Abstract
Completion of the Xenopus laevis genome sequence from inbred J strain animals has facilitated the generation of germline mutant X. laevis using targeted genome editing. In the last few years, numerous reports have demonstrated that TALENs are able to induce mutations in F0 Xenopus embryos, but none has demonstrated germline transmission of such mutations in X. laevis. In this report we used the oocyte host-transfer method to generate mutations in both tyrosinase homeologs and found highly-penetrant germline mutations; in contrast, embryonic injections yielded few germline mutations. We also compared the distribution of mutations in several F0 somatic tissues and germ cells and found that the majority of mutations in each tissue were different. These results establish that X. laevis J strain animals are very useful for generating germline mutations and that the oocyte host-transfer method is an efficient technique for generating mutations in both homeologs.
Keywords: Xenopus laevis, TALENs, J strain, tyrosinase, oocyte host-transfer, genome editing
Introduction
Xenopus laevis has a long history as a vertebrate model system in biomedical research, and has made important contributions to many fields, including cell cycle, somatic cell nuclear transfer, embryonic development, and cell fate specification. Due to its allotetraploid genome, the use of genetically heterogeneous animals, and its long generation time, X. laevis has not been utilized as a genetic model. In recent years, an inbred J strain line has become more widely available through the National Xenopus Resource (NXR), and this inbred strain was used in sequencing the X. laevis genome (Gantress et al., 2003; Pearl et al., 2012; Tochinai and Katagiri, 1975). As an allotetraploid, X. laevis is composed of two separate diploid genomes (2n=36) that segregate independently; there are nine pairs of homeologous chromosomes named Xla1S and Xla1L through Xla9S and Xla9L, where S (short) and L (long) refer to their relative lengths (Krylov and Tlapakova, 2015; Matsuda et al., 2015). With the recent sequencing of the X. laevis genome, combined with advances made in genome editing, X. laevis now offers additional benefits as a genetic model system.
The development of genome editing technologies (CRISPR/Cas and TALENs) has greatly facilitated the ability of investigators to create loss-of-function mutant alleles in virtually any model organism, including Xenopus (Harrison et al., 2014; Peng et al., 2014). Transcription activator-like effector nucleases (TALENs) are comprised of two modular arrays of DNA-binding domains coupled to opposing catalytic subunits of the heterodimeric FokI nuclease, and can be designed to target almost any region in the genome (Joung and Sander, 2013; H. Kim and J.-S. Kim, 2014). Simple microinjection of TALEN mRNAs into X. laevis embryos is sufficient to induce site-specific lesions and can be designed to target either one or both alloalleles (Nakade et al., 2015; Sakane et al., 2013; Suzuki et al., 2013). Although these studies demonstrated that TALENs are capable of inducing mutations in both homeologous genes, injections into embryos were not sufficient to generate a null phenotype. This result probably reflects the time required for the TALEN mRNAs to be translated during the rapid cell divisions of early Xenopus development.
A complementary approach that would provide more time for TALEN proteins to act on genomic DNA is to inject TALEN mRNAs into oocytes, rather than embryos. This well-established technique is known as the oocyte host-transfer (OHT) method, and has been commonly used to knock down maternally stored mRNAs to examine their function in early embryogenesis (Heasman et al., 1991; Olson et al., 2012). In the OHT method, the ovary is isolated and stage VI oocytes are defolliculated manually; they can then be injected and cultured for 24-48 hours in vitro, after which maturation is induced by addition of progesterone to the media. The next day, injected oocytes are implanted into an ovulating host female and the eggs are collected and fertilized in vitro the same day. Although laborious, in comparison to embryo injections, it is very effective in producing F0 TALEN-mediated gene knock-out in Xenopus laevis, with mutations present in both alloalleles (Miyamoto et al., 2015; Nakajima and Yaoita, 2015a). However, the TALEN-induced mutations were mosaic, with several different mutations identified in each alloallele. The distribution of mutations in different tissues was not described and it was not clear whether mutations were present in the germ cells.
With increased used of J strain animals, our goal was to develop an albino J strain X. laevis line that would benefit the wider Xenopus community. In this paper, we examined the efficiency of germline transmission of TALEN-induced mutations in F0 adult X. laevis J strain animals produced by oocyte host-transfer, and determined whether the same germ cell mutations were induced in somatic tissues. Tyrosinase is an essential component of melanin biosynthesis, the disruption of which produces an albino phenotype in Xenopus, allowing for easy phenotypic identification of a knockout phenotype (Blitz et al., 2013; Guo et al., 2014; Ishibashi et al., 2012; Nakajima et al., 2012; Nakajima and Yaoita, 2015b; Nakayama et al., 2013; Suzuki et al., 2013). We designed TALENs to target both tyrosinase homeologs present on the 2S and 2L chromosomes and examined the efficiency of germline transmission to the F1 generation, and genotyped five different tissues in the adult female F0 frog.
Materials and Methods
TALEN production
A single pair of TALENs were designed using the TAL Effector Targeter 2.0 (https://tale-nt.cac.cornell.edu/) to target the 3’ end of the first exons of both homeologs of X. laevis tyrosinase located on the 2S and 2L chromosomes based on the X. laevis genome data available from Xenbase (James-Zorn et al., 2015). The forward TALEN was designed to target GCCCATGAAGCTCCAG, and the reverse TALEN was designed to target CGGTACTTCTTGCTGC. The TALEN constructs were created using the Golden Gate and TAL Effector Kit 2.0 (Addgene) (Cermak et al., 2011) and cloned into the destination vectors pCS2-TAL3DDD and pCS2-TAL3RRR (provided Dr. David Grunwald) (Hu et al., 2013). The final constructs were confirmed by Sanger sequencing, and mRNAs were in vitro transcribed from the linearized constructs with a mMESSAGE mMACHINE SP6 Transcription kit (Life Technologies, Carlsbad, CA).
Oocyte Host-Transfer
700 pg of each TALEN mRNA were co-injected into manually-defolliculated, stage VI oocytes from a J strain female, and the oocytes were cultured in OCM (70% Leibovitz L-15 media, 0.4% BSA, 1X Pen-Strep, 0.5 μg/mL gentamicin, pH 7.8) for 24 hours. The oocytes were then matured by overnight application (16 hours) of 2 µM progesterone in OCM (Sigma, St Louis, MO); an albino host wild type female was induced to ovulate by injection with 500 IUs of human chorionic gonadotropin (HCG). The following day, matured J strain oocytes were transplanted into the wild type host female body cavity through a 16-gauge needle, and eggs were collected in 1X MMR and then fertilized in vitro with J strain sperm. The donor injected-embryos were pigmented and easily identifiable from the albino host eggs. For embryo injections, the same amount of each TALEN mRNA was injected into one-cell embryos that had been produced by in vitro fertilization with J strain sperm.
Husbandry
All animals were housed and handled in the National Xenopus Resource in accordance with animal care protocol 15-02B approved by the Marine Biology Laboratory IACUC. The Xenopus J strain animals were bred in house at the NXR from parents obtained from Jacques Robert (University of Rochester). Female frogs were primed with 50U PMSG 3-4 days prior to induction of ovulation. Ovulation was induced by injecting females with either 400 IU HCG or with 150 μg ovine luteinizing hormone (LH) (National Hormone and Peptide Program, Torrance, California). LH dose used was equivalent to 2 μg/g body weight. These albino J strain F0 frogs are deposited in the NXR and designated RRID:NXR_0.0041 and is designated Xla.tyrtm1NXR.
Genotyping
Genomic DNA was isolated from F0 tissue biopsies from the single OHT female and individual F1 embryos that were produced by backcrossing the F0 OHT frogs with wild type J strain using DNAzol reagent (Thermo Fisher, Waltham, MA) and DNeasy Blood and Tissue kits (Qiagen, Hilden, Germany). Homeolog-specific PCR of genomic DNA was performed using Taq polymerase with Thermopol buffer (New England Biolabs, Ipswich, MA) and the following primers: tyr.L fwd GTTTGCAATGGCCAGTTTCC, tyr.L rev TCATCCCACCACGAGCAA, tyr.S fwd GCTTGCAATGGCCAGTTCCC, tyr.S rev CCATCCCACCACCACCAC. PCR products were then cloned into pCR2.1 (Life Technologies, Carlsbad, CA).
Results
Recent sequencing of the X. laevis genome has shown that the homeologous tyrosinase (tyr) genes are located on the 2S and 2L chromosomes (Xenbase). Previous reports showed that injection of TALENs targeting both tyr homeologs (tyr.L and tyr.S) into one-cell stage X. laevis embryos results in mosaic pigmentation defects (Suzuki et al., 2013), whereas injection into oocytes produces more complete albinism (Miyamoto et al., 2015; Nakajima and Yaoita, 2015a). None of these studies however, examined the distribution of mutations within different adult organs. To determine how mosaic the mutations are in different adult organs, we injected one pair of TALEN mRNAs targeting the 3’ end of the first exon of both tyr.L and tyr.S into stage VI oocytes (Fig. 1A). After transferring the oocytes into a donor female, we obtained 16 viable embryos. At stage 45, 10 tadpoles were completely albino, two tadpoles showed partial albinism and four tadpoles had normal pigmentation (data not shown). We raised the resulting albino tadpoles to adulthood, and three survived past metamorphosis (two males and one female); all three were found to be primarily albino, except for a few patches of pigmentation (Fig. 1B). In contrast, embryos injected with the same TALEN mRNAs showed a higher level of mosaic pigmentation (Fig. 1C). Once these frogs reached sexual maturity we mated them to each other as well as to wild type J strain partners to determine the genotype and phenotype of F1 offspring and to create a line of J strain albino frogs with a defined genetic mutation in each alloallele. Mating the OHT female to OHT male #1 resulted in ~75% F1 albino tadpoles, whereas mating with OHT male #2 resulted in ~50% albino tadpoles (Fig. 1D,E and data not shown).
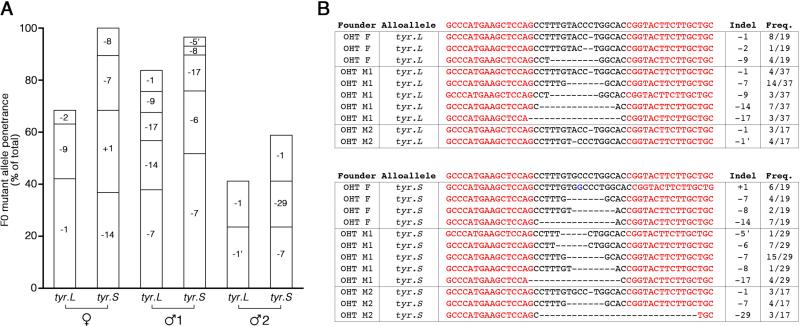
Figure 1.
Examples of F0 and F1 X. laevis tyr mutants. (A) Schematic diagram of tyrosinase locus, showing the five exons. The scissors show where the TALENs were designed to target the 3’ end of the first exon, and the arrows show the location of primers used for genotyping. (B) Picture of an 8-month-old tyr mutant F0 J strain frog generated by oocyte host transfer. Notice small patch of pigmentation in the body and eye (arrows). (C) Picture of an 8-month-old F0 tyr mutant frog generated by injection of TALEN mRNAs into one-cell stage embryos. (D) Picture of natural mating of two tyr mutant F0 frogs (OHT female and OHT male #1) at 14 months. (E) Picture of an F1 tyr mutant frog from mating of OHT female and OHT male #1 at 6 months of age.
To determine the exact mutations present in germ cells, we crossed them with wild type J strain frogs, and genotyped both homeologs in at least 17 F1 embryos from each cross; to identify homeolog-specific mutations, we used PCR primers specific to tyr.S or tyr.L. The number of different mutations inherited from a single founder F0 frog ranged from 2 to 5 (Fig. 2A). Analysis of genomic DNA from single F1 progeny confirmed a high level of germ line transmission of mutant tyr alleles: the female founder transmitted mutant alleles to ~85% of progeny, one male founder transmitted mutant alleles to ~90% of his progeny, and the other male founder transmitted mutant alleles to ~50% of his progeny (Fig. 2A). Both homeologs were similarly affected, and several identical deletions were observed independently on both chromosomes in separate founders (Fig. 2B). This demonstrates there are several different germline mutations in a single F0 adult, consistent with the fact that there are four presumptive primordial germ cells at the 32-cell stage (Houston et al., 1998; Ressom and Dixon, 1988). In contrast, we found almost no germline mutations in F1 embryos derived from F0 adult frogs that had been injected with tyr TALEN mRNAs at the one-cell stage (data not shown). Overall, these results demonstrate that the oocyte host-transfer method is an effective strategy for generating germ line mutations.
Figure 2.
Genotyping of F1 tyr mutant embryos. (A) Bar graph illustrating the frequency of tyr.L and tyr.S mutations in F1 embryos generated by mating F0 OHT frog with wild type J strain. Wild type alleles make up the remainder of each column. (B) Sequence of mutations identified in F1 embryos. Frequency refers to the number of F1 embryos that contained the mutation. Red font indicates TALEN target sites.
We anticipated that injecting TALENs into the oocyte would lead to the induction of mutations at an early stage, since the TALEN proteins would be present before fertilization. However, the presence of more than two mutations (and some wild type alleles) in the germ cells suggests that the TALENs were acting later in development. We wanted to determine if the same mutations found in the germ cells were also present in other tissues. If the TALENs induced mutations early in development then we expected to find identical mutations in somatic tissues as well. We therefore isolated genomic DNA from five different organs of the female OHT frog representing each germ layer, heart (mesoderm), skin (ectoderm), liver, lung and gut (endoderm), and genotyped the tyr.L and tyr.S loci.
Unexpectedly, we identified many additional mutations in each somatic tissue. For example, in F1 embryos derived from the F0 OHT female, we identified four different tyr.L mutations, −1, −2, −9, and wild type (Fig. 2A), yet in the F0 somatic tissues we recovered an additional sixteen unique mutant alleles (Fig. 3A). Furthermore, the most prevalent tyr.L germline mutation, −1, which was found in ~40% of F1 embryos (Fig. 2B), was only found in three of the five somatic tissues sampled (heart, skin and liver) and at much lower frequencies of 5-10% (Fig. 3A). Similar discrepancies were observed between F0 somatic tissues and F1 embryos for tyr.S mutations. The most prevalent germline mutations in the OHT F0 female, +1 and −14, which accounted for nearly 70% of all alleles transmitted to F1 progeny (Fig. 2B), were either entirely absent or found at a low frequency in only a subset of the somatic tissues (Fig. 3B). For both homeologs we found unique mutations in each somatic tissue that were not present in the other organs; many more mutations in somatic tissues that were not found in the germline, and drastically different frequencies for those mutations that occurred in both germline and somatic tissues. These results demonstrate that expressing TALENs in oocytes followed by host-transfer can lead to efficient bi-alloallelic mutagenesis in X. laevis, but that the mutations are still induced after fertilization, which results in mosaic tissues in the F0 frogs.
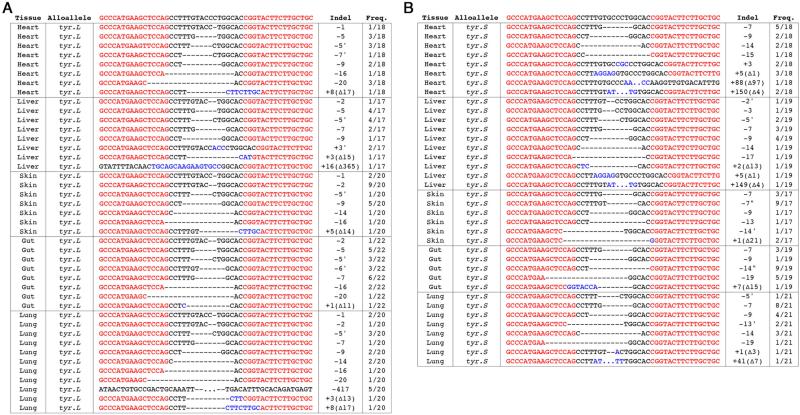
Figure 3.
Genotyping of F0 somatic tissue of female OHT tyr mutant frog. (A) Bar graph illustrating the frequency of tyr.L and tyr.S mutations in F0 organs. Wild type alleles make up the remainder of each column. *Only insertion numbers indicated; details of the deletion are found in panel B. (B) Sequence of mutations identified in heart, liver and skin of F0 female OHT frog. Frequency refers to the number of clones that contained the mutation. Red font indicates TALEN target sites.
The −9 mutation was identified in all three F0 somatic tissues at similar penetrance (19% vs 24% in germ cells), whereas the −1 mutation was only found in the skin (10%), which was much less than that observed in F1 embryos (47%). A similar discrepancy was observed between F0 somatic tissues and F1 embryos for the tyr.L −2 mutation; the −2 mutation was found in 6% of F1 embryos (Fig. 2), but present only in F0 skin in 66% of clones (Fig. 3). For the tyr.S alloallele there were five genotypes in F1 embryos, +1, −7, −8, −14 and wild type (Fig. 2), yet only the −7 mutation was found in each F0 somatic tissue (Fig. 3). Much like the tyr.L alloallele, we identified twenty two additional tyr.S mutations in F0 somatic tissues that were not present in F1 embryos (Fig. 3B). For both homeologs we found different mutations in each somatic tissue that were not present in the other organs. These results demonstrate that expressing TALENs in oocytes does lead to efficient mutagenesis in both homeologs, but that the mutations are induced later than desirable during embryogenesis creating mosaic mutations in the F0 frogs.
Discussion
Here we report the successful generation of highly-penetrant, germline-transmitted mutant alleles of both homeologs of the X. laevis tyrosinase (tyr) gene by injection of TALEN mRNAs into oocytes. Our results demonstrate that the oocyte host-transfer technique is a more effective method to generate mutations in X. laevis in both homeologs. Injection the tyrosinase TALEN mRNAs into oocytes results in almost complete albinism in F0 animals, whereas injection into embryos produced only patches of albinism. In raising these animals to adulthood, we show, for the first time in X. laevis, highly-efficient germline transmission of TALEN-induced mutations to the F1 generation. Although germline transmission of mutations have been demonstrated in X. tropicalis this is the first instance of efficient germline transmission of mutations in X. laevis (Guo et al., 2014; Ishibashi et al., 2012; Lei et al., 2012; Nakajima et al., 2012). These results demonstrate that germline mutants can now be produced in X. laevis, and opens up the experimental toolkit for X. laevis researchers. The development of this new J strain X. laevis line, designated Xla.tyrtm1NXR, will be of benefit to the wider Xenopus community.
Our injection and culture of oocytes with TALEN mRNAs followed by host-transfer aimed to effect the disruption of coding sequence as early in development as possible, either by induction of indels in the oocyte prior to fertilization, or by allowing sufficient time for translation of TALEN mRNAs prior to the rapid cell divisions during early embryogenesis. Although we did find that injection of TALEN mRNAs into oocytes produced highly-penetrant mutant alleles, the mosaicism observed in somatic and germ line tissues within the founders suggests that editing of genomic DNA occurred after fertilization, which is in agreement with previous reports (Miyamoto et al., 2015; Nakajima et al., 2012; Nakajima and Yaoita, 2015a). The disparate mutations observed in this study between somatic tissues of different germ-layer origins and those found in the F1 progeny suggests care should be taken in extrapolating genotype results from a single F0 embryo. This is in agreement with results obtained with zinc-finger nucleases targeting the tyrosinase locus in X. tropicalis, where they found mutations in different regions of the skin of adult frogs that were not present in sperm (Nakajima et al., 2012). The presence of unique mutations in different organs illustrates the difficulty of deciphering the genotype of specific cells in the F0 generation. Notably, the fact that we found wild type alleles in some F0 organs, derived from tadpoles that were completely albino, suggests that genotyping early F0 embryos will not reveal all of the mutations present in that animal.
Our results are in agreement with two recent papers that also expressed TALENs prior to fertilization to demonstrate efficient F0 knockout of tyr and pax6 in X. laevis (Miyamoto et al., 2015; Nakajima and Yaoita, 2015a). The fact that we found unique mutations in each somatic organ that differed from those found in F1 embryos, suggests that mutations were induced after the 16-cell stage, consistent with recent results where the timecourse of induction of mutations was examined in OHT F0 embryos (Nakajima and Yaoita, 2015a). They found 50% mutagenesis of the tyr loci at the 8-cell stage, rising to 75% at the 32-cell stage, and only reaching 100% at stage 8.
The identification of unique mutations in three somatic tissues is consistent with Xenopus fate maps that show that heart and liver both arise from the D2.1 dorso-vegetal blastomere in the 16-cell embryo (Moody, 1987a). By the 32-cell stage the heart and liver lineages have started to separate, with heart mesoderm coming from dorsal C blastomeres, and liver coming from the dorsal D blastomeres (Dale and Slack, 1987; Moody, 1987b). In contrast, the skin lineage is already separated from heart and liver at the 8-cell stage; at this stage trunk epidermis (skin) arises from the V1 blastomere, whereas heart and liver derive from D1 and D2 (Moody and Kline, 1990). In contrast to the large number of mutations found in somatic tissues, we only found 3-5 mutations in F1 embryos, consistent with the fact that presumptive primordial germ cells arise from approximately four cells at the blastula stage (Houston et al., 1998; Ressom and Dixon, 1988). Such tissue-specific mutations suggests that care should be taken in extrapolating results based on the genotyping an entire F0 embryo, since it will not be clear if the mutations detected are in the specific tissue being studied.
Highlights.
Generation of F1 null mutants in X. laevis
Oocyte host-transfer method effective method to induce mutations in both homeologs
Unique mutations identified in each organ of F0 adult
Highly-penetrant germline mutations in X. laevis using oocyte host-transfer
Acknowledgements
We wish to thank Sean McNamara, Brian Suh, Lynn Ware and Alyssa Arseneau for their care of the frogs and help in the breeding and to Esther Pearl for her help throughout this project. This work was supported by grants from the NIH (P40OD010997 and R01HD084409). We also thank Dr. Doug Houston for showing us the oocyte host-transfer technique.
References
- Blitz IL, Biesinger J, Xie X, Cho KWY. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. doi:10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. doi:10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale L, Slack JM. Fate map for the 32-cell stage of Xenopus laevis. Development. 1987;99:527–551. doi: 10.1242/dev.99.4.527. [DOI] [PubMed] [Google Scholar]
- Gantress J, Maniero GD, Cohen N, Robert J. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology. 2003;311:254–262. doi: 10.1016/s0042-6822(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. doi:10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes Dev. 2014;28:1859–1872. doi: 10.1101/gad.248252.114. doi:10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Holwill S, Wylie CC. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods in cell biology. 1991;36:213–230. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- Hu R, Wallace J, Dahlem TJ, Grunwald DJ, O'Connell RM. Targeting human microRNA genes using engineered Tal-effector nucleases (TALENs). PLoS ONE. 2013;8:e63074. doi: 10.1371/journal.pone.0063074. doi:10.1371/journal.pone.0063074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, Cliffe R, Amaya E. Highly efficient bi-allelic mutation rates using TALENs in Xenopus tropicalis. Biol Open. 2012;1:1273–1276. doi: 10.1242/bio.20123228. doi:10.1242/bio.20123228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Burns KA, Fortriede JD, Lotay VS, Liu Y, Brad Karpinka J, Karimi K, Zorn AM, Vize PD. Xenbase: Core features, data acquisition, and data processing. Genesis. 2015;53:486–497. doi: 10.1002/dvg.22873. doi:10.1002/dvg.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. doi:10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. doi:10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Krylov V, Tlapakova T. Xenopus Cytogenetics and Chromosomal Evolution. Cytogenet Genome Res. 2015;145:192–200. doi: 10.1159/000406550. doi:10.1159/000406550. [DOI] [PubMed] [Google Scholar]
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CHK, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci USA. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. doi:10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Uno Y, Kondo M, Gilchrist MJ, Zorn AM, Rokhsar DS, Schmid M, Taira M. A New Nomenclature of Xenopus laevis Chromosomes Based on the Phylogenetic Relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res. 2015;145:187–191. doi: 10.1159/000381292. doi:10.1159/000381292. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Suzuki K-IT, Suzuki M, Sakane Y, Sakuma T, Herberg S, Simeone A, Simpson D, Jullien J, Yamamoto T, Gurdon JB. The Expression of TALEN before Fertilization Provides a Rapid Knock-Out Phenotype in Xenopus laevis Founder Embryos. PLoS ONE. 2015;10:e0142946. doi: 10.1371/journal.pone.0142946. doi:10.1371/journal.pone.0142946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SA. Fates of the Blastomeres of the 16-Cell Stage Xenopus Embryo. Dev Biol. 1987a;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Moody SA. Fates of the Blastomeres of the 32-Cell-Stage Xenopus Embryo. Dev Biol. 1987b;122:300–319. doi: 10.1016/0012-1606(87)90296-x. [DOI] [PubMed] [Google Scholar]
- Moody SA, Kline MJ. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. 1990;182:347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
- Nakade S, Sakuma T, Sakane Y, Hara Y, Kurabayashi A, Kashiwagi K, Kashiwagi A, Yamamoto T, Obara M. Homeolog-specific targeted mutagenesis in Xenopus laevis using TALENs. In Vitro Cell.Dev.Biol.-Animal. 2015;51:879–884. doi: 10.1007/s11626-015-9912-0. doi:10.1007/s11626-015-9912-0. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Nakajima T, Takase M, Yaoita Y. Generation of albino Xenopus tropicalis using zinc-finger nucleases. Dev Growth Differ. 2012;54:777–784. doi: 10.1111/dgd.12006. doi:10.1111/dgd.12006. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Highly efficient gene knockout by injection of TALEN mRNAs into oocytes and host transfer in Xenopus laevis. Biol Open. 2015a;4:180–185. doi: 10.1242/bio.201410009. doi:10.1242/bio.201410009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Development of a new approach for targeted gene editing in primordial germ cells using TALENs in Xenopus. Biol Open. 2015b;4:259–266. doi: 10.1242/bio.201410926. doi:10.1242/bio.201410926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. doi:10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DJ, Hulstrand AM, Houston DW. Methods in Molecular Biology, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2012. Maternal mRNA Knock-down Studies: Antisense Experiments Using the Host-Transfer Technique in Xenopus laevis and Xenopus tropicalis; pp. 167–182. doi:10.1007/978-1-61779-992-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl EJ, Grainger RM, Guille M, Horb ME. Development of Xenopus resource centers: the National Xenopus Resource and the European Xenopus Resource Center. Genesis. 2012;50:155–163. doi: 10.1002/dvg.22013. doi:10.1002/dvg.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Clark KJ, Campbell JM, Panetta MR, Guo Y, Ekker SC. Making designer mutants in model organisms. Development. 2014;141:4042–4054. doi: 10.1242/dev.102186. doi:10.1242/dev.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressom RE, Dixon KE. Relocation and Reorganization of Germ Plasm in Xenopus Embryos After Fertilization. Development. 1988;103:507–518. doi: 10.1242/dev.103.3.507. [DOI] [PubMed] [Google Scholar]
- Sakane Y, Sakuma T, Kashiwagi K, Kashiwagi A, Yamamoto T, Suzuki K-IT. Targeted mutagenesis of multiple and paralogous genes in Xenopus laevisusing two pairs of transcription activator-like effector nucleases. Dev Growth Differ. 2013;56:108–114. doi: 10.1111/dgd.12105. doi:10.1111/dgd.12105. [DOI] [PubMed] [Google Scholar]
- Suzuki K-IT, Isoyama Y, Kashiwagi K, Sakuma T, Ochiai H, Sakamoto N, Furuno N, Kashiwagi A, Yamamoto T. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol Open. 2013;2:448–452. doi: 10.1242/bio.20133855. doi:10.1242/bio.20133855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochinai S, Katagiri C. Complete abrogation of immune response to skin allografts and rabbit erythrocytes in the early thymectomized Xenopus. Dev Growth Differ. 1975;17:383–394. doi: 10.1111/j.1440-169X.1975.00383.x. [DOI] [PubMed] [Google Scholar]