Abstract
Insect olfactory receptors (ORs) in the peripheral olfactory system play an important role detecting elements of information from the environment. At present, various approaches are used for deorphanizing of ORs in insect. In this study, we compared methods for functional analysis of ORs in vitro and in vivo taking the candidate pheromone receptor OR13 of Helicoverpa assulta (HassOR13) as the object of our experiments. We found that the natural system was more sensitive than those utilizing transgenic Drosophila. The two-electrode voltage-clamp recording is more suitable for functional screening of large numbers of ORs, while the in vivo transgenic Drosophila system could prove more accurate to further validate the function of a specific OR. We also found that, among the different solvents used to dissolve pheromones and odorants, hexane offered good reproducibility and high sensitivity. Finally, the function of ORs was indirectly confirmed in transgenic Drosophila, showing that odor-activation of ORs-expressing olfactory receptor neurons (ORNs) can mediate behavioral choices. In summary, our results compare advantages and drawbacks of different approaches, thus helping in the choice of the method most suitable, in each specific situation, for deorphanizing insect ORs.
The sense of smell in insects is of critical importance for every aspect of their life. Perception of odors and pheromones starts with detection of volatile molecules at the periphery of the sensory system, involving olfactory receptors (ORs) expressed on the membrane of olfactory receptor neurons (ORNs)1,2,3. Activation of ORs by odorants triggers generation of ORN action potentials, that converge to glomeruli of antennal lobes and eventually are integrated with other sensory inputs in the central nervous system (CNS)4,5. Since the first insect ORs from Drosophila melanogaster were identified using a bioinformatics-based approach6, a large amount of data on ORs has been accumulating from various insect taxa, thanks to recent simple and inexpensive methods of transcriptome sequencing7,8,9,10,11,12,13,14,15,16. Identification of the OR repertoire represents the first step towards understanding how the insect integrates and processes the huge diversity of chemical messages in the environment, originating from food, enemies and mates17. Consequentially, several methods for the functional characterization of insect ORs have been developed in which large numbers of biologically relevant odorants can be tested18,19,20,21,22,23,24,25,26,27.
Previous reports indicated that ORs cannot be properly folded when expressed in bacteria. Instead, they can be functionally characterized in eukaryotes, either in vitro or in vivo, along with well-established experimental strategies19,28,29,30,31,32,33,34,35,36,37,38. In vitro, heterologous expression systems, including Xenopus oocytes, have been adopted to probe the function of insect ORs19,28,32,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54. The first member of Drosophila was deorphanized using the Xenopus oocytes and two-electrode voltage-clamp system, this still being the most common technique adopted for heterologous expression28. Generally, Xenopus oocytes are injected with cRNAs encoding a specific OR and the odorant receptor co-receptor (Orco). The presence of Orco significantly increases the sensitivity and the specificity of individual ORs19.
Heterologous expression of odorant receptors can be also conducted in vivo using transgenic Drosophila techniques, which include two main paradigms, the “empty neuron” system29 and the Or67dGAL4 knock-in system33. The Drosophila “empty neuron” system, originally constructed for the deorphanization of Drosophila ORs in 2003, is a combination of a GAL4 driver line under the Or22a promoter in the Δ halo background and a fly line with UAS–‘OR gene’. In that way, the “Favorite” OR gene is inserted next to UAS-promoter to be expressed in the “empty/mutant” ab3A (basiconic sensilla) neuron. Another Or67dGAL4 knock-in system generates mutant alleles in which the open reading frame of Or67d is replaced with GAL4 and introduces independent UAS –‘OR gene’ transgene insertions into the Or67dGAL4 line, which allows for expression of the OR in the unique ORN of the antennal trichoid sensilla (at1). With both systems, the technique of single sensillum recording (SSR) is used to monitor the electrophysiological responses of OSNs expressing the exogenous candidate OR. The recent literature shows that in vivo Drosophila expression systems are used by several research groups for functional identification of odorant receptors from other insect species, because of close similarities with their natural cellular environment17,33,55,56,57,58,59,60,61,62,63.
A large number of insect ORs have been functionally investigated using the above mentioned methods17,18,19,23,24,25,26,27,28,29,30,31,55,56,58,59,64,65,66,67,68,69,70,71,72. In this study we investigate the strengths of some currently used method used to deorphanize insect ORs. Specifically, we have performed functional analysis of HassOR13 using both heterologous Xenopus expression and in vivo Or67dGAL4 knock-in Drosophila transgenic systems. Further, we compared the results obtained via approaches with the performance of ORNs expressing HassOR13 in the moth. Previous reports have shown that odorant receptors are responsible for the specificity of ORNs, thus relating the performance of an odorant receptor to electrophysiological recordings. We have also verified the function of ORs at the behavioral level. Finally, we have compared the performance of different solvents for in vivo electrophysiological recording. Our results highlight advantages and the drawbacks of the two main approaches for OR functional characterization and provide information guidelines to select a suitable method to deorphanize insect ORs.
Results
Comparison between in vitro and in vivo protocols
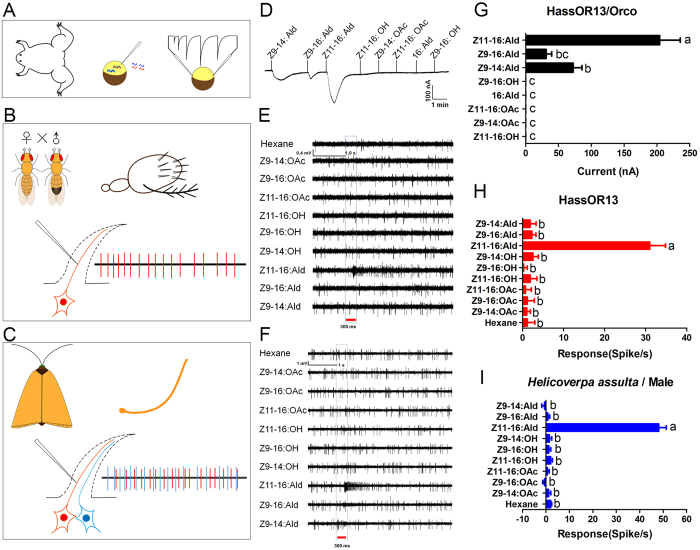
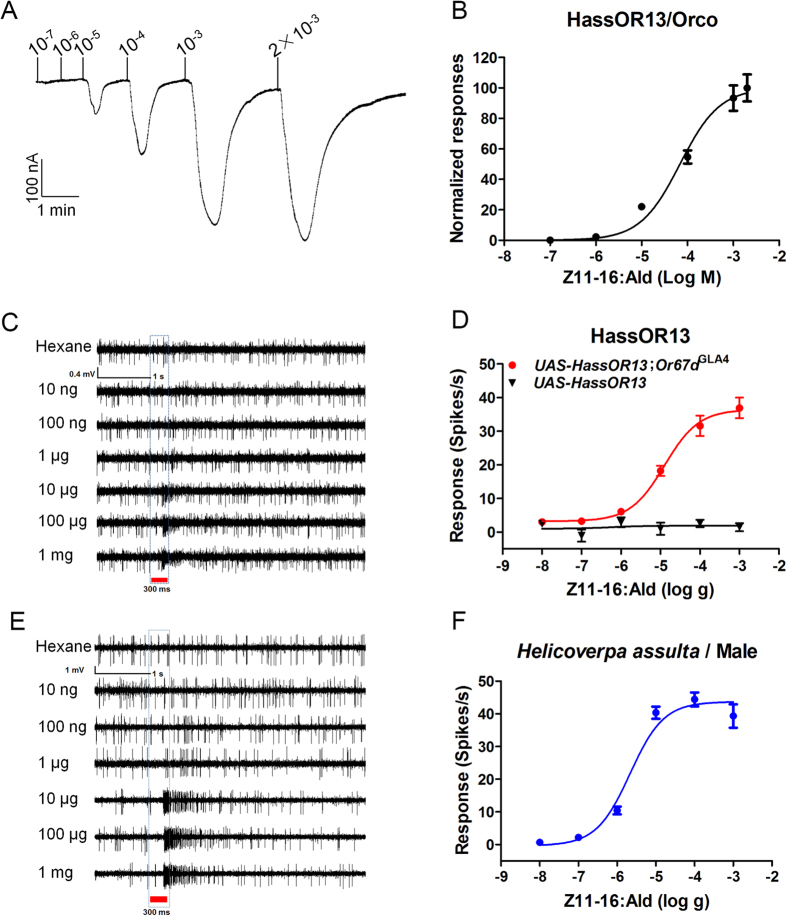
The receptor HassOR13 is tuned to the second sex pheromone component, (Z)-11-hexadecenal (Z11-16:Ald) in H. assulta66. Here, we used HassOR13 as a model to compare different methods of functional analysis. The first approach utilizes the in vitro heterologous expression systems in Xenopus oocytes (Fig. 1A). When co-expressed with Orco of H. assulta (HassOrco), HassOR13 robustly responded to Z11-16:Ald at a concentration of 10−4 M, but only weakly to the major pheromone component (Z)-9-hexadecenal (Z9-16:Ald) and to the non-specific pheromone (Z)-9-tetradecenal (Z9-14:Ald)73 (Fig. 1D). The signal evoked by Z11-16:Ald (204 ± 32 nA) was significantly larger than those produced by Z9-16:Ald and Z9-14:Ald, (72 ± 13 nA and 31 ± 8 nA, respectively, P < 0.01) (Fig. 1G). Dose–response experiments showed that the heterodimer HassOR13/HassOrco was sensitive to Z11-16:Ald with an EC50 value of 6.84 × 10−5 M (Fig. 2A,B and Table 1).
Figure 1. Functional comparison of HassOR13 responses between in vitro and in vivo methods.
(A) Schematic diagram of Xenopus oocyte system. The morphological schematic of Xenopus is shown on the left and cRNA microinjection is shown in the middle. Responses of ORs were recorded using two-electrode voltage-clamp technique, as shown on the right. (B) Schematic diagram of Or67dGAL4 knock-in system. The morphological schematic of fly (male and female) is shown at the top on the left, fly antennae covered by the sensilla, mostly trichoid sensilla (at1) are shown at the top on the right, and responses of ORN from single sensilla (SSR) are shown at the bottom of the figure. (C) Schematic diagram of the endogenous system in moths. The morphological schematic of a moth is shown at the top left and its antennae at the top right. Responses of ORNs (SSR) are shown at the bottom of the figure. (D) Inward currents from HassOR13/HassOrco Xenopus oocytes in response to 10−4 M solutions of pheromone compounds. (E) SSR traces from HassOR13-expressing neurons in at1 sensilla of Drosophila in response to pheromone compounds. (F) SSR traces of HassOR13-expressing neurons in type A sensilla of H. assulta in response to pheromone compounds. (G) Response profile of HassOR13/HassOrco Xenopus oocytes. The amplitude evoked by Z11-16:Ald was significantly larger than others (P < 0.01, one-way ANOVA followed Duncan’s multiple range test, 204 ± 32 nA, n = 6). (H) Average SSR responses of HassOR13-expressing neurons in at1 sensilla of Drosophila. The response was exclusively elicited by Z11-16:Ald (P < 0.01, one-way ANOVA followed Duncan’s multiple range test, 31 ± 4 spikes ⁄ s, n = 7). (I) Average SSR responses of HassOR13-expressing neurons in type A sensilla of H. assulta. The response was exclusively elicited by Z11-16:Ald (P < 0.01, one-way ANOVA followed Duncan’s multiple range test, 48 ± 3 spikes ⁄ s, n = 9). Bars labelled with different letters are significantly different. Error bars indicate SEM.
Figure 2. Dose-response relationships for in vitro and in vivo system.
(A) HassOR13/HassOrco expressed in Xenopus oocytes and stimulated with a concentration range of Z11-16:Ald. (B) Dose–response curve of HassOR13/HassOrco stimulated with Z11-16:Ald across a series of concentrations from 10−7 M to 2 × 10−3 M (n = 7). Data are reported as percent of maximal responses. Error bars indicate SEM. (C) SSR traces showing Z11-16:Ald-evoked activity of at1 neurons from UAS- HassOR13; Or67dGAL4 homozygous line. No response was evoked by hexane. (D) Z11-16:Ald-induced dose–response curves from at1 neurons expressing (UAS- HassOR13; Or67dGAL4, n = 13) and non-expressing (UAS-HassOR13, n = 11) HassOR13, stimulated with 10−3 g to 10−8 g. Error bars indicate SEM. (E) SSR traces showing Z11-16:Ald-evoked neuronal activities in H. assulta across a range of concentrations. No response was evoked by hexane. (F) Z11-16:Ald-induced dose–response curves in H. assulta (n = 12). Error bars indicate SEM.
Table 1. Comparison of dose-response values recorded with in vitro and in vivo system.
| Comparison in different systems | EC50 value | Standard error of EC50 | Test number | |
|---|---|---|---|---|
| In vitro | Xenopus oocyte system | 6.84 × 10−5 M | 7.93 × 10−6 | 7 |
| In vivo | Transgenic system in Drosophila | 1.26 × 10−5 g | 5.47 × 10−6 | 13 |
| The endogenous system in H. assulta | 2.15 × 10−6 g | 4.10 × 10−7 | 12 | |
Using an in vivo system, the HassOR13 gene was expressed in at1 neurons of Drosophila and the resulting UAS-HassOR13 flies were crossed with a mutant knock-in allele Or67dGAL4 driver line33. Then action potentials were recorded from the olfactory neurons within a single sensillum (Fig. 1B). The results showed that the HassOR13-expressing neurons in at1 specifically responded to the secondary sex pheromone component Z11-16:Ald at the dose of 1 mg loaded in the stimulus cartridge (P < 0.01) (Fig. 1E,H). In a dose–response experiment, neurons in at1 started firing at doses of Z11-16:Ald as low as 10 ng, with an EC50 value of 1.26 × 10−5 g (Fig. 2C,D, Table 1). For control lines UAS-HassOR13, no response to Z11-16:Ald was recorded at the same doses (Fig. 2D). We concluded that HassOR13 was selectively activated by Z11-16:Ald.
Recent in situ hybridization and single sensillum recording studies reported three types of trichoid sensilla on the antenna of H. assulta, with type A containing neurons responding only to Z11-16:Ald66,74. We directly recorded responses of trichoid sensilla type A from H. assulta antenna and compared the result with those obtained from transgenic fly lines (Fig. 1C). We first confirmed that neurons expressing HassOR13 gene were activated by Z11-16:Ald at a dose of 1 mg (P < 0.01) (Fig. 1F,I). Then, we measured the dose–response curve across a dose range from 10 ng to 1 mg (Fig. 2E,F) obtaining an EC50 value of 2.15 × 10−6 g (Table 1).
Effect of solvent
To evaluate the effects of different solvents used to dilute stimuli in single-sensillum experiments, we recorded responses of HassOR13 expressed in Drosophila at1 neurons to Z11-16:Ald dissolved in paraffin oil, hexane or methylene dichloride in a dose range from 10 ng to 1 mg (Fig. 3A). When the ligand was diluted in paraffin oil, the sensitivity (EC50 = 1.06 × 10−4 g) of HassOR13 to Z11-16:Ald in the system was markedly lower than when using methylene dichloride (EC50 = 1.31 × 10−5 g) or hexane (EC50 = 9.84 × 10−6 g) (Table 2). Figure 3 shows representative traces recorded at doses of 100 μg of the pheromone dissolved in the different solvents (Fig. 3B).
Figure 3. Functional comparison of HassOR13 expressed in Drosophila lines using different solvents.
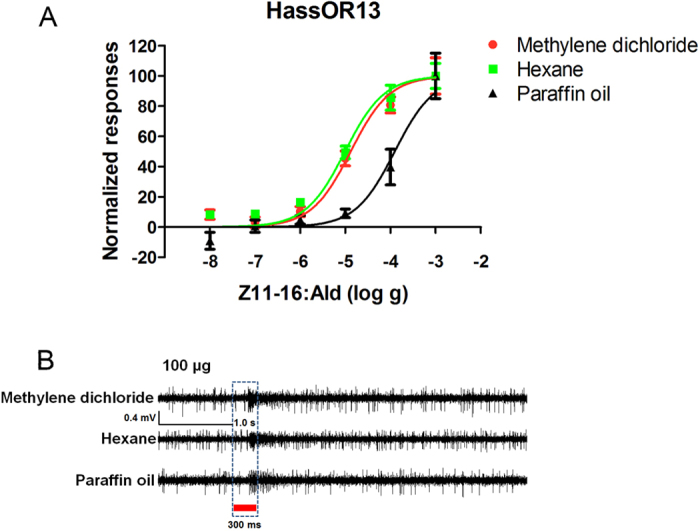
(A) Z11-16:Ald dose–response relationship for HassOR13 using paraffin oil, hexane or methylene dichloride as solvents. Z11-16:Ald was used at doses from 10−3 g to 10−8 g. Error bars indicate SEM. (B) SSR traces were recorded at the dose of 100 μg.
Table 2. Dose-response values in HassOR13-expressed in Drosophila lines using different solvents.
| Or67dGAL4 knock-in system, in vivo | EC50 value | Standard error of EC50 | Test number | |
|---|---|---|---|---|
| Different solvents | Methylene dichloride | 1. 31 × 10−5 g | 1.10 × 10−5 | 12 |
| Hexane | 9.84 × 10−6 g | 5.47 × 10−6 | 13 | |
| Paraffin oil | 1.06 × 10−4 g | 6.92 × 10−5 | 10 | |
Drosophila lines expressing OR13 are attracted to Z11-16:Ald
Next, we asked if HassOR13-expressing Drosophila would also exhibit a behavioral phenotype. Therefore, we performed behavior experiments using a two-choice bait trap assay62. Wild-type flies showed significant preference for 11-cis-vaccenyl acetate (cVA) compared to Z11-16:Ald and to paraffin oil (P < 0.05) (Fig. 4A). However, flies expressing HassOR13 were attracted to Z11-16:Ald (P < 0.05), but not to cVA (Fig. 4A) at a dose of 10 μg. In UAS-HassOR13; Or67dGAL4 lines, attraction to Z11-16:Ald was observed at doses from 10−7 g to 10−4 g. The attraction preference index (PI) of male flies gradually increased with the amount of Z11-16:Ald up to 10−4 g with an EC50 value of 3.7 × 10−7 g (Fig. 4B). In these experiments both male and female transgenic flies were attracted to the moth pheromone (Fig. 4C). Taken together, these data indicate that HassOR13 can mediate attraction to Z11-16:Ald in Drosophila by activating at1 neurons, thus confirming the function of this odorant receptor.
Figure 4. Attraction to Z11-16:Ald of Drosophila lines expressing HassOR13.
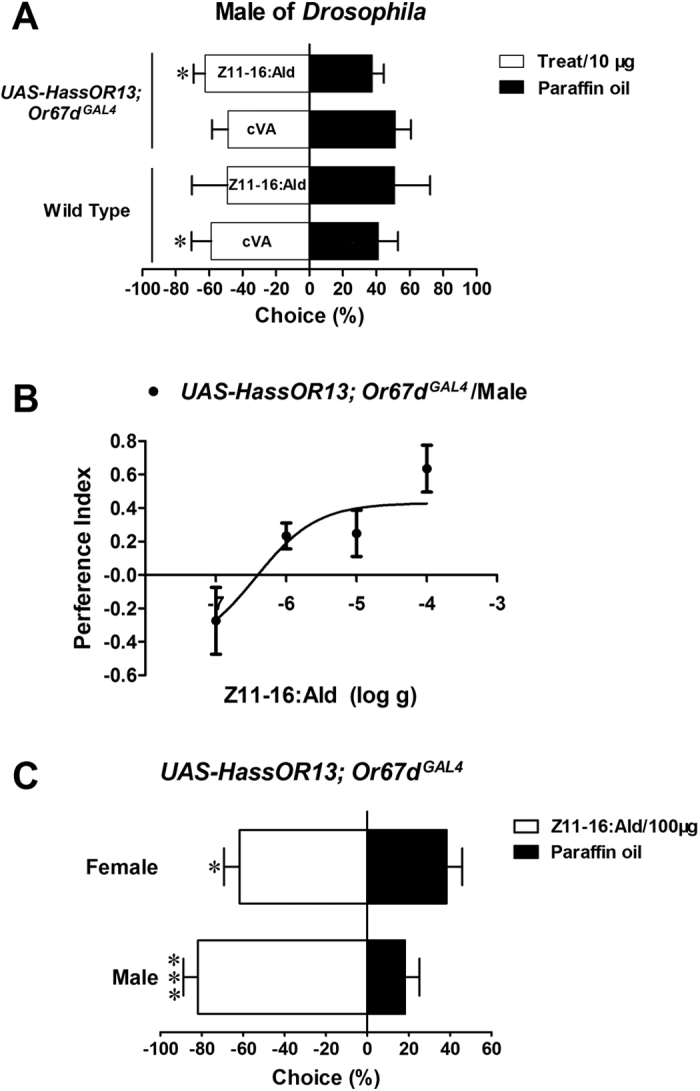
(A) Two-choice behavioral assay at the dose of 10 μg. Wild-type flies display strong attraction toward cVA (*P < 0.05, Chi-square test, χ2 = 5.59, df = 1), which is abolished in HassOR13-expressing fly lines (P = 0.55, Chi-square test, χ2 = 0.35, df = 1). The attraction preference of HassOR13-expressing fly lines (*P < 0.05, Chi-square test, χ2 = 5.77, df = 1) and wild-type flies (P = 0.56, Chi-square test, χ2 = 0.34, df = 1) toward Z11-16:Ald are shown. Error bars indicate SEM. (B) Attraction of HassOR13-expressing male fly lines to Z11-16:Ald gradually increased with the dose of pheromone up to 10−4 g (n = 3~6). (C) Two-choice behavioral assay of HassOR13-expressing fly lines at the dose of 100 μg. Both males and females were significantly attracted to Z11-16:Ald compared to paraffin oil (for male, ***P < 0.001, Chi-square test, χ2 = 49.87, df = 1; for female, *P < 0.05, Chi-square test, χ2 = 6.22, df = 1). Error bars indicate SEM.
Discussion
Rapidly and accurately deorphanizing OR genes is very important to elucidate how the insect converts external chemical signals into electrical signals through ORNs at the periphery of the olfactory system. Among the several methods developed during the last decade for the functional characterization of insect ORs, including the use of transgenic Drosophila and heterologous expression in Xenopus oocytes, it is sometimes difficult to choose the most suitable protocol for each research purpose. In this study we have compared in vitro and in vivo systems to study the function of HassOR13. In both cases, cells or neurons expressing HassOR13 were specifically activated by Z11-16:Ald. Of the two in vivo approaches, we found that the endogenous system was more sensitive (EC50 = 2.15 × 10−6 g) than that utilizing transgenic Drosophila (EC50 = 1.26 × 10−5 g). A comparison between in vivo and in vitro systems is not feasible, because we record electric currents in the two-electrode voltage-clamp technique used for heterologous expression systems, while we measure frequency of firing (spikes⁄s) when recording from single sensilla of transgenic flies. Each method presents its advantages and drawbacks. Sometimes, OR function cannot be properly reproduced in Xenopus expression system probably due to the absence of odorant binding proteins (OBPs)65,71. On the other hand, OR genes cannot always be expressed in transgenic fly lines17. Therefore, in vitro or in vivo protocols must be adopted depending on specific requirements. For example, the two-electrode voltage clamp recording is more practical in functional screenings of large numbers of ORs, while the in vivo transgenic Drosophila system is generally more accurate.
As for the choice of a solvent to dissolve odorant stimuli, we tested the three most used in the literature, paraffin oil, methylene dichloride and hexane20,55,56,58. The last two provided stronger responses compared to paraffin oil. This is due to the much lower volatility of the pheromone when dissolved in paraffin oil. Hexane remains probably the solvent of choice, offering a good reproducibility with a high sensitivity, while methylene dichloride can generate spontaneous firing of a specific neuron in some cases (Figure S1). However, paraffin oil has its advantage when testing a large number of odorants, since highly volatile compounds are likely to evaporate less from this solvent.
In our study, we used the Or67dGAL4 knock-in system to express HassOR13 gene in at1 sensilla of Drosophila. On the basis of the one-to-one relationship between ORs and ORNs, as well as on the odor-selective activation of ORs, we tested behavioral responses of transgenic fly lines to odorants. In general, OrcoGAL4 driven UAS-OR lines or the lines with odorant receptor promoter to drive expression of GAL4 have been adopted to monitor behavioral preference for specific odors matching ORs-expressing neurons within defined sensilla36,37,62. In our research we performed behavioral assays with flies expressing HassOR13 in their ORNs under control of the Or67dGAL4 driver background. We observed strong attraction of both male and female transgenic lines to Z11-16:Ald with a EC50 value in agreement with single sensillum recording. This indicates that odor-activation of ORs-expressing ORNs can mediate behavioral choice. At the same time, we can use behaviour assays to indirectly identify the function of specific ORs, without the need to perform electrophysiological recordings.
In summary, we have compared different approaches to study the function of ORs in vitro and in vivo, presenting the advantages and the drawbacks of each method. Studying the interactions of pheromones and odorants with their receptors still requires complex methodologies, as ORs cannot be expressed and isolated in their active forms.
Materials and Methods
Insect rearing
H. assulta individuals were reared in our laboratory with an artificial diet at the larval stage75 and 10% honey solution at the adult stage, at 26 ± 1 °C, 65% ± 5% relative humidity and under photoperiod of 16 h light: 8 h dark. Pupae were sexed and put into separate cages for eclosion. Drosophila stocks were fed on cornmeal-agar-molasses medium and maintained under a 12 h light: 12 h dark cycle at 25 °C and 60% relative humidity.
Pheromone components
(Z)-9-hexadecenol-1-ol (Z9-16:OH), (Z)-9-tetradecen-1-ol (Z9-14:OH), (Z)-9-tetradecenyl acetate (Z9-14:OAc), (Z)-9-hexadecadecenyl acetate (Z9-16:OAc), (Z)-11-hexadecenal (Z11-16:Ald) and (Z)-11-hexadecen-1-ol (Z11-16:OH) (both 95% minimum purity) were purchased from Nimrod Inc. (Changzhou, China). (Z)-9-tetradecenal (Z9-14:Ald), (Z)-9-hexadecenal (Z9-16:Ald), (Z)-11-hexadecenyl acetate (Z11-16:OAc) (all 93–95% minimum purity) were purchased from Bedoukian (Danbury, CT, USA). Paraffin oil, methylene chloride and hexane (96–98% minimum purity) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
cRNA synthesis and Electrophysiological recording with two-electrode voltage-clamp
HassOR13 and HassOrco genes were cloned into eukaryotic expression vector pT7Ts and stored as plasmids in our laboratory. cRNAs were synthesized using mMESSAGE mMACHINE T7 Ultra Kit (Ambion, Austin, TX, USA) following the manufacturer’s instructions. HassOR13 was expressed in Xenopus oocytes according to the following protocol. 27.6 ng of both HassOR13 and HassOrco cRNA were microinjected into mature oocytes (stage V–VII), that had been treated with 2 mg/mL collagenase in washing buffer (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, and 5 mM HEPES, pH 7.6) for 1–2 h at room temperature. Then, oocytes were cultured for 4–7 days at 18 °C in 1 × Ringer’s solution (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, 0.8 mM CaCl2, and 5 mM HEPES, pH 7.6) supplemented with 5% dialysed horse serum, 50 mg/mL tetracycline, 100 mg/mL streptomycin and 550 mg/mL sodium pyruvate. The recording methods of two-electrode voltage-clamp followed previously reported protocols25,27. Whole-cell currents were obtained from the injected Xenopus oocytes with a two-electrode voltage-clamp and recorded with an OC-725C oocyte clamp (Warner Instruments, Hamden, CT, USA) at a holding potential of −80 mV. Oocytes were exposed to compounds in ascending order of concentration with an interval between exposures that allowed the current to return to baseline. Data acquisition and analysis were carried out with Digidata 1440 A and PCLAMP 10.2 software (Axon Instruments Inc., Union City, CA, USA). GRAPHPAD PRISM 5.0 software (GraphPad Software Inc., San Diego, CA, USA) was used to analyze dose–response data.
Fly strains
Transgenic lines were generated according to standard procedures as described below. The open reading frame encoding HassOR13 was cloned into the pVALIUM20 vector76. Independent homozygous UAS-HassOR13 lines (with transgene insertions into chromosome II) were generated at Tsinghua Fly Center (Beijing, China). Driver mutant allele Or67dGAL4 stock was provided by Dr Barry J. Dickson33. The balancer w−/w−; sp/CyO; TM3/TM6B was used to cross with homozygous driver lines. Then, the driver line in Or67dGAL4 mutant background was crossed with UAS-HassOR13 balancer line to establish final homozygous stock w+/w+; UAS-HassOR13/UAS-HassOR13; Or67dGAL4/Or67dGAL4 which expressed HassOR13 in at1 sensilla neurons. Each HassOR13 insertion was confirmed by sequencing of genomic DNA prepared from mutant lines. Both final stock and wild-type Canton-S strain were used for electrophysiological experiments.
Single sensillum recordings
Extracellular electrophysiological recordings were performed on single at1 sensilla of 1- to 10-day-old flies. The antenna was fixed using standard procedures55,77. The reference electrode was placed in the fly eye, under a microscope (LEICA Z16 APO, Germany) at 920 × magnification. Action potentials were recorded by inserting a tungsten wire electrode in the base or in the shaft of a sensillum of the fly antenna. Signals were amplified 10 × by a high impedance pre-amplifier (IDAC-4 USB System, Syntech, Kirchzarten, Germany), sent to a PC via an analog-digital converter and analyzed off-line with AUTOSPIKE v. 3.9 software (Syntech, Kirchzarten, Germany). The filter was set with a 500 Hz low cutoff and a 3 kHz high cutoff. AC signals were recorded for 10 s, starting 1 s before stimulation. Responses were calculated by counting the number of action potentials one second after stimulation (with a delay of 200 ms to allow the odorant to travel down the airstream), and subtracting the number counted in the second before stimultion.
Odor stimulation
Aliquots of odorants were dissolved in paraffin oil, methylene chloride or hexane (vol/vol) and 10 μL of each solution were loaded onto a 0.5 × 40 mm filter paper strip (Whatman), which was placed inside a Pasteur pipette. Hexane, methylene chloride or paraffin oil alone were tested as negative controls. For dose-response relationships, serial dilutions were made in increasing doses of 0.001, 0.01, 0.1, 1, 10 and 100 μg/μL and loaded on filter paper strips. The preparation was held in a humidified continuous air flow delivered by the Syntech Stimulus controller (CS-55 model, Syntech) at 1.4 L/min. Stimulus pulses were added for 300 ms. During stimulation, the compensatory flow was switched off.
Behavioral assays
Attraction to odours was measured using a modified two choice trap assay62,78. Two to three day old female and male (1:1) adult fruit flies were starved for 40–42 h in collection cages containing 1% agarose gel. 40–60 flies per repeat were anaesthetized on ice, then placed into a 1 L glass beaker covered with a 150 mm Petri dish with three holes covered by nylon mesh for ventilation. Odor traps were made from 40 mL plastic vials with a 1 mL pipette tip inserted at the top, and placed in the glass beaker. Traps contained a filter paper strip soaked with either 10 μL of the odor (cVA or Z11-16:Ald) at different dilutions in paraffin oil or just paraffin oil was added. Behaviour tests were conducted for 24 h in the dark at room temperature. Each treatment was repeated three to six times. Dose–response curves (10−7 to 10−4) were used to calculate the preference index (PI), according to the formula PI = (#flies in odor vial − #flies in control vial)/total # of flies.
Statistical analysis
All data were presented as mean ± SEM. Data multiple comparison over three groups was assessed by one-way analysis of variance (ANOVA) following Duncan’s multiple range test for variable (α = 0.05), and two-sample analysis was performed using Student’s t-test (α = 0.05). Two choice trap assay results were compared using Chi-square test. All statistic comparison were assayed with SPSS Statistics 16.0 (SPSS Inc., Chicago, IL, USA).
Additional Information
How to cite this article: Wang, B. et al. Comparison of research methods for functional characterization of insect olfactory receptors. Sci. Rep. 6, 32806; doi: 10.1038/srep32806 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Anandasankar Ray for kindly providing the Or67dGAL4 mutant knock-in fly line under Dr. Barry Dickson’s permission, Dr. Jian-Quan Ni for kindly providing the pVALIUM20 vector, the Tsinghua Fly Center (Beijing, China) for the transformation service, and Dr. Jie Shen (China Agricultural University, Beijing, China) for kindly providing the balancer w-; sp/CyO; TM3/TM6B and wild-type Canton-S strain. Thanks to Prof. Paolo Pelosi for editorial assistance and comments on the manuscript. This project was supported by National Natural Science Foundation of China (31402023, 31230062, 31321004) and China Postdoctoral Science Foundation (2014M550905).
Footnotes
Author Contributions B.W., Y.L. and G.W. designed the experiments. B.W. performed the experiments. Y.L. and K.H. contributed reagents/materials/gene identification. B.W., Y.L. and G.W. analyzed the data. B.W and G.W. wrote and revised the paper.
References
- Vosshall L. B. & Stocker R. F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30, 505–533 (2007). [DOI] [PubMed] [Google Scholar]
- Leal W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391 (2013). [DOI] [PubMed] [Google Scholar]
- Vosshall L. B. Laying a controversial smell theory to rest. Proc. Natl. Acad. Sci. USA 112, 6525–6526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh E., Bohbot J. D. & Zwiebel L. J. Peripheral olfactory signaling in insects. Current Opinion in Insect Science 6, 86–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. D. & Löfstedt C. Moth pheromone receptors: gene sequences, function, and evolution. Front. Ecol. Evol. 3, 105 (2015). [Google Scholar]
- Clyne P. J. et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338 (1999). [DOI] [PubMed] [Google Scholar]
- Liu Y., Gu S. H., Zhang Y. J., Guo Y. Y. & Wang G. R. Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS One 7, e48260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. N. et al. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae). BMC Genomics 15, 501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. P., Liu Y., Walker W. B., Li J. H. & Wang G. R. Molecular characterization of the Aphis gossypii olfactory receptor gene families. PLoS One 9, e101187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D. P. et al. Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 10, 846–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Dippel S. et al. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15, 1141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. H. et al. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. PLoS One 9, e103420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges T. K. et al. Species-specific chemosensory gene expression in the olfactory organs of the malaria vector Anopheles gambiae. BMC Genomics 15, 1089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang S., Zhang Z., Wang H. & Kong X. Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (Lepidoptera: Lasiocampidae). Insect Biochem. Mol. Biol. 52, 69–81 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PloS One 10, e0117054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Ozaki K., Ishikawa Y. & Matsuo T. Identification of candidate odorant receptors in asian corn borer Ostrinia furnacalis. PLoS One 10, e0121261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fouchier A. et al. Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera. Front. Ecol. Evol. 3, 95 (2015). [Google Scholar]
- Hallem E. A., Ho M. G. & Carlson J. R. The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979 (2004). [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Sakurai T., Nishioka T. & Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642 (2005). [DOI] [PubMed] [Google Scholar]
- Hallem E. A. & Carlson J. R. Coding of odors by a receptor repertoire. Cell 125, 143–160 (2006). [DOI] [PubMed] [Google Scholar]
- Grosse-Wilde E., Gohl T., Bouche E., Breer H. & Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 25, 2364–2373 (2007). [DOI] [PubMed] [Google Scholar]
- Wanner K. W. et al. A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc.Natl. Acad. Sci. USA 104, 14383–14388 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuno H. et al. Identification of receptors of main sex-pheromone components of three lepidopteran species. Eur. J. Neurosci. 28, 893–902 (2008). [DOI] [PubMed] [Google Scholar]
- Carey A. F., Wang G., Su C. Y., Zwiebel L. J. & Carlson J. R. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. R., Carey A. F., Carlson J. R. & Zwiebel L. J. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 107, 4418–4423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner K. W. et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One 5, e8685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. R., Vasquez G. M., Schal C., Zwiebel L. J. & Gould F. Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol. 20, 125–133 (2011). [DOI] [PubMed] [Google Scholar]
- Wetzel C. H. et al. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proc. Natl. Acad. Sci. USA 98, 9377–9380 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. A., van Naters W., Warr C. G., Steinbrecht R. A. & Carlson J. R. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841 (2003). [DOI] [PubMed] [Google Scholar]
- Sakurai T. et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. USA 101, 16653–16658 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., Vannice K. S. & Vosshall L. B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293 (2007). [DOI] [PubMed] [Google Scholar]
- Kiely A., Authier A., Kralicek A. V., Warr C. G. & Newcomb R. D. Functional analysis of a Drosophila melanogaster olfactory receptor expressed in Sf9 cells. J. Neurosci. Methods 159, 189–194 (2007). [DOI] [PubMed] [Google Scholar]
- Kurtovic A., Widmer A. & Dickson B. J. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546 (2007). [DOI] [PubMed] [Google Scholar]
- Sakurai T. et al. A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genet. 7, e1002115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweck H. K., Ebrahim S. A., Farhan A., Hansson B. S. & Stensmyr M. C. Olfactory proxy detection of dietary antioxidants in Drosophila. Curr. Biol. 25, 455–466 (2015). [DOI] [PubMed] [Google Scholar]
- Dweck H. K. et al. Pheromones mediating copulation and attraction in Drosophila. Proc. Natl. Acad. Sci. USA 112, 2829–2835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. C. & Potter C. J. Re-classification of Drosophila melanogaster trichoid and intermediate sensilla using fluorescence-guided single sensillum recording. PLoS One 10, e0139675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T. et al. Targeted disruption of a single sex pheromone receptor gene completely abolishes in vivo pheromone response in the silkmoth. Sci. Rep. 5, 11001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. & Smart T. G. HEK293 cell line: a vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods 51, 187–200 (2005). [DOI] [PubMed] [Google Scholar]
- Grosse-Wilde E., Svatos A. & Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses 31, 547–555 (2006). [DOI] [PubMed] [Google Scholar]
- Lundin C. et al. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 581, 5601–5604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. R. et al. Molecular basis of female-specific odorant responses in Bombyx mori. Insect. Biochem. Mol. Biol. 39, 189–197 (2009). [DOI] [PubMed] [Google Scholar]
- Forstner M., Breer H. & Krieger J. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci. 5, 745–757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. D. et al. Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem. Senses 34, 383–394 (2009). [DOI] [PubMed] [Google Scholar]
- Schneider E. H. & Seifert R. Sf9 cells: a versatile model system to investigate the pharmacological properties of G protein-coupled receptors. Pharmacol. Ther. 128, 387–418 (2010). [DOI] [PubMed] [Google Scholar]
- Tsitoura P. et al. Expression and membrane topology of Anopheles gambiae odorant receptors in lepidopteron insect cells. PLoS One 5, e15428 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot J. D. et al. Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem. Senses 36, 149–160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. L., Pask G. M., Rinker D. C. & Zwiebel L. J. Functional agonism of insect odorant receptor ion channels. Proc. Natl. Acad. Sci. USA 108, 8821–8825 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregitzer P. et al. Plant odorants interfere with detection of sex pheromone signals by male Heliothis virescens. Front. Cell. Neurosci. 6, 42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- German P. F., van der Poel S., Carraher C., Kralicek A. V. & Newcomb R. D. Insights into subunit interactions within the insect olfactory receptor complex using FRET. Insect Biochem. Mol. Biol. 43, 138–145 (2013). [DOI] [PubMed] [Google Scholar]
- Kumar B. N. et al. A conserved aspartic acid is important for agonist (VUAA1) and odorant/tuning receptor-dependent activation of the insect odorant co-receptor (Orco). PLoS One 8, e70218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. & Luetje C. W. Trace amines inhibit insect odorant receptor function through antagonism of the co-receptor subunit. F1000Research 3, 84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudianos C. et al. Odor memories regulate olfactory receptor expression in the sensory periphery. Eur. J. Neurosci. 39, 1642–1654 (2014). [DOI] [PubMed] [Google Scholar]
- Corcoran J. A., Jordan M. D., Carraher C. & Newcomb R. D. A novel method to study insect olfactory receptor function using HEK293 cells. Insect Biochem. Mol. Biol. 54, 22–32 (2014). [DOI] [PubMed] [Google Scholar]
- Syed Z., Ishida Y., Taylor K., Kimbrell D. A. & Leal W. S. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc. Natl. Acad. Sci. USA 103, 16538–16543 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Z., Kopp A., Kimbrell D. A. & Leal W. S. Bombykol receptors in the silkworm moth and the fruit fly. Proc. Natl. Acad. Sci. USA 107, 9436–9439 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Z., Pelletier J., Flounders E., Chitolina R. F. & Leal W. S. Generic insect repellent detector from the fruit fly Drosophila melanogaster. PLoS One 6, e17705 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagné N. et al. Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596 (2012). [DOI] [PubMed] [Google Scholar]
- Vasquez G. M., Syed Z., Estes P. A., Leal W. S. & Gould F. Specificity of the receptor for the major sex pheromone component in Heliothis virescens. J. Insect Sci. 13, 160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. S., Xia R., Zhang H., Jin X. & Smith D. P. Lipid flippase modulates olfactory receptor expression and odorant sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA 111, 7831–7836 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., van Naters W. & Carlson J. R. Molecular determinants of odorant receptor function in insects. J. Bioscience. 39, 555–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronderos D. S., Lin C. C., Potter C. J. & Smith D. P. Farnesol-detecting olfactory neurons in Drosophila. J. Neurosci. 34, 3959–3968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueira-Vieira C., Kimbrell D. A., de Carvalho W. J. & Leal W. S. Facile functional analysis of insect odorant receptors expressed in the fruit fly: validation with receptors from taxonomically distant and closely related species. Cell. Mol. Life Sci. 71, 4675–4680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu C. C., Lin K. J. & Wang G. R. Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS One 8, e62094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M. et al. Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PloS one 8, e62098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. J. et al. Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem. Mol. Biol. 48, 63–74 (2014). [DOI] [PubMed] [Google Scholar]
- Liu C. C., Liu Y., Walker W. B., Dong S. L. & Wang G. R. Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hubner). Insect Biochem. Mol. Biol. 43, 747–754 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. An odorant receptor from the common cutworm (Spodoptera litura) exclusively tuned to the important plant volatile cis-3-hexenyl acetate. Insect Mol. Biol. 22, 424–432 (2013). [DOI] [PubMed] [Google Scholar]
- Liu C. C. et al. Narrow tuning of an odorant receptor to plant volatiles in Spodoptera exigua (Hubner). Insect Mol. Biol. 23, 487–496 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang Y. N. et al. Functional characterization of sex pheromone receptors in the purple stem borer, Sesamia inferens (Walker). Insect Mol. Biol. 23, 611–620 (2014). [DOI] [PubMed] [Google Scholar]
- Chang H. T. et al. Pheromone binding proteins enhance the sensitivity of olfactory receptors to sex pheromones in Chilo suppressalis. Sci. Rep. 5, 13093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem. senses 40, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- Berg B. G. & Mustaparta H. The significance of major pheromone components and interspecific signals as expressed by receptor neurons in the oriental tobacco budworm moth, Helicoverpa assulta. J. Comp. Physiol. A. 177, 683–694 (1995). [Google Scholar]
- Wu H. et al. Specific olfactory neurons and glomeruli are associated to differences in behavioral responses to pheromone components between two Helicoverpa species. Front. Behav. Neurosci. 9, 206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J. F. et al. Research on developing an improved artificial diet for Helicoverpa assulta. Chinese Journal of Applied Entomology 50, 261–267 (2013). [Google Scholar]
- Ni J. Q. et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. methods 8, 405–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M., Foster K. & Carlson J. R. Odor coding in the Drosophila antenna. Neuron 30, 537–552 (2001). [DOI] [PubMed] [Google Scholar]
- Potter C. J., Tasic B., Russler E. V., Liang L. & Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.