Abstract
Background: We have an incomplete understanding of the differences between cancer stem cells (CSCs) in human papillomavirus–positive (HPV-positive) and –negative (HPV-negative) head and neck squamous cell cancer (HNSCC). The PI3K pathway has the most frequent activating genetic events in HNSCC (especially HPV-positive driven), but the differential signaling between CSCs and non-CSCs is also unknown.
Methods: We addressed these unresolved questions using CSCs identified from 10 HNSCC patient-derived xenografts (PDXs). Sored populations were serially passaged in nude mice to evaluate tumorigenicity and tumor recapitulation. The transcription profile of HNSCC CSCs was characterized by mRNA sequencing, and the susceptibility of CSCs to therapy was investigated using an in vivo model. SOX2 transcriptional activity was used to follow the asymmetric division of PDX-derived CSCs. All statistical tests were two-sided.
Results: CSCs were enriched by high aldehyde dehydrogenase (ALDH) activity and CD44 expression and were similar between HPV-positive and HPV-negative cases (percent tumor formation injecting ≤ 1x103 cells: ALDH+CD44high = 65.8%, ALDH-CD44high = 33.1%, ALDH+CD44high = 20.0%; and injecting 1x105 cells: ALDH-CD44low = 4.4%). CSCs were resistant to conventional therapy and had PI3K/mTOR pathway overexpression (GSEA pathway enrichment, P < .001), and PI3K inhibition in vivo decreased their tumorigenicity (40.0%–100.0% across cases). PI3K/mTOR directly regulated SOX2 protein levels, and SOX2 in turn activated ALDH1A1 (P < .001 013C and 067C) expression and ALDH activity (ALDH+ [%] empty-control vs SOX2, 0.4% ± 0.4% vs 14.5% ± 9.8%, P = .03 for 013C and 1.7% ± 1.3% vs 3.6% ± 3.4%, P = .04 for 067C) in 013C and 067 cells. SOX2 enhanced sphere and tumor growth (spheres/well, 013C P < .001 and 067C P = .04) and therapy resistance. SOX2 expression prompted mesenchymal-to-epithelial transition (MET) by inducing CDH1 (013C P = .002, 067C P = .01), followed by asymmetric division and proliferation, which contributed to tumor formation.
Conclusions: The molecular link between PI3K activation and CSC properties found in this study provides insights into therapeutic strategies for HNSCC. Constitutive expression of SOX2 in HNSCC cells generates a CSC-like population that enables CSC studies.
Head and neck squamous cell carcinoma (HNSCC) incidence is increasing in the United States, mostly because of a rise in human papillomavirus (HPV)–positive oropharyngeal HNSCCs (45 780 vs 29 370 new cases in 2015 vs 2005) (1–4). Cancer stem cells (CSCs) maintain an undifferentiated phenotype, are resistant to therapy (5), and repopulate tumor heterogeneity upon recurrence or metastasis (6). These characteristics make CSCs an attractive yet challenging therapeutic target (7). CSCs of HPV-negative HNSCC are most consistently defined by aldehyde dehydrogenase (ALDH) activity (8) or CD44 expression (9), but prior investigations have relied heavily on in vitro studies or single markers to define CSC populations (10).
Phosphoinositide 3 kinase (PI3K) signaling is commonly activated in HNSCC by PIK3CA amplification or mutation (11–13); however, antitumor responses to PI3K inhibition can occur with or without these activating genetic events (14–16). PI3K signaling promotes tumor cell proliferation and survival (17) and is overexpressed in CSCs across different cancers (18,19). PIK3CA oncogenic alterations occur more frequently in HPV-positive compared with HPV-negative HNSCC (56% vs 34%, respectively) (20). The following unanswered questions may explain the differential clinical behavior of these HNSCC subtypes: 1) What markers define CSCs in HPV-positive HNSCC? 2) What are the differences or similarities in signaling pathways between HPV-positive and HPV-negative HNSCC CSCs?
Using HPV-negative and HPV-positive patient-derived xenografts (PDXs), we defined a consistent population of ALDH+CD44high CSCs across HNSCC subtypes. We characterized the core transcriptional profile of CSCs and their susceptibility to anticancer agents, showing that CSCs (HPV-positive and HPV-negative) are resistant to standard therapy but are particularly susceptible to PI3K inhibition. We report how PI3K and mechanistic target of rapamycin (mTOR) signaling through the eukaryotic translation initiation factor 4E (EIF4E) specifically upregulated the sex determining region Y box 2 (SOX2), a transcription factor linked to stemness in squamous cancers (21,22). SOX2 in turn increased the ALDH+ cell population by direct upregulation of ALDH1A1 and enhanced spheroid and tumor formation. This is the first report using a large panel of HNSCC PDX samples to elucidate how CSCs harness deregulated PI3K signaling to orchestrate sustained tumor growth.
Methods
PDX Generation
The protocol for studies involving human subjects was approved by the Colorado Multiple Institutional Review Board (COMIRB #08-0552), and informed written consent was obtained from all patients whose tissues were used for this study. The University of Colorado Institutional Animal Care and Use Committee approved all experiments involving mice. PDX generation and characterization was previously reported (14). Briefly, tumor pieces were implanted on both flanks of five six- to 10-week-old female Athymic Nude-Foxn1nu mice (Envigo, Denver, CO). Patient cases (F4-F8 generation) were expanded into cohorts of 10 to 20 mice for CSC isolation.
CSC Implantation In Vivo
CSCs were sorted into PBS +2% fetal bovine serum (FBS), suspended in 1:1, DMEM +10% FBS:Matrigel (Corning, Corning, NY) to 100 µL per injection, and injected (25 g needle) into the flanks of nude mice. Mice were monitored for tumor growth for up to one year.
SOX2/OCT4 Response Elements (SORE6) Reporter
The SORE6 assay was previously described (23) and consists of six SOX2/OCT4 response elements in front a minimal mCMVp promoter driving the expression of destabilized forms of GFP or mCherry. Lentiviral media containing the mCMVp-GFP (control), mCMVp-mCherry (control), SORE6-mCMVp-GFP, or SORE6-mCMV-mCherry reporters were generated using HEK293T cells and the pCMV-SSV-G and pHR-8.2ΔR packaging plasmids. Cell lines or PDX-derived CSCs were cultured in media containing virus for 24 hours. Cells containing the SORE6 reporter were selected using puromycin (1 µg/mL) for 7 days. Expression of GFP or mCherry was observed by fluorescence microscopy and flow cytometry (gating set at > 99.9% of control cells).
Time-Lapse Microscopy of Tumor-Derived CSCs
SORE6+ CSCs were seeded in chamber slides on a Matrigel layer overlaid with media +2% Matrigel, and imaged every 20 minutes using a Zeiss Axio Observer Z1 inverted microscope (Zeiss, Oberkochen, Germany) (Zeiss software Rel. 4.8) in a climate chamber.
Statistical Analysis
In vitro and in vivo (using ≥5 mice/group) experiments were compared with a two-group t test. Calculations were done using GraphPad Prism version 7.0 and SPSS version 11. Data are represented graphically as mean ± SD. GSEA estimates the statistical significance of the enrichment scores by a two-sided modified Kolmogorov-Smirnov permutation test. P values of less than .05 were statistically significant. All statistical tests were two-sided.
Results
Tumorigenicity of Tumor Populations Defined by ALDH Activity and CD44 Expression
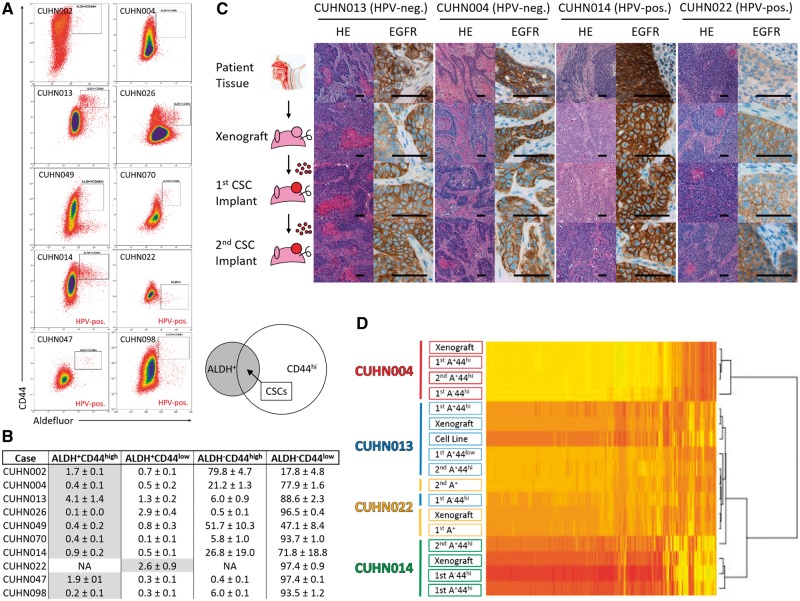
To identify common or distinct CSC populations across subtypes, we studied tumorigenicity in 10 (6 HPV-negative, 4 HPV-positive) HNSCC PDX cases (Supplementary Table 1, available online). Cells were sorted by CD44 expression and Aldefluor activity and implanted on nude mice. The CD44high population was typically larger than the ALDH+ fraction, while the ALDH+CD44high population ranged from 0.1% to 4.1% of tumor cells (Figure 1, A and B). Despite substantial population heterogeneity across cases, ALDH+CD44high defined the most tumorigenic population in nine of the 10 cases; CUHN022 did not express detectable CD44, thus ALDH activity alone defined the tumorigenic (CSC-enriched) population in this particular tumor. Average tumor formation rate (across 10 cases) was 65.8% when 1000 or fewer CSCs (ALDH+CD44high) were implanted, vs 33.1% and 20.0% with 1000 or fewer ALDH-CD44high and ALDH+CD44low cells, and 4.4% when implanting 100 000 negative cells (Table 1). ALDH+CD44high cells met the tumor-initiating criteria of CSCs in both HPV-positive and HPV-negative HNSCCs.
Figure 1.
Evaluation of tumor recapitulation following serial implantation of cancer stem cells (CSCs) in mice. A) Heterogeneous staining of CD44 cell surface protein and aldehyde dehydrogenase activity (Aldefluor assay) across 10 head and neck squamous cell carcinoma (HNSCC) patient-derived xenograft (PDX) cases. Gated regions are representative of the sorted CSC population for each PDX model defined in Table 1. Diagram depicting the CSC population falling within the overlapping region of the larger aldehyde dehydrogenase (ALDH)+ and CD44high populations. B) Tumor cell composition across PDX models measured as percentages (flow cytometry) for each of the four possible (ALDH-CD44low, ALDH-CD44high, ALDH+CD44low, ALDH+CD44high) tumor populations (average of 2+ sorted tumors). C) Tumors resulting from two consecutive CSC implantations for four HNSCC PDX show tumors retain characteristics of the originating tumor, including morphology (hematoxylin and eosin staining) and epidermal growth factor receptor (EGFR) expression with serial implantation. Scale bars = 100 µm. D) Tumors arising from the first or second/serial CSC implantation consistently cluster with the originating PDX tumor (clustering generated with FPKM values > 1). Tumors arising from CUHN004 and CUHN013 non-CSCs (ALDH-CD44high) were more distantly associated with the original PDX tumor. ALDH = aldehyde dehydrogenase; CSC = cancer stem cell; FPKM = fragments per kilobase of exon per million fragments mapped; HNSCC = head and neck squamous cell carcinoma; HPV-neg. = human papilloma virus–negative; HPV-pos. = human papilloma virus–positive; PDX = patient-derived xenograft.
Table 1.
CSC implantation results for 10 HNSCC PDX cases*
| ALDH- | ALDH- | ALDH+ | ALDH+ | Serial passage of CSC populations | |||||
|---|---|---|---|---|---|---|---|---|---|
| CD44low | CD44high | CD44low | CD44high | ||||||
| HPV status | Case | 105 cells | ≤103 cells | ≤103 cells | ≤103 cells | 101 cells | 102 cells | 103 cells | 104 cells |
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||
| HPV-negative | CUHN013 | 1/30 | 8/20 | 10/20 | 13/20 | – | 1/5 | 2/5 | 3/5 |
| (3.3) | (40.0) | (50.0) | (65.0)† | (20.0) | (40.0) | (60.0) | |||
| CUHN004 | 1/25 | 9/30 | 1/20 | 11/20 | – | 2/5 | 2/5 | – | |
| (4.0) | (30.0) | (5.0) | (55.0)† | (40.0) | (40.0) | ||||
| CUHN002 | 0/15 | 11/25 | 3/10 | 9/15 | |||||
| (0.0) | (44.0) | (30.0) | (60.0)† | ||||||
| CUHN026 | 0/10 | 3/10 | 4/10 | 6/10 | |||||
| (0.0) | (30.0) | (40.0) | (60.0)† | ||||||
| CUHN049 | 0/10 | 2/10 | 0/10 | 6/10 | |||||
| (0.0) | (10.0) | (0.0) | (60.0)† | ||||||
| CUHN070 | 0/10 | 0/10 | 0/10 | 7/10 | |||||
| (0.0) | (0.0) | (0.0) | (70.0)† | ||||||
| HPV-neg. | 2/100 | 33/105 | 18/80 | 58/95 | |||||
| (2.0) | (31.4) | (22.5) | (61.0) | ||||||
| HPV-positive | CUHN014 | 3/10 | 7/10 | 0/10 | 10/10 | 2/5 | 5/5 | 5/5 | – |
| (30.0) | (70.0) | (0.0) | (100.0) | (40.0) | (100.0) | (100) | |||
| CUHN022 | 0/10 | – | 6/20 | – | – | 1/5 | 2/5 | 4/5 | |
| (0.0) | (30.0)† | (20.0) | (40.0) | (80.0) | |||||
| CUHN047 | 0/10 | 1/10 | 0/5 | 6/10 | |||||
| (0.0) | (10.0) | (0.0) | (60.0)† | ||||||
| CUHN098 | 1/5 | 2/5 | 0/5 | 5/5 | |||||
| (20.0) | (40.0) | (0.0) | (100.0)† | ||||||
| HPV-pos. | 4/35 | 10/25 | 6/40 | 21/25 | |||||
| (11.4) | (40.0) | (15.0) | (84.0) | ||||||
| Combined | 6/135 | 43/130 | 24/120 | 79/120 | |||||
| (4.4) | (33.1) | (20.0) | (65.8) | ||||||
Single-positive populations (ALDH+ or CD44high) were capable of forming tumors at lower frequency. Tumor take rate and growth of the initial and second (serial) passage of CUHN004, CUHN013, CUHN0014, and CUHN022 tumor-generating populations were similar to that of the first passage. ALDH = aldehyde dehydrogenase; CSC = cancer stem cell; HPV-neg. = human papillomavirus–negative; HPV-pos. = human papillomavirus–positive; HNSCC = head and neck squamous cell carcinoma; PDX = patient-derived xenograft.
Identifies the CSC population for each case.
Morphological and Molecular Marker Comparison of Originating and CSC-Initiated Tumors
We examined morphology, epidermal growth factor (EGFR) expression, and transcriptome profiles over sequential CSC implantations. Hematoxylin and eosin staining showed that morphology of CSC-derived tumors was comparable with the originating tumor; EGFR membrane staining was consistent between patient tissue, PDX tumors, and CSC-initiated tumors (Figure 1C). Single positive cell (defined as ALDH+CD44low or ALDH-CD44high) implantations generated tumors with less differentiated squamous morphology than CSC-originated tumors (Supplementary Figure 1A, available online).
Tumors from CSCs, compared with single positive cells, had the most similar mRNA-seq profiles when compared with the original patient tumor (Figure 1D). To assess tumorigenic stability, we sorted and implanted CSCs from the first passage of CSC-derived tumors and found that tumor take and growth rates were consistent with the first implantation (Table 1; Supplementary Figure 1B, available online). Second passage CSC-derived tumors were also most similar to the original patient tumor (Figure 1, C and D).
Gene Expression Analysis of HNSCC CSCs
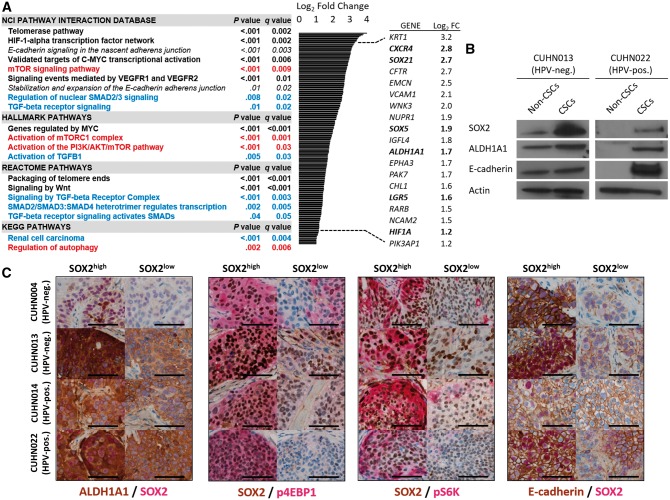
We compared the transcriptome profiles of CSCs and non-CSCs from HPV-negative (CUHN004, CUHN013) and HPV-positive (CUHN014, CUHN022) tumors. (Supplementary Figure 1C, available online). CUHN004 and CUHN022 were PIK3CA wild-type, and CUHN013 and CUHN014 had high-level PIK3CA copy gain. We observed statistically significant enrichment of signaling networks crucial in stem cell and CSC biology across HPV-positive and HPV-negative CSCs, including packaging of telomere ends (P < .001), PI3K/mTOR pathway (P < .001), E-cadherin signaling (P < .001), genes regulated by MYC (P < .001), signaling by wingless-related integration site (WNT; P < .001), and signaling by transforming growth factor beta (TGF-β; P < .001) (Figure 2A; Supplementary Table 2, available online). HPV-negative CSCs had lower p53 signaling compared with HPV-positive CSCs (P < .001), consistent with their respective TP53 mutant and wild-type status (Supplementary Table 3, available online). Upregulation of stemness-related genes in CSCs including SOX21 (P = .005), a mediator of SOX2 gene regulation (24), and ALDH1A1 (P = .003), a marker of CSCs (8,25), were statistically significant across HPV subtypes. SOX2 and ALDH1A1 protein levels were dramatically higher in CSCs (Figure 2B), and ALDH1A1 was associated with cells expressing SOX2 in patient tumors (Figure 2C).
Figure 2.
Transcriptome and protein expression profile of head and neck squamous cell carcinoma (HNSCC) cancer stem cells (CSCs). A) Gene set enrichment analysis (GSEA) pathway enrichment in HNSCC CSC populations compared with cells negative for CSC markers and relevant genes statistically significantly upregulated in CSCs across cases (bold = stem cell–related processes; red = phosphoinositide 3-kinase (PI3K)/ mechanistic target of rapamycin (mTOR)–related pathways; blue = transforming growth factor beta–related pathways). Statistical significance was calculated by a two-sided modified Kolmogorov-Smirnov permutation test. B) Immunoblot and reverse transcription polymerase chain reaction analysis of tumor-derived (CUHN013, CUHN022) CSCs and cells negative for CSC markers. C) Co-expression of SOX2 with aldehyde dehydrogenase (ALDH)1A1, markers of PI3K/mTOR pathway activation and E-cadherin in vivo. Scale bars = 100 µm. CSC = cancer stem cell; HNSCC = head and neck squamous cell carcinoma; HPV-neg. = human papilloma virus–negative; HPV-pos. = human papilloma virus–positive; mTOR = mechanistic target of rapamycin; PI3K = phosphoinositide 3-kinase; TGF-β = transforming growth factor beta.
SOX2-high cells have increased levels of both p4EBP1 and pS6K (effectors of PI3K/mTOR) when compared with SOX2-low cells (Figure 2C), especially in areas near vessels and stroma. E-cadherin showed consistent mRNA and protein enrichment in CSCs, as well as in vivo costaining distribution with SOX2 (Figure 2, A–C), suggesting a more epithelial phenotype.
Response of CSCs to a Panel of Therapies Using an In Vivo Re-implantation Assay
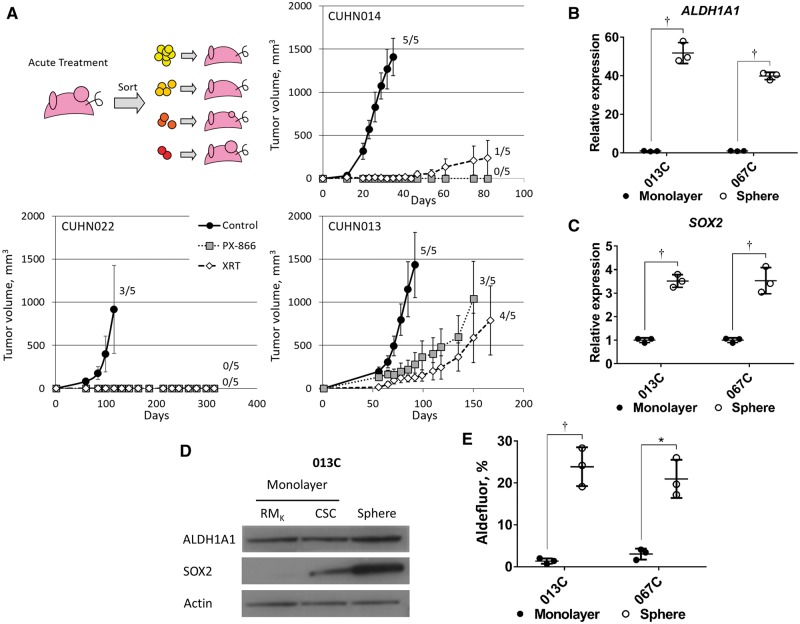
To assess CSC response to therapy in vivo, we treated CUHN013 (HPV-negative), CUHN014 (HPV-positive), and CUHN022 (HPV-positive) tumor-bearing mice with vehicle, radiation therapy (XRT), docetaxel, cetuximab, and the PI3K inhibitors ZSTK474 and PX-866. Tumors were then resected, sorted, and the resulting cell populations (see Table 1) were implanted in mice (Figure 3A). CSCs generated more tumors in control and treatment groups compared with non-CSC populations (Supplementary Table 4, available online), confirming that CSCs are more tumorigenic, with or without treatment, which impacts mouse survival (Supplementary Figure 1D, available online).
Figure 3.
Effects of phosphoinositide 3-kinase inhibition on cancer stem cells (CSCs) and SOX2 expression in cell lines. A) Schema for pretreatment of tumors prior to sorting of CSCs and tumor populations. Average tumor growth of pretreated (control, PX-866 and XRT) CSC populations (CUHN014 = 10 aldehyde dehydrogenase [ALDH]+CD44high cells, CUHN022 = 10 000 ALDH+ cells, CUHN013 = 10 000 ALDH+CD44high cells) was substantially decreased with PX-866 or XRT. Numbers represent tumor take rate for implantations in five mice (tumor volume > 500 mm3). B) Expression of ALDH1A1 and (C) SOX2 are upregulated in sphere culture (013C, 067C cells) when compared with cells grown in a monolayer. D) ALDH1A1 and SOX2 protein levels are also increased in sphere culture compared with monolayer cells grown in either RMK media (10% fetal bovine serum) or serum-free CSC media. E) The ALDH+ population is increased in sphere culture. Graphed results are presented as mean ± SD of three independent experiments. Statistical significance was calculated by the two-tailed Student’s t test (*P < .05; †P < .01). ALDH = aldehyde dehydrogenase; CSC = cancer stem cell; PI3K = phosphoinositide 3-kinase.
XRT decreased tumorigenicity in CUHN014 (100.0% control vs 20.0% XRT) and CUHN022 (60.0% control vs 0.0% XRT), but not CUHN013 (100.0% control vs 80.0% XRT) (Figure 3A). Both PI3K inhibitors delayed and/or blocked tumor formation by CSCs in all cases (control vs PX-866, ZSTK, 100.0% vs 0.0%, 40.0% for CUHN014; 60.0% vs 0.0%, 0.0% for CUHN022; 100.0% vs 60.0%, 20.0% for CUN013), whereas cetuximab and docetaxel had minimal impact in tumorigenicity (Figure 3A; Supplementary Figure 1E, available online). In summary, CSC tumorigenicity was associated with resistance to conventional therapy, whereas CSCs showed susceptibility to PI3K inhibition across etiologic subtypes.
Effects of PI3K Inhibition on SOX2 and the ALDH+ Population
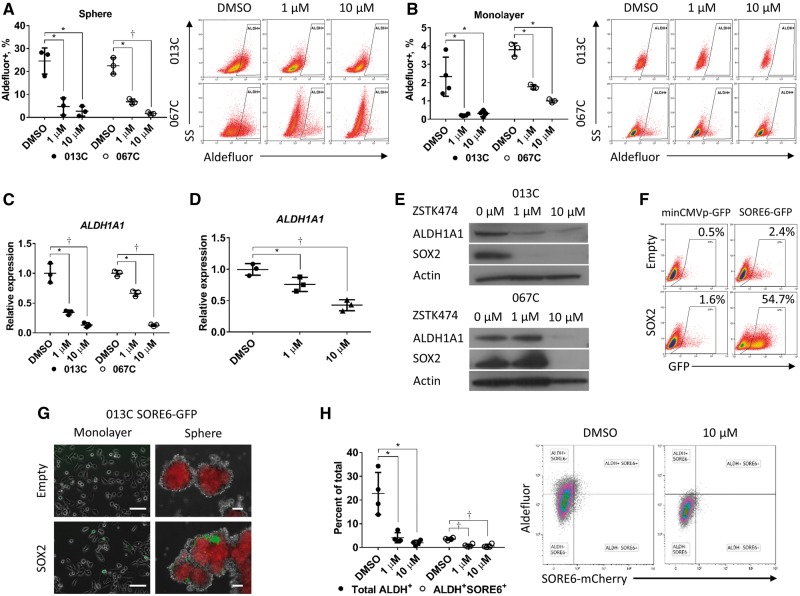
We generated early-passage cell lines (referred to as 013C, 036C, and 067C) that form tumors similar to the original tumor histologically (Supplementary Figure 2A, available online) and molecularly (Figure 1D), suggesting conservation of CSC-like populations and properties. Compared with monolayer culture, cells grown in CSC-enriching sphere culture had higher expression of ALDH1A1 (fold change ± SD, 51.7 ± 5.4, P = .004 for 013C, and 39.8 ± 1.9, P < .001 for 067C), SOX2 mRNA (fold change ± SD, 3.5 ± 0.3, P = .002 for 013C, and 3.6 ± 0.5, P = .009 for 067C) (Figure 3, B and C), increased ALDH1A1 and SOX2 protein levels (Figure 3D), and increased ALDH+ cell populations (monolayer vs sphere, 1.4% ± 0.7 vs 23.9% ± 4.6, P = .009 for 013C, and 3.0% ± 1.3 vs 21.0% ± 4.5, P = .02 for 067C) (Figure 3E). PI3K inhibition (ZSTK474) decreased the ALDH+ population in both sphere (DMSO vs 1 µM and 10 µM, 24.6% ± 5.7 vs 4.7% ± 3.7, P = .01, and 2.7% ± 2.2, P = .01 for 013C; 22.5% ± 3.6 vs 6.8% ± 1.2, P = .01, and 1.6% ± 1.7, P = .008 for 067C) and monolayer cultures (Figure 4, A and B). ZSTK474 decreased ALDH1A1 expression in both cell line sphere (1 µM and 10 µM relative to DMSO=1, 0.39 ± 0.04, P = .02, and 0.13 ± 0.04, P = .009 for 013C; 0.66 ± 0.05, P = .001, and 0.13 ± 0.02, P < .001 for 067C) (Figure 4C) and cell line monolayer (Supplementary Figure 2B, available online), as well as tumor-derived CSC sphere cultures of tumor-derived CSCs (1 µM and 10 µM relative to DMSO = 1, 0.76 ± 0.11, P = .04, and 0.43 ± 0.09, P = .002) (Figure 4D). SOX2 and ALDH1A1 protein levels decreased with ZSTK474 in a dose-dependent manner (Figure 4E; Supplementary Figure 2C, available online). Targeting EGFR directly or its downstream signaling with a MEK/ERK inhibitor did not suppress ALDH1A1 expression (Supplementary Figure 2D, available online). These results suggested that PI3K signaling regulates both SOX2 and ALDH1A1 and the ALDH+ population in HNSCC cells independently of EGFR.
Figure 4.
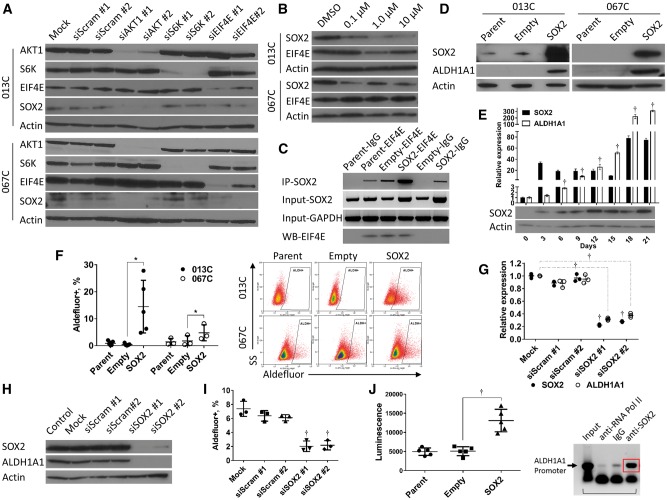
Effects of phosphoinositide 3-kinase (PI3K) inhibition on SOX2 transcriptional activity and the aldehyde dehydrogenase (ALDH)+ population. A) The ALDH+ population is statistically significantly reduced in sphere cultures and (B) monolayer cultures (013C, 067C cells) treated with the PI3K inhibitor ZSTK474. Gated regions are set at 0.1 positive of N,N-diethylaminobenzaldehyde-negative controls. C) PI3K inhibition (ZSTK474) suppresses ALDH1A1 gene expression in 013C cells grown as spheres and (D) patient-derived xenograft tumor-derived cancer stem cells (ALDH+CD44high) grown in sphere culture. E) Immunoblot showing that PI3K pathway inhibition by ZSTK474 decreases ALDH1A1 and SOX2 protein levels within 48 hours in 013C, 067C cells. F) Exogenous expression of SOX2 increases the SORE6-GFP+ population in 013C cells. Gated regions set as 0.5% of control GFP+ cells. G) Sphere culture of 013C-SOX2 cells has increased SORE6-GFP+ cells compared with control cells and monolayer culture. Scale bars = 100 µm. H) PI3K treatment decreased activation of the SORE6 reporter and the ALDH+ population in 013C spheres. Graphed results are presented as mean ± SD of three independent experiments. Statistical significance was calculated by the two-tailed Student’s t test (*P < .05; †P < .01). ALDH = aldehyde dehydrogenase; PI3K = phosphoinositide 3-kinase; SORE6 = SOX2/OCT4 response elements reporter.
Role of PI3K/mTOR Signaling in the Regulation of SOX2
We next measured SOX2 transcriptional activity using a SOX2/OCT4 response elements reporter (SORE6) (Supplementary Figure 2E, available online) (23). SORE6 was highly activated both in retroviral-mediated SOX2 expressing cell lines (Figure 4, F and G) and in unmodified CSCs indicating high baseline SOX2 (Supplementary Figure 2F, available online). PI3K inhibition of 013C spheres statistically significantly decreased the SORE6+ population (1 µM P = .009, 10 µM P = .006) (Supplementary Figure 2G, available online) and simultaneously decreased both the ALDH+ population (1 µM P = .02, 10 µM P = .02) and the ALDH+/SORE6+ population (DMSO vs 1 µM and 10 µM, 3.6% ± 0.5 vs 0.9% ± 0.6%, P < .001, and 0.6% ± 0.74%, P = .002) (Figure 4H). These findings establish that PI3K inhibition suppresses both SOX2 and ALDH activity within the same population.
SOX2 protein levels decrease following PI3K/mTOR inhibition in cancer cells (19,26), but the mechanism was not defined. We knocked down AKT1 (downstream of PI3K), and S6K and EIF4E (both downstream of TORC1) (Supplementary Figure 2H, available online). SOX2 levels decreased after silencing AKT1 (siAKT#1 P < .001, siAKT#2 P = .08 for 0.13C and siAKT#1 P = .008, siAKT#2 P = .10 for 067C) or EIF4E (siEIF4E#1 P = .06, siEIF4E#2 P = .005 for 013C and siEIF4E#1 P = .36, siEIF4E#2 P = .02 for 067C) (Figure 5A; Supplementary Figure 2I, available online). EIF4E regulates protein levels through both mRNA capping and initiation of translation downstream of mTOR signaling, which is negatively regulated by hypo-phosphorylated 4EBP1 (27). 4EGI-1 inhibited the interaction of EIF4E and EIF4G and decreased SOX2 levels and the ALDH+ fraction in HNSCC cells (Figure 5B; Supplementary Figure 3A, available online); ZSTK474 decreased levels of phosphorylated (Ser65) 4EBP1 in 013C cells (Supplementary Figure 3B, available online), allowing inactivation of EIF4E. We then performed RNA immunoprecipitation (RIP) and found that EIF4E physically bound SOX2 mRNA (Figure 5C), suggesting mRNA capping of SOX2 mRNA regulates translation. More SOX2 mRNA was bound by EIF4E when cells exogenously expressed more SOX2 mRNA (Supplementary Figure 3C, available online), though the same amount of EIF4E was immunoprecipitated (Figure 5C). Taken together, PI3K/mTOR signaling regulates EIF4E capping of SOX2 mRNA and translation of SOX2.
Figure 5.
Role of phosphoinositide 3-kinase/mechanistic target of rapamycin in the regulation of translation of SOX2 protein and aldehyde dehydrogenase (ALDH)1A1 expression. A) Silencing AKT1 or EIF4E statistically significantly reduces SOX2 protein levels in 013C and 067C cells. B) Treatment of 013C and 067C cells with the EIF4E inhibitor 4EGI-1 decreased SOX2 protein. C) The EIF4E protein physically binds SOX2 mRNA as measured by RNA immunoprecipitation using EIF4E specific antibodies. Western blot analysis showing successful precipitation of EIF4E protein. D) Exogenous expression of SOX2 (013C, 067C cells) increases ALDH1A1 protein. E) ALDH1A1 levels statistically significantly increase within six days following exogenous expression of SOX2. F) SOX2 expression increased the ALDH+ population in both 013C and 067C cells. G and H) Silencing of SOX2 in overexpression cells decreases ALDH1A1 expression and protein levels and (I) the ALDH+ population. J) Exogenous expression of SOX2 activates an ALDH1A1-promoter as measured by a luciferase assay. Chromatin immunoprecipitation using SOX2 antibodies suggests SOX2 associates with the ALDH1A1 promoter in 013C cells. Graphed results are presented as mean ± SD of three or more independent experiments. Statistical significance was calculated by the two-tailed Student’s t test (*P < .05; †P < .01). ALDH = aldehyde dehydrogenase; mTOR = mechanistic target of rapamycin; PI3K = phosphoinositide 3-kinase.
Role of SOX2 in the Maintenance of the ALDH+ Population
Retroviral-mediated expression of SOX2 increased ALDH1A1 mRNA (Supplementary Figure 3D, available online) and protein levels (Figure 5D; Supplementary Figure 3E, available online). ALDH1A1 upregulation occurred rapidly (<6 days) and led to a larger ALDH+ population (empty vs SOX2, 0.4% ± 0.4 vs 14.5% ± 9.8, P = .03 for 013C, and 1.7% ± 1.3 vs 3.6% ± 3.4, P = .04 for 067C) (Figure 5, E and F). PI3K inhibition or SOX2 expression had no effect on CD44 surface protein levels (Supplementary Figure 3F, available online). SOX2 knockdown suppressed ALDH1A1 transcripts (P < .001), protein levels, and the ALDH+ population (P < .001) (Figure 5, G–I) in 013C cells. Finally, retroviral-mediated SOX2 expression activated the ALDH1A1 promoter as measured by luciferase activity (P = .002) and chromatin immunoprecipitation (ChIP) using SOX2 antibodies, showing that SOX2 bound to the ALDH1A1 promoter (Figure 5J). These data confirm that SOX2 controls ALDH1A1 levels by direct regulation of the ALDH1A1 promoter, and ALDH1A1 is responsible for the ALDH+ population in HNSCCs.
Role of SOX2 in the Regulation of the CSC Phenotype
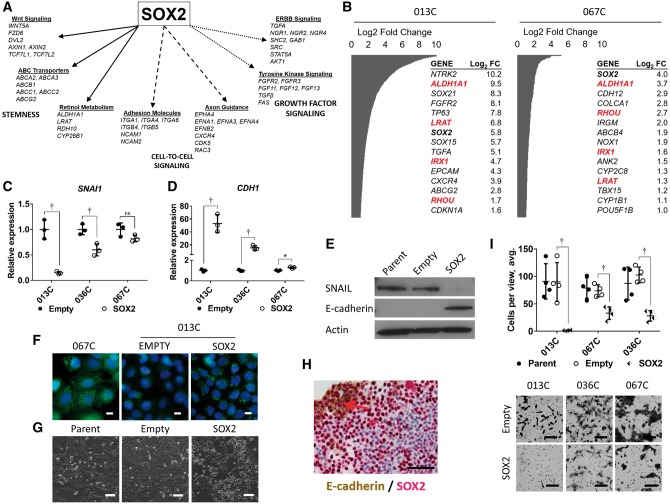
An unsupervised whole transcriptome comparison between control-vector and SOX2-overexpressing cells found that SOX2 expression increased stem cell, cell-to-cell, and growth factor–signaling pathways (013C), and genes key to CSCs like ALDH1A1, SOX2, and CXCR4 (013C, 067C) (Figure 6, A and B). These data are consistent with the signaling pattern associated with HNSCC CSCs (Figure 2A) and reveal that SOX2 expression induced CSC-like signaling.
Figure 6.
Effects of SOX2 expression on cellular morphology and gene expression. A) mRNA-sequencing analysis of 013C cells expressing SOX2 highlights increased expression of stemness pathways, cell-to-cell signaling and growth factor signaling. B) Selected cancer genes upregulated by SOX2 expressing 013C and 067C cells. Red = genes upregulated in both cell lines. C) Exogenous expression of SOX2 in 013C cells suppresses expression of SNAI1 mRNA while (D) increasing CDH1 expression. E) SOX2 expression decreases SNAIL protein levels while increasing E-cadherin in 013C cells. F) E-cadherin expression detected by immunocytochemistry. 067C cells demonstrate “typical” epithelial e-cadherin staining, while increased levels of e-cadherin in SOX2 expression 013C cells appears to be localized in the cytoplasm. This was confirmed by two different antibodies. Scale bars = 10 µm. G) Exogenous expression of SOX2 leads to a more epithelial phenotype in 013C cells. Scale bars = 100 µm. H) SOX2 (red) and E-cadherin (brown) expression are colocalized (red arrow) on the proliferating sphere surface, while SOX2 levels decrease towards the sphere interior (013C-SOX2 cells). Scale bar = 100 µm. I) SOX2 expression statistically significantly decreases invasiveness of 013C, 036C, 067C cells as measured in a matrigel-coated, 8 µm pore chamber assay. Scale bars = 100 µm. Graphed results are presented as mean ± SD of three or more independent experiments. Statistical significance was calculated by the two-tailed Student’s t test (*P < .05; †P < .01).
SOX2 expression suppressed the pro-EMT factor SNAI1 (013C P = .002, 036C P = .01, 067C P = .09) and increased expression of CDH1 (E-cadherin; 013C P = .002, 036C P = .002, 067C P = .01), generating an epithelial phenotype (Figure 6, C–G). E-cadherin was associated with high SOX2 expression at the outer growing edge of tumor spheres (Figure 6H), and retroviral-mediated SOX2 expression decreased invasiveness of HNSCC cells (invading cells/view empty vector vs SOX2, 90.0 ± 33.5 vs 1.4 ± 0.7, P = .01 for 013C, 73.7 ± 11.0 vs 33.0 ± 11.5, P = .002 for 036C, 102.6 ± 14.4 vs 28.1 ± 9.6, P < .001 for 067C) (Figure 6I). TGF-β signaling was enriched in CSCs (Figure 2A), but cell lines or tumor-derived CSCs treated with TGF-β inhibition (LY2109761 for 48 hours) did not have altered expression of SNAI1 or CDH1, even when phosphorylation of Smad3 was inhibited (Supplementary Figure 4, A–C, available online). However, treating these cells with extra TGF-β1 (10 ng/mL for 24 h) increased SNAI1 expression that was blocked by LY2109761 (Supplementary Figure 4C, available online). Neither TGF-β1 nor LY2109761 affected HNSCC cell invasion (Supplementary Figure 4D, available online). Together, these results suggest that SOX2 overcomes endogenous TGF-β effects on EMT, growth inhibition, and invasion and thus has a dominant effect on maintaining an epithelial phenotype (Figure 2, A–C) (28).
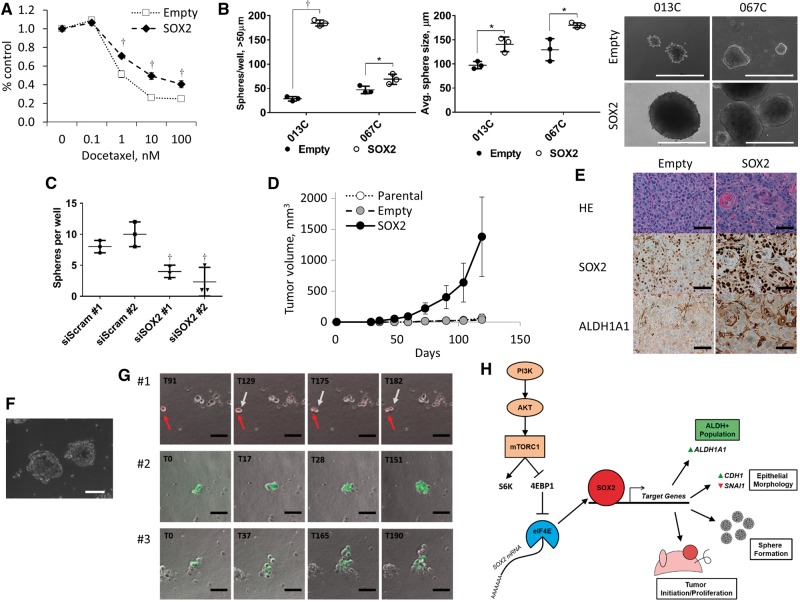
Transitions in cellular morphology are associated with changes in sensitivity to therapy (29), and we found that SOX2 expression decreased sensitivity to docetaxel (Figure 7A). Sphere formation (spheres/well, 013C P < .001 and 067C P = .04) and sphere size (average sphere diameter, 013C P = .02 and 067C P = .02) were also enhanced by SOX2 expression (Figure 7B). CUHN013 PDX CSCs readily form spheres compared with non-CSC cells (Supplementary Figure 4E, available online), and SOX2 knockdown decreases sphere formation (P = .002) (Figure 7C). SOX2-expressing 013C cells increased tumor initiation and growth in mice and continued to express higher levels of SOX2 and ALDH1A1 in vivo (Figure 7, D and E; Supplementary Figure 4F, available online). These findings confirm that SOX2 induces an epithelial phenotype, enables characteristics of CSCs, and increases the ALDH+ (CSC-like) population in HNSCC, adding to the armamentarium to study CSC biology (30).
Figure 7.
SOX2 expression promotes tumor formation. A) Exogenous expression of SOX2 increased resistance to docetaxel by the MTS assay (013C cells). B) SOX2 expression increases sphere number and sphere size in serum-free low attachment conditions (013C, 067C cells). Scale bar = 400 µm. C) Silencing of SOX2 in tumor- derived CUHN013 cancer stem cells (CSCs) decreases sphere number (sphere initiation) but not sphere size (proliferation) in culture. Graphed results are presented as mean ± SD of three or more independent experiments. Statistical significance was calculated by the two-tailed Student’s t test (*P < .05; †P < .01). D) SOX2 expression increased tumor initiation and growth of relative few (105) 013C cells implanted into five mice compared with parental and empty-vector controls, which had difficulty generating tumors within four months. E) Tumors resulting from SOX2 cells have higher levels of SOX2 and aldehyde dehydrogenase (ALDH)1A1 than control tumors. Scale bars = 100 µm. F) Sorted CUHN013 CSCs readily form spheroids in an anchorage-independent matrigel assay when cultured in serum-free CSC media. Scale bars = 200 µm. G) Single SORE6+ CSCs generate both SORE+ and SORE- daughter cells in anchorage-independent conditions. Three representative time-lapse cases where SORE6-mCherry+ or SORE6-GFP+ CSCs give rise to SORE6- cells. Scale bars = 100 µm. H) Proposed mechanism of the phosphoinositide 3-kinase/mechanistic target of rapamycin role in “stemness” maintenance by regulating SOX2 and ALDH1A1 expression. CSC = cancer stem cell; mTOR = mechanistic target of rapamycin; PI3K = phosphoinositide 3-kinase; SORE6 = SOX2/OCT4 response element.
SOX2 Transcriptional Activity in Daughter Cells of Single Tumor–Derived CSCs
We demonstrated that CSCs can recapitulate the originating tumor in vivo, giving rise to a heterogeneous cell population. We then examined asymmetric division assessing changes in SOX2 activity in vitro using an anchorage-independent, three-dimensional matrigel assay where CSCs formed spheres (Figure 7F). SORE6+ tumor-derived CSCs were individually seeded on a matrigel layer, and single cells were followed using time-lapse microscopy (120 hours). Single SORE6+ CSCs gave rise to multiple SORE6- daughter cells while maintaining fluorescence in others (Figure 7G), suggesting that daughter cells have different levels of SOX2 activity and likely diversified properties. Therefore, SOX2 activity may be used to detect and quantify asymmetric division, which is an early event in sphere/tumor initiation leading to tumor repopulation.
Discussion
CSCs are small tumor cell populations that perpetuate growth and recapitulate tumor heterogeneity by asymmetric division, are resistant to therapy, and enable phenotypic flexibility leading to invasion and metastasis (6). PDX models enable studying relevant cancer subtypes that have been underrepresented in conventional cell line panels like HPV-positive HNSCC. ALDH activity and CD44 expression defined CSCs regardless of HPV status. CSCs serially passaged over several generations faithfully recapitulated the histology and expression profiles observed in the originating tumor. Together, ALDH activity and CD44 expression are the primary markers for CSCs in HNSCC regardless of etiology.
Despite clinical and etiologic heterogeneity in our cohort, the transcriptome analysis of CSCs vs non-CSCs identified common signaling themes across subtypes (most importantly, HPV-negative and HPV-positive), including enrichment of stem cell maintenance pathways (telomere maintenance, Wnt, HIF1α, and MYC), PI3K/mTOR signaling, and TGF-β/SMAD. Lower p53 signaling in HPV-negative (vs HPV-positive) CSCs was expected as HPV-negative HNSCC tumors harbor inactivating TP53 mutations, making them more resistant to XRT (31). The use of core signaling pathways to drive oncogenesis has profound translational implications as it indicates common therapeutic avenues across HNSCC clinical subtypes and other cancer types (18,32,33).
CSCs were less susceptible to conventional therapies compared with non-CSCs. Because tumor-stroma interactions can dictate the efficacy of XRT and cytotoxic agents (34), we used in vivo treatment to model CSC behavior after therapy. PI3K inhibition and XRT targeted CSCs preferentially, compared with docetaxel or cetuximab, blocking tumor regrowth regardless of PIK3CA (or HPV) status. These findings support inhibiting PI3K signaling in combination with XRT to target HNSCC CSCs.
We identified a link between PI3K/mTOR signaling (the most commonly activating genetic event in HNSCC) and the regulation of key CSC genes. One of the primary properties of CSCs, ALDH activity, was regulated by PI3K signaling and SOX2. Retroviral-mediated SOX2 expression dramatically increased ALDH1A1 and the ALDH+ cell population by interacting directly with the ALDH1A1 promoter. SOX2 expression regulated other key CSC features in HNSCC, including sphere and tumor formation and drug resistance (Figure 7H). Constitutive expression of SOX2 in HNSCC cells generates a CSC-like population that enables CSC studies.
HNSCC CSCs have an epithelial phenotype by immunohistochemistry and have high E-cadherin mRNA and protein expression, which are distinct from descriptions of CSCs in other tumor types (33,35). Modeling of CSC by constitutive SOX2 expression unequivocally promoted an epithelial morphology in HNSCC cells by increasing E-cadherin and decreasing SNAIL. E-cadherin can be used as an alternative to OCT4 for the generation of iPSCs (36) and induces pluripotent stem cells (37), supporting the relevance of E-cadherin in CSCs. SCC CSCs transition between epithelial and mesenchymal states because of environmental stressors (38) and when metastasizing are able to gain a more mesenchymal phenotype without loss of E-cadherin (39). Recent reports have shown that metastasis can occur without EMT (40,41), and the data presented here establish that HNSCC CSCs derived from patient tissue have an epithelial phenotype.
SORE6+ CSCs are capable of asymmetric division, fulfilling another hallmark of CSCs (6). Using a 3D matrix model, we were able to track the asymmetric division of a single HNSCC CSC by the real-time binding of response elements by SOX2 (23). Individual SORE6+ HNSCC CSCs generated both positive and diminished/negative daughter cells while initiating tumor sphere formation, suggesting the SORE6+ daughter cells are the remaining CSC population, while the SORE6- (with low SOX2 activity) cells are likely a more differentiated lineage population. This supports that CSCs can recapitulate the heterogeneity of the original tumor via their ability to asymmetrically divide. To our knowledge, this is the first report tracking the asymmetric division of CSCs derived directly from patient tumors.
Our studies linking PI3K signaling with CSCs properties are not without limitations. A recent report demonstrates the dynamic ability of squamous tumor cells to reactivate PI3K/mTOR through EGFR/AXL/PKC following pharmacological inhibition of PI3Kα (42). Similarly, resistance to PI3K inhibition in human epidermal growth factor receptor (HER) 2–positive breast cancer occurs following rapid activation of HER3 (43), suggesting that multitargeted therapy may be required (44). Finally, we need to validate our results prospectively by correlating CSC profiles and outcomes in HNSCC.
We have characterized the molecular features and functional properties of HNSCC CSCs derived from PDXs representing tumors of both HPV and tobacco etiologies. PI3K signaling enhanced SOX2 translation, which in turn induced the expression of genes regulating the stemness characteristics of CSCs. These data support targeting PI3K therapeutically in HNSCC. Our findings have elucidated a mechanistic relationship linking the most common core genetic alterations in HNSCC with fundamental CSC features, thus identifying promising therapeutic avenues.
Funding
This work was supported by National Institutes of Health grants R01CA149456 (AJ), R21DE019712 (AJ), and R01DE024371 (XJW and AJ); and P30-CA046934 (University of Colorado Cancer Center Support Grant), P30-AR057212 (University of Colorado Skin Diseases Research Center Support Grant), DE020649 (XJW), U01AA021724 (VV), Ruth L. Kirschstein National Research Service Award T32CA17468 (XJW; PNL trainee), Training in Otolaryngology Research T32DC012280 (CN trainee), the Daniel and Janet Mordecai Foundation (AJ), and the Peter and Rhonda Grant Foundation (AJ).
Notes
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
The authors are indebted to the patients who donated their tissue, blood, and time and to the clinical teams who facilitated patient informed consent, as well as sample and data acquisition. The authors wish to thank Karen Helm for helpful discussions, Pamela Garl for her manuscript review, and Scott Peterson (Oncothyreon, Seattle, WA) for providing PX-866.
Dr. Jimeno has received laboratory and clinical trial research support from Oncothyreon Pharmaceuticals. The other authors declare no disclosures.
Materials will be shared per the University of Colorado’s Office for Technology Transfer policies and Institutional Review Board. Gene expression arrays are deposited in the National Center for Biotechnology Information Gene Expression Omnibus.
Authors: Stephen B. Keysar, Phuong N. Le, Bettina Miller, Brian C. Jackson, Justin R. Eagles, Cera Nieto, Jihye Kim, Binwu Tang, Magdalena J. Glogowska, J. Jason Morton, Nuria Padilla-Just, Karina Gomez, Emily Warnock, Julie Reisinger, John J. Arcaroli, Wells A. Messersmith, Lalage M. Wakefield, Dexiang Gao, Aik-Choon Tan, Hilary Serracino, Vasilis Vasiliou, Dennis R. Roop, Xiao-Jing Wang, Antonio Jimeno
Affiliations of authors: Division of Medical Oncology, Department of Medicine (SBK, PNL, BM, BCJ, JRE, CN, JK, MJG, JJM, NPJ, KG, EW, JR, JJA, WAM, ACT, AJ), Department of Biostatistics and Informatics (JK, DG, ACT), Department of Pathology (HS, XJW), Department of Dermatology (DRR), and Gates Center for Regenerative Medicine (DRR, XJW, AJ), University of Colorado School of Medicine, Aurora, CO; Laboratory of Cancer Biology and Genetics, National Cancer Institute, Bethesda, MD (BT, LMW); Department of Environmental Health Sciences, Yale School of Public Health, Yale School of Medicine, New Haven, CT (VV).
Supplementary Material
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55(1):10–30. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Engels EA, Anderson WF, et al. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26(4):612–619. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–319. [DOI] [PubMed] [Google Scholar]
- 6.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–9344. [DOI] [PubMed] [Google Scholar]
- 7.Winquist RJ, Boucher DM, Wood M, et al. Targeting cancer stem cells for more effective therapies: Taking out cancer's locomotive engine. Biochem Pharmacol. 2009;78(4):326–334. [DOI] [PubMed] [Google Scholar]
- 8.Chen YC, Chen YW, Hsu HS, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385(3):307–313. [DOI] [PubMed] [Google Scholar]
- 9.Prince ME, Ailles LE. Cancer stem cells in head and neck squamous cell cancer. J Clin Oncol. 2008;26(17):2871–2875. [DOI] [PubMed] [Google Scholar]
- 10.Bhaijee F, Pepper DJ, Pitman KT, et al. Cancer stem cells in head and neck squamous cell carcinoma: a review of current knowledge and future applications. Head Neck. 2012;34(6):894–899. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keysar SB, Astling DP, Anderson RT, et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Mol Oncol. 2013;7(4):776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hafner C, Landthaler M, Vogt T. Activation of the PI3K/AKT signalling pathway in non-melanoma skin cancer is not mediated by oncogenic PIK3CA and AKT1 hotspot mutations. Exp Dermatol. 2010;19(8):e222–e227. [DOI] [PubMed] [Google Scholar]
- 16.Lui VW, Hedberg ML, Li H, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fresno Vara JA, Casado E, de Castro J, et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. [DOI] [PubMed] [Google Scholar]
- 18.Hambardzumyan D, Becher OJ, Rosenblum MK, et al. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22(4):436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh S, Trevino J, Bora-Singhal N, et al. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012;11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegle JM, Basin A, Sastre-Perona A, et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun. 2014;5:4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boumahdi S, Driessens G, Lapouge G, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511(7508):246–250. [DOI] [PubMed] [Google Scholar]
- 23.Tang B, Raviv A, Esposito D, et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports. 2015;4(1):155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzmichev AN, Kim SK, D'Alessio AC, et al. Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr Biol. 2012;22(18):1705–1710. [DOI] [PubMed] [Google Scholar]
- 25.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Zhang Y, Zhang Y, et al. Downregulation of cancer stem cell properties via mTOR signaling pathway inhibition by rapamycin in nasopharyngeal carcinoma. Int J Oncol. 2015;47(3):909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mamane Y, Petroulakis E, Rong L, et al. eIF4E—from translation to transformation. Oncogene. 2004;23(18):3172–3179. [DOI] [PubMed] [Google Scholar]
- 28.Samavarchi-Tehrani P, Golipour A, David L, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. [DOI] [PubMed] [Google Scholar]
- 29.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. [DOI] [PubMed] [Google Scholar]
- 30.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimple RJ, Smith MA, Blitzer GC, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73(15):4791–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myant KB, Cammareri P, McGhee EJ, et al. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12(6):761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160(5):963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Campisi J, Higano C, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18(9):1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redmer T, Diecke S, Grigoryan T, et al. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12(7):720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer H, Keramari M, Ward CM. Using cadherin expression to assess spontaneous differentiation of embryonic stem cells. Methods Mol Biol. 2011;690:81–94. [DOI] [PubMed] [Google Scholar]
- 38.Biddle A, Liang X, Gammon L, et al. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71(15):5317–5326. [DOI] [PubMed] [Google Scholar]
- 39.White RA, Neiman JM, Reddi A, et al. Epithelial stem cell mutations that promote squamous cell carcinoma metastasis. J Clin Invest. 2013;123(10):4390–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer KR, Durrans A, Lee S, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elkabets M, Pazarentzos E, Juric D, et al. AXL mediates resistance to PI3Kalpha inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27(4):533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakrabarty A, Sanchez V, Kuba MG, et al. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A. 2012;109(8):2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrett JT, Chakrabarty A, Arteaga CL. Will PI3K pathway inhibitors be effective as single agents in patients with cancer? Oncotarget. 2011;2(12):1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.