Abstract
While the progression of mutated colonic cells is dependent upon interactions between the initiated epithelium and surrounding stroma, the nature of these interactions is poorly understood. Here the development of an ultra-sensitive laser-capture microdissection (LCM)/RNA-seq approach for studying the epithelial and stromal compartments of aberrant crypt foci (ACF) is described. ACF are the earliest identifiable pre-neoplastic lesion found within the human colon and are detected using high-definition endoscopy with contrast dye-spray. The current analysis focused on the epithelium of ACF with somatic mutations to either KRAS, BRAF, or APC, with expression patterns compared to normal mucosa from each patient. By comparing gene expression patterns between groups, an increase in a number of pro-inflammatory NF-κB target genes were identified that were specific to ACF epithelium, including TIMP1, RELA and RELB. Distinct transcriptional changes associated with each somatic mutation were observed and a subset display a BRAFV600E-mediated senescence-associated transcriptome characterized by increased expression of CDKN2A. Finally, LCM-captured ACF-associated stroma was found to be transcriptionally distinct from normal stroma, with an up-regulation of genes related to immune cell infiltration and fibroblast activation. Immunofluorescence confirmed increased CD3+ T cells within the stromal microenvironment of ACF and an abundance of activated fibroblasts. Collectively, these results provide new insight into the cellular interplay that occurs at the earliest stages of colonic neoplasia, highlighting the important role of NF-kB, activated stromal fibroblasts and lymphocyte infiltration.
Implications
Fibroblasts and immune cells in the stromal microenvironment play an important role during the earliest stages of colon carcinogenesis.
INTRODUCTION
Reciprocal interactions between transformed epithelial cells and their associated stroma are a critical determinant of neoplastic growth (1). Stromal cells constitute a large fraction of solid tumors and are important drivers of the carcinogenic process (2). The stroma that is in direct contact with transformed epithelium becomes “activated”, displaying an altered phenotype that produces growth-promoting factors that enhances cancer progression (3,4). However, little is known regarding the importance of epithelial-stromal interactions at the very early stages of neoplasia when critical steps in cancer progression occur and targeted interventions may elicit the greatest benefit.
Aberrant crypt foci (ACF) are the earliest morphologically identifiable mucosal abnormality found in the human colon (5), a subset of which may progress to colorectal cancer (CRC). While abundant in the distal colon, ACF in the proximal colon are relatively infrequent (<1 per colon), but are more likely to progress to malignancy. Proximal ACF share many molecular features with more advanced neoplasia (6), including somatic mutations to KRAS, BRAF, and APC (7), and may be precursor lesions of “interval” colon cancers that develop in the proximal colon between scheduled colonoscopies. The present study focuses on a detailed characterization of these diminutive (<5 mm) proximal lesions identified by high-definition (HD), magnifying chromoendoscopy. By developing a more comprehensive understanding of epithelial-stromal interactions occurring in these very early lesions, it may be possible to develop a better understanding of how their progression is regulated. This knowledge could ultimately be utilized to identify high-risk patient populations and to develop strategies for preventing cancer development, particularly interval cancers of the proximal colon.
MATERIALS AND METHODS
Clinical specimens
All colon specimens used in this study were obtained from patients enrolled in an ongoing ACF study at UConn Health-John Dempsey Hospital (Farmington, CT) between 2010 and 2014. Samples were freshly isolated during the colonoscopy procedure and immediately frozen in OCT-embedding media. Each subject provided written informed consent prior to inclusion in the study. HD-chromoendoscopy was performed in the distal 20-cm of the colorectum and throughout the entire proximal colon using 0.1% indigo-carmine dye-spray for contrast enhancement. The identification and histologic evaluation of ACF has been described previously (8,9). In addition, each subject had a histologically confirmed corresponding normal biopsy specimen removed from the same side of the colon, generally within 2-cm of the ACF biopsy. Mutational spectra of ACF were determined using DNA-MS analysis (Sequenom) (8). Only ACF with confirmed, non-overlapping somatic mutations to either KRAS, BRAF or APC were selected for further analysis. The study was approved by the University of Connecticut Health Center IRB (#IE-10-068OS-3) in accordance with NIH human research study guidelines.
Laser-capture microdissection and RNA extraction
Highly enriched epithelial and stromal RNA samples were obtained by laser-capture microdissection (LCM) using an ArcturusXT (ThermoFisher Scientific). Frozen sections were routinely cut at 9-μm thickness on PEN membrane slides and stored at −80 °C until use. After 15 seconds of air-drying, sections were rehydrated and dehydrated sequentially in 75% EtOH, ddH2O, ddH2O, 75% EtOH, 95% EtOH, and 100% EtOH for 30 seconds each. Each wash solution was treated with ProtectRNA RNase Inhibitor (Sigma-Aldrich). Following the final 100% EtOH wash, slides were washed in xylenes for 5 minutes. Serial sections were laser-captured following the manufacturers’ protocol. RNA was extracted using the Arcturus PicoPure Frozen RNA Isolation Kit and quantified using a Qubit 3.0 Fluorimeter. The quality of a subset of RNA samples was tested using a 2100 Bioanalyzer (Agilent Technologies).
Ion Personal Genome sequencing
Sequencing libraries were prepared using two panels: the Ion AmpliSeq RNA Apoptosis Panel, targeting 267 genes involved in the cellular apoptosis pathway, and a customized Ion AmpliSeq Senescence panel targeting 20 known senescence genes. The following genes were included in the senescence panel: CDKN2a (p16), CDKN2A (p14), MAPK12, MAP2K6, CCND1, BMI1, BRAF, SKP2, E2F1, CDKN1B, MAPKAPK5, MAPK14, MAP2K3, VHL, BACH1, CCNE1, MAPK11, MDM4, NPM1, MAPK13, and MKI67. This panel included specific primers capable of differentiating between the p16 and p14 alternate reading frames of CDKN2A (10). Genes common to both panels were used for normalization (11,12). Libraries were barcoded using the Ion Xpress Barcode Adapters (ThermoFisher). After barcoding, libraries were quantified using a Bioanalyzer and quantitative PCR, diluted to 25 pM and pooled in batches of three to four per 318 PGM chip (ThermoFisher). The libraries were incorporated during template preparation using the Ion PGM IC 200 Kit using an IonChef and sequenced, resulting in 800,000 to 1,200,000 reads per panel library with a mean read-depth of 8X for the apoptosis panel and over 100,000X for the senescence panel. Four samples from one batch belonging to three stromal pairs were excluded from the analysis due to poor library preparation (Supplementary Figure 1A and B). Six gene targets were removed prior to analysis due to possible off-target pseudo-gene amplification that was identified by GenBank (Supplementary Figure 1C and D). Raw and processed data was uploaded to GEO (GSE77312).
Data analysis
Data analysis was performed using the Torrent Suite (4.4.0) and the Coverage Analysis and AmpliseqRNA plug-ins. Genes were considered differentially expressed with a fold change >1.5 or <0.66. Gene set enrichment analysis (GSEA) software was downloaded from the Broad Institute GSEA portal (http://www.broadinstitute.org/gsea/index.jsp) and applied to the sample set using each of the genes from both the apoptosis and senescence panels. For the analysis, the GSE77312 data set was partitioned into the following four subgroups: ACF epithelial cells, ACF stromal cells, normal epithelial cells and normal stromal cells. GSEA was carried out for all gene lists using signal-to-noise ratios to rank genes. The significance of enrichment was estimated using 1,000 gene permutations, with a statistical cutoff of 0.05 for the nominal P-value. All other statistical analyses were performed with GraphPad Prism version 5.0 or in R (3.1.2). All P-values are two-sided. P-values <0.05 were considered statistically significant.
Immunofluorescence and quantification
Frozen tissue sections (9-μm thick) were collected on Superfrost (ThermoFisher) slides. All immunofluorescence reactions of the same type were performed in a single batch. The following primary antibodies were used: rabbit antibody to alpha smooth muscle actin (polyclonal; Abcam; Ab5694; 1:200 dilution), CD3+ (monoclonal; eBioscience; UCHT1, 1:200 dilution), and mouse antibody to vimentin [V9] (monoclonal IgG1; Santa Cruz Biotechnology; sc-6260; 1:200 dilution). The following secondary antibodies were used: goat anti-rabbit IgG conjugated to Alexa Fluor 568 (ThermoFisher; A-11036) and goat anti-mouse IgG conjugated to Alexa Fluor 488 (ThermoFisher; A-11001). Briefly, tissue sections were fixed for 30 min in acetone at −20°C and blocked in PBS containing 1% BSA (Sigma). Sections were washed with PBS and incubated overnight with primary antibody solution (in PBS with 0.1% BSA). Next, sections were washed and incubated with secondary antibodies (in PBS with 0.1% BSA). The nuclei counterstain was performed with DAPI (100 ng mL−1 in dH2O) for 10 min. Slides were washed and mounted in Vectashield Hardset Medium (Vector Laboratories; H–1400). Imaging was done on an Olympus fluorescence microscope with a 40x and 10x objective, using QCapture Pro 7 imaging software (Qimaging). Images were processed and combined using the ImageJ and Photoshop software programs. Activated fibroblasts are commonly identified by their expression of α-SMA (13,14). However, pericryptal myofibroblasts also express α-SMA and are normally present within the stromal cell population that line the base of colonic crypts (15). Although, pericryptal myofibroblasts are capable of becoming activated, only non-pericryptal α-SMA/vimentin fibroblasts were enumerated in the present study differentiated based on histological landmarks. To quantify activated fibroblasts within each sample, three independent 40x fields that consisted of more than 50% stroma area were taken from each slide. The number of activated fibroblasts were enumerated by counting the number of dual positive α-SMA and vimentin cells per field. The total area of stroma was calculated by creating an image mask of each field in Photoshop. Activated fibroblast density was calculated by dividing the number of activated fibroblasts by stromal area. Quantification of CD3+ cells followed an identical process, but used biopsy samples from different patients due to primary sample depletion.
RESULTS
ACF epithelium and stroma display distinct expression profiles
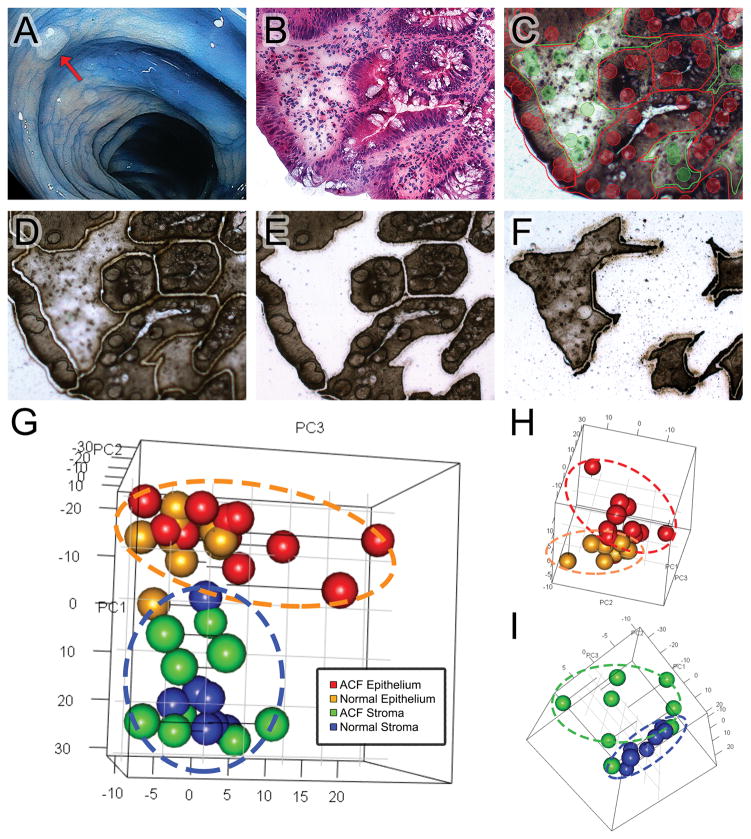
Our sample set is comprised of twelve ACF biopsies that were collected from the proximal colons of ten patients. A representative ACF identified in the proximal colon is shown in Figure 1A. ACF were histologically confirmed and then further selected based on their mutation status as determined by DNA-MS sequencing. Each biopsied ACF had only one detectable somatic mutation in either KRAS (KRASG12D, KRASG12V), BRAF (BRAFV600E), or APC (APCR876*, APCR1450*) (Table S1). In addition, a corresponding normal biopsy specimen was removed from the same side of the colon, generally within 2-cm of the ACF biopsy and confirmed as histologically normal. As shown in Figure 1 (panels B–F), colonic crypts and stroma were outlined by the LCM software and populations of stromal and epithelial cells were selectively laser-captured for analysis. Because the activation of apoptosis and senescence signaling represent critical barriers for early neoplastic progression, RNA extracted from laser-captured cells was subjected to targeted RNA-seq using a panel of 279-genes that cover these pathways. Targeted AmpliSeq panels were used for this study because of their increased sensitivity, providing the ability to quantify even low abundance transcripts. After completing the targeted transcriptomics analysis, principal component analysis (PCA) was used to evaluate broad transcriptional differences across all samples (Figure 1G). Epithelial and stromal samples were clearly delineated (dotted lines), clustering independently during sub-analyses (Figure 1H and 1I). These results demonstrate that the epithelium and stromal cells of an ACF are transcriptionally distinct from nearby normal mucosa.
Figure 1. LCM workflow for sample collection and analysis.
(A) A representative ACF identified in the proximal colon of a patient during HD-chromoendoscopy (red arrow), using 0.1% indigo Carmine dye-spray for contrast enhancement. (B) H&E slide of a dysplastic ACF harboring an APC r876* mutation. (C) Unstained PEN serial section of the same sample viewed from the internal camera of the Arcturus XT with slide mark-up indicated. Two capture groups were used: epithelial crypts (red) and the surrounding stroma (green). (D) An ultraviolet laser was used to cut the tissue from the slide and (E) epithelial and (F) stromal cells were captured onto separate caps for downstream RNA isolation, library preparation, and sequencing. (G) 3-D principal component analysis (PCA) of the RNA-seq data that characterizes the trends exhibited by the expression profiles of ACF epithelium (red), matched normal epithelium (orange), ACF- associated stroma (green), and matched normal stroma (blue). Each sample is represented by a single dot. In 3-D plot of epithelial (H) and stromal (I) samples. ACF separate from matched normal.
Proximal ACF activate anti-apoptotic pathways and up-regulate NF-κB family genes
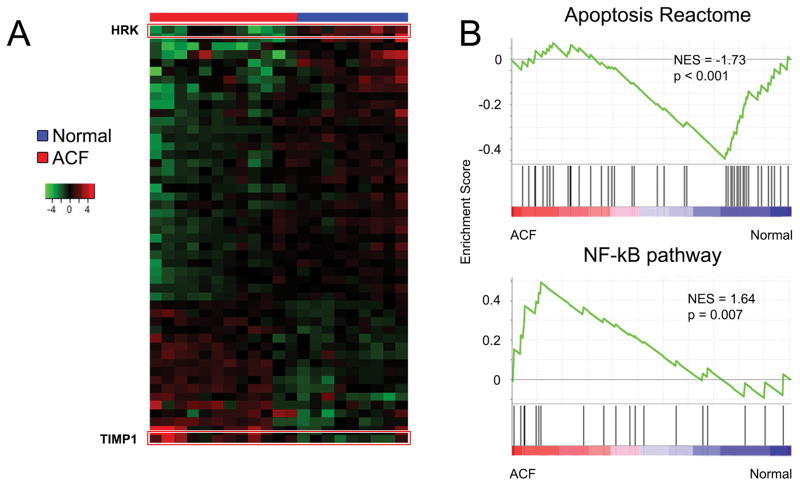
To further assess the signaling pathways responsible for the differential clustering observed in the PCA, gene expression patterns were evaluated in laser-captured epithelial cells from ACF and normal mucosa. Differential expression analysis identified 50 significantly altered genes between ACF and normal crypts (Figure 2A, Table S2). To evaluate these changes, Gene Set Enrichment Analysis (GSEA) was utilized to identify enriched signatures within the ACF epithelial cells compared to normal epithelia. In general, ACF epithelium demonstrate down-regulation of genes involved in apoptosis, referred to as the ‘apoptosis reactome’, and up-regulation of the NF-κB pathway (Figure 2B). Overall, fourteen pro-apoptotic genes were down-regulated in ACF. The most significantly down-regulated (6.9-fold, p=0.00009) gene was hara-kiri (HRK). HRK encodes a protein that is a potent activator of apoptosis that is frequently down-regulated in a variety of cancers and is hypermethylated in CRC. In addition, cytochrome C (CYCS) and caspase 3 (CASP3) expression were down-regulated relative to normal (1.5-fold; p=0.05 and 1.8-fold; p=0.004, respectively). Among the 10 anti-apoptotic genes that were up-regulated in ACF epithelial cells, two important NF-κB gene family members, RELA and RELB, were increased by ~2-fold (p=0.00001) and 1.8-fold (p=0.02), respectively. Several additional NF-kB activators and target genes were also up-regulated in ACF. Caspase recruitment domain family member 6 (CARD6) was increased by 3.7-fold in ACF relative to normal (p=0.02). The mRNA for receptor-interacting serine/threonine-protein kinase 2 (RIPK2), known to directly interact with CARD6 (16), was also increased (1.6-fold, p=0.01). The most significantly up-regulated gene present in proximal ACF was tissue inhibitor of metalloproteinase 1 (TIMP1), which was significantly increased (4.7-fold; p=0.0004). TIMP1 is a key NF-κB target gene that plays a central role in mediating NF-κB activity, as well as contributing to the maintenance of the extracellular matrix.
Figure 2. Comparison of normal and ACF epithelial gene expression patterns.
(A) Heat-map shows 50 significantly altered genes between ACF (red) and normal (blue) epithelium. The TIMP-1 gene shows the highest up-regulation (4.7-fold, p=0.0004), and the HRK gene is the most down-regulated (6.8 fold, p<0.01). (B) Gene set enrichment analysis (GSEA) identifies multiple differentially expressed gene sets. ACF are negatively enriched for genes within the apoptosis reactome, while positively enriched for genes in the NF-κB pathway.
Mutation-specific transcriptional patterns in ACF
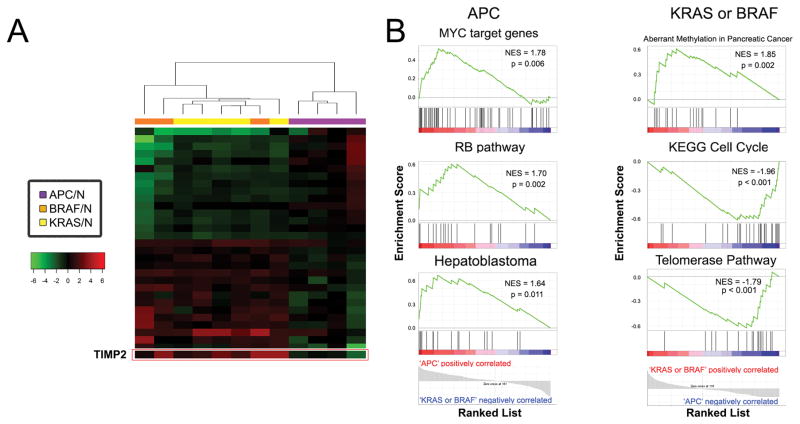
Inter-individual variation in gene expression patterns often complicates the results of expression profiling analyses. Thus to develop a quantitative assessment of mutation-specific effects, potential confounders were controlled for by normalizing each sample to its corresponding matched normal procured from the same patient. After data normalization, we then compared gene expression profiles in ACF between APC versus BRAF or KRAS to determine whether differentially expressed genes may be segregated based upon their mutation status. As shown by the heat-map results depicted in Figure 3A, a total of 31 genes were differentially expressed between the groups. GSEA (Figure 3B, left panel) identified a significant enrichment of MYC target genes (MYC is upregulated 3.5-fold, p = 0.03, Table S3), as well as genes within the RB-pathway and the hepatoblastoma gene set in ACF harboring APC mutations. In addition, the M2342 gene set was down-regulated in APC-mutant ACF (17). This finding is consistent with earlier studies reporting differential gene expression that is associated with β-catenin inactivation (NES = −2.03, p <0.001). Analyses of KRAS- and BRAF-mutant ACF epithelia showed up-regulation of gene sets involved in aberrant DNA methylation. In contrast, reduced gene expression was observed for gene sets associated with cell cycling and the telomerase pathway (Figure 3B, right panel). Collectively, these findings suggest that somatic mutations in ACF are capable of enforcing transcriptional changes that promote critical cellular processes associated with more advanced neoplasia. Several genes that did not fall into significantly enriched GSEA categories were associated with NF-κB signaling (Table S3). For example, the expression of RELB (2.9-fold, p=0.02) and its downstream effector, BIRC3 (2.9-fold, p=0.04) were both significantly altered in KRAS and BRAF-mutant ACF in comparison to APC-mutant epithelia. Interestingly, the most up-regulated gene (9.5-fold; p=0.002) in BRAF- and KRAS-mutant ACF epithelia was the secreted tissue inhibitor of metalloproteinase, TIMP2, which plays a key role in anti-tumor transcriptional activity within the tumor microenvironment.
Figure 3. Oncogenic mutations activate distinct transcriptional profiles.
ACF transcriptomes were normalized to patient controls to identify mutation-specific changes. (A) Heat-map identifies 32 genes differentially expressed genes between RAS-activated ACF and APC-mutant ACF. The TIMP-2 gene shows the highest up-regulation (9.5-fold, p=0.002) (B) Left panel; APC-mutant ACF were significantly enriched for gene sets including MYC target genes, RB pathway, and hepatoblastoma, a pediatric liver cancer that has abnormal Wnt/B-catenin signaling and is associated with individuals with familial adenomatous polyposis (FAP). Right panel; KRAS- and BRAF-mutant ACF up-regulate gene sets related to aberrant methylation. In addition, gene sets for cell cycle and telomerase pathway were significantly down-regulated, which may be indicative of a senescence phenotype.
BRAF-mutant ACF are associated with up-regulation of senescence-associated genes
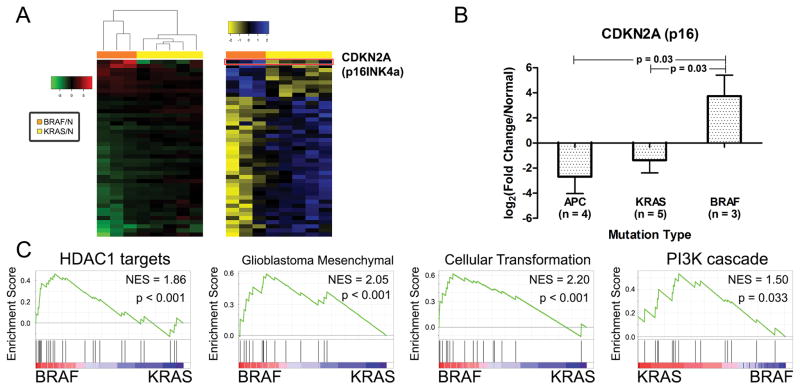
Distinct carcinogenic pathways dependent upon BRAF or KRAS activating mutations have been described in the colon. Thus the possibility was tested that KRAS and BRAF may activate different transcriptional networks that can be identified even at this early stage of neoplastic transformation. To illustrate these differences, a z-score transformation was applied to gene expression data from laser-captured epithelial cells. As shown in Figure 4A, a total of 43 differentially expressed genes were identified. Within this panel, 42 genes showed expression differences in the same direction when compared to normal epithelia (Table S4), although in each case the magnitude of change was higher in BRAF- compared to KRAS-mutant ACF. The one exception was p16 (CDKN2A), encoding the tumor suppressor protein (p16INK4A). p16 affords a critical barrier to cancer progression and has been implicated in the ‘alternative’ CRC pathway (18). Transcripts for p16 were significantly higher (3.5-fold; p=0.03) in BRAF-mutant ACF compared to both KRAS- and APC-mutant epithelial cells (Figure 4B). GSEA was also applied to the BRAF- and KRAS-mutant ACF. As shown in Figure 4C, three distinct gene sets were preferentially activated in BRAF-mutant ACF: histone deacetylase 1 (HDAC1), glioblastoma mesenchymal subtype and a ‘cellular transformation’ signature, whereas KRAS-mutant ACF were enriched for the PI3K cascade.
Figure 4. KRAS- and BRAF-mutant ACF are transcriptionally distinct.
(A) Left panel; heat-map compares normalized gene expression profiles between KRAS- and BRAF-mutant ACF. Forty-three genes were differentially expressed between the two groups. Right panel; z-score transformation of gene expression data. (B) p16INK4a mRNA is significantly up-regulated in BRAF-mutant ACF compared to both KRAS- and APC-mutant ACF. (C) GSEA for BRAF-mutant ACF identified enrichment in gene sets including histone deacetylase 1 (HDAC1) targets, glioblastoma mesenchymal subtype, a cellular transformation signature, and a negative enrichment for the PI3K cascade.
ACF-associated stroma is pro-inflammatory with the presence of activated stromal fibroblasts
Among the differentially expressed genes identified within laser-captured epithelial cells were a subset that encode secreted proteins. This observation suggests that early transformed epithelial cells may impart signals that directly contribute to the reorganization of the underlying stromal microenvironment. Thus LCM was used to capture populations of stromal cells in direct contact with the transformed epithelia. As shown by the heat-map depicted in Figure 5A, supervised clustering analysis revealed a total of 68 differentially expressed genes (Table S5). Six of 9 stromal samples (left side) laser-captured from ACF clustered independently from normal stromal samples.
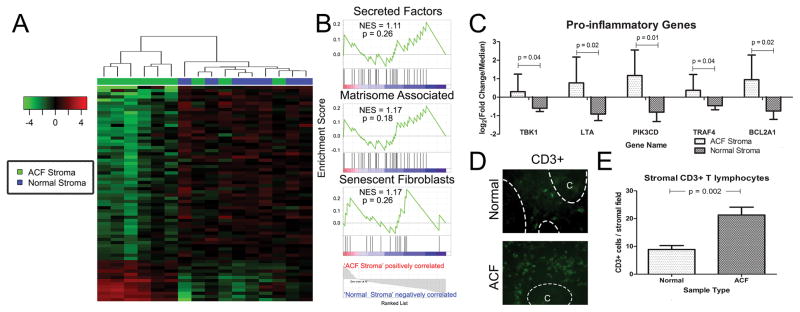
Figure 5. ACF associated stroma have an altered transcriptome with increase immune infiltration.
(A) Heat-map showing supervised clustering analysis. Sixty-eight genes were differentially expressed between ACF stromal samples and normal controls. Six of 9 samples demonstrate a distinct transcriptional profile from normal controls. (B) GSEA determined that cells isolated from the ACF stroma are enriched for gene sets for secreted factors, changes to the extracellular matrisome and enrichment for the senescent fibroblast phenotype. (C) Several pro-inflammatory genes are significantly upregulated in ACF-stroma and are leukocyte and lymphocyte specific. (D) Immunofluorescence of normal and ACF sections confirm increased abundance of CD3+ cells within ACF stroma. Epithelial crypt area is labelled ‘C’ and separated by dashed lines. (E) Quantification of CD3+ cells from 15 high magnification (40x) fields from five ACF and normal pairs
GSEA identified the enrichment of gene sets for the ‘extracellular matrisome’, ‘secreted factors’ and the ‘senescent fibroblast phenotype’ (Figure 5B). Although the changes were only modest, a number of genes involved in pro-inflammatory signaling and fibroblast activation were identified. For example, LTA (3.2-fold; p=0.012), PIK3CD (4.0-fold; p=0.011), BCL2A1 (3.2-fold; p=0.02), TBK1 (1.9-fold; p=0.04) and TRAF4 (1.8-fold; p=0.04) were each significantly up-regulated (Figure 5C). These results suggest an influx of pro-inflammatory cells and a release of potent cytokines into the ACF-associated stromal microenvironment. In fact, immunofluorescence analysis of ACF-associated stroma revealed an increase (2.4; p=0.002) in the number of CD3+ T lymphocytes compared to normal tissue (Figures 5D and 5E). The presence of CD3+ T lymphocytes was limited to adjacent stromal tissue with no observable infiltration within the epithelium.
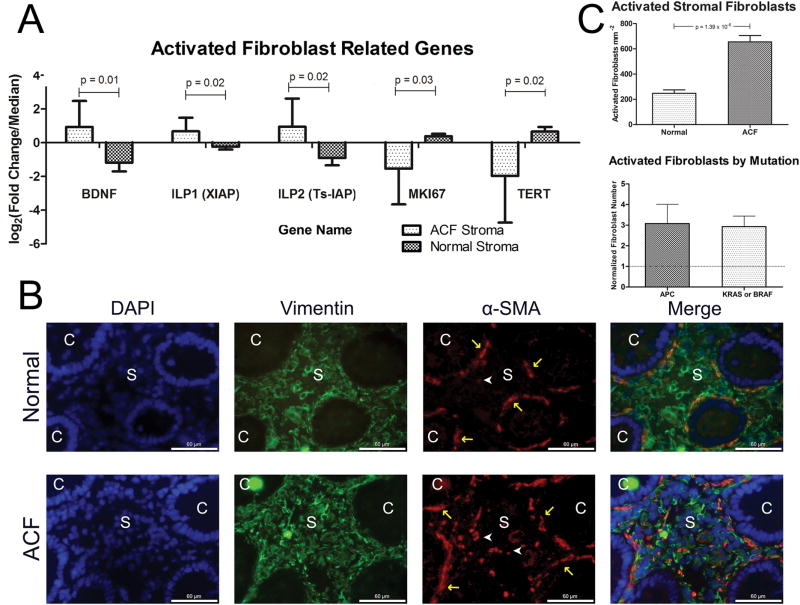
Another gene within the ACF stroma that was significantly differentially expressed was BDNF (4.3-fold; p=0.013). BDNF has been shown to be secreted by senescence-activated fibroblasts. In addition, several senescence-associated genes were down-regulated, including TERT (6.2-fold; p=0.03) and KI67 (3.8-fold; p=0.03). ACF-associated stroma also showed increased expression levels of the IAP proteins, X-IAP (1.9-fold; p=0.02) and Ts-IAP (3.6-fold; p-0.02) (Figure 6A). These genes are known to contribute to tumor progression via activation of NF-κB and contribute to fibronectin expression, which may increase fibroblast activation (19,20). Thus to confirm the presence of activated fibroblasts within the ACF-associated stroma, colon sections were stained for vimentin and α-SMA (Figure 6B). As shown in Figure 6C (top), quantification of serial sections identified a significant increase in the density of activated fibroblasts (3.0-fold; p=0.00001) compared to normal stroma. However, there were no detectable differences in the number of activated fibroblasts in ACF with distinct somatic mutations (KRAS, BRAF and APC) (Figure 6C, bottom).
Figure 6. Fibroblasts are activated within ACF stroma.
(A) Five genes related to activated senescent fibroblasts were up-regulated in ACF stroma. (B) Immunofluorescence analysis of normal (n = 10) and ACF (n = 11) sections confirm the presence of activated fibroblasts within the ACF-associated stroma. Epithelial crypt area is labeled ‘C’ and stromal area is labeled with an ‘S’. Dual-labeled normal appearing myofibroblasts, which line the base on the crypt, are marked with yellow arrows and activated fibroblasts are labeled with white arrowheads. White scale bar is 60 micrometers. (C) Left Panel; quantification of activated fibroblasts from ACF samples performed using 3 independent 40x fields containing greater than 50% stromal area. Number of non-myofibroblast dual co-stained cells were quantified and divided by their calculated stromal area to determine activated fibroblast density. Right panel; normalizing each ACF (n = 11) by its matched normal did not reveal any difference by mutation type.
DISCUSSION
Although considerable effort is presently being directed towards the development and application of targeted cancer therapies, preventing cancer remains the most promising approach for reducing overall cancer mortality rates. However, our understanding of the earliest events in cancer progression remains limited, particularly in humans. To gain further insight into the earliest stages of colon cancer development, we have focused on ACF. ACF are precancerous epithelial growths commonly found in the human colon. Although ACF are frequently observed in the distal colorectum, they are far less frequent in the proximal colon, averaging less than one lesion per subject. However, the molecular, histological and demographic features of proximal ACF indicate that they are more likely to advance to a significant neoplasm. In the present study we have taken advantage of the accessibility of these early proximal lesions to study molecular features of the epithelial-stromal interactions that regulate their progression. Understanding the nature of these interactions could ultimately uncover specific biomarkers and intercellular signaling pathways that could be monitored and targeted for highly effective cancer prevention. This information could be of central importance for proximal ACF since these lesions may progress rapidly and lead to the formation of interval cancers in the proximal colon.
The reciprocal interaction between transformed epithelial cells and their associated stroma is an important determinant of early neoplastic growth. The stromal microenvironment within the colon is comprised of extracellular matrix components, secreted factors, and stromal cells, which include fibroblasts as well as resident and infiltrating immune cells. Previous studies of premalignant changes associated with cancer, including head and neck (21), breast (22) and ovarian (23) cancers, have found evidence for a pro-inflammatory stromal microenvironment. Characteristic changes include increased cytokine signaling and immune cell infiltration. In addition, a mouse model of squamous skin carcinogenesis demonstrated that fibroblasts isolated from the stroma of dysplastic lesions become activated prior to invasion. These activated dermal fibroblasts promote inflammation, mediating immune cell recruitment and altering tumor angiogenesis in an NF-κB-dependent manner (20). Furthermore, stromal fibroblasts associated with advanced CRC display genomic signatures capable of accurately predicting cancer relapse (24). However, the role of the stromal environment during the earliest stages of neoplastic transformation is less well-defined in human. Using a combination of LCM and targeted RNA-seq against a total of 279 gene targets enriched for apoptotic and senescence signaling, we have characterized alterations to the transcriptome of epithelial and stromal cell populations obtained from the human colon. We have identified transcriptionally distinct gene signatures in ACF that cluster independently from normal mucosa and are further dependent upon underlying somatic mutations.
An important observation in the present study is that a single somatic mutation can activate distinct transcriptional changes within epithelial cells. For example, in ACF harboring an APC mutation (e.g. APCR876*, APCR1450*), gene sets for the RB and MYC pathways are significantly enriched (Figure 3B), which is consistent with activation of the WNT pathway (25). This in turn, leads to dysregulation of c-MYC with concomitant activation of the RB pathway (26,27). As further evidence of mutation-specific effects, KRAS-mutant ACF up-regulated a gene set for the PI3K/AKT/MTOR pathway more intensely than BRAF mutants, which is consistent with RAS being upstream of RAF in the growth factor receptor/MAPK signaling pathway. Furthermore, KRAS- and BRAF-mutant ACF displayed up-regulation of gene sets related to aberrant methylation, a characteristic of the ‘alternative’ pathway to CRC (28), where genome-wide hypermethylation may be a key driver to cancer progression. KRAS and BRAF mutations have been identified in both hyperplastic polyps and serrated adenomas with associated CpG island hypermethylation of promoter regions (CIMP-H) (29). Although several reports have described a limited constellation of DNA methylation changes in ACF, it would be interesting to examine the broader extent of epigenetic modifications in these early lesions. Finally, the presence of either KRAS or BRAF mutations were associated with a down-regulation of a set of genes involved in the cell cycle and telomerase pathway, suggesting that OIS programs may have been activated.
We further observed that senescence is particularly evident in BRAF-mutant ACF that expressed high levels of p16, an important mediator of OIS (Figure 4B). In fact, OIS may provide an important mechanism for limiting the growth of these early pre-neoplastic lesions, leading in most cases to their regression and elimination. However, it is likely that not all BRAF/KRAS ACF that undergo OIS are eliminated. For example, a subset of melanocytic nevi, which typically undergo mutant BRAFV600E-induced senescence, are able to bypass senescence and progress to melanoma (30). One method of senescence bypass is the induction of a secretory phenotype. Senescent cells can undergo changes in cellular metabolism, leading to increased production of secretory factors (31,32). Within the colonic mucosa, we believe that these secretory factors may exert a pro-tumorigenic influence upon the associated tissue microenvironment that promotes the progression of adjacent premalignant cells (33,34). In contrast, these senescence changes were not observed in APC-mutant ACF, suggesting a fundamental difference in how these senescence pathways are triggered (35), and possibly underlies the greater malignant potential of APC-mutant lesions.
Activation of the pro-inflammatory transcription factor NF-κB represents an important mechanism contributing to cancer promotion. NF-κB activation typically occurs during early stages of carcinogenesis (36) and exerts an anti-apoptotic effect while promoting angiogenesis. In CRC, the NF-κB pathway can become constitutively activated via oncogenic activation of either KRAS or BRAF, while APC can directly regulate NF-κB activation through beta-catenin (37). In the present study, we have found up-regulation of several pro-inflammatory NF-κB-related target genes in ACF (Figure 2B), indicating that the NF-kB/inflammation axis plays a pivotal role in regulating cancer development at the ACF stage.
Several studies have demonstrated that NF-κB signaling plays a fundamental role in OIS activation and the induction of a secretory phenotype. In the present study, we have observed a significant up-regulation of secreted TIMP family members in ACF samples examined regardless of their underlying somatic mutation (Figure 2A and 3A). TIMP-1 secretion by cancer cells induces trans-differentiation of normal fibroblasts into activated fibroblasts in xenograft mouse models and in co-culture systems of liver, prostate (38), and colon cancers (39). We also observed TIMP-2 upregulation in BRAF and KRAS-mutant ACF, although the effect of TIMP-2 on tumor progression is less well-defined (40). Some studies have correlated high levels of TIMP-2 with an adverse prognosis (41,42), while other studies have observed an opposite effect (43–45). The general consensus, however, is that overexpression of TIMP family proteins elicit effects on ECM turnover and facilitates formation of the tumor microenvironment. These findings suggest that ACF epithelium may interact with stromal cells and induce remodeling of the underlying of the supportive microenvironment (46,47).
Finally by analyzing directly adjacent stromal tissue, we have demonstrated for the first time that reactive stromal changes accompany the earliest detectable stages of human colonic neoplasia (Figure 5 and 6). We have observed a modest enrichment of genes reflective of a secretory milieu, with associated alterations to the extracellular matrix. Several genes within the secretory factor gene set were significantly elevated, characteristic of immune cell infiltration and activation of stromal fibroblasts. Immune cell infiltration into ACF tissue was further demonstrated by CD3+ lymphocyte immunofluorescence. In addition to immune cells, fibroblasts are a major constituent of the extracellular matrix and may become activated by a complex array of stimuli arising during tissue injury. Upon activation, fibroblasts alter their cellular phenotype, increasing expression of α-SMA protein (48). The activated fibroblasts acquire a senescent and highly secretory phenotype, promoting carcinogenesis by the secretion of growth factors and inflammatory cytokines (32,49). In our study, one key observation was the large up-regulation of BDNF (Table S5), which is secreted by activated fibroblasts (50). We confirmed the increased abundance of activated fibroblasts within ACF stroma by immunofluorescence of serial sections from the same samples. Taken together, our findings imply that inflammatory gene expression by mutated epithelium, and/or defects in the epithelial barrier related to altered colonic cryptal structure, may trigger this stromal reaction.
In summary, our results indicate that pre-neoplastic cells within the human colon undergo mutation-specific transcriptional reprogramming. These changes are associated with the induction of epithelial-stromal interactions that may influence the outcome of these early neoplastic changes. Pro-inflammatory NF-κB pathway signaling in initiated colonic epithelium may mediate the activation of underlying stromal fibroblasts and recruitment of immune cells within the stromal microenvironment. Our results provide new insights into the dynamic, complex interplay that occurs in early colorectal neoplasia, highlighting the role of activated stromal fibroblasts and potential therapeutic targeting of NF-κB for prevention of early neoplasia.
Supplementary Material
Acknowledgments
Financial support: This work was supported by the State of Connecticut Department of Public Health, Biomedical Research Applications #2012-0913 and #2015-0901 (to D.W. Rosenberg) and NIH 1RO1CA159976 (to D.W. Rosenberg).
The authors thank Dr. Jeoffrey Schageman (Thermo Fisher Scientific) for his bioinformatics expertise in 582 identifying potential pseudogenes.
Footnotes
Disclosures/Conflict of Interest: SJ, KV, and AC are employees of Thermo Fisher Scientific. The remaining authors (AM, CG, TJD, DWR) do not have any conflict of interest to declare.
Transcript Profiling: GSE77312, reviewer link provided in comments
Author Contributions: Conception and design (AM, DWR), Development of methodology (AM, SJ), DWR, Acquisition of data (AM, SJ, TJD), Analysis and interpretation of data (AM, SJ, DWR), Writing of manuscript (AM, CG, DWR), Review of the manuscript (AM, CG, DWR), Administrative support (DWR, KV, AC), Technical support (SJ), Material support (SJ, KV, AC), Study supervision (DWR)
References
- 1.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. Journal of cell science. 2005;118(Pt 3):485–96. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nature genetics. 2015;47(4):320–9. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 3.Rasanen K, Vaheri A. Activation of fibroblasts in cancer stroma. Experimental cell research. 2010;316(17):2713–22. doi: 10.1016/j.yexcr.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 4.De Wever O, Mareel M. Role of tissue stroma in cancer cell invasion. The Journal of pathology. 2003;200(4):429–47. doi: 10.1002/path.1398. [DOI] [PubMed] [Google Scholar]
- 5.Stevens RG, Swede H, Rosenberg DW. Epidemiology of colonic aberrant crypt foci: review and analysis of existing studies. Cancer letters. 2007;252(2):171–83. doi: 10.1016/j.canlet.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue A, Okamoto K, Fujino Y, Nakagawa T, Muguruma N, Sannomiya K, et al. B-RAF mutation and accumulated gene methylation in aberrant crypt foci (ACF), sessile serrated adenoma/polyp (SSA/P) and cancer in SSA/P. British journal of cancer. 2015;112(2):403–12. doi: 10.1038/bjc.2014.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg DW, Yang S, Pleau DC, Greenspan EJ, Stevens RG, Rajan TV, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer research. 2007;67(8):3551–4. doi: 10.1158/0008-5472.CAN-07-0343. [DOI] [PubMed] [Google Scholar]
- 8.Drew DA, Devers TJ, O’Brien MJ, Horelik NA, Levine J, Rosenberg DW. HD chromoendoscopy coupled with DNA mass spectrometry profiling identifies somatic mutations in microdissected human proximal aberrant crypt foci. Molecular cancer research: MCR. 2014;12(6):823–9. doi: 10.1158/1541-7786.MCR-13-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drew DA, Devers T, Horelik N, Yang S, O’Brien M, Wu R, et al. Nanoproteomic analysis of extracellular receptor kinase-1/2 post-translational activation in microdissected human hyperplastic colon lesions. Proteomics. 2013;13(9):1428–36. doi: 10.1002/pmic.201200430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laud K, Marian C, Avril MF, Barrois M, Chompret A, Goldstein AM, et al. Comprehensive analysis of CDKN2A (p16INK4A/p14ARF) and CDKN2B genes in 53 melanoma index cases considered to be at heightened risk of melanoma. Journal of medical genetics. 2006;43(1):39–47. doi: 10.1136/jmg.2005.033498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes & development. 2010;24(22):2463–79. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128(2):295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 13.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature reviews Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 14.Chuaysri C, Thuwajit P, Paupairoj A, Chau-In S, Suthiphongchai T, Thuwajit C. Alpha-smooth muscle actin-positive fibroblasts promote biliary cell proliferation and correlate with poor survival in cholangiocarcinoma. Oncology reports. 2009;21(4):957–69. doi: 10.3892/or_00000309. [DOI] [PubMed] [Google Scholar]
- 15.Khalil H, Nie W, Edwards RA, Yoo J. Isolation of primary myofibroblasts from mouse and human colon tissue. Journal of visualized experiments: JoVE. 2013;(80) doi: 10.3791/50611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufner A, Mak TW. CARD tricks: controlling the interactions of CARD6 with RICK and microtubules. Cell cycle (Georgetown, Tex) 2006;5(8):797–800. doi: 10.4161/cc.5.8.2635. [DOI] [PubMed] [Google Scholar]
- 17.Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Molecular and cellular biology. 2007;27(21):7551–9. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennecke M, Kriegl L, Bajbouj M, Retzlaff K, Robine S, Jung A, et al. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer cell. 2010;18(2):135–46. doi: 10.1016/j.ccr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer cell. 2010;17(1):53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer cell. 2010;17(2):135–47. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SD, De Costa AM, Young MR. Effect of the premalignant and tumor microenvironment on immune cell cytokine production in head and neck cancer. Cancers. 2014;6(2):756–70. doi: 10.3390/cancers6020756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast cancer research: BCR. 2009;11(1):R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saad AF, Hu W, Sood AK. Microenvironment and pathogenesis of epithelial ovarian cancer. Hormones & cancer. 2010;1(6):277–90. doi: 10.1007/s12672-010-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berdiel-Acer M, Cuadras D, Diaz-Maroto NG, Sanjuan X, Serrano T, Berenguer A, et al. A monotonic and prognostic genomic signature from fibroblasts for colorectal cancer initiation, progression, and metastasis. Molecular cancer research: MCR. 2014;12(9):1254–66. doi: 10.1158/1541-7786.MCR-14-0121. [DOI] [PubMed] [Google Scholar]
- 25.Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2000;18(9):1967–79. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- 26.Nevins JR. The Rb/E2F pathway and cancer. Human molecular genetics. 2001;10(7):699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 27.beta-catenin links the APC gene to MYC in colon cancer. Gastroenterology. 1998;115(5):1041c–42. doi: 10.1016/s0016-5085(98)70067-8. [DOI] [PubMed] [Google Scholar]
- 28.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–30. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang S, Farraye FA, Mack C, Posnik O, O’Brien MJ. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. The American journal of surgical pathology. 2004;28(11):1452–9. doi: 10.1097/01.pas.0000141404.56839.6a. [DOI] [PubMed] [Google Scholar]
- 30.Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene. 2004;23(16):2919–33. doi: 10.1038/sj.onc.1207518. [DOI] [PubMed] [Google Scholar]
- 31.Kozlowski M, Ladurner AG. ATM, MacroH2A.1, and SASP: The Checks and Balances of Cellular Senescence. Molecular cell. 2015;59(5):713–5. doi: 10.1016/j.molcel.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Salminen A, Kauppinen A, Kaarniranta K. Emerging role of NF-kappaB signaling in the induction of senescence-associated secretory phenotype (SASP) Cellular signalling. 2012;24(4):835–45. doi: 10.1016/j.cellsig.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Pazolli E, Alspach E, Milczarek A, Prior J, Piwnica-Worms D, Stewart SA. Chromatin remodeling underlies the senescence-associated secretory phenotype of tumor stromal fibroblasts that supports cancer progression. Cancer research. 2012;72(9):2251–61. doi: 10.1158/0008-5472.CAN-11-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer metastasis reviews. 2010;29(2):273–83. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole AM, Ridgway RA, Derkits SE, Parry L, Barker N, Clevers H, et al. p21 loss blocks senescence following Apc loss and provokes tumourigenesis in the renal but not the intestinal epithelium. EMBO molecular medicine. 2010;2(11):472–86. doi: 10.1002/emmm.201000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 37.Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22(56):8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 38.Song T, Dou C, Jia Y, Tu K, Zheng X. TIMP-1 activated carcinoma-associated fibroblasts inhibit tumor apoptosis by activating SDF1/CXCR4 signaling in hepatocellular carcinoma. Oncotarget. 2015;6(14):12061–79. doi: 10.18632/oncotarget.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong Y, Scott E, Lu R, Xu Y, Oh WK, Yu Q. TIMP-1 promotes accumulation of cancer associated fibroblasts and cancer progression. PloS one. 2013;8(10):e77366. doi: 10.1371/journal.pone.0077366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain Z, Khan MI, Shahid M, Almajhdi FN. S-adenosylmethionine, a methyl donor, up regulates tissue inhibitor of metalloproteinase-2 in colorectal cancer. Genetics and molecular research: GMR. 2013;12(2):1106–18. doi: 10.4238/2013.April.10.6. [DOI] [PubMed] [Google Scholar]
- 41.Clarke MR, Imhoff FM, Baird SK. Mesenchymal stem cells inhibit breast cancer cell migration and invasion through secretion of tissue inhibitor of metalloproteinase-1 and -2. Molecular carcinogenesis. 2015;54(10):1214–9. doi: 10.1002/mc.22178. [DOI] [PubMed] [Google Scholar]
- 42.Remacle A, McCarthy K, Noel A, Maguire T, McDermott E, O’Higgins N, et al. High levels of TIMP-2 correlate with adverse prognosis in breast cancer. International journal of cancer. 2000;89(2):118–21. doi: 10.1002/(sici)1097-0215(20000320)89:2<118::aid-ijc3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 43.Kapral M, Wawszczyk J, Jurzak M, Hollek A, Weglarz L. The effect of inositol hexaphosphate on the expression of selected metalloproteinases and their tissue inhibitors in IL-1beta-stimulated colon cancer cells. International journal of colorectal disease. 2012;27(11):1419–28. doi: 10.1007/s00384-012-1445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuda H, Aoki H. Host expression of matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 in normal colon tissue affects metastatic potential of colorectal cancer. Diseases of the colon and rectum. 1999;42(3):393–7. doi: 10.1007/BF02236360. [DOI] [PubMed] [Google Scholar]
- 45.Groblewska M, Mroczko B, Gryko M, Pryczynicz A, Guzinska-Ustymowicz K, Kedra B, et al. Serum levels and tissue expression of matrix metalloproteinase 2 (MMP-2) and tissue inhibitor of metalloproteinases 2 (TIMP-2) in colorectal cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(4):3793–802. doi: 10.1007/s13277-013-1502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remillard TC, Bratslavsky G, Jensen-Taubman S, Stetler-Stevenson WG, Bourboulia D. Molecular mechanisms of tissue inhibitor of metalloproteinase 2 in the tumor microenvironment. Molecular and cellular therapies. 2014;2:17. doi: 10.1186/2052-8426-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stetler-Stevenson WG, Gavil NV. Normalization of the tumor microenvironment: evidence for tissue inhibitor of metalloproteinase-2 as a cancer therapeutic. Connective tissue research. 2014;55(1):13–9. doi: 10.3109/03008207.2013.867339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Molecular biology of the cell. 2001;12(9):2730–41. doi: 10.1091/mbc.12.9.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual review of pathology. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dudas J, Bitsche M, Schartinger V, Falkeis C, Sprinzl GM, Riechelmann H. Fibroblasts produce brain-derived neurotrophic factor and induce mesenchymal transition of oral tumor cells. Oral oncology. 2011;47(2):98–103. doi: 10.1016/j.oraloncology.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.