Abstract
We previously reported reduced expression of Sox4 in metastatic melanoma and its role in suppression of cell migration and invasion through inhibition of NF-κB p50. Sox4 can also bind to the promoter sequence of Dicer, a miRNA biogenesis factor. Interestingly, altered expression of Dicer was also observed in cancers. However, the potential mechanisms which regulate Dicer expression and its potential significance in melanoma progression are unknown. Here we studied the regulation of Dicer expression by Sox4 and its role in suppression of melanoma invasion. Our data showed that Sox4 positively regulates Dicer expression by binding to its promoter sequences and enhancing its activity. We found that knockdown of Dicer enhances the matrigel invasion of melanoma cells by at least 2-fold. In addition, we revealed that overexpression of exogenous Dicer reverts the enhanced melanoma cell invasion upon Sox4 knockdown. Furthermore, we examined the expression of Dicer protein in a large set of melanocytic lesions (n=504) at different stages by tissue microarray and found that Dicer expression is inversely correlated with melanoma progression (P < 0.0001). Consistently, reduced Dicer expression was correlated with a poorer overall and disease-specific 5-year survival of patients (P = 0.015 and 0.0029, respectively). In addition, we found a significant correlation between expression of Sox4 and Dicer proteins in melanoma biopsies (P = 0.009), further indicating the regulation of Dicer expression by Sox4. Finally, we revealed that knockdown of Sox4 induces a major change in the expression pattern of miRNAs in melanoma cells, mainly due to reduced expression of Dicer. Our results pinpoint the regulation of Dicer expression by Sox4 in melanoma and the critical role of Dicer in suppression of melanoma invasion. Our findings on Sox4 regulated miRNA biogenesis pathway may aid toward the development of novel targeted therapeutic approaches for melanoma.
Keywords: Sox4, Dicer, Melanoma, Invasion, miRNA
Introduction
Melanoma is an aggressive disease due to its notorious potential to invade and metastasize to other organs (1). Altered expression of multiple genes is observed upon progression of melanoma toward metastasis. Some of these changes are required for melanocytes to adapt to the environments other than epidermis which host the secondary tumors, while many other factors are involved in tumor cell invasion, motility, angiogenesis, proliferation, and evasion of apoptosis (2).
microRNAs (miRNAs) are a class of non-coding RNA which regulate the expression of target genes at post-transcriptional level. Biogenesis of the majority of miRNA starts from transcription by RNA polymerase II, producing pri-miRNA. The pri-miRNA is cleaved by Drosha in conjunction with DGCR8 in the nucleus, generating the pre-miRNA which is then transferred to the cytoplasm where Dicer further processes the hairpin into the mature miRNA (3). Deregulated miRNA has been reported in many types of cancers, both promoting and suppressing the process of cancer initiation or progression depending on the type of the malignancy and the miRNA of interest (4–5). With exception of few types of malignancies, a general downregulation of miRNA expression in cancers has been observed, mainly due to chromosomal abnormality, epigenetic changes, or aberrant expression and function of the miRNA biogenesis factors such as Dicer (6). In addition, reduced expression of Dicer was also reported to promote tumorigenesis in animal models (7–9). Despite all these progresses, the expression status of most miRNAs and their biogenesis factors, as well as the mechanisms which regulate the expression of miRNAs in melanoma is largely unknown.
Altered expression of Sry-related high-mobility group (HMG) box4 (Sox4) transcription factors has been reported in several types of human malignancies (10–12). Sox4 was shown to suppress apoptotic cell death in prostate (13) and adenoid (14) cancer cell lines which suggest that it may positively contribute to the progression of certain types of cancer. On the other hand, others have shown that Sox4 expression in other human cancer cells induces apoptotic cell death (12,15–16). Sox4 can also promote cell cycle arrest and apoptosis to inhibit tumorigenesis in human colon cancer cell line (17), suggesting that it may have a potential tumor-suppressive function in other tissues. We recently reported a significant correlation between reduced Sox4 expression and melanoma metastasis (18). We observed a positive correlation between Sox4 expression and 5-year survival of melanoma patients. In addition, we showed that Sox4 suppresses melanoma cell migration and invasion by inhibiting nuclear factor (NF)-κB p50 expression (18). Sox4 has also been reported to bind to the promoter sequence of certain miRNA biogenesis factors and components of the RISC such as Dicer, Argonaute 1, and RNA Helicase A in prostate cancer cell lines (19). Nevertheless, the significance of the interaction between Sox4 and miRNA biogenesis factors in suppression of melanoma invasion is not understood.
Here we confirmed that Sox4 positively regulates Dicer expression at transcription level in melanoma and this regulation is critical for expression of a considerable number of cancer-related miRNAs and suppression of melanoma invasion. In addition, we found that Dicer expression is reduced in metastatic melanoma and Dicer expression is positively correlated with overall and disease-specific 5-year survival of melanoma patients, suggesting that Dicer expression may be used as a promising prognostic marker and therapeutic target for malignant melanoma.
Results
Regulation of Dicer expression by Sox4
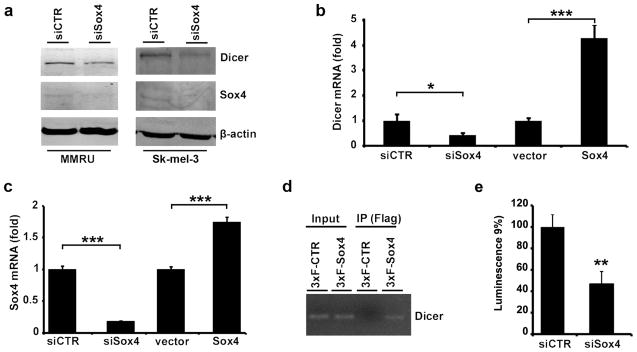
To identify whether Sox4 is able to regulate the expression of Dicer in melanoma cells, we knocked down Sox4 expression in two different melanoma cell lines and observed a marked reduction in Dicer expression at protein level (Figure 1a). To further investigate the mechanism by which Sox4 regulates Dicer expression, we used real-time qPCR and found that knockdown of Sox4 causes a marked reduction in Dicer mRNA level, while a modest increase in Sox4 expression mediated by transfection of MMRU cells with POTB7-Sox4 increased Dicer mRNA expression (Figure 1b and c). In addition, through ChIP assay using anti-Flag antibody mediated pull-down of 3×Flag-Sox4, we confirmed that Sox4 is able to bind to sequences about 2200bp upstream of Dicer transcription start site (Figure 1d). Consistently, using a luciferase reporter construct under control of Dicer promoter, we showed that Sox4 knockdown resulted in over 50% reduction in Dicer promoter activity (Figure 1e). These data indicate that Sox4 positively regulates the expression of Dicer at transcriptional level by binding to and activating its promoter region.
Figure 1.
Regulation of Dicer expression by Sox4. (a) Protein extracts were prepared 72 hours after transfection and analyzed for the expression of Dicer and Sox4 by western blot. β-actin was used as a loading control. (b, c) For quantitative reverse transcription PCR analysis, cells were transfected with siCTR, siSox4, empty vector or POTB7-Sox4 and lysed for total RNA extraction and reverse transcription (72 and 24 hours after transfection for siRNA and plasmid transfected cells, respectively). Expression of Dicer and Sox4 mRNAs was measured by real-time quantitative PCR and normalized with β-actin as loading control. *P < 0.05, ***P < 0.001, Student’s t-test. (d) Flag-Sox4 protein binds to the Dicer promoter sequence in MMRU cells in vivo, demonstrated by ChIP assay. (e) Dicer promoter luciferase reporter activity of control and Sox4 knockdown MMRU cells. Whole-cell extracts were prepared 243h after transfection, and luciferase levels were measured by luminometry. All values are expressed as mean±s.d. **P < 0.01.
Suppression of melanoma cell invasion by Sox4 in Dicer-dependent manner
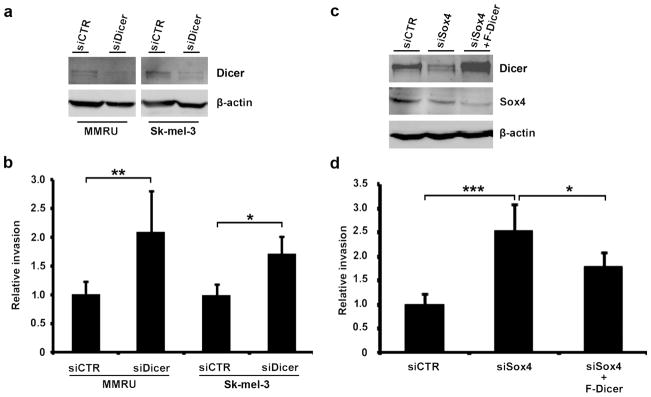
To study the role of Dicer in melanoma invasion, we examined the ability of MMRU and Sk-mel-3 cells to invade through matrigel using the Boyden chamber assay. We found that Dicer knockdown (Figure 2a) enhanced matrigel invasion of MMRU cells by 2-fold and Sk-mel-3 cells by 1.7-fold compared with the respective controls (Figure 2b), indicating that Dicer suppresses invasion of melanoma cells. It might be argued that the enhanced invasion by Dicer-KD might be due to increased cell growth rate and not enhanced matrigel invasion ability. To address this concern we used cell growth assay and found that Dicer-KD MMRU and Sk-mel-3 cells grow slightly but significantly slower than their siCTR transfected counterparts (Supplementary Fig. S1), excluding the possibility that the augmented invasion after Dicer-KD was due to higher cell growth rate.
Figure 2.
Sox4-mediated enhancement of Dicer expression is required for suppression of melanoma cell invasion. (a) Western blot confirms reduction of Dicer protein in MMRU and Sk-mel-3 cells 72 h after transfection with siDicer. (b) For Boyden chamber assay, MMRU and Sk-mel-3 cells were suspended in serum-free medium and seeded on matrigel, incubated at 37°C for 24 hours, stained by crystal violet, and quantified. *P < 0.05, **P < 0.01, Student’s t-test. (c) Western blot confirms rescued expression of Dicer after transfection of Sox4-KD MMRU cells with Flag-Dicer. (d) Overexpression of Flag-Dicer can revert Sox4-KD enhanced MMRU cell matrigel invasion. *P < 0.05, ***p < 0.001, Student’s t-test.
We previously reported that knockdown of Sox4 enhanced MMRU cell invasion by 3-fold (18). To understand the significance of Sox4-regulate Dicer expression in suppression of melanoma invasion we overexpressed Flag-Dicer in Sox4-KD cells (Figure 2c) and observed that overexpression of Flag-Dicer reduces invasion in Sox4-KD MMRU cell by 30 percents (Figure 2d), indicating that Flag-Dicer is able to at least partially revert the Sox4-KD phenotype.
Reduced cytoplasmic Dicer expression is correlated with melanoma progression
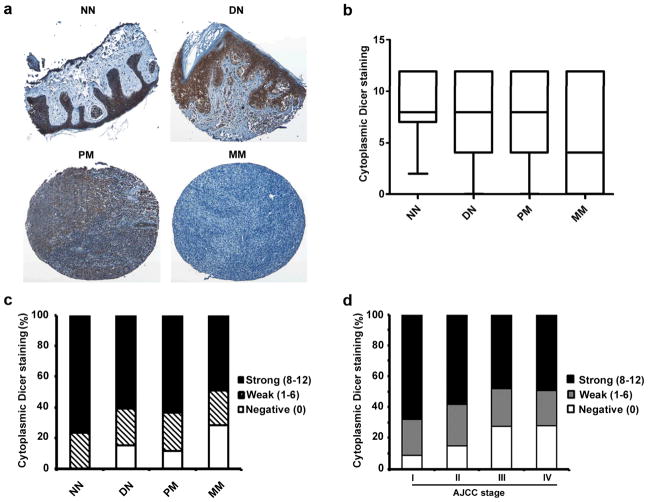
We used TMA technology to investigate the expression pattern of Dicer in melanoma biopsies and its possible correlation with melanoma progression. Immunohistochemical staining with Dicer-specific antibody (Sigma) revealed a predominant cytoplasmic staining of Dicer (Figure 3a). Notably, a significant difference in cytoplasmic Dicer staining was observed between different stages of melanoma. Kruskal-Wallis test revealed a clear reduction in the expression of Dicer in metastatic melanoma compared with other stages of melanocytic lesions (P = 0.003; Figure 3b), but not among normal nevi dysplastic nevi and primary melanomas. In addition, when we grouped the samples into negative, weak, or strong Dicer staining, we found a significant difference between primary melanomas and metastatic melanomas (P = 0.0001, χ2 test; Figure 3c), although the differences among normal nevi, dysplastic nevi, and primary melanomas were not significant (P > 0.05). Our data further demonstrated that Dicer expression is also inversely correlated with American Joint Committee on Cancer (AJCC) stages (P =0.003, χ2 test; Figure 3d).
Figure 3.
Reduced expression of cytoplasmic Dicer correlates with melanoma progression. (a) Representative images of normal nevi (NN) and dysplastic nevi (DN) with strong cytoplasmic staining, primary melanoma (PM) with moderate staining, and metastatic melanoma (MM) with negative cytoplasmic Dicer staining. (b) Kruskal-Wallis test for differences in Dicer staining among NN, DN, PM, and MM. The median is depicted as a horizontal line inside each box (P = 0.003). (c) Chi square test for differences in Dicer staining in NN, DN, PM, and MM. Significant difference was found between PM and MM (P = 0.0001). (d) Cytoplasmic Dicer expression is negatively associated with AJCC stage of melanoma cases (P = 0.003, χ2 test).
In addition, we found that expression of cytoplasmic Dicer is positively correlated with lymphocytic response in primary melanoma cases (P = 0.036, χ2 test; Table 1). Furthermore, primary melanoma samples from male individuals show less Dicer expression compared to their female counterparts (P = 0.009, χ2 test; Table 1). We did not find any significant correlation between cytoplasmic Dicer expression and patient age, tumor thickness, location, subtype, or ulceration status (Table 1).
Table 1.
Cytoplasmic Dicer staining and clinicopathologic characteristics of 262 cases of primary melanomas
| Cytoplasmic Dicer staining
|
Total | p value* | |||
|---|---|---|---|---|---|
| Negative | Weak | Strong | |||
| Age | |||||
| ≤62 | 15 (11.2%) | 31 (23.1%) | 88 (65.7%) | 134 | 0.702 |
| >62 | 15 (11.7%) | 35 (27.3%) | 78 (61.0%) | 128 | |
| Sex | |||||
| Male | 23 (16.7%) | 32 (23.2%) | 83 (60.1%) | 138 | 0.009 |
| Female | 6 (4.8%) | 35 (28.2%) | 83 (67.0%) | 124 | |
| Tumor thickness (mm) | |||||
| ≤2 mm | 12 (8.2%) | 34 (23.1%) | 101 (68.7%) | 147 | 0.073 |
| >2mm | 18 (15.7%) | 32 (27.8%) | 65 (56.5%) | 115 | |
| Ulceration | |||||
| Present | 8 (14.8%) | 14 (25.9%) | 32 (59.3%) | 54 | 0.648 |
| Absent | 22 (10.6%) | 52 (25.0%) | 134 (64.4%) | 208 | |
| Lymphocytic response | |||||
| Present | 12 (12.8%) | 15 (15.9%) | 67 (71.3%) | 94 | 0.036 |
| Absent | 18 (10.7%) | 51 (30.4%) | 99 (58.9%) | 168 | |
| Tumor subtype | |||||
| Lentigo maligna | 10 (21.3%) | 9 (19.1%) | 28 (59.6%) | 47 | 0.256 |
| Superficial spreading | 8 (8.8%) | 24 (26.4%) | 59 (64.8%) | 91 | |
| Nodular | 5 (10.9%) | 9 (19.6%) | 32 (69.5%) | 46 | |
| Other† | 7 (9.0%) | 24 (30.8%) | 47 (60.2%) | 78 | |
| Site‡ | |||||
| Sun-exposed | 13 (13.8%) | 22 (23.4%) | 59 (62.8%) | 94 | 0.632 |
| Sun-protected | 17 (10.1%) | 44 (26.2%) | 107 (63.7%) | 168 | |
χ2 test.
Other: unspecified subtype.
Sun-protected sites: trunk, arm, leg and feet; Sun-exposed sites: head and neck.
To further validate these findings we used a separate anti-Dicer antibody (Clonegene) to probe for the expression of Dicer in a smaller TMA construct, containing 31 cases of dysplastic nevi, 71 cases of primary and 46 cases of metastatic melanomas (Supplementary Figure S2a). Statistical analysis showed that expression of cytoplasmic Dicer is significantly lost in metastatic melanoma compared with dysplastic nevi and primary melanoma (P = 0.001, χ2 test; Supplementary Figure S2b). We did not observe any significant correlation between the expression of cytoplasmic Dicer and clinicopathologic parameters using this antibody (Supplementary Table S1).
Cytoplasmic Dicer staining positively correlates with better 5-year patient survival
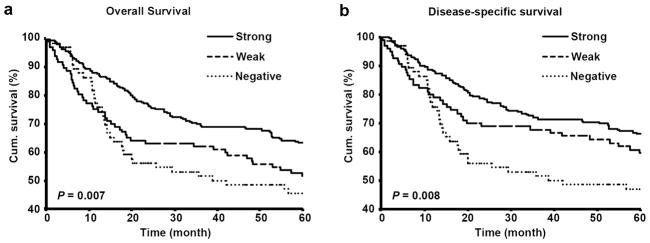
We evaluated the correlation between cytoplasmic Dicer expression and 5-year survival of primary and metastatic melanoma patients by constructing Kaplan-Meier survival curves. Overall survival in strong Dicer staining group was 63.6% compared with 52.1% in weak and 46.2% in negative Dicer staining group (P = 0.007, log rank test; Figure 4a). In addition, the disease-specific 5-year survival of the patients was significantly reduced from 67.1% in strong Dicer staining group to 62.5% in weak and 47.7% in negative Dicer staining group (P = 0.008, log rank test; Figure 4b), indicating a significant positive correlation between Dicer expression and melanoma patients survival. We also performed multivariate Cox regression analysis including cytoplasmic Dicer staining, age and sex. The results revealed that Dicer staining is able to predict both overall and disease-specific patient survival (P = 0.002 and 0.007, respectively; Supplementary Table S2) independent of age or sex of the patients. Using data obtained with the second anti-Dicer antibody, we also observed a clear positive correlation between Dicer expression and 5-year patient survival. Although the correlation between Dicer expression and overall survival was not significant (P = 0.077, log rank test; Supplementary Figure S3a), Dicer expression was significantly correlated with disease-specific survival (P = 0.037, log rank test; Supplementary Figure S3b).
Figure 4.
Dicer expression positively correlates with 5-year survival of melanoma patients. (a) Overall and (b) disease-specific 5-year survival of all primary and metastatic melanoma patients.
Positive correlation between expression of Sox4 and Dicer in melanoma patients
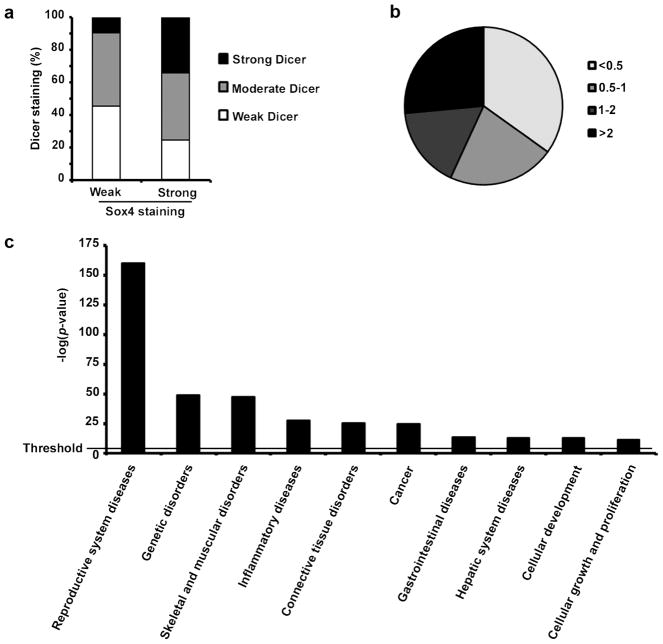
We used our previous data for Sox4 TMA (18) and obtained the cytoplasmic Dicer expression for the corresponding cases from the current TMA data set to compare the expression pattern of both Sox4 and Dicer in the same melanocytic lesions. We grouped all cases based on the expression pattern of Sox4 into weak Sox4 (42 cases) or strong Sox4 group (102 cases) and found that expression of Dicer is considerably higher in cases with strong Sox4 staining than those with weak Sox4 staining (P = 0.004, χ2 test; Figure 5a). Accordingly, the percentage of cases with strong Dicer staining increased from 9% in weak Sox4 group to 34% in strong Sox4 group while the percentage of cases with negative Dicer staining decreases from 45% in weak Sox4 group to 24% in strong Sox4 group.
Figure 5.
Regulation of miRNAs expression by Sox4 in melanoma. (a) positive correlation between Sox4 and Dicer expression in 144 melanocytic lesions at different stages. Dicer expression was compared between weak or strong Sox4 expressing cases (P = 0.004, χ2 test). (b) Total RNA from siCTR or siSox4 transfected MMRU cells was prepared with Qiazol extraction followed by Poly-A Tailing reactions and miRNA cDNA synthesis. miRNAs were grouped by pair-wise comparison of our expression data sets (siSox4 vs siCTR) based on their expression fold-change. (c) Functional analysis of the miRNA profiling data was performed using the Ingenuity Pathway Analysis. The horizontal line indicate the p-value = 0.05 threshold.
Sox4 regulates the expression of a large subset of miRNAs
Considering the regulation of Dicer expression and to investigate whether Sox4 has a role in biogenesis of miRNAs in melanoma cells we used a qPCR based miRNA profiling method to detect the changes in expression of all validated and predicted mature human miRNAs after Sox4 knockdown in MMRU cells. Our results showed at least 2-fold downregulation of 34.9% of miRNAs in Sox4-KD cells compared with siCTR transfected control cells. Sox4-KD also increased the expression of 26.5% of miRNAs by at least 2-fold and had no significant effect on the other 38.6% miRNAs (Figure 5b). These results indicated a major change in the expression pattern of more than 60% of miRNAs after Sox4-KD in melanoma cells, implying the critical role of Sox4 in this process. Next, to determine the most important biological functions that are affected by Sox4-KD mediated change in miRNA expression we used Ingenuity Pathway Analysis (IPA) software and analyzed the miRNAs with at least 2-fold up- or down-regulation after Sox4-KD. Our results demonstrated that reproductive system diseases-related miRNAs show the most significant change after Sox4 knockdown in melanoma cells (Figure 5c). Interestingly, cancer related miRNAs also showed a very significant change upon Sox4-KD (Figure 5c). According to IPA’s database, 61 cancer related miRNAs were affected by Sox4-KD, 53 of which were at least 2-fold downregulated (Supplementary Table S3). In addition, between 21 metastasis-related miRNAs which were affected by Sox4-KD, 19 miRNAs showed at least 2-fold downregulation (Supplementary Table S4). Moreover, 26 out of 28 melanoma-related miRNAs identified by this software were at least 2-fold downregulated after Sox4-KD (Supplementary Table S5).
We also screened for expression profile of miRNAs in Sox4-KD MMRU cells after rescuing Dicer expression by exogenous Flag-Dicer. Consistent with a downstream role of Dicer in Sox4-regulated miRNA expression, we found a reversal in the expression of majority miRNAs which showed either reduced or increased expression after Sox4-KD with overexpression of Flag-Dicer (GEO database; accession# GSE36715). For instance, of 307 miRNAs with more than two-fold reduced expression after Sox4-KD, expression of 198 miRNAs were either comparable or more than control group after rescue of Dicer expression (Supplementary Table S6). Interestingly, rescue of Dicer expression also revert the Sox4-KD induced change in the expression of majority miRNAs involved in cancer, metastasis or melanoma (Supplementary Tables S3, S4 & S5).
Discussion
High tendency to invade other tissues is the leading cause of melanoma-related death. Therefore, identifying the factors which are involved in regulation of melanoma invasion and metastasis is of outstanding priorities. We previously reported that expression of Sox4 is reduced in metastatic melanoma and revealed that Sox4 suppressed melanoma cell migration and invasion at least partially through suppression of (NF)-κB p50 expression. Different groups investigated the pattern of Sox4 binding to the promoters or regulation of expression of its target genes in human genome (19–21) which created a wealth of knowledge regarding Sox4’s target network. However, these high-throughput screenings did not return consistent results. For instance, Sox4 was shown to bind to promoter of Dicer in prostate carcinoma cell line (19), while the same observation was not made in small cell lung cancer (21), nor its knockdown was shown to change Dicer expression in hepatocellular carcinoma cell lines (20). Although the actual reason for this discrepancy is not known, different sensitivity in the methods and variations in the biology of different tissues might play a role.
In this study, we investigated the regulation of Dicer expression by Sox4 in melanoma and its significance in Sox4-mediated suppression of melanoma invasion. We observed that Sox4 is able to regulate the expression of Dicer at both mRNA and protein levels (Figure 1a and b). Using overexpression of Flag-Sox4 construct and pull-down with anti-Flag antibody, we were able to confirm by ChIP assay that Sox4 binds to Dicer promoter sequences in melanoma cells (Figure 1d). Interestingly, the region which showed the highest degree of amplification after pull-down of 3×Flag-Sox4, contains two adjacent consensus Sox4 binding sites (AACAAAG and AACAATA) (22–23), which may explain the significance of Sox4 binding to this region. Reduction of the Dicer promoter activity upon Sox4 knockdown as shown by luciferase reporter assay (Figure 1e) further confirmed the Sox4-regulated regulation of Dicer transcription. To understand the role of Dicer in regulation of melanoma invasion, we used matrigel invasion assay and for the first time revealed that knockdown of Dicer augments invasion ability of two different melanoma cells (Figure 2b). Consistently, Dicer expression had been previously proven critical in suppression of metastasis in multiple types of cancer, downregulation of which would enhance tumor metastasis (24–25). Excitingly, we also observed that overexpression of Dicer in melanoma cells can revert the Sox4-KD phenotype (Figure 2d), indicating the downstream role of Dicer in Sox4-mediated suppression of melanoma invasion.
By using immunohistochemistry (Figure 3a), we observed a significant reduction in cytoplasmic Dicer expression in metastatic melanoma compared with primary melanoma, dysplastic nevi and normal nevi (Figure 3b and c), suggesting that reduced or lost expression of Dicer may be relevant to the process of melanoma metastasis. Interestingly, we also observed an inverse correlation between Dicer expression and AJCC staging of the melanoma cases (Figure 3d). Accordingly, the main and only significant difference in cytoplasmic staining was between AJCC stage II and III, which corresponds to transition from primary melanoma to lymph node metastasis. This observation is consistent with our Kruskal-Wallis and χ2 tests (Figure 3b & c), in which Dicer expression is mainly reduced in metastatic melanomas compared with the earlier stages of melanocytic lesions. Altogether, these data suggest that Dicer expression is reduced during the course of melanoma progression, implying an inhibitory role for Dicer in this process.
Aberrant expression of Dicer has been reported in multiple types of tumors. For instance, reduced expression of Dicer was shown in ovarian, lung, hepatocellular and basal cell carcinomas (26–30). On the other hand, overexpression of Dicer was reported in prostate cancer, colon and squamous cell carcinomas (30–32). This obvious discrepancy between different reports highlights the possibility of tissue specific function for Dicer in cancer development. In a recent report, Ma et al observed that Dicer protein is upregulated in melanoma compared to other skin cancers such as carcinomas and sarcomas (33). When compared among all examined cutaneous malignancies, they found Dicer upregulation in a tumor-type specific manner, namely in melanoma compared with melanocytic nevi (33). We believe that this discrepancy might be due to differences in sample size (521 cases in our study compared with 223 cases used by Ma et al) which could significantly affect the power of the study, inherent differences in the source populations from which the samples were collected or possible differences in the immunoreactivity of the antibodies. To further address this concern, we performed an immunohistochemistry test on a TMA construct containing 31 cases of dysplastic nevi, 71 cases of primary melanomas and 46 cases of metastatic melanomas using the same antibody mentioned in Ma et al report (Supplementary Figure S2). We observed that cytoplasmic Dicer staining using this antibody closely resembles our original TMA analysis (Figure 3), as opposed to what Ma et al observed. This further verifies our original findings that Dicer expression is decreased during the course of melanoma progression. Furthermore, we found that Dicer expression positively correlate with 5-year overall and disease-specific survival of primary and metastatic melanoma patients (Figure 4a and b), confirmed by a second anti-Dicer antibody (Supplementary Figure S3).
The survival analysis using data produced by both antibodies revealed a positive correlation between Dicer expression and disease specific 5-year survival (Figure 4a and Supplementary Figure S3). However, despite an observed significant correlation between cytoplasmic Dicer expression and overall patient survival (P = 0.007; Figure 4a) using the first antibody, we found no significant correlation between overall survival and Dicer expression using the second antibody (P = 0.077; Supplementary Figure S3b). We believe this is due to smaller sample size with the second antibody(n=117) compared with the analysis with the first antibody (n=397).
This is the first study on the correlation between Dicer expression and melanoma patient survival. As shown in Figure 2b, Dicer-KD significantly stimulated melanoma cell invasion which is one of the most critical events in the process of cancer progression toward metastasis (34), suggesting that Dicer functions as an inhibitor of melanoma metastasis, that is by far the most important cause of melanoma related death. Moreover, we found a positive correlation between expression of Dicer and lymphocytic response in primary melanomas (Table 1). Lymphocytic response in melanoma is known to be responsible for killing tumor cells and may induce spontaneous regression. In addition, lymphocytic response in melanoma vertical growth phase is a prognostic factor associated with better patient survival (35). Suppression of melanoma invasion and enhancement of lymphocytic response may explain the observed positive correlation between Dicer expression and higher survival rate in patients.
Interestingly, we also observed less expression of cytoplasmic Dicer in male samples compared with female individual (P = 0.009, Table 1). Using the second anti-Dicer antibody, we found a similar trend, although not statistically significant possibly due to smaller sample size (Supplementary Table S1). Although we are not aware of the functional significance or the mechanism responsible for this differential pattern of Dicer expression, it may be interesting to investigate possible gender-specific differences in miRNAs expression and/or maturation, especially during the course of cancer development.
By comparing the expression status of these Sox4 and Dicer proteins in our TMA database, we found that there is a significant positive correlation between their expression (Figure 5a), further confirming our observation that Dicer expression is regulated by Sox4 in melanoma cells. As a matter of fact, this phenomenon may not be exclusive to melanoma. For instance, independent studies demonstrated overexpression of both Dicer (32) and Sox4 (11) in prostate cancer. Considering the observed upregulation of Dicer by Sox4 in prostate cancer cell lines (19), it would be interesting to study the correlation between their expressions in this type of malignancy.
Finally, we investigated the role of Sox4 in biogenesis of mature miRNAs in melanoma cells in vitro. Our data demonstrated that knockdown of Sox4 alters the expression of the majority of miRNAs expressed by these cells (Figure 5b) implying involvement of Sox4 in the process of miRNA biogenesis. Consistent with our other observations, we found that rescued Dicer expression can revert the Sox4-KD induced changes in the expression of majority miRNAs. Our analysis showed a clear reversion in expression of those miRNAs which were either up- or down-regulated upon Sox4 knockdown toward normalcy after rescue of Dicer expression indicating the requirement for Dicer in regulation of miRNAs expression by Sox4 (Supplementary Table S3, S4, S5, S6). It is noteworthy that, although expression of majority miRNAs shifted upon overexpression of Dicer in Sox4 knockdown cells, a considerable number of miRNAs did not show a significant change which indicates existence of mechanisms other than Dicer upregulation by which Sox4 can modify the expression of these miRNAs. Regulating the expression of other miRNA biogenesis factors such as Drosha and Ago1 by Sox4, direct effect of Sox4 on promoters which control the expression of some pri-miRNAs and indirect effect of Sox4 on transcription of pri-miRNAs through downstream transcription factors such as p53, the expression of which has been shown to be regulated by Sox4 (36), are a few possible scenarios which need verification in future studies. We also studied the cellular functions that are regulated by those miRNAs, expression of which were affected by Sox4, using Ingenuity Pathway Analysis software. Our data revealed that reproductive system diseases represented the most relevant function in our analysis. This observation is not surprising, as Sox4 has been previously shown to be involved in reproductive system (37–38). Cancer related miRNAs were also one of the most significantly changed groups in our study (Figure 5c). Further analysis revealed that Sox4-KD affects the expression of cancer, metastasis and melanoma-related miRNAs, the majority of which are more than 2-fold down-regulated (Supplementary Table S3–S5). To our knowledge, this was the first study to investigate the role of Sox4 in regulation of miRNAs and demonstrated the significant effect of this gene in the expression profile of miRNAs in melanoma cells.
Although we showed that Sox4 and Dicer regulates invasion of melanoma cells in vitro and that their reduced expression levels are exhibited in advanced stages of melanoma, it is not known whether low expression levels of Sox4 and or Dicer would promote melanoma metastasis in vivo. We believe metastasis assays in mice, either using xenografts or tail vein injection of melanoma cells with Sox4 and Dicer knockdown would address this question in the future.
In conclusion, our data suggest a reduction in the expression of Dicer in melanoma which could be due to a downregulation of its upstream regulator Sox4, resulting in perturbation of the basic machinery controlling miRNA biogenesis in melanoma cells. We found that similar to Sox4, Dicer expression is positively correlated with melanoma patient survival, revealing the prognostic value of Dicer in melanoma. We further proved that alterations in the expression of Sox4 and Dicer significantly affect the invasive potential of melanoma cells and may play a key role in the molecular pathogenesis of melanoma. Our findings on Sox4 regulated miRNA biogenesis pathway may aid toward development of novel targeted therapeutic approaches for melanoma.
Materials and Methods
Cell culture, plasmid construction and siRNA transfection
Human melanoma cell lines, MMRU and Sk-mel-3 were cultured as described previously (18). The Sox4 open reading frame was subcloned from pOTB7-Sox4 (ImaGegen, Berlin, Germany) into 3×Flag-CMV-7.1 as previously described (18). pCDNA3-Flag-Dicer was kindly provided by Dr. Ian J. MacRae (The Scripps Research Institute). Plasmids were transfected into 50% confluent cells using Effectene transfection reagent (Qiagen, Valencia, CA). For RNAi experiments, cells were grown to 50% confluency before transfection of either nonspecific control siRNA (siCTR), Sox4 specific siRNA (sc-38412, Santa Cruz Biotechnology, Santa Cruz, CA) or Dicer siRNA (siDicer, Qiagen SI00300006) using siLentFect transfection reagent (Bio-Rad, Mississauga, Ontario, Canada). The cells were harvested at 72 hours after transfection for indicated analysis.
Western blot
Proteins extraction and Western blot was performed as described previously (39). The following antibodies were used for Western blot: rabbit anti-Sox4 (1:250; Santa Cruz Biotechnology), rabbit anti-Dicer (1:1000; Sigma, St Louis, MO), mouse anti-Flag (1:1000; Applied Biological Materials, Richmond, BC, Canada), and mouse anti-β-actin (1:10,000; Sigma). Infrared-dye-labelled secondary antibody was applied to the blots and signals were detected with Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln, NE).
Real-time reverse transcription PCR
Total RNA was prepared by Qiazol extraction (Qiagen) and reverse transcribed into cDNA with the Transcriptor cDNA Synthesis System (Roche, Indianapolis, IN, USA). Real-time qPCR was performed with SYBR Green Master mix system (Roche). The sequences of human Sox4 primers were 5′-GGTCTCTAGTTCTTGCACGCTC-3′ (forward) and 5′-CGGAATCGGCACTAAGGAG- 3′ (reverse). The primers for human Dicer were 5′-GTACGACTACCACAAGTACTT-3′ (forward) and 5′-ATAGTACACCTGCCAGACTGT-3′ (reverse). The primers for human β-actin were 5′-CCCTGAGGCACTCTTC-3′ (forward) and 5′-AGGTCTTTGCGGATGT-3′ (reverse).
Chromatin immunoprecipitation assay
MMRU cells were transfected with 3×Flag-Sox4 construct using the Effectene transfection reagent (Qiagen). Twenty four hours after transfection, formaldehyde-fixed cells were immunoprecipitated overnight with mouse anti-Flag (1:1000; Applied Biological Materials) and the associated genomic DNA was analyzed by PCR and agarose gel electrophoresis. We designed four sets of primers, spanning a distance from −3kb to +1000bp of the Dicer transcription start site. A list of the sequences of primers and the site of their corresponding amplicons is shown in Supplementary Table S7.
Luciferase reporter assay
The human Dicer promoter reporter gene construct (pGL3-DICER-Prom, (40) was kindly provided by Dr. D.E. Fisher (Harvard Medical School, MA) through Addgene (plasmid 25851). siCTR or siSox4 treated MMRU cells were plated in 12-well plates and transfected with pGL3-DICER-Prom. Firefly luciferase activities in the lysates were determined after 243h using the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA) and were normalized with Renilla pRL-CMV plasmid transfection.
Cell invasion assay
Cell invasion analysis was done using the Boyden chamber assay as previously described (41). In brief, 40 μl of 5 mg/ml matrigel (BD Biosciences, Mississauga, Canada) in serum-free medium was added to the upper compartment of 24-well Transwell culture chambers (with 8.0 μm pore size polycarbonate membrane). 5×104 cells suspended in 250 μl of serum-free medium were seeded on the upper compartment, and 750 μl of complete medium was added to the lower compartment and incubated for 24 hours. Invaded cells on the lower side of the filter were stained with 0.5% crystal violet and the retained dye was extracted by 30% acetic acid followed by reading the absorbance at 590 nm.
Sulforhodamine B (SRB) cell growth assay
Twenty four hours after transfection with siDicer and siCTR, MMRU and Sk-mel-3 cells were seeded in 24-well plates. Cells were fixed and stained as previously described (18). The initial time point (0 hours) was measured 6 hours after seeding. Subsequent time points were measured by fixing cells 24, 48, and 72 hours later.
miRNA profiling assay
The miRNA profiling array was carried out using Applied Biological Materials (ABM)’s miRNA profiling service (ABM C201). Total RNA from siCTR, siCTR/Flag-vector, siSox4, or siSox4/Flag-Dicer transfected MMRU cells was prepared with Qiazol extraction followed by Poly-A Tailing reactions and miRNA cDNA synthesis (ABM C204). Real-time qPCR reactions and instrumental analysis was performed using Roche LightCycler480. The expression profile of miRNAs in all four groups is deposited and publically available in GEO database (accession number: GSE36715). Functional analysis of the miRNA profiling data was performed using the Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA). Lists of miRNAs were generated by pair-wise comparison of our expression data sets (siSox4 vs siCTR and siSox4/F-Dicer vs CTR). miRNAs with 2-fold up- or down-regulation after Sox4-knockdown were subjected to the Ingenuity knowledge proprietary database to identify the biological functions that were most significant to the data sets.
Construction of TMA
Formalin-fixed paraffin-embedded tissues from 30 normal nevi, 87 dysplastic nevi, 262 primary melanomas, and 135 metastatic melanomas were used in the present study. All specimens were obtained from the 1990 to 1998 archives of the Department of Pathology, Vancouver General Hospital. The use of human skin tissues in this study was approved by the Clinical Research Ethics Board of the University of British Columbia. TMA slides were prepared as described previously (18).
Immunohistochemistry
Immunohistochemistry procedure was performed as described previously (18) using polyclonal Rabbit anti-Dicer antibody (1:100, Sigma HPA000694) or a monoclonal Rabbit anti-Dicer antibody (1:100; CloneGene, Hartford, CT).
Evaluation of immunostaining
The evaluation of cytoplasmic Dicer expression was made blindly by two independent observers (including one dermatopathologist) simultaneously. The cytoplasmic Dicer staining was scored into four grades according to the following staining intensities: 0, 1+, 2+, and 3+. Percentages of Dicer-positive cells were also scored into five categories: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The immunoreactive score, which is calculated by multiplying the scores of staining intensity and the percentage of positive cells, was used as the final staining score, defined as follows: 0, negative; 1–6, Weak and 8–12, strong.
Statistical analyses
The Kruskal-Wallis test was applied to compare the Dicer cytoplasmic staining between normal nevi, dysplastic nevi, primary melanoma and metastatic melanoma using the GraphPad prism4 software. Other statistical analyses were performed with the SPSS 11.5 software. The correlations between nuclear Sox4 expression and clinicopathologic variables, including age, gender, tumor thickness, location, and ulceration were analyzed byχ2 test. Fisher’s exact test was used to estimate the significance of the incidence of biological functions by calculating a P value determining the probability that the association between the miRNA dataset and these biological functions is significant. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Ian J. MacRae for providing the pCDNA3-Flag-Dicer construct, Stephanie Smith, Kate Orchard, Ronald Wong, Ladan Fazli, Larry Tan, Cecilia Sjoestroem, Liang L. Liu and Yabin Cheng for technical assistance and Drs. Michael Cox, Aziz Ghahary and Vincent Duronio for helpful discussions. This work was supported by grants from Canadian Institutes of Health Research (MOP-93810, MOP-110974 and CCI-117958) and Canadian Dermatology Foundation to G.L. S.M.J. is a recipient of University of British Columbia Graduate Fellowship. S.M.J. and G.S.A are recipients of Canadian Institutes of Health Research - Skin Research Training Centre Trainee Awards.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 4.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Gene Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315. e2301–2304. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, Luider TM, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10:648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu PB, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Research. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 12.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 13.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer research. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 14.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 15.Ahn SG, Kim HS, Jeong SW, Kim BE, Rhim H, Shim JY, et al. Sox-4 is a positive regulator of Hep3B and HepG2 cells’ apoptosis induced by prostaglandin (PG)A(2) and delta(12)-PGJ(2) Experimental & molecular medicine. 2002;34:243–249. doi: 10.1038/emm.2002.34. [DOI] [PubMed] [Google Scholar]
- 16.Hur EH, Hur W, Choi JY, Kim IK, Kim HY, Yoon SK, et al. Functional identification of the pro-apoptotic effector domain in human Sox4. Biochemical and biophysical research communications. 2004;325:59–67. doi: 10.1016/j.bbrc.2004.09.215. [DOI] [PubMed] [Google Scholar]
- 17.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, et al. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci USA. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jafarnejad SM, Wani AA, Martinka M, Li G. Prognostic significance of Sox4 expression in human cutaneous melanoma and its role in cell migration and invasion. Am J Pathol. 2010;177:2741–2752. doi: 10.2353/ajpath.2010.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 21.Castillo SD, Matheu A, Mariani N, Carretero J, Lopez-Rios F, Lovell-Badge R, et al. Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-3506. [DOI] [PubMed] [Google Scholar]
- 22.Wotton D, Lake RA, Farr CJ, Owen MJ. The high mobility group transcription factor, SOX4, transactivates the human CD2 enhancer. J Biol Chem. 1995;270:7515–7522. doi: 10.1074/jbc.270.13.7515. [DOI] [PubMed] [Google Scholar]
- 23.Vandewetering M, Oosterwegel M, Vannorren K, Clevers H. Sox-4, an Sry-Like Hmg Box Protein, Is a Transcriptional Activator in Lymphocytes. Embo J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su XH, Chakravarti D, Cho MS, Liu LZ, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–U168. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faggad A, Budczies J, Tchernitsa O, Darb-Esfahani S, Sehouli J, Muller BM, et al. Prognostic significance of Dicer expression in ovarian cancer-link to global microRNA changes and oestrogen receptor expression. J Pathol. 2010;220:382–391. doi: 10.1002/path.2658. [DOI] [PubMed] [Google Scholar]
- 28.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JF, Shen W, Liu NZ, Zeng GL, Yang M, Zuo GQ, et al. Down-regulation of Dicer in hepatocellular carcinoma. Med Oncol. 2011;28:804–809. doi: 10.1007/s12032-010-9520-5. [DOI] [PubMed] [Google Scholar]
- 30.Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, et al. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest. 2010;28:649–653. doi: 10.3109/07357901003630918. [DOI] [PubMed] [Google Scholar]
- 31.Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Z, Swede H, Cassarino D, Fleming E, Fire A, Dadras SS. Up-regulated Dicer expression in patients with cutaneous melanoma. PLoS One. 2011;6:e20494. doi: 10.1371/journal.pone.0020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Spatz A, Batist G, Eggermont AMM. The biology behind prognostic factors of cutaneous melanoma. Curr Opin Oncol. 2010;22:163–168. doi: 10.1097/CCO.0b013e328337fe8f. [DOI] [PubMed] [Google Scholar]
- 36.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, et al. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. P Natl Acad Sci USA. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt SM, Clarke CL. Expression and hormonal regulation of the Sox4 gene in mouse female reproductive tissues. Biol Reprod. 1999;61:476–481. doi: 10.1095/biolreprod61.2.476. [DOI] [PubMed] [Google Scholar]
- 38.Graham JD, Hunt SM, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol. 1999;22:295–304. doi: 10.1677/jme.0.0220295. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Li G. ING3 promotes UV-induced apoptosis via Fas/caspase-8 pathway in melanoma cells. J Biol Chem. 2006;281:11887–11893. doi: 10.1074/jbc.M511309200. [DOI] [PubMed] [Google Scholar]
- 40.Levy C, Khaled M, Robinson KC, Veguilla RA, Chen PH, Yokoyama S, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong RP, Ng P, Dedhar S, Li G. The role of integrin-linked kinase in melanoma cell migration, invasion, and tumor growth. Mol Cancer Ther. 2007;6:1692–1700. doi: 10.1158/1535-7163.MCT-07-0134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.