Abstract
African trypanosomes and related kinetoplastid parasites selectively traffic specific membrane proteins to the flagellar membrane, but the mechanisms for this trafficking are poorly understood. We show here that KHARON, a protein originally identified in Leishmania parasites, interacts with a putative trypanosome calcium channel and is required for its targeting to the flagellar membrane. KHARON is located at the base of the flagellar axoneme, where it likely mediates targeting of flagellar membrane proteins, but is also on the subpellicular microtubules and the mitotic spindle. Hence, KHARON is probably a multifunctional protein that associates with several components of the trypanosome cytoskeleton. RNA interference-mediated knockdown of KHARON mRNA results in failure of the calcium channel to enter the flagellar membrane, detachment of the flagellum from the cell body, and disruption of mitotic spindles. Furthermore, knockdown of KHARON mRNA induces a lethal failure of cytokinesis in both bloodstream (mammalian host) and procyclic (insect vector) life cycle stages, and KHARON is thus critical for parasite viability.
Keywords: calcium channel, cytoskeleton, membrane protein, mitotic spindle, trafficking, Trypanosoma brucei
Introduction
African trypanosomes such as Trypanosoma brucei and related subspecies are parasitic protists that cause sleeping sickness in humans and the wasting disease nagana in cattle. Between 2010 and 2013 there were ∼7,000 new cases reported per year (1), although considerably larger numbers of people are at risk for infection (2), and disease in domestic livestock represents a major economic burden in impoverished regions of Africa. In addition to the medical and veterinary impact of these parasites, they also exhibit many fascinating and unusual biological features, and their study has led to novel discoveries such as glycosylphosphatidylinositol anchors for membrane proteins, trans-splicing of mRNAs, extensive editing of mitochondrial mRNAs, and global polycistronic transcription of gene arrays.
These parasites are flagellated in all life cycle stages, including the bloodstream forms (BFs)4 that infect the mammalian host and in all of the developmental stages that infect the tsetse fly vector. Given the multiple important roles that flagella play for trypanosomes, these cellular appendages have been studied extensively (3, 4). Although studies on axonemal and luminal proteins predominated originally, considerable work has been done more recently on proteins of the trypanosome flagellar membrane (5). One of the first such proteins identified was a receptor type adenylate cyclase (6) that is likely involved in sensing the extracellular environment and relaying signals via synthesis of cAMP, and recent work on this family of proteins has uncovered roles for evasion of host innate immunity (7) and in social motility in insect stage procyclic forms (PFs) (8–10). Furthermore, a variety of putative transporters and channels are present in the flagellar membrane (5), where they may play roles in nutrient or ion uptake and potentially in nutrient or solute sensing (11). Other important roles for the flagellar membrane are attachment to the salivary gland epithelium in the tsetse fly (12), flagellar adherence of gametes during mating (13), and attachment of the tip of the new flagellum to the body of the old flagellum during parasite replication, where attachment of flagella is required for cleavage furrow formation and proper cell division (14–16).
Although a host of proteins are known that are targeted selectively to the flagellar membrane of trypanosomes or related parasites such as Trypanosoma cruzi or Leishmania (17–20), the mechanisms for this selective surface targeting are largely obscure. In studies of the flagellar specific glucose transporter 1 from Leishmania mexicana, LmxGT1, our laboratory has identified a novel protein KHARON (KH),5 which interacts with the flagellar targeting signal and mediates trafficking of this permease to the flagellar membrane (21). Although this ∼57-kDa protein did not exhibit detectable sequence similarity to proteins outside the kinetoplastid parasites nor did it possess any recognizable conserved sequence motifs (21), the existence of orthologs of KH in all kinetoplastid parasites whose genomes have been sequenced suggested that this protein might be important for flagellar targeting of multiple membrane proteins across this family of parasites. However, to date the role of KH in other parasites has not been investigated. Furthermore, various genetic advantages of T. brucei, such as the ability to readily knock down mRNAs by inducible RNAi, as well as the existence of a flagellar membrane proteome (5), suggested that this parasite might represent a particularly facile system with which to study targeting of flagellar membrane proteins. In the present study, we have investigated KH from T. brucei (TbKH) and established that it is involved in flagellar trafficking of a putative calcium channel, TbCaCh (also called FS179) (5). Notably, RNAi-mediated knockdown of KH mRNA not only prevented flagellar trafficking of TbCaCh but was also lethal to both BF and insect stage PF parasites, resulting in detachment of flagella from the cell body via the flagellar attachment zone and failure of the parasites to undergo cytokinesis. Subcellular localization of KH demonstrated that this protein is located at the base of the flagellar axoneme, where it is likely to mediate import of integral membrane proteins into the flagellar membrane, but also on the subpellicular microtubule cytoskeleton and unexpectedly on the mitotic spindle. Hence KH is a central component of flagellar membrane transport machinery and also associates with multiple components of the trypanosome cytoskeleton, where it is likely to serve diverse functions in parasite cell biology. The results reported here establish T. brucei as an ideal experimental system in which to study the multiple functions of KH and in particular to dissect the mechanism whereby it mediates trafficking of integral membrane proteins to the flagellar membrane.
Results
Identification of TbKH and Subcellular Localization in BF Trypanosomes
Recent studies by our laboratory identified the KHARON protein of L. mexicana, LmxKH, as a cytoskeleton-associated protein that mediates the translocation of the LmxGT1 glucose transporter to the flagellar surface membrane (21). Because a BLAST search employing GenBankTM and TriTrypDB revealed the presence of KH orthologs in kinetoplastid parasites but not in other organisms, we hypothesized that KH might play a fundamental biological role in this entire family of protists. Protein sequence alignment of LmxKH (gene ID in TriTrypDB: LmxM.36.5850) and TbKH (gene ID: Tb927.10.8940) revealed 27% (112/411) identity and an additional 11% similarity, and several segments of sequence that are present in LmxKH (520 amino acids) are absent from TbKH (411 amino acids). Hence, whereas KH exists in trypanosomes, it is relatively divergent in sequence between these two related parasites, thus underscoring the need to test the biological functions in both organisms. A conserved sequence signature encompassing amino acids 227–235 of TbKH (NESVDVLNL) is the largest block of contiguous sequence present in both KH orthologs.
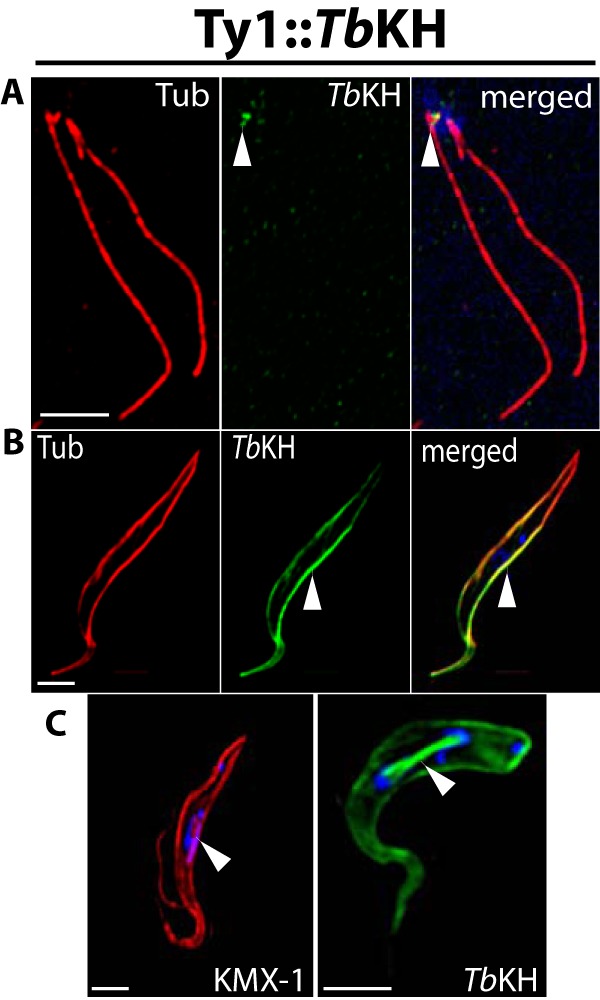
To determine the subcellular localization of TbKH in BF parasites, transgenic parasites were generated in which the TbKH open reading frame was endogenously tagged with the Ty1 epitope on the N terminus or with the HA3 epitope on the C terminus. To facilitate isolation of separated flagella for some experiments, these transgenic lines were also prepared in a parasite genetic background, the TbFLA1RNAi line, allowing inducible RNAi (22, 23) against the TbFLA1 gene (24). Inducing degradation of TbFLA1 mRNA results in separation of flagellar attachment to the cell body by disrupting the flagellar attachment zone, the interface that encompasses parts of both the flagellar and cell body membranes and associated structures where the two membranes adhere (25). This disruption allows the non-adherent flagella to be sheared off the cell body and purified by differential centrifugation (26). Immunofluorescence of the transgenic BFs demonstrated that epitope-tagged TbKH was located in three discrete compartments: 1) In purified flagella, Ty1::TbKH localizes at the base of the flagella (Fig. 1A). 2) In whole cell images, Ty1::TbKH localizes in the subpellicular microtubule cytoskeleton (Fig. 1B) as indicated by the overlap of the TbKH signal with α-tubulin signal; in addition, these two fluorescence signals also overlap in detergent-extracted cytoskeletons from BF parasites (Fig. 1C), consistent with TbKH being a cytoskeleton-associated protein. 3) Unexpectedly, Ty1::TbKH also localizes in the mitotic spindle in dividing BF parasites (Fig. 1D), where it exhibits intense fluorescence on a tubule that connects two replicated nuclei. To confirm that this localization represents the mitotic spindle, parasites were co-stained with the KMX-1 antibody (Fig. 1E) that preferentially recognizes β-tubulin associated with the spindle (27). These images show strong overlap of the Ty1::TbKH- and KMX-1-associated signals at the tubule that connects separating nuclei. In addition, spindle morphology has been shown to change during the cell cycle (28), with the rhomboid shaped spindle (Fig. 1E) observed early in mitosis developing into a dense central spindle (Fig. 1D) later in the nuclear replication process, similar to the morphologies observed for Ty1::TbKH fluorescence.
FIGURE 1.
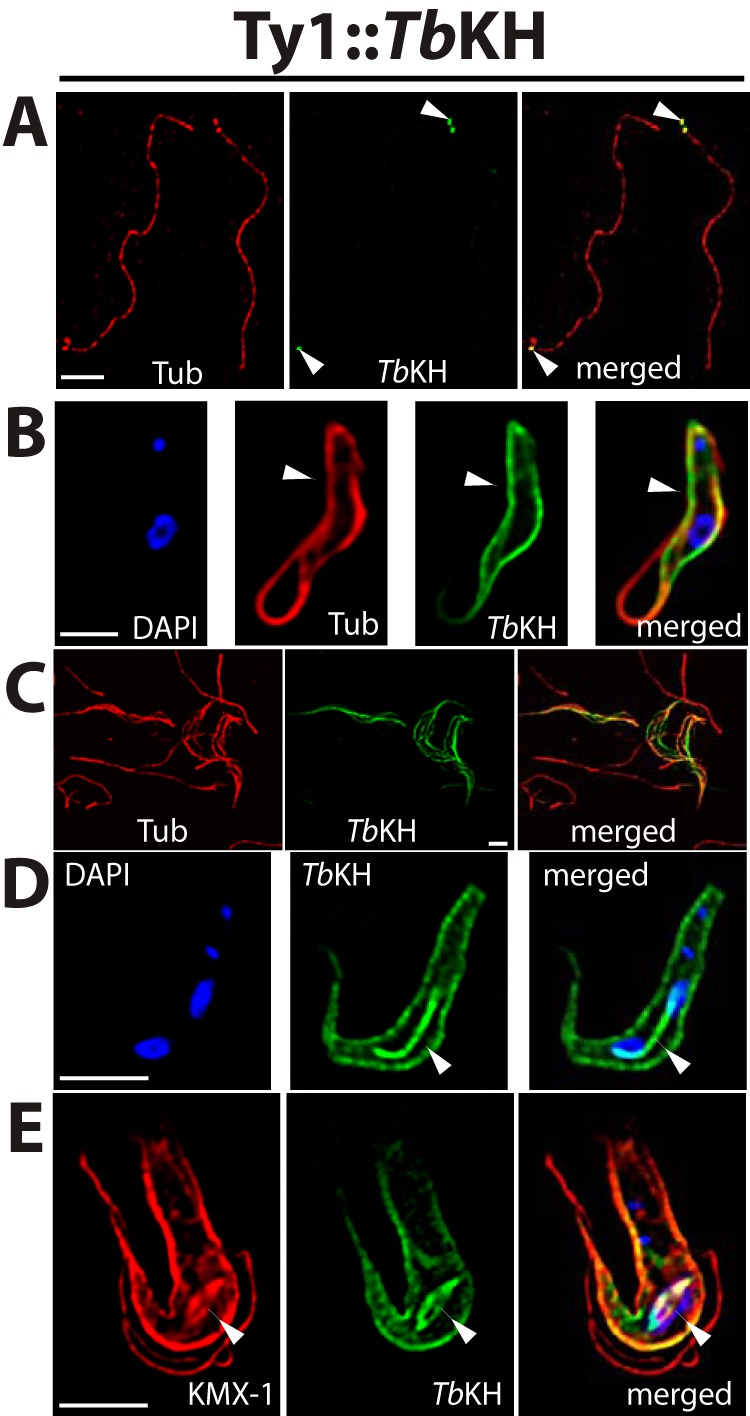
Subcellular localization of TbKH in BF parasites using an N-terminal Ty1 epitope tag. A, flagellar axonemes prepared from Ty1::TbKH expressing BF trypanosomes by selective depolymerization of subpellicular microtubules using high salt. The sample was immunostained with anti-α-tubulin pAb (Tub, red) and anti-Ty1 (BB2) mAb (Ty1::TbKH, green). White arrowheads denote localization of Ty1::TbKH at the base of the flagellum. Scale bars (white) in all panels represent 4 μm. B, the Ty1::TbKH cell line was immunostained with anti-Ty1 (BB2) mAb (Ty1::TbKH, green) and anti-α-tubulin pAb (Tub, red) and demonstrates staining of Ty1::TbKH on the subpellicular microtubules, indicated by white arrowheads. C, detergent-extracted cytoskeletons from BF parasites expressing Ty1::TbKH were stained with BB2 mAb (TbKH, green) and with anti-α-tubulin antibody (Tub, red). D, the Ty1::TbKH cell line was stained with BB2 mAb (TbKH, green). This image displays a parasite in which Ty1:TbKH staining is associated with both the subpellicular microtubules and the mitotic spindle that connects the two nuclei late during karyokinesis. E, another Ty1::TbKH-expressing parasite captured earlier during karyokinesis than the cell shown in D. In this image, the KMX-1 anti-β-tubulin antibody (red) was employed, and it stained spindle microtubules as well as other microtubules (61) and displayed the rhomboid spindle that converges at two poles at opposite ends of dividing nuclei (62). All preparations were stained with DAPI that detects both nuclear and kinetoplast DNA (blue). White arrowheads in D and E point to the mitotic spindle.
To confirm that the localizations observed above did not represent epitope tag-mediated mistargeting, similar images were also obtained from parasite lines expressing TbKH1 with a complementary epitope tag, the trimeric hemagglutinin epitope HA3, fused to the alternate C terminus of KH. These images confirmed localization at the base of the flagellum (Fig. 2A), where the tagged protein is immediately adjacent to DAPI fluorescence signal that is associated with kinetoplast DNA, the highly catenated mitochondrial DNA that is physically attached to the basal body (29), the subpellicular microtubules (Fig. 2B), and the mitotic spindle (Fig. 2C) for both Ty1::TbKH and TbKH::HA3 (compare Figs. 1 and 2). Whole parasites that are expressing TbKH::HA3 do, however, also exhibit punctate staining in the interior of the cell in addition to staining on the subpellicular microtubules (Fig. 2B) or the spindle (Fig. 2C). The absence of cross-reacting bands on Western blots of lysates from wild type BF parasites (see Fig. 6G, lane 1) developed with anti-HA monoclonal antibody or immunofluorescence signal from wild type BF parasites stained with the same antibody (not shown) suggests that this punctate staining does not represent endogenous cross-reacting material. The additional puncta could represent a proportion of TbKH::HA3 that is mistargeted, indicating that addition of HA3 to the C terminus may represent a less optimal tag than addition of Ty1 to the N terminus.
FIGURE 2.
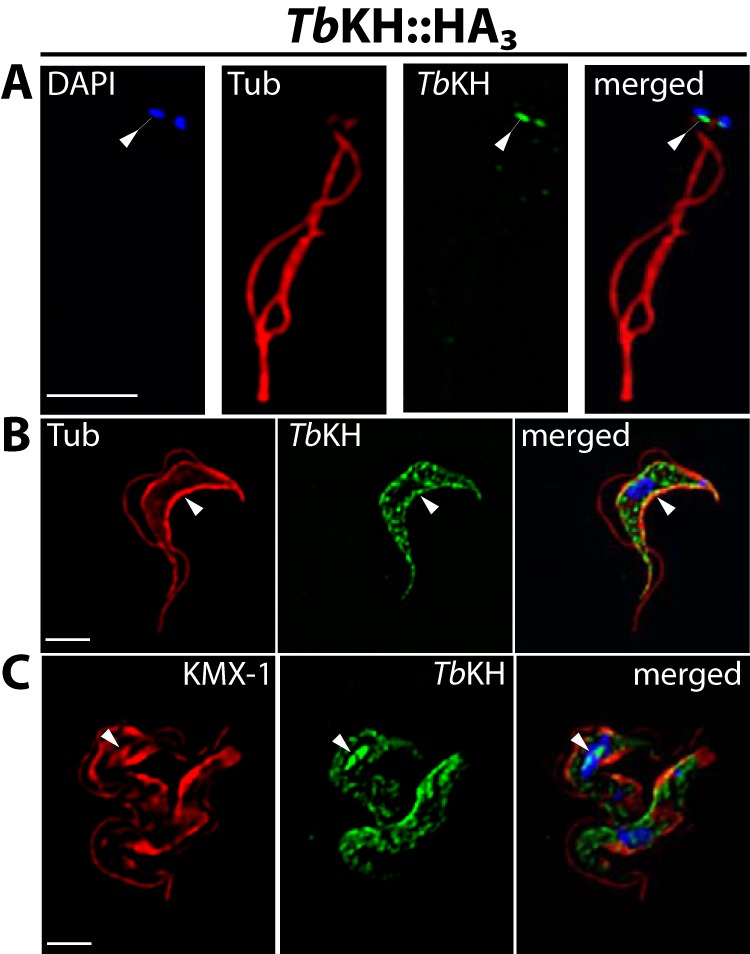
Subcellular localization of TbKH in BF parasites using a C-terminal HA3 epitope tag. Localization of TbKH::HA3 in isolated flagella (A), subpellicular microtubules (B), and mitotic spindles and subpellicular microtubules (C), prepared as described for Fig. 1 but employing anti-HA mAb for immunostaining. Tub, α-tubulin.
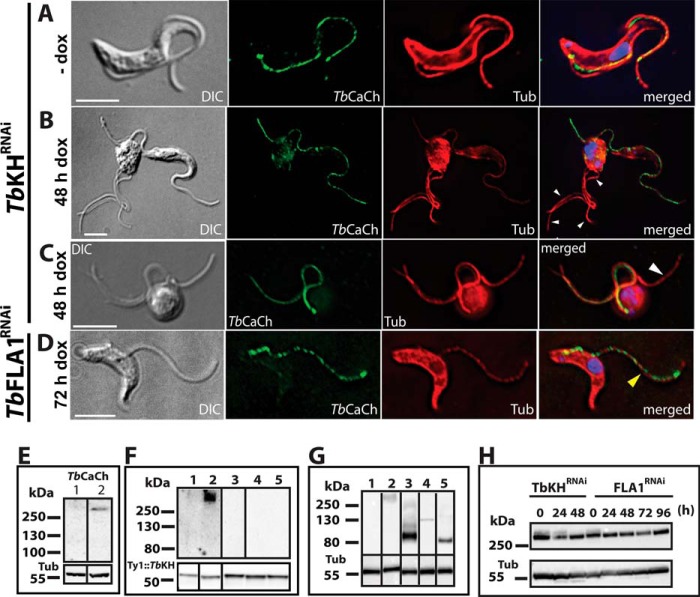
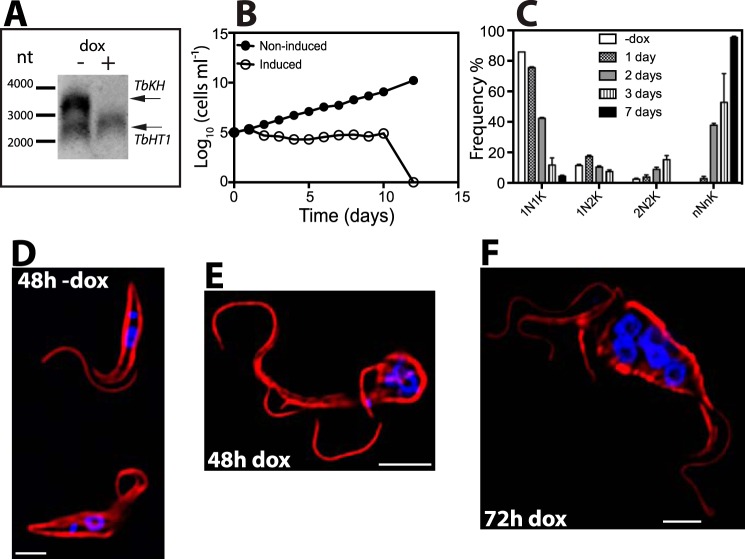
FIGURE 6.
TbCaCh depends on TbKH for translocation to the flagellar membrane in BF parasites. TbCaCh::HA3/TbKHRNAi or TbCaCh::HA3/TbFLA1RNAi cell lines were induced with 1 μg ml−1 doxycycline (dox). Parasites were stained with DAPI and immunostained with anti-HA mAb (TbCaCh, green) and anti-α-tubulin pAb (Tub, red). A, non-induced TbCaCh::HA3/TbKHRNAi parasites (−dox). B and C, TbCaCh::HA3/TbKHRNAi parasites induced for 48 h (48 h dox). D, TbCaCh::HA3/TbFLA1RNAi parasites induced for 72 h (72 h dox). White arrowheads in B and C indicate flagella where TbCaCh is absent, and the yellow arrowhead in D indicates a flagellum where TbCaCh is present. E, immunoblot of isolated flagella from TbCaCh::HA3/TbKHRNAi (lane 1) and TbCaCh::HA3/TbFLA1RNAi (lane 2) cells, induced for 48 and 72 h, respectively, and probed with anti-HA mAb (TbCaCh, upper panel), and anti-α-tubulin mAb (Tub, lower panel) as loading control. F, immunoblot of co-immunoprecipitated proteins from Ty1::TbKH (lane 1), TbCaCh::HA3/Ty1::TbKH (lane 2), TbCaATPase::HA3/Ty1::TbKH (lane 3), FS33::HA3/Ty1::TbK1 (lane 4), and FS133::HA3/Ty1::TbKH (lane 5) expressing parasites. Immunoprecipitation was performed with BB2 mAb directed against the Ty1 epitope to precipitate Ty1::TbKH, and the blot of the immunoprecipitated material was probed (upper panel) with antibody directed against the HA epitope to detect co-immunoprecipitated proteins. The lower panel shows the blot probed with BB2 mAb to detect the immunoprecipitated Ty1::TbKH. G, immunoblot of input total protein lysates from Ty1::TbKH expressing lines used for co-immunoprecipitation experiments in F. The lanes were spliced from different sections of the blots in F and G, according to the order in F, and delineated by black bars. H, immunoblot of total protein lysates (20 μg) from TbCaCh::HA3/TbKHRNAi or TbCaCh::HA3/TbFLA1RNAi cell lines induced with 1 μg ml−1 doxycycline (dox) at various time points, to determine the level of TbCaCh::HA3 following induction of RNAi. The protein blots were probed with anti-HA mAb (TbCaCh, upper panel) and anti-α-tubulin mAb (Tub, lower panel) as loading controls.
Although spindle localization had not been detected for LmxKH in the original studies on L. mexicana (21), subsequent images of hydroxyurea synchronized promastigotes (Fig. 3) demonstrate that LmxKH is also associated with the mitotic spindle in that parasite and that this newly discovered localization is thus likely universal. Hydroxyurea synchronization was employed to increase the percentage of parasites captured in mitosis and thus facilitate the detection of mitotic spindles that are associated with LmxKH, but similar images were also observed with lower frequency using non-synchronized parasites (data not shown). L. mexicana promastigotes assumed an oval morphology following exposure to hydroxyurea rather than the elongated shape of untreated promastigotes, but LmxKH was nonetheless apparent on the mitotic spindle that separates dividing nuclei.
FIGURE 3.
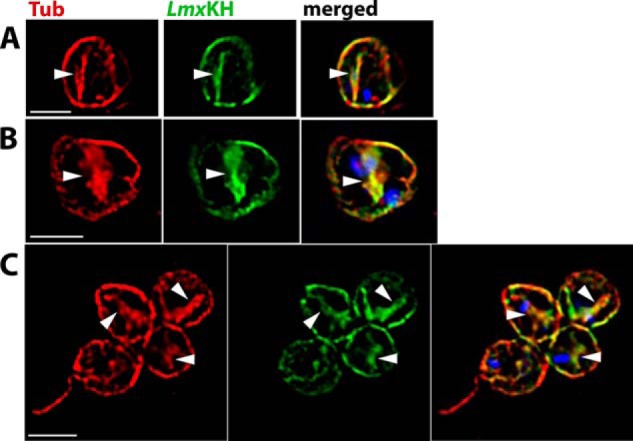
Overlap of LmxKH with the mitotic spindle. Promastigotes of L. mexicana expressing LmxKH::HA3 were synchronized with hydroxyurea, released from the cell cycle block by washing out hydroxyurea, and cultured in hydroxyurea-free RPMI medium for 7 h. A–C show images of parasites that were stained with anti-α-tubulin pAb (red) and anti-HA antibody pAb (green) and images with the red, green, and blue (DAPI-stained) images merged (merged). White arrowheads mark mitotic spindles that stain with both anti-α-tubulin and anti-HA antibodies.
TbKHARON Is Essential for Viability of the BF Parasites
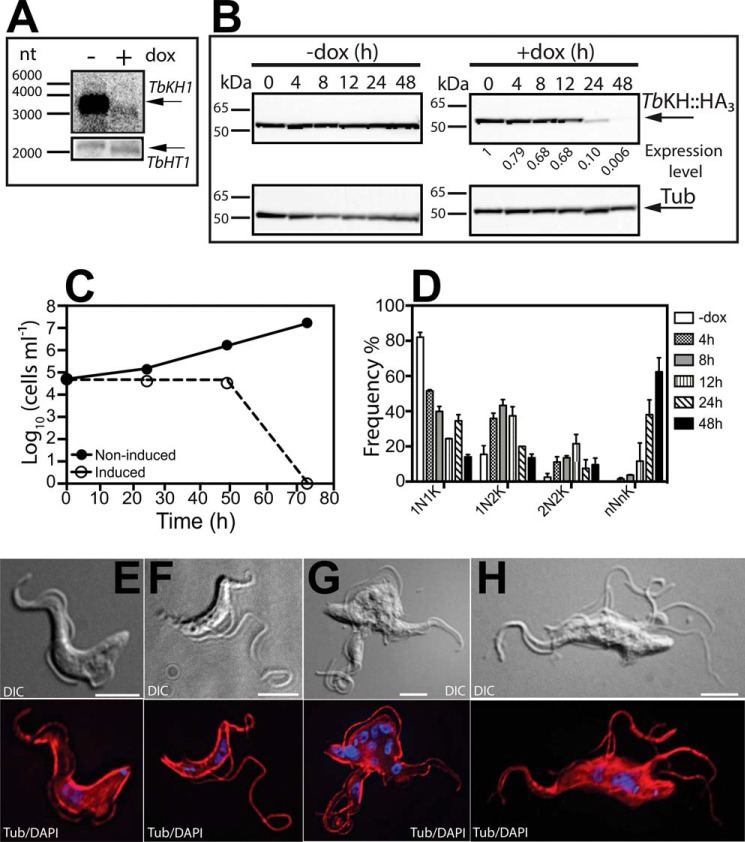
To determine whether TbKH is essential for BF parasites, we inhibited the expression of TbKH by RNAi. The transgenic BF/pHD1313 line (30), which expresses the tetracycline repressor, was transfected with the TbKHRNAi construct employing the p3666 vector (22) that transcribes the hairpin loop RNA driven by the EP procyclin promoter fused to four tetracycline operators. Induction of expression of this hairpin loop RNA with doxycycline for 24 h reduced the level of TbKH mRNA by at least 6-fold (Fig. 4A, upper panel), as determined by densitometry, whereas T. brucei hexose transporter 1 (TbHT1) mRNA remained unchanged (Fig. 4A, bottom panel). A time course for expression of TbKH::HA3 following induction of RNAi against TbKH mRNA (Fig. 4B) showed that the protein was almost completely eliminated by 48 h after induction. Upon induction of RNAi, parasite growth was severely compromised (Fig. 4C, open circles), resulting in death of BFs by 72 h. Whereas uninduced parasites exhibited normal morphology upon staining with anti-α-tubulin antibody and DAPI (Fig. 4E), parasites where RNAi had been induced for 12 h (Fig. 4F), 24 h (Fig. 4G), or 48 h (Fig. 4H) continued to replicate nuclear and kinetoplast DNA, as determined by the multiple DAPI staining foci, but failed to undergo cell division, resulting in cells with multiple nuclei, kinetoplasts, and flagella. These parasites are similar to the “monster” cell phenotype originally described for RNAi directed against mRNAs encoding various flagellar axoneme proteins (31). Microscopic quantification of parasites with a normal complement of kinetoplasts and nuclei (1K1N) or with altered numbers of both genomes (e.g. 2K1N, 2K2N, etc.) revealed that 1N1K parasites are reduced rapidly following induction of RNAi and that monster cells like those in Fig. 4G began to dominate the population increasingly from 0 to 48 h (Fig. 4D). Additionally, the flagella became detached from the cell body along their length as early as 12 h. Microscopic observation of live cells revealed that detached flagella remained motile even after 48 h of RNAi induction, similar to the phenotype reported upon RNAi of TbFLA1 (24, 28).
FIGURE 4.
Depletion of TbKH is lethal for BF parasites. A, Northern blot of total RNA from TbKHRNAi parasites grown in the presence (+) and absence (−) of doxycycline (dox) for 24 h was hybridized against a probe recognizing TbKH (upper panel) or the hexose transporter TbHT1 (lower panel). Arrows indicate the positions of the relevant mRNAs. Molecular weight markers are indicated in nucleotides (nt). B, time course for loss of TbKH::HA3 protein following induction of RNAi against TbKH mRNA. BF parasites expressing TbKH::HA3 and carrying the TbKHRNAi construct were induced (+dox) or not induced (−dox) with doxycycline for the times indicated (h). Cell lysates (20 μg) were resolved by SDS-PAGE, blotted, and probed with anti-HA (top panels) or anti-α-tubulin (bottom panels, Tub) antibodies. Molecular weight markers are indicated at the left of each panel in kDa units. The numbers below the upper right panel indicate the relative signal intensity for TbKH::HA3 (Expression level), first normalized to the α-tubulin signal at each time point and then normalized to the signal at 0 h (set to 1). C, growth curve of induced (empty circles) and non-induced (filled circles) TbKHRNAi cell line. Parasite density was quantified by phase contrast microscopy using a hemacytometer. The points represent the means of two biological replicates, but the error bars are too small to be visible on the logarithmic scale employed for the y axis. D, frequency (%) of cells with different numbers of nuclei (N) and kinetoplasts (K) at different times following induction of RNAi against TbKH mRNA. The results represent the average and range of two independent experiments. E–H, representative TbKHRNAi cells undergoing RNAi were stained with DAPI (blue) and immunostained with anti-α-tubulin mAb (Tub, red) at (E) 0 h, (F) 12 h, (G) 24 h, and (H) 48 h after RNAi induction. DIC indicates images collected by differential interference contrast microscopy.
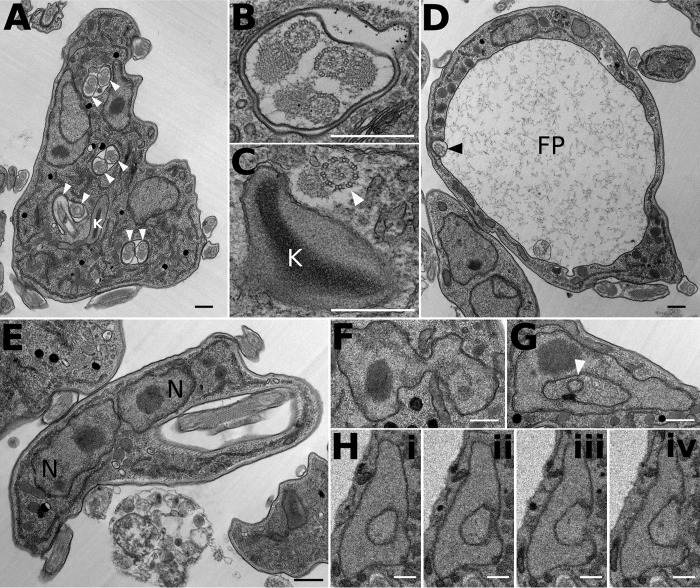
BF trypanosomes induced for RNAi against TbKH for 24 or 48 h were also examined by electron microscopy. Images show a number of abnormalities, including multiple flagellar pockets, multiple axonemes within single flagellar membranes (Fig. 5, A and B), axonemes without flagellar membranes (Fig. 5C), enlarged flagellar pockets (Fig. 5D), multiple nuclei in close apposition (Fig. 5E), and nuclei with apparent membranous inclusions, which represent invaginations of the nuclear envelope (Fig. 5, F–H).
FIGURE 5.
Abnormal subcellular structures in BF T. brucei following induction of RNAi against TbKH mRNA. Transmission electron micrographs (A–G) and serial block face scanning electron micrographs (H, panels i–iv) show multiple flagella (white arrowheads) in multiple flagellar pockets (A); note the presence of paraflagellar rods in several profiles. B, multiple flagellar axonemes within a single flagellar membrane. C, flagellar axoneme (white arrowhead) with associated PFR but without a flagellar membrane within the cell body. D, enlarged flagellar pocket (FP) with a single flagellum (black arrowhead). E, two nuclei (N) abnormally close together. F, nucleus with several lobes. G, nucleus with apparent membranous inclusion (white arrowhead). H, panels i–iv, sequential sections through a nucleus showing that apparent membranous inclusions represent invaginations of the nuclear envelope. All scale bars represent 500 nm. Induction of RNAi against TbKH mRNA was as follows: 24 h (A), 18 h (B), 24 h (C), 48 h (D), 24 h (E), 24 h (F), 25 h (G), and 48 h (H).
Role of TbKHARON in Trafficking of Flagellar Membrane Proteins
To determine whether TbKH could be involved in trafficking of flagellar membrane proteins, we tested the potential role of this protein in localization of several flagellar membrane proteins that had been identified in the flagellar membrane proteome and localized by immunofluorescence in BF parasites (5): a putative Ca2+ channel (TbCaCh, FS179, Tb927.10.2880), a bona fide (32) Ca2+ ATPase, (TbCaATPase, FS60, Tb927.8.1200), a membrane-bound adenylate cyclase (FS33, Tb927.8.7940), and an integral membrane protein of unknown function (FS133, Tb927.2.1700). Upon knockdown of TbKH mRNA, normal flagellar trafficking of the TbCaCh to the region of the flagellum that adheres to the cell body (Ref. 5 and Fig. 6A) was blocked, resulting in BF parasites with TbCaCh in only a few (Fig. 6, B and C) of the multiple flagella, as indicated by white arrowheads that mark flagella that stain with anti-α-tubulin but not with antibody directed against the HA3 epitope on TbCaCh::HA3. Isolation of sheared flagella followed by immunoblotting using antibody against the HA3 tag also confirmed that TbCaCh was largely absent from the flagella under TbKH RNAi conditions (Fig. 6E, lane 1) but not when RNAi was directed against TbFLA1 mRNA (Fig. 6, D and E, lane 2), a different knockdown that also results in detachment of flagella from the cell body (24). Hence, mistargeting of TbCaCh is a specific consequence of down-regulating TbKH and not simply an outcome of flagellar detachment.
To examine whether TbKH may interact with flagellar membrane proteins that it targets to the flagellar membrane, we monitored possible co-immunoprecipitation of Ty1::TbKH and HA-tagged flagellar membrane proteins. In BFs expressing Ty1::TbKH, cells were cross-linked with formaldehyde and sonicated to disrupt cytoskeletons, followed by immunoprecipitation with BB2 monoclonal antibody directed against the Ty1 epitope. This procedure was carried out employing BF clones that also expressed TbCaCh::HA3. As negative controls, similar experiments were done in BF parasites expressing TbCaATPase::HA3, FS33::HA3, and FS133::HA3, proteins that do not depend on TbKH for trafficking to the flagellar membrane (data not shown). TbCaCh::HA3 co-immunoprecipitated along with Ty1::TbKH (Fig. 6F, upper panel, lane 2), whereas none of the other tested flagellar membrane proteins did so (Fig. 6F, upper panel, lanes 1 and 3–5). Control immunoblots of input lysates before immunoprecipitation (Fig. 6G) demonstrated that these control lines did express the relevant tagged proteins, supporting the notion that the absence of signals in the blot in Fig. 6F was due to failure or any of these proteins to interact with TbKH and not simply to failure of the cell line to express the tagged flagellar membrane protein. Immunoblots of the immunoprecipitates were also probed with BB2 antibody (Fig. 6F, lower panel) to demonstrate that similar amounts of Ty1::KH had been captured in the immunoprecipitation step.
Loss of TbCaCh from flagella following induction of TbKHRNAi was due to failure to target this channel to flagellar membranes and not simply to degradation of the channel. Thus, immunoblots (Fig. 6H, TbKHRNAi) of lysates from parasites that had been induced for RNAi against TbKH mRNA for 24 or 48 h showed that TbCaCh was still present at levels similar to that seen before induction of RNAi (0 h). Similar results were obtained for parasites where RNAi had been induced against TbFLA1 mRNA (Fig. 6H, TbFLA1RNAi).
Localization of TbCaATPase, a protein that traffics to puncta in both the flagellar membrane and the cell body (Fig. 7A), was also determined following induction of RNAi against TbKH mRNA. This flagellar membrane protein continued to traffic to the flagellar membrane when RNAi was induced against either TbKH (Fig. 7B) or TbFLA1 (Fig. 7C) mRNA. Immunoblots of isolated flagella probed with antibody against the HA3 tag also demonstrated that TbCaATPase was still present in flagella following induction of RNAi against either TbKH or TbFLA1 mRNA (Fig. 7D, upper panel, lanes 1 and 2, respectively). This result demonstrates that induction of RNAi against TbKH mRNA does not cause global disintegration of the flagellar membrane and/or nonspecific removal of all flagellar membrane proteins but rather specifically impairs flagellar trafficking of TbCaCh.
FIGURE 7.
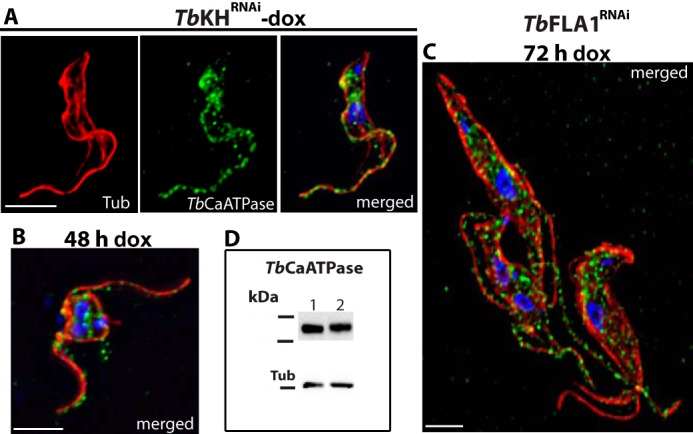
TbCaATPase translocation to the flagellar membrane is TbKH-independent. TbCaATPase::HA3/TbKHRNAi and TbCaATPase::HA3/TbFLA1RNAiclones were induced with doxycycline for 48 and 72 h, respectively; stained with DAPI (blue); and immunostained with anti-HA mAb (TbCaATPase, green) and anti-α-tubulin pAb (Tub, red). A, non-induced TbCaATPase::HA3/TbKHRNAi clone (−dox). B, TbCaATPase::HA3/TbKHRNAi clone induced for 48 h (48 h dox). C, TbCaATPase::HA3/TbFLA1RNAi clone induced for 72 h (72 h dox). D, immunoblot of isolated flagella from TbCaATPase::HA3/TbKHRNAi (lane 1) and TbCaATPase::HA3/TbFLA1RNAi (lane 2) cells induced for 48 and 72 h, respectively, was immunodetected with anti-HA mAb (TbCaATPase, upper panel) and anti-α-tubulin mAb (Tub, lower panel) as loading control.
TbKHARON Is a Functionally Important Component of the Mitotic Spindle in BF Parasites
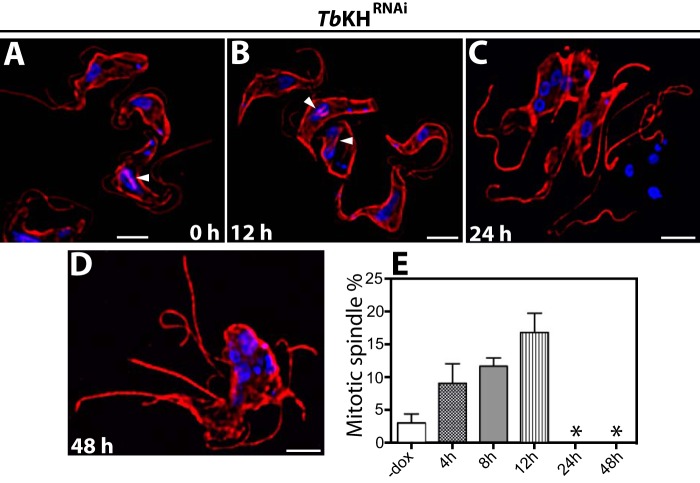
To investigate the potential role of TbKH in the mitotic spindle of BF parasites, the mitotic spindle fate was followed by immunofluorescence microscopy, employing KMX-1 antibodies, following the induction of RNAi against TbKH mRNA for 12, 24, or 48 h (Fig. 8, A–D). Prior to induction of RNAi, 3 ± 1% (n = 2, 100 cells counted per sample) of BF parasites had observable spindles (Fig. 8E). Following induction of RNAi against TbKH, the percentage of BF parasites with spindles first increased to 17 ± 2% at 12 h after induction, but at 24 and 48 h, no spindles were observed (Fig. 8, C and D, and asterisks in E). This observation suggests that TbKH not only localizes to the mitotic spindles but is also necessary for their integrity and/or stability. Moreover, 24 h RNAi-induced parasites have multiple nuclei that appeared to be well separated (Fig. 8C), whereas parasites induced for 48 h had multiple clumped nuclei (Fig. 8D), suggesting that nuclear segregation may be impaired following sufficient reduction in the level of TbKH mRNA. In addition, synchronization of Ty1::TbKH parasites using hydroxyurea (33) resulted in an increase in the number of parasites showing specific Ty1::TbKH fluorescence signal on the mitotic spindle of dividing cells, from ∼5% of total cells in asynchronous to ∼20% in synchronous parasites, as determined by examining 100 cells by immunofluorescence microscopy. This observation further confirms the association of TbKH with the mitotic spindle.
FIGURE 8.
Loss of mitotic spindle in BF parasites upon induction of RNAi against TbKH. A–D, TbKHRNAi cells were grown in the presence of doxycycline for times between 0 and 48 h, stained with DAPI (blue), and immunostained with KMX-1 mAb (red). The white arrowheads point to mitotic spindles that were visible in uninduced cells and in parasites for which RNAi against TbKH had been induced for 12 h, but spindles were not visible in cells that had been induced for 24 or 48 h. E, percentage of BF parasites showing mitotic spindles prior to induction (−dox) and 4, 8, 12, 24, or 48 h after induction with doxycycline. Numbers represent the means and ranges of two independent experiments for which 100 cells were counted. No mitotic spindles were detected at 24 and 48 h, as indicated by asterisks.
TbKHARON Is a Cytoskeletal Protein Essential for Viability of the PF Parasites
To establish the subcellular localization of TbKH in PF parasites, a PF line was generated that carried a copy of TbKH ORF endogenously tagged on the N terminus with Ty1. TbKH localization in PFs was essentially the same as that observed in BFs. In isolated flagellar cytoskeletons (31), Ty1::TbKH was detected at the base of the flagella adjacent to the DAPI-stained kinetoplast DNA (Fig. 9A). In whole PF parasites, Ty1::TbKH was observed in the subpellicular microtubule cytoskeleton by overlapping fluorescence with the α-tubulin signal (Fig. 9B) and in the mitotic spindles of dividing PF (Fig. 9C, right panel) similar to the signal observed in parasites stained with KMX-1 antibody (Fig. 9C, left panel). In Fig. 9C, separate cells were stained with each primary antiserum, because the same secondary antiserum is employed to detect both primary monoclonal antibodies. Induction of RNAi resulted in loss of detectable TbKH mRNA within 48 h (Fig. 10A), and whereas parasite growth arrested by this time, PF cell death was not complete until 12.5 days (Fig. 10B), much longer than for BF parasites. Induction of RNAi also resulted in formation of polynucleated and polyflagellated PF parasites with disrupted attachment of the flagella to the cell body (Fig. 10, C, E, and F), unlike uninduced PFs (Fig. 10D). Hence, the phenotypes upon induction of RNAi against TbKH mRNA are similar in BF and PF parasites. This observation is notably distinct from the monster cell phenotype originally observed using RNAi directed against components of the flagellar axoneme (31), where the cytokinesis defect was only observed for BF but not PF trypanosomes. Of note, both BF and PF parasites undergoing RNAi against TbKH were still motile, as determined by phase contrast microscopy, even after these dramatic morphological changes.
FIGURE 9.
s Subcellular localization of TbKH in PF parasites. A, flagellar axonemes were isolated from Ty1::TbKH-expressing PF parasites by detergent lysis and incubation with NaCl (“Experimental Procedures”) to disrupt the subpellicular microtubules, and preparations were stained with antibodies against α-tubulin (Tub, red) and Ty1 (BB2 mAb, green) and with DAPI to visualize kDNA (blue). The white arrowhead indicates the green fluorescence representing Ty1::TbKH at the base of the flagella. B, Ty1::TbKH PF parasites were immunostained with BB2 mAb (TbKH, green) and anti-α-tubulin (Tub, red) to show localization of TbKH that partially overlaps with the subpellicular microtubules (white arrowhead). C, Ty1::TbKH PF parasites were stained with DAPI (blue) and immunostained with BB2 mAb (TbKH, green) or KMX-1 mAb (Tub, red). The white arrowhead indicates KMX-1 antibody staining on the spindle between two nuclei (left image) and TbKH on the mitotic spindle (right image) in parasites undergoing karyokinesis.
FIGURE 10.
Effect of TbKH depletion on PF parasites. A, Northern blot of total RNA from parasites grown in the presence (+) and absence (−) of 1 μg ml−1 doxycycline (dox) for 48 h was hybridized against probes for TbKH (upper band) or TbHT1 (lower band). Molecular weight markers are indicated in nucleotides (nt), and the positions of the relevant mRNAs are indicated by arrows. B, growth curve of induced (empty circles) and non-induced (filled circles) TbKHRNAi PF parasites. The data represent the averages of two independent experiments; the error bars are too small to see on the logarithmic scale employed for the y axis. C, frequency (%) of cells with different numbers of nuclei (N) and kinetoplasts (K) following induction of RNAi for 1–7 days. The data represent the means and ranges of two independent experiments for which 100 cells were counted. The absence of the black-filled bar for 1N1K and 2N2K samples indicates that no parasites with these characteristics were observed on day 7. D–F, TbKH1RNAi PF parasites grown in the presence and absence of doxycycline were stained with DAPI (blue) and immunostained with anti-α-tubulin mAb (red). Parasite samples were processed at different time points, 48 h minus doxycycline (48h −dox) (D), 48 h plus doxycycline (48h dox) (E), and 72 h plus doxycycline (72h dox) (F).
Rescue of L. mexicana Δlmxkh[pGT1::GFP] Phenotype by TbKH
To determine whether TbKH could substitute in L. mexicana for LmxKH regarding trafficking of LmxGT1 to the flagellar membrane and support growth of intracellular amastigotes, the Δlmxkh[pGT1::GFP] knock-out line was complemented with the TbKH ORF expressed from an episome. Partial restoration of LmxGT1 flagellar membrane targeting (Fig. 11, A and B) and growth of intracellular amastigotes (Fig. 11C) was observed. These results indicate that TbKH can function, albeit suboptimally, in trafficking of flagellar membrane proteins in L. mexicana, despite the fact that sequence identity for the KH proteins is relatively low between the two species of parasite.
FIGURE 11.
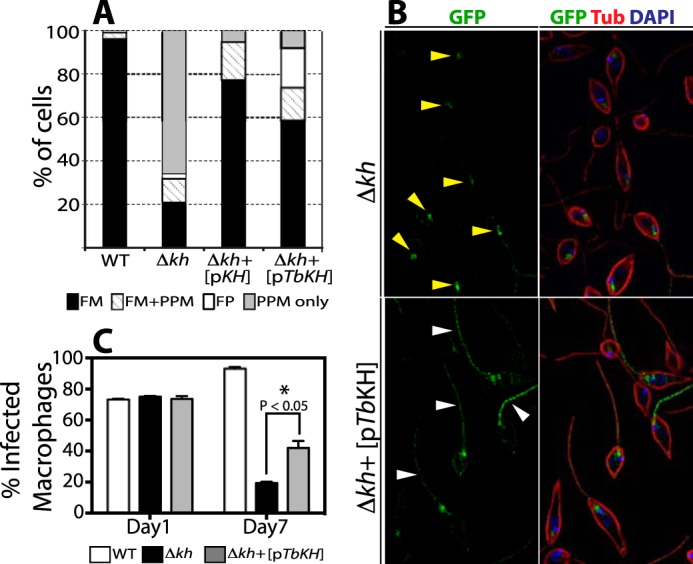
Functional complementation of Leishmania kharon null mutant by TbKH. L. mexicana Δlmxkh null mutants (Δkh) expressing the LmxGT1 flagellar glucose transporter tagged with GFP at it C terminus [pGT1::GFP] were transfected with an episomal expression vector encoding the TbKH gene [pTbKH]. Δlmxkh[pGT1::GFP] (Δkh) and Δlmxkh[pGT1::GFP]/pTbKH (Δkh+[pTbKH]) were stained with DAPI (blue) and immunostained with anti-GFP pAb (GFP, green) and anti-α-tubulin mAb (Tub, red). A, quantification of LmxGT1::GFP localization (%) in L. mexicana lines. The data for WT L. mexicana, Δkh, and Δkh[pKh] (Δkh null mutants complimented with an episomal expression vector encompassing the LmxKH gene) were previously published by Tran et al. (21) and are reproduced here for comparison. The key at the bottom shows the labeling scheme for quantification of parasites that contain GT1::GFP in the flagellar membrane (FM), flagellar membrane plus pellicular plasma membrane (FM+PPM), flagellar pocket (FP), and pellicular plasma membrane only (PPM only). The number of parasites examined in each quantification was: WT = 210, Δkh = 241, Δkh+[pKH] = 139, and Δkh+[pTbKH] = 249. B, examples of LmxGT1::GFP localization in non-complemented (Δkh, upper panel) and complemented (Δkh+[pTbKH]) mutants. Yellow arrowheads in the upper panel indicate GT1::GFP localized in the flagellar pocket. White arrowheads in the lower panel indicate GT1::GFP in the flagellum. C, quantification of THP-1 infection with WT, Δkh mutant, and Δkh complemented with an episomal copy of TbKH Δkh+[pTbKH] promastigotes. Day 1 and Day 7 indicate the time elapsed following infection. The data represent the averages and standard deviations of triplicate infections (n = 3), each counted in triplicate and averaged. The statistical significance for comparison of the null mutant and complemented null mutant on day 7 was determined by two-tailed Student's t test (p < 0.05).
Discussion
Cilia and flagella are multifunctional organelles that are represented broadly in biology, from unicellular eukaryotes to differentiated mammalian cells. Over the past decade or so, the role of cilia and flagella as sensors of the extracellular environment has emerged, with a corresponding emphasis on the membrane proteins that mediate these sensory processes (34). In parallel, a burgeoning interest has developed in the mechanisms whereby membrane proteins are selectively targeted to cilia and flagella (35). Recent studies on African trypanosomes have uncovered novel unexpected functions for flagella in these parasites. Szempruch et al. (36) have reported that BF trypanosomes generate extracellular vesicles that originate from the flagellar membrane, contain flagellar proteins and virulence determinants such as serum resistance-associated protein, and deliver these vesicles to host cells including erythrocytes. This observation further underscores the importance of flagellar membrane proteins in the biology and virulence of African trypanosomes. Furthermore, flagellar membranes of PF trypanosomes can fuse with each other and transfer both membrane-bound and cytosolic proteins between the two flagellar-attached parasites (37), suggesting that flagella may be organelles involved in cell-cell communication. In addition, flagellar adenylate cyclases are involved in the process of social motility (8–10), a coordinated motility process that is important for migration of parasites within the tsetse fly and for successful colonization of this insect vector (38). Hence, understanding the function of parasite flagella and the processes for targeting integral membrane proteins to these organelles is central to the biology of these parasites and the diseases they cause.
The current characterization of TbKH has resulted in a number of novel discoveries that extend beyond the initial discovery of KH in L. mexicana. First, KH is essential for viability of both BF and PF trypanosomes, establishing the critical role of this protein in a second kinetoplastid parasite, in this case in both mammalian and insect life cycle stages. Second, a novel putative Ca2+ channel is dependent upon TbKH for flagellar targeting, demonstrating that this conserved trafficking machinery mediates flagellar targeting of at least two distinct membrane proteins in diverse parasites. Third, a new location for KH has emerged, namely its association with the mitotic spindle.
TbKH is involved in flagellar targeting of the putative Ca2+ channel TbCaCh, and this function is likely mediated by TbKH located at the base of the flagellum. During biosynthesis, integral membrane proteins are first incorporated into the surface membrane at the flagellar pocket (39), from where they migrate to the plasma membrane surrounding the cell body or the flagellar membrane. The presence of KH in close apposition to the flagellar pocket membrane may thus mediate, by a still unknown mechanism, the entry of TbCaCh and LmxGT1 into the adjacent flagellar membrane. This function of TbKH in targeting flagellar membrane proteins is likely to be essential for viability of BF and PF parasites. Thus an early phenotype accompanying induction of RNAi against TbKH RNA is detachment of the flagellum from the cell body. Previous studies have also demonstrated that induction of RNAi against TbCaCh mRNA results in detachment of the flagella from the cell body and parasite death (5, 40). This observation suggests that failure of TbKHRNAi knockdown parasites to target TbCaCh to its normal location in the component of the flagellar membrane that associates with the cell body would be sufficient to disrupt this junction between the flagellar and cell body membranes. Failure of BF parasites to divide, resulting in multinucleate, multiflagellated monster cells, was previously observed by Gull and colleagues (31) following induction of RNAi against mRNAs encoding proteins of the flagellar axoneme. These authors suggested that proper flagellar function is required for establishment of the cleavage furrow during mitosis, thus establishing an intimate link between flagella and cytokinesis. A number of other studies (41–44) have also underscored a role for the flagellum in cytokinesis and parasite viability. Hence, it is not surprising that disruption of targeting for flagellar membrane proteins, such as TbCaCh and possibly others, following RNAi against TbKH mRNA would induce failure of cytokinesis and a lethal phenotype in BF parasites. It is, however, notable that the essentiality of flagellar function noted in the previous studies on other flagellar proteins was typically reserved to the BF stage, but induction of RNAi in the PF forms was not lethal. The role of TbKH is distinct in this regard, because RNAi induced against TbKH mRNA is lethal in both life cycle stages.
If TbCaCh is a bona fide calcium channel, it may also play an important role in flagellar physiology that is dependent upon flagellar localization. One example of such an organelle-dependent function for Ca2+ channels is provided by the cilia of mammalian kidney tubule cells, where several of these channels are thought to be involved in sensing fluid flow within this organ (45). We anticipate that some other flagellar membrane proteins will likely be dependent upon TbKH for targeting to this organelle. It should be possible to identify other potential cargo for TbKH by performing quantitative proteomics to detect proteins whose relative abundance in isolated flagella decreases after induction of RNAi directed against TbKH mRNA.
Although the specific functions of TbKH at the mitotic spindle and the subpellicular microtubules are not yet clear, this protein could also be required for critical processes at those loci. RNAi targeting of TbKH mRNA impairs mitotic spindle formation, a deficiency that would be expected to be lethal. However, the effect of TbKH RNAi on spindle integrity is only detected at 24 h, after the detachment of flagella and failure of cytokinesis, which are already detected at 12 h. Hence, loss of spindles is probably not the primary lethal defect of knocking down TbKH mRNA. Similarly, association of TbKH with the subpellicular microtubules might mediate critical functions for this extensive cytoskeletal network. In principle, lethality could be due to disrupted KHARON function at the base of the flagellum, on the mitotic spindle, or in the subpellicular cytoskeleton, or loss of KHARON at all three loci may contribute to the lethal phenotype, potentially with different kinetics. Hence one challenge will be to dissect the potentially distinct functions of TbKH at each of its subcellular loci. Experiments employing biotin proximity labeling (46)6 suggest that both TbKH and LmxKH may associate with multiple additional subunits to form KHARON Complexes, and this possibility is being further investigated. These potential separate complexes may share KH and other subunits, but they may also incorporate other distinct subunits that are unique to the subcellular localization and specific function of each complex. If this model of KH structure and function is correct, then targeting complex-specific subunits using RNAi may allow dissection of the distinct functions of each complex and insight into the role of KH and its associated subunits at each of its three subcellular locations.
Experimental Procedures
Growth and Transfection of T. brucei Cell Lines
BF Lister 427 and PF TREU927 T. brucei cell lines were grown as described previously (30). For T7 RNA polymerase-independent driven expression (22), BF or PF parasites transfected with the pHD1313 plasmid expressing the tetracycline repressor, kindly provided by Dr. Christine Clayton (47), were generated and grown in 2.5 μg ml−1 phleomycin. Subsequently, 5–10 μg of linearized plasmid DNA was used to transfect mid-log phase parasites as described (48). Transfectants were isolated by limiting dilution according to published protocols (49, 50).
Hydroxyurea Synchronization
Synchronization of BF Ty1::TbKH cell line with hydroxyurea was performed as described by Forsythe et al. (33). Briefly, parasites were incubated in HMI-9 (51) culture medium containing 10 μg ml−1 of hydroxyurea for 6 h at 37 °C in 5% CO2. Later, parasites were pelleted by centrifugation at 1600 × g for 10 min at room temperature to remove hydroxyurea and resuspended in fresh HMI-9 culture medium, and centrifugation was repeated. Finally, parasites were resuspended in 10 ml of fresh HMI-9 and incubated for 3 h at 37 °C in 5% CO2. Then parasites were processed for fluorescence microscopy as indicated.
RNA Isolation and Northern Blotting
Total RNA from T. brucei parasites was isolated using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Northern blotting analysis was performed as described elsewhere (52). Non-radioactive detection employing DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) was used according to the manufacturer's instructions, and the Image Quant LAS 400 (GE Healthcare) was employed for image acquisition. ImageJ software (National Institutes of Health) was used for densitometry analysis and Adobe Photoshop CS3 and Adobe Illustrator CS3 to create figures.
Primary Amino Acid Sequence Analysis
For DNA sequence analysis of TbKH and amino acid sequence alignment with the LmxKH ORF, MacVector software (Intelligenetics) was employed. Sequence similarity was analyzed using the BLAST search and GenBankTM or TritrypDB.
Inhibition of Gene Expression by RNAi
To inhibit the expression of TbKH in both BF and PF parasites, a construct was generated based on the p3666 vector harboring the blasticidin (BSD) resistance marker (22) that integrates into the rRNA gene repeat locus and generates a hairpin RNA derived from the TbKH ORF. A 521-bp gene fragment corresponding to nucleotides 340–860 of the TbKH ORF (Tb927.10.8940) was employed, identified as an optimal fragment for RNAi using RNAit software. The TbKHRNAi construct was linearized and transfected into BF or PF/pHD1313 cell lines as described (30) to generate BF or PF/TbKHRNAi clonal lines. RNAi clones were selected by resistance to 2.5 μg ml−1 phleomycin and 5 μg ml−1 (10 μg ml−1 for PF) blasticidin, and expression of hairpin loop RNAi was induced by addition of 1 μg ml−1 doxycycline. Similarly, a TbFLA1RNAi hairpin loop construct was generated using a 539-bp gene fragment corresponding to nucleotides 692–1230 bp of the TbFLA1 ORF (Tb927.8.4010), identified by RNAit software, and the p3666 plasmid as described above. A BF/TbFLA1RNAi clonal line was produced as described for the TbKHRNAi clones. RNAi was induced by adding 1 μg ml−1 doxycycline to the medium.
Endogenous Epitope Tagging
For endogenous tagging of TbKH at the N terminus with the Ty1 epitope tag (53), a DNA segment representing sequence 500 bp upstream of the start codon of the TbKH ORF was ligated to the BSD/BB2 module (54) followed by another segment representing the first 170 bp corresponding to the N-terminal open reading frame. For endogenous tagging of TbKH at the C terminus with a trimeric hemagglutinin (HA3) tag, 500 bp corresponding to C-terminal coding region of the TbKH ORF was ligated to the pMOTag2H plasmid (55), followed by a 500-bp segment downstream of the stop codon. Constructs for expression of flagellar membrane proteins FS33, FS60, FS133, and FS179 tagged with a HA3 epitope at the C terminus were kindly provided by Dr. Kent Hill (5). Parasites were transfected with 5 μg of linear construct as described (48) and selected using 5 μg ml−1 blasticidin or 0.1 μg ml−1 puromycin and cloned by limiting dilution.
Immunofluorescence Microscopy
For immunofluorescence microscopy, parasites (5 × 106) were centrifuged at 700 × g for 4 min and washed twice at room temperature with PBS, pH 7.2, containing 10 mm glucose. The cell pellet was resuspended in 4% paraformaldehyde in PBS, pH 7.2, and incubated for 15 min at room temperature; cells were centrifuged as described above, washed once with 10 mm glycine in PBS and once with PBS, resuspended in 100 μl PBS, spotted onto poly-l-lysine-coated coverslips, blocked with 2% goat serum, 0.01% sodium azide, 0.01% saponin in PBS (blocking solution) for 1 h at room temperature, rinsed three times with PBS, and incubated with primary antibodies for 1 h at room temperature. The following primary antibodies were employed: 1:500 dilution anti-HA.11 clone 16B12 mAb (BioLegend, catalog no. MMS-101R, lot no. D13FF01558), 1:1000 dilution anti-HA polyclonal antibody (pAb) (Sigma-Aldrich, catalog no. H6908, batch no. 083M4837V), 1:500 dilution anti-Ty1 (BB2) mAb (53), 1:1000 dilution anti-α-tubulin mAb (Sigma-Aldrich, catalog no. T5168, batch no. 104M4790V), 1:1000 dilution anti-α-tubulin pAb (Sigma-Aldrich, catalog no. SAB3501071, lot no. 75971302), 1:1000 anti-β-tubulin clone KMX-1 mAb (Millipore, catalog no. MAB3408, lot no. 2646950), and 1:1000 anti-GFP pAb (Molecular Probes, catalog no. A11122, lot no. 847818). Subsequently, the cells were rinsed as before and incubated with a 1:1000 dilution of secondary antibodies coupled to Alexa Fluor dyes (Molecular Probes) as follows: Alexa Fluor® 488 goat anti-mouse IgG (H+L) (catalog no. A11001, lot no. 1572559), Alexa Fluor® 595 goat anti-mouse IgG (H+L) (catalog no. A11005, lot no. 1524907), Alexa Fluor® 594 goat anti-rabbit IgG (H+L) (catalog no. A11012, lot no. 84B2-1), and Alexa Fluor® 488 goat anti-rabbit IgG (H+L) (catalog no. A11008, lot no. 84B1-1), as indicated, in blocking solution for 1 h at room temperature in the dark. Coverslips were rinsed three times with PBS and mounted onto slides using DAPI Fluoromount-G (SouthernBiotech). Fluorescence images were acquired using a wide field deconvolution system (Applied Precision Instruments, Inc.) consisting of an inverted Nikon TE 200 Eclipse microscope, a Kodak CH350 CCD camera and the Deltavision operating system. Images were acquired using a 60× objective in a 1024 × 1024 format and deconvolved with nine iterations using SoftWoRx software. Adobe Photoshop CS3 and Adobe Illustrator CS3 (Adobe Systems Inc.) were used to create image compositions.
Images that included differential interference contrast (DIC) were acquired on a high resolution wide field Core DV system (Applied PrecisionTM). This system is an Olympus IX71 inverted microscope with a proprietary XYZ stage enclosed in a controlled environment chamber; DIC transmitted light and a solid state module for fluorescence. The camera is a Nikon Coolsnap ES2 HQ. Each image was acquired as Z stacks in a 1024 × 1024 format with a 60× 1.42 NA PlanApo lens in three colors: Alexa Fluor® 488, Alexa Fluor® 594, and DAPI. The pixel size in XYZ was 0.107 × 0.107 × 0.2 microns. The images were deconvolved with the appropriate optical transfer function using an iterative algorithm of 10 iterations. The histogram was optimized for the most positive image and applied to all the other images for consistency before saving the images as 24-bit merged TIFF. A reference DIC image was acquired from the middle of the Z stack.
Flagellar Protein Purification, Formaldehyde Cross-linking, Co-immunoprecipitation, and Western Blotting Assay
Flagellar purification was performed as described (26) with slight changes. Briefly, BF/TbKHRNAi clones or BF/TbFLA1RNAi clones were grown in 1 μg ml−1 doxycycline for 48 or 72 h, respectively, and pelleted at 420 × g for 20 min. Cell pellets were washed with buffer A (25 mm Na+-Tricine, pH 7, 1% BSA, 0.1 mm CaCl2, 0.2 mm EDTA, 5 mm MgCl2, and 12 mm β-mercaptoethanol) containing 0.32 m sucrose and centrifuged at 420 × g for 10 min. Cell pellets were gently resuspended at 3 × 108 parasites ml−1 in buffer A plus 0.3 m sucrose, transferred into Eppendorf tubes and vortexed for 5 min or until microscopic verification of flagellum detachment, followed by centrifugation at 420 × g for 10 min. Supernatants were recovered and centrifuged at 1600 × g for 10 min at 4 °C. Pellets containing the isolated flagella were resuspended in 25 μl of PBS and used for Western blotting assays. Also, flagellar cytoskeleton purification from wild type BF and PF clones was performed by isolation of cytoskeletons using Nonidet P-40 extraction, followed by 1 m NaCl treatment according to Broadhead et al. (31) to dissociate the subpellicular cytoskeleton. The remaining flagellar cytoskeletons were processed for immunofluorescence microscopy as indicated.
For formaldehyde cross-linking and co-immunoprecipitation, cell lines were prepared that expressed TbKH endogenously tagged with the Ty1 epitope at the N terminus (Ty1::TbKH) and also co-expressed a flagellar membrane protein of interest endogenously tagged at its C terminus with an HA3 epitope (FS33::HA3, FS60::HA3, FA133:HA3, and FS179::HA3) (5). Parasites were washed once with PBS and pelleted at 1000 × g for 10 min, and cell pellets were resuspended in 9.37 ml of PBS plus 0.63 ml of 16% formaldehyde-EM grade (Polysciences, Inc.) and incubated at 37 °C for 10 min. Subsequently, 1 ml of 2.5 m glycine in PBS was added to the cross-linked samples and incubated at 37 °C for 5 min to terminate cross-linking, and cross-linked parasites were pelleted at 1000 × g for 10 min and washed twice with PBS. Finally, cell pellets were resuspended in 500 μl of Tween 20/PBS buffer (0.2% Tween 20 in 1× PBS, pH 7.2) containing 1× of HaltTM protease inhibitor single-use mixture (Thermo Fisher). Cross-linked samples were sonicated on ice using two 15-s pulses at maximum output (Branson Sonifer 150). After disruption, samples were centrifuged at 16,000 × g for 20 min at 4 °C. Supernatants were transferred to new Eppendorf tubes and incubated overnight with 1:50 dilution of BB2 mAb at 4 °C on a rocker. Later, 30 μl of PureProteomeTM protein A/G mix magnetic beads (Millipore) were incubated for 30 min at room temperature on a rocker to capture Ty1::TbKH proteins that had been cross-linked to other partner proteins. Two final washes of the magnetic beads for 30 s at room temperature with radioimmune precipitation assay buffer were performed to increase stringency and reduce background. Co-immunoprecipitated proteins were eluted, and cross-linking was reversed by adding 30 μl of 1× NUPAGE®-LDS sample buffer (Life Technologies) containing 10 mm DTT followed by heating at 70 °C for 30 min. Eluted protein samples were used for Western blotting analysis.
Flagellar protein extracts or co-immunoprecipitated protein extracts from 2.5 × 107 parasites per lane were resolved on NUPAGE® 4–12% Bis-Tris Mini Gels using the XCell SureLock® Mini-Cell system following the manufacturer's instructions (Life Technologies), and proteins were transferred onto PVDF membranes (Millipore). Anti-HA.11 clone 16B12 mAb (BioLegend, catalog no. MMS-101R, lot no. D13FF01558) 1:2500 dilution, BB2 mAb (53) 1:500 dilution, and anti-α-tubulin mAb (Sigma-Aldrich, catalog no. T5168, batch no. 104M4790V) 1:10000 dilution were used as primary antibodies, and goat anti-mouse-HRP-conjugated (Jackson ImmunoResearch Laboratories, catalog no. 115-03-174, lot no. 117119) 1:15000 dilution was employed as secondary antibody. SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher) was used for detection, and an Image Quant LAS 400 (GE Healthcare) scanner was employed to acquire luminescent images. Adobe Photoshop CS3 and Adobe Illustrator CS3 (Adobe Systems Inc.) were used to create image compositions. Immunoblots of total BF cell lysates, such as those in Figs. 4B and 6H, were performed by the same method, and ImageJ was employed for quantification of band densities.
Functional Complementation of L. mexicana kh Null Mutant (Δlmxkh) with TbKH, and Macrophage Infections
Transgenic Δlmxkh overexpressing LmxGT1::GFP, previously generated (21), was transfected with the TbKH ORF cloned into the pX63HYG expression vector (56) for episomal expression. Stable transfectants were selected on bleomycin (50 μg ml−1')/puromycin (50 μg ml−1)/neomycin (100 μg ml−1)/hygromycin (80 μg ml−1) and tested for their ability to translocate LmxGT1::GFP to the flagellar membrane as monitored by immunofluorescence microscopy as indicated above.
Macrophage infections were performed by infecting differentiated macrophages from the human acute leukemia monocyte cell line (THP-1) in triplicate samples, as described (57). Briefly, 3 × 105 differentiated THP-1 cells were infected with stationary phase L. mexicana (10:1 promastigotes:macrophages ratio) for 4 h. Infected macrophages were then washed exhaustively to remove extracellular promastigotes. Cultures were incubated at 37 °C in a 5% CO2 environment, and infected macrophages were stained using the HEMA 3 STAT PACK staining kit as described by the manufacture (Fisher Scientific) at 1 and 7 days after infection. Stained infected macrophages were examined using a Nikon Eclipse 50i microscope equipped with a 100s× 1.25 NA oil objective (Nikon Instruments). The number of parasites per 100 macrophages was determined by counting 100 cells in each of the triplicate experiments.
Electron Microscopy
BF/TbKHRNAi clone was grown in the presence of 1 μg ml−1 doxycycline to induce RNAi against TbKH mRNA, and parasite samples were harvested at 0, 18, 24, and 48 h postinduction. Parasites were fixed in 2.5% glutaraldehyde by addition of 1/10 volume of 25% glutaraldehyde directly to the culture and incubated at room temperature for 5 min. Fixed cells were subsequently collected and centrifuged for 10 min at 800 × g. The cells were resuspended in 1 ml of 0.1 m sodium phosphate buffer, pH 7.4, containing 50 mm sucrose and pelleted for 10 min at 800 × g. The cells were then resuspended in 1 ml of buffered fixative (2.5% glutaraldehyde, 2% formaldehyde, and 50 mm sucrose in 0.1 m phosphate buffer, pH 7.4), pelleted for 1 min at 21,000 × g in a microcentrifuge, and stored at 4 °C.
Fixed samples were prepared for transmission electron microscopy and serial block face scanning electron microscopy. Transmission electron microscopy samples were prepared and imaged as previously described (57). Serial block face scanning electron microscopy samples were washed in 0.1 m PIPES buffer and then embedded in a mix of 4% low melting point agarose and 4% gelatin from porcine skin. Excess agarose/gelatin was trimmed away, and samples were prepared according to Wilke et al. (58) with minor modifications: samples were incubated at 4 °C for 1 h in a 1:1 mix of 3% potassium ferricyanide in 0.2 m PIPES buffer and 4% aqueous osmium tetroxide. Samples were washed with double distilled (dd) H2O between each of the subsequent heavy metal staining steps, which consisted of 1% thiocarbohydrazide in ddH2O for 20 min at room temperature, 2% aqueous osmium tetroxide for 30 min at 4 °C, 1% uranyl acetate overnight at 4 °C, and lead aspartate (0.02 m lead nitrate and 0.03 m aspartic acid) for 30 min at room temperature. Final washes in ddH2O were performed prior to dehydration through a graded series of ethanol and acetone incubations. The samples were then infiltrated with and embedded in Durcupan ACM epoxy resin. Serial block face images were captured with a Zeiss Merlin Compact field emission gun scanning electron microscope with variable pressure mode and a Gatan 3View system using digital micrograph with an accelerating voltage of 3 kV, variable pressure of 45 Pa, aperture size of 30 μm, 4.7-nm lateral resolution, and 50-nm axial resolution and processed using ImageJ (59).
Synchronization of L. mexicana with Hydroxyurea
Promastigotes of L. mexicana were synchronized with hydroxyurea as described by da Silva et al. (60). Briefly, mid-log phase promastigotes were incubated in RPMI culture medium containing 5 mm hydroxyurea for 14 h at 26 °C. Later, parasites were pelleted by centrifugation at 1600 × g for 10 min at room temperature to remove hydroxyurea, resuspended in fresh RPMI, and incubated for 7 h at 26 °C. Subsequently, parasite samples were processed for fluorescence microscopy as indicated.
Author Contributions
M. A. S. designed, performed, and analyzed the experiments in Figs. 1–4 and 6–11 and contributed to writing the paper. K. D. T. designed, preformed, and analyzed the experiments in Fig. 11. J. V., E. J., and E. G. designed, performed, and analyzed the experiments in Fig. 5. S. H. performed and analyzed the experiments in Fig. 3. S. M. L. designed and analyzed experiments in Figs. 1–4 and 6–9 and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Kent Hill for providing constructs encoding the FS179, FS60, FS33, and FS133 proteins fused to HA3 epitopes tags at their C termini. We also thank Brooke Morriswood for thoughtful comments on the manuscript. We acknowledge the Advanced Light Microscopy Core at the Jungers Center of the Oregon Health & Science University for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R21-AI114822 and R01-AI121160 (to S. M. L.). This work was also supported by Biotechnology and Biological Sciences Research Council Grant BB/C014122/1 and Shared Instrumentation Grant S10-RR023432 from the National Center for Research Resources, a component of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The L. mexicana protein was originally designated KHARON1, or KH1, to allow subsequent numbering of additional subunits in a KHARON complex. However, for simplicity, we now refer to KHARON1 as KHARON or KH for both L. mexicana and T. brucei, and associated proteins, currently under investigation, will be designated KHARON-associated proteins, or KHAPs.
K. D. Tran and M. A. Sanchez, unpublished data.
- BF
- bloodstream form
- PF
- procyclic form
- DIC
- differential interference contrast
- ddH2O
- double distilled H2O.
References
- 1. Franco J. R., Simarro P. P., Diarra A., and Jannin J. G. (2014) Epidemiology of human African trypanosomiasis. Clin. Epidemiol. 6, 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brun R., Blum J., Chappuis F., and Burri C. (2010) Human African trypanosomiasis. Lancet 375, 148–159 [DOI] [PubMed] [Google Scholar]
- 3. Ralston K. S., Kabututu Z. P., Melehani J. H., Oberholzer M., and Hill K. L. (2009) The Trypanosoma brucei flagellum: moving parasites in new directions. Annu. Rev. Microbiol. 63, 335–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Langousis G., and Hill K. (2014) Motility and more: the flagellum of Trypanosoma brucei. Nat. Rev. Microbiol. 12, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oberholzer M., Langousis G., Nguyen H., Saada E., Shimogawa M., Jonsson Z., Nguyen S., Wohlschlegel J., and Hill K. (2011) Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol. Cell. Proteomics 10, M111.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alexandre S., Paindavoine P., Hanocq-Quertier J., Paturiaux-Hanocq F., Tebabi P., and Pays E. (1996) Families of adenylate cyclase genes in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 173–182 [DOI] [PubMed] [Google Scholar]
- 7. Salmon D., Vanwalleghem G., Morias Y., Denoeud J., Krumbholz C., Lhommé F., Bachmaier S., Kador M., Gossmann J., Dias F. B., De Muylder G., Uzureau P., Magez S., Moser M., De Baetselier P., et al. (2012) Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science 337, 463–466 [DOI] [PubMed] [Google Scholar]
- 8. Lopez M. A., Saada E. A., and Hill K. L. (2015) Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes. Eukaryot. Cell 14, 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saada E. A., Kabututu Z. P., Lopez M., Shimogawa M. M., Langousis G., Oberholzer M., Riestra A., Jonsson Z. O., Wohlschlegel J. A., and Hill K. L. (2014) Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei flagellar membrane. Eukaryot. Cell 13, 1064–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oberholzer M., Saada E., and Hill K. (2015) Cyclic AMP regulates social behavior in African trypanosomes. MBio 6, e01954–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bassarak B., Uzcátegui N. L., Schönfeld C., and Duszenko M. (2011) Functional characterization of three aquaglyceroporins from Trypanosoma brucei in osmoregulation and glycerol transport. Cell Physiol. Biochem. 27, 411–420 [DOI] [PubMed] [Google Scholar]
- 12. Rotureau B., Subota I., Buisson J., and Bastin P. (2012) A new asymmetric division contributes to the continuous production of infective trypanosomes in the tsetse fly. Development 139, 1842–1850 [DOI] [PubMed] [Google Scholar]
- 13. Peacock L., Ferris V., Bailey M., and Gibson W. (2014) Mating compatibility in the parasitic protist Trypanosoma brucei. Parasit. Vectors 7, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farr H., and Gull K. (2012) Cytokinesis in trypanosomes. Cytoskeleton 69, 931–941 [DOI] [PubMed] [Google Scholar]
- 15. Hughes L., Towers K., Starborg T., Gull K., and Vaughan S. (2013) A cell-body groove housing the new flagellum tip suggests an adaptation of cellular morphogenesis for parasitism in the bloodstream form of Trypanosoma brucei. J. Cell Sci. 126, 5748–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wheeler R. J., Scheumann N., Wickstead B., Gull K., and Vaughan S. (2013) Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: implications for microtubule-based morphogenesis and mutant analysis. Mol. Microbiol. 90, 1339–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wingard J. N., Ladner J., Vanarotti M., Fisher A. J., Robinson H., Buchanan K. T., Engman D. M., and Ames J. B. (2008) Structural insights into membrane targeting by the flagellar calcium-binding protein (FCaBP), a myristoylated and palmitoylated calcium sensor in Trypanosoma cruzi. J. Biol. Chem. 283, 23388–23396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maric D., Olson C. L., Xu X., Ames J. B., and Engman D. M. (2015) Calcium-dependent membrane association of a flagellar calcium sensor does not require calcium binding. Mol. Biochem. Parasitol. 201, 72–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Denny P. W., Gokool S., Russell D. G., Field M. C., and Smith D. F. (2000) Acylation-dependent protein export in Leishmania. J. Biol. Chem. 275, 11017–11025 [DOI] [PubMed] [Google Scholar]
- 20. Mills E., Price H. P., Johner A., Emerson J. E., and Smith D. F. (2007) Kinetoplastid PPEF phosphatases: dual acylated proteins expressed in the endomembrane system of Leishmania. Mol. Biochem. Parasitol. 152, 22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tran K. D., Rodriguez-Contreras D., Vieira D. P., Yates P. A., David L., Beatty W., Elferich J., and Landfear S. M. (2013) KHARON1 mediates flagellar targeting of a glucose transporter in Leishmania mexicana and is critical for viability of infectious intracellular amastigotes. J. Biol. Chem. 288, 22721–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sunter J., Wickstead B., Gull K., and Carrington M. (2012) A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PLoS One 7, e35167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolev N. G., Tschudi C., and Ullu E. (2011) RNA interference in protozoan parasites: achievements and challenges. Eukaryot. Cell 10, 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LaCount D. J., Barrett B., and Donelson J. E. (2002) Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277, 17580–17588 [DOI] [PubMed] [Google Scholar]
- 25. Sunter J. D., Benz C., Andre J., Whipple S., McKean P. G., Gull K., Ginger M. L., and Lukes J. (2015) Modulation of flagellum attachment zone protein FLAM3 and regulation of the cell shape in Trypanosoma brucei life cycle transitions. J. Cell Sci. 128, 3117–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subota I., Julkowska D., Vincensini L., Reeg N., Buisson J., Blisnick T., Huet D., Perrot S., Santi-Rocca J., Duchateau M., Hourdel V., Rousselle J. C., Cayet N., Namane A., Chamot-Rooke J., i (2014) Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localisation and dynamics. Mol. Cell. Proteomics 13, 1769–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sasse R., and Gull K. (1988) Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell Sci. 90, 577–589 [DOI] [PubMed] [Google Scholar]
- 28. Sun S. Y., Wang C., Yuan Y. A., and He C. Y. (2013) An intracellular membrane junction consisting of flagellum adhesion glycoproteins links flagellum biogenesis to cell morphogenesis in Trypanosoma brucei. J. Cell Sci. 126, 520–531 [DOI] [PubMed] [Google Scholar]
- 29. Gluenz E., Povelones M. L., Englund P. T., and Gull K. (2011) The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Mol. Cell Biol. 31, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanchez M. (2013) Molecular identification and characterization of an essential pyruvate transporter from Trypanosoma brucei. J. Biol. Chem. 288, 14428–14437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Broadhead R., Dawe H. R., Farr H., Griffiths S., Hart S. R., Portman N., Shaw M. K., Ginger M. L., Gaskell S. J., McKean P. G., and Gull K. (2006) Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440, 224–227 [DOI] [PubMed] [Google Scholar]
- 32. Luo S., Rohloff P., Cox J., Uyemura S. A., and Docampo R. (2004) Trypanosoma brucei plasma membrane-type Ca2+-ATPase 1 (TbPMC1) and 2 (TbPMC2) genes encode functional Ca2+-ATPases localized to the acidocalcisomes and plasma membrane, and essential for Ca2+ homeostasis and growth. J. Biol. Chem. 279, 14427–14439 [DOI] [PubMed] [Google Scholar]
- 33. Forsythe G. R., McCulloch R., and Hammarton T. C. (2009) Hydroxyurea-induced synchronisation of bloodstream stage Trypanosoma brucei. Mol. Biochem. Parasitol. 164, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berbari N. F., O'Connor A. K., Haycraft C. J., and Yoder B. K. (2009) The primary cilium as a complex signaling center. Curr. Biol. 19, R526–R535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nachury M. V., Seeley E. S., and Jin H. (2010) Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier. Annu. Rev. Cell Dev. Biol. 26, 59–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szempruch A. J., Sykes S. E., Kieft R., Dennison L., Becker A. C., Gartrell A., Martin W. J., Nakayasu E. S., Almeida I. C., Hajduk S. L., and Harrington J. M. (2016) Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell 164, 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imhof S., Fragoso C., Hemphill A., Von Schubert C., Li D., Leganty W., Betzig E., and Roditi I. (2016) Flagellar membrane fusion and protein exchange in trypanosomes: a new form cell-cell comunication? F1000Res. 5, 682–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Imhof S., Vu X. L., Bütikofer P., and Roditi I. (2015) A glycosylation mutant of Trypanosoma brucei links social motility defects in vitro to impaired colonization of tsetse flies in vivo. Eukaryot. Cell 14, 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webster P., and Russell D. (1993) The flagellar pocket of trypanosomatids. Parasitol. Today 9, 201–206 [DOI] [PubMed] [Google Scholar]
- 40. Serricchio M., Schmid A. W., Steinmann M. E., Sigel E., Rauch M., Julkowska D., Bonnefoy S., Fort C., Bastin P., and Bütikofer P. (2015) Flagellar membranes are rich in raft-forming phospholipids. Biol. Open 4, 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kohl L., Robinson D., and Bastin P. (2003) Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22, 5336–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Branche C., Kohl L., Toutirais G., Buisson J., Cosson J., and Bastin P. (2006) Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 119, 3443–3455 [DOI] [PubMed] [Google Scholar]
- 43. Ralston K. S., Lerner A. G., Diener D. R., and Hill K. L. (2006) Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell 5, 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baron D. M., Ralston K. S., Kabututu Z. P., and Hill K. L. (2007) Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J. Cell Sci. 120, 478–491 [DOI] [PubMed] [Google Scholar]
- 45. Patel A. (2015) The primary cilium calcium channels and their role in flow sensing. Pflugers Arch. 467, 157–165 [DOI] [PubMed] [Google Scholar]
- 46. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Deursen F. J., Shahi S. K., Turner C. M., Hartmann C., Guerra-Giraldez C., Matthews K. R., and Clayton C. E. (2001) Characterisation of the growth and differentiation in vivo and in vitro-of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasitol. 112, 163–171 [DOI] [PubMed] [Google Scholar]
- 48. Ortiz D., Sanchez M. A., Quecke P., and Landfear S. M. (2009) Two novel nucleobase/pentamidine transporters from Trypanosoma brucei. Mol. Biochem. Parasitol. 163, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burkard G., Fragoso C. M., and Roditi I. (2007) Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 153, 220–223 [DOI] [PubMed] [Google Scholar]
- 50. McCulloch R., Vassella E., Burton P., Boshart M., and Barry J. (2004) Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262, 53–86 [DOI] [PubMed] [Google Scholar]
- 51. Hirumi H., and Hirumi K. (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75, 985–989 [PubMed] [Google Scholar]
- 52. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratoty, Cold Spring Harbor, NY [Google Scholar]
- 53. Bastin P., Bagherzadeh Z., Matthews K. R., and Gull K. (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235–239 [DOI] [PubMed] [Google Scholar]
- 54. Shen S., Arhin G. K., Ullu E., and Tschudi C. (2001) In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113, 171–173 [DOI] [PubMed] [Google Scholar]
- 55. Oberholzer M., Morand S., Kunz S., and Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 56. Cruz A., Coburn C. M., and Beverley S. M. (1991) Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. U.S.A. 88, 7170–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tran K. D., Vieira D. P., Sanchez M. A., Valli J., Gluenz E., and Landfear S. M. (2015) Kharon1 null mutants of Leishmania mexicana are avirulent in mice and exhibit a cytokinesis defect within macrophages. PLoS One 10, e0134432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wilke S. A., Antonios J. K., Bushong E. A., Badkoobehi A., Malek E., Hwang M., Terada M., Ellisman M. H., and Ghosh A. (2013) Deconstructing complexity: serial block-face electron microscopic analysis of the hippocampal mossy fiber synapse. J. Neurosci. 33, 507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abramoff M. D., Magalhães P. J., and Ram Sunanda J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 60. da Silva M. S., Monteiro J. P., Nunes V. S., Vasconcelos E. J., Perez A. M., Freitas-Júnior Lde H., Elias M. C., and Cano M. I. (2013) Leishmania amazonensis promastigotes present two distinct modes of nucleus and kinetoplast segregation during cell cycle. PLoS One 8, e81397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Birkett C. R., Foster K. E., Johnson L., and Gull K. (1985) Use of monoclonal antibodies to analyse the expression of a multi-tubulin family. FEBS Lett. 187, 211–218 [DOI] [PubMed] [Google Scholar]
- 62. Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., and Gull K. (2000) Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma 108, 501–513 [DOI] [PubMed] [Google Scholar]