Abstract
Intravenous (IV) busulfan doses are often personalized to a concentration at steady state (Css) using the patient’s clearance, which is estimated with therapeutic drug monitoring. We sought to identify biomarkers of IV busulfan clearance using a targeted pharmacometabonomics approach. A total of 200 metabolites were quantitated in 106 plasma samples, each obtained before IV busulfan administration in hematopoietic cell transplant (HCT) recipients. Both univariate linear regression with false discovery rate (FDR), and pathway enrichment analyses using the Global test were performed. In the univariate analysis, glycine, N-acetylglycine, 2-hydroxyisovaleric acid, creatine, serine, and tyrosine and were statistically significantly associated with IV busulfan clearance at P<0.05, with the first three satisfying the FDR of q<0.1. Using pathway enrichment analysis, the glycine, serine, and threonine metabolism pathway was the only pathway statistically significantly associated with IV busulfan clearance at P<0.05 and q<0.1, and a pathway impact >0.1. Glycine is a component of glutathione, which is conjugated with busulfan via glutathione transferase enzymes. These results demonstrate the potential utility of pharmacometabonomics to inform IV busulfan dosing. Future studies are required to validate these findings.
Keywords: pharmacometabonomics, metabolomics, busulfan, hematopoietic cell transplant, pharmacokinetics
Graphical Abstract
Introduction
Allogeneic hematopoietic cell transplantation (HCT) offers a curative treatment for a variety of malignant and nonmalignant disorders.1, 2 The alkylating agent busulfan is often part of an HCT conditioning regimen. Recent data has shown improved overall survival in HCT recipients conditioned with intravenous (IV) busulfan compared to total body irradiation (TBI); however, toxicities of the HCT conditioning treatments persist.1–3 Busulfan has a narrow therapeutic index, and plasma exposure, expressed as concentration at steady state (Css) or area under the plasma concentration-time curve (AUC), forecasts the efficacy of busulfan-containing conditioning regimens.4, 5 Low busulfan Css, caused by rapid clearance, is associated with reduced efficacy, e.g., increased risk of relapse6 or rejection,7, 8 while high busulfan Css is associated with hepatotoxicity7, 9–12 and non-relapse mortality (NRM).13 Personalizing busulfan doses to target plasma Css, using a patient-specific busulfan clearance, improves these clinical outcomes.9, 14–17 This process of pharmacokinetic sampling and modeling to personalize busulfan dosing is termed targeted busulfan (TBU), therapeutic drug monitoring or pharmacokinetic (PK)-guided dosing; the latter will be used for the remainder of the manuscript. However, this approach is insufficient as relapse and NRM persist even with PK-guided IV busulfan dosing. Novel biomarkers that can be used to predict IV busulfan clearance before treatment begins, and potentially improve overall survival in HCT recipients, are therefore needed. Identification of such markers could lead to estimating a patient-specific IV busulfan clearance and their personalized dose before IV busulfan administration, reducing the resource intensity of PK-guided dosing of busulfan in HCT conditioning. Furthermore, such analyses could identify novel biomarkers associated with NRM or overall survival.
Substantial insight regarding drug metabolism and response has been gained through the use of metabolomics – the profiling of a broad range of small molecules present in biological fluids.18–24 For example, recent metabolomics studies have provided insight regarding potential biomarkers for graft versus host disease (GVHD) in allogeneic HCT recipients.25, 26 More specifically, Clayton, et al., introduced the concept of personalized drug treatment using pre-dose metabolite profiling to predict drug response in individual subjects, which the authors termed, “pharmacometabonomics”. 22, 23 Against this background, we sought to identify biomarkers predictive of IV busulfan clearance using a targeted pharmacometabonomics approach consisting of 200 metabolites in plasma prior to IV busulfan administration in 106 allogeneic HCT recipients.
Methods
Study population
This was an ancillary retrospective study of 108 subjects who received HCT conditioning with IV busulfan and PK-guided dosing from April 2006 to November 2012 under the aegis of protocols approved by the Fred Hutchinson Cancer Research Center (Fred Hutch) Institutional Review Board. Of the 108 subjects, insufficient sample available for pharmacometabonomics analysis for two subjects, leaving a total of 106 samples for analysis. All subjects were diagnosed with hematologic disorders and had adequate renal (i.e., serum creatinine < 1.5 mg/dl, and creatinine clearance or radioisotope glomerular filtration rate > 60 ml/min/1.73 m2) and liver (i.e., total bilirubin < 1.5 mg/dl and alanine aminotransferase < 300 units/l) function. Demographic data were taken from the subjects’ medical charts [age, sex, height, total body weight (i.e., actual; TBW), dosing weight (calculated as previously described;27), body surface area, and clinical information (disease, conditioning regimen)].
All subjects underwent PK-guided dosing of IV busulfan as part of HCT conditioning. Other conditioning agents included cyclophosphamide (n=70) or fludarabine monophosphate (fludarabine; n=36). Standard practice for prophylaxis of busulfan-induced seizures was phenytoin. Similar antiemetics, antibiotics and antifungals were given per institutional Standard Practice Guidelines. All subjects provided written informed consent before participating in the treatment protocols. The Fred Hutch Institutional Review Board approved both the treatment protocol and the retrospective analysis of samples to identify biomarkers of IV busulfan clearance using pharmacometabonomics.
IV busulfan dosing
Clearance after the first busulfan dose (dose 1), administered in the morning for all subjects, was the primary outcome of interest. The busulfan dose 1 was calculated using TBW if it was less than ideal body weight, or adjusted ideal body weight (AIBW, which equals 0.25 (TBW – ideal weight) + ideal weight) if it was greater than IBW. The IBW in adults was calculated as follows: for males = 50 kg + (2.3 kg for each inch over 5 feet); for females = 45.5 kg + (2.3 kg for each inch over 5 feet). Subsequent IV busulfan doses were personalized to achieve the desired target busulfan Css, chosen for the treatment protocol by the attending physician (i.e., clinician-chosen).
Blood samples (3 ml/sample) were collected in sodium heparin tubes before the morning doses of days 1, 2, and 3 of IV busulfan administration. For those subjects receiving daily IV busulfan, pharmacokinetic samples were drawn at the end of the 3-hour infusion, and at 3.25, 4.5, 6, 8, 11, and 24 hours (i.e., prior to subsequent dose) after the beginning of the infusion.27 For those subjects receiving IV busulfan every 6 hours (Q6h), PK samples were drawn at the end of the 2-hour infusion, and at 2.25, 2.5, 3, 4, 5, and 6, hours (i.e., prior to subsequent dose) after the beginning of the infusion. All samples were stored on wet-ice or refrigerated, and transported to the Seattle Cancer Care Alliance Busulfan Laboratory, a College of American Pathology-certified laboratory that has focused exclusively on PK-guided dosing of busulfan since 1996. Plasma busulfan concentrations were analyzed by gas chromatography with mass-selective detection as previously described.27 The dynamic range was from 1.97 to 4.54 ml/min/kg normal fat mass (NFM; see Supporting Methods for further details)28 and the intraday and interday coefficients of variation were less than 5% and 8%, respectively. After quantitation of busulfan samples, the individual subject’s concentration-time data underwent pharmacokinetic modeling using Phoenix WinNonlin (Certara USA, Princeton, NJ) to obtain each individual’s busulfan area under the curve (AUC) from time 0 to infinity (AUC0 to ∞). After dose 1, the patient-specific clearance and Css were calculated based on the following equations: clearance = dose divided by AUC0-∞ and Css = AUC0-∞ divided by the IV busulfan dosing frequency. After calculation of the patient-specific clearance, the personalized dose was calculated linearly to achieve the target Css and this personalized dose was administered for subsequent doses. The patient-specific clearance was measured after the morning doses of 2 and 3 of IV busulfan administration as well; these data were not used in this analysis. For the present dataset, the concentration-time data underwent noncompartmental analysis and all clearances are reported based on NFM, the optimal body metric for IV busulfan clearance over a population of pediatric to adult allogeneic HCT recipients.28
Targeted pharmacometabonomics sample collection
All samples for pharmacometabonomics analyses were conducted on baseline plasma samples collected prior to IV busulfan administration. Each subject had one sample (i.e., one sample per subject). In 59 subjects, the blood sample was obtained before any conditioning agents were administered (i.e., no conditioning) and blood had been drawn into citrate blood collection tubes (BCTs). In the remaining 47 subjects, samples were drawn after administration of other conditioning agents (n=40 cyclophosphamide/busulfan27 and n=7 fludarabine/busulfan/thymoglobulin)29. For these subjects, the pre-transplant pharmacometabonomics sample had been drawn into an EDTA BCT, refrigerated shortly thereafter at a target temperature of 4°C until transport (within 12 hrs) to the University of Washington/Fred Hutch Pharmacokinetics Laboratory. The sample was subsequently centrifuged and the resultant plasma frozen at −80°C for either cyclophosphamide or fludarabine pharmacokinetic analysis as previously reported.27, 29 The samples underwent two freeze-thaw cycles before the targeted pharmacometabonomics analysis (i.e., the analysis was conducted with the second thaw).
Pharmacometabonomics analysis
Metabolite profiling of plasma was completed at the University of Washington’s Northwest Metabolomics Research Center. Targeted pharmacometabonomics analysis was carried out using a liquid chromatography tandem mass spectrometry (LC-MS/MS) platform in both positive and negative ion modes against 200 standard metabolites (see Supporting Information Table S1) from numerous Kyoto Encyclopedia of Genes and Genomes (KEGG)-defined metabolic pathways30 (e.g., glycolysis, TCA cycle, amino acid metabolism, glutathione, etc.) of potential significance to monitor diet effects, along with 24 internal standards for concentration determinations.31–33 All plasma samples were prepared in batches of 30 samples. A standard protocol was used34–37,33 where 25 μL plasma and 150 μL high performance liquid chromatography (HPLC) grade methanol were combined in an Eppendorf vial and vortexed for 2 min. After 20 min storage at −20 °C the samples were centrifuged at 18,000 g for 10 min. A fixed volume of 150 μL supernatant was collected and placed in a new Eppendorf vial. The protein pellets were mixed with another 300 μL HPLC grade methanol, then vortexed for 10 min and centrifuged for 10 min at 18,000 g. 250 μL was collected and combined with the previous 150 μL sample. Samples were then dried at 30 °C in a SpeedVac for 3 h.
Prior to each LC run, samples were reconstituted with 100 μL 5 mM ammonium acetate in 95% water/5% acetonitrile + 0.5% acetic acid, and filtered through 0.45 μm PVDF filters (Phenomenex, Torrance, CA) prior to analysis on an AB Sciex QTrap 5500 LC-MS/MS system (AB Sciex, Toronto, ON, Canada).35–37 The LC system was composed of two Agilent 1260 binary pumps, an Agilent 1260 auto-sampler and Agilent 1290 column compartment containing a column-switching valve (Agilent Technologies, Santa Clara, CA). Each sample was injected twice, 10 μL for analysis using negative ionization mode and 2 μL for analysis using positive ionization mode. Both chromatographic separations were performed in reverse phase (RP) on Thermo Accucore PFP columns (150 × 2.1 mm, 2.6 μm particle size, Thermo Fisher Scientific Inc., Waltham, MA). The flow rate was 0.250 mL/min, auto-sampler temperature was kept at 4 °C, and the column compartment was set at 40 °C. The mobile phase was composed of Solvents A (5 mM ammonium acetate in water + 0.5% acetic acid + 0.5% acetonitrile) and B (acetonitrile + 0.5% acetic acid + 0.5% water). After chromatographic separation, MS ionization and data acquisition was performed using AB Sciex QTrap 5500 mass spectrometer (AB Sciex, Toronto, ON, Canada) with electrospray ionization (ESI) source. The collision gas was 99.99% pure nitrogen. The data gathered through the multiple reaction monitoring were integrated using MultiQuant 2.1 software (AB Sciex, Toronto, ON, Canada).33 A pooled QC sample was run for every 10 biological samples to assess instrument performance. The intra-assay average CV was 7.8% across all samples.
Statistical analysis
Of the 200 metabolites measured, 118 had detectable signal in all samples and were retained for analysis. The majority of metabolites were skewed to higher values and were therefore log-transformed using the natural logarithm to approximate a normal distribution. A univariate linear regression model was used to assess marginal associations of each metabolite individually on IV busulfan clearance (continuous) after dose 1. Because two different BCTs were used, the effect of BCT type was assessed. Although the results were similar with and without adjustment (data not shown), BCT type was included in the univariate analyses. In our previous evaluation of 1,610 HCT recipients (n=904 male and n=689 female), used to inform the present analysis, we found no effect of gender or weight on IV busulfan clearance, the endpoint of interest.28 However, to ensure that gender or weight did not confound our results, both were tested and found not to affect significance of the metabolites evaluated with the exception of a single metabolite. Homovanilate became significant with adjustment for gender (data not shown), but this would be expected by chance due to the high number of individual tests performed. Therefore, results without adjustment for gender and weight are presented. To determine the percent of variance explained in our model, R2 was calculated including all significant metabolites in a single regression model. Benjamini-Hochberg methods were used to control for false discovery rate (FDR).38 Individual metabolites were considered for both P<0.05 and q<0.1.
To consider metabolites that coordinately predict IV busulfan clearance, pathway analyses using all metabolites were carried out using MetaboAnalyst 3.0 (see Supporting Information Table S2),39, 40 integrating pathway enrichment analysis and pathway topology analysis for visualization. Within the pathway analysis module, metabolites were auto-scaled (mean-centered and divided by the standard deviation of each variable), and IV busulfan clearance was evaluated as a continuous outcome. Four metabolites from our panel, aminoisobutyric acid, cystamine, inositol, and N-acetylneuraminate, were not present in the MetaboAnalyst compound library [derived from KEGG,30 Small Molecule Pathway Database41 (SMPDB)], and Human Metabolome Database42 (HMDB)], and thus were not included in the pathway analyses. The Global test,43 which evaluates changes among groups of metabolites, was used for statistical significance of pathway enrichment analysis, with FDR of q<0.1 for multiple comparisons. While 56 pathways contained at least one metabolite from our panel, only 26 pathways contained 4 or more metabolites, sufficient for meaningful pathway analysis (see Supporting Information Table S2). Betweeness centrality (shortest path between nodes), based on metabolite centrality in a given metabolic network, was used to calculate metabolite importance.44 Pathway impact was calculated as the sum of the importance measures of the pathway-specific metabolites, normalized by the sum of the importance measures of all metabolites in each pathway.45
A post-hoc univariate receiver operating characteristic (ROC) curve was performed on the most significant metabolite, glycine, to evaluate performance as a predictive biomarker using the ROCR package in R.46 For this ROC analysis we defined two groups of subjects based on their IV busulfan clearance being above or below the median. The area under the ROC (AUROC) curve and 95% confidence interval (95% CI) are also reported.
Results
Patient characteristics
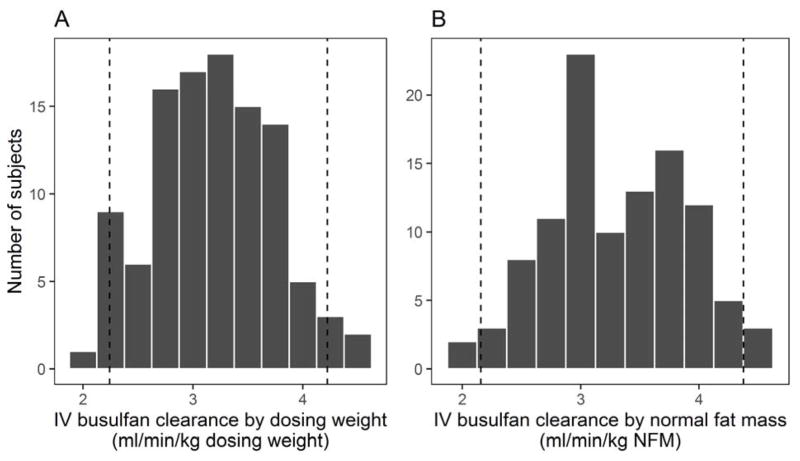
Pre-transplant characteristics and diagnoses of the 106 HCT subjects are given in Table 1. Mean age was 50.4 y (range 22–66), and BMI was 20.0 ± 2.2 kg/m2. Slightly more subjects were male (60%). All samples were collected prior to IV busulfan administration; however, 47 subjects (44%) began other conditioning regimens prior to sample collection. Mean IV busulfan clearance after dose 1 was 3.2 ± 0.5 ml/min/kg of dosing weight and 3.3 ± 0.6 ml/min/kg NFM (Figure 1), which is in agreement with previous studies.10
Table 1.
Demographic and clinical data for the HCT study population (N=106)
| Parameter | Na |
|---|---|
| Age (y) | 50.4 (21.6–65.8) |
| Male sex | 64 (60%) |
| Dosing weight (kg)b | 69.4 ± 11.1 |
| Body mass index [BMI; weight (kg)/height (m)2] | 20.0 ± 2.2 |
| Normal fat mass (NFM; kg) | 67.6 ± 13.9 |
| HCT Conditioningc | |
| Cyclophosphamide/Busulfan | 67 (63%) |
| Busulfan/Cyclophosphamide | 3 (3%) |
| Fludarabine/Busulfan | 27 (25%) |
| Fludarabine/Busulfan/Thymoglobulin | 9 (8%) |
| Busulfan dosing frequency | |
| Every 6 hours | 11 (10%) |
| Every 24 hours | 95 (90%) |
| IV busulfan clearance (ml/min/kg NFM) | 3.33 ± 0.59 |
| Diagnosis | |
| Aplastic anemia | 1 (1%) |
| Acute lymphoblastic leukemia | 1 (1%) |
| Acute myeloid leukemia | 45 (42%) |
| Chronic myeloid leukemia (CML) | 4 (4%) |
| Chronic myelomonocytic leukemia | 2 (2%) |
| Myelodysplastic syndrome (MDS) | 21 (20%) |
| MDS/CML | 1 (1%) |
| Myelofibrosis | 26 (25%) |
| Myeloproliferative disease | 5 (5%) |
| Blood collection tube | |
| Citrate | 59 (56%) |
| EDTA | 47 (44%) |
| Drugs present in sample | |
| None | 59 (56%) |
| Cyclophosphamide | 40 (38%) |
| Fludarabine | 7 (7%) |
Data presented as: number (%) or mean ± standard deviation; percentages may not add up to 100 due to rounding
Total body weight was used for busulfan dosing if total body weight was less than ideal body weight, whereas adjusted ideal body weight was used if total body weight was greater than ideal body weight.
Listed in administration order; all subjects received PK-guided dosing of busulfan, in which the IV busulfan dose was personalized based on clearance
Figure 1.
IV busulfan clearance of the population, shown by dosing weight (A) and normal fat mass (NFM; B). Dashed vertical lines border 95% of the observed values.
Pharmacometabonomics
In the univariate analysis, six metabolites were statistically significantly associated with IV busulfan clearance at P<0.05: glycine, N-acetylglycine, creatine, and serine, were positively associated with clearance, and tyrosine and 2-hydroxyisovaleric acid, were negatively associated. Three of these metabolites satisfied the FDR of q<0.1: glycine, N-acetylglycine and 2-hydroxyisovaleric acid (Table 2). Inclusion of all six nominally significant metabolites in a single regression model explained ~16% of the variability (R2 =0.16).
Table 2.
Endogenous metabolites significantly associated with IV busulfan clearance by univariate analysis
| Metabolite | Functiona | Directionb | P-value | q-valuec |
|---|---|---|---|---|
| Glycine | Amino acid; involved in biosynthesis of proteins, including glutathione, involved the metabolism of busulfan, as well as purines, heme, bile salts, and creatine; inhibitory neurotrasmitter | + | 0.0006 | 0.08 |
| N-Acetylglycine | Acetylated glycine; important for synthesis, stability and localization of proteins | + | 0.002 | 0.09 |
| 2-Hydroxyisovaleric Acid | Fatty acid; derived from metabolism of valine, leucine and isoleucine; may originate from ketogenesis | − | 0.002 | 0.09 |
| Creatine | Involved in energy production, primarily in muscle; produced from glycine and arginine | + | 0.006 | 0.23 |
| Serine | Amino acid; involved in production of purines and pyrimidines; inhibitory neurotransmitter | + | 0.03 | 0.80 |
| Tyrosine | Amino acid; biosynthesis of proteins; essential component of neurotransmitters; receptor kinases involved in immune signaling | − | 0.03 | 0.80 |
Information pertaining to function is derived from Kyoto Encyclopedia of Genes and Genomes, Human Metabolome Database or PubChem unless otherwise noted
Indicates whether the metabolite was positively or negatively associated with IV busulfan clearance
False discovery rate (Benjamini-Hochberg)
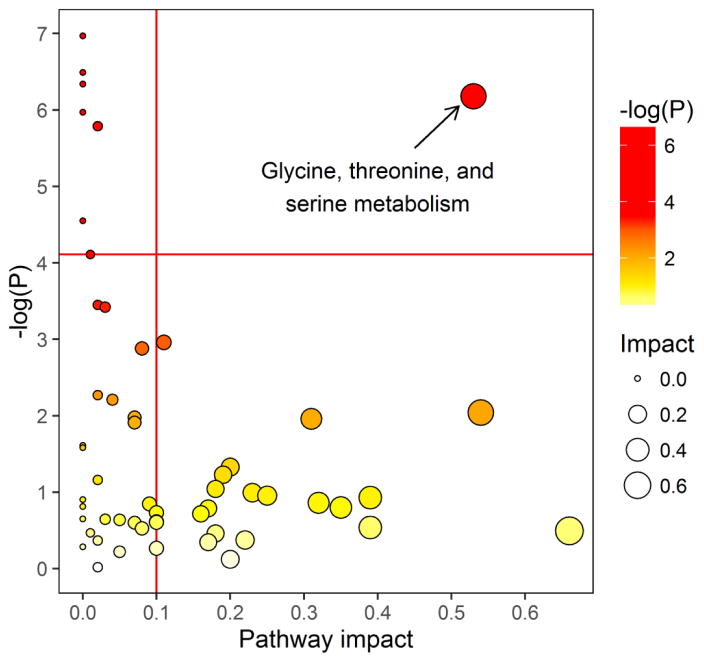
In the pathway enrichment analysis, considering all metabolites together, the only pathway meeting both a statistical significance level of P<0.05 and q<0.1 and any measureable pathway impact factor (e.g., <0.1) was the glycine, serine, and threonine metabolism pathway, which was significantly associated with IV busulfan clearance (P=0.002, FDR=0.028, impact=0.53; Figures 2, 3 and Table 3). Other pathways that were statistically significant at P<0.05 and q<0.1 were thiamine, porphyrin, cyanoamino acid, methane, and glutathione metabolism, but all had pathway low impact values (<0.1). The post-hoc univariate AUROC curve for glycine was 0.66 (95% CI: 0.55, 0.75, Figure 4).
Figure 2.
Overview of pathway enrichment analysis. All dots represent matched pathways from topology pathway analysis. Pathways are colored according to their significance values from pathway enrichment analysis, with gradations from yellow, having the least significance, to red having the highest significance (exact P values are given in Tables 3 and S2). Pathways above the horizontal red line correspond to q<0.1. Pathway impact is indicated on the x-axis. Pathways to the right of the vertical red line on the x-axis have an impact score >0.1. Glycine, threonine and serine metabolism was the only significant pathway with q<0.1 and a pathway impact value >0.1. Other pathways that were significant but had a negligible pathway impact (e.g., <1: thiamin, porphyrin, cyanoamino acid, glutathione, methane and sphingolipid metabolism; all values are given in Table 3) all included glycine.
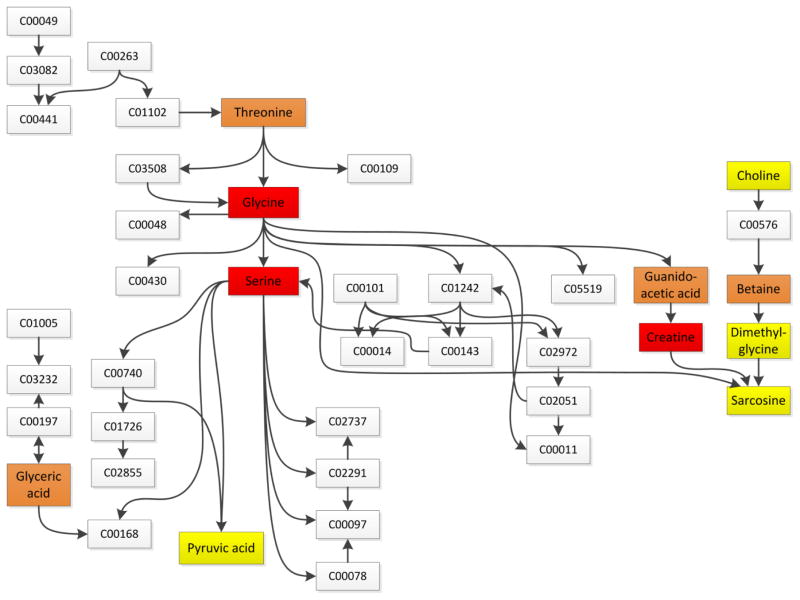
Figure 3.
Glycine, Serine, and Threonine Metabolism pathway. Named metabolites were included in the pharmacometabonomics panel; boxed metabolites shown by letter and number combination show metabolites in the pathway but not included in the metabolite panel. Metabolites are colored according to their significance values from pathway enrichment analysis, with yellow having the least significance (P=0.2–0.8), orange having moderate significance (P=0.03–0.12), and red having the highest significance (P=0.01–0.0006). C00049: aspartic acid; C03082: aspartyl-4-phosphate; C00441: aspartate-semialdehyde; C00197: 3-phospho-D-glycerate; C00168: hydroxypyruvic acid; C03508: 2-amino-3-oxobutanoic acid; C03232: phosphohydroxypyruvic acid; C00740: D-serine; C01726: lombricine; C02855: N-phospho-D-lombricine; C00048: glycoxylic acid; C00263: homoserine; C01102: O-phosphohomoserine; C00430: 5-aminolevulinic acid; C02737: phosphatidylserine; C00014: ammonia; C00078: tryptophan; C00109: 2-ketobutyric acid; C00101: tetrahydrofolic acid; C00143: 5,10-methylene-THF; C02291: cystathionine; C00097: cysteine; S-aminomethyldihydrolipoylprotein; C02972: dihydrolipoylprotein; C02051: lipoylprotein; C00011: carbon dioxide; C05519: allothreonine; C00576: betaine aldehyde; C01242: S-aminomethylidihydrolipoyl-protein; Nine metabolites are omitted from the figure for ease of presentation: C03283: 2,4-diaminobutanoate; C06442: N-gamma-acetyldiaminobuturate; C06231: ectoine; C01005: phosphoserine; C16432: 5-hydroxyectoine; C01888: aminoacetone; C00546: pyruvaldehyde; C03194: 1-aminopropan-2-ol; C05235: hydroxyacetone.
Table 3.
Top pathways, significance and impact from pathway enrichment analyses, sorted by increasing P-values.
| Pathway Name | Total Metabolitesa | Matched Metabolitesb | P-value | −log(P)c | q-valued | Impacte |
|---|---|---|---|---|---|---|
| Thiamine metabolism | 24 | 2 | 0.0009 | 6.97 | 0.028 | 0.0 |
| Porphyrin and chlorophyll metabolism | 104 | 2 | 0.002 | 6.49 | 0.028 | 0.0 |
| Cyanoamino acid metabolism | 16 | 4 | 0.002 | 6.34 | 0.028 | 0.0 |
| Glycine, serine and threonine metabolism | 48 | 11 | 0.002 | 6.18 | 0.028 | 0.53 |
| Glutathione metabolism | 38 | 5 | 0.003 | 5.97 | 0.028 | 0.0 |
| Methane metabolism | 34 | 4 | 0.003 | 5.79 | 0.028 | 0.02 |
| Sphingolipid metabolism | 25 | 1 | 0.01 | 4.55 | 0.07 | 0.0 |
| Sulfur metabolism | 18 | 1 | 0.01 | 4.55 | 0.07 | 0.0 |
| Nitrogen metabolism | 39 | 8 | 0.02 | 4.11 | 0.10 | 0.01 |
| Glycerolipid metabolism | 32 | 1 | 0.03 | 3.45 | 0.17 | 0.02 |
| Primary bile acid biosynthesis | 47 | 4 | 0.03 | 3.42 | 0.17 | 0.03 |
Total number of metabolites in the pathway
Number of matched metabolites, explained in Statistical Analysis section
−log(P) is the negative natural log of the P value for each pathway shown in Figure 2
False Discovery Rate (Benjamini-Hochberg)
Impact is the pathway impact value on IV busulfan clearance calculated from pathway topology analysis
Figure 4.
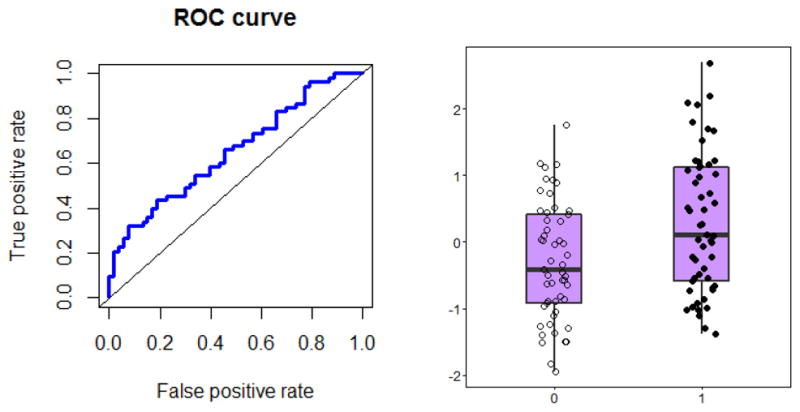
Evaluation of glycine as a predictor for IV busulfan clearance. Area Under the Receiver Operator Characteristic (AUROC) curve for the most promising metabolite, glycine = 0.66 (95% CI: 0.55, 0.75). Box plots represent mean (interquartile range) of low busulfan clearance (0; below the median) and high busulfan clearance (1; above the median).
Discussion
The key findings of this analysis are: 1) glycine, N-acetylglycine, creatine, serine, tyrosine and 2-hydroxyisovaleric acid plasma concentrations were associated with IV busulfan clearance, with the AUROC curve for glycine alone being 0.66; and 2) of the 26 pathways with sufficient metabolites for analysis, the glycine, serine, and threonine metabolism pathway was most highly associated with IV busulfan clearance. In this analysis, we took a first step towards identifying endogenous plasma metabolites associated with IV busulfan clearance with the long-range goal of personalizing IV busulfan doses using biomarkers identified via pharmacometabonomics.
Busulfan is a widely-used alternative to TBI in preparation for HCT.47 A bi-functional anti-neoplastic alkylating agent, busulfan hydrolyzes in aqueous solutions to release methanesulfonate moieties. The resulting reactive carbonium ions alkylate DNA, destroying existing blood cells and remaining cancer cells. While overall survival after HCT is improved with busulfan conditioning, increased efficacy and reduced toxicity of busulfan-based conditioning is needed. It is well known that busulfan has a narrow therapeutic index, with many HCT centers obtaining busulfan pharmacokinetic data in allogeneic HCT recipients.
The patient-specific IV busulfan clearance, not busulfan Css, is the relevant endpoint because the goal with PK-guided dosing is to obtain the patient-specific IV busulfan clearance to be used for dose personalization to achieve the clinician-chosen target busulfan Css. Currently, weight-based dosing of the initial busulfan dose achieves the clinician-chosen target busulfan Css in 22.6% of patients.48 After the initial busulfan dose, serial pharmacokinetic samples are obtained to determine the plasma exposure and the patient-specific IV busulfan clearance (as AUC = dose/clearance). That patient-specific clearance is then used with the clinician-chosen target Css (as Css=AUC/dosing frequency) to personalize the subsequent IV busulfan doses with the intent of achieving the clinican-chosen target Css. This process is referred to as PK-guided dosing or therapeutic drug monitoring (TDM). Personalizing IV busulfan doses using PK-guided dosing results in over 85% of patients achieving the busulfan clinician-chosen target Css at the end of 4-days of busulfan therapy.48 Thus, busulfan Css was not the endpoint of interest because it is confounded by the personalized IV busulfan dose adjustments–made using the patient-specific IV busulfan clearance– to achieve the clinician-chosen target busulfan Css. PK-guided dosing improves clinical outcomes, but it cannot be conducted with the shorter (i.e., <4 day) busulfan courses included in reduced intensity conditioning regimens. Even with the traditional four days of busulfan conditioning, PK-guided dosing is time sensitive and resource-intensive. It is anticipated that ‘omics techniques can improve –or ideally replace–PK-guided busulfan dosing to decrease its resource intensity.
Numerous small studies have evaluated the association of busulfan pharmacokinetics with the constitutional pharmacogenomics of genes regulating the enzymes involved in busulfan disposition.49 The major elimination pathway of busulfan is through conjugation with glutathione to form an unstable S-glutathione sulfonium conjugate γ-glutamyl-β-(S-tetrahydrothiophenium)-alanyl-glycine (GS+THT).50 This reaction is mainly catalyzed by glutathione transferase (GST) isoenzymes A1-1, with GSTM1-1 and GSTP1-1 having minor roles.51, 52 While highly polymorphic, variants in GSTA1 and GSTM1 are not associated with IV busulfan clearance53–55 possibly due to redundancy in function across GST enzymes.56 Therefore, other methods or biomarkers for personalization are needed.
Metabolomics is a commonly used approach for biomarker discovery.57–59 While there are no other studies evaluating IV busulfan clearance in HCT subjects, a few studies have examined the metabolome to identify subsequent clinical outcomes among allogeneic HCT recipients. Reikvam, et al.,26 used metabolite profiling of 766 analytes to evaluate whether pre-transplant metabolic status in 75 HCT subjects was associated with GVHD. Altered pre-transplant levels of several immunoregulatory metabolites, including BCAA and tyrosine derivatives, were found among subjects who later developed GVHD. The authors hypothesized that these metabolites may be involved in the development of GVHD. Another study evaluated 40 thiol/redox metabolites associated with early stages of GVHD between syngeneic and allogeneic HCT recipients. Reduced glutathione was significantly decreased while oxidized glutathione was increased among allogeneic compared to syngeneic recipients as well as non-transplant controls, indicating early shifts in oxidative stress.25 Further, an accumulation of cysteine, cystathione and cysteinylglycine was associated with early GVHD among the allogeneic HCT subjects.25 These studies, as well as our own, highlight the opportunity pharmacometabonomics could offer to improve clinical outcomes in HCT recipients.
In the present study, we analyzed 200 metabolites representing over 25 pathways. Six metabolites measured pre-administration were associated with subsequent IV busulfan clearance. Glycine, N-acetylglycine and 2-hydroxyisovaleric acid remained significant with FDR <0.1. In addition, glycine, serine, and threonine metabolism was significant in pathway enrichment analyses. Other pathways were statistically significant, but were driven mainly by glycine, and contained few metabolites from our panel, such that pathway impact values were negligible (e.g., < 0.02). Glycine is a non-essential amino acid that can be endogenously synthesized from serine, threonine or choline. In addition to roles in the production of purines, bile acids, creatine and heme, glycine is a component of glutathione— which is involved in the metabolism of busulfan.60
It is tempting to speculate that more substrate for glutathione production may be driving the association between glycine and IV busulfan clearance. In fact, glutathione was one of the pathways significantly associated with increased IV busulfan clearance. However, our panel contained only five of the 38 metabolites in the pathway: glycine, pyroglutamic acid, ornithine, glutamic acid and cadaverine, with glycine being the only metabolite to reach statistical significance individually. Further, the other four metabolites were either inversely associated with IV busulfan clearance or only slightly positive. Other components of glutathione consist of the amino acids cysteine and glutamate (see Supporting Information Figure S1). While cysteine was not included in our panel due to difficulty in measuring it by mass spectrometry, glutamate was inversely associated with IV busulfan clearance. This would suggest that increased substrate for the production may not explain the relation between glycine and increased IV busulfan clearance. However, whereas we had 11 of the 48 metabolites represented in the glycine, serine and threonine pathway, only five metabolites were included in the glutathione pathway, which may have been insufficient for a complete evaluation. Nonetheless, we cannot rule out another mode of action contributing to increased IV busulfan clearance, e.g., an altered amino acid pool pre-administration, as all metabolites were amino acids or their derivatives. In addition to amino acids in the glycine, serine, and threonine pathway (glycine, serine and creatine), tyrosine and 2-hydroxyisovaleric acid were negatively associated with IV busulfan clearance. Tyrosine is a non-essential amino acid which can be synthesized from phenylalanine, while 2-hydroxyisovaleric acid is a fatty acid derivative of leucine, a branched chain amino acid (BCAA). Although a decrease in 2-hydroxyisovaleric acid was observed, no associations were found in other BCAA, including isoleucine and valine, or their metabolites. These observations may also reflect dietary or other exposures at the time of measurement.
Strengths of this work include the large population of over 100 HCT subjects, a well characterized IV busulfan pharmacokinetic database, and the targeted panel providing high accuracy of metabolite identification and relative abundances. As with all studies, there were some limitations worth noting. Importantly, the current dataset had insufficient coverage of the glutathione pathway involved in busulfan metabolism. While 13 metabolites in the glutathione pathway were measured, only five had detectable signal in our plasma samples. Thus, greater sensitivity of the eight that did not have detectable signals (i.e., putrescine, spermidine, spermine, cysteine, cysteinylglycine, glutathione, oxidized glutathione, and ascorbic acid) is needed. Having more information about this pathway pre-administration would have provided a more complete picture. Future studies should further focus the targeted analysis on this pathway. In addition not all 118 metabolites had matches in MetaboAnalyst. Four metabolites (i.e., aminoisobutyric acid, cystamine, inositol, N-acetylneuraminate) were therefore not included in pathway enrichment analyses, although none of these metabolites were significant on their own in univariate analyses. Another potential limitation is the two types of BCTs used for sample collection. However, we evaluated associations both with and without adjustment of BCT type and found no differences among the significant metabolites; thus, it is unlikely that this factor had any effect on the results. An analysis of the targeted metabolome with clinical outcomes is also needed. Finally, these finding should be replicated in an independent cohort.
Conclusion
In conclusion, this work demonstrates that glycine, and potentially other metabolites in the glycine, serine, and threonine metabolism pathway predict IV busulfan clearance in HCT subjects. Further studies, including greater interrogation of the glutathione pathway, are needed to validate these results which may have the prospect of personalizing IV busulfan dosing and potentially improve clinical outcomes.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Cancer Institute grants: CA182963, CA18029, and 5P30CA015704. We are also grateful to the physicians, nurses, physician assistants, nurse practitioners, pharmacists, and support staff caring for our patients, and to the patients who participated in this study.
Abbreviations
- HCT
hematopoietic cell transplant
- BMI
body mass index
- NFM
normal fat mass
- TBU
targeted busulfan (PK-guided IV busulfan dosing)
- CY
cyclophosphamide
- FLU
fludarabine monophosphate
- THY
thymoglobulin
Footnotes
Contribution: J.S.M. collected the data; D.R. oversaw the targeted pharmacometabonomics quantitation and edited the manuscript; S.L.N., T.W.R., L.M.S., J.S.M. analyzed the data and made the figures; J.S.M. designed the research; and S.L.N. and J.S.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Supporting Information. Methods: Normal fat mass equations; Table S1. Full list of measured metabolites; Table S2. All 56 metabolic pathways, significance and impact from pathway enrichment analyses; Figure S1. Glutathione synthesis.
References
- 1.Bredeson C, Lerademacher J, Kato K, Dipersio JF, Agura E, Devine SM, Appelbaum FR, Tomblyn MR, Laport GG, Zhu X, McCarthy PL, Ho VT, Cooke KR, Armstrong E, Smith A, Rizzo JD, Burkart JM, Pasquini MC. Prospective cohort study comparing intravenous busulfan to total body irradiation in hematopoietic cell transplantation. Blood. 2013;122(24):3871–8. doi: 10.1182/blood-2013-08-519009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Copelan EA, Hamilton BK, Avalos B, Ahn KW, Bolwell BJ, Zhu X, Aljurf M, van Besien K, Bredeson CN, Cahn JY, Costa LJ, de Lima M, Gale RP, Hale GA, Halter J, Hamadani M, Inamoto Y, Kamble RT, Litzow MR, Loren AW, Marks DI, Olavarria E, Roy V, Sabloff M, Savani BN, Seftel M, Schouten HC, Ustun C, Waller EK, Weisdorf DJ, Wirk B, Horowitz MM, Arora M, Szer J, Cortes J, Kalaycio ME, Maziarz RT, Saber W. Better leukemia-free and overall survival in AML in first remission following cyclophosphamide in combination with busulfan compared to TBI. Blood. 2013;122(24):3863–70. doi: 10.1182/blood-2013-07-514448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieder ML, McDonald GB, Kida A, Hingorani S, Armenian SH, Cooke KR, Pulsipher MA, Baker KS National Cancer Institute-National Heart. Lung and Blood Institute/pediatric Blood and Marrow Transplant Consortium First International Consensus Conference on late effects after pediatric hematopoietic cell transplantation: long-term organ damage and dysfunction. Biol Blood Marrow Transplant. 2011;17(11):1573–84. doi: 10.1016/j.bbmt.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813–26. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 5.McCune JS, Holmberg LA. Busulfan in hematopoietic stem cell transplant setting. Expert Opin Drug Metab Toxicol. 2009;5(8):957–69. doi: 10.1517/17425250903107764. [DOI] [PubMed] [Google Scholar]
- 6.Slattery JT, Clift RA, Buckner CD, Radich J, Storer B, Bensinger WI, Soll E, Anasetti C, Bowden R, Bryant E, Chauncey T, Deeg HJ, Doney KC, Flowers M, Gooley T, Hansen JA, Martin PJ, McDonald GB, Nash R, Petersdorf EW, Sanders JE, Schoch G, Stewart P, Storb R, Appelbaum FR, et al. Marrow transplantation for chronic myeloid leukemia: the influence of plasma busulfan levels on the outcome of transplantation. Blood. 1997;89(8):3055–60. [PubMed] [Google Scholar]
- 7.Slattery JT, Sanders JE, Buckner CD, Schaffer RL, Lambert KW, Langer FP, Anasetti C, Bensinger WI, Fisher LD, Appelbaum FR, et al. Graft-rejection and toxicity following bone marrow transplantation in relation to busulfan pharmacokinetics. Bone Marrow Transplant. 1995;16(1):31–42. [PubMed] [Google Scholar]
- 8.Bolinger AM, Zangwill AB, Slattery JT, Glidden D, DeSantes K, Heyn L, Risler LJ, Bostrom B, Cowan MJ. An evaluation of engraftment, toxicity and busulfan concentration in children receiving bone marrow transplantation for leukemia or genetic disease. Bone Marrow Transplant. 2000;25(2):925–30. doi: 10.1038/sj.bmt.1702371. [DOI] [PubMed] [Google Scholar]
- 9.Grochow LB, Jones RJ, Brundrett RB, Braine HG, Chen TL, Saral R, Santos GW, Colvin OM. Pharmacokinetics of busulfan: correlation with veno-occlusive disease in patients undergoing bone marrow transplantation. Cancer Chemother Pharmacol. 1989;25(1):55–61. doi: 10.1007/BF00694339. [DOI] [PubMed] [Google Scholar]
- 10.Perkins JB, Kim J, Anasetti C, Fernandez HF, Perez LE, Ayala E, Kharfan-Dabaja MA, Tomblyn MR, Sullivan DM, Pidala JA, Field TL. Maximally tolerated busulfan systemic exposure in combination with fludarabine as conditioning before allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2012;18(7):1099–107. doi: 10.1016/j.bbmt.2011.12.584. [DOI] [PubMed] [Google Scholar]
- 11.Dix SP, Wingard JR, Mullins RE, Jerkunica I, Davidson TG, Gilmore CE, York RC, Lin LS, Devine SM, Geller RB, Heffner LT, Hillyer CD, Holland HK, Winton EF, Saral R. Association of busulfan area under the curve with veno-occlusive disease following BMT. Bone Marrow Transplant. 1996;17(2):225–30. [PubMed] [Google Scholar]
- 12.Veal GJ, Nguyen L, Paci A, Riggi M, Amiel M, Valteau-Couanet D, Brock P, Ladenstein R, Vassal G. Busulfan pharmacokinetics following intravenous and oral dosing regimens in children receiving high-dose myeloablative chemotherapy for high-risk neuroblastoma as part of the HR-NBL-1/SIOPEN trial. Eur J Cancer. 2012;48(16):3063–72. doi: 10.1016/j.ejca.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 13.Geddes M, Kangarloo SB, Naveed F, Quinlan D, Chaudhry MA, Stewart D, Savoie ML, Bahlis NJ, Brown C, Storek J, Andersson BS, Russell JA. High busulfan exposure is associated with worse outcomes in a daily i.v. busulfan and fludarabine allogeneic transplant regimen. Biol Blood Marrow Transplant. 2008;14(2):220–8. doi: 10.1016/j.bbmt.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 14.Bolinger AM, Zangwill AB, Slattery JT, Risler LJ, Sultan DH, Glidden DV, Norstad D, Cowan MJ. Target dose adjustment of busulfan in pediatric patients undergoing bone marrow transplantation. Bone Marrow Transplant. 2001;28(11):1013–8. doi: 10.1038/sj.bmt.1703264. [DOI] [PubMed] [Google Scholar]
- 15.Chang C, Storer BE, Scott BL, Bryant EM, Shulman HM, Flowers ME, Sandmaier BM, Witherspoon RP, Nash RA, Sanders JE, Bedalov A, Hansen JA, Clurman BE, Storb R, Appelbaum FR, Deeg HJ. Hematopoietic cell transplantation in patients with myelodysplastic syndrome or acute myeloid leukemia arising from myelodysplastic syndrome: similar outcomes in patients with de novo disease and disease following prior therapy or antecedent hematologic disorders. Blood. 2007;110(4):1379–87. doi: 10.1182/blood-2007-02-076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deeg HJ, Storer B, Slattery JT, Anasetti C, Doney KC, Hansen JA, Kiem HP, Martin PJ, Petersdorf E, Radich JP, Sanders JE, Shulman HM, Warren EH, Witherspoon RP, Bryant EM, Chauncey TR, Getzendaner L, Storb R, Appelbaum FR. Conditioning with targeted busulfan and cyclophosphamide for hemopoietic stem cell transplantation from related and unrelated donors in patients with myelodysplastic syndrome. Blood. 2002;100(4):1201–7. doi: 10.1182/blood-2002-02-0527. [DOI] [PubMed] [Google Scholar]
- 17.Radich JP, Gooley T, Bensinger W, Chauncey T, Clift R, Flowers M, Martin P, Slattery J, Sultan D, Appelbaum FR. HLA-matched related hematopoietic cell transplantation for chronic-phase CML using a targeted busulfan and cyclophosphamide preparative regimen. Blood. 2003;102(1):31–35. doi: 10.1182/blood-2002-08-2619. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham K, Claus SP, Lindon JC, Holmes E, Everett JR, Nicholson JK, Coen M. Pharmacometabonomic characterization of xenobiotic and endogenous metabolic phenotypes that account for inter-individual variation in isoniazid-induced toxicological response. J Proteome Res. 2012;11(9):4630–42. doi: 10.1021/pr300430u. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Krausz KW, Idle JR, Gonzalez FJ. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J Biol Chem. 2008;283(8):4543–59. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao D, Shi X, Wang L, Gosnell BA, Chen C. Characterization of differential cocaine metabolism in mouse and rat through metabolomics-guided metabolite profiling. Drug Metab Dispos. 2013;41(1):79–88. doi: 10.1124/dmd.112.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1(2):153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 22.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ, Everett JR, Nicholson JK. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440(7087):1073–7. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 23.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106(34):14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phapale PB, Kim SD, Lee HW, Lim M, Kale DD, Kim YL, Cho JH, Hwang D, Yoon YR. An integrative approach for identifying a metabolic phenotype predictive of individualized pharmacokinetics of tacrolimus. Clin Pharmacol Ther. 2010;87(4):426–36. doi: 10.1038/clpt.2009.296. [DOI] [PubMed] [Google Scholar]
- 25.Suh JH, Kanathezhath B, Shenvi S, Guo H, Zhou A, Tiwana A, Kuypers F, Ames BN, Walters MC. Thiol/redox metabolomic profiling implicates GSH dysregulation in early experimental graft versus host disease (GVHD) PLoS One. 2014;9(2):e88868. doi: 10.1371/journal.pone.0088868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reikvam H, Hatfield K, Bruserud O. The pre-transplant systemic metabolic profile reflects a risk of acute graft versus host disease after allogeneic stem cell transplantation. Metabolomics. 2015;12(12) doi: 10.1007/s11306-015-0880-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezvani AR, McCune JS, Storer BE, Batchelder A, Kida A, Deeg HJ, McDonald GB. Cyclophosphamide followed by intravenous targeted busulfan for allogeneic hematopoietic cell transplantation: pharmacokinetics and clinical outcomes. Biol Blood Marrow Transplant. 2013;19(7):1033–9. doi: 10.1016/j.bbmt.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCune JS, Bemer MJ, Barrett JS, Scott Baker K, Gamis AS, Holford NH. Busulfan in infant to adult hematopoietic cell transplant recipients: a population pharmacokinetic model for initial and Bayesian dose personalization. Clin Cancer Res. 2014;20(3):754–63. doi: 10.1158/1078-0432.CCR-13-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCune JS, Woodahl EL, Furlong T, Storer B, Wang J, Heimfeld S, Deeg HJ, O’Donnell PV. A pilot pharmacologic biomarker study of busulfan and fludarabine in hematopoietic cell transplant recipients. Cancer Chemother Pharmacol. 2012;69(1):263–72. doi: 10.1007/s00280-011-1736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei R, Li G, Seymour AB. High-throughput and multiplexed LC/MS/MRM method for targeted metabolomics. Anal Chem. 2010;82(13):5527–33. doi: 10.1021/ac100331b. [DOI] [PubMed] [Google Scholar]
- 32.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125(1):76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res. 2014;13:4120–30. doi: 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- 34.Barton S, Navarro SL, Buas MF, Schwarz Y, Gu H, Djukovic D, Raftery D, Kratz M, Neuhouser ML, Lampe JW. Targeted plasma metabolome response to variations in dietary glycemic load in a randomized, controlled, crossover feeding trial in healthy adults. Food Funct. 2015;6(9):2949–56. doi: 10.1039/c5fo00287g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13(3):529–39. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H, Du J, Carnevale Neto F, Carroll PA, Turner SJ, Chiorean EG, Eisenman RN, Raftery D. Metabolomics method to comprehensively analyze amino acids in different domains. Analyst. 2015;140(8):2726–34. doi: 10.1039/c4an02386b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, Cheng PF, Anderson S, Ulrich M, Hurley JB, Raftery D, Ayer DE, Eisenman RN. Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell. 2015;27(2):271–85. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Royal Statist Soc Serial B. 1995;57(1):289–300. [Google Scholar]
- 39.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40(Web Server issue):W127–33. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, Djoumbou Y, Liu Y, Deng L, Guo AC, Han B, Pon A, Wilson M, Rafatnia S, Liu P, Wishart DS. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42(Database issue):D478–84. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(D1):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20(1):93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 44.Aittokallio T, Schwikowski B. Graph-based methods for analysing networks in cell biology. Brief Bioinform. 2006;7(3):243–55. doi: 10.1093/bib/bbl022. [DOI] [PubMed] [Google Scholar]
- 45.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26(18):2342–4. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 46.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 47.Ciurea SO, Andersson BS. Busulfan in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2009;15(5):523–36. doi: 10.1016/j.bbmt.2008.12.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh RF, Pawlikowski MA, Blough DK, McDonald GB, O’Donnell PV, Rezvani A, Deeg HJ, McCune JS. Accurate targeting of daily intravenous busulfan with 8-hour blood sampling in hospitalized adult hematopoietic cell transplant recipients. Biol Blood Marrow Transplant. 2012;18(2):265–72. doi: 10.1016/j.bbmt.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaziev J, Nguyen L, Puozzo C, Mozzi AF, Casella M, Perrone Donnorso M, Gravina P, Sodani P, Marziali M, Isgro A, Simone MD, Andreani M, Formosa A, Testi M, Federici G, Bernardini S, Lucarelli G. Novel pharmacokinetic behavior of intravenous busulfan in children with thalassemia undergoing hematopoietic stem cell transplantation: a prospective evaluation of pharmacokinetic and pharmacodynamic profile with therapeutic drug monitoring. Blood. 2010;115(22):4597–604. doi: 10.1182/blood-2010-01-265405. [DOI] [PubMed] [Google Scholar]
- 50.Younis IR, Elliott M, Peer CJ, Cooper AJ, Pinto JT, Konat GW, Kraszpulski M, Petros WP, Callery PS. Dehydroalanine analog of glutathione: an electrophilic busulfan metabolite that binds to human glutathione S-transferase A1-1. J Pharmacol Exp Ther. 2008;327(3):770–6. doi: 10.1124/jpet.108.142208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Czerwinski M, Gibbs JP, Slattery JT. Busulfan conjugation by glutathione S-transferases alpha, mu, and pi. Drug Metab Dispos. 1996;24(9):1015–9. [PubMed] [Google Scholar]
- 52.Ritter CA, Bohnenstengel F, Hofmann U, Kroemer HK, Sperker B. Determination of tetrahydrothiophene formation as a probe of in vitro busulfan metabolism by human glutathione S-transferase A1-1: use of a highly sensitive gas chromatographic-mass spectrometric method. J Chromatogr B Biomed Sci Appl. 1999;730(1):25–31. doi: 10.1016/s0378-4347(99)00170-x. [DOI] [PubMed] [Google Scholar]
- 53.Zwaveling J, Press RR, Bredius RG, van Derstraaten TR, den Hartigh J, Bartelink IH, Boelens JJ, Guchelaar HJ. Glutathione S-transferase polymorphisms are not associated with population pharmacokinetic parameters of busulfan in pediatric patients. Ther Drug Monit. 2008;30(4):504–10. doi: 10.1097/FTD.0b013e3181817428. [DOI] [PubMed] [Google Scholar]
- 54.Ansari M, Lauzon-Joset JF, Vachon MF, Duval M, Theoret Y, Champagne MA, Krajinovic M. Influence of GST gene polymorphisms on busulfan pharmacokinetics in children. Bone Marrow Transplant. 2010;45(2):261–7. doi: 10.1038/bmt.2009.143. [DOI] [PubMed] [Google Scholar]
- 55.Abbasi N, Vadnais B, Knutson JA, Blough DK, Kelly EJ, O’Donnell PV, Deeg HJ, Pawlikowski MA, Ho RJ, McCune JS. Pharmacogenetics of intravenous and oral busulfan in hematopoietic cell transplant recipients. J Clin Pharmacol. 2011;51(10):1429–38. doi: 10.1177/0091270010382915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830(5):3217–66. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 57.Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP. Targeted metabolomics for biomarker discovery. Angew Chem Int Ed Engl. 2010;49(32):5426–45. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- 58.Mikami T, Aoki M, Kimura T. The application of mass spectrometry to proteomics and metabolomics in biomarker discovery and drug development. Curr Mol Pharmacol. 2012;5(2):301–16. doi: 10.2174/1874467211205020301. [DOI] [PubMed] [Google Scholar]
- 59.Monteiro MS, Carvalho M, Bastos ML, Guedes de Pinho P. Metabolomics analysis for biomarker discovery: advances and challenges. Curr Med Chem. 2013;20(2):257–71. doi: 10.2174/092986713804806621. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Wu Z, Dai Z, Yang Y, Wang J, Wu G. Glycine metabolism in animals and humans: implications for nutrition and health. Amino Acids. 2013;45(3):463–77. doi: 10.1007/s00726-013-1493-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.