Abstract
Werner syndrome (WS) is a prototypical segmental progeroid syndrome characterized by multiple features consistent with accelerated aging. It is caused by null mutations of the WRN gene, which encodes a member of the RECQ family of DNA helicases. A unique feature of the WRN helicase is the presence of an exonuclease domain in its N-terminal region. Biochemical and cell biological studies during the past decade have demonstrated involvements of the WRN protein in multiple DNA transactions, including DNA repair, recombination, replication and transcription. A role of the WRN protein in telomere maintenance could explain many of the WS phenotypes. Recent discoveries of new progeroid loci found in atypical Werner cases continue to support the concept of genomic instability as a major mechanism of biological aging. Based on these biological insights, efforts are underway to develop therapeutic interventions for WS and related progeroid syndromes.
1. Introduction
Werner syndrome (WS; OMIM# 277700) is a rare genetic disorder that displays clinical features suggestive of accelerated aging. WS was originally described by a German medical student, Otto Werner, in 1904 (Werner, 1985). Werner reported a family of four siblings, ages 31 to 40, who presented with “Cataracts in Connection with Scleroderma” as well as short stature and premature graying of hair. The term, “Werner’s syndrome” was first used in 1934 by Oppenheimer and Kugel who described a new case of WS (Oppenheimer and Kugel, 1934) and subsequently by Thannhauser in 1945, who provided a comprehensive review of “Werner’s syndrome (Progeria of the Adults)” (Thannhauser, 1945). The gene responsible for WS was discovered in 1996 through then-ground breaking positional cloning method (Yu et al., 1996). This review summarizes our current understandings of clinical phenotypes, normal functions of the WRN gene product and potential therapeutic approaches.
2. Classical Aspects of the Werner syndrome
WS is a rare autosomal recessive disorder characterized by an array of features consistent with accelerated aging (Fig 1)(Oshima et al., 2014; Takemoto et al., 2013). This is one of the few adult-onset syndromes of accelerated aging in which patients generally develop normally until they reach adolescence. The first sign, often recognized retrospectively, is a lack of a growth spurt and a relatively short stature as adults. Beginning in the early third decade of life patients begin to develop an aged appearance that includes skin atrophy, loss of subcutaneous fat and graying and loss of hair. Bilateral cataracts requiring surgery are seen in virtually all cases by the late 20s or early 30s (Huang et al., 2006; Takemoto et al., 2013). This is accompanied by a series of common age-related diseases that appear during middle age. These disorders include type 2 diabetes mellitus, hypogonadism, osteoporosis, atherosclerosis and malignancies. Several studies report that 30-40% of WS cases had children before gonadal atrophy leading to early loss of fertility in their 30s (Goto, 1997; Takemoto et al., 2013). Indolent deep ulcerations around Achilles tendons and, less frequently, at elbows, are almost pathognomonic to WS. These are associated with extensive subcutaneous calcifications and often lead to amputation of feet or lower extremities (Takemoto et al., 2013). Other features frequently seen in WS include a high pitched hoarse voice (recognizable over the phone), characteristic facial features (a “pinched” facial appearance), thin limbs, truncal obesity, and flat feet. The most common causes of death are cancer and myocardial infarction, at a median age of 54 (Huang et al., 2006). This is 7 years older than the median age of death reported in 1996 (Epstein et al., 1966), likely owing to improvements of medical care, as the median age for the extraction of cataracts (at age 31) were comparable in both eras.
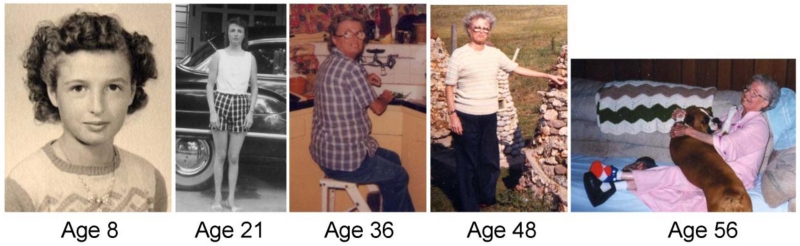
Fig. 1.
Werner syndrome patient with homozygous null WRN mutations. Although apparently normal at age 8, cataracts were removed at age 36 and severe ankle ulcerations were recorded at age 56. (Registry# SANAN1010) (Hisama et al., 2006)
There are clinical discordances in the presentation of age-related disorders between WS and normal aging. For example, systematic reviews of cancer in WS patients revealed a much higher incidence of sarcomas than expected for an age-matched control cohort (Goto et al., 1996; Lauper et al., 2013). The most common neoplasms in WS are thyroid follicular carcinomas, followed by malignant melanoma, meningioma, soft tissue sarcomas, primary bone tumors and leukemia/myelodysplasia. The elevated risk of these neoplasms ranges from 2 to 60-fold higher than population controls (Goto et al., 1996; Lauper et al., 2013). The arteriosclerosis of WS patients includes premature and severe forms of atherosclerosis, arteriolosclerosis and medial calcinosis. Hypertension, however, is not a common feature of WS. The cataracts seen in WS are almost always posterior sub-capsular, in contrast to those seen in normal aged people, which are typically nuclear cataracts. Osteoporosis in WS is more severe in distal limb bones than in the vertebral column, the opposite of what is seen in normal aged individuals. In addition, osteosclerosis of distal phalanges is highly characteristic of WS, though rarely observed during normal aging. There is no evidence for increased deposition of a variety of amyloids in WS, and dementias of the Alzheimer type are not a common feature of WS (Martin et al., 1999). Mental retardation, dysmorphology, skeletal anomalies and other developmental abnormalities are not features of WS; when present, they are likely due to co-existing disorders. The discordances above may be attributed to a number of factors, such as differential expressions and regulations of the WRN protein in various cell types and tissues, rates of cell turnover, and variations in the replicative potentials of various types of stem cells. The presence or absence of the compensatory enzymes or signal transduction pathways among various tissues may also play important roles. It is clear, however, that further studies are needed to explain the characteristic distributions of phenotypes.
Clinical criteria can be used to facilitate a diagnosis of WS. These are detailed at the International Registry of Werner Syndrome (Table 1) (www.wernersyndrome.org)(Oshima et al., 2014). Cardinal signs include bilateral cataracts (present in 99% of WS cases), premature graying and/or thinning of scalp hair (100%), characteristic dermatologic changes (96%) and short stature (95%)(Huang et al., 2006). Over 91% of affected individuals had all four cardinal signs (Huang et al., 2006). A related set of diagnostic criteria based on a national survey of 146 clinically diagnosed Japanese patients with WS lists progeroid changes of hair, cataracts, scleroderma-like changes of skin, intractable skin ulcers, soft-tissue calcifications, bird-like facies, and abnormal voice can serve as cardinal signs (Takemoto et al., 2013). Confirmation of a clinical diagnosis requires WRN gene testing.
Table 1.
Diagnostic criteria of Werner syndrome
| International Registry of Werner Syndrome (Oshima et al., 2014) | Japanese Registry (Takemoto et al., 2013) |
|---|---|
I. Cardinal signs and symptoms (onset over 10 years old)
|
I. Cardinal signs and symptoms (onset over 10 until 40years-of-age)
|
II. Further signs and symptoms
|
II. Other signs and symptoms
|
| III. Genetic testing | |
| Addendum: Mental retardation is seldom found in WS and cognitive function is often appropriate for the age |
|
| Definite: All the cardinal signs and two others. | Confirmed: All cardinal signs are present or a gene mutation in addition to at least three cardinal signs. |
| Probable: The first three cardinal signs and any two others | |
| Possible: Either cataracts or dermatological alterations and any four others. |
Suspected: Two or more cardinal signs or 1–2 cardinal signs in addition to other signs. |
| Exclusion: Onset of signs and symptoms before adolescence (except stature) |
3. WRN gene product and WRN mutations
Classical WS is caused by homozygous or compound heterozygous loss of function mutations in the WRN gene (Fig 2) (Yu et al., 1996). WRN is the only known gene in which mutations cause classical WS, and WS is the only known genetic disorder caused by null mutations of the WRN gene. The WRN locus is located on human chromosome 8p12, and consists of 34 coding exons spanning 140kb (Yu et al., 1996). The encoded WRN protein is a 1,432-amino acid, 160 kDa multifunctional nuclear protein with a 3’→5’ exonuclease domain in its N-terminal region (Huang et al., 1998), an ATP-dependent 3’→5’ helicase in its central region (Gray et al., 1997) and a nuclear localization signal in its C-terminal region (Matsumoto et al., 1997; Suzuki et al., 2001). There are two other consensus domains: the RecQ helicase conserved region (RQC) and the “helicase, RNaseD, C-terminal conserved region” (HRDC). Structural analysis and biochemical studies demonstrated that RQC, with its winged-helix (WH) domain, is critical for substrate-specific DNA binding to initiate unwinding (Kitano et al., 2010; Tadokoro et al., 2012; von Kobbe et al., 2003). The HRDC domain also plays a role in DNA binding, particularly for recruiting WRN protein to dsDNA breaks (DSBs)(Kitano et al., 2007; Lan et al., 2005; von Kobbe et al., 2003). The region between RQC and HRDC was also shown to possess single strand-DNA annealing activity and may influence oligomerization of the WRN protein (Muftuoglu et al., 2008).
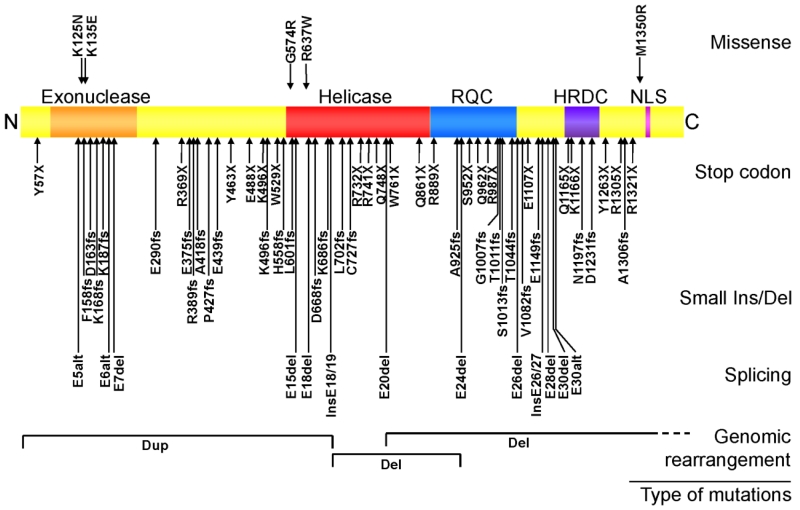
Fig. 2.
WRN disease mutations in classical WS patients. The rectangular box shows the WRN protein. Known functional domains are: exonuclease region (Exo), helicase region, RecQ C-terminus consensus region (RQC), helicase and RnaseD consensus region (HRDC) and the nuclear localization signal (NLS). Disease mutations are grouped based on the types of mutations. Splicing mutations that result in identical exon skipping are combined and indicated by the number of unique mutations as in (2). Splicing mutations that create new splice sites are indicated as “nss”. Modified from (Friedrich et al., 2010).
More than 70 different disease mutations have been identified in classical WS patients from all over the world (Friedrich et al., 2010; Uhrhammer et al., 2006). A majority of the disease mutations are stop codons, small indels or splicing mutations which cause truncations with loss of the nuclear localization signal at the C-terminus of the WRN protein and/or promote nonsense-mediated mutant mRNA decay. Two amino acid substitution mutations within the exonuclease domain, p.Lys125Asn and Lys135Glu, render the WRN protein unstable and are functionally null mutations as well (Huang et al., 2006). Genomic rearrangements leading to loss of function with or without involvement of neighboring loci have been identified by array comparative genome hybridization (array CGH)(Friedrich et al., 2010). Thus, virtually all WRN mutations in clinically ascertained patients are functional null alleles. There has been one report of possible genotype-phenotype correlation of the type of thyroid carcinoma with the location of WRN mutations in Japanese WS patients: follicular carcinomas were associated with C-terminal WRN mutations and papillary carcinoma with WRN N-terminal mutations (Ishikawa et al., 1999). Further studies are needed to clarify the effect of residual mutant WRN proteins in WS patients.
Two WRN amino acid substitution mutations have been identified within the helicase domain, p.Gly574Arg and p.Arg637Trp, in compound heterozygotes in association with null mutations (Friedrich et al., 2010; Uhrhammer et al., 2006). Protein studies confirmed that the p.Gly574Arg change abolishes helicase activity, and that the p.Arg637Trp is predicted to inactivate helicase function (Tadokoro et al., 2013). The phenotypes of patients carrying p. Gly574Arg or p.Arg637Trp appear to be indistinguishable from those with WRN null mutations. (Tadokoro et al., 2013; Uhrhammer et al., 2006)These data support the idea that the helicase activity is a crucial function of the WRN protein. Consistent with this idea, a mouse model carrying a homozygous helicase domain deletion mutation (Wrn^Δhel/Δhel) showed various signs of genomic instability and metabolic abnormalities resembling those seen in WS patients, and had 10-15% shorter median lifespan compared to controls (Labbe et al., 2011; Lebel et al., 2003). In contrast, a mouse model with a homozygous null mutation (Wrn^−/−) appeared to live beyond two years of age without developing an obvious phenotype under normal dietary and husbandry conditions (Lombard et al., 2000; Moore et al., 2008). These findings raise the possibility that the presence of helicase-deficient WRN protein may render subtle but deleterious dominant negative effects that are not present in individuals with null mutations.
Ethnicity-specific WRN mutations have been reported in Japanese, Sardinian, Indian/Pakistani, Moroccan, Turkish and Dutch patients (Friedrich et al., 2010; Saha et al., 2013a). The prevalence of specific WRN mutations varies depending on the level of consanguinity. In the Japanese population, the heterozygote frequency is estimated to be 1/166 (Satoh et al., 1999). Similarly, in the Sardinian population, a heterozygote frequency is estimated be of the order of 1/120 (Masala et al., 2007). It is likely that other founder mutations and populations with high prevalence will be identified as the awareness of WS increases, and as genetic testing become more common worldwide.
4. Cellular functions of the WRN gene product
Extensive biochemical studies of the substrate preference of WRN protein have identified complex DNA structures such as G4 quadruplexes, Holiday junctions and bubble structures as preferred substrates (Brosh et al., 2006; Compton et al., 2008; Kamath-Loeb et al., 2012a; Kamath-Loeb et al., 1998; Shen et al., 1998). This suggests a role of WRN in resolving complex intermediate DNA structures generated either normally or accidentally during DNA metabolism. Two major characteristics of cells derived from WS patients are genomic instability and very limited replicative capacity (Oshima et al., 1995; Salk et al., 1985a; Salk et al., 1985b). At the chromosomal level, genomic instability of WS fibroblasts has been described as variegated translocation mosaicism, with characteristic multiple, variable, predominantly stable chromosomal aberrations (Melcher et al., 2000). Biochemical and cell biological studies demonstrated that the roles of WRN protein in various DNA transactions, including DNA repair, replication, transcription and telomere maintenance, maintain genomic stability.
Double-strand breaks (DBSs) are potentially lethal DNA damages that can be generated by exogenous causes as well as by endogenous processes. DBSs are repaired through either homologous recombination (HR) or error-prone non-homologous end joining (NHEJ). It has been proposed that the choice between HR and NHEJ is determined in part by heterochromatin status and the chemical nature of the DSB, and that canonical NHEJ may be preferred for exogenous DSB repair during the G0 and G1 phases of the cell cycle (Kakarougkas and Jeggo, 2014; Keijzers et al., 2014). WRN physically interacts with a number of players in NHEJ such as Ku70/80, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and DNA ligase IV/XRCC4 (Kusumoto-Matsuo et al., 2014; Kusumoto et al., 2008). The potential physiologic significance of WRN in NHEJ repair is suggested by the demonstration of extensive deletions of reporter plasmids, particularly at 3’ overhang ends, in cells lacking WRN protein (Oshima et al., 2002). Ku70/80 and Ligase IV/XRCC4 stimulate WRN exonuclease but not WRN helicase activity, again suggesting a specific requirement of WRN in NHEJ (Cooper et al., 2000; Li and Comai, 2001). HR repair of DSBs may predominate during late S and G2 phases of the cell cycle, when a sister chromatid is available to serve as a repair template (Johnson and Jasin, 2000). Integrated recombination reporter substrates have been used to identify a role for the WRN helicase and exonuclease catalytic activities in promoting the resolution of recombinant DNA molecules during the postsynaptic phase of HR (Saintigny et al., 2002).
More recently, WRN was shown to participate in 5’→3’ DNA end resection in human cells, a step during HR that creates a substrate for RAD51 binding and subsequent D-loop formation and strand exchange (Sturzenegger et al., 2014). WRN carries this function in cooperation with the DNA2 endonuclease, likely using its helicase activity to generate a substrate for DNA2. In this role, WRN is redundant with its close homolog, BLM, though it is conceivable that different cell lineages may preferentially utilize one or the other RECQ helicase. In fact, a recently identified heterozygous mutation in RAD51, p.T131P, which elicits a Fanconi anemia-like phenotype, revealed that WRN/DNA2 and not BLM/DNA2 is recruited to resect DNA at mitomycin C crosslinks (Wang et al., 2015). Together, these findings establish a role for WRN early in the HR pathway.
HR also plays a major role in the protection or resolution and repair of stalled or broken replication forks during S phase in eukaryotic cells (Kakarougkas and Jeggo, 2014; Petermann and Helleday, 2010). Stalled replication forks may exist in a dynamic, regulated equilibrium between a regressed form (a “chicken foot”) and its reversal. A RECQ helicase, RECQ1 (or RECQL1), reverses regressed forks in a manner that is blocked by PARP1 activation (Berti et al., 2013), and RAD51 may protect forks from regression (Zellweger et al., 2015), as well as limit resection of DNA at forks (see below)(Hashimoto et al., 2010). Both regressed and un-regressed forks may become substrates for nascent DNA end resection that may differ in its extent and in the identities of the proteins involved, but is, in principle, similar to that occurring during HR (Neelsen and Lopes, 2015; Schlacher et al., 2011; Schlacher et al., 2012).
WRN and DNA2 are thought to execute productive, limited resection at stalled forks that facilitates fork restart when conditions become permissive for resumption of DNA synthesis. These functions of WRN have been uncovered using DNA fiber technologies that quantify replication fork progression and can demonstrate, for example, that in hydroxyurea-arrested WRN-deficient human cells both fork resection during arrest and fork restart after hydroxyurea removal, are reduced (Ammazzalorso et al., 2010; Franchitto et al., 2008; Iannascoli et al., 2015; Sidorova et al., 2013; Sidorova et al., 2008; Thangavel et al., 2015). Both WRN helicase and exonuclease activities are implicated in preserving replication forks for restart. WRN may also have an additional role in facilitating DNA synthesis for the first moments after fork restart (Sidorova et al., 2013; Sidorova et al., 2008), which is thought to involve its functional interaction with specialized polymerases such as polymerase eta (Kamath-Loeb et al., 2007). To add complexity to this already elaborate orchestra of activities surrounding stalled forks, it has recently been shown that WRN may have a non-enzymatic role in preventing unproductive nascent DNA resection at forks that developed double strand breaks due to an encounter with a camptothecin-induced topoisomerase I-DNA adduct (Su et al., 2014). Lastly, replication fork phenotypes of WRN deficiency are not limited to cases where replication progression is severely disrupted by genotoxic agents. An early study demonstrated increased stalling of nascent forks during an unperturbed S phase in primary fibroblasts deficient in WRN (Rodriguez-Lopez et al., 2002), and later work noted a subtle reduction in fork progression rate during a normal S phase in WRN-depleted transformed human fibroblasts (Sidorova et al., 2013). Chronic low-grade destabilization of forks that can drive the genomic instability phenotype of WRN deficiency, is also suggested by the increased common fragile site expression in WRN-deficient cells (Murfuni et al., 2012; Pirzio et al., 2008).
WRN may also participate in base excision repair through the interactions with APE endonuclease and DNA polymerase β (Sidorova and Monnat, 2015). There is a single publication showing the interaction of WRN and XPG, suggesting an involvement of WRN in nucleotide excision repair (Trego et al., 2011).
5. Telomere maintenance and Werner syndrome
Telomeres are DNA-protein structures that cap and protect the ends of chromosomes. Telomere complexes consist of TTAGGG repeats with 3’ single strand overhangs which can strand-invade within the repeat region to form a T-loop. Telomeric DNA is then bound by several proteins to form the shelterin complex. These proteins include TRF2, TRF1, POT1, TPP1, Rap 1 and TIN2 (O’Sullivan and Karlseder, 2010; Palm and de Lange, 2008). In normal cells, the shelterin components TRF2, TRF1 and POT1 directly bind to WRN protein and modulate its enzymatic activities (Edwards et al., 2014; Opresko, 2008). Several lines of evidence argue that telomeres are physiological substrates of WRN protein, and that aberrant telomere maintenance in the absence of WRN may be an important disease mechanism leading to the progeroid phenotypes of WS patients. Tandem repeats of TTAGGG sequences can form G4 quadruplexes, which are preferred substrates of WRN helicase in vitro (Brosh et al., 2006). In the absence of WRN protein, persistent G4 structures could lead to accelerated loss of telomere repeats if not resolved by other RECQ helicases in a timely fashion (Damerla et al., 2012). Replication of telomeres requires progressive resolution of four way junctions formed at the crosses of D- and T-loops, another preferred substrate of WRN (Opresko et al., 2009). Alteration of WRN function through expression of a dominant-negative WRN helicase mutant resulted in substantial loss of telomeric DNA that was associated with replication of the G-rich strand (Arnoult et al., 2009; Crabbe et al., 2004), consistent with the notion that cells lacking WRN helicase activity suffer delayed replication of the G4-forming telomere strand and subsequent shortening of telomeres from single sister chromatids. The increased vulnerability of telomeric G-rich DNA to WRN loss was explained by the consensus that these strands are replicated by the lagging mechanism only and thus spend more time in a single-stranded state, which facilitates G4 formation. Interestingly however, recent studies that combined telomere FISH and DNA fiber analysis of replication, showed that replication initiation events can in fact be detected within telomeric repeats, suggesting that a subset of G-rich strands are replicated by the leading strand mechanism (Drosopoulos et al., 2012; Sfeir et al., 2009). In mouse cells, where initiations within telomeric repeats are more prevalent than in human cells, it was possible to observe that the impact of WRN as well as BLM deficiency on fork progression was more pronounced in those DNA molecules where a G-rich strand was in fact copied by the leading strand mechanism (Drosopoulos et al., 2015). The same study confirmed that telomeric G4 signals, as visualized by a G4-specific antibody, were indeed elevated in WRN or BLM-deficient cells.
Whether or not WRN counteracts G4 formation in leading or lagging strand replication, it is clear that failure to replicate telomeric repeats properly will eventually lead to critically short telomeres, which are no longer end-protected, and can initiate a persistent DNA damage response and promote catastrophic chromosomal end fusions (Crabbe et al., 2007). Of note, in a WRN null mutant mouse model of WS, disease phenotypes were observed only in 3rd generation or later telomerase-deficient mice (mTerc^−/− Wrn^−/−) having short telomeres (Chang et al., 2004). Interestingly, hTERT immortalization of WS fibroblasts abolishes both the proliferative defect and some of the genotoxic hypersensitivity displayed by WS cells (Hisama et al., 2000). Although the degree of damage accumulation at non-telomeric DNA was not assessed in these WS hTERT fibroblasts, this observation is consistent with the idea that telomeres are particularly vulnerable to a loss of WRN function and that telomere phenotypes may be important in WS disease pathogenesis.
The roles of WRN protein at telomeres described above may in part explain the characteristic tumor spectrum of WS patients. The development of malignant neoplasms require a mechanism of telomere elongation, generally through re-activation of hTERT expression or through acquisition of the alternative lengthening of telomeres, referred to as ALT (an “alternative” mechanism). The ALT is mediated by homologous recombination of telomere sequences and is more frequently adopted by sarcomas (Bryan et al., 1997). Mouse embryonic fibroblasts (MEF) derived from the 5th generation (G5) of mTerc^−/− Wrn^−/− mice showed elevated numbers of telomere sister chromatid exchanges (T-SCE). Tumors derived from the G5 mTerc^−/− Wrn^−/− MEF showed the characteristics of ALT cells (Laud et al., 2005). Elevated SCEs, mainly localized to telomeres, were also observed in in human WRN deficient cells (Gocha et al., 2014; Hagelstrom et al., 2010). For example, the widely used AG11395 human WS cell line is ALT-positive (Fasching et al., 2005). These findings support the hypothesis that WRN normally suppresses T-SCEs, and that the absence of WRN facilitates the activation of ALT as a telomere maintenance mechanism.
6. Translational approaches to WS
Currently there is no cure for WS. Clinical management has focused on treating manifestations, preventing secondary complications and screening for acquired diseases common to WS. Treatment of WS patients is similar to that of the general population, with the exception of neoplasia, where the use of DNA-damaging chemotherapeutic agents may be modified to reflect the sensitivity of WS cells to several classes of chemotherapeutic agents (Mao et al., 2010).
Several novel therapeutic approaches are also being explored that may more directly influence WS disease progression. One example involves mTOR inhibitors. The mTOR pathway is a key modulator of aging and age-related diseases across wide varieties of species. Important biological roles of the mTOR pathway include promoting cell proliferation in nutrient-rich environments (high mTOR activity), and the diversion of metabolic resources for stem cell maintenance in nutrient-poor environments (low mTOR activity)(Johnson et al., 2013). We reported that mTOR signaling and basal autophagy are upregulated in WS cells, and that long-term rapamycin treatment resulted in improved growth rate, reduced accumulation of DNA damage foci and improved nuclear morphology. Autophagy markers were also reduced to near-normal levels, possibly due to the clearance of damaged proteins (Saha et al., 2014). DNA damage accumulation in WS cells may compromise protein homeostasis, which in turn further contributes to impaired DNA repair. Support for this hypothesis comes from the independent observation that hydrogen sulfide can abrogate a protein aggregation phenotype and attenuate oxidative damage in WS cells via modulation of the mTOR pathway (Talaei et al., 2013).
Selective inhibitors of p38 mitogen-activated protein kinase (MAPK) have also been investigated as a potential pharmaceutical intervention for WS and other genomic instability syndromes (Davis et al., 2005; Tivey et al., 2013). The MAPK mediates stress-signaling, which activates two tumor suppressor pathways, p53/p21WAF1 and pRb/p16INK4A and induces premature senescence of primary fibroblasts (Yaswen and Campisi, 2007). A small molecule that targets p38 MAPK was shown to suppress accumulation of p21WAF1 and restore the replicative potential of WS fibroblasts (Davis et al., 2005; Tivey et al., 2013). Neither mTOR inhibitors nor p38 MAPK inhibitors have as yet been tested in WS patients.
There is a single case report of astaxanthin-treatment of a WS patient, in which administration of this keto-cartinoid markedly improved the patient’s fatty liver possibly, via its anti-inflammatory and anti-oxidative capabilities (Takemoto et al., 2015).
The availability of human induced pluripotent cells (hiPSCs) has provided a new approach to disease modeling and treatment (Zhu and Huangfu, 2013). The hiPSCs are derived through the reprogramming of human somatic cells using ectopic expression of transcription factors, including OCT3/4, SOX2, KLF4, MYC, NANOG and LIN28 (Shimamoto et al., 2014; Takahashi and Yamanaka, 2006). There have been two reports of hiPSC derived from WS patient fibroblasts (Cheung et al., 2014; Shimamoto et al., 2014), both of which showed reversion of senescence-related cellular phenotypes observed in fibroblasts. Comparison of the gene expression profiles of WS and control fibroblasts revealed numerous differences, most of which are believed to be involved in cellular aging. The gene expression profiles of WS hiPSCs were indistinguishable from those of control hiPSCs fibroblasts (Cheung et al., 2014; Shimamoto et al., 2014). While introduction of hTERT was shown to reverse some of the WS cellular phenotypes (Cheung et al., 2014; Hisama et al., 2000), the reversion of gene expression profiles of WS hiPSCs cannot be solely attributed to activation of hTERT (Shimamoto et al., 2014). Moreover WS hiPSCs were karyotypically stable (Shimamoto et al., 2014). The evidence of significant degrees of genomic stabilization in these WS hiPSCs is consistent with a hypothesis of early development gene actions that compensate, or at least partially compensate, for null mutations at the WRN locus. These precedents provide the basis for determining whether stem cell-based therapies may eventually be applied to WS patients to, e.g., promote the healing of the leg ulcers that often leads to amputation in many WS patients (Shimamoto et al., 2015). The first human iPSC-based therapy has already begun, in September 2014, for age-related macular degeneration (Kamao et al., 2014).
WS human embryonic stem cells (hESCs) have also been generated by WRN knockout in hESCs. The analysis of WRN−/− hESCs and derived MSCs (mesenchymal stem cells) compared to WRN+/+ cells again showed cellular phenotypes associated with accelerated aging (Zhang et al., 2015). Epigenomic and chromatin analyses of WRN−/− MSCs revealed reduced levels of H3K9me3 as well as reduced expression of heterochromatin protein 1α (HP1α), suppressor of variegation 3-9 homolog 1 (SUV39H1), and lamina-associated polypeptide 2β (LAP2β). Interestingly, knockdown of SUV39H1 or HP1α in wild-type MSCs led to accelerated cellular senescence without induction of DNA damage makers, raising a possibility that chromatin disorganization, not DNA damage, may be responsible for the pathogenesis of WS (Zhang et al., 2015). In fact, loss of heterochromatin and the resulting transcriptional “leakage” have now been implicated as a factor in cellular aging, providing support for this novel understanding of WS etiology (O’Sullivan and Karlseder, 2012).
7. International Registry of Werner Syndrome and atypical Werner syndrome
The International Registry of Werner Syndrome (Department of Pathology, University of Washington, Seattle, WA) (www.wernersyndrome.org) was established in 1988 to recruit WS patients from all over the world for the positional cloning of the WRN gene (Yu et al., 1996). Following the cloning of the WRN gene, our Registry has expanded its scope to search for causative mutations and mechanisms responsible for a broader range of progeroid syndromes. One prominent example is our work to better define patients with “atypical Werner syndrome (AWS)”. These patients were suspected to have WS on clinical grounds, but were found on subsequent analysis to lack WRN mutations. As of 2015, our International Registry has enrolled 71 AWS cases, in addition to 149 WS patients with documented WRN mutations.
Our analyses of AWS patients have revealed several potential candidate genes. Among the first candidate genes examined in AWS was the LMNA gene, which encodes nuclear intermediate filaments, lamin A/C, and is known to be mutated in the Hutchinson-Gilford Progeria syndrome (HGPS) and other laminopathies (Eriksson et al., 2003). LMNA mutations identified in AWS are either dominant heterozygous amino acid substitutions, or are splicing mutations which result in the generation of amounts of progerin that are much smaller than those responsible for HGPS (Chen et al., 2003; Hisama et al., 2011). LMNA mutant cells show evidence of genomic instability such as accumulations of double strand breaks, redistribution of ATM/ATR, and accelerated telomere shortening (Benson et al., 2010; Liu et al., 2006; Saha et al., 2013b). Another gene mutated in a small subset of AWS is POLD1, which encodes the catalytic subunit of the replication DNA polymerase δ, a known WRN-interacting protein complex (Kamath-Loeb et al., 2012b; Lessel et al., 2015; Szekely et al., 2000). The primary role of POLD1 is in lagging strand synthesis, and in translesion synthesis (TLS) of lagging strands as well as leading strand synthesis (Johnson et al., 2015). The POLD1 mutations found in AWS are dominant heterozygous mutations, p.Ser605del and p.Arg507Cys, which perturb polymerase activity, unlike cancer-disposing germline mutations that abolish its exonuclease activity (Lessel et al., 2015; Palles et al., 2013). The p.Ser605del was previously described in patients with a multisystem disorder known as the MDPL syndrome (mandibular hypoplasia, deafness, progeroid features, lipodystrophy) (Weedon et al., 2013). Despite the obvious connection with genomic instability, malignancies leading to death are not reported among LMNA mutant and POLD1 mutant AWS in our Registry.
Next generation sequencing of members of two AWS pedigrees have also revealed homozygous and compound heterozygous mutations in SPRTN (SprT-like N-terminal domain), a gene previously associated with Ruijs-Aalfs syndrome (RAS)(Lessel et al., 2014b; Ruijs et al., 2003). RAS is characterized by developmental retardation, skeletal abnormalities and a progeroid appearance. Of note, SPRTN is an adaptor protein that binds to a TLS polymerase η (encoded by POLH) that functions in post-replication repair of DNA leading strands (Lessel et al., 2014b). Both of the SPRTN mutant individuals we studied developed hepatomas. We have also identified homozygous SAMHD1 (SAM domain- and HD domain-containing protein 1) mutation in a patient with progeroid feature and progressive ataxia (Lessel et al., 2014a). SAMHD1 encodes a dNTP pool regulator with 3′→5′ exonuclease activity, has been implicated in DNA damage response (Beloglazova et al., 2013), and when mutant has been linked to a subset of Aicardi-Goutieres syndrome (AGS). AGS is a genetically heterogeneous neurological disorder known for pronounced phenotypic variability. Our SAMHD1 mutant AWS patient carried a heterozygous WRN mutation which might have synergistic effects with SAMHD1 mutation (Lessel et al., 2014a). Searches for novel genetic alterations in additional AWS cases are currently in progress. The Registry now welcomes inquiries from clinicians who believe they have identified patients with either classical WS, or those with atypical features. We particularly welcome cases for which members of nuclear pedigrees are available for genetic analysis.
8. Conclusions
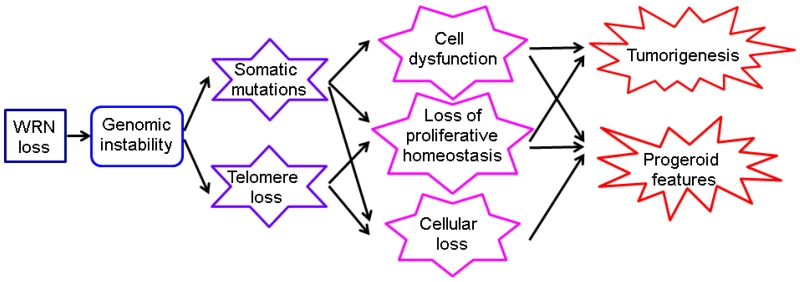
A combination of clinical and biological analyses of WS as a disease phenotype and the consequences of WRN mutations have led us to propose a simple model for the pathogenesis of WS (Fig 3)(Monnat, 2006). In this model, lack of WRN protein causes genomic instability, with the accumulation of somatic mutations, accelerated telomere attrition and cell dysfunction and cell loss in many cell lineages. These cellular consequences result in either atrophic or hyperplastic changes in many tissues or cell lineages, leading to progressive progeroid features accompanied by the emergence of tumors. Mutations of the WRN gene and various AWS genes appear to lead to overlapping yet distinct disease phenotypes, particularly as regards patterns of neoplasia. One can anticipate that further discoveries of novel segmental progeroid syndromes will also share features of genomic instability as a common pathogenetic mechanism underlying the accelerated appearances of a range of geriatric disorders, including a distinct disease- or syndrome-specific spectrum of neoplasia.
Fig. 3.
Model for the pathogenesis of WS. Modified from (Monnat, 2006). See text.
Acknowledgements
Authors thank Dr. George M. Martin for his critical comments. This was supported by the NIH grants, R24AG42328 (JO) and P01CA77852 (RJM, Jr).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. Embo J. 2010;29:3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Saintome C, Ourliac-Garnier I, Riou JF, Londono-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson EK, Lee SW, Aaronson SA. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J Cell Sci. 2010;123:2605–2612. doi: 10.1242/jcs.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Jr., Lopes M, Vindigni A. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr., Opresko PL, Bohr VA. Enzymatic mechanism of the WRN helicase/nuclease. Methods Enzymol. 2006;409:52–85. doi: 10.1016/S0076-6879(05)09004-X. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chen L, Lee L, Kudlow BA, Dos Santos HG, Sletvold O, Shafeghati Y, Botha EG, Garg A, Hanson NB, Martin GM, Mian IS, Kennedy BK, Oshima J. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- Cheung HH, Liu X, Canterel-Thouennon L, Li L, Edmonson C, Rennert OM. Telomerase protects werner syndrome lineage-specific stem cells from premature aging. Stem Cell Reports. 2014;2:534–546. doi: 10.1016/j.stemcr.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J Biol Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc Natl Acad Sci U S A. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Damerla RR, Knickelbein KE, Strutt S, Liu FJ, Wang H, Opresko PL. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012;11:3036–3044. doi: 10.4161/cc.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T, Baird DM, Haughton MF, Jones CJ, Kipling D. Prevention of accelerated cell aging in Werner syndrome using a p38 mitogen-activated protein kinase inhibitor. J Gerontol A Biol Sci Med Sci. 2005;60:1386–1393. doi: 10.1093/gerona/60.11.1386. [DOI] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210:191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J Cell Biol. 2012;197:253–266. doi: 10.1083/jcb.201112083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DN, Orren DK, Machwe A. Strand exchange of telomeric DNA catalyzed by the Werner syndrome protein (WRN) is specifically stimulated by TRF2. Nucleic Acids Res. 2014;42:7748–7761. doi: 10.1093/nar/gku454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Bower K, Reddel RR. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 2005;65:2722–2729. doi: 10.1158/0008-5472.CAN-04-2881. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich K, Lee L, Leistritz DF, Nurnberg G, Saha B, Hisama FM, Eyman DK, Lessel D, Nurnberg P, Li C, Garcia FVMJ, Kets CM, Schmidtke J, Cruz VT, Van den Akker PC, Boak J, Peter D, Compoginis G, Cefle K, Ozturk S, Lopez N, Wessel T, Poot M, Ippel PF, Groff-Kellermann B, Hoehn H, Martin GM, Kubisch C, Oshima J. WRN mutations in Werner syndrome patients: genomic rearrangements, unusual intronic mutations and ethnic-specific alterations. Hum Genet. 2010;128:103–111. doi: 10.1007/s00439-010-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocha AR, Acharya S, Groden J. WRN loss induces switching of telomerase-independent mechanisms of telomere elongation. PLoS One. 2014;9:e93991. doi: 10.1371/journal.pone.0093991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Hagelstrom RT, Blagoev KB, Niedernhofer LJ, Goodwin EH, Bailey SM. Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc Natl Acad Sci U S A. 2010;107:15768–15773. doi: 10.1073/pnas.1006338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisama FM, Bohr VA, Oshima J. WRN’s tenth anniversary. Sci Aging Knowledge Environ. 2006;2006:pe18. doi: 10.1126/sageke.2006.10.pe18. [DOI] [PubMed] [Google Scholar]
- Hisama FM, Chen YH, Meyn MS, Oshima J, Weissman SM. WRN or telomerase constructs reverse 4-nitroquinoline 1-oxide sensitivity in transformed Werner syndrome fibroblasts. Cancer Res. 2000;60:2372–2376. [PubMed] [Google Scholar]
- Hisama FM, Lessel D, Leistritz D, Friedrich K, McBride KL, Pastore MT, Gottesman GS, Saha B, Martin GM, Kubisch C, Oshima J. Coronary artery disease in a Werner syndrome-like form of progeria characterized by low levels of progerin, a splice variant of lamin A. Am J Med Genet A. 2011;155A:3002–3006. doi: 10.1002/ajmg.a.34336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, Dorn T, Spiegel R, Oral EA, Abid M, Battisti C, Lucci-Cordisco E, Neri G, Steed EH, Kidd A, Isley W, Showalter D, Vittone JL, Konstantinow A, Ring J, Meyer P, Wenger SL, von Herbay A, Wollina U, Schuelke M, Huizenga CR, Leistritz DF, Martin GM, Mian IS, Oshima J. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannascoli C, Palermo V, Murfuni I, Franchitto A, Pichierri P. The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic Acids Res. 2015;43:9788–9803. doi: 10.1093/nar/gkv836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Sugano H, Matsumoto T, Furuichi Y, Miller RW, Goto M. Unusual features of thyroid carcinomas in Japanese patients with Werner syndrome and possible genotype-phenotype relations to cell type and race. Cancer. 1999;85:1345–1352. [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerase delta in Replication of Both the Leading and Lagging DNA Strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, Oh K, Wasko BM, Ramos FJ, Palmiter RD, Rabinovitch PS, Morgan PG, Sedensky MM, Kaeberlein M. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamao H, Mandai M, Okamoto S, Sakai N, Suga A, Sugita S, Kiryu J, Takahashi M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb A, Loeb LA, Fry M. The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures. PLoS One. 2012a;7:e30189. doi: 10.1371/journal.pone.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein. II. Characterization of the integral 3′ --> 5′ DNA exonuclease. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Shen JC, Schmitt MW, Loeb LA. The Werner syndrome exonuclease facilitates DNA degradation and high fidelity DNA polymerization by human DNA polymerase delta. J Biol Chem. 2012b;287:12480–12490. doi: 10.1074/jbc.M111.332577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzers G, Maynard S, Shamanna RA, Rasmussen LJ, Croteau DL, Bohr VA. The role of RecQ helicases in non-homologous end-joining. Crit Rev Biochem Mol Biol. 2014;49:463–472. doi: 10.3109/10409238.2014.942450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18:177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Kitano K, Yoshihara N, Hakoshima T. Crystal structure of the HRDC domain of human Werner syndrome protein, WRN. J Biol Chem. 2007;282:2717–2728. doi: 10.1074/jbc.M610142200. [DOI] [PubMed] [Google Scholar]
- Kusumoto-Matsuo R, Ghosh D, Karmakar P, May A, Ramsden D, Bohr VA. Serines 440 and 467 in the Werner syndrome protein are phosphorylated by DNA-PK and affects its dynamics in response to DNA double strand breaks. Aging (Albany NY) 2014;6:70–81. doi: 10.18632/aging.100629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto R, Dawut L, Marchetti C, Wan Lee J, Vindigni A, Ramsden D, Bohr VA. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47:7548–7556. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe A, Garand C, Cogger VC, Paquet ER, Desbiens M, Le Couteur DG, Lebel M. Resveratrol improves insulin resistance hyperglycemia and hepatosteatosis but not hypertriglyceridemia, inflammation, and life span in a mouse model for Werner syndrome. J Gerontol A Biol Sci Med Sci. 2011;66:264–278. doi: 10.1093/gerona/glq184. [DOI] [PubMed] [Google Scholar]
- Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- Laud PR, Multani AS, Bailey SM, Wu L, Ma J, Kingsley C, Lebel M, Pathak S, DePinho RA, Chang S. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauper JM, Krause A, Vaughan TL, Monnat RJ., Jr. Spectrum and risk of neoplasia in Werner syndrome: a systematic review. PLoS One. 2013;8:e59709. doi: 10.1371/journal.pone.0059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am J Pathol. 2003;162:1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Hisama FM, Szakszon K, Saha B, Sanjuanelo AB, Salbert BA, Steele PD, Baldwin J, Brown WT, Piussan C, Plauchu H, Szilvassy J, Horkay E, Hogel J, Martin GM, Herr AJ, Oshima J, Kubisch C. POLD1 Germline Mutations in Patients Initially Diagnosed with Werner Syndrome. Hum Mutat. 2015;36:1070–1079. doi: 10.1002/humu.22833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Saha B, Hisama F, Kaymakamzade B, Nurlu G, Gursoy-Ozdemir Y, Thiele H, Nurnberg P, Martin GM, Kubisch C, Oshima J. Atypical Aicardi-Goutieres syndrome: is the WRN locus a modifier? Am J Med Genet A. 2014a;164A:2510–2513. doi: 10.1002/ajmg.a.36664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JC, Smith KR, Oehler J, Cabrera E, Freire R, Pope K, Nahid A, Norris F, Leventer RJ, Delatycki MB, Barbi G, von Ameln S, Hogel J, Degoricija M, Fertig R, Burkhalter MD, Hofmann K, Thiele H, Altmuller J, Nurnberg G, Nurnberg P, Bahlo M, Martin GM, Aalfs CM, Oshima J, Terzic J, Amor DJ, Dikic I, Ramadan K, Kubisch C. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nat Genet. 2014b;46:1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Comai L. Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J Biol Chem. 2001;276:9896–9902. doi: 10.1074/jbc.M008575200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rusinol A, Sinensky M, Wang Y, Zou Y. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J Cell Sci. 2006;119:4644–4649. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Beard C, Johnson B, Marciniak RA, Dausman J, Bronson R, Buhlmann JE, Lipman R, Curry R, Sharpe A, Jaenisch R, Guarente L. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol Cell Biol. 2000;20:3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Oshima J, Gray MD, Poot M. What geriatricians should know about the Werner syndrome. J Am Geriatr Soc. 1999;47:1136–1144. doi: 10.1111/j.1532-5415.1999.tb05240.x. [DOI] [PubMed] [Google Scholar]
- Masala MV, Scapaticci S, Olivieri C, Pirodda C, Montesu MA, Cuccuru MA, Pruneddu S, Danesino C, Cerimele D. Epidemiology and clinical aspects of Werner’s syndrome in North Sardinia: description of a cluster. Eur J Dermatol. 2007;17:213–216. doi: 10.1684/ejd.2007.0155. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Shimamoto A, Goto M, Furuichi Y. Impaired nuclear localization of defective DNA helicases in Werner’s syndrome. Nat Genet. 1997;16:335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- Melcher R, von Golitschek R, Steinlein C, Schindler D, Neitzel H, Kainer K, Schmid M, Hoehn H. Spectral karyotyping of Werner syndrome fibroblast cultures. Cytogenet Cell Genet. 2000;91:180–185. doi: 10.1159/000056841. [DOI] [PubMed] [Google Scholar]
- Monnat RJ., Jr. Werner syndrome as a model of human aging. In: Conn PM, editor. Handbook of models for human aging. Elsevier; 2006. pp. 961–976. [Google Scholar]
- Moore G, Knoblaugh S, Gollahon K, Rabinovitch P, Ladiges W. Hyperinsulinemia and insulin resistance in Wrn null mice fed a diabetogenic diet. Mechanisms of ageing and development. 2008;129:201–206. doi: 10.1016/j.mad.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I, De Santis A, Federico M, Bignami M, Pichierri P, Franchitto A. Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis. 2012;33:1655–1663. doi: 10.1093/carcin/bgs206. [DOI] [PubMed] [Google Scholar]
- Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. The great unravelling: chromatin as a modulator of the aging process. Trends Biochem Sci. 2012;37:466–476. doi: 10.1016/j.tibs.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer BS, Kugel VH. Werner’s syndrome - a heredo-familial disorder with scleroderma, bilateral juvenile cataracts, precocious graying of hair and endocrine stigmatization. Trans Assoc Amer Physicians. 1934;49:358–370. [Google Scholar]
- Opresko PL. Telomere ResQue and preservation--roles for the Werner syndrome protein and other RecQ helicases. Mechanisms of ageing and development. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Sowd G, Wang H. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PLoS One. 2009;4:e4825. doi: 10.1371/journal.pone.0004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima J, Campisi J, Tannock TC, Martin GM. Regulation of c-fos expression in senescing Werner syndrome fibroblasts differs from that observed in senescing fibroblasts from normal donors. J Cell Physiol. 1995;162:277–283. doi: 10.1002/jcp.1041620213. [DOI] [PubMed] [Google Scholar]
- Oshima J, Huang S, Pae C, Campisi J, Schiestl RH. Lack of WRN results in extensive deletion at nonhomologous joining ends. Cancer Res. 2002;62:547–551. [PubMed] [Google Scholar]
- Oshima J, Martin GM, Hisama FM. In: Werner Syndrome. Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews; Seattle (WA): 2014. [Google Scholar]
- Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- Ruijs MW, van Andel RN, Oshima J, Madan K, Nieuwint AW, Aalfs CM. Atypical progeroid syndrome: an unknown helicase gene defect? Am J Med Genet A. 2003;116A:295–299. doi: 10.1002/ajmg.a.10730. [DOI] [PubMed] [Google Scholar]
- Saha B, Cypro A, Martin GM, Oshima J. Rapamycin decreases DNA damage accumulation and enhances cell growth of WRN-deficient human fibroblasts. Aging Cell. 2014;13:573–575. doi: 10.1111/acel.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B, Lessel D, Nampoothiri S, Rao AS, Hisama FM, Peter D, Bennett C, Nurnberg G, Nurnberg P, Martin GM, Kubisch C, Oshima J. Ethnic-Specific WRN Mutations in South Asian Werner Syndrome Patients: Potential Founder Effect in Patients with Indian or Pakistani Ancestry. Mol Genet Genomic Med. 2013a;1:7–14. doi: 10.1002/mgg3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B, Zitnik G, Johnson S, Nguyen Q, Risques RA, Martin GM, Oshima J. DNA damage accumulation and TRF2 degradation in atypical Werner syndrome fibroblasts with LMNA mutations. Front Genet. 2013b;4:129. doi: 10.3389/fgene.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr. Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salk D, Au K, Hoehn H, Martin GM. Cytogenetic aspects of Werner syndrome. Adv Exp Med Biol. 1985a;190:541–546. doi: 10.1007/978-1-4684-7853-2_27. [DOI] [PubMed] [Google Scholar]
- Salk D, Bryant E, Hoehn H, Johnston P, Martin GM. Growth characteristics of Werner syndrome cells in vitro. Adv Exp Med Biol. 1985b;190:305–311. doi: 10.1007/978-1-4684-7853-2_14. [DOI] [PubMed] [Google Scholar]
- Satoh M, Imai M, Sugimoto M, Goto M, Furuichi Y. Prevalence of Werner’s syndrome heterozygotes in Japan. Lancet. 1999;353:1766. doi: 10.1016/S0140-6736(98)05869-3. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Kagawa H, Zensho K, Sera Y, Kazuki Y, Osaki M, Oshimura M, Ishigaki Y, Hamasaki K, Kodama Y, Yuasa S, Fukuda K, Hirashima K, Seimiya H, Koyama H, Shimizu T, Takemoto M, Yokote K, Goto M, Tahara H. Reprogramming suppresses premature senescence phenotypes of Werner syndrome cells and maintains chromosomal stability over long-term culture. PLoS One. 2014;9:e112900. doi: 10.1371/journal.pone.0112900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Yokote K, Tahara H. Werner Syndrome-specific induced pluripotent stem cells: recovery of telomere function by reprogramming. Front Genet. 2015;6:10. doi: 10.3389/fgene.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Kehrli K, Mao F, Monnat R., Jr. Distinct functions of human RECQ helicases WRN and BLM in replication fork recovery and progression after hydroxyurea-induced stalling. DNA Repair (Amst) 2013;12:128–139. doi: 10.1016/j.dnarep.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Li N, Folch A, Monnat RJ., Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Monnat RJ., Jr. Human RECQ helicases: roles in cancer, aging and inherited disease. Adv Genomics Genet. 2015;5:19–33. [Google Scholar]
- Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, Janscak P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J Biol Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Mukherjee S, Yang Y, Mori E, Bhattacharya S, Kobayashi J, Yannone SM, Chen DJ, Asaithamby A. Nonenzymatic role for WRN in preserving nascent DNA strands after replication stress. Cell Rep. 2014;9:1387–1401. doi: 10.1016/j.celrep.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shiratori M, Furuichi Y, Matsumoto T. Diverged nuclear localization of Werner helicase in human and mouse cells. Oncogene. 2001;20:2551–2558. doi: 10.1038/sj.onc.1204344. [DOI] [PubMed] [Google Scholar]
- Szekely AM, Chen YH, Zhang C, Oshima J, Weissman SM. Werner protein recruits DNA polymerase delta to the nucleolus. Proc Natl Acad Sci U S A. 2000;97:11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Kulikowicz T, Dawut L, Croteau DL, Bohr VA. DNA binding residues in the RQC domain of Werner protein are critical for its catalytic activities. Aging (Albany NY) 2012;4:417–429. doi: 10.18632/aging.100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Rybanska-Spaeder I, Kulikowicz T, Dawut L, Oshima J, Croteau DL, Bohr VA. Functional deficit associated with a missense Werner syndrome mutation. DNA Repair (Amst) 2013;12:414–421. doi: 10.1016/j.dnarep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takemoto M, Mori S, Kuzuya M, Yoshimoto S, Shimamoto A, Igarashi M, Tanaka Y, Miki T, Yokote K. Diagnostic criteria for Werner syndrome based on Japanese nationwide epidemiological survey. Geriatr Gerontol Int. 2013;13:475–481. doi: 10.1111/j.1447-0594.2012.00913.x. [DOI] [PubMed] [Google Scholar]
- Takemoto M, Yamaga M, Furuichi Y, Yokote K. Astaxanthin Improves Nonalcoholic Fatty Liver Disease in Werner Syndrome with Diabetes Mellitus. J Am Geriatr Soc. 2015;63:1271–1273. doi: 10.1111/jgs.13505. [DOI] [PubMed] [Google Scholar]
- Talaei F, van Praag VM, Henning RH. Hydrogen sulfide restores a normal morphological phenotype in Werner syndrome fibroblasts, attenuates oxidative damage and modulates mTOR pathway. Pharmacol Res. 2013;74:34–44. doi: 10.1016/j.phrs.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, Cejka P, Stewart S, Lopes M, Vindigni A. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannhauser SJ. Werner’s syndrome (progeria of the adults) and Rothmund’s syndrome: Two types of closely related hederofamilial atrophic dermatitis with juvenile cataracts and endocrine features; a critical study of five new cases. Ann Intern Med. 1945;23:599–626. [Google Scholar]
- Tivey HS, Brook AJ, Rokicki MJ, Kipling D, Davis T. p38 (MAPK) stress signalling in replicative senescence in fibroblasts from progeroid and genomic instability syndromes. Biogerontology. 2013;14:47–62. doi: 10.1007/s10522-012-9407-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trego KS, Chernikova SB, Davalos AR, Perry JJ, Finger LD, Ng C, Tsai MS, Yannone SM, Tainer JA, Campisi J, Cooper PK. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle. 2011;10:1998–2007. doi: 10.4161/cc.10.12.15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrhammer NA, Lafarge L, Dos Santos L, Domaszewska A, Lange M, Yang Y, Aractingi S, Bessis D, Bignon YJ. Werner syndrome and mutations of the WRN and LMNA genes in France. Human mutation. 2006;27:718–719. doi: 10.1002/humu.9435. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, Abhyankar A, Sougnez C, Gabriel SB, Auerbach AD, Kowalczykowski SC, Smogorzewska A. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Ellard S, Prindle MJ, Caswell R, Lango Allen H, Oram R, Godbole K, Yajnik CS, Sbraccia P, Novelli G, Turnpenny P, McCann E, Goh KJ, Wang Y, Fulford J, McCulloch LJ, Savage DB, O’Rahilly S, Kos K, Loeb LA, Semple RK, Hattersley AT. An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat Genet. 2013;45:947–950. doi: 10.1038/ng.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner O. On cataract in conjunction with scleroderma (translated by H. Hoehn) In: Salk D, Fujiwara Y, Martin GM, editors. Werner’s Syndrome and Human Aging, Vol. 190, Advances in Experimental Medicine and Biology. Plenum Press; New York: 1985. pp. 1–14. [Google Scholar]
- Yaswen P, Campisi J. Oncogene-induced senescence pathways weave an intricate tapestry. Cell. 2007;128:233–234. doi: 10.1016/j.cell.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Li M, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu GH, Belmonte JC. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Huangfu D. Human pluripotent stem cells: an emerging model in developmental biology. Development. 2013;140:705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]