Abstract
Physical interaction between organelles is a flexible event and essential for cells to adapt rapidly to environmental stimuli. Germinating plants utilize oil bodies and peroxisomes to mobilize storage lipids for the generation of sucrose as the main energy source. Although membrane interaction between oil bodies and peroxisomes has been widely observed, its underlying molecular mechanism is largely unknown. Here we present genetic evidence for control of the physical interaction between oil bodies and peroxisomes. We identified alleles of the sdp1 mutant altered in oil body morphology. This mutant accumulates bigger and more oil body aggregates compared with the wild type and showed defects in lipid mobilization during germination. SUGAR DEPENDENT 1 (SDP1) encodes major triacylglycerol lipase in Arabidopsis. Interestingly, sdp1 seedlings show enhanced physical interaction between oil bodies and peroxisomes compared with the wild type, whereas exogenous sucrose supplementation greatly suppresses the interaction. The same phenomenon occurs in the peroxisomal defective 1 (ped1) mutant, defective in lipid mobilization because of impaired peroxisomal β-oxidation, indicating that sucrose production is a key factor for oil body-peroxisomal dissociation. Peroxisomal dissociation and subsequent release from oil bodies is dependent on actin filaments. We also show that a peroxisomal ATP binding cassette transporter, PED3, is the potential anchor protein to the membranes of these organelles. Our results provide novel components linking lipid metabolism and oil body-peroxisome interaction whereby sucrose may act as a negative signal for the interaction of oil bodies and peroxisomes to fine-tune lipolysis.
Keywords: ABC transporter, lipid droplet, lipolysis, membrane, organelle, peroxisome, oil body, sucrose production
Introduction
Membrane-delimited organelles perform specialized subcellular functions. They communicate with each other through physical interaction, exerting functions as a community to benefit the cells, such as energy transduction and lipid delivery (1, 2). Those interactions are dynamic and rapid upon intracellular and extracellular signals. Notably, organelle interactions are highly selective, attributed to specific membrane protein-protein interaction, which, in some cases, is mediated through cytoplasmic linker proteins. For example, several mitochondrion- or ER-bound2 proteins are reported to promote mitochondrion-ER interaction, which results in apoptosis as well as rapid transmission of Ca2+ from the ER to mitochondria (3, 4).
Oil bodies, also referred to as lipid bodies or lipid droplets, have been widely found in eukaryotic cells and are the only place in cells that stores lipids for later usage. The hydrophobic core of oil bodies is composed of neutral lipids, mainly triacylglycerols (TAGs) and sterol esters (5, 6). In plant, TAGs stored in the seeds are the main energy reservoir during seed germination. Accumulation of those storage lipids occurs mainly in the cotyledon tissues during plant embryogenesis (7). This process is accompanied by oil body biogenesis, featuring an increased number and size of cellular oil bodies (8–10). Investigation of oil body membrane-associated proteins suggests that the feature of oil body membranes varies between animals and plants (11–13). Oleosins, caleosins, and steroleosins have been identified in plant as the major constituents of oil body membrane proteins, of which oleosin is the most abundant (14). For example, oleosins represent 80–90% of total proteins isolated from oil body membranes in sesame (15). Suppression of oleosins in Arabidopsis resulted in bigger oil bodies in embryonic tissues, suggesting that oleosins are involved in the regulation of oil body morphology (16).
Mobilization of TAGs is essential for germinating plants to generate sucrose as the main energy source during the nonautotrophic stage before photosynthesis. Degradation of TAGs mainly occurs in cotyledons. Sucrose generated from TAG metabolism is transported to nonautotrophic organs such as roots from the cotyledon through the phloem (17). Oil bodies and peroxisomes are central organelles for storage lipid degradation during seed germination. Sugar-dependent 1 (SDP1) is a patatin-like TAG lipase that resides on the oil body membrane and mobilizes TAGs up to 90% upon germination in Arabidopsis (18). SDP1 hydrolyzes TAG to produce fatty acids. Fatty acids are then translocated into peroxisomes by a peroxisomal membrane protein ATP binding cassette transporter, PED3 (also known as COMATOSE (CTS) and PXA1) (19–21), followed by CoA attachment by long chain acyl-CoA synthetases (LACS) 6 and 7 (22, 23). They are further degraded by peroxisomal matrix enzymes of peroxisomal fatty acid β-oxidation and the glyoxylate cycle, resulting in the generation of succinate, which is finally converted to sucrose through mitochondria (24). PED1, also known as KAT2, encodes 3-ketoacyl-CoA thiolase and catalyzes the last step of fatty acid β-oxidation, leading to the production of acetyl-CoA (25, 26). Arabidopsis mutants defective in the functions of either oil bodies (i.e. sdp1) or peroxisomes (i.e. ped1, ped3) display impaired storage lipid degradation and thus show sucrose-dependent germination or post-germinative growth (18, 24).
Compared with oil bodies, which are generally thought to be immobile in plant cells (27), peroxisomes are largely mobile and occasionally move across the cytoplasm in a microtubule- and actin filament-dependent manner in animals and plants, respectively (28–30). Direct membrane associations between oil bodies and peroxisomes have been detected in mammalian cells, yeast, and plants (31–33). For example, an electron microscopic study in cotyledon cells of the Arabidopsis ped1 mutant showed a direct membrane interaction between oil bodies and peroxisomes, where peroxisomal membrane invagination from membrane interaction sites was detected (33). Except for this study, to our knowledge, characterization of their interaction in planta is largely missing. Here we mainly performed microscopic observation and focused on the oil body-peroxisome interaction in Arabidopsis, including the newly identified allele of the sdp1 mutant as well as ped1 and ped3, all of which are defective in lipid mobilization and, thus, in sucrose production. We show an unanticipated negative correlation between lipolysis-mediated sucrose production and the oil body-peroxisome interaction that relies on actin filaments. Our data reveal novel factors for the regulation of physical interaction between oil bodies and peroxisomes and provide molecular and genetic links between lipid metabolism and the oil body-peroxisome interaction in plants.
Results
Identification of the sdp1 Mutant That Accumulates Oil Bodies during Seed Germination
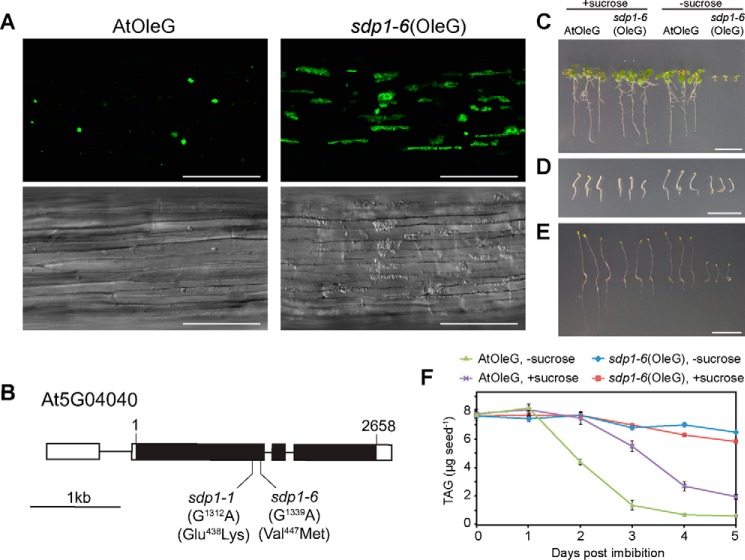
The morphology of oil bodies in plants is associated with oil body biogenesis, lipolysis, or biotic/abiotic stresses (16, 34, 35). To visualize oil bodies in living cells, we established transgenic Arabidopsis plants (AtOleG) expressing an oil body marker, a chimeric OLE1 gene that encodes oleosin, fused to GFP under the control of the 35S promoter (OleG) (36). Our previous report has shown that oil bodies in AtOleG aggregate with each other and are displayed as green spots under a fluorescent microscope (Fig. 1A) (36). The size of these aggregates and the amounts of storage lipids in the seedlings gradually decrease along germination. With the aim of identifying the mutants with altered morphology of oil bodies or/and oil body aggregates, we mutagenized AtOleG seeds by ethyl methane sulfonate treatment and conducted a forward genetic screening. As a result, we identified two mutants that accumulate more oil bodies in germinating seedlings (Fig. 1A). The diameter of oil body aggregates in the hypocotyl of 3-day-old AtOleG was 3∼10 μm (Fig. 1A, AtOleG). The oil body aggregates in the mutants appeared significantly larger (Fig. 1A, sdp1–6(OleG)); the size of those aggregates was 206 ± 141 μm2 versus 22 ± 16 μm2 in the wild type. The morphology of oil body aggregates in the mutants varied. Many of them appeared as tubular structures that extended up to 100 μm in length. The elongated morphology is probably due to the physical pressure caused by cell vacuolation. Big oil body aggregates were maintained in the mutants during the later stage of germination, whereas the aggregates became largely limited in the wild type, indicating higher accumulation of oil bodies in the mutants.
FIGURE 1.
Identification of the sdp1 mutant that shows oil body accumulation and defect of lipid mobilization during seed germination. A, oil bodies accumulate in the sdp1–6 mutant during postgerminative growth. Hypocotyl tissues of 3-day-old wild-type (AtOleG) and sdp1–6 (sdp1–6(OleG)) plants overexpressing OLE1 fused with GFP (OleG). Green particles represent oil body aggregates. Bottom panels, difference interference contrast images. Scale bars = 100 μm. B, gene structure of SDP1 with intron (line) and exon (closed boxes) organization and the positions of single nucleotide substitutions from guanine to adenine in the sdp1–1 and sdp1–6 mutants. The corresponding amino acid mutations are shown at the bottom. Open boxes indicate the 5′ and 3′ UTR, respectively. 1 and 2658 at the top indicate the nucleotide residue of the start and stop codons, respectively. C—E, 10-day-old (C and E) and 3-day-old (D) AtOleG and sdp1–6(OleG) seedlings grown under constant light (C) or dark conditions (D and E) in the absence or presence of 2% sucrose. Scale bars = 1 cm. F, TAG levels in AtOleG and sdp1–6(OleG) seedlings grown under dark conditions in the absence or presence of 2% sucrose. The data represent mean ± S.E. of three independent experiments (n = 150).
High-resolution mapping of the mutants revealed that the mutations in both mutants were located 3.9 centimorgans away from At5g01060, 1.0 centimorgans away from At5g05100, and close to At5g03730. We carefully analyzed the DNA sequence in this region and found that the mutants bear mutations in a gene, At5g04040, that is annotated as SDP1 (Fig. 1B), encoding a TAG lipase located on the oil body membrane. One of the mutants is identical to the previously reported sdp1–1 (18), whereas the other mutant contains a single nucleotide substitution in nucleotide 1339 (guanine to adenine), changing amino acid 447 from Val to Met in the encoded gene product (Fig. 1B). We designated this allele sdp1–6 and called the mutant sdp1–6(OleG). SDP1 is a major lipase in Arabidopsis, with higher lipase activity against TAG than diacylglycerol and monoacylglycerol and shares the highest sequence similarity with yeast TGL3 (18, 37). Like previously identified sdp1 mutant alleles (18), sdp1–6(OleG) exhibited sucrose-dependent post-germinative growth (Fig. 1, C and E) and a defect in lipid degradation (Fig. 1F). Interestingly, sucrose-dependent growth in the sdp1 mutant was not significant during early seedling germination (3 days, Fig. 1D) but became evident at a later stage (10 days, Fig. 1E) under dark conditions. We further analyzed the oil body-peroxisome interaction by using 3-day-old dark-grown seedlings in the presence or absence of sucrose to minimize the effects of the tissue stage on organelle appearance, such as differences in cell volume.
Sucrose Deficiency Caused by the Mutation in SDP1 Enhances the Physical Interaction between Oil Bodies and Peroxisomes
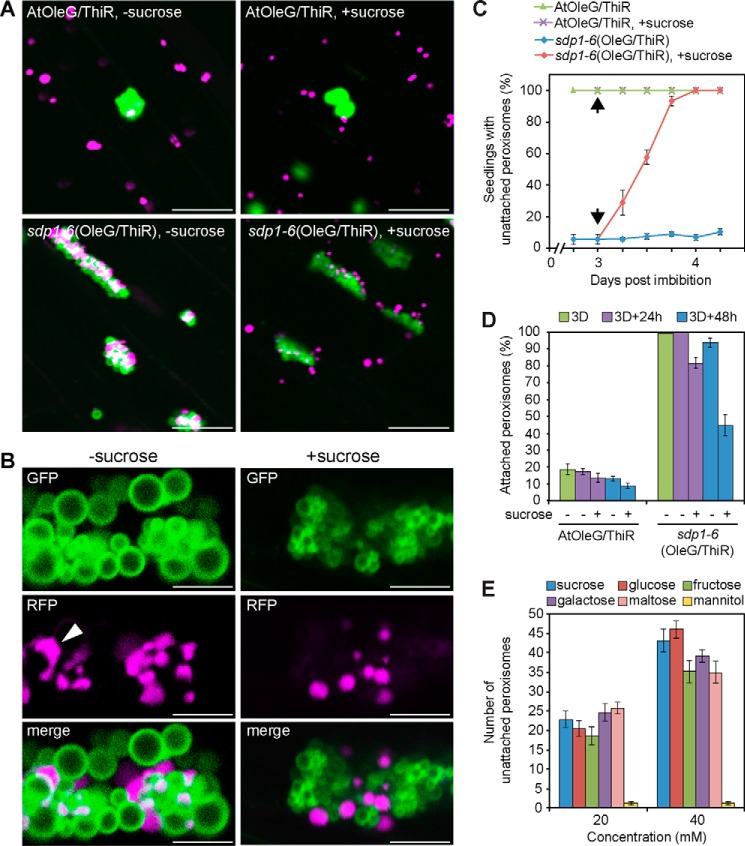
Both oil bodies and peroxisomes are involved in lipid metabolism, during which oil bodies deliver fatty acids to peroxisomes (20). Therefore, to elucidate the correlation between lipid metabolism and the dynamic behavior of oil bodies and peroxisomes, we established transgenic Arabidopsis, AtThiR, containing a chimeric gene encoding a fusion protein, ThiR, composed of the N terminus of peroxisomal 3-ketoacyl-CoA thiolase with peroxisomal targeting signal 2 and N-terminal red fluorescence protein (RFP) (25). Peroxisomes in AtThiR were visualized as red fluorescence. AtThiR was crossed with AtOleG and sdp1–6(OleG). In these plants, oil bodies and peroxisomes were visualized at the same time. They were designated AtOleG/ThiR and sdp1–6(OleG/ThiR). In AtOleG/ThiR seedlings, most peroxisomes were found in the cytosol (Fig. 2A, AtOleG/ThiR), and less than 20% of peroxisomes were attached to oil bodies (Fig. 2D). No significant difference was observed between AtOleG/ThiR seedlings grown in the presence or absence of sucrose (Fig. 2A). On the other hand, in sdp1–6(OleG/ThiR) seedlings grown without sucrose, nearly all peroxisomes were attached to oil bodies, forming oil body-peroxisome clusters (Fig. 2A). Interestingly, exogenous sucrose supplementation greatly reduced this attachment (Fig. 2, A–C). Sucrose did not affect the morphology of oil body aggregates in sdp1–6(OleG/ThiR) seedlings (Fig. 2A) at this stage, except that each oil body was generally smaller in the presence of sucrose (Fig. 2B). Oil body-peroxisome clusters in sdp1–6(OleG/ThiR) seedlings were observed in the presence or absence of sucrose (Fig. 2A); however, the direct membrane attachment between oil bodies and peroxisomes seemed largely reduced in the presence of sucrose (Fig. 2B). It is noted that peroxisomes were uniformly spherical in the presence of sucrose, whereas, in the absence of sucrose, those in clusters with oil bodies showed irregular and, in some cases, tubular shapes. Those elongated peroxisomes were frequently observed to surround oil body surfaces (Fig. 2B, arrowhead), forming a larger interaction interface.
FIGURE 2.
The enhanced physical interaction between oil bodies and peroxisomes in the sdp1 mutant was restored by exogenous sucrose application. A, oil bodies (green) and peroxisomes (magenta) were observed in the hypocotyl tissues of 3-day-old wild-type (AtOleG/ThiR) and sdp1–6 (sdp1–6(OleG/ThiR)) seedlings overexpressing OleG and ThiR grown in the absence (left panels) or presence (right panels) of sucrose. Scale bars = 20 μm. B, enlarged images of oil body-peroxisome clusters in sdp1–6(OleG/ThiR) that were grown under the same condition as in A in the absence (left panels) or presence (right panels) of sucrose. The arrowhead indicates an elongated peroxisome. Scale bars = 5 μm. C, quantification of seedlings exhibiting unattached peroxisomes in the hypocotyl. Unattached peroxisomes are represented by the appearance of peroxisomes in the cytosol. AtOleG/ThiR and sdp1–6(OleG/ThiR) seedlings grown in the absence of sucrose for 3 days were transferred to growth medium containing 2% sucrose (arrows) and observed under a microscope at 6-h intervals. Data represent the mean ± S.E. of three biological replicates (n = 60). D, percentage of oil body-attached peroxisomes in AtOleG and sdp1–6(OleG) before and after sucrose induction. The seedlings were grown without sucrose for 3 days (3D) and transferred to growth medium without (−) or with (+) 2% sucrose for 24 and 48 h. Data represent the mean ± S.E. of two biological replicates (n = 15). E, the effect of various sugars on the interaction. Shown is the quantification of peroxisomes that are dispersed in the cytosol from 3-day-old sdp1–6 (OleG/ThiR) seedlings grown on growth medium containing various sugars, including the metabolizable sugars sucrose, glucose, fructose, galactose, and maltose, and an unmetabolizable sugar, mannitol, under dark conditions. Data represents the mean ± S.E. of 20 samples.
To quantify the response of sdp1–6 seedlings against sucrose in relation to oil body-peroxisomal interaction, 3-day-old sdp1–6(OleG/ThiR) seedlings grown without sucrose were transferred to growth medium containing 2% sucrose. After induction, peroxisomal separation from oil bodies occurred in 20% of seedlings within 6 h and was eventually observed in all seedlings within 18–24 h (Fig. 2C). Almost no peroxisomes were observed in the cytosol in sdp1–6(OleG/ThiR) seedlings initially grown without sucrose (Fig. 2D). A total of 20% and 45% of peroxisomes appeared in the cytosol after 24 and 48 h of sucrose incubation, respectively (Fig. 2D). In contrast, all AtOleG/ThiR seedlings grown without sucrose showed the presence of unattached peroxisomes in the cytosol (Fig. 2C). Those unattached peroxisomes dispersed in the cytosol constituted about 80% of total peroxisomes (Fig. 2D). To rule out the possibility that sucrose-induced dissociation between oil bodies and peroxisomes might be a result of osmotic stress, we also tested the effect of various sugars, including mannitol, which is a reduced form of glucose and is used ordinarily as a control for osmotic pressure (Fig. 2E). sdp1–6(OleG/ThiR) seedlings were germinated on various sugars for 3 days under dark conditions, followed by quantification of peroxisomes. sdp1–6(OleG/ThiR) seedlings grown on metabolizable sugars showed dispersed peroxisomes in the cytosol with a similar level as those grown on sucrose, except that fructose, galactose, and maltose exhibited slightly lower levels than sucrose and glucose at 40 mm. By contrast, seedlings grown on an equivalent concentration of mannitol were deficient in those unattached peroxisomes and showed all peroxisomes clustering with oil bodies, indicating that the effect of sugars on the dissociation of peroxisomes from oil bodies is not due to the osmotic pressure but by the metabolizable sugars. These results show that both endogenous sucrose production in plants and exogenous sucrose application suppress physical interaction between oil bodies and peroxisomes.
To check whether there is a physical tethering between oil bodies and peroxisomes, we isolated oil bodies from sdp1–6(OleG/ThiR) seedlings and tested for the presence of peroxisomes. It has been known that oil bodies could be easily isolated by centrifugation of the crude extract prepared from etiolated cotyledons. As a result, oil bodies isolated from sdp1–6(OleG/ThiR) seedlings grown without sucrose contained significantly higher numbers of peroxisomes compared with those grown in the presence of sucrose (Fig. 3A). A Western blotting experiment showed the presence of a larger quantity of peroxisomal membrane (ascorbate peroxidase) and matrix (3-ketoacyl CoA thiolase) proteins in the oil body fraction isolated from sdp1–6(OleG/ThiR) seedlings grown without sucrose (Fig. 3B). These results indicate the presence of physical tethering between oil bodies and peroxisomes in seedlings grown without sucrose.
FIGURE 3.
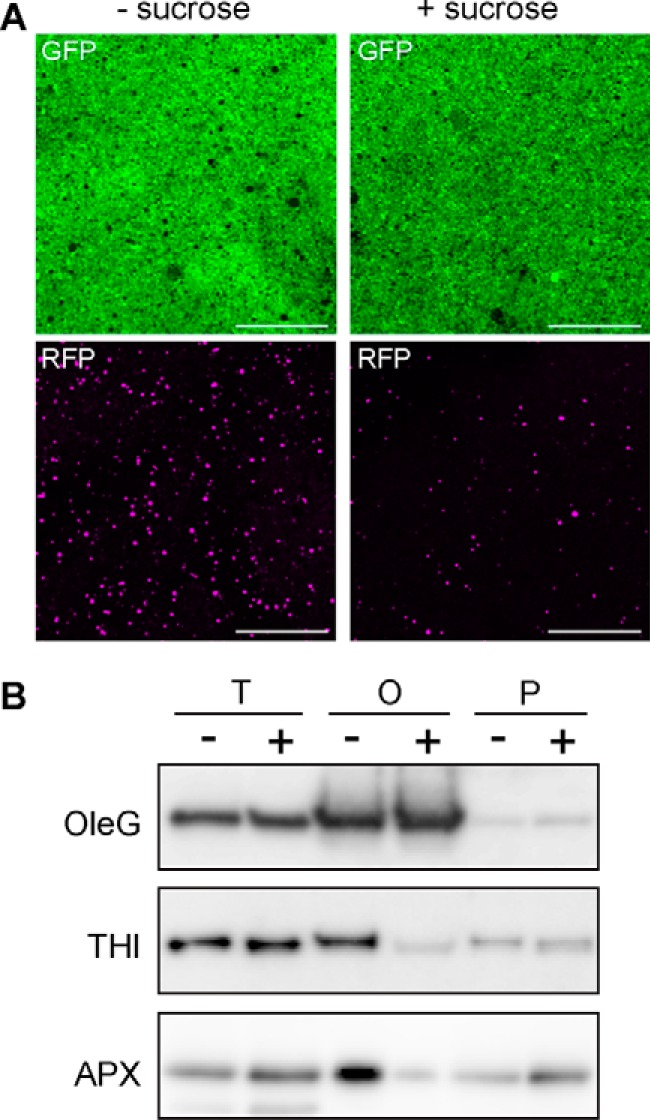
Quantification of peroxisomes in the isolated oil body fraction of the sdp1 mutant. A, oil bodies were isolated from 3-day-old sdp1–6(OleG/ThiR) seedlings grown with or without 2% sucrose and subjected to microscopic observation. Note that more peroxisomes were co-isolated with oil bodies from sdp1–6(OleG/ThiR) mutants grown without sucrose. Scale bars = 50 μm. B, detection of peroxisomal proteins by immunoblot analysis in the oil body fraction isolated from sdp1–6(OleG/ThiR) seedlings grown in the absence (−) or presence (+) of 2% sucrose. T, O, and P represent cell lysate, oil body, and pellet fraction, respectively. OleG, thiolase (THI), and ascorbate peroxidase (APX) were used as protein markers for the oil body membrane, peroxisomal matrix, and membrane, respectively.
Actin Filament-dependent Dissociation of Oil Bodies and Peroxisomes
Actin filaments drive the movement of various organelles, including peroxisomes in plants (28). Therefore, we investigated whether actin filaments are involved in the sucrose-induced dissociation of peroxisomes from oil bodies. Three-day-old sdp1–6(OleG/ThiR) seedlings initially grown without sucrose were transferred to sucrose medium supplemented with or without latrunculin B (LATB), an inhibitor of actin polymerization. As shown in Fig. 4A, LATB treatment negatively affected sucrose-induced peroxisomal dissociation from oil bodies in a concentration-dependent manner. Notably, 0.1 μm LATB completely abolished the sucrose-induced appearance of peroxisomes in the cytosol (Fig. 4B), indicating that peroxisomal dissociation from oil bodies requires peroxisome mobility mediated by actin filaments.
FIGURE 4.
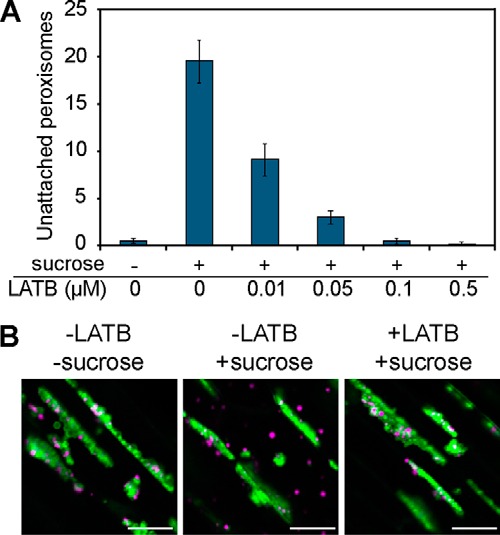
Sucrose-induced dissociation of peroxisomes from oil bodies requires actin filaments. A, 3-day old sdp1–6(OleG/ThiR) seedlings grown without sucrose were subjected to sucrose induction (+) for 24 h with various concentrations of LATB. The y axis indicates the number of peroxisomes detected in the cytosol of hypocotyl tissue. As a negative control, sucrose (−) was removed during induction. Data represent the mean ± S.D. (n = 14). B, microscopic images of oil bodies and peroxisomes after the sucrose induction experiment in A with or without 0.1 μm LATB in the sdp1–6(OleG/ThiR) mutant. Scale bars = 20 μm.
Exogenous Sucrose Reduces Oil Body-Peroxisome Interaction and Delays Storage Lipid Breakdown in Wild-type Germinating Seedlings
Next we tested whether the suppressive effect of sucrose on the oil body-peroxisome interaction observed in sdp1 also occurs in the wild type. Because oil bodies were reduced too rapidly in the hypocotyl tissues of the wild type during germination, we instead observed cotyledon tissues. Wild-type AtOleG/ThiR seeds were germinated on medium in the dark in the presence or absence of 2% sucrose, and epidermal cells were investigated. 2 days after germination, nearly all peroxisomes were attached to oil bodies in seedlings grown without sucrose (Fig. 5, A and C). Peroxisomes gradually disassociated from oil bodies from 3 days post-germination (Fig. 5, B and C). In contrast, large numbers of peroxisomes were observed in the cytosol, remaining unattached to oil bodies in the presence of sucrose (Fig. 5, A and C). The population of peroxisomes in physical association with oil bodies was increased 3 days post-germination compared with that 2 days post-germination and decreased 4 days after germination (Fig. 5, B and C). In general, the interaction rate between oil bodies and peroxisomes in the presence of sucrose was lower than in the absence of sucrose along germination, with a significant difference occurring 2 days post-germination. These data indicate that sucrose negatively regulates the physical interaction between oil bodies and peroxisomes in wild-type plants.
FIGURE 5.
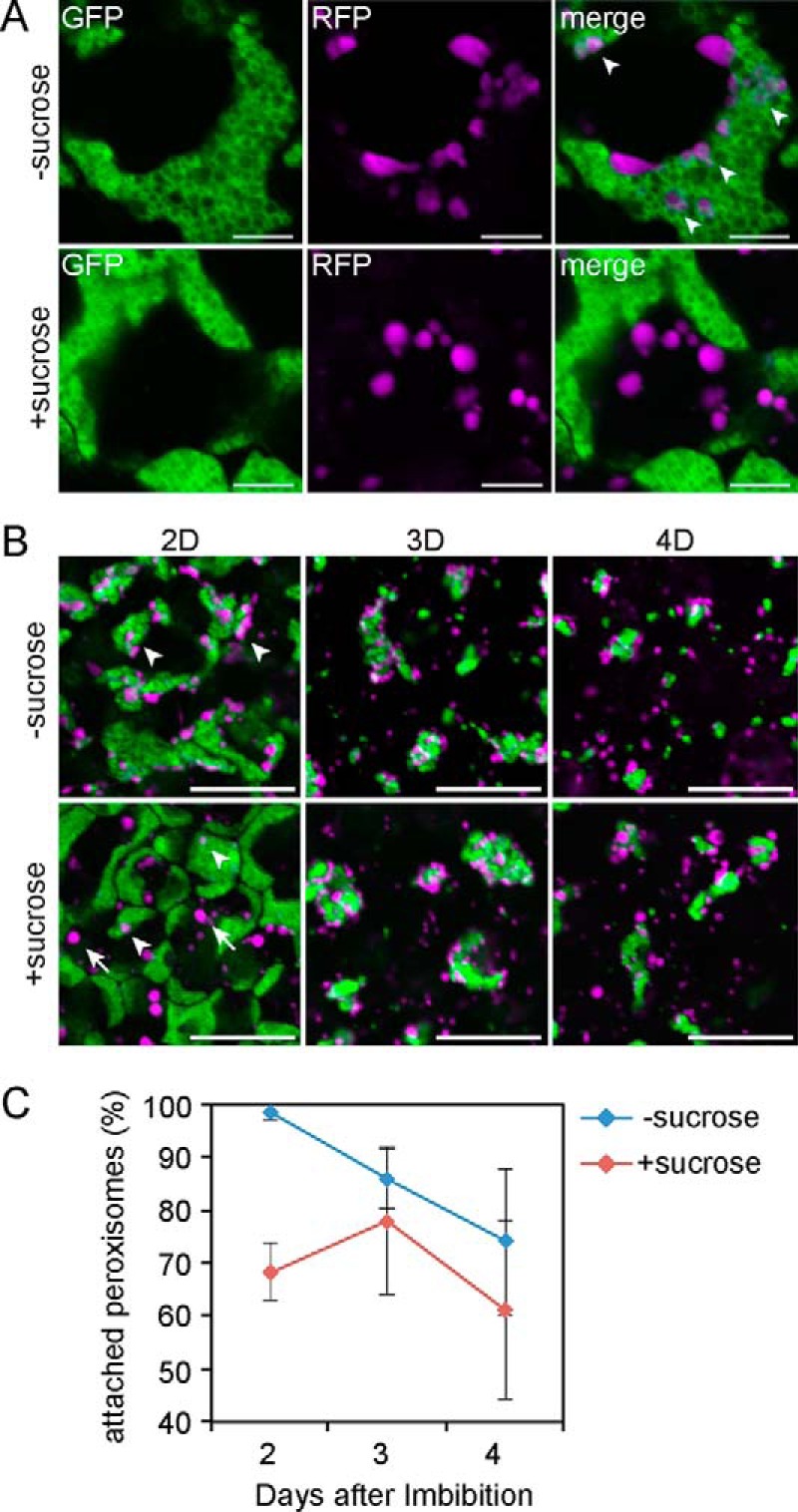
Sucrose effect on the interaction between oil bodies and peroxisomes in wild-type germinating seedlings. A, fluorescent images of cotyledon tissues from 2-day-old AtOleG/ThiR seedlings grown in the dark in the absence (top panels) or presence (bottom panels) of 2% sucrose. Scale bars = 5 μm. B, time-laps fluorescent images of cotyledon tissues of AtOleG/ThiR seedlings grown in the dark in the absence (top panels) or presence (bottom panels) of 2% sucrose for 2 (2D), 3 (3D), and 4 days (4D). Scale bars = 20 μm. C, percentage of oil body-attached peroxisomes among total peroxisomes in the cotyledon tissues of AtOleG/ThiR seedlings grown under the same conditions as in B. Data represent the mean ± S.E. of 15 samples. Arrowheads and arrows indicate oil body-attached and -unattached peroxisomes, respectively.
Enhanced Oil Body-Peroxisome Interaction Is Also Observed in the Sucrose-defective ped1 Mutant but Not in the ped3 Mutant
Like the sdp1 mutant, peroxisomal mutants defective in peroxisome biogenesis or oxidation of fatty acids also exhibit a defect in lipid breakdown (38). Therefore, we examined whether two peroxisomal mutants, ped1 and ped3 (20, 24), would, like sdp1, reveal a link between lipolysis and oil body-peroxisome attachment. The PED3 gene, also known as CTS/PXA1, which shares homology with human adrenoleukodystrophy (ALD) genes (39), encodes an ATP-binding cassette transporter on the peroxisomal membrane and transfers fatty acids into peroxisomes (20). PED1 encodes 3-ketoacyl CoA thiolase, which catalyzes the last step of peroxisomal fatty acid β-oxidation inside peroxisomes (26). Previous reports showed that both mutants display sucrose-dependent germination because of defects in their ability to break down TAGs during germination (24).
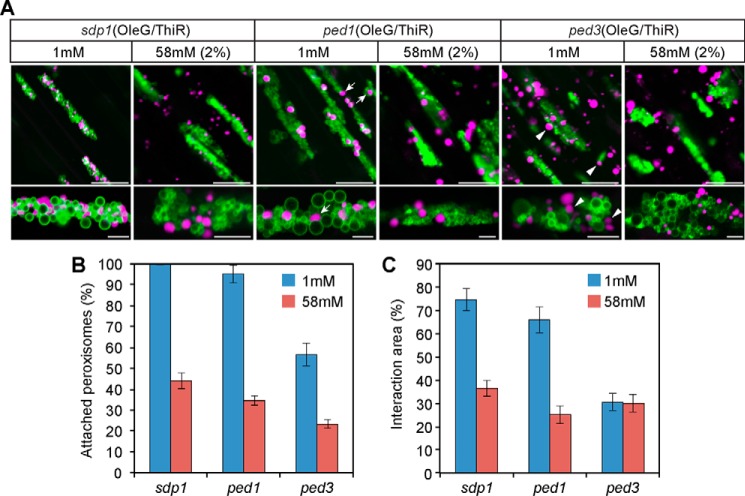
To visualize oil bodies and peroxisomes in these mutants, we crossed each loss-of-function mutant with AtOleG/ThiR, and homozygous progenies were designated ped1(OleG/ThiR) and ped3(OleG/ThiR). Because ped1(OleG/ThiR) and ped3(OleG/ThiR) did not germinate without sucrose, we added 1 mm sucrose to the growth medium to support germination of these seedlings. Under this condition, all peroxisomes were attached to oil bodies in sdp1–6(OleG/ThiR) (Fig. 6, A and B). Enlarged peroxisomes were observed in ped1/kat2 (Fig. 6A), which is consistent with our previous report (33). In ped1(OleG/ThiR), nearly all peroxisomes were attached to oil bodies under 1 mm sucrose (Fig. 6, A and B). Exogenous supplementation of 58 mm (2%) sucrose also reduced the physical interaction between oil bodies and peroxisomes (Fig. 6, A and B), which is similar to the findings for sdp1–6(OleG/ThiR). It should be noted that the peroxisomes in clusters with oil bodies in ped1(OleG/ThiR) under 1 mm (Fig. 6A, arrowheads) were more spherical than those in sdp1–6(OleG/ThiR) under the same condition. The membrane interaction interface was quantified in the oil body-peroxisome clusters between two sucrose conditions (Fig. 6C). Both ped1(OleG/ThiR) and sdp1–6(OleG/ThiR) showed a significantly reduced interaction interface between oil bodies and peroxisomes under 58 mm sucrose compared with 1 mm (Fig. 6C). Although the level of interaction was slightly lower in ped1(OleG/ThiR) than in sdp1–6(OleG/ThiR) under 1 mm sucrose (Fig. 6, B and C), our data show that both SDP1 and PED1 suppress oil body-peroxisomal interaction through sucrose production.
FIGURE 6.
PED3, but not PED1, is required for physical attachment between oil bodies and peroxisomes under sucrose-limited conditions. A, oil bodies and peroxisomes in the hypocotyl tissues of 3-day-old sdp1–6(OleG/ThiR), ped1(OleG/ThiR), and ped3(OleG/ThiR) seedlings grown in the presence of 1 or 58 mm (corresponds to 2%) sucrose. Bottom panels, magnified images of oil body-peroxisome aggregates for each top panel. Arrows and arrowheads indicate peroxisomes that are loosely bound to oil body membranes in ped1(OleG/ThiR) and ped3(OleG/ThiR), respectively. Scale bars = 20 μm (top panels) and 5 μm (bottom panels). B, quantitative analysis of oil body-attached peroxisomes in the hypocotyl tissues of each mutant. The same growth condition was used as in A. Data represent the mean ± S.E. (n = 15). C, quantitative analysis of the interaction area between oil bodies and peroxisomes in hypocotyl tissues. The same growth condition was used as in A. Data represent the mean ± S.E. (n = 15).
Interestingly, ped3(OleG/ThiR) exhibited significantly reduced oil body-peroxisome attachment compared with sdp1–6(OleG/ThiR) and ped1(OleG/ThiR). Under 1 mm sucrose, unlike sdp1–6(OleG/ThiR) and ped1(OleG/ThiR), half of the peroxisomes remained unattached to oil bodies in ped3(OleG/ThiR) (Fig. 6, A and B). In addition, peroxisomes in clusters with oil bodies in ped3(OleG/ThiR) under 1 mm sucrose (Fig. 6A, arrowheads) remained largely unattached to oil bodies, as did those in plants grown in the presence of 58 mm (2%) sucrose (Fig. 6). These results indicate that PED3 is involved in oil body-peroxisome tethering under sucrose-limited conditions.
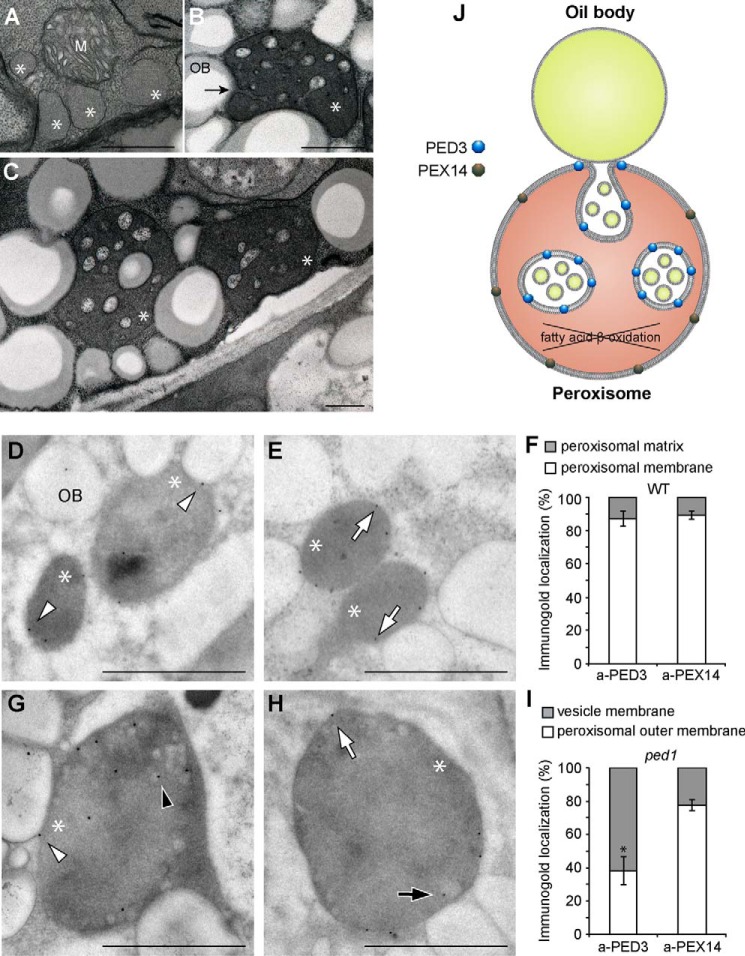
PED3 Localization on Specialized Membranes of Peroxisomes in the ped1 Mutant
We previously found that the enlarged peroxisomes in ped1 contain small vesicle-like structures (Fig. 7, B, C, G, and H) (33). Those structures result from peroxisomal membrane invagination from membrane interaction sites with oil bodies (Fig. 7B, arrow) (33). Ultramicroscopic observation of serial sections revealed that the vesicle-like structures are connected with each other, forming membrane continuity inside peroxisomes. Closer investigation of the content of vesicle-like structures showed that they encompass tiny vesicles that encircle materials with a similar election density as oil body-located storage lipids (Fig. 7, B, C, and J) (33). The membranes of these tiny vesicles showed a structural resemblance to those of oil bodies that are surrounded by phospholipid monolayer membranes (33), suggesting that the tiny vesicles within small vesicle-like structures might originate from oil bodies. An immunoelectron microscopic experiment using antibody raised against PED3 showed that PED3 was localized both on the membranes of vesicle-like structures (Fig. 7G, black arrowhead) and the peroxisomal outer membranes (Fig. 7G, white arrowhead) in the ped1 mutant. Quantification analysis on the location of gold particles shows that PED3 was localized significantly more on the membranes of vesicle-like structures than on peroxisomal outer membranes (62% versus 38%) (Fig. 7I and supplemental Table 1). We also tested the membrane localization of PEX14 that functions in peroxisomal protein import machinery (40). We previously reported the subcellular localization of PEX14 on the peroxisomal membrane using a biochemical experiment (40). In the ped1 mutant, we detected PEX14 on both the membranes of internalized vesicle-like structures and peroxisomal outer membranes (Fig. 7H). However, in contrast to PED3, a significant amount of PEX14 (78%) was detected on the peroxisomal outer membranes (Fig. 7, H, white arrow, and I, and supplemental Table 1), suggesting different targeting behavior of PED3 and PEX14 to the two types of peroxisomal membranes.
FIGURE 7.
Membrane localization of PED3 in the wild type and the ped1 mutant. A–C, peroxisomes in the etiolated cotyledons of 5-day-old wild-type (A) and ped1 mutant (B and C) seedlings. The arrow indicates membrane invagination of the enlarged peroxisomes at the interaction site with an oil body. Scale bars = 0.5 μm. M and OB indicate mitochondrion and oil body, respectively. Asterisks indicate the peroxisomes. D, E, G, and H, immunogold labeling of ultrathin sections of etiolated cotyledons from the wild type (D and E) and the ped1 mutant (G and H). Immunogold particles were coated with antibodies raised against PED3 (D and G) and PEX14 (E and H). Scale bars = 0.5 μm. Arrowheads and arrows indicate PED3- and PEX14-coated gold particles, respectively, on peroxisomal outer membranes (white) and vesicle membranes (black). F and I, quantitative analysis of the membrane localization of PED3 and PEX14 in the wild type (F) and ped1 mutant (I) based on immunogold labeling. The y axis represents the percentage of immunogold localization on different parts of peroxisomes. Data are derived from supplemental Table 1 and represent the mean ± S.D. of 10 sections (see also supplemental Table 1). * indicates a statistically significant difference between PED3 and PEX14 localization (p < 0.01, Student's t test). J, schematic showing the enrichment of PED3 and PEX14 on different membranes of enlarged peroxisomes in ped1.
Compared with the ped1 mutant, wild-type seedlings contain smaller peroxisomes and rarely show internal vesicle-like structures (Fig. 7, A, D, and E) (28, 33). Those internalized peroxisomal membrane structures are probably too transient to be observed in the wild type. We also performed immunogold electron microscopic experiment to detect the localization of PED3 and PEX14 in wild-type plants (Fig. 7, D–F). In this case, both PED3 and PEX14 were exclusively detected on the peroxisomal outer membranes (87% and 89%, respectively) (Fig. 7, D–F, and supplemental Table 1), consistent with our previous reports on the peroxisomal membrane localization of PED3 and PEX14 (19, 40). These data show that PED3 and PEX14 have a similar localization in the wild type but not in the ped1 mutant. It also indicates that the membrane property of vesicle-like structures and outer peroxisomal membranes might be not completely the same. The specific enrichment of PED3 on the membranes of vesicle-like structures that are derived from peroxisomal membrane invagination suggests that PED3 may initially act at the oil body-peroxisomal interaction site.
Discussion
Mutant screening aimed at the identification of the oil body regulator allowed us to identify a major TAG lipase, SDP1, in Arabidopsis. Here we report that oil bodies are highly accumulated in the identified sdp1 mutant because of the defect in TAG breakdown by sdp1 mutation. More importantly, we found abnormal organelle interaction of oil bodies and peroxisomes in the sdp1 mutant and thus revealed the unanticipated biological role of SDP1 in the physical interaction of oil bodies and peroxisomes.
Peroxisomes have been reported to interact with various organelles, including the ER, chloroplasts, and oil bodies (33, 41, 42). In addition, expression of chimeric peroxisomal membrane-bound ascorbate peroxidase in BY-2 cells resulted in oligomerization of peroxisomal membranes, leading to the formation of peroxisomal aggregates, a process known as membrane “zippering” (43, 44). It seems that energy starvation in cells signals peroxisomal movement to oil bodies in yeasts. It was shown that, when yeast cells were starved on oleate medium, more peroxisomes attached to oil bodies (31), which was accompanied by oil bodies being enwrapped and/or penetrated by peroxisomal membranes. Direct membrane interaction was also observed in the Arabidopsis ped1 mutant lacking peroxisomal fatty acid β-oxidation, whereas, in this case, in contrast to yeasts, invasion of peroxisomal and oil body membranes was observed (Fig. 7, B, C, and J) (33). A higher tendency of oil body-peroxisome interaction upon sucrose deprivation is clearly observed in our study of the Arabidopsis wild type and sucrose-defective mutants, indicating a common linkage of cellular sucrose homeostasis with the oil body-peroxisome interaction between plants and yeast. Although peroxisomal membrane proteins, including PEXs, are widely conserved among organisms (45), oil body membrane proteins largely vary between plants and mammals (11–13). Therefore, the conservation and divergence of molecular mechanisms underlying the oil body-peroxisome interaction remain to be elucidated.
Peroxisomes become largely mobile in the cytosol following germination (46), when Arabidopsis seedlings initiate mobilization of cellular storage lipids (18, 47), a process dependent on SPD1 function. Strikingly, our data showed that all peroxisomes were clustered to oil bodies and thus immobilized in the sdp1 mutant. Blockage of lipid mobilization by a defect in peroxisomal fatty acid β-oxidation, revealed by the ped1 mutant, similarly affected the interaction. This clearly shows that lipid mobilization mediated by SDP1 and PED1 suppresses physical interaction between oil bodies and peroxisomes. It is thus assumed that two organelles were initially clustered in seeds that lack significant lipid mobilization activity and gradually separate during seedling growth. Indeed, we observed the gradual decrease of the interaction along germination in Arabidopsis cotyledons (Fig. 5). The biological significance of oil body-peroxisome clusters is unclear. A generally accepted hypothesis is that the interaction is involved in direct fatty acid delivery from oil bodies to peroxisomes, benefitting lipid mobilization for seedling growth and development. In our observations, exogenous supplementation of 58 mm sucrose to the seedlings largely inhibited the clustering of oil bodies and peroxisomes and almost completely blocked storage lipid breakdown on 2 day after germination (Figs. 1F and 5). Interestingly, the lipid breakdown was recovered 1 day later, together with the increase in oil body-peroxisome clusters (Fig. 1F and 5), suggesting that the interaction might be important for the lipid degradation process.
Notably, our study clearly shows that sucrose, either produced in plants or exogenously supplied, leads peroxisomes to separate from oil bodies. The exogenously supplied sugars suppressing the interaction are limited to metabolizable sugars but not a non-metabolizable sugar, mannitol, suggesting that sugars act either directly as a signaling molecule or a carbon metabolite. Previous reports showed that the sugars act as signaling molecules to suppress storage lipid mobilization and starch degradation (48–50). For example, 140 mm glucose, but not sorbitol, blocked the degradation of storage lipids along germination. In addition, sucrose application on young seedlings of Arabidopsis had little impact on the expression level of genes involved in fatty acid β-oxidation and the glyoxylate cycle (51), suggesting that sugar-induced suppression of lipid degradation occurs through a still unknown mechanism. In our experiment, 58 mm (equivalent to 2%) sucrose also delayed the degradation process of storage lipids in wild-type germinating seedlings (Fig. 1F). It should be noted that the overall physical interaction between oil bodies and peroxisomes was also reduced by sucrose application at this stage (Fig. 5). By taking advantage of the inhibitory effect of a high concentration of sugars on seedling growth, forward genetic screening was carried out and identified a number of sucrose-insensitive mutants showing insensitivity toward the growth-inhibitory effect of sucrose or glucose, such as sis and gin mutants (52, 53). One of those identified mutants that is insensitive to abscisic acid, sis5, was shown to be resistant against glucose-induced inhibition of storage lipid mobilization (49). Whether sucrose-induced separation of oil bodies and peroxisomes acts through these sucrose signaling components remains to be tested. Such analyses may also be useful to understand the direct link between sucrose signaling and membrane communication between oil bodies and peroxisomes.
Identification of membrane anchor proteins is the key to find out the biological significance of the oil body-peroxisomal interaction. We show that, unlike sdp1 and ped1, the ped3 mutant has a deficiency in sucrose deprivation-induced oil body-peroxisome interaction (Fig. 6), indicating the positive role of PED3 on the oil body-peroxisome interaction. Subcellular localization of PED3 on specialized peroxisomal membranes that originate from the oil body-peroxisome membrane junction in the ped1 mutant (Fig. 7J) suggests that PED3 may initially function on the membrane interaction site. This raises the possibility that PED3 may act as an anchor protein on the membrane junction between oil bodies and peroxisomes. Alternatively, although we have not investigated it, the cytosolic accumulation of TAG-derived fatty acid derivatives that resulted from ped3 mutation may act as a direct or indirect signal to suppress the interaction. Nevertheless, if PED3 is an anchor protein, then the ped3 mutant would be an ideal genetic tool to investigate the biological importance of the interaction of the organelles. However, because of the biological role of PED3 as an ATP-binding cassette transporter (19), it is impossible in the ped3 mutant to dissect the importance of the oil body-peroxisome interaction from the fatty acid delivery function of PED3 in the context of fatty acid transport. Therefore, the identification of additional compounds that exclusively target the membrane-membrane interaction of oil bodies and peroxisomes is necessary to elicit the molecular mechanism and biological significance underlying the physical interaction between oil bodies and peroxisomes.
Removal of peroxisomes from oil bodies enables peroxisomes to actively interact with chloroplasts, a step that is involved in photorespiration to generate carbohydrates for plant growth after plants run out of energy reserves upon germination (42, 54). During this process, the mobilization of leaf peroxisomes to chloroplasts requires actin filaments. Our data suggest that detachment of peroxisomes from oil bodies and the subsequent release into the cytosol are also dependent on the activity of actin filaments, indicating an important role of actin filaments for peroxisomal relocation during lipolysis. Meanwhile, trapping peroxisomes to oil bodies, either by inhibiting actin filaments or by limiting sucrose production in sdp1 and ped1 mutants, apparently limits peroxisomal mobility and, thus, may delay the subsequent process of photorespiration requiring peroxisome-chloroplast interaction. It will be interesting to test whether and how the oil body-stacked peroxisomes in sdp1 (or ped1) affect plant photorespiration after light exposure.
Delaying lipid breakdown is of great importance for practical applications for increasing oil production in plants (55), and most mutants impaired in lipolysis exhibit severe germination problems. Modulating the anchor protein of peroxisomes and oil bodies can potentially manipulate the oil degradation process. Therefore, our results increase the number of potential targets for future biological approaches aimed at increasing plant oil yields.
Experimental Procedures
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia was used as the wild type. The generation of transgenic plants overexpressing OleG was described previously (36). Peroxisomes in Arabidopsis were visualized by stably expressing a chimeric gene, ThiR, encoding a fusion protein composed of the N terminus extrapeptide of Arabidopsis 3-ketoacyl-CoA thiolase (PED1) and monomeric red fluorescent protein 1 (mRFP1) (25). The N terminus of PED1 was amplified from the cDNA (25) by PCR using two primers: B1-AtThi (5′-AAAAAGCAGGCTCAATGGAGAAAGCGATCGAGAGA-3′) and PFRm-ihTtA (5′-AGTGCTGCATATCAGAGGATGGCCTCCTCCGAGGAC-3′). A DNA fragment derived from mRFP1 was amplified by PCR using two primers: B1-AtThi-mRFP (5′-AGTGCTGCATATCAGAGGATGGCCTCCTCCGAGGAC-3′) and mRFP-B2 (5′-AGAAAGCTGGGTTTTAGGCGCCGGTGGAGTGGCG-3′). The chimeric gene, ThiR, was constructed by PCR using these two DNA fragments and two primers: B1-AtThi and mRFP-B2. The ThiR DNA fragment was cloned in pDONR221 and transferred into Ti plasmid pH7WG2 (purchased from Gent University). The resulting Ti plasmid was introduced into Agrobacterium (strain C58C1rif) by electroporation. Transgenic plants overexpressing ThiR were generated by the floral dip method, an Agrobacterium-mediated transformation system (56). Peroxisomes and oil bodies were visualized in wild-type and sdp1 plants by crossing the wild type and sdp1 overexpressing OleG to ThiR-expressing plants. The ped1 and ped3–3 mutants were described previously (19, 24). To visualize oil bodies and peroxisomes in ped1 and ped3–3, these plants were crossed with transgenic plants expressing OleG and ThiR, and homozygous lines were selected from the F3 generation showing both sucrose dependence and green and red fluorescence in all seedlings.
Seeds were sown on growth medium (0.8% (w/v) agar, half-strength Murashige-Skoog salts) with or without 2% (w/v) sucrose. To test the effects of other sugars, various sugars were added to the growth medium to a final concentration of 20 or 40 mm. Germination was induced under continuous light conditions for 12 h at 22 °C following incubation in the dark for 48 h at 4 °C. In the experiment using actin filament inhibitor, 3-day-old sdp1 seedlings overexpressing OleG and ThiR grown on sucrose-free growth medium under dark conditions were transferred to liquid growth medium (half-strength Murashige-Skoog salts) with 100 mm sucrose containing LATB at the final concentrations indicated in the figures, followed by incubation for 24 h. Because LATB was dissolved in DMSO, the control plants were treated with the same amount of DMSO in liquid growth medium.
Isolation of sdp1 Mutants
Seeds from transgenic plants overexpressing the OleG fusion gene were mutagenized by ethyl methane sulfonate as described previously (36). M2 seeds were grown vertically on growth medium without sucrose for 3 days under dark conditions. GFP signals were observed in hypocotyl tissues under an epifluorescent binocular microscope (SteREO Lumar, Carl Zeiss) to enable the morphology of oil bodies to be compared with that of the wild type.
Map-based Cloning
To identify the responsible gene in the mutant, the F2 generation was analyzed by backcrossing the mutant with the wild type (Landsberg erecta accession). Homozygous mutant plants were isolated by GFP observation, which revealed oil body accumulation. Map-based cloning was conducted as described previously (40). Two simple sequence length polymorphism (SSLP) markers and one cleaved amplified polymorphic sequence (CAPS) marker were used in this study. The primer sequences used for the SSLP markers were 5′-TCTCCCCACTAGTTTTGTGTCC-3′ and 5′-GAAATCCAAATCCCAGAGAGG-3′ for At5g05100 and 5′-CCACTTGTTTCTCTCTCTAG-3′ and 5′-TATCAACAGAAACGCACCGAG-3′ for At5g03730. The primer sequences and restriction enzyme used for the CAPS marker for At5g01060 were TTGATATCTATCAATGGTGGAC and GCTAAGAATCAGTCCAAAATGC and EcoRV, respectively.
Fluorescence Microscopy
Fluorescent images were taken under an LSM510 laser-scanning confocal microscope (Carl Zeiss). Excitation wavelengths for GFP and RFP were 488 and 543 nm, respectively. For GFP and RFP signal detection, band pass filters of 505–530 and 560–615 nm were used, respectively.
Quantification of Peroxisomes
The number of peroxisomes was quantified using confocal fluorescence microscopic images. A fixed area of 15,000 μm2 was cropped in each image, and the number of peroxisomes, including those attached and unattached to oil bodies, was scored manually using ZEN 2010 software (Carl Zeiss). The interaction area of peroxisomes with oil bodies was quantified according to a method described previously (42). Images of clustered oil bodies and peroxisomes were derived from confocal microscopic images. A total of 15 aggregates were analyzed in each genotype. In each aggregate, the area of overlap between oil bodies and peroxisomes, which appeared as a white color because of overlap of the GFP and RFP signal in the image, and the total area of peroxisomes were scored using ImageJ (http://imagej.nih.gov/ij/). The interaction area (percent) was then calculated by dividing the area of overlap by the total area of peroxisomes.
Measurements of the size of peroxisomal aggregates were obtained from confocal fluorescence microscopic images of 3-day-old dark-grown seedlings. Images were derived from three independent experiments. The size of peroxisomal aggregates (n > 36) were automatically calculated using the Analyze Particles tool of ImageJ following application of the threshold for the GFP signal.
TAG Measurement
Seeds were sterilized, sown on growth medium, and incubated in the dark at 4 °C for 2 days. The seedlings were then transferred to a growth chamber under dark conditions at 22 °C and collected 0, 1, 2, 3, 4, and 5 days after germination. A total of 50 seedlings were collected in an Eppendorf tube. Then 100 μl of 5% Triton X-100 buffer was added, and the seeds were incubated at 80 °C for 3 min, followed by homogenization. An additional 200 μl of 5% Triton X-100 buffer was added to the sample, which was subjected to heat treatment at 80 °C for 2 min and cooled to room temperature. The heat treatment was repeated twice. After thorough mixing, each 20-μl sample was mixed with the reagent provided in the triglyceride E-test kit (Wako Pure Chemical, Osaka, Japan) and incubated at 37 °C for 3 min, followed by centrifugation at 15,000 × g for 5 min at 25 °C. The supernatant was subjected to the TAG assay according to the instructions of the manufacturer.
Immunoblot Analysis
Three-day-old seedlings were homogenized in sample buffer (0.4 m mannitol, 100 mm HEPES-KOH (pH 7.5), 10 mm KCl, 1 mm MgCl2, and 1 mm EDTA), followed by centrifugation at 100,000 × g for 20 min to separate the cell lysate into the oil body fraction (suspended on the surface of the solution) and the pellet fraction (at the bottom). Equal amounts of protein between samples in the same layer were separated by SDS-PAGE and transferred to a GVHP membrane (0.22 μm, Millipore), followed by immunoblot analysis as described previously (57). Antibodies raised against GFP (58), 3-ketoacyl-CoA thiolase (25), and ascorbate peroxidase (59) were used as primary antibodies at a 3:20,000 (v/v) dilution. Secondary antibody (horseradish peroxidase-conjugated anti-rabbit IgG, GE Healthcare) was applied at a 1:2000 (v/v) dilution. The reactive polypeptides were detected with Chemi-Lumi One reagent (Nacalai Tesque).
Electron Microscopic Analysis
The methods for sample preparation and detection by electron microscopic analysis were described previously (33). For immunoelectron microscopy, cotyledons from 5-day-old seedlings grown under dark conditions were frozen with a high-pressure freezing machine (HPM-010, Bal-Tec, Balzer, FL), followed by sectioning with a diamond knife ultramicrotome (EM UC7, Leica). Proteins in the samples were immunoreacted with primary antibodies against Arabidopsis PEX14 (1:500 (v/v)) (40) and PED3 (1:500 (v/v)) (19) for 1 h. The samples were then treated with protein A-gold (15 nm, BBInternational) as a secondary antibody for 30 min, washed with distilled water, and dried. Sections were stained with 4% uranyl acetate for 10 min at room temperature and examined under a transmission electron microscope (H-7650, Hitachi High-Tech Co.) at 80 kV. Measurements for the localization of gold particles were obtained from 10 EM pictures in each genotype with different antibodies, as shown in supplemental Table 1.
Author Contributions
S. C., M. H., and M. N. conceived and coordinated the study. M. H. performed the mutant screening. Y. H. and M. O. performed the immunoelectron microscopy experiments. K. O. helped with the image quantification analysis. S. C. performed the rest of the experiments. S. C., S. M., M. H., and M. N. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the staff of the Core Research Facilities and Model Plant Resource Facilities at the National Institute for Basic Biology for plant maintenance and technical support.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 25891029 to S. C. and 22120007 to M. N.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table 1.
- ER
- endoplasmic reticulum
- TAG
- triacylglycerol
- RFP
- red fluorescent protein
- LATB
- latrunculin B
- PEX
- peroxin.
References
- 1. Toulmay A., and Prinz W. A. (2011) Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 23, 458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hayashi T., Rizzuto R., Hajnoczky G., and Su T. P. (2009) MAM: more than just a housekeeper. Trends Cell Biol. 19, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K. F., Balla T., Mannella C. A., and Hajnóczky G. (2006) Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simmen T., Aslan J. E., Blagoveshchenskaya A. D., Thomas L., Wan L., Xiang Y., Feliciangeli S. F., Hung C. H., Crump C. M., and Thomas G. (2005) PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., and Daum G. (2008) Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283, 17065–17074 [DOI] [PubMed] [Google Scholar]
- 6. Tzen J., Cao Y., Laurent P., Ratnayake C., and Huang A. (1993) Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 101, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mansfield S. G., and Briarty L. G. (1992) Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can. J. Bot. 70, 151–164 [Google Scholar]
- 8. Brown L. A., Larson T. R., Graham I. A., Hawes C., Paudyal R., Warriner S. L., and Baker A. (2013) An inhibitor of oil body mobilization in Arabidopsis. New Phytol. 200, 641–649 [DOI] [PubMed] [Google Scholar]
- 9. Eastmond P. J. (2007) MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19, 1376–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lehner A., Corbineau F., and Bailly C. (2006) Changes in lipid status and glass properties in cotyledons of developing sunflower seeds. Plant Cell Physiol. 47, 818–828 [DOI] [PubMed] [Google Scholar]
- 11. Horn P. J., James C. N., Gidda S. K., Kilaru A., Dyer J. M., Mullen R. T., Ohlrogge J. B., and Chapman K. D. (2013) Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 162, 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R. V. Jr, and Walther T. C. (2013) Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell Proteomics 12, 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gidda S. K., Watt S., Collins-Silva J., Kilaru A., Arondel V., Yurchenko O., Horn P. J., James C. N., Shintani D., Ohlrogge J. B., Chapman K. D., Mullen R. T., and Dyer J. M. (2013) Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant Signal Behav. 8, e27141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tzen J. T. C. (2012) Integral proteins in plant oil bodies. ISRN Botany 2012, 1–16 [Google Scholar]
- 15. Tzen J. T., Peng C. C., Cheng D. J., Chen E. C., and Chiu J. M. (1997) A new method for seed oil body purification and examination of oil body integrity following germination. J. Biochem. 121, 762–768 [DOI] [PubMed] [Google Scholar]
- 16. Siloto R. M., Findlay K., Lopez-Villalobos A., Yeung E. C., Nykiforuk C. L., and Moloney M. M. (2006) The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell 18, 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayre B. G., Keller F., and Turgeon R. (2003) Symplastic continuity between companion cells and the translocation stream: long-distance transport is controlled by retention and retrieval mechanisms in the phloem. Plant Physiol. 131, 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eastmond P. J. (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayashi M., Nito K., Takei-Hoshi R., Yagi M., Kondo M., Suenaga A., Yamaya T., and Nishimura M. (2002) Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiol. 43, 1–11 [DOI] [PubMed] [Google Scholar]
- 20. Zolman B. K., Silva I. D., and Bartel B. (2001) The Arabidopsis pxa1 mutant is defective in an ATP-Binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127, 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- 21. Footitt S., Slocombe S. P., Larner V., Kurup S., Wu Y., Larson T., Graham I., Baker A., and Holdsworth M. (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 21, 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fulda M., Shockey J., Werber M., Wolter F. P., and Heinz E. (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant J. 32, 93–103 [DOI] [PubMed] [Google Scholar]
- 23. Hayashi H., De Bellis L., Hayashi Y., Nito K., Kato A., Hayashi M., Hara-Nishimura I., and Nishimura M. (2002) Molecular characterization of an Arabidopsis acyl-coenzyme a synthetase localized on glyoxysomal membranes. Plant Physiol. 130, 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayashi M., Toriyama K., Kondo M., and Nishimura M. (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kato A., Hayashi M., Takeuchi Y., and Nishimura M. (1996) cDNA cloning and expression of a gene for 3-ketoacyl-CoA thiolase in pumpkin cotyledons. Plant Mol. Biol. 31, 843–852 [DOI] [PubMed] [Google Scholar]
- 26. Germain V., Rylott E. L., Larson T. R., Sherson S. M., Bechtold N., Carde J. P., Bryce J. H., Graham I. A., and Smith S. M. (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J. 28, 1–12 [DOI] [PubMed] [Google Scholar]
- 27. Peng C. C., Lin I. P., Lin C. K., and Tzen J. T. (2003) Size and stability of reconstituted sesame oil bodies. Biotechnol. Prog. 19, 1623–1626 [DOI] [PubMed] [Google Scholar]
- 28. Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., and Nishimura M. (2002) Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43, 331–341 [DOI] [PubMed] [Google Scholar]
- 29. Jedd G., and Chua N. H. (2002) Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 43, 384–392 [DOI] [PubMed] [Google Scholar]
- 30. Rapp S., Saffrich R., Anton M., Jäkle U., Ansorge W., Gorgas K., and Just W. W. (1996) Microtubule-based peroxisome movement. J. Cell Sci. 109, 837–849 [DOI] [PubMed] [Google Scholar]
- 31. Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., and Goodman J. M. (2006) An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schrader M. (2001) Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J. Histochem. Cytochem. 49, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 33. Hayashi Y., Hayashi M., Hayashi H., Hara-Nishimura I., and Nishimura M. (2001) Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma 218, 83–94 [DOI] [PubMed] [Google Scholar]
- 34. Shimada T. L., Takano Y., Shimada T., Fujiwara M., Fukao Y., Mori M., Okazaki Y., Saito K., Sasaki R., Aoki K., and Hara-Nishimura I. (2014) Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 164, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shimada T. L., Shimada T., Takahashi H., Fukao Y., and Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55, 798–809 [DOI] [PubMed] [Google Scholar]
- 36. Hayashi M., Nanba C., Saito M., Kondo M., Takeda A., Watanabe Y., and Nishimura M. (2012) Loss of XRN4 function can trigger cosuppression in a sequence-dependent manner. Plant Cell Physiol. 53, 1310–1321 [DOI] [PubMed] [Google Scholar]
- 37. Athenstaedt K., and Daum G. (2003) YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278, 23317–23323 [DOI] [PubMed] [Google Scholar]
- 38. Nuttall J. M., Motley A., and Hettema E. H. (2011) Peroxisome biogenesis: recent advances. Curr. Opin. Cell Biol. 23, 421–426 [DOI] [PubMed] [Google Scholar]
- 39. Mosser J., Douar A. M., Sarde C. O., Kioschis P., Feil R., Moser H., Poustka A. M., Mandel J. L., and Aubourg P. (1993) Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature 361, 726–730 [DOI] [PubMed] [Google Scholar]
- 40. Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., and Nishimura M. (2000) AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19, 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R. L., and Rachubinski R. A. (2013) An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32, 2439–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oikawa K., Matsunaga S., Mano S., Kondo M., Yamada K., Hayashi M., Kagawa T., Kadota A., Sakamoto W., Higashi S., Watanabe M., Mitsui T., Shigemasa A., Iino T., Hosokawa Y., and Nishimura M. (2015) Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nature Plants 1, 15035. [DOI] [PubMed] [Google Scholar]
- 43. Mullen R. T., Lisenbee C. S., Flynn C. R., and Trelease R. N. (2001) Stable and transient expression of chimeric peroxisomal membrane proteins induces an independent “zippering” of peroxisomes and an endoplasmic reticulum subdomain. Planta 213, 849–863 [DOI] [PubMed] [Google Scholar]
- 44. Lisenbee C. S., Karnik S. K., and Trelease R. N. (2003) Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic 4, 491–501 [DOI] [PubMed] [Google Scholar]
- 45. Reumann S. (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135, 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mano S., Nakamori C., Nito K., Kondo M., and Nishimura M. (2006) The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 47, 604–618 [DOI] [PubMed] [Google Scholar]
- 47. Hernández M. L., Whitehead L., He Z., Gazda V., Gilday A., Kozhevnikova E., Vaistij F. E., Larson T. R., and Graham I. A. (2012) A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 160, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas B. R., and Rodriguez R. L. (1994) Metabolite signals regulate gene expression and source/sink relations in cereal seedlings. Plant Physiol. 106, 1235–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. To J. P., Reiter W. D., and Gibson S. I. (2002) Mobilization of seed storage lipid by Arabidopsis seedlings is retarded in the presence of exogenous sugars. BMC Plant Biol. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rognoni S., Teng S., Arru L., Smeekens S. C. M., and Perata P. (2007) Sugar effects on early seedling development in Arabidopsis. Plant Growth Regul. 52, 217–228 [Google Scholar]
- 51. Rylott E. L., Hooks M. A., and Graham I. A. (2001) Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana. Biochem. Soc. Trans. 29, 283–287 [DOI] [PubMed] [Google Scholar]
- 52. Zhou L., Jang J. C., Jones T. L., and Sheen J. (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. U.S.A. 95, 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gibson S. I., Laby R. J., and Kim D. (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem. Biophys. Res. Commun. 280, 196–203 [DOI] [PubMed] [Google Scholar]
- 54. Nishimura M., Yamaguchi J., Mori H., Akazawa T., and Yokota S. (1986) Immunocytochemical analysis shows that glyoxysomes are directly transformed to leaf peroxisomes during greening of pumpkin cotyledons. Plant Physiol. 81, 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fan J., Yan C., Roston R., Shanklin J., and Xu C. (2014) Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 26, 4119–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bechtold N., Ellis J., and Pelletier G. (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, Life Sciences 316, 1194–1199 [Google Scholar]
- 57. Cui S., Fukao Y., Mano S., Yamada K., Hayashi M., and Nishimura M. (2013) Proteomic analysis reveals that the Rab GTPase RabE1c is involved in the degradation of the peroxisomal protein receptor PEX7 (peroxin 7). J. Biol. Chem. 288, 6014–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mitsuhashi N., Shimada T., Mano S., Nishimura M., and Hara-Nishimura I. (2000) Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41, 993–1001 [DOI] [PubMed] [Google Scholar]
- 59. Yamaguchi K., Mori H., and Nishimura M. (1995) A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol. 36, 1157–1162 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.