Abstract
Heat shock cognate protein 70 (Hsc70) regulates protein homeostasis through its reversible interactions with client proteins. Hsc70 has two major domains: a nucleotide-binding domain (NBD), that hydrolyzes ATP, and a substrate-binding domain (SBD), where clients are bound. Members of the BAG family of co-chaperones, including Bag1 and Bag3, are known to accelerate release of both ADP and client from Hsc70. The release of nucleotide is known to be mediated by interactions between the conserved BAG domain and the Hsc70 NBD. However, less is known about the regions required for client release, and it is often assumed that this activity also requires the BAG domain. It is important to better understand this step because it determines how long clients remain in the inactive, bound state. Here, we report the surprising observation that truncated versions of either human Bag1 or Bag3, comprised only the BAG domain, promoted rapid release of nucleotide, but not client, in vitro. Rather, we found that a non-canonical interaction between Bag1/3 and the Hsc70 SBD is sufficient for accelerating this step. Moreover, client release did not seem to require the BAG domain or Hsc70 NBD. These results suggest that Bag1 and Bag3 control the stability of the Hsc70-client complex using at least two distinct protein-protein contacts, providing a previously under-appreciated layer of molecular regulation in the human Hsc70 system.
Keywords: 70 kilodalton heat shock protein (Hsp70), molecular chaperone, nuclear magnetic resonance (NMR), protein complex, protein-protein interaction
Introduction
Heat shock cognate protein 70 (Hsc70, HSPA8)2 is a molecular chaperone that regulates the folding, trafficking, function, stability, and turnover of its client proteins (1, 2). The versatility of the Hsc70 system is thought to arise from its association with co-chaperones. Co-chaperones bind to Hsc70 and, among other activities, help shuttle clients to cellular systems dedicated to folding, turnover, and other fates (3, 4). Accordingly, there is an interest in understanding the protein-protein interactions between Hsc70 and its co-chaperones because these contacts are important for “fine tuning” protein quality control and homeostasis.
Hsc70 is a 70-kDa chaperone that is composed of a nucleotide-binding domain (NBD) and a substrate-binding domain (SBD) (5). The SBD is further divided into a β-sandwich subdomain that contains a hydrophobic cleft for client binding (6), a helical “lid” subdomain that controls access to the cleft and a disordered C-terminal domain (CTD), which is a site of post-translational modifications and protein-protein interactions (7, 8). On the other side of the chaperone, the NBD is subdivided into two lobes (lobe I and lobe II) that each contributes to a nucleotide-binding cleft in which ATP is bound and hydrolyzed. Much of our knowledge of Hsc70 structure and dynamics has come from studies on the prokaryotic ortholog, DnaK. However, the high sequence conservation of this family of proteins (often ∼50% identity) suggests that similar mechanisms might be relevant across orthologs and paralogs (9). Despite this high level of homology, there also appears to be some differences in the regulation of the prokaryotic and eukaryotic family members (10, 11), including in the way that they interact with co-chaperones (12).
One of the most striking structural features of Hsc70/DnaK is the large motions of the NBD that accompany ATP binding and conversion to the ADP-bound state (13–15). These motions are, in turn, propagated into the SBD through a short, conserved linker and an allosteric network (16–19). Briefly, binding to ATP favors an “open” configuration of the lid subdomain (20), which occurs concurrent with docking of the NBD to the SBD. This ATP-bound conformer has relatively weak affinity for clients, as the client's off-rate is increased when the lid is open (21, 22). To continue the cycle, hydrolysis of nucleotide reorganizes lobes I and II, which “closes” the lid subdomain and strengthens the affinity for clients. In this ADP-bound state, the NBD and SBD are no longer docked; rather, they move relatively independently in solution (23). Because of these dramatic conformational changes, Hsc70/DnaK is often used as a model for intra- and inter-domain allostery in multi-domain proteins (5, 15).
Each step of Hsc70's conformational cycle is regulated by protein-protein interactions with co-chaperones. In eukaryotes, the BAG family of co-chaperones, including Bag1, Bag2, Bag3, Bag4, and Bag5, act as nucleotide-exchange factors (NEFs): promoting the release of ADP and client from the Hsc70 complex (4, 24, 25). This activity is important because it resolves a potentially rate-limiting step in the hydrolysis cycle, allowing Hsc70 to re-enter the ATP-bound state (26). The NEFs also govern the persistence and stability of the client complex (27). Most clients cannot fully fold when bound to Hsc70, such that dissociation is likely required for them to mature and function. Indeed, recent studies have shown that members of the Hsc70 family inhibit ligand binding to glucocorticoid receptor (28). In addition, this interaction keeps aggregation-prone clients, such as human telomerase repeat binding factor (hTRF1), on the folding trajectory by allowing proper secondary structures to form (29). Thus, Hsc70-client complexes seem to represent a “timing” mechanism for client folding, function and turnover, possibly helping to ensure proper quality control. These observations have also focused attention on the protein-protein interaction between Hsc70 and BAG family proteins as potential targets for drug discovery (3). Indeed, allosteric inhibitors of this interaction trigger degradation of a subset of Dengue viral proteins (30) and some oncogenes (31).
Each member of the BAG family of co-chaperones contains a conserved BAG domain that is important for binding to Hsc70. The BAG domain folds into a three-helix bundle that interacts with the NBD of human paralogs, including Hsc70 (HSPA8) and the closely related Hsp72 (HSPA1A). Co-crystal structures of Hsc70NBD bound to BAG domains have shown that the protein-protein interaction involves a large, polar surface that spans the “top” of lobes I and II (32–35). These co-crystal structures have also suggested a mechanism in which BAGs “open” the lobes of the NBD, favoring release of ADP and re-binding to ATP. This mechanism is reminiscent of that used by structurally unrelated NEFs, such as prokaryotic GrpE (36), although the details of the interaction and the surfaces involved can be different. Regardless of the identity of the NEF, conversion to the ATP-bound state would be expected to indirectly favor the release of clients from Hsc70, because this conformer has relatively weaker affinity (37). Consistent with that idea, full-length Bag1, Bag2, and Bag3 accelerate the release of a fluorescent client peptide from human Hsp72 and Hsc70 in vitro (38). Also, NEFs are known to release cholera toxin from inactive, chaperone-bound complexes in cells (39). However, other observations suggest that NEFs might release clients through a mechanism that is more complex. For example, Hsc70NBD binds to full-length Bag1 with an affinity of ∼12 nm in the ATP-bound state by isothermal calorimetry (ITC), but it only binds with an affinity of 95 nm to a truncated BAG domain under the same conditions (38). This result suggests that a secondary binding site might also contribute.
While studying the interaction between human Hsc70 and Bag1/3 in more detail, we made the surprising discovery that truncated variants, composed of only the BAG domain, are poor at promoting release of bound clients. This result suggested the possibility that, in the human system, release of ADP and client may not necessarily be linked. Using in vitro assays for nucleotide and client release, we characterized a non-canonical interaction between Bag1/3 and the Hsc70SBD that is essential for client release. This secondary contact was confirmed using an NMR-based binding assay and a truncated form of Bag3 that lacks the BAG domain. Together, these results support a revised model in which members of the BAG family of co-chaperones use two separate domains to promote release of nucleotide and clients from Hsc70.
Results
Isolated BAG Domains Promote Release of Nucleotide, but Not Model Client
In this study, we focused on the interaction of human Hsc70 (HSPA8) with Bag1 and Bag3. These systems were chosen because the Hsc70-Bag1 complex has been implicated in directing clients to the ubiquitin-proteasome system (UPS) (40), while the Hsc70-Bag3 pair links some clients to the macroautophagy pathway (41). Thus, these co-chaperones are some of the best-characterized BAG family members and they seem to be important in guiding client fate decisions under some conditions, such as antigen presentation (42). Further, the affinities of both protein-protein interactions are similar (Bag1 Kd ∼12 nm; Bag3 Kd ∼10 nm; binding to Hsc70NBD in the ATP-bound form by ITC) (38). To understand if nucleotide and client release activities are separable, we used two fluorescence polarization (FP) assays that have been previously developed (38). In the first, we saturated Hsc70 with a fluorescent nucleotide (N6-(6-amino)hexyl-ATP-5-FAM; ATP-FAM) and then measured nucleotide release after addition of BAG protein. ATP and ADP have similar Ki values in this platform (38), so it is often used to measure nucleotide release. In the second assay, a fluorescent client based on the well-characterized HLA peptide (FAM-RENLRIALRY; FAM-HLA) (43) was loaded into Hsc70 and its release by BAG proteins was measured. To make this assay especially sensitive to NEF activity, binding is carried out under conditions of high ADP (1 mm), such that the tight-binding conformer of Hsc70 is purposefully favored. In the first experiments, we compared the activities of full-length Bag1 and Bag3 to that of truncated versions composed of only the BAG domain (termed Bag1C (residues 107–219) and Bag3C (residues 421–499); Fig. 1A). In the nucleotide release assay, all of the proteins had approximately equivalent activity (EC50 values ∼0.4 to 0.7 μm; Fig. 1B), consistent with the reported importance of the BAG domain in ADP release (38). However, we found that Bag1C and Bag3C had only very weak ability to promote client release (EC50 > 10 μm; Fig. 1B). This was an unexpected result, which suggested that regions outside the BAG domain might be essential for this process.
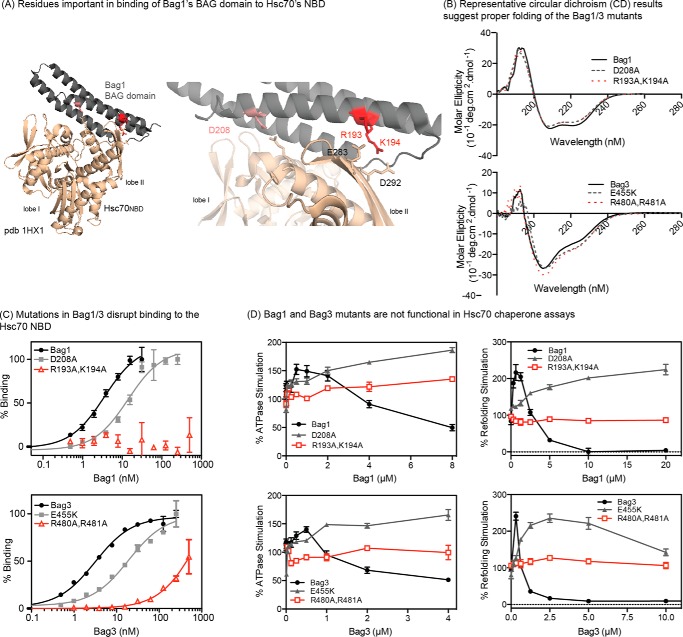
FIGURE 1.

Isolated BAG domains promote release of nucleotide, but not client, from Hsc70. A, partial domain architecture of human Bag1 and Bag3, highlighting the location of the BAG domain. Some known domains are not indicated for clarity. Bag1C and Bag3C are the isolated BAG domains. B, full-length Bag1/3, but not Bag1C or Bag3C, strongly promote release of a fluorescent client from Hsc70. The results are the average of four independent experiments performed in triplicate each. The error bars represent S.D. All of the NEFs were able to promote nucleotide release, as previously reported (see text).
Point Mutations in the BAG Domain Prevent Nucleotide Release, but Not Client Release
To disrupt binding of the BAG domains to Hsc70 with greater precision, we examined the co-crystal structure of Bag1's BAG domain bound to Hsc70NBD (PDB = 1HX1) (34). Residues in Bag1 that are predicted to be involved in key contacts were then selected for mutation, including D208A, R193A, and K194A (Fig. 2A). There is no reported co-crystal structure of Hsc70NBD with the BAG domain of Bag3, so we used primary sequence alignment (supplemental Fig. S1), intuition and homology models to select E455K, R480A, and R481A for mutation. We found that the single point mutations had relatively modest effects on affinity by themselves (see below), so we also expressed and purified the double mutants: Bag1 (R193A,K194A) and Bag3 (R480A,R481A). The mutant proteins were all folded normally, as judged by circular dichroism (Fig. 2B). Using a flow cytometry protein interaction assay (FCPIA), we found that the mutants, especially Bag1 (R193A,K194A) and Bag3 (R480A, R481A), had significantly reduced affinity for Hsc70NBD (Fig. 2C). These results suggested that the double mutants would be good tools for selectively disrupting the BAG-Hsc70NBD contact. To confirm this idea, we tested them in ATP turnover and luciferase-refolding assays, which are methods that are commonly used to characterize the effects of NEFs on Hsc70's enzyme functions in vitro (44). As expected, the double mutants were unable to promote Hsc70's steady state ATP turnover or firefly luciferase refolding activity (Fig. 2D). In these assays, NEFs, such as Bag1 or Bag3, are typically able to stimulate Hsc70 at low concentrations and then inhibit at higher concentrations. Activation occurs when the NEFs act on the rate-limiting step in the functional cycle. The apparent inhibitory effect is less intuitive, but is interpreted as an overstabilization of discrete conformers, such as the apo-state, during nucleotide cycling. Consistent with an intermediate loss of affinity, the single mutants, Bag1 D208A and Bag3 E455K, were still able to stimulate Hsc70, but only at high concentrations and the inhibitory effect was largely abolished (at least under the concentrations attainable in these platforms). The double mutants were not able to impact Hsc70 functions at all, consistent with the idea that they no longer have functional BAG domains.
FIGURE 2.
Design of point mutations in Bag1 and Bag3 that disrupt binding of the BAG domain to Hsc70. A, binding interface between Bag1/3 and Hsc70's NBD, highlighting the polar contacts. B, CD spectra of Bag1 and Bag3 proteins, showing that the mutations do not disrupt the overall structure. C, mutations in Bag1 or Bag3 interrupt binding to the Hsc70 NBD, as measured by FCPIA. D, mutations in Bag1/3 disrupt effects on the Hsc70 ATPase activity (left) and the refolding of denatured firefly luciferase (right). NEFs, such as Bag1 and Bag3, typically stimulate ATPase and luciferase refolding activity at low concentrations and then inhibit at higher ratios. In C and D, the results are shown as the average of at least three experiments performed in triplicate. The error bars represent S.D.
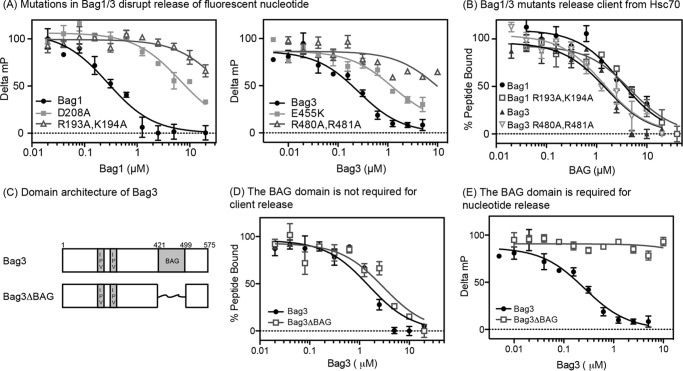
Next, we asked whether the mutants could release nucleotide and/or client from the Hsc70-client complex using the FP assays. As expected, Bag1 (R193A,K194A) and Bag3 (R480A, R481A) were only able to weakly promote release of nucleotide (EC50 > 10 μm; Fig. 3A), likely because their BAG domains were compromised. However, these mutants were normal in their ability to release client (EC50 ∼1 to 2 μm; Fig. 3B). The results with Bag1 (R193A,K194A) were particularly important because this double mutant had no measurable affinity for Hsc70NBD (KD > 500 μm; see Fig. 2C) and; thus, any remaining activity in client release assays must originate from a non-canonical interaction (meaning an interaction that does not involve the BAG domain). To further explore this model in Bag3, we found that deletion of the BAG domain created a co-chaperone (Bag3ΔBAG; Fig. 3C) that had normal activity in the client release assay (EC50 ∼2 μm; Fig. 3D), but not nucleotide release assay (EC50 > 10 μm; Fig. 3E). Together, these results suggested that independent regions might be responsible for the ability of Bag1 and Bag3 to promote nucleotide and client release.
FIGURE 3.
Mutation or deletion of the BAG domain blocks the ability to promote ADP release, but not client release. A, Bag1/3 mutants have decreased ability to release nucleotide from Hsp70. B, Bag1/3 mutants have normal activity in the client release assay. C, domain structure of the Bag3ΔBAG construct used in these studies is shown. D, deletion of the BAG domain from Bag3 has no effect on client release. E, deletion of the BAG domain from Bag3 abolishes nucleotide release. For all experiments, the results are the average of at least three experiments performed in triplicate each, and the error bars represent S.D.
The Non-canonical Interaction of Bag3 with Hsc70 Occurs in the SBD
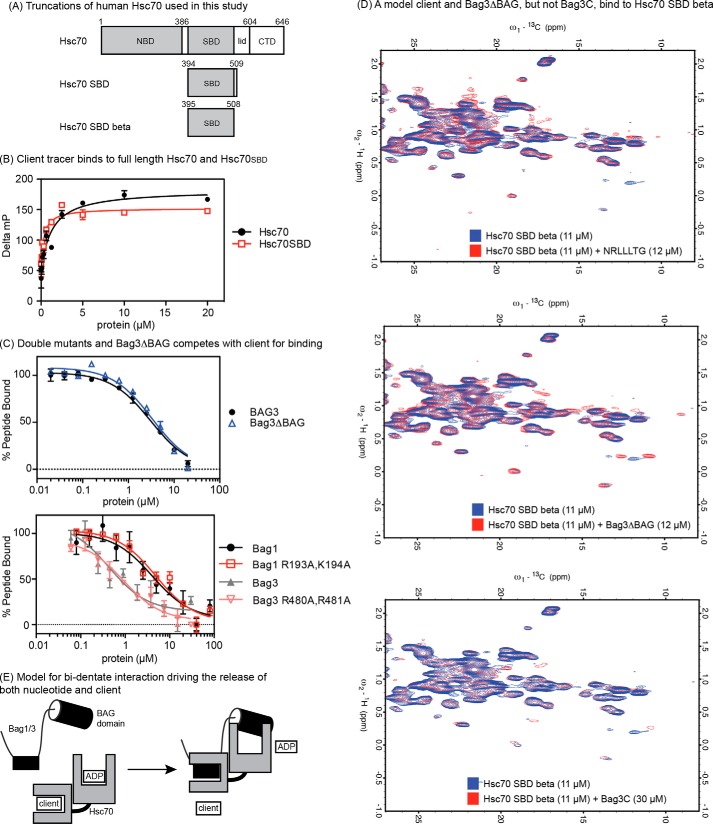
To understand which domain of Hsc70 might be required for this putative, non-canonical interaction, we created a truncated Hsc70 construct that lacked the NBD (Hsc70SBD; Fig. 4A). We found that the FAM-HLA tracer had a relatively poor signal (< 50 mP) upon binding to Hsc70SBD in the FP assay (data not shown), so we switched to a related tracer, FAM-LVEALY that is based on the well-known client peptide from microtubule-binding protein tau (10, 45). As expected, the FAM-LVEAVY tracer bound both full-length Hsc70 and Hsc70SBD with similar apparent affinity (Fig. 4B). Using this assay, we found that Bag1 and Bag3 accelerate release of the FAM-LVEAVY client from Hsc70SBD (Bag 1 EC50 ∼3 μm and Bag3 EC50 ∼1 μm; Fig. 4C). Moreover, we found that Bag1 (R193A,K194A), Bag3 (R480A, R481A), and Bag3ΔBAG all had similar activity as the wild type, full-length versions. Together, these results suggest that the non-canonical interaction does not require the NBD.
FIGURE 4.
The non-canonical interaction between Bag3 and Hsp72 occurs in the SBD. A, truncations of Hsc70 used in this study. B, client tracer interacts with both full-length Hsc70 and Hsc70SBD. C, Bag1, 3, double mutants and Bag3ΔBAG compete with client for binding to the Hsc70SBD. Results are the average of experiments performed in triplicate. Error is S.E. D, 600 MHz 13C-1H HSQC spectra of 11 μm 13C 15N Hsc70 (395–508) showing binding to a model client (NRLLLTG), to Bag3ΔBAG and to Bag3C. In each overlay, the apo-protein spectrum is in blue, and the spectrum of the Hsc70 SBDβ domain in the presence of ligand is shown in red. The model client and Bag3ΔBAG cause chemical shift perturbations and appearances of new resonances in slow exchange at ∼1:1, whereas Bag3C only causes minor changes at ∼3:1. E, model for the bi-dentate mechanism of human Bag1 and Bag3. The identity and location of the SBD-binding site is not yet clear (see supplemental Fig. S1).
To further understand whether the non-canonical interaction occurs in the SBD, we wanted to develop a distinct biophysical assay using NMR. Toward that goal, we expressed and purified a 13C 15N-labeled truncation of Hsc70 that was composed of only the β sandwich subdomain (Hsc70 SBD β; residues 395–508; see “Materials and Methods” for a description of how this construct was chosen and see supplemental Figs. S2, S3, and S4 for additional spectra and controls). After denaturation and refolding, we found that this protein was monomeric and had the expected affinity for the model client peptide, NRLLLTG (KD ∼10 μm; see supplemental Figs. S2, S3, and S4). We decided to carry out the binding studies at relatively low concentrations (11 μm) to favor biologically relevant interactions and to stay within the limits of Bag1/Bag3 solubility. However, under these conditions, the 15N-1H TROSY and/or HSQC spectra of Hsc70 SBDβ, especially in the apo-state, were not sufficient sensitive to be useful for ligand binding studies (data not shown). Hence, we opted for the more sensitive 13C-1H HSQC spectra of the methyl groups, which yielded good spectra at low concentrations (11 μm; Fig. 4D).
As an initial control, we worked with a sample containing 80 μm 13C 15N Hsc70 SBD β and confirmed that addition of a model client peptide, NRLLLTG (200 μm) caused extensive changes in the methyl spectra (supplemental Fig. S4). Specifically, extensive chemical shift changes were observed, including the appearance of new cross peaks in slow exchange. The appearance of new resonances, just as was seen in the 15N-1H TROSY spectra (supplemental Fig. S2) suggest that the Hsc70 apo-SBD is dynamically disordered and that it rigidifies upon peptide binding. Similar, but less dramatic changes have been reported for the SBD of DnaK, the Escherichia coli Hsc70 homologue (6). The 13C-1H HSQC spectra of Hsc70 SBDβ at 11 μm were, of course, less sensitive and we could not discern as many changes as seen in the spectra at 80 μm. However, NRLLLTG still caused significant chemical changes in a discrete number of methyl cross peaks (Fig. 4D) indicating binding with ∼10 μm affinity. This peptide also induced a small number of new peaks to appear in slow exchange (for example, ω1 ∼18 ppm and ω2 ∼1 ppm).
Using this assay platform, we asked whether Bag3ΔBAG or Bag3C could bind to Hsc70 SBDβ. We found that addition of Bag3ΔBAG (12 μm) induced new peaks (for example, ω1 ∼11 to 13 and ω2 ∼0.25) and strong CSPs, while nearly 3× more Bag3C (30 μm) was required to observe even modest and incomplete changes in the spectra (Fig. 4D). This result shows that Bag3ΔBAG, but not Bag3C, binds to Hsc70 SBDβ with an affinity of 10 μm or stronger. By analogy with the NRLLLTG peptide spectra, we assume that the chemical shift changes occur as a product of “rigidification” of Hsc70 SBDβ by binding to Bag3ΔBAG. Extensive characterization of the binding site or the changes will await future studies. For now, the NMR binding data serves to confirm that a region of Bag3 outside of the BAG domain binds to the Hsc70SBD.
Discussion
Bag1 and Bag3 Interact with Hsc70 through a Bi-dentate Mechanism and the Second Interaction Is Crucial for Client Release
Here, we report that a non-canonical interaction outside the BAG domain is essential for promoting client release in vitro. Most strikingly, isolated Bag1C or Bag3C were unable to promote client release, despite their normal activity in nucleotide release assays (see Fig. 1B). These results suggest a model in which Bag1 and Bag3 use at least two, distinct protein-protein interactions when binding to Hsc70 (Fig. 4E). Why are the activities of nucleotide release and client release separated in human Bag1 and Bag3? We speculate that, at least for some BAG family proteins, this bi-dentate interaction may provide an additional layer of regulation. In the canonical model, the interaction of the BAG domain with Hsc70NBD directly favors release of ADP and only indirectly triggers the release of clients after ATP re-binding and allosteric switching of the SBD. Indeed, even Bag1C is capable of promoting release of client from Hsp72 at high enough concentrations (EC50 ∼76 μm) in vitro (38). However, we speculate that the rate or extent of this allosteric program may not be sufficient to drive function of the NEF-Hsc70 system under some conditions in vivo. Instead, the secondary, non-canonical interaction in the Hsc70SBD might allow for greater control over the kinetics and timing of client “hand-off”, perhaps coordinating the release to coincide with delivery to other pathways. This multi-pronged interaction might even position the other regions of Bag1 or Bag3 (e.g. the PXXP motif of Bag3) so that they can recruit effectors of macroautophagy or the ubiquitin-proteasome system. Another possibility is that separating the two activities makes the system more responsive to cellular stress conditions. Under normal conditions, the levels of ATP are high compared with ADP, but this situation reverses in response to some types of stress or metabolic conditions. Thus, NEF function might sometimes be partly frustrated by re-binding of Hsc70 to ADP, disrupting progression through the nucleotide cycle. Under those conditions, the non-canonical contact might provide a salvage pathway to ensure dissociation of clients.
Is the Bi-dentate Interaction Conserved in Other NEFs?
It seems possible that other families of NEFs, such as GrpE and Hsp110, might also take advantage of bi-dentate binding mechanisms. For example, the crystal structure of GrpE bound to the NBD of DnaK shows a long helical coil extending from the site of the protein-protein interaction (46) that could possibly project toward the putative location of the SBD (which was truncated in the construct used for crystallography). While not directly parallel to the proposed mechanism of Bag1 and Bag3, the eukaryotic NEF, Hsp110, has spatially separated domains that are dedicated to binding Hsc70NBD and client (47, 48). It will be interesting to learn if other family members use bi-dentate mechanisms to coordinate nucleotide and client release. Such a model could create new opportunities for creating more selective inhibitors of NEF function. This possibility is important, given the diversity of functions played by NEFs (49). For example, it is not yet clear if this mechanism is conserved among the other members of the BAG family. Bag2 is important in linking some Hsc70 clients to the proteasome (50), while Bag5 has been implicated in alpha-synuclein homeostasis (51). Future work will be needed to see if these members of the co-chaperone family also bind Hsc70 through bi-dentate interactions.
Where on Bag1 and Bag3 Does the Non-canonical Interaction Occur?
Bag1 and Bag3 have little sequence identity (or similarity) beyond their conserved BAG domains (see supplemental Fig. S1). The NMR binding results suggested that the interaction could possibly involve a sequence that resembles a client because the changes in the 13C-1H HSQC NMR spectra are somewhat similar after addition of either NRLLLTG or Bag3ΔBAG (see Fig. 4D). In that putative model, a “client-like” sequence in Bag1 or Bag3 might directly bind to the SBD's cleft and release client by a competitive interaction. Along these lines, the fact that significant effects are observed by titration of 13C 15N Hsc70 SBDβ (11 μm) with approximately equivalent levels of Bag3ΔBAG (12 μm), suggests that the interaction is of relatively good affinity (estimated < 10 μm). Such an affinity would mean that the non-canonical interaction could potentially compete with many Hsc70 clients, which often have KD values in the low micromolar range, especially if the BAG domain interaction with Hsc70NBD was able to increase local concentration. However, Bag1 and Bag3 only have a limited number of predicted, consensus Hsc70 binding sites (see supplemental Fig. S1). Each protein has one predicted site within the BAG domains, but these regions are expected to be partly occluded in the folded, three-helix bundle and; more importantly, Bag1C and Bag3C had poor activity in the client release assays (see Fig. 1). Outside the BAG domain, Bag3 only has one predicted consensus Hsc70 binding motif and Bag1 has none (see supplemental Fig. S1), suggesting that an alternative explanation may be required. One possibility is that the putative, second site does not have sufficient sequence similarity to known clients, so it is not detected by prediction algorithms (52), Alternatively, it seems possible that the non-canonical interaction might occur, at least in part, outside of the classic client-binding cleft rather than being directly competitive. In theory, this interaction could reposition the lid subdomain and/or the CTD to indirectly regulate client off-rates and accessibility of the binding cleft. At this point, the location of the binding site on Bag1 and Bag3 and where it binds on the SBD are unclear.
The Thermodynamics of the Bi-dentate Interaction Remains Uncertain
Another issue that remains to be resolved is the role of avidity in the interactions of full-length Hsc70 with Bag1 and Bag3 in cells. The apparent affinity of Bag3C for Hsc70 is ∼30 nm (in the apo state), as measured by either ITC or FCPIA, while the affinity for full-length Bag3 is 10-fold better (∼3 nm) (38). That observation first suggested that there might be “missing” binding energy in the protein-protein complex. However, the apparent affinity of the Bag3ΔBAG protein for Hsc70 SBDβ by NMR was ∼10 μm or less (see Fig. 4), which is significantly tighter than what would be minimally needed to account for the “missing” affinity. We were not able to measure the binding of Bag1C, Bag3C, or Bag3ΔBAG to Hsc70SBD by ITC or FCPIA (data not shown). Indeed, technical challenges, such as solubility, persistent client retention and signal:noise, have prevented collection of the affinity values for all of the comparisons (e.g. Hsc70 binding to Bag3ΔBAG) that would be needed for a rigorous analysis of the thermodynamic contributions of the two sites to overall binding free energy. Regardless, the results reported here provide a strong impetus for the continued study of the non-canonical interactions between Hsc70 and BAG family members, as this contact appears to be necessary and sufficient for client release under some conditions.
Materials and Methods
Protein Expression and Purification
Human BAG1 (also called BAG1S), BAG3, BAG1C, BAG3C, Bag1, and Bag3 mutants, BAG3ΔBAG, Hsc70 (HSPA8), Hsc70 SBD (residues 394–509) were expressed in BL21(DE3) cells as previously described (38). Following purification on Ni-NTA resin, overnight TEV cleavage to remove His tags, monoQ (GE Healthcare) and size exclusion (SuperDex S200) columns, the stock proteins were dialyzed into HEPES buffer (25 mm HEPES, 5 mm MgCl2, 150 mm KCl pH 7.5). Hsc70 was also subject to 3 days of extensive dialysis to remove bound nucleotide and residual client.
Nucleotide Release
Fluorescence polarization (FP) experiments were carried out as previously described (38). Briefly, ATP-FAM (20 nm; Jena Bioscience) dissociation from Hsc70 (1 μm) was measured by incubating BAGs for 10 min in black, round-bottom, low-volume 384-well plates (Corning) in assay buffer (100 mm Tris, 20 mm KCl, 6 mm MgCl2 pH 7.4). After incubation, FP was measured (Ex 485 nm/Em 535 nm) using a SpectraMax M5 plate reader. Controls included wells lacking Hsc70. It has previously been reported that ATP and ADP have similar IC50 values for the ATP-FAM tracer (38).
Client Release
Release of fluorescent FAM-HLA or FAM-LVEAVY from Hsc70 was measured by FP, as described (38, 53). Briefly, a mixture of Hsc70 (1 μm) and FAM-HLA (25 nm; Anaspec) or FAM-LVEALY (25 nm, Genscript) was incubated with BAG proteins for 30 min at room temperature in assay buffer (100 mm Tris, 20 mm KCl, 6 mm MgCl2, 1 mm ADP, pH 7.4). FP was measured (Ex 485 nm/Em 535 nm) using a SpectraMax M5 plate reader. Pilot studies showed that 30 min was sufficient to achieve equilibrium.
Flow Cytometry Protein Interaction Assay (FCPIA)
The assay procedure was adopted from previous reports (38). Briefly, Hsc70 biotinylated on lysine residues was immobilized (1h at room temperature) on streptavidin-coated polystyrene beads (Spherotech), in the presence of nucleotide (1 mm). After immobilization, beads were washed and then incubated with fluorescently labeled NEF protein. Binding was detected using a flow cytometer to measure median bead-associated fluorescence. Beads capped with biocytin were used as a negative control, and nonspecific binding to beads was subtracted.
Chaperone Assays
The effects of BAG proteins on steady state ATPase activity were measured by malachite green assays, as previously described (38, 54). Effects on denatured firefly luciferase refolding were measured by luminescence assays, as previously described (38). Hsc70 was used at 1 μm and DnaJA2 was added to 0.5 μm.
NMR Spectroscopy
In initial studies, we found that the NMR spectra of Hsc70 residues 395–646 to be unusable for binding studies because residues 600–646 are dynamically disordered and obscure/dominate much of the NMR spectrum (55). We next tried the truncation 395–604, which contains only the β sandwich domain and alpha helical “lid”. However, we found that this construct, while monomeric, bound NRLLLTG peptide with a KD value above 100 μm (compared with the expected value of ∼1 μm for full-length Hsc70) perhaps suggesting that part of the lid might partially occlude the SBD cleft in this truncation. Finally, the lid was removed, yielding the constructs 394–509 (Hsc70 SBD) and 395–508 (Hsc70 SBDβ), which were used interchangeably. We found 395–508 bound client peptides with a KD < 10 μm, in addition to giving a well resolved NMR spectra (see supplemental Figs. S2–S4). This construct also yielded interpretable spectra (see below). Therefore, we elected to use 13C 15N Hsc70 SBDβ (residues 395–508 G3H6) for the NMR binding studies. No significant differences are expected between Hsc70 SBD and Hsc70 SBDβ, but both are included here for transparency.
13C 15N Hsc70 SBDβ (residues 395–508) G3H6 was expressed in BL21 cells in M9 minimal medium supplemented with isotopically labeled amino acids. Previous studies indicated that the protein was co-purified with bound clients, which need to be removed using an extensive purification protocol (see legend of supplemental Fig. S2). The SBD was bound to Ni-NTA and washed with 10 column volumes of 50 mm KH2PO4, 200 mm NaCl, 50 mm imidazole, pH 7.3 (buffer A). Next, the protein-bound beads were washed with 10 column volumes of buffer A containing 0.1% CAS36 detergent, followed by 10 column volumes buffer A without detergent. Subsequently, the protein was denatured on the column using 4 m guanidinium HCl (GdHCl) in buffer A and washed with 10 column volumes of the same. The protein was refolded on the resin using a linear gradient from 4 to 0 m GdHCl over 20 column volumes, and subsequently washed again with 10 column volumes of buffer A. Finally, the protein was eluted with 300 mm imidazole in buffer A. Eluants were exchanged into NH4HCO3 using G25 and lyophilized in glass tubes. Prior to NMR, protein was resuspended in 25 mm Tris, 50 mm NaCl, 1 mm EDTA, 2 mm DTT, 0.02% sodium azide, 5% D2O, SIGMAFAST protease inhibitors, pH 7.2, not exceeding 80 μm in protein concentration.
In its apo form, the domain remained monomeric up to a concentration of ∼80 μm as judged from a rotational correlation time of 8 ns (measured from one-dimensional 15N relaxation data), but it started to aggregate above that concentration. The TROSY NMR spectrum of the APO protein is quite different from the spectrum with bound peptide and many resonances are missing and/or broad (see supplemental Fig. S2). Upon addition of peptide, many peaks (re) appear in slow exchange and the spectrum becomes similar to that of an identical sample that was not refolded (see supplemental Figs. S2 and S3). These observations were taken to indicate that purification and refolding of the apo-β domain produces properly folded and functional protein. Moreover, we found that peptide binding at a concentration of 10 μm is stoichiometric. Resonance assignments of the domain with bound peptide are in progress. We decided to carry out the NMR studies at ∼ 11 μm protein concentration to ensure that only biologically relevant interactions would be monitored. However, at such low concentrations 15N-1H TROSY and/or HSQC spectra of especially the apo-state are of insufficient sensitivity to be useful for careful ligand binding studies. Hence, we opted for the much more sensitive 13C-1H HSQC spectra of the methyl groups. Several 13C-1H HSQC spectra of 11 μm Hsc70 SBDβ domain with different concentrations of ligands were recorded at 25 °C in 1 h each, using a Bruker 600 MHz NMR spectrometer with cryoprobe. Water suppression was accomplished with the Watergate technique.
Author Contributions
J. N. R. and E. R. P. Z. designed, performed, and interpreted experiments. J. E. G. designed and interpreted experiments. J. N. R., E. R. P. Z., and J. E. G. wrote the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grant NS059690. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S4.
- Hsc70
- heat shock cognate protein 70
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- CTD
- C-terminal domain
- BAG
- Bcl2-associated anthanogene
- ITC
- isothermal calorimetry
- NEF
- nucleotide-exchange factor
- FP
- fluorescence polarization
- FCPIA
- flow cytometry protein interaction assay.
References
- 1. Mayer M. P., and Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 3. Assimon V. A., Gillies A. T., Rauch J. N., and Gestwicki J. E. (2013) Hsp70 protein complexes as drug targets. Curr. Pharm. Des. 19, 404–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rampelt H., Mayer M. P., and Bukau B. (2011) Nucleotide exchange factors for Hsp70 chaperones. Methods Mol. Biol. 787, 83–91 [DOI] [PubMed] [Google Scholar]
- 5. Zuiderweg E. R., Bertelsen E. B., Rousaki A., Mayer M. P., Gestwicki J. E., and Ahmad A.. (2013) Allostery in the Hsp70 chaperone proteins. Topics in Current Chemistry 328, 99–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pellecchia M., Montgomery D. L., Stevens S. Y., Vander Kooi C. W., Feng H. P., Gierasch L. M., and Zuiderweg E. R. (2000) Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303 [DOI] [PubMed] [Google Scholar]
- 7. Muller P., Ruckova E., Halada P., Coates P. J., Hrstka R., Lane D. P., and Vojtesek B. (2013) C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene 32, 3101–3110 [DOI] [PubMed] [Google Scholar]
- 8. Assimon V. A., Southworth D. R., and Gestwicki J. E. (2015) Specific binding of tetratricopeptide repeat proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) is regulated by affinity and phosphorylation. Biochemistry 54, 7120–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hageman J., van Waarde M. A., Zylicz A., Walerych D., and Kampinga H. H. (2011) The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 435, 127–142 [DOI] [PubMed] [Google Scholar]
- 10. Jinwal U. K., Akoury E., Abisambra J. F., O'Leary J. C. 3rd, Thompson A. D., Blair L. J., Jin Y., Bacon J., Nordhues B. A., Cockman M., Zhang J., Li P., Zhang B., Borysov S., Uversky V. N., Biernat J., Mandelkow E., Gestwicki J. E., Zweckstetter M., and Dickey C. A.. (2013) Imbalance of Hsp70 family variants fosters tau accumulation. FASEB J. 27, 1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amick J., Schlanger S. E., Wachnowsky C., Moseng M. A., Emerson C. C., Dare M., Luo W. I., Ithychanda S. S., Nix J. C., Cowan J. A., Page R. C., and Misra S. (2014) Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone. Protein Sci. 23, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brehmer D., Rüdiger S., Gässler C. S., Klostermeier D., Packschies L., Reinstein J., Mayer M. P., and Bukau B. (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8, 427–432 [DOI] [PubMed] [Google Scholar]
- 13. Revington M., Holder T. M., and Zuiderweg E. R. (2004) NMR study of nucleotide-induced changes in the nucleotide binding domain of Thermus thermophilus Hsp70 chaperone DnaK: implications for the allosteric mechanism. J. Biol. Chem. 279, 33958–33967. [DOI] [PubMed] [Google Scholar]
- 14. Liu Q., and Hendrickson W. A. (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kityk R., Vogel M., Schlecht R., Bukau B., and Mayer M. P. (2015) Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 6, 8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vogel M., Bukau B., and Mayer M. P. (2006) Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell 21, 359–367 [DOI] [PubMed] [Google Scholar]
- 17. General I. J., Liu Y., Blackburn M. E., Mao W., Gierasch L. M., and Bahar I. (2014) ATPase subdomain IA is a mediator of interdomain allostery in Hsp70 molecular chaperones. PLoS Comp. Biol. 10, e1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., and Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smock R. G., Rivoire O., Russ W. P., Swain J. F., Leibler S., Ranganathan R., Gierasch L. M. (2010) An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol. Syst. Biol. 6, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., and Liu Q. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palleros D. R., Reid K. L., Shi L., Welch W. J., and Fink A. L. (1993) ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature 365, 664–666 [DOI] [PubMed] [Google Scholar]
- 22. Slepenkov S. V., and Witt S. N. (1998) Peptide-induced conformational changes in the molecular chaperone DnaK. Biochemistry 37, 16749–16756 [DOI] [PubMed] [Google Scholar]
- 23. Bertelsen E. B., Chang L., Gestwicki J. E., and Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabbage M., and Dickman M. B. (2008) The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol. Life Sci. 65, 1390–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Höhfeld J., and Jentsch S. (1997) GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16, 6209–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siegenthaler R. K., and Christen P. (2006) Tuning of DnaK chaperone action by nonnative protein sensor DnaJ and thermosensor GrpE. J. Biol. Chem. 281, 34448–34456 [DOI] [PubMed] [Google Scholar]
- 27. Groemping Y., Klostermeier D., Herrmann C., Veit T., Seidel R., and Reinstein J. (2001) Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE. J. Mol. Biol. 305, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 28. Kirschke E., Goswami D., Southworth D., Griffin P. R., and Agard D. A. (2014) Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sekhar A., Rosenzweig R., Bouvignies G., and Kay L. E. (2016) Hsp70 biases the folding pathways of client proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E2794–E2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taguwa S., Maringer K., Li X., Bernal-Rubio D., Rauch J. N., Gestwicki J. E., Andino R., Fernandez-Sesma A., and Frydman J. (2015) Defining Hsp70 Subnetworks in Dengue Virus replication reveals key vulnerability in flavivirus infection. Cell 163, 1108–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X., Srinivasan S. R., Connarn J., Ahmad A., Young Z. T., Kabza A. M., Zuiderweg E. R. P., Sun D., and Gestwicki J. E. (2015) Validation of the Hsp70-Bag3 Protein-Protein Interaction as a Potential Therapeutic Target in Cancer. Mol. Cancer Ther. 16, 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., and Moarefi I. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 33. Arakawa A., Handa N., Ohsawa N., Shida M., Kigawa T., Hayashi F., Shirouzu M., and Yokoyama S. (2010) The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange. Structure 18, 309–319 [DOI] [PubMed] [Google Scholar]
- 34. Briknarová K., Takayama S., Brive L., Havert M. L., Knee D. A., Velasco J., Homma S., Cabezas E., Stuart J., Hoyt D. W., Satterthwait A. C., Llinás M., Reed J. C., and Ely K. R. (2001) Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 8, 349–352 [DOI] [PubMed] [Google Scholar]
- 35. Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., and Misra S. (2008) Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Melero R., Moro F., Pérez-Calvo M. Á., Perales-Calvo J., Quintana-Gallardo L., Llorca O., Muga A., and Valpuesta J. M. (2015) Modulation of the chaperone DnaK allosterism by the nucleotide exchange factor GrpE. J. Biol. Chem. 290, 10083–10092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buczynski G., Slepenkov S. V., Sehorn M. G., and Witt S. N. (2001) Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-rate constants. J. Biol. Chem. 276, 27231–27236 [DOI] [PubMed] [Google Scholar]
- 38. Rauch J. N., and Gestwicki J. E. (2014) Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 289, 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams J. M., Inoue T., Chen G., and Tsai B. (2015) The nucleotide exchange factors Grp170 and Sil1 induce cholera toxin release from BiP to enable retrotranslocation. Mol. Biol. Cell 26, 2181–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsukahara F., and Maru Y. (2010) Bag1 directly routes immature BCR-ABL for proteasomal degradation. Blood 116, 3582–3592 [DOI] [PubMed] [Google Scholar]
- 41. Gamerdinger M., Hajieva P., Kaya A. M., Wolfrum U., Hartl F. U., and Behl C. (2009) Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 28, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kettern N., Rogon C., Limmer A., Schild H., and Höhfeld J. (2011) The Hsc/Hsp70 co-chaperone network controls antigen aggregation and presentation during maturation of professional antigen presenting cells. PLoS ONE 6, e16398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ricci L., and Williams K. P. (2008) Development of fluorescence polarization assays for the molecular chaperone Hsp70 family members: Hsp72 and DnaK. Curr. Chem. Genomics 2, 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schröder H., Langer T., Hartl F. U., and Bukau B. (1993) DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson A. D., Scaglione K. M, Prensner J., Gillies A. T., Chinnaiyan A., Paulson H. L., Jinwal U. K., Dickey C. A., and Gestwicki J. E. (2012) Analysis of the tau-associated proteome reveals that exchange of hsp70 for hsp90 is involved in tau degradation. ACS Chem. Biol. 7, 1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., and Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 [DOI] [PubMed] [Google Scholar]
- 47. Andréasson C., Fiaux J., Rampelt H., Mayer M. P., and Bukau B. (2008) Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 48. Mattoo R. U., Sharma S. K., Priya S., Finka A., and Goloubinoff P. (2013) Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J. Biol. Chem. 288, 21399–21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abrams J. L., Verghese J., Gibney P. A., and Morano K. A. (2014) Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289, 13155–13167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carrettiero D. C., Hernandez I., Neveu P., Papagiannakopoulos T., and Kosik K. S. (2009) The cochaperone BAG2 sweeps paired helical filament- insoluble tau from the microtubule. J. Neurosci. 29, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kalia L. V., Kalia S. K., Chau H., Lozano A. M., Hyman B. T., and McLean P. J. (2011) Ubiquitinylation of α-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2-associated athanogene 5 (BAG5). PLoS ONE 6, e14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Durme J., Maurer-Stroh S., Galladro R., Wilkinson H., Rousseau F., and Schymkowitz J. (2009) Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comp. Biol. 5, e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cesa L. C., Patury S., Komiyama T., Ahmad A., Zuiderweg E. R., and Gestwicki J. E. (2013) Inhibitors of difficult protein-protein interactions identified by high-throughput screening of multiprotein complexes. ACS Chem. Biol. 8, 1988–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang L., Miyata Y., Ung P. M., Bertelsen E. B., McQuade T. J., Carlson H. A., Zuiderweg E. R., and Gestwicki J. E. (2011) Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem. Biol. 18, 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith M. C., Scaglione K. M., Assimon V. A., Patury S., Thompson A. D., Dickey C. A., Southworth D. R., Paulson H. L., Gestwicki J. E., and Zuiderweg E. R. (2013) The E3 ubiquitin ligase CHIP and the molecular chaperone Hsc70 form a dynamic tethered complex. Biochemistry 52, 5354–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.