Abstract
Purpose
Neuroblastoma is a childhood cancer of the sympathetic nervous system with embryonic origins. Previous epidemiologic studies suggest maternal vitamin supplementation during pregnancy reduces the risk of neuroblastoma. We hypothesized offspring and maternal genetic variants in folate-related and choline-related genes are associated with neuroblastoma and modify the effects of maternal intake of folate, choline and folic acid.
Methods
The Neuroblastoma Epidemiology in North America (NENA) study recruited 563 affected children and their parents through the Children’s Oncology Group’s Childhood Cancer Research Network. We used questionnaires to ascertain pre-pregnancy supplementation and estimate usual maternal dietary intake of folate, choline and folic acid. We genotyped 955 genetic variants related to folate or choline using DNA extracted from saliva samples and used a log-linear model to estimate both child and maternal risk ratios and stratum-specific risk ratios for gene-environment interactions.
Results
Overall, no maternal or offspring genotypic results met criteria for a false discovery rate (FDR) Q-value <0.2. Associations were also null for gene-environment interaction with pre-pregnancy vitamin supplementation, dietary folic acid and folate. FDR significant gene-choline interactions were found for offspring SNPs rs10489810 and rs9966612 located in MTHFD1L and TYMS, respectively, with maternal choline dietary intake dichotomized at the first quartile.
Conclusion
These results suggest that variants related to one-carbon metabolism are not strongly associated with neuroblastoma. Choline-related variants may play a role; however, the functional consequences of the interacting variants are unknown and require independent replication.
Keywords: Neuroblastoma, genetics, folate, choline, case-parent triad
Introduction
Neuroblastoma is an embryonal tumor of the neural crest portion of the sympathetic nervous system and usually presents in children less than 1 year of age [1, 2]. Each year approximately 770 children in North America are diagnosed with neuroblastoma, with incidence rates slightly higher in males (7.7 per million) than females (6.9 per million) [2–5]. Familial cases of neuroblastoma have been associated with specific mutations in the PHOX2B and ALK genes and among non-familial cases, recent genome-wide association (GWA) studies have identified several common variants of interest [6–8].
Due to the embryonic origins of neuroblastoma, pre-pregnancy and early pregnancy exposures are crucial for its development. Epidemiologic studies have found evidence of an inverse association between maternal prenatal vitamin use and risk of neuroblastoma [9, 10]. One study reported a 60% reduction in risk for daily vitamin use in the month before or during pregnancy. Although these studies did not indicate which vitamin(s) may underlie the association with neuroblastoma, folate and choline may be important. Folate is essential for one-carbon metabolism and is important in cell proliferation and differentiation of neural crest cells [11, 12]. Choline is also involved in one-carbon metabolism and an essential building block for membrane development [13].
Due to the key role of folate and choline in fetal and neuronal development and the suggestive epidemiological evidence, we hypothesized that genetically-based alterations in the levels of folate and choline during development, acting jointly with maternal nutrition, may impact the risk of neuroblastoma. This study is the first to examine the risk of neuroblastoma with maternal and offspring single nucleotide polymorphisms (SNPs) as well as gene-environment interactions with maternal folate and choline dietary intake and pre-pregnancy maternal vitamin supplementation.
Methods
Study sample
The Neuroblastoma Epidemiology in North America (NENA) study used a case-parent triad design. Cases were identified from the Childhood Cancer Research Network (CCRN) – a registry system of cases maintained by the Children’s Oncology Group (COG) [14]. NENA approached families registered in the CCRN registry who agreed to be contacted for future research. Eligible cases had a primary diagnosis of neuroblastoma, including ganglioneuroblastoma but excluding ganglioneuroma. Cases were diagnosed before 6 years of age at a U.S. or Canadian COG institution from December 24, 2007 to July 31, 2013. The biologic mother had to be alive and willing to participate. The University of North Carolina at Chapel Hill (UNC) Institutional Review Board approved this study.
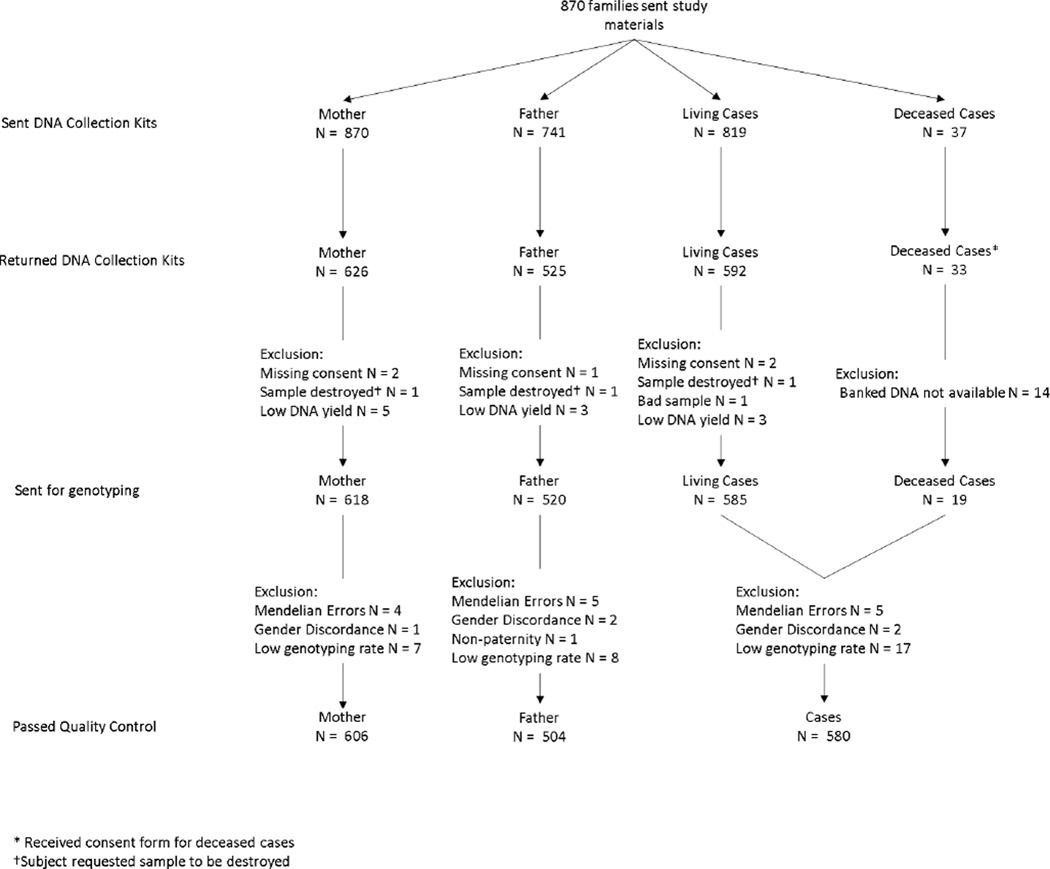
After the cases were identified through the CCRN, we sent a recruitment packet to 1347 families and 870 families agreed to enroll. Study materials sent included a consent form, questionnaire to be filled out by the mother, an Oragene saliva tube collection kit for the parents, and an Oragene saliva sponge/disc kit for the child. If the child was deceased, we delayed communication by 15 months after date of death and obtained a previously collected blood DNA sample from the COG Neuroblastoma Bio-repository at the Children’s Hospital of Philadelphia (CHOP).
Saliva samples were collected for 626 biological mothers, 592 living children, and 525 biological fathers. Blood samples were obtained for 19 deceased children (Supplemental Figure S1). Of the 630 maternal questionnaires received, two did not have a corresponding signed consent form and two were incomplete, resulting in 626 completed questionnaires for analysis (Figure 1).
Figure 1.
Flowchart for genetic and questionnaire quality control for triads and dyads.
Candidate genes and DNA collection
Genes were selected based on their role in the transport and metabolism of folate and choline as well as one-carbon metabolism. Since most of the mothers self-identified as white, TAGster with the greedy algorithm was used to capture haplotype tagging SNPs (minor allele frequency≥5%) that tag SNPs in high linkage disequilibrium (LD; r2≥0.8) for Hapmap 3 release III CEU population, located between 20kb upstream to 10kb downstream from the gene [15, 16]. The case-parent triad design is not subject to confounding by population stratification, thus ancestry-informative markers were not included [17]. A total of 693 SNPs in 38 folate-related and 302 SNPs in 19 choline-related genes were selected for genotyping.
DNA extraction and amplification was completed by the UNC Biospecimen Processing Facility. DNA was extracted using the Perkin-Elmer's Chemagic MSMI magnetic-bead extraction robotic system. Saliva samples from parents were collected in DNA Genotek’s OGR-500 collection kits. Saliva from the child was also collected by the parents using swabs. DNA quality was assessed with Nanodrop Optical Density and quantitated with Applied Biosystems® Taqman® RNase P detection kit. A total of 498 triads, 99 mother-child dyads, 5 father-child dyads and 27 other sets (mother-father dyads and singleton cases) with DNA yields greater than 2 µg were sent for genotyping.
Genotyping was performed by UNC’s Mammalian Genotyping Core Facility using the GoldenGate Assay with the Illumina BeadStation 500GX Genetic Analysis System. Allelic discrimination was based on allele-specific primer extension followed by ligation.
Genotyping Quality Control
For quality control purposes, a Centre de l'Étude du Polymorphisme Humain family triad and blinded duplicates were included on each plate. SNPs with a genotyping call rate less than 95% and showing a lack of defined clusters in the raw genetic intensity data were excluded (N=119). In total, 599 folate-related SNPs and 277 choline-related SNPs passed quality control. We assessed Hardy-Weinberg (HWE) equilibrium among parents who self-identified as white, using chi-square tests in PLINK (v1.07) and flagged (N=5), but did not exclude, SNPs that failed HWE at a false discovery rate (FDR) significance level of <0.2 [18, 19].
Individuals with genotyping rates <95% or gender discrepancies were excluded (N=36; Supplemental Figure S1). Relatedness was confirmed for each triad through measures of identity by descent. Triads and individuals with unexpected relatedness were excluded. For example, for non-paternity the paternal data was excluded (N=1). A total of 465 triads, 94 mother-child dyads, 4 father-child dyads and 61 others (13 mother-father dyads and 48 singletons) passed genetic quality control (Figure 1).
Biological and Clinical Variables
We obtained clinical and biologic characteristics of the tumor, such as tumor genetics and stage, from the COG Statistical and Data Center for all cases enrolled in a COG clinical protocol except 89 cases who were not enrolled. We used the COG prognostic risk schema that defined three prognostic risk-classifications: low-risk, intermediate-risk and high-risk. These risk-classifications are based on tumor characteristics, including stage and MYCN amplification, ploidy and patient age [20].
Maternal Vitamin Use
We ascertained the current and usual maternal diet during the preceding year using the Dietary History Questionnaire, a self-administered semi-quantified food frequency questionnaire (FFQ). We assumed maternal usual diet in the last year approximates pre-pregnancy diet. Completed FFQs were processed in Diet*Calc (version 1.5.0) to derive usual nutrient intake per day for the previous year. The nutrient and food group database was based on a compilation of national 24-hour dietary recall data from the National Health and Nutrition Examination Surveys (NHANES) conducted in 2001–2002, 2003–2004, and 2005–2006 (http://riskfactor.cancer.gov/dhq2/database). Certain foods not included in the original database were added by NENA staff in 100 gram amounts using the USDA database, standard release 24.
Mothers were also questioned about maternal dietary supplementation, including single vitamins and prenatal or multivitamin use 1 month pre-pregnancy and within each trimester of pregnancy. To aid in recall, an estimated conception date was provided; calculated by subtracting gestational age at delivery from infant birthdate. Since we are interested in pre-pregnancy and early pregnancy exposures, we focused on prenatal vitamin, including multivitamin, use 1 month pre-pregnancy.
Diet and Nutrient classification
We excluded questionnaires that reported calories per day below the 5th percentile (N=31; <854.47 calories) or above the 97th percentile (N=18; >4508.75 calories) (Figure 1). We focused on folate, folic acid and choline for gene-environment interaction. To take into account the different bioavailability of food folate and folic acid, dietary folate equivalent (DFE) was used to estimate total folate. To explore different dietary cutoffs, nutrients from the FFQ were dichotomized at the 25th percentile (<209.70 mg for choline; <389.83 µg DFE; and <100.69 µg folic acid) and dichotomized at the current daily recommendation for adult women. We used the recommended dietary allowance (RDA) for folate of 600 µg DFE for pregnant women [21]. Additionally, the Public Health Service Task Force recommends used 400 µg folic acid/day for prevention of birth defects for women trying to get pregnant [22]. We used the choline Adequate Intake of 425 mg/day for women [23].
We conducted an analysis combining prenatal vitamin use and folic acid and folate from diet. Maternal total exposure was dichotomized as low intake and sufficient intake. Women with intake in the lowest tertile of micronutrients from diet and with no prenatal supplementation 1 month pre-pregnancy were defined as “low intake”. Women were classified as “sufficient intake” otherwise. We only combined vitamin use with folic acid and folate from diet since choline is not commonly found in prenatal vitamins [24].
Statistical Analysis
There were three main analytic goals: 1) estimate the genotypic maternal and offspring risk ratios (RRs); 2) estimate stratum-specific RRs by neuroblastoma prognostic risk-classification and offspring age at diagnosis; and 3) assess multiplicative maternal and offspring gene-environment interactions with maternal choline, folate and folic acid intake. We used a log-linear model to simultaneously assess the offspring and maternal log-additive genetic main effects and gene-environment interaction [25].
Since there are no study controls in this analysis, the null background is discerned from the parental genotypes under the assumption of Mendelian transmission in the source population [26]. For assessing a maternally-mediated genetic association, the maternal genotype frequencies are compared to the paternal genotype frequencies under a further assumption of mating symmetry in the source population [17]. The maternal and offspring log-additive RRs were calculated simultaneously and thus are mutually adjusted. Missing parent genotypes can be accounted for with the expectation-maximization algorithm, which maximizes the observed-data likelihood by fractionally assigning incomplete triads into their data-compatible cells on the basis of the current parameter estimates, and then repeating the calculations iteratively up to convergence and maximization of the likelihood [27].
For the stratified analysis, the offspring and maternal genetic models were separately fit for each prognostic COG risk-classification and offspring age at diagnosis dichotomized at 1 year with separate mating types allowed within each stratum. This age dichotomy represents the two peaks in the neuroblastoma age at diagnosis distribution, and potentially corresponds to differences in etiology.
The gene-environment interaction model is an extension of the genetic only model with an additional term for the interaction of the offspring or maternal genotype and maternal vitamin intake [25]. This model allows genotypic RRs to differ across levels of vitamin intake. The main genotype effects were coded co-dominantly, while the interaction term is fit additively to enhance power. If interaction terms were significant after multiple testing correction, then the interaction model was refit co-dominantly to characterize the interaction in a less constrained way.
All p-values were corrected for the number of tests by false discovery rate (FDR) [28]. Results were considered significant if the FDR-corrected Q-value was less than 0.2. All estimated RRs are presented in relation to the minor allele at the specified SNP.
COG/CHOP Replication Study
We were able to provide replication of our findings for offspring genotypes using genotyping data from an ongoing GWA study. [29] Here, we conducted a GWA case-control study with 2101 neuroblastoma cases and 4202 healthy controls, with both sets of European-American ancestry. Information on this study has been described elsewhere [29]. Briefly, the cases were diagnosed with neuroblastoma and identified through the Neuroblastoma Bio-repository maintained by the COG, which collects germline and tumor specimens at the time of diagnosis. Controls with no known medical disorder were recruited from multiple sites within the CHOP Health Care Network. Cases enrolled in NENA were excluded from this cohort, resulting in 2,052 cases and 4,104 matched controls.
Imputation was performed on all case-control GWA data with IMPUTE2 using the world-wide 1000 Genomes Project Phase Release 3 data as reference [30]. SNPs with an info score less than 0.8 were excluded. The same SNPs used for the NENA case-parent analysis (N=783) were tested for case-control association with neuroblastoma using SNPTEST under an additive model [31]. About a third (n=264) of the NENA SNPs required imputation for the COG/CHOP replication study. Odds ratios (ORs) were FDR-corrected and compared with the RRs from NENA study.
Sensitivity Analysis
Since many women increase dairy consumption and decrease fish consumption during pregnancy, and both are rich in choline, we performed sensitivity analyses. The questionnaire asked if women during that pregnancy had changed their intake of foods prone to change, including dairy and fish [32]. The mothers were asked if during pregnancy intake had been “Much less than it is now”, “Somewhat less than it is now”, “Same as it is now”, “Somewhat more than it is now”, and “Much more than it is now”.
Choline levels were manually changed for women who reported having increased dairy consumption and decreased fish consumption during pregnancy. We calculated the average amount fish and dairy contributes to choline in NENA, 8.55 mg. Choline levels for mothers who reported their fish consumption during the pregnancy had been “much less than it is now” were decreased by 8.55 mg, resetting negative values to 0. For mothers who reported their dairy consumption during the pregnancy had been “much more than it is now”, their choline levels were increased by 73.87 mg, the average amount of choline. After this choline intake adjustment, choline was then dichotomized at the 25th percentile and the gene-environment model was fit again.
Since women who breastfeed are advised to consume more calories, which alters current nutrient intake, additional sensitivity analyses were done excluding breastfeeding women.
Results
Descriptive Statistics
Table 1 describes the demographics of our analytic sample of families (465 triads and 98 dyads). The mean age at diagnosis for the offspring was 1.7 years. As expected, the age at diagnosis differed across COG risk-classifications (p-value<0.001); the high-risk classification had the oldest mean age at diagnosis (2.6 years). This study included more male cases (53.6%) than female cases. This male excess was similar across COG risk-classification groups except for the low-risk classification (52.4% females). The predominant maternal race was white (84.8%). Almost 60% of mothers (N=349) reported using prenatal vitamin supplementation 1 month before conception (Table 2).
Table 1.
Descriptive statistics for triads with genetic data
| Total | Low-risk | Intermediate-risk | High-risk | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean(Std) | n | Mean(Std) | n | Mean(Std) | n | Mean(Std) | p-value* | ||
| Maternal Age (Yrs) | 606 | 29.7 (5.30) | 186 | 29.4 (5.14) | 146 | 29.5 (5.38) | 204 | 30.0 (5.34) | 0.591 | |
| Age at diagnosis (Yrs) | 618 | 1.7 (1.43) | 181 | 1.4 (1.40) | 149 | 0.9 (0.87) | 204 | 2.6 (1.20) | <0.001 | |
| N | % | N | % | N | % | N | % | |||
| Offspring gender | ||||||||||
| Female | 285 | 45.7 | 94 | 51.9 | 71 | 47.7 | 88 | 43.6 | 0.078 | |
| Male | 339 | 54.3 | 87 | 48.1 | 78 | 52.3 | 114 | 56.4 | ||
| Maternal race | ||||||||||
| White | 513 | 84.7 | 140 | 79.6 | 120 | 82.2 | 174 | 87.9 | 0.042 | |
| Black | 24 | 4.0 | 12 | 6.8 | 3 | 2.1 | 7 | 3.5 | ||
| Hispanic | 36 | 5.9 | 16 | 9.1 | 11 | 7.5 | 7 | 3.5 | ||
| Other | 33 | 5.5 | 8 | 4.6 | 12 | 8.2 | 10 | 5.1 | ||
| Missing | 18 | -- | 5 | -- | 3 | -- | 4 | -- | ||
Std: Standard Deviation; Yrs: Years
p-value is comparing the distribution across risk-classifications
Table 2.
Descriptive statistics of maternal usual dietary nutrient levels and supplemental pre-pregnancy vitamin consumption
| N | % | |
| Vitamin use 1 month pre-pregnancy | ||
| Yes | 349 | 59.4 |
| No | 239 | 40.6 |
| Missing | 36 | -- |
| N | Median (IQR) | |
| Choline (mg) | 559 | 279.78 (208.28–372.39) |
| Folate (µg Dietary Folate Equivalent) | 559 | 511.29 (389.71–698.35) |
| Folic Acid (µg) | 559 | 162.11 (100.69–233.79) |
IQR: Interquartile Range
Folate
We found no significant associations between folate-related maternal and offspring SNPs and neuroblastoma overall, or when stratified by COG risk-classification or offspring age at diagnosis (Supplemental table S1 and S2).
We observed no significant gene-environment interaction in relation to maternal or offspring genotypes for maternal vitamin supplementation 1 month pre-pregnancy or for maternal dietary folic acid or total folate intake. Results from the total exposure analysis combining prenatal vitamins and diet were also non-significant.
Choline
We found no significant associations for maternal or offspring choline SNPs, either overall or stratified by risk-classification or offspring age at diagnosis.
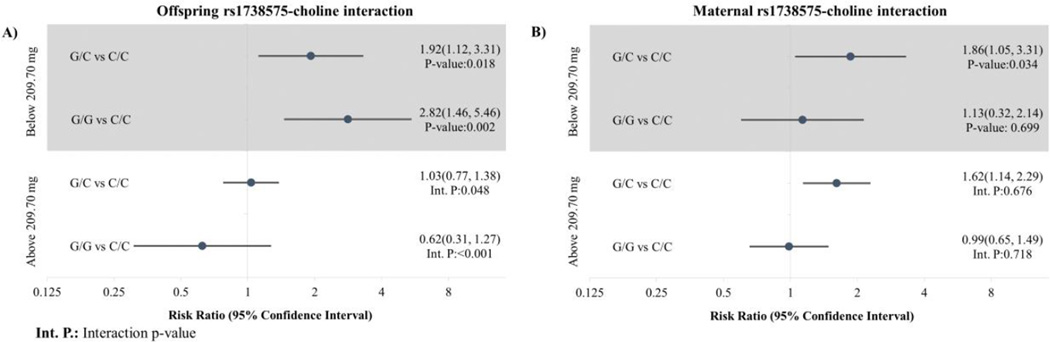
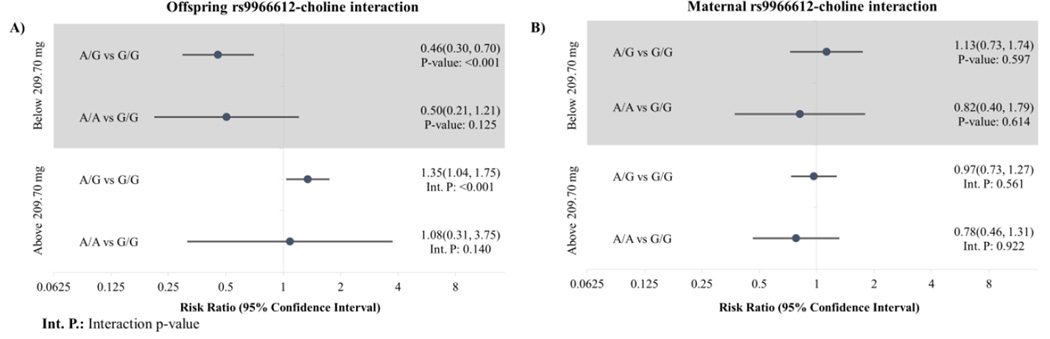
For the gene-choline interaction, we observed two significant log-additive interaction p-values for the 25th percentile in maternal choline consumption with the offspring SNP rs1738575 in MTHFD1L (interaction p-value<0.001; Q-value=0.076), and with the offspring SNP rs9966612 near TYMS (interaction p-value<0.001; Q-value=0.140). We refit the interaction model co-dominantly to provide allele-count-specific point estimates, resulting in wider confidence intervals due to the rarity of homozygotes. For mothers below the 25th percentile of choline consumption (Figure 2), offspring with the G allele of rs1738575 (MTHFD1L) were at increased risk of neuroblastoma (RR for G/C vs. C/C: 1.92, 95% CI: 1.12–3.31; RR for G/G vs. C/C: 2.82, 95% CI: 1.46–5.46), compared to a null association when maternal choline was above 25th percentile (RR for G/C vs. C/C: 1.03, 95% CI: 0.77–1.38; RR for G/G vs. C/C: 0.62, 95% CI: 0.31–1.27). We found offspring with the A allele in the SNP rs9966612 (TYMS) had lower risk of neuroblastoma (Figure 3) when maternal choline consumption was below the 25th percentile (RR for A/G versus G/G: 0.46, 95% CI: 0.30–0.70; RR for A/A versus G/G: 0.5, 95% CI: 0.21–1.21), with both lower than the relative risks among triads with maternal choline greater than the 25th percentile (RR for A/G versus G/G: 1.35, 95% CI: 1.04–1.75; RR for A/A versus G/G: 1.08, 95% CI: 0.31–3.75).
Figure 2.
A) Offspring and B) maternal MTHFD1L (rs1738575)-choline interaction modeled co-dominantly with maternal choline consumption dichotomized at the 25th percentile
Figure 3.
A) Offspring and B) maternal TYMS (rs9966612)-choline interaction modeled co-dominantly with maternal choline consumption dichotomized at the 25th percentile
When choline was dichotomized at the Adequate Intake level (425 mg), the log-additive interaction was significant for the offspring SNP rs10489810 located in SLC44A3 (interaction p-value<0.001; Q-value=0.173). Among mothers with below Adequate Intake of choline consumption, we found offspring with one T allele had little evidence for association (RR T/A vs. A/A: 0.91, 95% CI: 0.71–1.17) while those with 2 T alleles had an inverse association (RR T/T vs. A/A: 0.43, 95% CI: 0.26–0.70). Among mothers with above Adequate Intake level of choline, offspring with 1 T allele and those with 2 T alleles had an increased risk (RR T/A vs. A/A: 2.00, 95% CI: 1.11–3.60; RR T/T vs. A/A: 2.85, 95% CI: 0.98–8.30) of neuroblastoma.
Replication Study
Maternal genotyping and questionnaire data were not available from the replication study, thus only offspring genetic results were compared. No offspring variants in NENA were significant. A few COG/CHOP replication study results were significant (Supplemental table S2). However, the results from NENA for these SNPs were not also significant and the RRs were not directionally consistent between studies.
Sensitivity Analysis
Among women who were below the 25th percentile for choline, 10 mothers increased dairy consumption and 2 increased fish consumption during pregnancy. Among women with greater than the 25th percentile for choline consumption, only 8 decreased dairy consumption but 83 decreased fish consumption during pregnancy. After changing the choline intake for these families, both alleles for rs10489810 and rs9966612 remained significant (Supplemental table S3) and the point estimates changed little. We also found a significant interaction with offspring alleles in rs9478157 and rs1052751, neither of which had previously been significant.
We found no new significant results when women who were breastfeeding were excluded (N=46). The previously identified gene-choline interactions for offspring SNPs rs10489810 and rs9966612 remained nominally significant and were directionally unchanged.
Discussion
These analyses were motivated by prior epidemiologic evidence suggesting inadequate maternal consumption of folate, folic acid, and choline during pregnancy increases the risk of the unborn child subsequently developing neuroblastoma. Although SNPs within the one-carbon metabolism pathway have been previously associated with birth defects and childhood cancers, our study suggests SNPs from either choline or folate-related genes were not associated with neuroblastoma overall, within COG risk-classification categories, or by categories defined by age at diagnosis [33–36]. While significant SNPs were found in the COG/CHOP case-control replication study, those SNPs were not significant and were not directionally consistent with NENA results. The gene-environment interaction results suggest gene variants in choline pathways be modified by choline intake; however, since the identified SNPs lie within non-coding regions, the exact implications of these associations are unclear.
We found no offspring or maternal associations for the SNPs that were selected because they had previously been associated with cancer or birth defects. MTHFR 667C>T (rs1801133), one of the most highly studied variants with known functional effects on one-carbon metabolism, [37–39] had a null offspring association (RR: 0.99, 95% CI: 0.84–1.19) and a weak maternal association (RR: 1.16, 95% CI: 0.97–1.38). Two previous studies of candidate SNPs from folate-related genes identified SLC19A1 80G>A (rs1051266) as positively associated with neuroblastoma in Brazil [40, 41]. Montalvão-de-Azevedo et al. found maternal carriers of the G had 3 times the risk of offspring with neuroblastoma and offspring carriers had approximately 2.5 times the risk, which was replicated by de Miranda et al. [40, 41]. In NENA, we found no association (Maternal RR: 1.12, 95% CI: 0.96–1.32; offspring log-additive RR: 0.94, 95% CI: 0.79–1.11). The inconsistent findings may be due to chance, differences in ancestry and the associated linkage with a causative SNP, confounding by maternal genotype, or possibly different dietary and vitamin supplementation intake patterns in Brazil.
We found significant gene-choline interaction for two offspring SNPs, rs1738575 and rs9966612, respectively located in an intron of MTHFD1L and upstream from TYMS. MTHFD1L is involved in tetrahydrofolate conversion in the mitochondria during one-carbon metabolism [11]. SNP rs9966612 is about 8 kbp upstream from TYMS but within the intron of CLUL1 and 500 bp downstream from TYMSOS. However, there is no compelling evidence either TYMSOS or CLUL1 is related to neuroblastoma development [42]. Since we used haplotype tagging, these SNPs could be in LD with the casual SNP. To further explore correlated SNPs, we used SNP Annotation and Proxy Search developed by the Broad Institute to find SNPs in high LD (r2>0.8) based on the 1000 Genome CEU population [43]. SNPs in high LD with rs1738575 and rs9966612 have not previously been associated with disease. Choline is involved with one-carbon metabolism and both TYMS and MTHFD1L encode proteins essential to one-carbon metabolism [44]. The functionality of these variants is known.
When choline was dichotomized at the Adequate Intake level, we found one additional interacting offspring SNP, which appeared to increase the risk of neuroblastoma among those above the Adequate Intake level but decrease risk among offspring below. The offspring SNP rs10489810 is located within an intron of SLC44A3, a choline transporter. However, SNPs in SLC44A3 and those in high LD with rs10489810 have an unknown functional impact and have not previously been associated with any disease.
The present study has some limitations. Neuroblastoma is too rare to be studied prospectively and our assessment of pre-pregnancy maternal diet was necessarily retrospective. Studies have demonstrated that maternal preconception nutritional status is critical for early fetal development but the critical etiologic window specific to neuroblastoma is nonetheless unknown [45]. The mothers in NENA completed the FFQ shortly after enrollment (2 months to 9 years after the offspring birth date). This assessment of diet more likely mirrors pre-pregnancy diet rather than early pregnancy, when mothers may have changed diet due to morning sickness [46]. Moreover, the FFQ occurred during a time when their child was suffering with a critical illness or may have died, leading to the potential for substantial disruption of their routine eating patterns. However, nutrient levels of folic acid, folate and choline from diet did not significantly differ by risk-classification or vital status, suggesting that nutrient levels do not differ by severity of disease. Furthermore, our sensitivity analysis revealed the FDR-significant SNPs for gene-choline interaction were stable to differences in the estimation of choline levels related to reported changes in fish and dairy consumption during pregnancy. The sample studied in NENA were mostly white and highly educated (over 50% graduated college), and thus had greater rates of vitamin consumption and nutrient intake compared to the general population in the United States [47]. Although the nature of our study sample does not affect the validity of the study, it could reduce generalizability and introduces the possibility we are not capturing the “high-risk” population that could benefit the most from intervention.
The study has multiple strengths. This is the largest study conducted with both genetic and maternal questionnaire data to allow for the study of gene-environment interaction for candidate genes with neuroblastoma. The case-parent triad approach eliminates the need for a control group, a logistical and validity challenge for a North America-wide study. Additionally, the case-parent triad design is robust against bias due to population stratification and self-selection based on ethnicity. The case-parent triad approach also allows for the estimation of maternal RRs, which is especially important for diseases that can originate in utero. Mating symmetry (i.e. a genotype is over represented in either the mother or father not due to the disease state of the offspring) could be violated when assessing maternal associations. However, the null findings for maternal effects argue for symmetry for the studied SNPs. We also employed the use of an independent replication study that provided additional evidence for the robustness of our null results for offspring SNPs and neuroblastoma. Although a few of the CHOP/COG replication results were significant, they were not directionally consistent with NENA results. This is most likely to chance, but could also be due to differences LD patterns [48].
This study suggests that maternal and offspring SNPs in folate and choline-related genes are not strongly associated with neuroblastoma. Further, gene-environment interactions were not identified for maternal vitamin supplementation or total folate or folic acid intake from diet, suggesting there is no appreciable modification of effects of SNPs near folate genes by maternal diet or vitamin supplementation. Our suggestive diet-by-SNP interactions for SNPs related to the choline pathway warrant further study.
Supplementary Material
Acknowledgments
This research was supported in part by two grants from the National Cancer Institute (R01 CA 132887 and R01 CA124709) and the Lineberger Cancer Control Education Program (R25 CA57726)
Footnotes
There are no conflicts of interest to disclose.
References
- 1.Maris JM. Recent advances in neuroblastoma. N. Engl. J. Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hallett A, Traunecker H. A review and update on neuroblastoma. Paediatrics and Child Health. 2012;22:103–107. [Google Scholar]
- 3.Spix C, Pastore G, Sankila R, Stiller CA, Steliarova-Foucher E. Neuroblastoma incidence and survival in European children (1978–1997): report from the Automated Childhood Cancer Information System project. Eur. J. Cancer. 2006;42:2081–2091. doi: 10.1016/j.ejca.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Canadian Cancer Society’s Steering Committee on Cancer Statistics. Canadian Cancer Statistics 2011. 2011 [Google Scholar]
- 5.Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992–2004) Cancer. 2008;112:416–432. doi: 10.1002/cncr.23169. [DOI] [PubMed] [Google Scholar]
- 6.Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2010;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat. Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olshan AF, Smith JC, Bondy ML, Neglia JP, Pollock BH. Maternal vitamin use and reduced risk of neuroblastoma. Epidemiology. 2002;13:575–580. doi: 10.1097/00001648-200209000-00014. [DOI] [PubMed] [Google Scholar]
- 10.Michalek AM, Buck GM, Nasca PC, Freedman AN, Baptiste MS, Mahoney MC. Gravid health status, medication use, and risk of neuroblastoma. Am. J. Epidemiol. 1996;143:996–1001. doi: 10.1093/oxfordjournals.aje.a008682. [DOI] [PubMed] [Google Scholar]
- 11.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boot MJ, Steegers-Theunissen RP, Poelmann RE, Van Iperen L, Lindemans J, Gittenberger-de Groot AC. Folic acid and homocysteine affect neural crest and neuroepithelial cell outgrowth and differentiation in vitro. Dev. Dyn. 2003;227:301–308. doi: 10.1002/dvdy.10303. [DOI] [PubMed] [Google Scholar]
- 13.Zeisel SH, da Costa KA. Choline: an essential nutrient for public health. Nutr. Rev. 2009;67:615–623. doi: 10.1111/j.1753-4887.2009.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musselman JR, Spector LG, Krailo MD, et al. The Children's Oncology Group Childhood Cancer Research Network (CCRN): case catchment in the United States. Cancer. 2014;120:3007–3015. doi: 10.1002/cncr.28813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, Kaplan NL, Taylor JA. TAGster: efficient selection of LD tag SNPs in single or multiple populations. Bioinformatics. 2007;23:3254–3255. doi: 10.1093/bioinformatics/btm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilcox AJ, Weinberg CR, Lie RT. Distinguishing the effects of maternal and offspring genes through studies of "case-parent triads". Am. J. Epidemiol. 1998;148:893–901. doi: 10.1093/oxfordjournals.aje.a009715. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S. PLINK (v1.07) [Google Scholar]
- 20.Pizzo PA, Poplack DG. Principles and practice of pediatric oncology. Lippincott Williams & Wilkins; 2015. [Google Scholar]
- 21.Simpson JL, Bailey LB, Pietrzik K, Shane B, Holzgreve W. Micronutrients and women of reproductive potential: required dietary intake and consequences of dietary deficiency or excess. Part I--Folate, Vitamin B12, Vitamin B6. J. Matern. Fetal Neonatal Med. 2010;23:1323–1343. doi: 10.3109/14767051003678234. [DOI] [PubMed] [Google Scholar]
- 22.U. S. Preventive Services Task Force. Folic acid for the prevention of neural tube defects: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2009;150:626–631. doi: 10.7326/0003-4819-150-9-200905050-00009. [DOI] [PubMed] [Google Scholar]
- 23.Zeisel SH, Caudill MA. Choline. Advances in nutrition (Bethesda, Md.) 2010;1:46–48. doi: 10.1093/advances/nmx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masih SP, Plumptre L, Ly A, et al. Pregnant Canadian Women Achieve Recommended Intakes of One-Carbon Nutrients through Prenatal Supplementation but the Supplement Composition, Including Choline, Requires Reconsideration. J. Nutr. 2015;145:1824–1834. doi: 10.3945/jn.115.211300. [DOI] [PubMed] [Google Scholar]
- 25.Umbach DM, Weinberg CR. The use of case-parent triads to study joint effects of genotype and exposure. Am. J. Hum. Genet. 2000;66:251–261. doi: 10.1086/302707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am. J. Hum. Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinberg C. Allowing for missing parents in genetic studies of case-parent triads. Am. J. Hum. Genet. 1999;64:1186–1193. doi: 10.1086/302337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 29.Diskin SJ, Capasso M, Schnepp RW, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 31.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Sotres-Alvarez D, Herring AH, Siega-Riz AM. Latent transition models to study women's changing of dietary patterns from pregnancy to 1 year postpartum. Am. J. Epidemiol. 2013;177:852–861. doi: 10.1093/aje/kws303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parle-McDermott A, Pangilinan F, O'Brien KK, et al. A common variant in MTHFD1L is associated with neural tube defects and mRNA splicing efficiency. Hum. Mutat. 2009;30:1650–1656. doi: 10.1002/humu.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deak KL, Dickerson ME, Linney E, et al. Analysis of ALDH1A2, CYP26A1, CYP26B1, CRABP1, and CRABP2 in human neural tube defects suggests a possible association with alleles in ALDH1A2. Birth defects research. Part A, Clinical and molecular teratology. 2005;73:868–875. doi: 10.1002/bdra.20183. [DOI] [PubMed] [Google Scholar]
- 35.Etheredge AJ, Finnell RH, Carmichael SL, et al. Maternal and infant gene-folate interactions and the risk of neural tube defects. Am J Med Genet A. 2012;158A:2439–2446. doi: 10.1002/ajmg.a.35552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Wang J, Zhao L, Liu X, Mi W. Methylenetetrahydrofolate reductase polymorphisms and risk of acute lymphoblastic leukemia-evidence from an updated meta-analysis including 35 studies. BMC medical genetics. 2012;13:77. doi: 10.1186/1471-2350-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo YL, Cheng YL, Ye P, Wang W, Gao XH, Chen Q. Association between MTHFR polymorphisms and orofacial clefts risk: a meta-analysis. Birth defects research. Part A, Clinical and molecular teratology. 2012;94:237–244. doi: 10.1002/bdra.23005. [DOI] [PubMed] [Google Scholar]
- 38.Zhao M, Li X, Xing C, Zhou B. Association of methylenetetrahydrofolate reductase C677T and A1298C polymorphisms with colorectal cancer risk: A meta-analysis. Biomedical reports. 2013;1:781–791. doi: 10.3892/br.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J, Yin M, Dreyer ZE, et al. A meta-analysis of MTHFR C677T and A1298C polymorphisms and risk of acute lymphoblastic leukemia in children. Pediatric blood & cancer. 2012;58:513–518. doi: 10.1002/pbc.23137. [DOI] [PubMed] [Google Scholar]
- 40.de Miranda DO, Barros JE, Vieira MM, et al. Reduced folate carrier-1 G80a gene polymorphism is associated with neuroblastoma's development. Mol. Biol. Rep. 2014;41:5069–5075. doi: 10.1007/s11033-014-3372-6. [DOI] [PubMed] [Google Scholar]
- 41.Montalvao-de-Azevedo R, Vasconcelos GM, Vargas FR, et al. RFC-1 80G>A polymorphism in case-mother/control-mother dyads is associated with risk of nephroblastoma and neuroblastoma. Genet. Test. Mol. Biomarkers. 2015;19:75–81. doi: 10.1089/gtmb.2014.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturgill GM, Pauer GJ, Bala E, et al. Mutation screen of the cone-specific gene, CLUL1, in 376 patients with age-related macular degeneration. Ophthalmic Genet. 2006;27:151–155. doi: 10.1080/13816810600976871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu. Rev. Nutr. 2001;21:255–282. doi: 10.1146/annurev.nutr.21.1.255. [DOI] [PubMed] [Google Scholar]
- 45.King JC. Physiology of pregnancy and nutrient metabolism. Am. J. Clin. Nutr. 2000;71:1218S–1225S. doi: 10.1093/ajcn/71.5.1218s. [DOI] [PubMed] [Google Scholar]
- 46.Anderson AS. Symposium on 'nutritional adaptation to pregnancy and lactation'. Pregnancy as a time for dietary change? Proc. Nutr. Soc. 2001;60:497–504. doi: 10.1079/pns2001113. [DOI] [PubMed] [Google Scholar]
- 47.Yang Q, Cogswell ME, Hamner HC, et al. Folic acid source, usual intake, and folate and vitamin B-12 status in US adults: National Health and Nutrition Examination Survey (NHANES) 2003–2006. Am. J. Clin. Nutr. 2010;91:64–72. doi: 10.3945/ajcn.2009.28401. [DOI] [PubMed] [Google Scholar]
- 48.Kraft P, Zeggini E, Ioannidis JPA. Replication in genome-wide association studies. Statistical science : a review journal of the Institute of Mathematical Statistics. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.