Abstract
Radiation therapy can be an effective way to kill cancer cells using ionizing radiation, but some tumors are resistant to radiation therapy and the underlying mechanism still remains elusive. It is therefore necessary to establish an appropriate working model to study and monitor radiation-mediated cancer therapy. In response to cellular stress, the metabolome is the integrated profiling of changes in all metabolites in cells, which can be used to investigate radiation tolerance mechanisms and identify targets for cancer radiation sensibilization. In this study, using 1H nuclear magnetic resonance for untargeted metabolic profiling in radiation-tolerant mouse melanoma cell line B16, we comprehensively investigated changes in metabolites and metabolic network in B16 cells in response to radiation. Principal component analysis and partial least squares discriminant analysis indicated the difference in cellular metabolites between the untreated cells and X-ray radiated cells. In radiated cells, the content of alanine, glutamate, glycine and choline was increased, while the content of leucine, lactate, creatine and creatine phosphate was decreased. Enrichment analysis of metabolic pathway showed that the changes in metabolites were related to multiple metabolic pathways including the metabolism of glycine, arginine, taurine, glycolysis, and gluconeogenesis. Taken together, with cellular metabolome study followed by bioinformatic analysis to profile specific metabolic pathways in response to radiation, we deepened our understanding of radiation-resistant mechanisms and radiation sensibilization in cancer, which may further provide a theoretical and practical basis for personalized cancer therapy.
Introduction
Ionizing radiation is the main cause of death for cancer cells in radiation therapy, but many cancers for example melanoma are not sensitive to radiation therapy, resulting in poor clinic effects [1, 2]. Radiotherapy is one of the most important methods of cancer treatment along with surgery and chemotherapy. Radiotherapy was administered to shrink tumor in advanced melanoma, or prevent tumor relapse after surgical treatment [3, 4].
In addition to killing cancer cells, radiation therapy also leads to damage of normal cells and tissues. It is therefore an urgent medical concern to protect the normal cells in addition to killing cancer cells as many as possible. To this end, studies on radiosensitization are receiving more and more attention in radiobiology [5, 6]. As a comprehensively used reagent, a radiosensitizer can facilitate the sensitivity of cancer cells in response to radiation, which accordingly promotes the effects of therapy by increasing radiation-mediated cancer cell death [7, 8]. Initially, radiosensitizers were used in radiation-resistant anaerobic cells in solid tumors, but this application has now extended to other cell types in cancers [9]. Though a radiosensitizer is effective in increasing the effectiveness of radiation therapy for cancer, most radiosensitizers are chemotherapy drugs, with unavoidable toxicity for normal cells. Therefore, the search for low-toxic and highly effective radiosensitization is an urgent need in tumor radiation therapy.
The nuclear magnetic resonance (NMR) spectrum–based cellular metabolome analysis is a novel method to assist radiosensitization study in tumor radiation therapy. 1H NMR analysis is an ideal high throughput method to detect small-metabolites in biological samples [10, 11]. With pattern recognition, 1H-NMR-based metabolome analysis screens differentially expressed metabolites between different samples [12]. Followed by bioinformatics analysis on the basis of a public database, the changed metabolic pathways can be further identified.
In this study, using radiation-tolerant mouse melanoma cell line B16 as research model [13], cells were first radiated with sublethal doses of X-ray radiation; then cellular metabolites were collected, which were further analyzed by 600 MHz 1H NMR to screen differentially expressed metabolites. Following bioinformatic analysis for those quantified metabolites, potential targets of disturbed metabolic pathway can be used for radiosensitization study to discover high efficacy drugs for sensitization in radiotherapy. The underlying mechanism of metabolome changes and radiation resistance in B16 cells was discussed.
Experimental Procedures
Cell line and cell culture
Mouse melanoma cell line B16 (obtained from China Center for Type Culture Collection, Wuhan University, Wuhan, China) was maintained in a complete RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% Penicillin-Streptomycin. After a sublethal dose of X-ray radiation (18 Gy) using RS2000 X-ray irradiator (Rad Source Technologies Inc., GA, U.S.), 1×106 cells were subcultured in 25 cm2 plates in 5% CO2 at 37°C. Untreated cells cultured in the same conditions were used as control cells. Cells were harvested after 48 h culture for further experiments. The experiment was repeated 3 times.
Cellular metabolites collection
One-step quenching and extraction were applied as previously described to cell samples [14, 15]. Briefly, after the removal of cell culture medium, the cells were washed twice with ice-cold PBS, and then the pellets were quenched by 2 mL −20°C methanol (HPLC grade, Kermel Chemical Reagent, Tianjin, China). Afterwards, the cells were scraped and transferred into a 15-mL centrifugation tube, followed by adding 2 mL −20°C chloroform and ice-cold ddH2O (V/V/V 1:1:0.7). Cell lysates were mixed by vortex for 5mins and left stand for 15 mins, and then centrifuged with 14,000 g for 30 min at 4°C, generating two-phase extraction. The aqueous phase was lyophilized and then dissolved into 450 μL D2O with 50 μL buffer (1.5 M K2HPO4, 0.375 M NaH2PO4, 0.1% TSP, 0.2% NaN3, pH 7.4) [16]. After being mixed with vortex, undissolved substances were removed by 14,000 × g centrifugation for 5 mins at 4°C and the supernatant was transferred into a 5-mm nuclear magnetic tube for NMR analysis.
1H Nuclear magnetic spectra collection
1H NMR analysis was performed by a Bruker AVANCE III 600 MHz NMR spectrometer at 298.15K. The samples were analyzed by pre-saturated pressed water peak pulse sequence noesypr1d ([RD-90°-t1-90°-tM-90°-ACQ]). Relaxation delay was set as 3 s, t1 4 μs, tM 120 ms, sampling time 1.64 s. Two hundred fifty-six free induction decays (FIDs) were collected by TOPSPIN software. The data point at 32 K was kept and adjusted to 64 K with spectral width 10 kHz. Spectral data were acquired from Fourier transform, by a window from all FIDs multiplying exponential function of line width 1 Hz.
Spectral data preprocessing
Phase and baseline of all NMR spectra were adjusted by MestReNova software, and the TMSP (Trimethylsilylpropanoic acid) peak was set as 0 ppm. The text was derived after calculating piecewise integration, with 0.002 ppm as the interval. Data preprocessing was performed with self-prepared MATLAB script. Data points within 0.6–9.5 ppm were kept and the peak water interval (4.5–5.2 ppm) was removed. For each spectrum, 4450 bins were kept. To reduce the difference due to different sample concentration, the dilution factor of each sample was calculated according to a control spectrum after probabilistic quotient normalization (PQN) [17]
Pseudo two-dimensional spectra were drawn, based on preprocessing the data of statistical total correlation spectroscopy (STOCSY), which indicated correlation factors among each chemical shift [18]. To reduce the ratio of false positives, the threshold of the correlation factor r was calculated by an adjusted p value and number of varieties by Bonferroni.
Dimensionality reduction and pattern recognition analysis
Dimensionality reduction and pattern recognition analysis was carried out by imputing preprocessed data into the software SIMCA-P+ (Ver. 12.0, Umetrics, Umeå, Sweden). To reduce the difference due to different sample concentrations, data were converted by Pareto [19].
Firstly, principal component analysis (PCA) was applied to perform data-dimensionality reduction and get a data preview [20]. Afterwards, we use partial least squares discriminant analysis (PLS-DA), a supervised pattern recognition method, to determine the modeling that generated the greatest difference between radiated sample and untreated sample. Grouping information used as response variable Y in calculating PLS-DA model. Q2 and R2were generated by software calculation to evaluate the degree of fit and forecasting ability of model. To avoid over-fitting, 7-fold cross-validation was performed for 400 repeats [21–23].
The validated model was formally established by orthogonal projections to latent structures discriminant analysis (OPLS-DA), significant components were separated into one predictive component, t1, to describe the differences and one or more orthogonal components to filtering the irrelevant noise, the two datasets may be the most distinguished on score chart [24]. After exporting the model correlation factor p(corr) and variable importance projection (VIP) as well as the load value, the loading diagram was drawn after backtracking transformation, and then differential metabolites were identified.
Univariate analysis
Univariate analysis was performed on those differential metabolites identified by pattern recognition and substances identified by STOCSY. The relative concentration of differential metabolites was acquired by calculating the integration of spectra peaks. According to the p value calculated by the Mann–Whitney test in GraphPad Prism 6 (Ver. 6.01, GraphPad Software, Inc., CA, U.S.), the statistical significance of the differential metabolite was further verified.
Pathway enrichment analysis
The names of the metabolites and their KEGG IDs were matched using MetaboAnalyst (http://www.metaboanalyst.ca/) [25]. Pathway enrichment analysis was performed using MBRole [26]. The relevant pathways were acquired by inputting metabolite KEGG ID and running a Hypergeometric Test. The metabolic pathway illustration was generated by Cytoscape and MetScape [27].
Results
Radiation changed the metabolic profiling of B16 cells
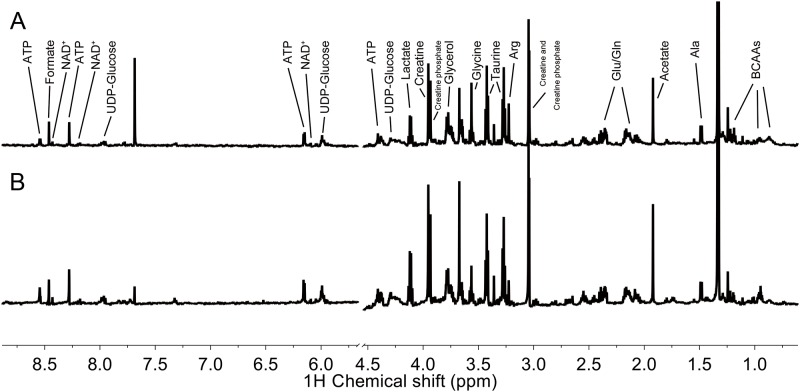
To identify the representative metabolites in B16 cells in response to radiation, we firstly compared 1H NMR spectra from X-ray radiated B16 cells and untreated control cells. The representative spectrum of cellular soluble metabolite from X-ray radiated B16 cells and untreated control cells was indicated in Fig 1A and 1B, where the spectrum peak with δ0.5–8.6 ppm chemical shift was kept and water peak was removed. With ChenomxNMR Suite software analysis and statistical total correlation spectroscopy (STOCSY), major metabolites in the spectrum were confirmed. Chemical shifts and corresponding groups were summarized in Table 1.
Fig 1. Representative spectra and peak assignment.
A: NMR spectra of irradiated B16 cells; B: NMR spectra of untreated B16 cells. Spectrum peak with chemical shift δ 0.5–8.6 ppm was kept and water peak as removed. The main metabolites indicated in the spectra were identified by Chenomx NMR Suite software and statistical total correlation spectroscopy (STOCSY).
Table 1. Peak assignments and integral regions.
| Metabolite | Assignment | δ (ppm) | Untreated | Irradiated | |||
|---|---|---|---|---|---|---|---|
| From | To | Mean | SD | Mean | SD | ||
| Acetate | α-CH3 (d) | 1.93 | 1.91 | 448.79 | 50.31 | 341.49 | 61.04 |
| Ala | β-CH3 (d) | 1.50 | 1.47 | 155.38 | 27.86 | 177.45 | 7.02 |
| Asp | α-CH (dd) | 2.69 | 2.64 | 146.49 | 17.89 | 141.64 | 20.93 |
| β-CH2 (dd) | 2.71 | 2.69 | 40.42 | 12.33 | 37.25 | 10.26 | |
| β-CH2 (dd) | 2.84 | 2.79 | 49.80 | 7.61 | 47.41 | 3.81 | |
| ATP | 5’-CH2 (m) | 4.44 | 4.40 | 284.96 | 30.21 | 207.76 | 16.93 |
| 1’-CH (d) | 6.17 | 6.14 | 205.79 | 9.59 | 170.23 | 18.21 | |
| 2-CH (s) | 8.29 | 8.27 | 143.62 | 14.75 | 117.74 | 9.66 | |
| Choline | N-CH3 (s) | 3.23 | 3.22 | 100.98 | 9.95 | 203.64 | 9.29 |
| Creatine | N-CH3 (s) | 3.04 | 3.03 | 503.70 | 37.06 | 435.52 | 58.59 |
| α-CH2 (s) | 3.94 | 3.92 | 387.04 | 17.13 | 312.79 | 25.32 | |
| CP | N-CH3 (s) | 3.06 | 3.04 | 927.33 | 103.48 | 740.98 | 68.27 |
| α-CH2 (s) | 3.97 | 3.94 | 624.26 | 33.22 | 509.38 | 41.90 | |
| Formate | H (s) | 8.47 | 8.45 | 113.10 | 17.25 | 96.75 | 16.06 |
| Glu | β-CH2 (m) | 2.10 | 2.02 | 329.62 | 43.42 | 327.81 | 37.01 |
| γ-CH2 (m) | 2.38 | 2.32 | 402.98 | 14.69 | 429.76 | 22.17 | |
| Glucose | 1-CH (d) | 5.25 | 5.23 | 28.29 | 9.19 | 11.23 | 9.49 |
| Glutamine | γ-CH2 (m) | 2.48 | 2.43 | 150.35 | 21.21 | 148.91 | 20.88 |
| Glutathione | Glu-γ-CH2 (m) | 2.59 | 2.54 | 220.70 | 33.58 | 214.76 | 28.23 |
| Cys-β-CH2 (m) | 3.01 | 2.95 | 104.20 | 22.41 | 115.49 | 26.55 | |
| Glycerol | CH2 (m) | 3.56 | 3.54 | 121.56 | 19.39 | 139.28 | 29.26 |
| CH2 (m) | 3.59 | 3.57 | 124.95 | 5.70 | 144.05 | 24.38 | |
| CH2 (m) | 3.67 | 3.64 | 337.51 | 44.32 | 394.20 | 32.42 | |
| Glycine | α-CH2 (s) | 3.57 | 3.56 | 135.84 | 3.69 | 240.80 | 11.82 |
| Ile | β-CH3 (d) | 1.03 | 1.00 | 43.81 | 8.75 | 36.68 | 13.68 |
| Lactate | CH3 (d) | 1.35 | 1.31 | 2737.81 | 98.25 | 1950.38 | 238.82 |
| CH (q) | 4.14 | 4.09 | 527.75 | 36.47 | 390.44 | 49.93 | |
| Leu | γ-CH3 (dd) | 0.98 | 0.95 | 176.43 | 9.18 | 105.66 | 9.56 |
| CH2 and CH (m) | 1.77 | 1.71 | 149.40 | 20.52 | 54.86 | 27.83 | |
| Methanol | CH3 (s) | 3.37 | 3.35 | 262.02 | 145.77 | 92.50 | 28.61 |
| Pyruvate | CH3 (s) | 2.40 | 2.39 | 95.97 | 14.72 | 84.79 | 14.17 |
| Succinate | 2 × CH2 (s) | 2.41 | 2.40 | 43.69 | 10.14 | 40.02 | 5.73 |
| Taurine | N-CH2 (t) | 3.29 | 3.25 | 844.63 | 35.21 | 851.09 | 33.52 |
| S-CH2 (t) | 3.44 | 3.41 | 842.21 | 26.76 | 828.75 | 35.74 | |
| UDP-glucose | Glucose 4-CH (m) | 3.50 | 3.45 | 127.82 | 33.61 | 73.28 | 19.22 |
| Glucose 6-CH2 (m) | 3.90 | 3.88 | 46.02 | 13.02 | 42.22 | 9.58 | |
(s) singlet, (d) doublet, (t) triplet, (q) quartet, (dd) doublet of doublet, (m) multiplet
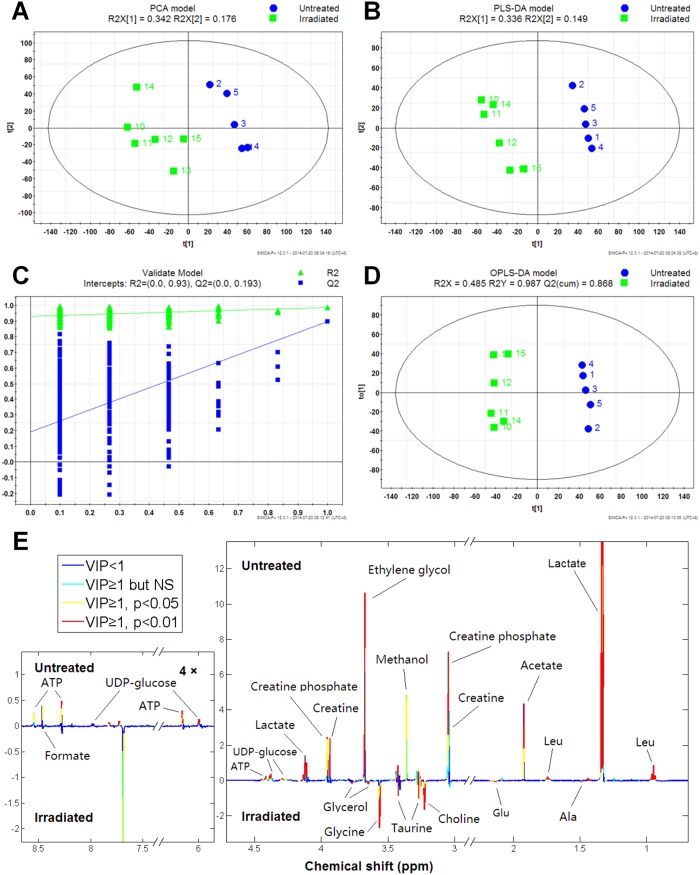
Principal component analysis (PCA) is an unsupervised pattern recognition algorithm, which is able to effectively eliminate the interference of artificial factors and indicates the sample distribution in principal component space. Taking advantage of this, all data were analyzed by PCA. A score plot of the first two principal components are shown in Fig 2A, radiated B16 cells and untreated B16 cells can be distinguished by first two principal components (PC1 and PC2), suggesting a metabolic profiling difference between radiated B16 cells and untreated B16 cells.
Fig 2. Multivariate analysis and back-scaled loading plot.
Differential “metabogram” plotted according to correlation coefficient and VIP value. A: Radiated and untreated B16 cells were distinguished in principal component score chart, which indicated that the metabolic profiling of radiated B16 cells was different with that of untreated cells, t[1] and t[2] are scores on PC1 and PC2, respectively. B: Score chart of partial least squares discriminant analysis model distinguished radiated B16 cells and untreated B16 cells. C: 400 cross validations indicated little over-fitting and a reliable model. D:OPLS-DA model distinguished radiated and untreated B16 cells. E: Differential metabolites were distinguished in radiated and untreated cells according to VIP, relevant index r and loading value.
Next, a supervised pattern recognition algorithm was applied using PLS-DA. As shown in Fig 2B, radiated and untreated cells were able to be distinguished in the principal component in the score chart. After 400 cross-validations, little over-fitting was observed and the data presented good predictive feature in the model (Fig 2C). On the basis of a reliable PLS-DA model, orthogonal partial least squares discriminant analysis (OPLS-DA) model was established by rotated principal–component projection to filter irrelevant information, and the predication of principal components in the samples of two individual groups showed the greatest distinction (Fig 2D). Furthermore, differentially expressed metabolites between the two group samples were distinguished by analyzing the VIP of each variety, the correlation coefficient r, and the loading value. According to VIP and the correlation coefficient r, a metabogram was presented that clearly indicated changes in metabolites in different treatments. In summary, in response to radiation, the contents of leucine, lactate, acetate, creatine, creatine phosphate, methanol, ethylene glycol, UDP-glucose, ATP, and formate were decreased in cells, while the contents of alanine, glutamate, taurine, choline, glycerol, and glycine were increased in cells.
Radiation induced the content change in metabolites in B16 cells
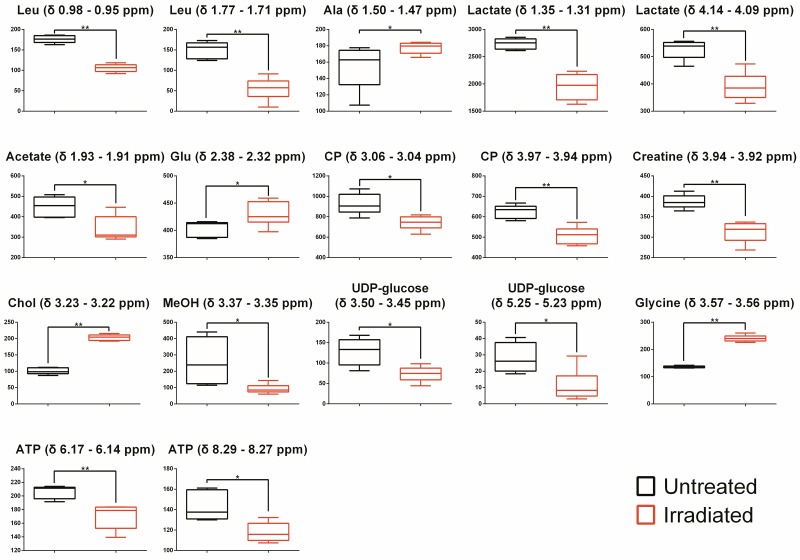
By calculating the spectra peaks of corresponding metabolites, the relative concentration of metabolites was acquired. Single-variable analysis was performed to validate the statistical difference. For each metabolite, 1 or 2 non-overlapping or little-overlapping spectrum peaks were selected as specific spectrum peaks to calculate the integral area by which the relative concentration of each metabolite was acquired (Table 1). The relative concentration of differentially expressed metabolites was performed in a non-parametric statistical test of single variable to generate a p value by the Mann–Whitney test, and the differentially expressed metabolites with statistical significance were indicated in Fig 3 (* p < 0.05, ** p < 0.01). Consistent with the results of multivariable analysis, in response to radiation, the contents of alanine, glutamate, choline and glycine were increased, while the content of leucine, lactate, acetate, creatine, creatine phosphate, methanol, UDP-glucose, and ATP were decreased.
Fig 3. Relative concentration determined by the area under the peaks.
Leu, Leucine; Ala, Alanine; CP, Creatine phosphate; Chol, Choline; MeOH, Methanol. Calculation ofp values were done by the Mann–Whitney test, * p < 0.05, ** p < 0.01.
Radiation changed the metabolic pathways and network in B16 cells
Through pattern recognition analysis and single variable analysis, in total 12 differentially expressed metabolites in B16 cells were identified upon radiation, including 4 content increased metabolites (alanine, glutamate, choline, and glycine) and 8 content-decreased metabolites (leucine, lactate, acetate, creatine, creatine phosphate, methanol, UDP-glucose and ATP). Correspondingly, for those content-changeable metabolites, their KEGG IDs and changes were listed in Table 2.
Table 2. Radiation related metabolites in B16 cells.
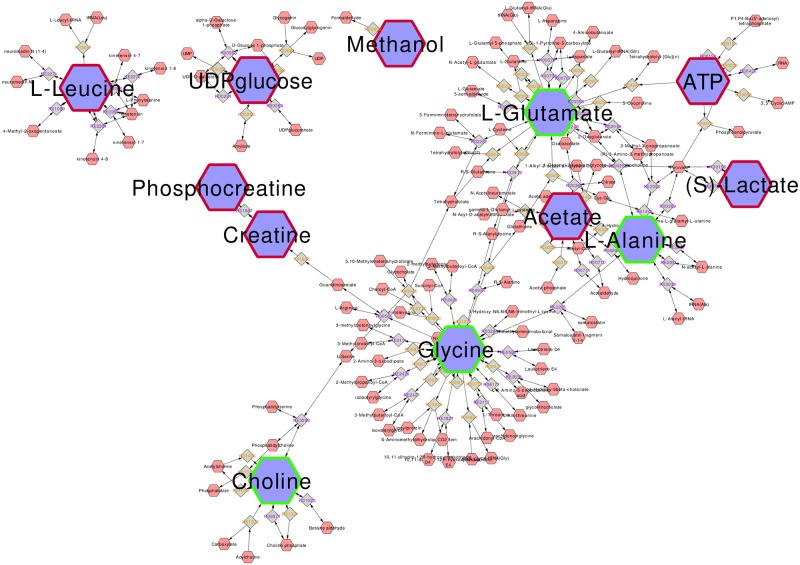
In MBRole, pathway enrichment analysis indicated that differentially expressed metabolites were involved in multiple metabolic pathways, including the metabolism of glycine, taurine, arginine and alanine (Table 3). Using MetScape, metabolic network illustration shows the connection of these metabolites (Fig 4), where hexagons represente metabolites, red frames indicate the decreased metabolites, green frames indicate increased metabolites, squares indicate KEGG ID, rounded rectangles represent enzymes, and blue represents regulative genes.
Table 3. Influenced pathways generated from MBRole by enrichment analysis (adjusted p values< 0.05).
| id | label | pval | adj. pval | in bckgnd | in set | % |
|---|---|---|---|---|---|---|
| mmu02010 | ABC transporters | 6.16E-06 | 3.26E-04 | 90 | 5 | 41.7 |
| mmu01100 | Metabolic pathways | 1.95E-05 | 5.17E-04 | 1455 | 12 | 100 |
| mmu00970 | Aminoacyl-tRNA biosynthesis | 7.71E-05 | 1.36E-03 | 75 | 4 | 33.3 |
| mmu00260 | Glycine, serine and threonine metabolism | 4.85E-04 | 6.42E-03 | 49 | 3 | 25 |
| mmu00430 | Taurine and hypotaurine metabolism | 1.89E-03 | 1.93E-02 | 20 | 2 | 16.7 |
| mmu00330 | Arginine and proline metabolism | 2.19E-03 | 1.93E-02 | 82 | 3 | 25 |
| mmu00250 | Alanine, aspartate and glutamate metabolism | 2.72E-03 | 2.06E-02 | 24 | 2 | 16.7 |
| mmu00910 | Nitrogen metabolism | 3.19E-03 | 2.12E-02 | 26 | 2 | 16.7 |
| mmu00450 | Selenoamino acid metabolism | 4.24E-03 | 2.32E-02 | 30 | 2 | 16.7 |
| mmu00010 | Glycolysis / Gluconeogenesis | 4.53E-03 | 2.32E-02 | 31 | 2 | 16.7 |
| mmu00620 | Pyruvate metabolism | 4.82E-03 | 2.32E-02 | 32 | 2 | 16.7 |
| mmu00680 | Methane metabolism | 5.43E-03 | 2.40E-02 | 34 | 2 | 16.7 |
| mmu00480 | Glutathione metabolism | 6.76E-03 | 2.76E-02 | 38 | 2 | 16.7 |
| mmu04080 | Neuroactive ligand-receptor interaction | 7.73E-03 | 2.93E-02 | 128 | 3 | 25 |
| mmu01110 | Biosynthesis of secondary metabolites | 2.94E-02 | 1.04E-01 | 1023 | 7 | 58.3 |
| mmu00230 | Purine metabolism | 3.64E-02 | 1.21E-01 | 92 | 2 | 16.7 |
Fig 4. Radiation-associated metabolic network generated by MetScape.
Red frames indicate increased metabolites in radiated B16 cells and green frames indicate decreased ones.
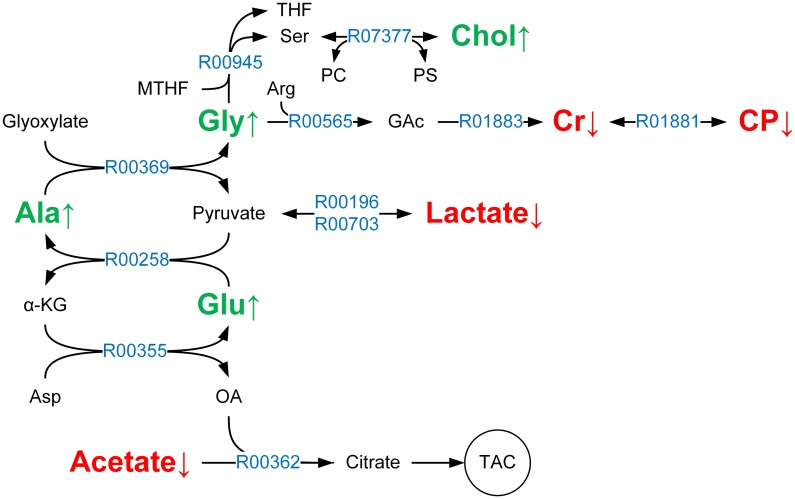
From metabolic pathway and network analysis, we observed that glutamate and pyruvate were converted into alanine and α-ketoglutarate by catalysis of alanine aminotransferase (ALT); Alanine and glyoxalic acid were converted into glycine and pyruvate by catalysis of Alanine-glyoxylate transaminase (AGT) (Fig 5). For those catalytic reactions, key enzymes and the EC number were listed in Table 4.
Fig 5. Altered pathways in radiated B16 cells.
PC, Phosphatidylcholines; PS, Phosphatidylserine; MTHF, 5,10-methylenetetrahydrofolate; THF, Tetrahydrofolate; GAc, Guanidinoacetate; Arg, Arginine; α-KG, α-ketoglutarate, also known as oxoglutarate; Asp, aspartate; OA, Oxaloacetate; TAC, tricarboxylic acid cycle, also known as citrate cycle. Red represents increased metabolites and green represents decreased metabolites in response to radiation.
Table 4. Key enzymes in altered pathways.
| Reaction | Enzyme | Abbr. | EC |
|---|---|---|---|
| R00703 | L-lactate dehydrogenase | LDH | 1.1.1.27 |
| R00196 | L-lactate dehydrogenase (cytochrome) | 1.1.2.3 | |
| R01883 | Guanidinoacetate N-methyltransferase | GAMT | 2.1.1.2 |
| R00945 | Glycine hydroxymethyltransferase | 2.1.2.1 | |
| R00565 | Glycine amidinotransferase | AGAT | 2.1.4.1 |
| R00355 | Aspartate transaminase | AST | 2.6.1.1 |
| R00258 | Alanine transaminase | ALT | 2.6.1.2 |
| R00369 | Alanine-glyoxylate transaminase | AGT | 2.6.1.44 |
| R01881 | Creatine kinase | CK | 2.7.3.2 |
| R07377 | Phosphatidylserine synthase 1 | 2.7.8.- | |
| R00362 | Citrate (pro-3S)-lyase | 4.1.3.6 |
Discussion
To investigate the impacts of radiation, the response of the metabolome in B16 cells to radiation were analyzed. Though metabolome analysis was performed in many clinic tissues or biofluids [28, 29], using cultured cell samples as a metabolic research model has multiple advantages, as not only can one exclude individual differences in clinic samples and tumor tissue heterogeneity, but also they are stable and have a high controllability; therefore, cell samples are appropriate for preliminary studies [30].
Radiation therapy can generate free radicals and reactive oxygen species (ROS) from X-ray radiation, which induces DNA damage in M-phase and G2-phase cells and subsequently blocks cells in the G2/M phase. By inducing cell cycle arrest–mediated apoptosis, cancer cells are killed with radiation therapy [31–33]. Nevertheless, it has been shown that multiple mechanisms lead to cancer tolerance of radiation therapy (e.g., anti-apoptosis [34], DNA damage repair [35], cell cycle regulation [36]). Previous studies indicated that radiation had little impact on the B16 cell cycle, and the observation of γH2AX foci showed that the ratio of DNA double strand break in radiated B16 cells was higher than that of untreated cells [37]. Impaired DNA double-strand break repair by chemical castration in prostate cancer had an improved response to radiotherapy [38]. In this study, the content of glycine in radiated B16 cells was increased. It is known that glycine can react with 5,10-methylenetetrahydrofolate by catalysis of glycine hydroxymethyltransferase to generate serine and tetrahydrofolic acid, and the latter is an important coenzyme of DNA synthesis process. This conclusion suggested that radiation tolerance in B16 cells may arise from DNA damage repair. Accordingly, by activating specific metabolic pathways to block DNA damage repair, radiation-tolerant cancer cells can be killed, which may possibly be an effective way of radiotherapy sensitization.
In response to radiation in B16 cells, the contents of glutamate, alanine, glycine, and choline were increased, which indicated that the activity of aspartate transaminase (AST), alanine transaminase (ALT), and alanine-glyoxylate transaminase (AGT) was elevated. Therefore, we speculate that, with a series of biochemical reactions upon the catalysis of those enzymes, more glycine may be synthesized to participate in DNA damage repair. On the other hand, with the catalysis of phosphatidylcholine (PC) synthase, serine can react with PC to generate choline and phosphatidylserine (PS). Though we did not observe PC and PS in cellular soluble metabolites upon radiation, due to their poor water solubility (no corresponding signal was observed in the hydrogen spectrum), the elevated content of choline in response to radiation indicated that the catalytic reaction occurred.
Though it has been reported that cancer cells, specially cancer stem cells, can synthesize antioxidant substances and reduce the generation of ROS to acquire radiation tolerance [39, 40], in this study, the content of the two antioxidant substances taurine and glutathione did not increase significantly in response to radiation, suggesting that radiation tolerance in B16 cells may have different mechanisms.
Conclusions
With NMR analysis, we observed a series of change in metabolites in response to radiation in B16 cells. Based on previous studies, we concluded that these metabolites are involved in radiation tolerance in B16 cells. Taken together, our results suggested that radiation tolerance in B16 cells may result from the repair of radiation induced DNA damage.
With intensive bioinformatic analysis, NMR based metabolome analysis can be used to identify the specific metabolic pathways in response to radiation, which may provide potential targets for radiotherapy sensitization and furthermore offers technical support and theoretical evidence for personalized radiation therapy.
Supporting Information
Dataset A. NMR spectra using MestReNova software. Dataset B. Associated network generated by MetScape, a Cytoscape network file type of Fig 4. Table A. All pathways generated from MBRole by enrichment analysis, using metabolites in Table 2.
(ZIP)
Acknowledgments
The authors are grateful to Dr. Donghai Lin and Dr. Caihua Huang (College of Chemistry and Chemical Engineering, Xiamen University) for instructions and assistance in data analysis. The authors also want to thank Jinping Gu (College of Chemistry and Chemical Engineering, Xiamen University) for MATLAB script writing. This work was supported, in part, by grants from the National Natural Scientific Foundation of China (Nos. 81430055, 81372452); Programs for Changjiang Scholars and Innovative Research Team in University (No.IRT_15R13); International Cooperation Project of the Ministry of Science and Technology of China (No. 2015DFA31320); Project for Innovative Research Team in Guangxi Natural Science Foundation (2015GXNSFFA139001); Project of Science and Technology of Guangxi (Nos. 14125008-2-12, 1599005-2-10).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The present study was supported, in part, by grants from the National Natural Scientific Foundation of China (nos. 81430055 and 81372452), Programs for Changjiang Scholars and Innovative Research Team in University (No.IRT_15R13), the International Cooperation Project of the Ministry of Science and Technology of China (no. 2015DFA31320), the Project for Innovative Research Team in Guangxi Natural Science Foundation (2015GX NSFFA139001), and the Project of Science and Technology of Guangxi (nos. 14125008-2-12 and 1599005-2-10).
References
- 1.Huber SM, Butz L, Stegen B, Klumpp L, Klumpp D, Eckert F. Role of ion channels in ionizing radiation-induced cell death. Biochim Biophys Acta. 2015;1848(10 Pt B):2657–64. 10.1016/j.bbamem.2014.11.004 . [DOI] [PubMed] [Google Scholar]
- 2.Werthmoller N, Frey B, Ruckert M, Lotter M, Fietkau R, Gaipl US. Combination of ionising radiation with hyperthermia increases the immunogenic potential of B16-F10 melanoma cells in vitro and in vivo. Int J Hyperthermia. 2016:1–8. 10.3109/02656736.2015.1106011 . [DOI] [PubMed] [Google Scholar]
- 3.Hong A, Fogarty G, Izard MA. The role of radiation therapy in the management of metastatic melanoma in the brain. Int J Surg Oncol. 2012;2012:294735 10.1155/2012/294735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibault JE, Dewas S, Mirabel X, Mortier L, Penel N, Vanseymortier L, et al. Adjuvant radiation therapy in metastatic lymph nodes from melanoma. Radiat Oncol. 2011;6:12 10.1186/1748-717X-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams TM, Galban S, Li F, Heist KA, Galban CJ, Lawrence TS, et al. DW-MRI as a Predictive Biomarker of Radiosensitization of GBM through Targeted Inhibition of Checkpoint Kinases. Transl Oncol. 2013;6(2):133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao SL, Akhtar AJ, McKenna KA, Bedi GC, Sidransky D, Mabry M, et al. Selective radiosensitization of p53-deficient cells by caffeine-mediated activation of p34cdc2 kinase. Nat Med. 1996;2(10):1140–3. .8837615 [Google Scholar]
- 7.Purnell PR, Mack PC, Tepper CG, Evans CP, Green TP, Gumerlock PH, et al. The Src inhibitor AZD0530 blocks invasion and may act as a radiosensitizer in lung cancer cells. J Thorac Oncol. 2009;4(4):448–54. 10.1097/JTO.0b013e31819c78fb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biaglow JE, Clark EP, Epp ER, Morse-Guadio M, Varnes ME, Mitchell JB. Nonprotein thiols and the radiation response of A549 human lung carcinoma cells. Int J Radiat Biol Relat Stud Phys Chem Med. 1983;44(5):489–95. . [DOI] [PubMed] [Google Scholar]
- 9.Ni X, Zhang Y, Zennami K, Castanares M, Mukherjee A, Raval RR, et al. Systemic Administration and Targeted Radiosensitization via Chemically Synthetic Aptamer-siRNA Chimeras in Human Tumor Xenografts. Mol Cancer Ther. 2015;14(12):2797–804. 10.1158/1535-7163.MCT-15-0291-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiziani S, Lopes V, Gunther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11(3):269–76, 4p following [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15(2):431–40. 10.1158/1078-0432.CCR-08-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diao C, Zhao L, Guan M, Zheng Y, Chen M, Yang Y, et al. Systemic and characteristic metabolites in the serum of streptozotocin-induced diabetic rats at different stages as revealed by a (1)H-NMR based metabonomic approach. Mol Biosyst. 2014;10(3):686–93. 10.1039/c3mb70609e . [DOI] [PubMed] [Google Scholar]
- 13.Bernhard EJ, Maity A, Muschel RJ, McKenna WG. Effects of ionizing radiation on cell cycle progression. A review. Radiat Environ Biophys. 1995;34(2):79–83. . [DOI] [PubMed] [Google Scholar]
- 14.Teng Q, Huang W, Collette TW, Ekman DR, Tan C. A direct cell quenching method for cell-culture based metabolomics. Metabolomics. 2009;5(2):199–208. [Google Scholar]
- 15.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical chemistry. 2011;83(9):3406–14. 10.1021/ac103313x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao C, Hao F, Qin X, Wang Y, Tang H. An optimized buffer system for NMR-based urinary metabonomics with effective pH control, chemical shift consistency and dilution minimization. The Analyst. 2009;134(5):916–25. 10.1039/b818802e . [DOI] [PubMed] [Google Scholar]
- 17.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Analytical chemistry. 2006;78(13):4281–90. 10.1021/ac051632c . [DOI] [PubMed] [Google Scholar]
- 18.Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, et al. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Analytical chemistry. 2005;77(5):1282–9. 10.1021/ac048630x . [DOI] [PubMed] [Google Scholar]
- 19.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC genomics. 2006;7:142 10.1186/1471-2164-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindon JC, Holmes E, Nicholson JK. Metabonomics techniques and applications to pharmaceutical research & development. Pharmaceutical research. 2006;23(6):1075–88. 10.1007/s11095-006-0025-z . [DOI] [PubMed] [Google Scholar]
- 21.Westerhuis JA, Hoefsloot HC, Smit S, Vis DJ, Smilde AK, van Velzen EJ, et al. Assessment of PLSDA cross validation. Metabolomics. 2008;4(1):81–9. [Google Scholar]
- 22.Szymanska E, Saccenti E, Smilde AK, Westerhuis JA. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics. 2012;8(Suppl 1):3–16. 10.1007/s11306-011-0330-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel J, Krumsiek J, Theis FJ. Statistical methods for the analysis of high-throughput metabolomics data. Computational and structural biotechnology journal. 2013;4:e201301009 10.5936/csbj.201301009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boccard J, Rutledge DN. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Analytica chimica acta. 2013;769:30–9. 10.1016/j.aca.2013.01.022 . [DOI] [PubMed] [Google Scholar]
- 25.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic acids research. 2012;40(Web Server issue):W127–33. 10.1093/nar/gks374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27(5):730–1. 10.1093/bioinformatics/btr001 . [DOI] [PubMed] [Google Scholar]
- 27.Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–80. 10.1093/bioinformatics/btr661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11 10.1186/s40170-016-0151-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duarte IF, Rocha CM, Gil AM. Metabolic profiling of biofluids: potential in lung cancer screening and diagnosis. Expert Rev Mol Diagn. 2013;13(7):737–48. 10.1586/14737159.2013.835570 . [DOI] [PubMed] [Google Scholar]
- 30.Tan J, Hong Y, Su J, Hu N, Lai Z, Lu X, et al. Advances in Detection Technologies of Early cancer. Cell Communications. 2015;2(2):36–45. [Google Scholar]
- 31.Gillette EL, Gillette SM. Principles of radiation therapy. Seminars in veterinary medicine and surgery. 1995;10(3):129–34. . [PubMed] [Google Scholar]
- 32.Borek C. Antioxidants and radiation therapy. The Journal of nutrition. 2004;134(11):3207S–9S. . [DOI] [PubMed] [Google Scholar]
- 33.Lomax ME, Folkes LK, O'Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clinical oncology. 2013;25(10):578–85. 10.1016/j.clon.2013.06.007 . [DOI] [PubMed] [Google Scholar]
- 34.Kruger M, Amort J, Wilgenbus P, Helmstadter JP, Grechowa I, Ebert J, et al. The anti-apoptotic PON2 protein is Wnt/beta-catenin-regulated and correlates with radiotherapy resistance in OSCC patients. Oncotarget. 2016. 10.18632/oncotarget.9013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Zhu T, Gao YF, Zheng W, Wang CJ, Xiao L, et al. Targeting DNA Damage Response in the Radio(Chemo)therapy of Non-Small Cell Lung Cancer. Int J Mol Sci. 2016;17(6). 10.3390/ijms17060839 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen XY, Wang Z, Li B, Zhang YJ, Li YY. Pim-3 contributes to radioresistance through regulation of the cell cycle and DNA damage repair in pancreatic cancer cells. Biochem Biophys Res Commun. 2016;473(1):296–302. 10.1016/j.bbrc.2016.03.099 . [DOI] [PubMed] [Google Scholar]
- 37.Matchuk O, Zamulaeva I, Kovalev O, Saenko A. Radioresistance mechanisms of side population cells in mouse melanoma cell line B16. Cell and Tissue Biology. 2013;7(6):556–62. [PubMed] [Google Scholar]
- 38.Tarish FL, Schultz N, Tanoglidi A, Hamberg H, Letocha H, Karaszi K, et al. Castration radiosensitizes prostate cancer tissue by impairing DNA double-strand break repair. Sci Transl Med. 2015;7(312):312re11 10.1126/scitranslmed.aac5671 . [DOI] [PubMed] [Google Scholar]
- 39.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–3. 10.1038/nature07733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer journal. 2013;19(3):200–7. 10.1097/PPO.0b013e318292e4e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset A. NMR spectra using MestReNova software. Dataset B. Associated network generated by MetScape, a Cytoscape network file type of Fig 4. Table A. All pathways generated from MBRole by enrichment analysis, using metabolites in Table 2.
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.