Abstract
Transformation-associated recombination (TAR) cloning represents a unique tool for isolation and manipulation of large DNA molecules. The technique exploits a high level of homologous recombination in the yeast Sacharomyces cerevisiae. So far, TAR cloning is the only method available to selectively recover chromosomal segments up to 300 kb in length from complex and simple genomes. In addition, TAR cloning allows the assembly and cloning of entire microbe genomes up to several Mb as well as engineering of large metabolic pathways. In this review, we summarize applications of TAR cloning for functional/structural genomics and synthetic biology.
Keywords: transformation-associated recombination, TAR cloning, human artificial chromosome, HAC, synthetic biology
Introduction
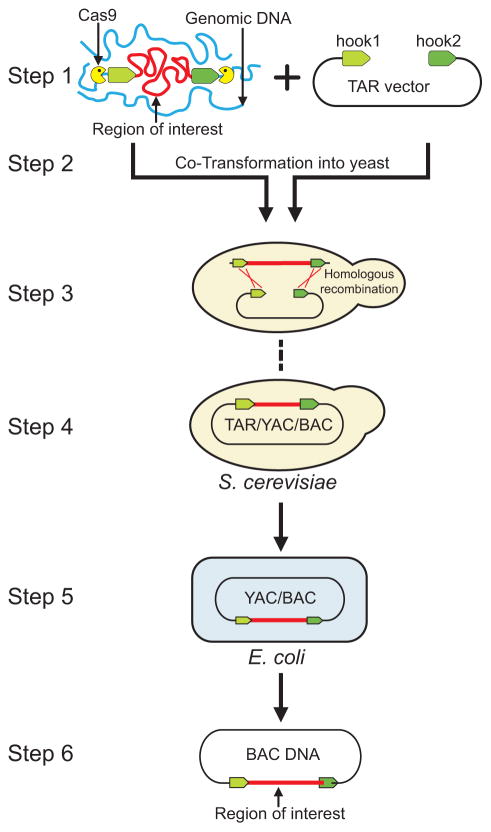
Transformation-associated recombination (TAR) cloning is based on the well-known property of free DNA ends as efficient substrates for homologous recombination in yeast (Orr-Weawer et al. 1981). Assembly of two DNA molecules by recombination in yeast was first demonstrated by Botstein and colleagues (Kunes et al. 1985; Ma et al. 1987). This team reported that a yeast vector DNA, which is broken in a sequence absent in the yeast genome, is efficiently rescued by recombination with a homologous restriction fragment included during yeast transformation (Kunes et al. 1985). A couple of years later, the authors suggested using this in vivo bi-molecular reaction as a novel method for plasmid construction as an alternative to the more common method of in vitro DNA ligation (Ma et al. 1987). The breakthrough step was the demonstration that at specific conditions, transformation-associated recombination in yeast can be utilized for selective isolation of large genomic regions from total mammalian genomic DNA as linear or circular yeast artificial chromosomes (YACs) (Larionov et al. 1996; Larionov et al. 1997; Kouprina et al. 1998; Kouprina and Larionov 2003). While the principles of this technology were developed in the late 1990s, the main parameters of TAR cloning have been modified during the past few years, which has allowed for highly efficient and reproducible gene targeting. Under such optimized conditions, any desirable chromosomal fragment up to 300 kb can be isolated in yeast from multiple samples within 2 weeks with a yield of gene/genomic fragment positive clones as high as 32% (Kouprina and Larionov 2006; 2008; Lee et al. 2015). For comparison, the frequency of gene-positive clones in genomic BAC libraries that are generated by DNA ligation is less than 0.003% (Asakawa et al. 1997). The selectivity of gene isolation by TAR cloning from complex genomes was achieved by use of highly competent yeast spheroplasts and exclusion of a yeast origin of replication (ARS element) from a TAR vector. Propagation of TAR-generated YACs in yeast cells absolutely depends on acquisition of mammalian DNA fragments with ARS-like sequences that can function as an origin of replication in yeast. Because these sequences are common in mammalian DNA, with approximately one ARS-like sequence per 20–30 kb (Stinchcomb et al. 1980), most mammalian genes can be isolated by TAR cloning. For chromosomal regions with multiple repetitive elements (such as the centromere and telomere) for GC-rich regions that are poor in ARS-like sequences, and for very simple genomes, the ARS frequency might be reduced, which precludes their isolation by the standard method. For such cases, a modified TAR method has been developed (Noskov et al. 2003). In this system, the TAR vector contains an ARS and a counter-selectable marker. Negative genetic selection eliminates the background caused by vector recircularization that results from end-joining during yeast transformation. It is worth noting that cloning of large GC-rich bacterial DNA fragments may sometimes be challenging even in the presence of an ARS-containing vector, and in this case, only fragments of approximately 100 kb can be recovered (Noskov et al. 2012). A general scheme of TAR cloning of a single copy gene from total genomic DNA is presented in Figure 1.
Fig. 1.
The consecutive experimental steps of selective gene isolation. Step1: The diagram shows TAR cloning of a gene of interest from total genomic DNA with a TAR vector containing YAC and BAC cassettes and two unique targeting sequences (hook1 and hook2) (in green) homologous to the 5′ and 3′ end of a gene of interest. The size of hooks may be as small as 60 bp (Noskov et al. 2001). The hook sequences may either be both unique, or one of the hook sequences may be a common repeat (for example, an Alu repeat for cloning from human genomic DNA). The TAR vector DNA is linearized by a unique endonuclease located between the hooks to expose targeting sequences. If necessary, genomic DNA may be treated by CRISPR/Cas9 endonuclease before yeast transformation, which will greatly increase the yield of gene/region-positive TAR clones. Step 2: Genomic DNA and a linearized TAR vector are co-transformed into yeast Saccharomyces cerevisiae cells. Steps 3 and 4: Recombination between targeting sequences in the vector and the targeted sequences in the genomic DNA fragment leads to rescue of the fragment (or gene) as a circular TAR/YAC/BAC molecule. Step 5: Transferring TAR-isolated molecules containing a region of interest from yeast cells to bacterial cells by electroporation. Step 6: BAC DNA is isolated by a standard procedure for further sequencing or functional analysis.
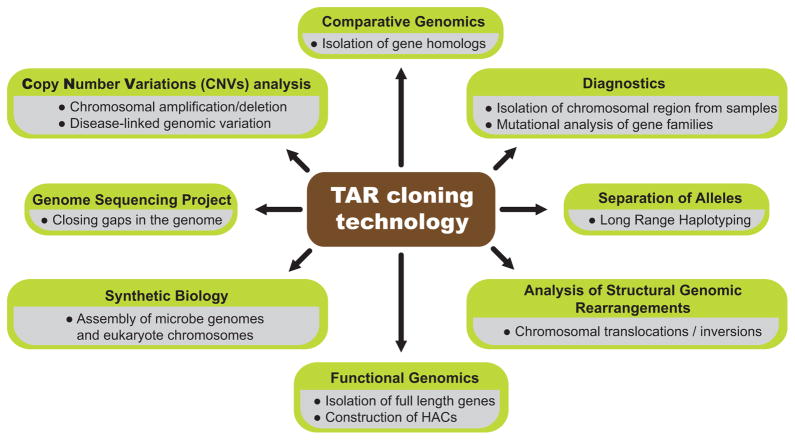
TAR cloning has found many applications in the post genomic era (Fig. 2). For example, it serves as a tool for selective isolation of a specific chromosomal segment or gene from an individual. It can also be used to isolate rearranged chromosomal regions, such as translocations and inversions, from patients and model organisms. Here, we provide an overview of the recent applications of TAR cloning for structural and functional genomics as well as for synthetic biology. We also discuss the role of TAR cloning in construction of gene delivery vectors based on human artificial chromosomes (HACs), along with the benefits of coupling TAR gene cloning technology with the HAC gene delivery and expression system.
Fig. 2.
Multiple applications of the TAR cloning technology.
Applications of TAR cloning
Selective isolation of full-size single copy genes and gene clusters from total genomes
More and more publications have appeared describing complex mechanisms regulating gene or gene cluster expression by means of alternative splicing, alternative promoter-enhancer usage, and expression of non-coding RNAs from intronic regions. Therefore, full-size genes containing all of the necessary regulatory regions become preferable for gene functional studies because only in such configurations can “physiological” expression be achieved. The efficient homologous recombination machinery of the budding yeast host allows for the selective isolation of any full-length gene or gene cluster from entire complex genomes through recombination between a TAR cloning vector, containing targeting sequences homologous to a region/gene of interes and homologous sequences in the co-transformed genomic DNA (Fig. 1). This results in rescue of the desired chromosomal fragment or gene in a circular or linear form that is able to propagate, segregate, and be selected in yeast cells.
For complex genomes such as human, the basic TAR cloning protocol yields between 0.5 and 5 % of the target gene-positive clones (Kouprina and Larionov 2006), while for simple microbe genomes such as the protist Trypanosoma brucei, Trypanosoma equiperdum, or Plasmodium falciparum, the yield is much higher. For example, the yield of the TAR-cloned Plasmodium falciparum var genes reached up to 48% (Gaida et al. 2011), while TAR cloning of the repertoires of VSG expression site containing telomeres isolated from Trypanosoma equiperdum resulted in a yield as high as 18% (Young et al. 2008). Recently, we significantly increased the fraction of gene-positive clones isolated from complex genomes up to 32% by using Cas9 nucleases that create specific double strand breaks bracketing the target genomic DNA sequence, which makes the ends of the targeted region highly recombinogenic (Lee et al. 2015) (Fig. 1).
TAR cloning of genes and gene clusters from simple genomes
At present, TAR cloning is routinely used for isolation of genes and gene clusters from simple genomes such as bacteria, protist and virus. The method was also adopted for rapid construction of biochemical pathways. Examples of direct capture of genes or gene clusters for functional studies or commercial purposes include cloning of the repertoire of individual Plasmodium falciparum var genes up to 30 kb in size (Gaida et al. 2011), the Pseudoalteromonas alterochromide gene cluster of 34 kb in size controlling the production of alterochromide lipopeptides (Ross et al. 2015), the nataxazole biosynthesis gene cluster for heterologous expression into S. albus J1074 and S. lividans JT46 (Cano-Prieto et al. 2015), the lipopeptide gene cluster of 73 kb in size from the marine actinomycete Saccharomonospora sp. to yield the antibiotic taromycin A (Yamanaka et al. 2014), the Salinispora pacifica natural product gene cluster of 21.3 kb in size for the biosynthesis of enterocin (Bonet et al. 2015), the amicoumacin biosynthetic gene cluster from the marine Bacillus subtilis 1779 for antibiotic production in Bacillus host (Li et al. 2015a), as well as identification of thiotetronic acid antibiotic biosynthetic pathways in Salinispora (Tang et al. 2015), isolation of genes controlling production of colibactin in E. coli to clarify the biosynthetic pathway to (pre)colibactin (Li et al. 2015b) and isolation of the repertoire of VSG expression site containing telomeres from Trypanosoma brucei gambiense, T. b. brucei and T. equiperdum (Becker et al. 2004; Young et al. 2008). One very interesting work presents a general experimental framework that permits the recovery of large natural product biosynthetic gene clusters on overlapping soil-derived eDNA (environmental) clones and the resulting reassembly of these large gene clusters (~90 kb) using transformation-associated recombination (TAR) in yeast (Kim et al. 2010; Feng et al. 2011). The application of TAR cloning for rapid cloning and assembly of biosynthetic gene clusters from collections of overlapping eDNA clones is an important step toward being able to functionally study larger natural product gene clusters from uncultured bacteria. Large-scale functional cloning followed by screening of resulting eDNA clones should be a productive strategy for generating previously structurally uncharacterized chemical entities for use in future drug development efforts. TAR cloning has also been adapted to serve as a general platform for the engineering of large metabolic pathways from basic parts, which is summarized in the referenced publications (Shao et al 2009; Ongley et al. 2013; Agarwal et al. 2014; Mizutani 2015; Yuan et al. 2016). To summarize, TAR cloning from microbial genomes helps to better understand the evolutionary and physiological role of natural products, their structural/functional features, mechanisms of action and biosynthesis, which is can be used in the future to address fundamental questions in environmental evolution and biotechnology (Giordano et al. 2015).
TAR cloning of full-size genes and entire loci from complex genomes
Complex genomes include animal and plant genomes. Because TAR cloning is applicable for multiple genomic samples and clinical material, it is becoming a powerful tool for functional studies in complex genomes. Over the past few years, TAR cloning has been used to isolate unique regions, full-size genes, and gene clusters up to 250 kb from genomes of the human, nonhuman primate, and mouse (Kouprina and Larionov 2006; Kim et al. 2011; Kononenko et al. 2014). Physical analysis has demonstrated a high fidelity of the material isolated by TAR cloning. For example, by transfecting the isolates into mammalian cells, the functionality of several genes was confirmed, including the breast cancer gene BRCA1 (Annab et al. 2000; Kononenko et al. 2014), the human HPRT gene (Kouprina et al. 1998), the tumor suppressor gene KAI1 (Kouprina and Larionov 2006), the metastasis suppressor gene TEY1 (Nihei et al. 2002), the human TERT gene coding for a catalytic subunit of human telomerase (Leem et al. 2002), the cancer-associated genes VHL (mutated in von Hippel–Lindau syndrome) and NBS1 (mutated in Nijmegen breakage syndrome (Kim et al. 2011). Further evidence of the accuracy of the TAR cloning was obtained by isolation and shot-gun-sequencing of the genomic regions containing the SPANX (Sperm Protein Associated with the Nucleus on the X chromosome) genes (Kouprina et al. 2004a; 2005a; 2007a; 2007b; 2012). The sequences of one of the SPANX gene family members, SPANX-C, isolated from several individuals differed only by a single nucleotide substitution due to natural polymorphism (Kouprina et al. 2005a). TAR-isolated genes can also be successfully used in transgenesis. For example, transgenic mice carrying the entire 50 kb hTERT locus were constructed and used to show that in vivo expression of human and mouse TERT genes differ significantly, raising a concern about the use of mouse models for human cancer and aging (Horikawa et al. 2005).
Separation of gene alleles and long-range molecular haplotyping
Haplotype analysis is a common tool used in association studies. The joint distribution of adjacent markers in a population, which is the haplotype frequency, represents the correlation between those markers. A major difficulty in using haplotype markers lies in determining the haplotype phase for individuals heterozygous for more than one marker. Classically, haplotypes are resolved by pedigree analysis, but this approach is limited by having to collect DNA samples from appropriate family members and by meiotic recombination. Separation of individual chromosomes by construction of hybrid cell lines, microdissection of chromosomes, or amplification of spermatocyte DNA are also laborious and time-consuming and cannot be used for a large number of individuals. At present, many computer programs are available for haplotype analysis of samples of unrelated individuals (Salem et al. 2005; Kuleshov et al. 2014; Wang et al. 2015), though all of them have limitations and are difficult in resolving the phase of maternal and paternal chromosomes. TAR cloning can be used to unambiguously resolve the “haplotype problem”. Both parental alleles of a gene can be quickly isolated in one TAR cloning experiment because homologous recombination between targeting hooks and the target sequence occurs at equal frequencies in both chromosomes (Fig. 3).
Fig. 3.
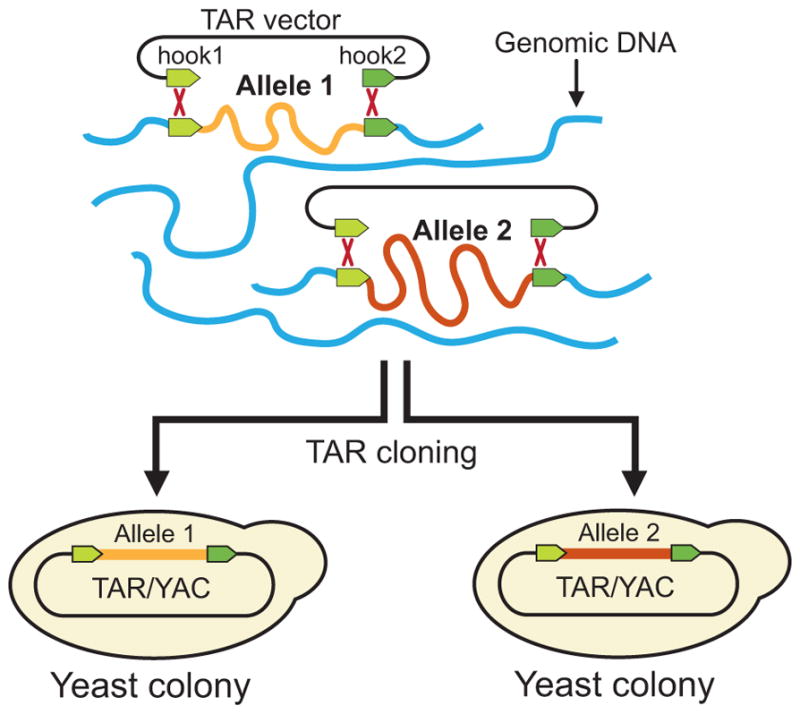
TAR cloning for separation of alleles and long-range haplotyping. TAR cloning experiments result in isolation of multiple gene-positive clones. Thus, the genomic region corresponding to different alleles becomes separated into different yeast cells, allowing for independent analysis.
One good example of the application of TAR cloning for this purpose is separation of the alleles of the hTERT gene (Kim et al. 2003). To prove that different alleles were indeed isolated, we examined the VNTR (a variable number of tandem repeats) blocks of the TAR isolates. As known, the hTERT gene contains four VNTR blocks – two in intron 2 and two in intron 6. Earlier analysis of the segregation of these VNTRs in families revealed that all were transmitted through meiosis and followed a Mendelian inheritance pattern. Analysis of TAR isolates containing parental hTERT alleles determined the specific combination of minisatellites at each of the polymorphic sites.
An example of the application of TAR cloning for long-range molecular haplotypying is isolation of the SPANX genes from different individuals (Kouprina et al. 2007a; 2007b; 2012). This case is very complicated because sequences of these gene members have a high level of homology and reside within large segmental duplications with >95% identity. Therefore, their mutational analysis could not be carried out by a routine PCR analysis or by the next-generation sequencing. We applied a TAR-cloning-based approach by cloning each member of this SPANX gene family (SPANX-A1, SPANX-A2, SPANX-B, SPANX-C, and SPANX-D). Analysis of the TAR clones revealed that roughly two thirds of SPANX sequence variants resulted from gene conversion suggesting a high frequency of recombination between the genes. Based on traces of gene conversion detected in the SPANX-C locus, it was concluded that recombinational interaction operates over a long distance (SPANX-D and SPANX-C genes are separated by more than 500 kb). In most cases, reports of gene conversion in the human genome are based on comparison of DNA sequences obtained from different individuals. In the case of SPANX genes, donor and targeted sequences were isolated from the same individual. Therefore, our data provided a unique opportunity to analyze the mechanism of gene conversion in humans and build long-range SPANX haplotypes in individuals. Analogously, TAR cloning may be applicable to other gene families and large segmental duplications (SDs), whose analysis is prohibited by a high level of homology between family members.
For a long time, the cluster of SPANX genes was considered to play a role in prostate cancer. Direct isolation of a set of overlapping genomic segments of the 750 kb region in X-linked families with predisposition to prostate cancer revealed no disease-specific alterations within the SPANX gene cluster (Kouprina et al. 2012). Thus, TAR cloning allowed exclusion of the 750-kb genetically unstable region at Xq27 as a candidate locus for prostate malignancy.
To summarize, TAR cloning is well suited for separation of alleles and for a large-scale analysis of long-range haplotypes in multiply heterozygous individuals, and can also be used to identify haplotypes that may contribute to disease.
Isolation of gene homologues for evolutionary studies
Since up to 15% divergence in DNA sequences does not prevent recombination during transformation in yeast (Noskov et al. 2003b), TAR cloning can be used to isolate gene homologues. This technique was used to reconstruct the evolutionary history of several disease-associated genes: the breast cancer gene BRCA1 (Pavlicek et al. 2004), the microcephaly gene ASPM (Kouprina et al. 2004b), the cancer/testis-specific antigen gene family SPANX (Kouprina et al 2004a; 2005a), and the ATM and NBS1 genes involved in DNA repair (NK, unpublished observations). Complete genomic copies of these genes from human, chimpanzee, gorilla, bonobo, orangutan, and rhesus macaque genomes were isolated using TAR vectors containing human-specific targeting sequences developed from 5′ and 3′ gene-flanking regions. Analysis of cloned sequence regions revealed that some genes have evolved under strong positive selection. For example, ASPM, which encodes a centrosomal protein (Kouprina et al. 2005b), showed accelerated evolution in the African hominoid clade, preceding hominid brain expansion by several million years (Kouprina et al. 2004b). These data suggest that the evolutionary selection of specific segments of the ASPM sequence strongly relates to differences in cerebral cortical size. Analysis of the non-synonymous/synonymous ratio in the BRCA1 gene revealed that most of the internal BRCA1 sequence is variable between primates and has evolved under positive selection. In contrast, the terminal regions of BRCA1, which encode the RING finger and BRCT domains, experienced negative selection, which left them almost identical between the compared primates (Pavlicek et al. 2004). Finally, on the basis of protein sequence conservation, we have been able to identify missense changes that are likely to compromise BRCA1 function.
Comparison of TAR-isolated, full-length gene homologues also provide information on the evolution of noncoding regions. An increasing body of evidence indicates the importance of noncoding regions in regulating gene expression and their involvement in genomic rearrangements resulting in gene inactivation. For example, a significant fraction of germ-line BRCA1 mutations, which cause hereditary predisposition to breast and ovarian cancers, are deletions and duplications that involve one or more exons. Most of them are caused by recombination between Alu repeats, which are particularly numerous in BRCA1. Sequence analysis of full-size BRCA1 homologues isolated by TAR cloning from a representative group of nonhuman primates revealed that Alu-mediated rearrangements, including Alu transpositions and Alu-associated deletions, are the major forces of evolutionary changes in noncoding BRCA1 sequences (Pavlicek et al. 2004). Moreover, analysis of BRCA1 TAR-isolated clones indicated that the structural instability of the locus might be an intrinsic feature of anthropoids. Most of the Alu repeats involved in disease-associated genomic rearrangements have been retained in nonhuman primates, suggesting that the repeats are of functional significance.
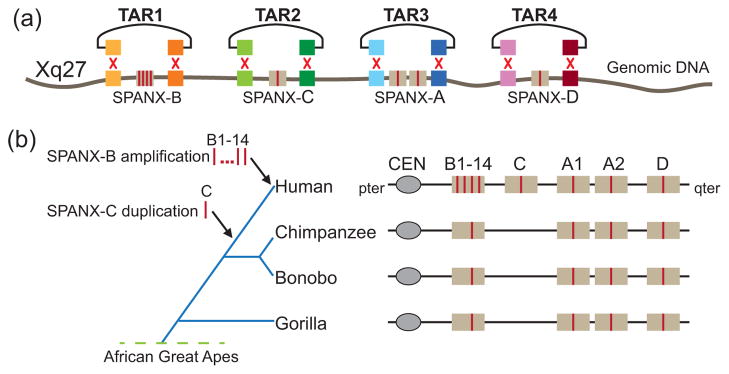
A good example of the application of TAR cloning for evolutionary analysis is reconstruction of evolutionary history of the SPANX-A/D gene family in primates (Kouprina et al. 2004a; 2005a). Rapid evolution of these genes and their location within segmental duplications (SDs) impede a routine PCR or next generation sequencing analyses of syntenic chromosomal segments that is required to detect lineage-specific amplification of these genes. To overcome this problem, we selectively isolated syntenic segments from human, chimpanzee, bonobo, gorilla, orangutan, and macaque. Sequence comparison revealed that TAR clones from syntenic regions of chimpanzee, bonobo, and gorilla genomes do not contain the SPANX-C gene, meaning that the SPANX-C gene is human specific. Analysis of the SPANX-B locus revealed a variable number of gene copies ranging from 1 to 14 due to the tandem duplication of a 12-kb DNA segment carrying the SPANX-B gene present only in humans. Moreover, the analysis revealed that the SPANX-A/D gene family is absent in orangutan and macaque (Fig. 4).
Fig. 4.
Organization of the SPANX genes in primates. Syntenic genomic fragments containing different members of the SPANX gene family from the human, chimpanzee, bonobo, gorilla, orangutan, and macaque were isolated by TAR cloning. Unique targeting hooks in the TAR vectors were chosen from the available human genome sequences. Comparison of the sequences showed that TAR clones from chimpanzee, bonobo, and gorilla do not contain SPANX-C along with the duplication. In addition, the amplification of SPANX-B from one to 14 copies in different individuals is also human specific. The SPANX-A/D gene family is absent in the orangutan and macaque.
Isolation of chromosomal regions that are unclonable in bacterial vectors
It is well known that some genomic regions are missing in the existing BAC libraries because they cannot be cloned efficiently, if at all, in E. coli cells. Such regions may include long inverted repeats and sequences with Z-DNA-like structures that are extremely unstable in E. coli. Poorly clonable human sequences include both non-coding and coding regions. Human DNA sequences that are unstable or unclonable in E. coli could be cloned and propagated in yeast using TAR-generated YACs. Two genes that are toxic for bacterial cells (MUC2 and KAI1) have been TAR-cloned in yeast and then sequenced. Re-sequencing of these regions showed that the errors in the draft genome sequence were the results of both missassembly and loss of specific DNA sequences during cloning in E. coli (Kouprina et al. 2003a).
TAR cloning was put to use in the final phase of the human genome sequencing and significantly contributed to closing the gaps. As an example, several gap regions were selectively recovered from human chromosome 19 (Leem et al. 2004; Grimwood et al. 2004) by TAR cloning using sequence information of the flanking contigs. Analysis of the gap sequences revealed that they contain several abnormalities that could result in instability of the sequences in microbe hosts, including large blocks of micro- and mini-satellites and a high density of Alu repeats. In addition, sequencing of these gap regions allowed us to generate a complete sequence of four genes, including the neuronal cell signaling gene SCK1/SLI that contains a record number of mini-satellites, most of which are polymorphic and transmitted through meiosis in a Mendelian fashion (Leem et al. 2004). The same recombinational cloning approach may be applied for isolation of genomic fragments from other genomes that are not clonable in E. coli vectors.
Construction of human artificial chromosomes (HACs) for gene functional studies
Progress in functional genomics depends on both the availability of full-length genes and a suitable system for gene delivery. TAR cloning solved the problem of full-length gene isolation. During the past few years, there has also been great progress in construction of new gene delivery vectors, i.e. human artificial chromosomes or HACs (Harrington et al. 1997; Ikeno et al. 1998; Henning et al. 1999; Ebersole et al. 2000; Mejía et al. 2002; Ikeno et al. 2002; Kouprina et al. 2003a; Katoh et al. 2004; Kazuki and Oshimura 2011; Kouprina et al. 2013; Kouprina et al. 2014; Oshimura et al. 2015). HACs are extra chromosomes carrying all of the required components of a functional kinetochore assembled on a large block of centromeric alphoid DNA repeats. Therefore, they are maintained stably as an additional 47th chromosome in human cells, thus avoiding insertional mutagenesis due to integration into host chromosomes. They do not cause severe immunogenic responses, which have been a serious problem with viral-based vectors (Lufino et al. 2008; Buchholz et al. 2009; Wanisch et al. 2009; Epstein 2009; Maier et al. 2010; Banasik et al. 2010; Mátrai et al. 2010; Mingozzi et al. 2011; Zhang et al. 2014). HACs have essentially unlimited cloning capacity and may carry one or several therapeutic genes surrounded by their long range controlling elements that should confer physiological levels of fully regulated gene expression (Tedesco et al. 2011; Kim et al. 2011; Kononenko et al. 2014). When regulatory elements such as enhancers, for example, are too far away from the gene or even on a different chromosome, recapitulation of physiological regulation of endogenous loci can be achieved using a HAC vector containing several gene acceptor sites. A gene of interest may be inserted into one acceptor site, while the enhancer element is inserted into another (Suzuki et al. 2014). Several studies have demonstrated the efficacy of de novo assembled HACs for delivery and expression of full-size genes, including those isolated by TAR cloning, in human or embryonic cells with their further differentiated progeny in mice (Kim et al. 2011; Kononenko et al. 2014; Liskovykh et al. 2015; Oshimura et al. 2015).
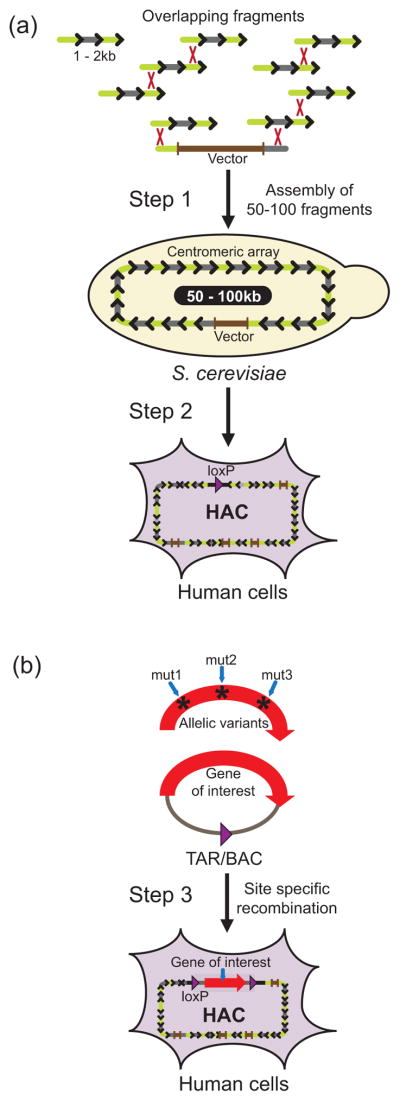
Existing strategies for HAC construction can be divided into two classes: top down, based on the truncation of an existing chromosome into a much smaller mini-chromosome suitable for further manipulation, and bottom up, based on transfection of large blocks of centromeric alphoid DNA into human cells with resulting HAC formation (Oshimura et al. 2015; Katona 2015; Moralli and Monaco 2015). TAR cloning allows for the construction of synthetic arrays of alphoid DNA with a predetermined structure, which may be then used as substrates for HAC formation (Ebersole et al. 2005). This approach starts with a short alphoid DNA multimer (e.g., a dimer) that is assembled by in vivo homologous recombination in yeast (TAR cloning) into alphoid arrays up to 150 kb in size (Fig. 5a) (Ebersole et al. 2005). The minimum size of the array that is required for de novo HAC formation is ~40 kb. After transfection of such synthetic arrays [or natural alphoid arrays cloned by TAR cloning (Kouprina et al 2003b)] into human cells, de novo HACs are generated, ranging in size from 1 Mb to 10 Mb due to amplification of the input alphoid DNA (Fig. 5a). Thus, the use of TAR cloning advances construction of HACs with a predetermined structure, which is very useful for analysis of the human kinetochore (Bergmann et al. 2012; Ohzeki et al 2015).
Fig. 5.
Coupling of TAR gene cloning technology with the HAC gene delivery and expression system. (a) Amplification of synthetic alphoid DNA repeats for HAC construction by TAR cloning. Step 1: Amplification involves two sub-steps: rolling-circle amplification (RCA) of 340 bp alphoid DNA dimers to 1–2 kb tandem repeats (direction of repeats is shown by arrows) and co-transformation of the TAR vector and a mixture of the overlapping tandem repeats followed by end-to-end recombination through interaction of the recombined fragments with the TAR vector hooks that results in the rescue of large alphoid arrays up to 100 kb in size as circular molecules. The vector contains a yeast cassette, a mammalian selectable marker (the Neo or BS gene), and a BAC replicon. Step 2: After transfection into human cells, both the synthetic and natural arrays are capable of de novo HAC formation. To convert a HAC into a gene delivery vector, a unique loxP gene loading site is inserted. (b) Step 3: Loading of a TAR-isolated gene allele (mut1, mut2 or mut3) of a gene of interest into the loxP site of the HAC gene delivery vector by Cre-loxP mediated recombination.
The most advanced de novo constructed HAC is the alphoidtetO-HAC engineered from a ~50 kb synthetic alphoid DNA array, in which the tetracycline operator (tet-O) sequences were embedded (Nakano et al. 2008). Therefore, this HAC contains a conditional kinetochore that can be inactivated by expression of tet-repressor fusion proteins, resulting in loss of the HAC from populations of dividing cells. It was subsequently demonstrated that the alphoidtetO-HAC may carry a genomic copy of a TAR-cloned gene (either VHL, NBS1, BRCA1 or HPRT) (Fig. 5b) (Kim et al. 2011; Kononenko et al. 2014), which if necessary may be successfully eliminated from cells by loss of the HAC following kinetochore inactivation.
In the recent study, the HAC vector containing a regulated centromere was used for delivery and expression of the TAR-cloned 90 kb genomic copy of the BRCA1 gene into the gene-deficient human cells (Kononenko et al. 2014). The use of this system demonstrated that deficiency in BRCA1 results in an elevated level of transcription of diverged pericentromeric repeats, forming constitutive heterochromatin, as well as higher-order alphoid DNA repeats (HORs), forming a functional kinetochore. These data support the hypothesis that epigenetic alterations of these regions initiated in the absence of BRCA1 could result in kinetochore disassembly, leading to chromosome instability and cell transformation.
Thus, the alphoidtetO-HAC vector has significant advantages because it provides a mechanism to compare the phenotype of human target cells with or without a functional copy of any cloned gene of interest, a feature that provides a control for phenotypic changes attributed to the expression of HAC-encoded genes. This generation of human artificial chromosomes from synthetic alphoid arrays seems to be the most suitable for studies of gene function and therapeutic applications.
TAR cloning for synthetic biology
The recent progress in synthetic genomics was determined to a certain extend by the use of TAR cloning. A few years ago the J Craig Venter Institute had attempted complete synthesis and assembly of a whole bacterial (M. genitalium) genome (582,970 bp), a culmination of about 10 years of work (Gibson et al. 2008a). The M. genitalium genome was assembled using a combination of in vitro enzymatic and in vivo TAR cloning methods. In the early stage, an in vitro recombination method was utilized to assemble 25 DNA cassettes with an average length of 24 kb into eight 72 kb assemblies and subsequently assembled into four 144 kb assemblies. It was found that the efficiency of the in vitro procedure greatly declined as the assemblies became larger, and the half-genomes, each of 290 kb in size, were unable to be assembled at all. Therefore, the in vivo recombination in yeast was exploited to complete the final whole genome assembly. In the meantime, it was also found possible to directly assemble 25 DNA cassettes from the earliest stages into a complete genome in a single step using TAR cloning in yeast (Gibson et al. 2008b). Thus, use of TAR cloning greatly simplifies the assembly of large DNA molecules from both synthetic and natural fragments.
Subsequently, Gibson et al. (2010) reported the creation of a bacterial cell controlled by a chemically synthesized genome. A 1.1 Mb M. mycoides genome was assembled by TAR cloning from eleven ~100 kb DNA segments in yeast (Lartigue et al. 2009); it was then transplanted into a closely related M. capricolum recipient cell to form a new M. mycoides cell that was controlled solely by the synthetic genome. The new cells have expected phenotypic properties and are capable of continuous self-replication. Thus, the full circle starting from de novo synthesis of a whole mycoplasma genome and ending with its transfer into a bacterial cell has been accomplished. These organisms were first selected for genome cloning/replacements because of their small genome size and also because of the expectation that the genomes were unlikely to produce toxic gene products in yeast as a result of their alternative genetic code.
So far, cloning in yeast of whole genomes or individual chromosomes was successfully accomplished for species such as M. pneumoniae (0.8 Mb) (Benders et al. 2010), Acholeplasma laidlawii (1.5 Mb) (Karas et al. 2012), Prochlorococcus marinus MED4 (1.6 Mb) (Tagwerker et al. 2012), and eukaryotic algal Phaeodactylum tricornutum (27.4 Mb) (Karas et al. 2013). In the last case, two individual chromosomes with a size of ~500 kb were assembled by TAR cloning in yeast. At present, propagation in yeast has been hampered for the Acholeplasma laidlawii genome due to the presence of a single gene that is toxic to yeast cells. Also, a high G+C content (55%) prevents propagation in yeast of bacterial fragments larger than 200 kb. It becomes a possibility if an additional ARS sequence(s) is added (Noskov et al. 2012; Karas et al. 2013).
In contrast to most other eukaryotic organisms, the introduction of targeted and specific modifications such as gene deletions, insertions, or replacements into the genome of Saccharomyces cerevisiae has been standard for decades (Hinnen et al. 1978; Scherer and Davis 1979). Therefore, the reported assembly of microbe genomes in yeast provides an unprecedented opportunity for their further modifications in order to address key questions of synthetic biology.
The yeast Saccharomyces cerevisiae is the most intensively studied unicellular eukaryote and one of the main industrial microorganisms used in the production of biochemicals. The potential of yeast as a powerful host for synthetic biology has already been successfully demonstrated by both basic research, namely the de novo synthesis of a complete chromosome (Annaluru et al. 2014), and the application-oriented engineering of complex pathways, such as the synthesis of amorphadiene and vanillin (Brochado et al. 2013; Westfall et al. 2012). Recently, the TAR cloning strategy has been applied for assembling genetic pathways for expression in Saccharomyces cerevisiae (Mitchell et al. 2015). The authors demonstrated the assembly of four-, five- and six-gene pathways to generate S. cerevisiae cells synthesizing β-carotene and violacein.
Currently, the most ambitious project in yeast synthetic biology is the complete de novo synthesis of all 16 chromosomes, Sc2.0. Although this project is at its very beginning, with one synthetic chromosome (chromosome III) completed (Annaluru et al. 2014), the consortium plans to finish all additional chromosomes by 2019. It is worth noting that the assembly of overlapping DNA fragments by TAR cloning was a critical step in de novo synthesis of the first yeast chromosomes (Annaluru et al. 2014).
To conclude, TAR cloning has emerged an essential method for the assembly of new microbe genomes and synthetic biology. Additional information about strategies for cloning and manipulation of natural and synthetic chromosomes can be found in recent publications (Gibson 2014; Karas et al. 2015).
Acknowledgments
We thank Dr. Nicolas Lee for helping in preparation of figures.
Funding This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, USA (V.L. and N.K).
Footnotes
Conflict of interest The authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Agarwal V, El Gamal AA, Yamanaka K, Poth D, Kersten RD, Schorn M, Allen EE, Moore BS. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol. 2014;10:640–U182. doi: 10.1038/nchembio.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H, Lemire S, Pires DP, Lu TK. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015;1:187–196. doi: 10.1016/j.cels.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annab L, Kouprina N, Solomon G, Cable L, Hill D, Barrett JC, Larionov V, Afshari C. Isolation of functional copy of the human BRCA1 gene by TAR cloning in yeast. Gene. 2000;250:201–208. doi: 10.1016/s0378-1119(00)00180-3. [DOI] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell La, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, Cai Y, Zeller K, Agmon N, Han JS, Hadjithomas M, Tullman J, Caravelli K, Cirelli K, Guo Z, London V, Yeluru A, Murugan S, Kandavelou K, Agier N, Fischer G, Yang K, Martin JA, Bilgel M, Bohutski P, Boulier KM, Capaldo BJ, Chang J, Charoen K, Choi WJ, Deng P, DiCarlo JE, Doong J, Dunn J, Feinberg JI, Fernandez C, Floria CE, Gladowski D, Hadidi P, Ishizuka I, Jabbari J, Lau CY, Lee PA, Li S, Lin D, Linder ME, Ling J, Liu J, Liu J, London M, Ma H, Mao J, McDade JE, McMillan A, Moore AM, Oh WC, Ouyang Y, Patel R, Paul M, Paulsen LC, Qiu J, Rhee A, Rubashkin MG, Soh IY, Sotuyo NE, Srinivas V, Suarez A, Wong A, Wong R, Xie WR, Xu Y, Yu AT, Koszul R, Bader JS, Boeke JD, Chandrasegaran S. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa S, Abe I, Kudoh Y, Kishi N, Wang Y, Kubota R, Kudoh J, Kawasaki K, Minoshima S, Shimizu N. Human BAC library: construction and rapid screening. Gene. 1997;191:69–79. doi: 10.1016/s0378-1119(97)00044-9. [DOI] [PubMed] [Google Scholar]
- Becker M, Aitcheson N, Byles E, Wickstead B, Louis E, Rudenko G. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 2004;14:2319–2329. doi: 10.1101/gr.2955304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benders GA, Noskov VN, Denisova EA, Lartigue C, Gibson DG, Assad-Garcia N, Chuang RY, Carrera W, Moodie M, Algire MA, Phan Q, Alperovich N, Vashee S, Merryman C, Venter JC, Smith HO, Glass JI, Hutchison CA., 3rd Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 2010;38:2558–2569. doi: 10.1093/nar/gkq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Martins NM, Larionov V, Masumoto H, Earnshaw WC. HACking the centromere chromatin code: insights from human artificial chromosomes. Chromosome Res. 2012;20:505–519. doi: 10.1007/s10577-012-9293-0. [DOI] [PubMed] [Google Scholar]
- Bonet B, Teufel R, Crüsemann M, Ziemert N, Moore BS. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J Nat Prod. 2015;78:539–542. doi: 10.1021/np500664q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochado AR, Patil KR. Overexpression of O-methyltransferase leads to improved vanillin production in baker’s yeast only when complemented with model-guided network engineering. Biotechnol Bioeng. 2013;110:656–659. doi: 10.1002/bit.24731. [DOI] [PubMed] [Google Scholar]
- Buchholz CJ, Mühlebach MD, Cichutek K. Lentiviral vectors with measles virus glycoproteins - dream team for gene transfer? Trends Biotechnol. 2009;27:259–265. doi: 10.1016/j.tibtech.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Cano-Prieto C, García-Salcedo R, Sánchez-Hidalgo M, Braña AF, Fiedler HP, Méndez C, Salas JA, Olano C. Genome Mining of Streptomyces sp. Tü 6176: Characterization of the Nataxazole Biosynthesis Pathway. Chembiochem. 2015;16:1461–1473. doi: 10.1002/cbic.201500153. [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- Epstein AL. Progress and prospects: biological properties and technological advances of herpes simplex virus type 1-based amplicon vectors. Gene Ther. 2009;16:709–715. doi: 10.1038/gt.2009.42. [DOI] [PubMed] [Google Scholar]
- Ebersole T, Okamoto Y, Noskov VN, Kouprina N, Kim JH, Leem SH, Barrett JC, Masumoto H, Larionov V. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Kallifidas D, Brady SF. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc Natl Acad Sci U S A. 2011;108:12629–12634. doi: 10.1073/pnas.1103921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida A, Becker MM, Schmid CD, Bühlmann T, Louis EJ, Beck HP. Cloning of the repertoire of individual Plasmodium falciparum var genes using transformation associated recombination (TAR) PLoS One. 2011;6:e17782. doi: 10.1371/journal.pone.0017782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, 3rd, Smith HO. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008a;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., 3rd One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A. 2008b;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, 3rd, Smith HO, Venter JC. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Gibson DG. Programming biological operating systems: genome design, assembly and activation. Nat Methods. 2014;11:521–526. doi: 10.1038/nmeth.2894. [DOI] [PubMed] [Google Scholar]
- Giordano D, Coppola D, Russo R, Denaro R, Giuliano L, Lauro FM, di Prisco G, Verde C. Marine Microbial Secondary Metabolites: Pathways, Evolution and Physiological. Adv Microb Physiol. 2015;66:357–428. doi: 10.1016/bs.ampbs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O, Tran-Gyamfi M, Aerts A, Altherr M, Ashworth L, Bajorek E, Black S, Branscomb E, Caenepeel S, Carrano A, Caoile C, Chan YM, Christensen M, Cleland CA, Copeland A, Dalin E, Dehal P, Denys M, Detter JC, Escobar J, Flowers D, Fotopulos D, Garcia C, Georgescu AM, Glavina T, Gomez M, Gonzales E, Groza M, Hammon N, Hawkins T, Haydu L, Ho I, Huang W, Israni S, Jett J, Kadner K, Kimball H, Kobayashi A, Larionov V, Leem SH, Lopez F, Lou Y, Lowry S, Malfatti S, Martinez D, McCready P, Medina C, Morgan J, Nelson K, Nolan M, Ovcharenko I, Pitluck S, Pollard M, Popkie AP, Predki P, Quan G, Ramirez L, Rash S, Retterer J, Rodriguez A, Rogers S, Salamov A, Salazar A, She X, Smith D, Slezak T, Solovyev V, Thayer N, Tice H, Tsai M, Ustaszewska A, Vo N, Wagner M, Wheeler J, Wu K, Xie G, Yang J, Dubchak I, Furey TS, DeJong P, Dickson M, Gordon D, Eichler EE, Pennacchio LA, Richardson P, Stubbs L, Rokhsar DS, Myers RM, Rubin EM, Lucas SM. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Henning KA, Novotny EA, Compton ST, Guan XY, Liu PP, Ashlock MA. Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc Natl Acad Sci USA. 1999;96:592–597. doi: 10.1073/pnas.96.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A, Hicks JB, Fink GR. Transformation of yeast. Proc Natl Acad Sci U S A. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa I, Chiang JC, Patterson T, Feigenbaum L, Leem SH, Michishita E, Larionov V, Hodes RJ, Barrett JC. Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repression element. Proc Natl Acad Sci USA. 2005;102:18437–18442. doi: 10.1073/pnas.0508964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- Ikeno M, Inagaki H, Nagata K, Morita M, Ichinose H, Okazaki T. Generation of human artificial chromosomes expressing naturally controlled guanosine triphosphate cyclohydrolase I gene. Genes Cells. 2002;7:1021–1032. doi: 10.1046/j.1365-2443.2002.00580.x. [DOI] [PubMed] [Google Scholar]
- Karas BJ, Tagwerker C, Yonemoto IT, Hutchison CA, 3rd, Smith HO. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth Biol. 2012;1:22–28. doi: 10.1021/sb200013j. [DOI] [PubMed] [Google Scholar]
- Karas BJ, Molparia B, Jablanovic J, Hermann WJ, Lin YC, Dupont CL, Tagwerker C, Yonemoto IT, Noskov VN, Chuang RY, Allen AE, Glass JI, Hutchison CA, 3rd, Smith HO, Venter JC, Weyman PD. Assembly of eukaryotic algal chromosomes in yeast. J Biol Eng. 2013;7:30. doi: 10.1186/1754-1611-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas BJ, Suzuki Y, Weyman PD. Strategies for cloning and manipulating natural and synthetic chromosomes. Chromosome Res. 2015;23:57–68. doi: 10.1007/s10577-014-9455-3. [DOI] [PubMed] [Google Scholar]
- Katoh M, Ayabe F, Norikane S, Okada T, Masumoto H, Horike S, Shirayoshi Y, Oshimura M. Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun. 2004;321:280–290. doi: 10.1016/j.bbrc.2004.06.145. [DOI] [PubMed] [Google Scholar]
- Katona RL. De novo formed satellite DNA-based mammalian artificial chromosomes and their possible applications. Chromosome Res. 2015;23:143–157. doi: 10.1007/s10577-014-9458-0. [DOI] [PubMed] [Google Scholar]
- Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol Ther. 2011;19:1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Feng Z, Bauer JD, Kallifidas D, Calle PY, Brady SF. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers. 2010;93:833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Leem SH, Sunwoo Y, Kouprina N. Separation of long-range human TERT gene haplotypes by transformation-associated recombination cloning in yeast. Oncogene. 2003;22:2443–2456. doi: 10.1038/sj.onc.1206316. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kononenko A, Erliandri I, Kim TA, Nakano M, Iida Y, Barrett JC, Oshimura M, Masumoto H, Earnshaw WC, Larionov V, Kouprina N. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc Natl Acad Sci U S A. 2011;108:20048–20053. doi: 10.1073/pnas.1114483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononenko AV, Bansal R, Lee NC, Grimes BR, Masumoto H, Earnshaw WC, Larionov V, Kouprina N. A portable BRCA1-HAC (human artificial chromosome) module for analysis of BRCA1 tumor suppressor function. Nucleic Acids Res. 2014;42(21) doi: 10.1093/nar/gku870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Annab L, Graves J, Afshari C, Barrett JC, Resnick MA, Larionov V. Functional copies of a human gene can be directly isolated by TAR cloning with a small 3′ end target sequence. Proc Natl Acad Sci USA. 1998;95:4469–4474. doi: 10.1073/pnas.95.8.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Leem SH, Solomon G, Ly A, Koriabine M, Otstot J, Pak E, Dutra A, Zhao S, Barrett JC, Larionov V. Segments missing from the draft human genome sequence can be isolated by TAR cloning in yeast. EMBO Rep. 2003a;4:257–262. doi: 10.1038/sj.embor.embor766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Ebersole T, Koriabine M, Pak E, Rogozin IB, Katoh M, Oshimura M, Ogi K, Peredelchuk M, Solomon G, Brown W, Barrett JC, Larionov V. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 2003b;31:922–934. doi: 10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Larionov V. Exploiting the yeast Saccharomyces cerevisiae for the study of the organization and evolution of complex genomes. FEMS Microbiol Rev. 2003;27:629–649. doi: 10.1016/S0168-6445(03)00070-6. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Mullokandov M, Rogozin IB, Collins NK, Solomon G, Otstot J, Risinger JI, Koonin EV, Barrett JC, Larionov V. The SPANX gene family of cancer/testis-specific antigens: rapid evolution and amplification in African great apes and hominids. Proc Natl Acad Sci U S A. 2004a;101:3077–3082. doi: 10.1073/pnas.0308532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon YH, Collura R, Ruvolo M, Barrett JC, Woods CG, Walsh CA, Jurka J, Larionov V. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004b;2:653–663. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Noskov VN, Solomon G, Otstot J, Isaacs W, Carpten JD, Trent JM, Schleutker J, Barrett JC, Jurka J, Larionov V. Dynamic structure of the SPANX gene cluster mapped to the prostate cancer susceptibility locus HPCX at Xq27. Genome Res. 2005a;15:1477–1486. doi: 10.1101/gr.4212705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki JI, Mochida GH, Risinger JI, Goldsmitt P, Gunsior M, Solomon G, Gersch W, Kim JH, Barrett JC, Walsh CA, Jurka J, Masumoto H, Larionov V. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genetics. 2005b;14:2155–2165. doi: 10.1093/hmg/ddi220. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Larionov V. TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat Rev Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Noskov VN, Pavlicek A, Collins NK, Schoppee Bortz PD, Ottolenghi C, Loukinov D, Goldsmith P, Risinger JI, Kim JH, Westbrook VA, Solomon G, Sounders H, Herr JC, Jurka J, Lobanenkov V, Schlessinger D, Larionov V. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PLoS One. 2007a;2:e359. doi: 10.1371/journal.pone.0000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Noskov VN, Solomon G, Otstot J, Isaacs W, Xu J, Schleutker J, Larionov V. Mutational analysis of SPANX genes in families with X-linked prostate cancer. Prostate. 2007b;67:820–828. doi: 10.1002/pros.20561. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Lee NC, Pavlicek A, Samoshkin A, Kim JH, Lee HS, Varma S, Reinhold WC, Otstot J, Solomon G, Davis S, Meltzer PS, Schleutker J, Larionov V. Exclusion of the 750-kb genetically unstable region at Xq27 as a candidate locus for prostate malignancy in HPCX1-linked families. Genes Chromosomes Cancer. 2012;51:933–948. doi: 10.1002/gcc.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Earnshaw WC, Masumoto H, Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell Mol Life Sci. 2013;70:1135–1148. doi: 10.1007/s00018-012-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Tomilin AN, Masumoto H, Earnshaw WC, Larionov V. Human artificial chromosome-based gene delivery vectors for biomedicine and biotechnology. Expert Opin Drug Deliv. 2014;11:517–535. doi: 10.1517/17425247.2014.882314. [DOI] [PubMed] [Google Scholar]
- Kuleshov V, Xie D, Chen R, Pushkarev D, Ma Z, Blauwkamp T, Kertesz M, Snyder M. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S, Botstein D, Fox MS. Transformation of yeast with linearized plasmid DNA. Formation of inverted dimers and recombinant plasmid products. J Mol Biol. 1985;184:375–387. doi: 10.1016/0022-2836(85)90288-8. [DOI] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci U S A. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Solomon G, Barrett JC, Resnick MA. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc Natl Acad Sci USA. 1997;94:7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue C, Vashee S, Algire MA, Chuang RY, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA, 3rd, Smith HO, Venter JC, Glass JI. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- Lee NC, Larionov V, Kouprina N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015;43:e55. doi: 10.1093/nar/gkv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem SH, Londono-Vallejo JA, Kim JH, Bui H, Tubacher E, Solomon G, Park JE, Horikawa I, Kouprina N, Barrett JC, Larionov V. Cloning of the human telomerase gene: Complete genomic sequence and analysis of tandem repeats polymorphisms in intronic regions. Oncogene. 2002;21:769–777. doi: 10.1038/sj.onc.1205122. [DOI] [PubMed] [Google Scholar]
- Leem SH, Kouprina N, Grimwood J, Kim JH, Mullokandov M, Yoon YH, Chae JY, Morgan J, Lucas S, Richardson P, Detter C, Glavina T, Rubin E, Barrett JC, Larionov V. Closing the gaps on human chromosome 19 revealed genes with a high density of repetitive tandemly arrayed elements. Genome Res. 2004;14:239–246. doi: 10.1101/gr.1929904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li Z, Yamanaka K, Xu Y, Zhang W, Vlamakis H, Kolter R, Moore BS, Qian PY. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci Rep. 2015a;5:9383. doi: 10.1038/srep09383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZR, Li Y, Lai JY, Tang J, Wang B, Lu L, Zhu G, Wu X, Xu Y, Qian PY. Critical Intermediates Reveal New Biosynthetic Events in the Enigmatic Colibactin Pathway. Chembiochem. 2015b;16:1715–1719. doi: 10.1002/cbic.201500239. [DOI] [PubMed] [Google Scholar]
- Liskovykh M, Ponomartsev S, Popova E, Bader M, Kouprina N, Larionov V, Alenina N, Tomilin A. Stable maintenance of de novo assembled human artificial chromosomes in embryonic stem cells and their differentiated progeny in mice. Cell Cycle. 2015;14:1268–1273. doi: 10.1080/15384101.2015.1014151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufino MM, Edser PA, Wade-Martins R. Advances in high-capacity extrachromosomal vector technology: episomal maintenance, vector delivery, and transgene expression. Mol Ther. 2008;16:1525–1538. doi: 10.1038/mt.2008.156. [DOI] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Maier P, von Kalle C, Laufs S. Retroviral vectors for gene therapy. Future Microbiol. 2010;5:1507–1523. doi: 10.2217/fmb.10.100. [DOI] [PubMed] [Google Scholar]
- Mejía JE, Alazami A, Willmott A, Marschall P, Levy E, Earnshaw WC, Larin Z. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics. 2002;79:297–304. doi: 10.1006/geno.2002.6704. [DOI] [PubMed] [Google Scholar]
- Mingozzi F, Katherine A, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–356. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Mitchell LA, Chuang J, Agmon N, Khunsriraksakul C, Phillips NA, Cai Y, Truong DM, Veerakumar A, Wang Y, Mayorga M, Blomquist P, Sadda P, Trueheart J, Boeke JD. Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 2015;43:6620–6630. doi: 10.1093/nar/gkv466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K. High-throughput plasmid construction using homologous recombination in yeast: its mechanisms and application to protein production for X-ray crystallography. Bioscience Biotechnology and Biochemistry. 2015;79:1–10. doi: 10.1080/09168451.2014.952614. [DOI] [PubMed] [Google Scholar]
- Moralli D, Monaco ZL. Developing de novo human artificial chromosomes in embryonic stem cells using HSV-1 amplicon technology. Chromosome Res. 2015;23:105–110. doi: 10.1007/s10577-014-9456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassman R, Vagnarelli P, Kandels-Lewis S, Earnshaw WC, Larionov V, Masumoto H. Inactivation of a human kinetochore by a specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihei N, Kouprina N, Larionov V, Oshima J, Martin GM, Ichikawa T, Barrett JC. Functional Evidence for a Metastasis Suppressor Gene for Rat Prostate Cancer within a 60 kb Region on Human Chromosome 8p21-p12. Cancer Res. 2002;62:367–370. [PubMed] [Google Scholar]
- Noskov V, Koriabine M, Solomon G, Randolph M, Barrett JC, Leem SH, Stubbs L, Kouprina N, Larionov V. Defining the minimal length of sequence homology required for selective gene isolation by TAR cloning. Nucleic Acids Res. 2001;29:E62. doi: 10.1093/nar/29.6.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov VN, Kouprina N, Leem SH, Ouspenski I, Barrett JC, Larionov V. A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics. 2003a;4:16. doi: 10.1186/1471-2164-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov VN, Leem SH, Solomon G, Mullokandov M, Chae JY, Yoon YH, Shin YS, Kouprina N, Larionov V. A novel strategy for analysis of gene homologues and segmental genome duplications. J Mol Evol. 2003b;56:702–710. doi: 10.1007/s00239-002-2442-x. [DOI] [PubMed] [Google Scholar]
- Noskov VN, Karas BJ, Young L, Chuang RY, Gibson DG, Lin YC, Stam J, Yonemoto IT, Suzuki Y, Andrews-Pfannkoch C, Glass JI, Smith HO, Hutchison CA, 3rd, Venter JC, Weyman PD. Assembly of large, high G+C bacterial DNA fragments in yeast. ACS Synth Biol. 2012;1:267–273. doi: 10.1021/sb3000194. [DOI] [PubMed] [Google Scholar]
- Ongley SE, Bian X, Neilan BA, Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep. 2013;30:1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Larionov V, Earnshaw WC, Masumoto H. Genetic and epigenetic regulation of centromeres: a look at HAC formation. Chromosome Res. 2015;23:87–103. doi: 10.1007/s10577-015-9470-z. [DOI] [PubMed] [Google Scholar]
- Oshimura M, Uno N, Kazuki Y, Katoh M, Inoue T. A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges. Chromosome Res. 2015;23:111–133. doi: 10.1007/s10577-014-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek A, Noskov N, Kouprina N, Barrett JC, Jurka J, Larionov V. Evolution of the tumor suppressor BRCA1 locus in primates: implications for cancer predisposition. Hum Mol Genet. 2004;13:1–15. doi: 10.1093/hmg/ddh301. [DOI] [PubMed] [Google Scholar]
- Salem RM, Wessel J, Schork NJ. A comprehensive literature review of haplotyping software and methods for use with unrelated individuals. Hum Genomics. 2005;2:39–66. doi: 10.1186/1479-7364-2-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosomal segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Thomas M, Kelly J, Selker E, Davis RW. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci U S A. 1980;77:4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kazuki Y, Oshimura M, Hara T. A novel system for simultaneous or sequential integration of multiple gene-loading vectors into a defined site of a human artificial chromosome. PLoS One. 2014;9(10):e110404. doi: 10.1371/journal.pone.0110404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Li J, Millán-Aguiñaga N, Zhang JJ, O’Neill EC, Ugalde JA, Jensen PR, Mantovani SM, Moore BS. Identification of Thiotetronic Acid Antibiotic Biosynthetic Pathways by Target-directed Genome Mining. ACS Chem Biol. 2015;10:2841–2849. doi: 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C, Dupont CL, Karas BJ, Ma L, Chuang RY, Benders GA, Ramon A, Novotny M, Montague MG, Venepally P, Brami D, Schwartz A, Andrews-Pfannkoch C, Gibson DG, Glass JI, Smith HO, Venter JC, Hutchison CA., 3rd Sequence analysis of a complete 1.66 Mb Prochlorococcus marinus MED4 genome cloned in yeast. Nucleic Acids Res. 2012;40:10375–10383. doi: 10.1093/nar/gks823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco FS, Hoshiya H, D’Antona G, Gerli MF, Messina G, Antonini S, Tonlorenzi R, Benedetti S, Berghella L, Torrente Y, Kazuki Y, Bottinelli R, Oshimura M, Cossu G. Stem cell-mediated transfer of a human artificial chromosome ameliorates muscular dystrophy. Sci Transl Med. 2011;3:96ra78. doi: 10.1126/scitranslmed.3002342. [DOI] [PubMed] [Google Scholar]
- Ross AC, Gulland LE, Dorrestein PC, Moore BS. Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synth Biol. 2015;4:414–420. doi: 10.1021/sb500280q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Beck CR, English AC, Meng Q, Buhay C, Han Y, Doddapaneni HV, Yu F, Boerwinkle E, Lupski JR, Muzny DM, Gibbs RA. PacBio-LITS: a large-insert targeted sequencing method for characterization of human disease-associated chromosomal structural variations. BMC Genomics. 2015;16:214. doi: 10.1186/s12864-015-1370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanisch K, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109:E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R, Taylor JE, Kurioka A, Becker M, Louis EJ, Rudenko G. Isolation and analysis of the genetic diversity of repertoires of VSG expression site containing telomeres from Trypanosoma brucei gambiense, T. b. brucei and T. equiperdum. BMC Genomics. 2008;9:385. doi: 10.1186/1471-2164-9-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Andersen E, Zhao H. Flexible and Versatile Strategy for the Construction of Large Biochemical Pathways. ACS Synth Biol. 2016;5:46–52. doi: 10.1021/acssynbio.5b00117. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hagedorn C, Schulz E, Lipps HJ, Ehrhardt A. Viral hybrid-vectors for delivery of autonomous replicons. Curr Gene Ther. 2014;14:10–23. doi: 10.2174/1566523213666131223130024. [DOI] [PubMed] [Google Scholar]