertion site. A qPCR of the DNA-PolIII gene was used as a control. For each strain, a dilution of the culture was also plated on BHI. Eight clones per strain were re-isolated on BHI plates, and DNA was extracted from each using the Instagene kit (Biorad). To detect the presence of skinCd into the chromosome, the 5’ junction of the skinCd element was PCR-amplified using primers OBD0742 (in CD1231) and QRTBD326 (in sigK).
Construction of a ΔskinCd mutant
A derivative of strain 630Δerm lacking the skinCd was obtained as follows. Strain 630Δerm (pDIA6103-CD1234) was grown 4 h in TY. After inducing expression of CD1234 expression, the cells were serially diluted, plated on BHI and DNA extracted for several clones. To verify skinCd excision, PCR was performed with primers flanking the site of skinCd insertion (QRTBD326 and IMV833) or the 5’ junction of the skinCd element (OBD0742 in CD1231 and QRTBD326 in sigK). One clone in which sigK was amplified and lacking the 5’ junction of skinCd skin was selected. To cure the selected clone of pDIA6103-CD1234, cells were diluted 6-fold in TY medium before plating on BHI. About 70% of the clones obtained after plating were TmS and had lost pDIA6103-CD1234 as checked by PCR.
RNA extraction and quantitative RT-PCR
Total RNA was isolated from strains 630Δerm and 630Δerm Δskin, from the CD1231 and CD1234 mutants and from the complementation strains. These strains were grown in SM medium, the cells were collected by centrifugation, resuspended in RNApro™ solution and RNA extracted using the FastRNA Pro Blue Kit, according to the manufacturer’s instructions (MP Biomedicals). Quantitative real-time PCR (qRT-PCR) analysis was performed as previously described [57]. The primers used for each marker are listed in S1 Table. In each sample, the quantity of cDNAs of a gene was normalized to the quantity of cDNAs of the DNApolIII gene. The relative change in gene expression was recorded as the ratio of normalized target concentrations (ΔΔCt) [58].
Assay for recombinase activity and identification of attP
The CD1234 coding region was PCR-amplified using primers CD1234-Fw and -Rev. The resulting 252 bp DNA fragment was cleaved with NdeI and XhoI and cloned between the same sites of pET33b (Novagen), creating pMS484. CD1234 fused to the His6-tag was PCR-amplified from pMS484 with primers CD1234 Fw and T7ter, cleaved with NdeI and cloned between the NdeI and EcoRV sites of pETDuet-1, to create pMS494. The CD1231 coding region was amplified using primers CD1231-Fw and -Rev. The resulting 1518 bp fragment was digested with NcoI and SalI and ligated to pFN127 (a pET16b derivative carrying a Strep-tag) to yield pMS486. CD1231 fused to the Strep-tag was removed from pMS486 by digestion with NcoI and XhoI and cloned into pETDuet-1 producing pMS499, or in pMS494 to give pMS500. We used pMS499 and pMS500 (see above) and CD1231-specific primers to convert the serine codon at position 10 to an alanine codon producing pMS512 and pMS513, respectively. Derivatives of BL21(DE3) were constructed bearing pFT74 and pMS494 and either pMS499, pMS500, pMS512, or pMS513. The resulting strains were grown to an OD600nm of about 0.6, induced with 1 mM isopropyl-D-thiogalactopyranoside (IPTG), and incubated for 3 h before the cells were harvested. Plasmid DNA was extracted and analyzed by cleavage with NotI and BamHI. pFT74R results from miniskinCd excision, by recombination, from pFT74.
The attP site was obtained by PCR using IMV825 and IMV824 and DNA extracted from 630Δerm after 24 h of growth in SM. The PCR product was first cloned into pGEM-T-easy (Promega) and the attP site was then released with NotI and inserted into pFT74R, in both directions, to produced pMS510 and pMS511. Derivatives of BL21(DE3) were constructed, bearing pMS510 or pMS511 and either pMS494, pMS499, pMS500, pMS512, or pMS513. The various strains were grown to an OD600nm of about 0.6, induced with 1 mM IPTG, and incubated for 3 h before the cells were harvested. Plasmid DNA was extracted and cleaved with BamHI and NotI for the detection of recombination events.
Pull-down assays
Derivatives of BL21 (DE3) bearing plasmids for the production of CD1231-Strep, CD1234-His6 or Tgl-His6, were grown to mid-log phase (OD600nm≈0.6) in LB, and induced with 1 mM IPTG for 3 hours before the cells were harvested by centrifugation. The cell sediment was resuspended in 1-ml portions of buffer A [100 mM NaCl, 10 mM Tris-HCl (pH 8.0), 10% glycerol] per 50 ml of induced culture and lysed in a French pressure cell (18,000 lb/in2). In one set of assays, the lysates containing CD1231-Strep, CD1234-His6, or both, were cleared by centrifugation and 1 ml of each lysate was independently incubated with 50 μl of a 50% slurry of Ni2+-NTA agarose (Qiagen) at room temperature for 30 min. The Ni2+-NTA agarose was washed three times in buffer B (same as A but with 200 mM NaCl). In another set of assays, lysates containing either CD1234-His6 or Tgl-His6 were incubated with the Ni2+-NTA beads as above, and then incubated with a CD1231-Strep-containing extract for a further 30 min at room temperature. The beads were washed as described above. In all cases, the washed beads with bound proteins were resuspended in a final volume of 30 μl. The samples were resolved on 15% SDS-PAGE and subject to immunoblotting. Anti-His6 or anti-Strep antibodies were used at a 1:1000 dilution, and a rabbit secondary antibody conjugated to horseradish peroxidase (from Sigma) was used at a 1:10000 dilution. The immunoblots were developed with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Spore production, purification and spore coat extraction
For spore production, 5 ml of BHI was inoculated with of C. difficile and grown for 12 h at 37°C in anaerobic conditions. Fresh BHI (125 ml) was then inoculated with 1.25 ml of the pre-inoculum and the cultures incubated at 37°C under anaerobic conditions for 7 days. Cells were collected by centrifugation (for 10 min at 4800xg), resuspended in cold water and stored overnight at 4°C. Spores were then purified with a 42% Gastrografin (Schering) step gradient, as previously described [59]. The pellet was washed 10 times with cold water, and stored at -20°C. To analyze the profile of spore coat proteins, the spores were resuspended in extraction buffer (0.125 mM Tris-HCl, 5% β-mercaptoetanol, 2% SDS, 0.025% bromophenol blue, 0.5 mM DTT, 5% glycerol, pH 6.8) to a final OD580 of 4, and boiled. The extracted proteins were resolved by 15% SDS-PAGE and visualized by Coomassie brilliant blue R-250 staining. For identification, protein bands were excised and digested with trypsin, before analysis by matrix-assisted laser desorption ionization mass spectrometry.
Microscopy and image analysis
Samples (1 ml) were withdrawn from SM cultures at the desired times following inoculation, and the cells collected by centrifugation (4000 xg, for 10 min, at 4°C). The cells were washed with 1ml of PBS and resuspended in 0.1 ml of PBS supplemented with the membrane dye Mitotracker Green (MTG) at 0.5 μg.ml-1 and the DNA stain DAPI (4',6-diamidino-2-phenylindole; 50 μg.ml-1) (Invitrogen). For SNAP staining, culture samples of 1 ml were stained for 30 min with 250 nM SNAP-Cell TMR-Star (New England Biolabs) as described before [17]. Cells were washed four times by centrifugation (4000 g, 5 min) and ressupended in 1ml of PBS. The cells were resuspended in 1 ml of PBS containing 0.5 μg.ml-1 of MTG. Cells were mounted on 1.7% agarose coated glass slides and imaged as previously described [60]. Images were analyzed using the Metamorph software suite version 5.8 (Universal Imaging). For quantification of the SNAPCd-TMR Star signal, 6x6 pixel regions were defined in the desired cell and the average pixel intensity was calculated, and corrected by subtracting the average pixel intensity of the background. Small fluctuations of fluorescence among different fields were corrected by normalizing to the average pixel intensity obtained for the intrinsic autofluorescence of C. difficile cells [61].
Transmission Electron Microscopy
For thin sectioning transmission electron microscopy (TEM) analysis, C. difficile were purified by density gradient centrifugation as described above. Samples were processed for TEM as described previously [62].
Supporting Information
The alignment was generated using T-coffee (www.tcoffee.org). The color code indicates conservation, as indicated by the scale on the lower right corner. The conserved motifs are indicated, as well as the boundaries between the N- (NTD) and C-terminal (CTD) domains of the recombinase. The black dot indicates the catalytic nucleophile (Ser10) and the brown dots additional catalytic residues in CD1231. The position homologous to residue Glu449 in the ϕC31 recombinase is indicated by a red triangle. The E449K substitution in the ϕ C31 protein results in a “hyperactive” recombinase able to recombine attL and attR in the absence of the RDF.
(TIF)
A: schematic representation of gene inactivation by a type II Intron with an associated Retro-transposon-Activated Marker (RAM) [54]. The group II intron (bracket), originally in pMTL007 (top), carries a RAM element interrupting an ermB determinant (white). The intron was retargeted to CD1231 or CD1234 (blue) by altering the IBS, EBS1 and EBS2 sequences (grey and white stripes; top) by overlapping PCR. Splicing out of the td group I intron from the ermB gene in the RAM restores a functional marker allowing positive selection of mutants following intron integration. Primers used to confirm the integration and orientation of the type II intron are also indicated (bottom). B: chromosomal DNA of EmR C. difficile conjugants obtained during CD1231 inactivation by Clostron and of strain 630Δerm were screened by PCR using primer pairs RAM-F/R to confirm splicing out of the group I intron in the mutant (lane 1 and 2). We also performed PCR using chromosomal DNA of strain 630Δerm and of the CD1231 mutant (lane 3 and 4) with the intron primer EBSu and with a primer in CD1231 (IMV737). To verify the integration of the Ll.LtrB intron into the right gene targets, we further performed PCR using chromosomal DNA of strain 630Δerm and of the CD1231 mutant (lane 5 and 6) with two primers flanking the insertion site in CD1231 (IMV737-IMV736). Chromosomal DNA from the 630Δerm strain corresponded to lane 2, 3 and 6 while chromosomal DNA of the CD1231 mutant corresponded to lane 1, 3 and 5. The smart ladder (Eurogentec) was used as a molecular weight marker. C: chromosomal DNA of EmR C. difficile conjugants obtained during CD1234 inactivation by Clostron and of strain 630Δerm were screened by PCR using primer pairs RAM-F/R to confirm splicing out of the group I intron in the mutant (lane 1 and 2). To verify the integration of the Ll.LtrB intron into the right gene targets, we further performed PCR using chromosomal DNA of strain 630Δerm and of the CD1234 mutant with the intron primer EBSu and with a primer in CD1234 (IMV696) (lane 3 and 4) or with two primers flanking the insertion site in CD1234 (IMV695-IMV696) (lane 5 and 6). Chromosomal DNA from the 630Δerm strain corresponded to lane 2, 4 and 6 while chromosomal DNA of each mutant corresponded to lane 1, 3 and 5. The smart ladder (Eurogentec) was used as a molecular weight marker. D: southern blot analysis of genomic DNA from C. difficile 630Δerm, CDIP526 (CD1231::erm) and CDIP396 (CD1234::erm) mutant strains with an intron probe. Chromosomal DNA (6 μg in each reaction) was digested with HindIII. Southern blot analyses were performed using Amersham ECL Direct Nucleic Acid labelling and detection reagents, in accordance with the manufacturer's guidelines and visualised using Super Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific). The probe was produced by PCR using OBD522 and OBD523 primers (S1 Table), designed within the group II intron sequence.
(TIF)
Panel A: The DNA sequence immediately uptream of the coding region of spoIIID is presented. The transcriptional start site (+1, red) as previously mapped [34], the -10 and -35 promoter elements (green) that match the consensus for σE recognition (represented below the sequence), SpoIIID boxes (yellow), and the start codon of spoIIID, are indicated. Panel B corresponds to an alignment of the SpoIIID box identified upstream of CD1234, sigK and spoIIID. The sequence logo was created from on the WebLogo website (http://weblogo.berkeley.edu) using SpoIIID boxes identified in this study. The asterisk means that the sequence of the SpoIIID2 box is in opposite orientation. Panel C shows the SpoIIID box of B. subtilis as described in Eichenberger et al [19].
(TIF)
A: whole cell extracts were made from E. coli BL21(DE3) derivatives induced with IPTG to produce CD1231-Strep (expected size 59 kDa), CD1234-His6 (15 kDa), or both (as indicated by the «+ » signs). B: the proteins were detected in the extracts by immunoblotting (first three lanes) using anti-Strep (top) or anti-His6 antibodies (bottom). CD1231-Strep was detected as a species migrating around 60 kDa as indicated (the asterisks represent likely degradation products). CD1234-His6 was detected as a species of about 12 kDa, as indicated. The last three lanes of panel B represent the results of a pull down assay, in which CD1231-Strep (lane 4), CD1234-His6 (lane 5) or the two co-produced proteins (lane 6) were incubated with Ni2+-NTA agarose beads. CD1234-His6 alone (lane 5) binds to the beads. Residual binding of CD1231-Strep to the beads (lane 4) was also detected, but retention of the protein was greatly increased in the presence of CD1234-His6 (lane 6). C: the left side of the panel shows the immnoblot analysis of whole cell extracts prepared from E. coli BL21(DE3) strains producing the indicated proteins (« + » signs) individually. The extracts containing Tgl-His6 [39,40] or CD1234-His6 were incubated with the Ni2+-NTA agarose beads after which the extract containing CD1231-Strep was added. Proteins were eluted, resolved by SDS-PAGE and subject to immunoblot analysis with anti-Strep (top) or anti-His6 antibodies (bottom). The asterisks represent likely degradation products. D: diagram of the domain organization of CD1231 (as in Fig 1B) showing the location of a protease-sensitive site at the end of the NTD, in the recombinases from phages C31 and Bxb1 and from transposon TnpX. The calculated size of the full-lenght protein and of two fragments containing the C-terminal end of the protein (CTD and C-terminal extension) is shown. Fragments of about 37 and 40 kDa most likely containing the C-terminal end of CD1231-Strep may interact with CD1234-His6 (see main text for details).
(TIF)
E. coli cells were transformed with plasmids pFT74 and pMS510 and pMS511 corresponding to pFT74R+attP [carrying a recombined sigK gene and containing the attP site in both possible orientations (attP or attP´; see also Fig 7)]. Each E. coli strain containing one of these plasmids was then transformed with plasmids expressing either CD1231-Strep or CD1231S10A-Strep, and CD1234-His6 or both under the control of an IPTG inducible promoter. Plasmids were recovered from E. coli strains and analyzed by digestion with NotI and BamHI. Plasmid DNA from pFT74 and pFT74R+attP were used as controls for non-recombined and recombined sigK, respectively. The size of molecular size marker (in bp) is indicated on the left side of the panels. The “+” sign identifies clones where recombination has occurred (a total of 8 clones were analyzed).
(TIF)
A: strategy of construction of the 630Δerm ΔskinCd strain. A plasmid (pDIA6103-CD1234) carrying the gene CD1234 expressed under the control of a Ptet promoter was transferred into strain 630Δerm by conjugation. After 4 h of growth, CD1234 expression was induced with ATc for 2 h and cells were plated on BHI. The loss of the plasmid was obtained and verified as outlined in the Materials and Methods section. B: verification of the skinCd excision into the chromosome of the strain 630Δerm ΔskinCd after 5 h of growth in TY medium (vegetative cells). Lane 1: PCR recombined sigK (qRTBD326-IMV833); lane 2: PCR skinCd junction (OBD742-qRTBD326); lane 3: PCR excised form (IMV824-IMV825). C: detection of skinCd in strain 630Δerm during vegetative growth in TY medium (4 h). The gel shows the PCR products obtained with the following primer pairs: lane 1, PCR recombined sigK (qRTBD326-IMV833); lane 2, PCR skinCd junction (OBD742-qRTBD326); lane 3, PCR excised form (IMV824-IMV825).
(TIF)
The spores from both strains were purified as described for Fig 8B. The figure shows whole spores from which the panels of Fig 8B were derived. The yellow arrows point to the lamellar inner coat layers, whereas the red arrows point to the more electrondense spore surface layers.
(TIF)
(DOCX)
Total RNAs were extracted from C. difficile 630Δerm and 630Δerm ΔskinCd strains, after 10 h, 14 h, 18 h, 20 h and 24 h of growth in SM medium. After reverse transcription, specific cDNAs were quantified by qRT-PCR using the DNApolIII gene for normalization.
(DOCX)
Acknowledgments
We are thankful to Marc Monot for helpful discussions and to Olga Soutourina for her help with the kinetics of sporulation.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by “Fundação para a Ciência e a Tecnologia” [grant Pest-C/EQB/LA0006/2011 to AOH], by program IF of “Fundação para a Ciência e a Tecnologia” [IF/00268/2013/CP1173/CT0006 to MS], by the “Institut Pasteur”, by the “Université Paris 7” and by the European Union Marie Sklodowska Curie Innovative Training Networks [grant number 642068 to AOH and IMV]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–41. [DOI] [PubMed] [Google Scholar]
- 2.Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6(3):212–25. 10.1111/1758-2229.12130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends in microbiology. 2014;22(7):406–16. 10.1016/j.tim.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart GC. The Exosporium Layer of Bacterial Spores: a Connection to the Environment and the Infected Host. Microbiol Mol Biol Rev. 2015;79(4):437–57. 10.1128/MMBR.00050-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190(7):2505–12. 10.1128/JB.01765-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll KC, Bartlett JG. Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annu Rev Microbiol. 2011;65:501–21. 10.1146/annurev-micro-090110-102824 [DOI] [PubMed] [Google Scholar]
- 7.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80(8):2704–11. 10.1128/IAI.00147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janoir C, Deneve C, Bouttier S, Barbut F, Hoys S, Caleechum L, et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun. 2013;81(10):3757–69. 10.1128/IAI.00515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS microbiology reviews. 2012;36(1):131–48. 10.1111/j.1574-6976.2011.00310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68(2):234–62. 68/2/234 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abecasis AB, Serrano M, Alves R, Quintais L, Pereira-Leal JB, Henriques AO. A genomic signature and the identification of new sporulation genes. J Bacteriol. 2013;195(9):2101–15. 10.1128/JB.02110-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol. 2012;14(11):2870–90. 10.1111/j.1462-2920.2012.02841.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hinai MA, Jones SW, Papoutsakis ET. sigmaK of Clostridium acetobutylicum Is the First Known Sporulation-Specific Sigma Factor with Two Developmentally Separated Roles, One Early and One Late in Sporulation. J Bacteriol. 2014;196(2):287–99. 10.1128/JB.01103-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fimlaid KA, Shen A. Diverse mechanisms regulate sporulation sigma factor activity in the Firmicutes. Curr Opin Microbiol. 2015;24:88–95. 10.1016/j.mib.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saujet L, Pereira FC, Henriques AO, Martin-Verstraete I. The regulatory network controlling spore formation in Clostridium difficile. FEMS Microbiol Lett. 2014;358(1):1–10. 10.1111/1574-6968.12540 [DOI] [PubMed] [Google Scholar]
- 16.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, et al. Global analysis of the sporulation pathway of Clostridium difficile. PLoS genetics. 2013;9(8):e1003660 10.1371/journal.pgen.1003660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, Couture-Tosi E, et al. The Spore Differentiation Pathway in the Enteric Pathogen Clostridium difficile. PLoS genetics. 2013;9(10):e1003782 10.1371/journal.pgen.1003782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, et al. Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in Clostridium difficile. PLoS genetics. 2013;9(10):e1003756 10.1371/journal.pgen.1003756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS biology. 2004;2(10):e328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fimlaid KA, Jensen O, Donnelly ML, Siegrist MS, Shen A. Regulation of Clostridium difficile Spore Formation by the SpoIIQ and SpoIIIA Proteins. PLoS genetics. 2015;11(10):e1005562 10.1371/journal.pgen.1005562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano M, Crawshaw AD, Dembek M, Monteiro JM, Pereira FC, de Pinho MG, et al. The SpoIIQ-SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis. Mol microbiol. 2016;100:204–28. 10.1111/mmi.13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MC. Phage-encoded Serine Integrases and Other Large Serine Recombinases. Microbiol Spectr. 2015;3(4). [DOI] [PubMed] [Google Scholar]
- 23.Halberg R, Kroos L. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J Mol Biol. 1994;243(3):425–36. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel B, Kroos L, Poth H, Youngman P, Losick R. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes & development. 1989;3(11):1735–44. [DOI] [PubMed] [Google Scholar]
- 25.de Hoon MJ, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr Biol. 2010;20(17):R735–45. 10.1016/j.cub.2010.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrano M, Gao J, Bota J, Bate AR, Meisner J, Eichenberger P, et al. Dual-specificity anti-sigma factor reinforces control of cell-type specific gene expression in Bacillus subtilis. PLoS genetics. 2015;11(4):e1005104 10.1371/journal.pgen.1005104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haraldsen JD, Sonenshein AL. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol microbiol. 2003;48(3):811–21. [DOI] [PubMed] [Google Scholar]
- 28.Brouwer MS, Allan E, Mullany P, Roberts AP. Draft genome sequence of the nontoxigenic Clostridium difficile strain CD37. J Bacteriol. 2012;194(8):2125–6. 10.1128/JB.00122-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes & development. 1990;4(4):525–35. [DOI] [PubMed] [Google Scholar]
- 30.Pishdadian K, Fimlaid KA, Shen A. SpoIIID-mediated regulation of sigmaK function during Clostridium difficile sporulation. Mol microbiol. 2015;95(2):189–208. 10.1111/mmi.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato T, Samori Y, Kobayashi Y. The cisA cistron of Bacillus subtilis sporulation gene spoIVC encodes a protein homologous to a site-specific recombinase. J Bacteriol. 1990;172(2):1092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe K, Kawano Y, Iwamoto K, Arai K, Maruyama Y, Eichenberger P, et al. Developmentally-regulated excision of the SPbeta prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS genetics. 2014;10(10):e1004636 10.1371/journal.pgen.1004636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Harada K, Ohta Y, Kobayashi Y. Expression of the Bacillus subtilis spoIVCA gene, which encodes a site-specific recombinase, depends on the spoIIGB product. J Bacteriol. 1994;176(3):935–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, et al. Genome-Wide Identification of Regulatory RNAs in the Human Pathogen Clostridium difficile. PLoS genetics. 2013;9(5):e1003493 10.1371/journal.pgen.1003493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popham DL, Stragier P. Binding of the Bacillus subtilis spoIVCA product to the recombination sites of the element interrupting the sigma K-encoding gene. Proc Natl Acad Sci U S A. 1992;89(13):5991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic acids research. 2001;29(11):2205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutherford K, Van Duyne GD. The ins and outs of serine integrase site-specific recombination. Curr Opin Struct Biol. 2014;24:125–31. 10.1016/j.sbi.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MC, Brown WR, McEwan AR, Rowley PA. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans. 2010;38(2):388–94. 10.1042/BST0380388 [DOI] [PubMed] [Google Scholar]
- 39.Fernandes CG, Placido D, Lousa D, Brito JA, Isidro A, Soares CM, et al. Structural and Functional Characterization of an Ancient Bacterial Transglutaminase Sheds Light on the Minimal Requirements for Protein Cross-Linking. Biochemistry. 2015;54(37):5723–34. 10.1021/acs.biochem.5b00661 [DOI] [PubMed] [Google Scholar]
- 40.Zilhao R, Isticato R, Martins LO, Steil L, Volker U, Ricca E, et al. Assembly and function of a spore coat-associated transglutaminase of Bacillus subtilis. J Bacteriol. 2005;187(22):7753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62(2):239–50. [DOI] [PubMed] [Google Scholar]
- 42.Oke V, Losick R. Multilevel regulation of the sporulation transcription factor sigma K in Bacillus subtilis. J Bacteriol. 1993;175(22):7341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs Pro-sigma K processing in Bacillus subtilis. Genes & development. 1991;5(3):456–66. [DOI] [PubMed] [Google Scholar]
- 44.Dekel E, Mangan S, Alon U. Environmental selection of the feed-forward loop circuit in gene-regulation networks. Phys Biol. 2005;2(2):81–8. [DOI] [PubMed] [Google Scholar]
- 45.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15(10):2631–41. 10.1111/1462-2920.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, et al. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci U S A. 2003;100(3):1316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KP, Born Y, Lurz R, Eichenseher F, Zimmer M, Loessner MJ, et al. Inducible Clostridium perfringens bacteriophages PhiS9 and PhiS63: Different genome structures and a fully functional sigK intervening element. Bacteriophage. 2012;2(2):89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Hinai MA, Jones SW, Papoutsakis ET. The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol Mol Biol Rev. 2015;79(1):19–37. 10.1128/MMBR.00025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurka H, Ehrenreich A, Ludwig W, Monot M, Rupnik M, Barbut F, et al. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLoS One. 2014;9(1):e86535 10.1371/journal.pone.0086535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abe K, Yoshinari A, Aoyagi T, Hirota Y, Iwamoto K, Sato T. Regulated DNA rearrangement during sporulation in Bacillus weihenstephanensis KBAB4. Mol microbiol. 2013;90(2):415–27. 10.1111/mmi.12375 [DOI] [PubMed] [Google Scholar]
- 51.Ramaswamy KS, Carrasco CD, Fatma T, Golden JW. Cell-type specificity of the Anabaena fdxN-element rearrangement requires xisH and xisI. Mol microbiol. 1997;23(6):1241–9. [DOI] [PubMed] [Google Scholar]
- 52.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012;150(4):792–802. 10.1016/j.cell.2012.06.036 [DOI] [PubMed] [Google Scholar]
- 53.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nature rev Microbiol. 2015;13(10):641–50. [DOI] [PubMed] [Google Scholar]
- 54.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15(3):443–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods. 2007;70(3):452–64. 10.1016/j.mimet.2007.05.021 . [DOI] [PubMed] [Google Scholar]
- 56.Heap JT, Pennington OJ, Cartman ST, Minton NP. A modular system for Clostridium shuttle plasmids. J Microbiol Methods. 2009;78(1):79–85. 10.1016/j.mimet.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 57.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol. 2011;193(13):3186–96. 10.1128/JB.00272-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 59.Henriques AO, Beall BW, Roland K, Moran CP Jr. Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J Bacteriol. 1995;177(12):3394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Serrano M, Real G, Santos J, Carneiro J, Moran CP Jr., Henriques AO. A negative feedback loop that limits the ectopic activation of a cell type-specific sporulation sigma factor of Bacillus subtilis. PLoS genetics. 2011;7(9):e1002220 PGENETICS-D-10-00494 [pii]. 10.1371/journal.pgen.1002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.George WL, Sutter VL, Citron D, Finegold SM. Selective and differential medium for isolation of Clostridium difficile. J Clin Microbiol. 1979;9(2):214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henriques AO, Melsen LR, Moran CP Jr. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J Bacteriol. 1998;180(9):2285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The alignment was generated using T-coffee (www.tcoffee.org). The color code indicates conservation, as indicated by the scale on the lower right corner. The conserved motifs are indicated, as well as the boundaries between the N- (NTD) and C-terminal (CTD) domains of the recombinase. The black dot indicates the catalytic nucleophile (Ser10) and the brown dots additional catalytic residues in CD1231. The position homologous to residue Glu449 in the ϕC31 recombinase is indicated by a red triangle. The E449K substitution in the ϕ C31 protein results in a “hyperactive” recombinase able to recombine attL and attR in the absence of the RDF.
(TIF)
A: schematic representation of gene inactivation by a type II Intron with an associated Retro-transposon-Activated Marker (RAM) [54]. The group II intron (bracket), originally in pMTL007 (top), carries a RAM element interrupting an ermB determinant (white). The intron was retargeted to CD1231 or CD1234 (blue) by altering the IBS, EBS1 and EBS2 sequences (grey and white stripes; top) by overlapping PCR. Splicing out of the td group I intron from the ermB gene in the RAM restores a functional marker allowing positive selection of mutants following intron integration. Primers used to confirm the integration and orientation of the type II intron are also indicated (bottom). B: chromosomal DNA of EmR C. difficile conjugants obtained during CD1231 inactivation by Clostron and of strain 630Δerm were screened by PCR using primer pairs RAM-F/R to confirm splicing out of the group I intron in the mutant (lane 1 and 2). We also performed PCR using chromosomal DNA of strain 630Δerm and of the CD1231 mutant (lane 3 and 4) with the intron primer EBSu and with a primer in CD1231 (IMV737). To verify the integration of the Ll.LtrB intron into the right gene targets, we further performed PCR using chromosomal DNA of strain 630Δerm and of the CD1231 mutant (lane 5 and 6) with two primers flanking the insertion site in CD1231 (IMV737-IMV736). Chromosomal DNA from the 630Δerm strain corresponded to lane 2, 3 and 6 while chromosomal DNA of the CD1231 mutant corresponded to lane 1, 3 and 5. The smart ladder (Eurogentec) was used as a molecular weight marker. C: chromosomal DNA of EmR C. difficile conjugants obtained during CD1234 inactivation by Clostron and of strain 630Δerm were screened by PCR using primer pairs RAM-F/R to confirm splicing out of the group I intron in the mutant (lane 1 and 2). To verify the integration of the Ll.LtrB intron into the right gene targets, we further performed PCR using chromosomal DNA of strain 630Δerm and of the CD1234 mutant with the intron primer EBSu and with a primer in CD1234 (IMV696) (lane 3 and 4) or with two primers flanking the insertion site in CD1234 (IMV695-IMV696) (lane 5 and 6). Chromosomal DNA from the 630Δerm strain corresponded to lane 2, 4 and 6 while chromosomal DNA of each mutant corresponded to lane 1, 3 and 5. The smart ladder (Eurogentec) was used as a molecular weight marker. D: southern blot analysis of genomic DNA from C. difficile 630Δerm, CDIP526 (CD1231::erm) and CDIP396 (CD1234::erm) mutant strains with an intron probe. Chromosomal DNA (6 μg in each reaction) was digested with HindIII. Southern blot analyses were performed using Amersham ECL Direct Nucleic Acid labelling and detection reagents, in accordance with the manufacturer's guidelines and visualised using Super Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific). The probe was produced by PCR using OBD522 and OBD523 primers (S1 Table), designed within the group II intron sequence.
(TIF)
Panel A: The DNA sequence immediately uptream of the coding region of spoIIID is presented. The transcriptional start site (+1, red) as previously mapped [34], the -10 and -35 promoter elements (green) that match the consensus for σE recognition (represented below the sequence), SpoIIID boxes (yellow), and the start codon of spoIIID, are indicated. Panel B corresponds to an alignment of the SpoIIID box identified upstream of CD1234, sigK and spoIIID. The sequence logo was created from on the WebLogo website (http://weblogo.berkeley.edu) using SpoIIID boxes identified in this study. The asterisk means that the sequence of the SpoIIID2 box is in opposite orientation. Panel C shows the SpoIIID box of B. subtilis as described in Eichenberger et al [19].
(TIF)
A: whole cell extracts were made from E. coli BL21(DE3) derivatives induced with IPTG to produce CD1231-Strep (expected size 59 kDa), CD1234-His6 (15 kDa), or both (as indicated by the «+ » signs). B: the proteins were detected in the extracts by immunoblotting (first three lanes) using anti-Strep (top) or anti-His6 antibodies (bottom). CD1231-Strep was detected as a species migrating around 60 kDa as indicated (the asterisks represent likely degradation products). CD1234-His6 was detected as a species of about 12 kDa, as indicated. The last three lanes of panel B represent the results of a pull down assay, in which CD1231-Strep (lane 4), CD1234-His6 (lane 5) or the two co-produced proteins (lane 6) were incubated with Ni2+-NTA agarose beads. CD1234-His6 alone (lane 5) binds to the beads. Residual binding of CD1231-Strep to the beads (lane 4) was also detected, but retention of the protein was greatly increased in the presence of CD1234-His6 (lane 6). C: the left side of the panel shows the immnoblot analysis of whole cell extracts prepared from E. coli BL21(DE3) strains producing the indicated proteins (« + » signs) individually. The extracts containing Tgl-His6 [39,40] or CD1234-His6 were incubated with the Ni2+-NTA agarose beads after which the extract containing CD1231-Strep was added. Proteins were eluted, resolved by SDS-PAGE and subject to immunoblot analysis with anti-Strep (top) or anti-His6 antibodies (bottom). The asterisks represent likely degradation products. D: diagram of the domain organization of CD1231 (as in Fig 1B) showing the location of a protease-sensitive site at the end of the NTD, in the recombinases from phages C31 and Bxb1 and from transposon TnpX. The calculated size of the full-lenght protein and of two fragments containing the C-terminal end of the protein (CTD and C-terminal extension) is shown. Fragments of about 37 and 40 kDa most likely containing the C-terminal end of CD1231-Strep may interact with CD1234-His6 (see main text for details).
(TIF)
E. coli cells were transformed with plasmids pFT74 and pMS510 and pMS511 corresponding to pFT74R+attP [carrying a recombined sigK gene and containing the attP site in both possible orientations (attP or attP´; see also Fig 7)]. Each E. coli strain containing one of these plasmids was then transformed with plasmids expressing either CD1231-Strep or CD1231S10A-Strep, and CD1234-His6 or both under the control of an IPTG inducible promoter. Plasmids were recovered from E. coli strains and analyzed by digestion with NotI and BamHI. Plasmid DNA from pFT74 and pFT74R+attP were used as controls for non-recombined and recombined sigK, respectively. The size of molecular size marker (in bp) is indicated on the left side of the panels. The “+” sign identifies clones where recombination has occurred (a total of 8 clones were analyzed).
(TIF)
A: strategy of construction of the 630Δerm ΔskinCd strain. A plasmid (pDIA6103-CD1234) carrying the gene CD1234 expressed under the control of a Ptet promoter was transferred into strain 630Δerm by conjugation. After 4 h of growth, CD1234 expression was induced with ATc for 2 h and cells were plated on BHI. The loss of the plasmid was obtained and verified as outlined in the Materials and Methods section. B: verification of the skinCd excision into the chromosome of the strain 630Δerm ΔskinCd after 5 h of growth in TY medium (vegetative cells). Lane 1: PCR recombined sigK (qRTBD326-IMV833); lane 2: PCR skinCd junction (OBD742-qRTBD326); lane 3: PCR excised form (IMV824-IMV825). C: detection of skinCd in strain 630Δerm during vegetative growth in TY medium (4 h). The gel shows the PCR products obtained with the following primer pairs: lane 1, PCR recombined sigK (qRTBD326-IMV833); lane 2, PCR skinCd junction (OBD742-qRTBD326); lane 3, PCR excised form (IMV824-IMV825).
(TIF)
The spores from both strains were purified as described for Fig 8B. The figure shows whole spores from which the panels of Fig 8B were derived. The yellow arrows point to the lamellar inner coat layers, whereas the red arrows point to the more electrondense spore surface layers.
(TIF)
(DOCX)
Total RNAs were extracted from C. difficile 630Δerm and 630Δerm ΔskinCd strains, after 10 h, 14 h, 18 h, 20 h and 24 h of growth in SM medium. After reverse transcription, specific cDNAs were quantified by qRT-PCR using the DNApolIII gene for normalization.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.