Abstract
Development of a Plasmodium falciparum (Pf) transmission blocking vaccine (TBV) has the potential to significantly impact malaria control. Antibodies elicited against sexual stage proteins in the human bloodstream are taken up with the blood meal of the mosquitoes and inactivate parasite development in the mosquito. In a phase 1 trial, a leading TBV identified as Pfs25-EPA/Alhydrogel® appeared safe and immunogenic, however, the level of Pfs25-specific antibodies were likely too low for an effective vaccine. Pfs230, a 230-kDa sexual stage protein expressed in gametocytes is an alternative vaccine candidate. A unique 6-cysteine-rich domain structure within Pfs230 have thwarted its recombinant expression and characterization for clinical evaluation for nearly a quarter of a century. Here, we report on the identification, biochemical, biophysical, and immunological characterization of recombinant Pfs230 domains. Rabbit antibodies generated against recombinant Pfs230 domains blocked mosquito transmission of a laboratory strain and two field isolates using an ex vivo assay. A planned clinical trial of the Pfs230 vaccine is a significant step toward the potential development of a transmission blocking vaccine to eliminate malaria.
Keywords: malaria, protein structure, recombinant protein expression, vaccine, yeast, Pfs230, Pichia pastoris, transmission blocking
Introduction
Development of a malaria vaccine that effectively protects against parasite infection of both the natural host, Anopheles mosquitoes, and its secondary host, man, would effectively disrupt transmission and clinical disease. The most well known investigational vaccine against Plasmodium falciparum malaria that recently received a positive scientific opinion from the Committee for Medicinal Products for Human Use of the European Medicines Agency in July 2015, is identified as RTS,S (MosquirixTM). This vaccine targets the circumsporozoite protein that is present on the surface of the sporozoite, the parasite stage that infects man (1). RTS,S, a virus like-particle-based vaccine, is protective against clinical disease in about 30% of the young children who participated in a phase 3 trial (1, 2).
Efforts toward development of a vaccine to disrupt parasite infection of the mosquito host, also identified as a transmission blocking vaccine have to date only been able to evaluate a sexual stage-specific protein, Pfs25,2 which is a 25-kDa protein expressed on the zygote and ookinete surfaces. A phase 1 study demonstrated that human antibodies raised against a recombinant Pfs25 (Pfs25H) protein formulated in Montanide ISA 51, a water-in-oil adjuvant formulation, were biologically active in an ex vivo feeding assay (3), however, this formulation was not deemed suitable for a public health vaccine. More recently, in preclinical studies, Pfs25H has been shown to have enhanced immunogenic properties when chemically conjugated to a carrier molecule such as Neisseria meningitis outer membrane protein complex (4), or Pseudomonas aeruginosa ExoProtein A (EPA) (5, 6). In particular, the chemically conjugated Pfs25-EPA has the biophysical features of a nanoparticle with a diameter of about 25 μm in solution, similar in size to that of the RTS,S (7). In a phase 1 human trial, the Pfs25-EPA/Alhydrogel® formulation was shown to be safe and immunogenic, and generated human antibodies that reduced transmission by greater than 50% using an ex vivo standard membrane feeding assay (SMFA).3 The levels of Pfs25-specific antibodies, however, appeared insufficient for complete disruption of the mosquito infection. At a minimum, the Pfs25-EPA-conjugated nanoparticle may require a more potent adjuvant formulation, the inclusion of other sexual stage transmission-blocking vaccine candidates or both.
Other transmission-blocking vaccine candidates have been identified (9–13). Nearly a quarter of a century ago, Quakyi et al. (14) recognized the potential of the P. falciparum sexual stage 230-kDa protein (Pfs230) as a candidate for a transmission blocking vaccine. Pfs230 forms a complex with Pfs48/45, a glycosylphosphatidylinositol anchored protein (15) on the surface of sexual stage parasites (16, 17). The complex is involved in fertilization of male and female gametes. Pfs230 is initially a 360-kDa gametocyte surface protein that is proteolytically processed during development at the amino terminus (18). Polyclonal (19, 20) and monoclonal (14, 21) antibodies have been shown to block parasite transmission using the ex vivo SMFA in a complement-dependent manner. Pfs230 has been speculated to have a male gamete function because parasites with a truncated form of Pfs230 were shown to adhere to uninfected erythrocytes, even though their capacity to infect mosquitoes was severely decreased by greater than 95% (22). Various forms or domains of Pfs230 have been expressed in several recombinant expression systems, including Saccharomyces cerevisiae, Escherichia coli, plants, and wheat germ (9, 12, 13, 23) but to date none of the recombinant forms of Pfs230 have been suitable for extensive biochemical and biophysical characterization to effectively characterize a functional antibody response. A primary bottleneck in recombinant protein production has been the presence of an unique disulfide bond structure initially modeled by Gerloff et al. (24). More recent characterization of a poorly expressed, E. coli recombinant P. falciparum asexual parasite protein Pf12 by NMR led to the confirmation that the 6-cysteine protein-folds within Pf12 are similar to a Toxoplasma protein identified as SAG1 (25).
Given the difficulty to produce recombinant proteins with a 6-cysteine motif in a manner suitable for full biochemical and biophysical characterization, we aimed to develop a process and recombinant protein product of suitable quality, quantity, and purity for human clinical testing. Two scalable and commercial production platforms were assessed. Here we report on the identification, production, and characterization of an E. coli expressed and refolded Pfs230 protein (Pfs230D1–2) and a Pichia pastoris-expressed Pfs230 protein (Pfs230D1H) comprised of domains 1–2 or domain 1, respectively. Both forms of purified recombinant Pfs230 induced transmission blocking antibodies in rabbits, however, based on comparative studies the P. pastoris Pfs230D1H protein was more effective at inducing transmission blocking antibodies. As a result, a new Pfs230D1 protein without any heterologous amino acids, identified as Pfs230D1M, was developed and fully characterized. Recombinant Pfs230D1M is a promising component of a transmission blocking vaccine.
Results
Production and Characterization of Recombinant Pfs230 Proteins
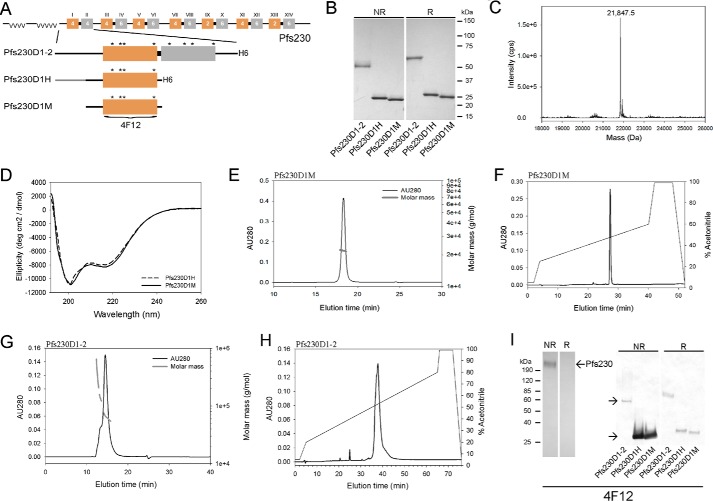
Early work by Williamson et al. (23) identified a natural cleavage site upstream of Pfs230 domain 1, which was used as the basis for the N-terminal boundary. The boundary for the C terminus varied depending on the expression system. In P. pastoris, the recombinant protein boundaries included the non-structured domain through domain 1 (Pfs230D1H) and in E. coli the boundaries included the non-structured domain through domain 2 (Pfs230D1–2) (see schematic Fig. 1A). Both recombinant Pfs230D1H and Pfs230D1–2 proteins contained a His6 C-terminal fusion tag to facilitate purification. A third recombinant form of Pfs230, Pfs230D1M, was homologous to the Pfs230D1H protein except for the removal of the His6 fusion tag. The E. coli expressed Pfs230D1–2 protein was refolded and purified as described under “Experimental Procedures.” Recombinant Pfs230D1–2 had the expected N termini (MEYVDEK) with the non-native M retained as determined by Edman degradation. Recombinant Pfs230D1–2 demonstrated the presence of disulfide bond formation as observed by a mobility shift by Coomassie Blue staining following SDS-PAGE under non-reduced versus reduced conditions (Fig. 1B). Additional analyses by analytical size exclusion chromatography with online multiangle light scattering (SEC-MALS-HPLC) and reversed-phase (RP) HPLC demonstrated that the protein was principally monomeric in solution (85%) with an observed molar mass of 64.5 kDa and by RP-HPLC was resolved as a single asymmetric peak (Fig. 1, G and H, respectively).
FIGURE 1.
Production of recombinant Pfs230 proteins. A, schematic of native Pfs230 based on a report by Gerloff et al. (24) and recombinant Pfs230D1–2, Pfs230D1H, and Pfs230D1M, as well as, for B-H, the biochemical and biophysical characterizations of the different recombinant forms. B, Coomassie Blue-stained SDS-PAGE analysis of recombinant forms of Pfs230. C, analysis of the Pfs230D1M intact mass by LC/MS. D, comparative analysis of the ellipticity of Pfs230D1M and Pfs230D1H by far-UV circular dichroism. E and F, analysis of Pfs230D1M by SEC-MALS-HPLC and RP-HPLC, respectively. G and H, analysis of Pfs230D1–2 by SEC-MALS-HPLC and RP-HPLC, respectively. I, Western blotting analysis with Pfs230-specific mAb 4F12 against native gametocyte lysate (left panel) or recombinant forms of Pfs230 (right panel) under both non-reduced and reducing conditions, respectively. Notations in panel A include Roman numerals showing 14 distinct cysteine domains, alpha numeric numbers represent the number of cysteines in each domain, and the gold-gray color represents a double domain as defined by Gerloff et al. (24). Asterisks denote position of cysteines within domains 1 and 2. Boundary for the epitope recognized by the Pfs230D1-specific mAb 4F12 is shown in panel A.
Even though the initial P. pastoris expression plasmid was designed to secrete a recombinant protein that contained the non-structured domain through domain 1 (amino acids Tyr444-Gly736), the Pfs230D1H shown in Fig. 1B by Coomassie Blue staining was consistently cleaved at Ser542 based on N-terminal sequencing and intact electrospray ionization mass spectroscopy (observed mass: 21,847.5 Da was consistent with an expected mass with cleavage at Ser542). This constituted a loss of a 98-amino acid N-terminal fragment. Based on these empirical results, a second production clone was developed such that the designed synthetic gene and translated protein were consistent with the N terminus observed for the Pfs230D1H. The new purified recombinant protein, identified as Pfs230D1M, had the expected N terminus (SVLQSGAL) and a native C terminus due to the removal of the His6 fusion tag. The expected intact mass (21,849.8 Da) was consistent with the observed intact mass (21,847.5 Da) (Fig. 1C). An expected shift in the intact mass of approximately 4 daltons was observed upon reduction of Pfs230D1M consistent with the presence of two disulfide bonds. The far-UV CD spectrum demonstrated that Pfs230D1H and Pfs230D1M had similar secondary structures (Fig. 1D). The secondary structure of Pfs230D1M using DICHROWEB was determined to be α-helix: 8 ± 2%, β-sheet: 33 ± 5%, β-turn: 19 ± 7%, and unordered: 40 ± 5%. SEC-MALS-HPLC showed that Pfs230D1M and Pfs230D1H were monomeric in solution with molar masses of 21.9 and 23.1 kDa, respectively (Fig. 1E and data not shown, respectively). An analysis of their integrity by reversed-phase HPLC (RP-HPLC) showed a single symmetrical peak for each recombinant protein (Fig. 1F and data not shown, respectively). Finally, we evaluated the thermal stability of Pfs230D1M following a temperature ramp and a change in the secondary structure using far-UV CD spectroscopy. A major change in the ellipticity was observed between 55 and 60 °C and after complete denaturation at 80 °C and cooling to 20 °C, the secondary structure appeared similar to the original material (Fig. 2) indicating the thermal stability is suitable for a vaccine component.
FIGURE 2.
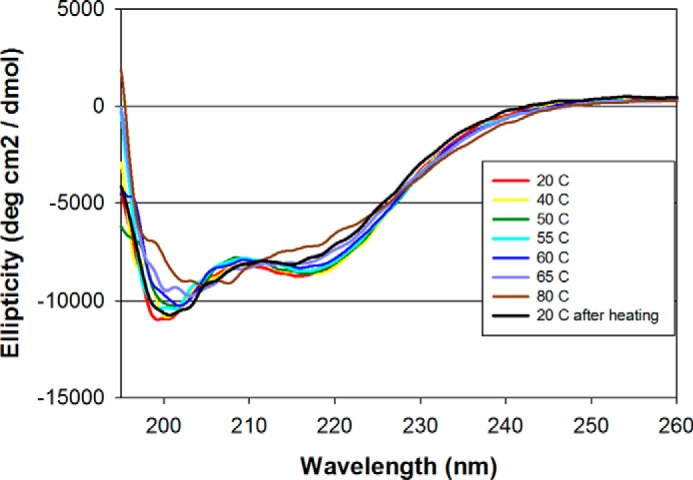
Circular dichroism analysis of Pfs230D1M in aqueous solution. The ellipticity (deg cm2/dmol) was plotted as a function of wavelength (nm). Raw data measured in millidegrees was converted into ellipticity (deg cm2/dmol). Spectra were obtained at increments from 20 to 80 °C and then at 20 °C following cooling for protein renaturation.
To accurately assess the immunogenicity of these recombinant proteins, the endotoxin and host cell protein content were also determined. The endotoxin levels were all below 5 endotoxin units/mg of protein and the concentration of host cell proteins, in particular for the P. pastoris expressed proteins using a process specific immunoblot assay were less than 0.5% (w/w) (26).
Evaluation of Recombinant Pfs230 Protein Conformation
To assess the recombinant proteins' conformation, a panel of monoclonal antibodies (mAbs) were produced against P. falciparum-enriched gametes and then screened for reactivity against native and recombinant forms of Pfs230. One mAb, identified as 4F12, recognized both native and recombinant forms of Pfs230 by Western blotting (Fig. 1I, left and right panels, respectively). The reactivity was observed primarily under non-reduced conditions indicating mAb 4F12 recognized a conformational epitope. Affinity purification of native gamete lysates with mAb 4F12 followed by in-gel tryptic digestion and mass spectroscopy identified tryptic peptide fragments enriched for Pfs230 (data not shown). mAb 4F12 also recognized Pfs230 on the surface of unfixed gametes by live IFA (Fig. 3A). A Pfs25-specific mAb 4B7 was included in the live IFA study as a positive control for the assay (Fig. 3A). Secondary antibody reagents were negative for parasite staining (data not shown). mAb 4F12, isotype IgG1, inhibited mosquito infectivity in the ex vivo SMFA by 99.6 to 100% at 1.0 mg/ml along with a corresponding reduction in parasite prevalence (as defined as the percentage of uninfected mosquitoes compared with the total number of mosquitoes fed) with or without human complement (Table 1). The Pfs25-specific mAb 4B7, which blocked mosquito transmission by greater than 85% at ∼0.5 mg/ml (Table 1), was included as a positive control (27).
FIGURE 3.
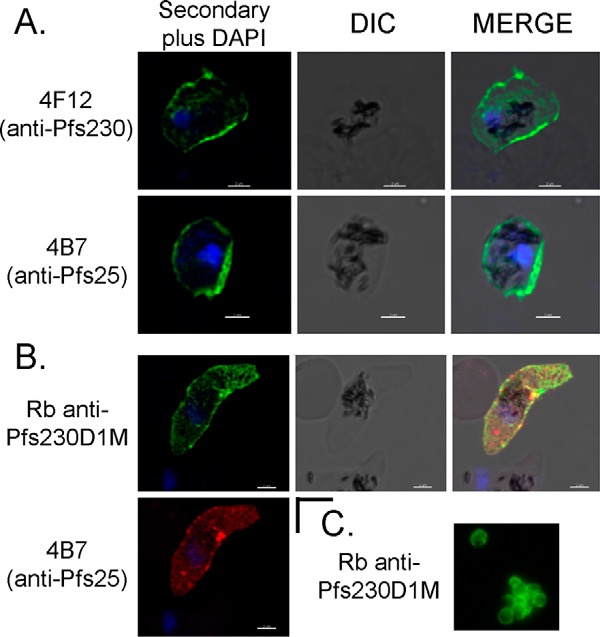
Surface reactivity of antibodies against Pfs230 and Pfs25 on live gametes. A, modified surface immunofluorescence assay analysis using Pfs230- and Pfs25-specific mAbs 4F12 and 4B7, and B, rabbit Pfs230D1M antiserum alone or in combination with Pfs25-specific mAb 4B7. C, live surface immunofluorescence assay analysis is shown for rabbit Pfs230D1M antiserum. Scale bar is equal to 2 μm.
TABLE 1.
Standard membrane feeding assay results for Pfs230D1 and Pfs25-specific mAbs 4F12 and 4B7
| Antigen | mAba | Experiment No. | Inf/Dissb | Avg No. oocysts | Median No. oocysts (range) | Reduction oocysts | Reduction prevalence |
|---|---|---|---|---|---|---|---|
| % | |||||||
| TBA without human complement | |||||||
| Pfs25 | 4B7 | 1 | 10/20 | 1.0 | 0.5 (0–3) | 95.2 | 50 |
| 2 | 18/20 | 4.45 | 3 (0–17) | 86.9 | 10 | ||
| Pfs230 | 4F12 | 1 | 0/20 | 0 | 0 (0–0) | 100 | 100 |
| 2 | 0/20 | 0 | 0 (0–0) | 100 | 100 | ||
| Controlc | 1 | 19/20 | 16.1 | 14.5 (0–45) | 0 | NAd | |
| 2 | 18/20 | 40.2 | 35.5 (0–94) | 0 | NA | ||
| TBA with human complement | |||||||
| Pfs25 | 4B7 | 3 | 19/20 | 4.4 | 3 (0–15) | 86.5 | 5 |
| 4 | 15/20 | 2.5 | 1.5 (0–7) | 95.1 | 25 | ||
| Pfs230 | 4F12 | 1 | 1/20 | 0.05 | 0 (0–1) | 99.8 | 95 |
| 2 | 3/20 | 0.15 | 0 (0–1) | 99.6 | 85 | ||
| Controlc | 1 | 20/20 | 28.5 | 29.5 (1–59) | 0 | NA | |
| 2 | 20/20 | 41.0 | 33 (11–83) | 0 | NA | ||
| Controlc | 3 | 20/20 | 24.4 | 23 (4–51) | 0 | NA | |
| 4 | 20/20 | 51.4 | 46 (18–98) | 0 | NA | ||
a Pfs25-specific mAb 4B7 is shown as an assay control using a fixed number of ELISA units (3250 EU), which equates to approximately 0.5 mg/ml of IgG; 4F12 was tested at 1.0 mg/ml of IgG.
b Inf., infected; Diss., dissected.
c Control: PBS with or without human complement.
d NA, not applicable.
Immunogenicity and Functional Activity of Rabbit Antiserum Generated against Recombinant Pfs230 Proteins
Paired rabbits were immunized with the three recombinant forms of Pfs230 formulated in a water-in-oil adjuvant at different stages during development. The rabbit antisera generated against each of the three Pfs230 proteins recognized native Pfs230 on the surface of live gametes by IFA using a modified-live IFA approach (Fig. 3 or data not shown) or a standard approach as shown for rabbit Pfs230D1M-specific antisera (Fig. 3C). During the modified live IFA, the rabbit Pfs230D1M-specific antiserum was co-incubated with the Pfs25-specific mAb 4B7 (Fig. 3B). In this analysis, Pfs230 was shown to be co-expressed with Pfs25 on the surface of a live sexual stage parasite, likely a young zygote. These antisera were evaluated for transmission blocking activity using the ex vivo SMFA. The SMFA results along with the antibody titers as determined by ELISA and represented as a reciprocal dilution at an OD1 are summarized in Table 2. Antisera from all three sets of paired rabbits generated transmission blocking activity (TBA). Most importantly, a head to head comparison to assess the TBA of Pfs230D1–2 and Pfs230D1H rabbit serum at a 1:16 dilution showed that Pfs230D1H induced an enhanced level of blocking activity (0–4.7 versus 99–100%, p < 0.001) (Table 2). This surprising result was confirmed by additional studies in which antibodies against Pfs230D1–2 consistently required a higher concentration of antisera or IgG to achieve similar levels of TBA (data not shown). Subsequently, the functional activity and ELISA titers induced by Pfs230D1M were shown to be similar for Pfs230D1H (Table 2). As anticipated (20), human complement was important for optimum blocking using sera or purified rabbit IgG because SMFA studies performed in the absence of human complement showed a decreased blocking activity (data not shown). Given the significant enhancement in TBA generated by the recombinant Pfs230D1H/M proteins, the recombinant Pfs230D1M protein was selected for further development.
TABLE 2.
Standard membrane feeding assay results for rabbit antisera generated against Pfs230D1–2, Pfs230D1H, and Pfs230D1M with human complement
| Antigen | Rabbit No. | Serum dilution | ELISA titer OD1a |
Infected/disssected | Average No. oocysts | Median No. oocysts (range) | Reduction oocysts | Reduction prevalence | ||
|---|---|---|---|---|---|---|---|---|---|---|
| D1–2 | D1H | D1M | ||||||||
| % | ||||||||||
| D1–2 | 1.1 | 1:4.5 | 81K | 37K | NDb | 7/20 | 0.85 | 0 (0–5) | 96.9c | 65 |
| 1.2 | 117K | 25K | ND | 13/20 | 2.30 | 1 (1–11) | 91.5c | 35 | ||
| 1.1 | 1:16 | 20/20 | 67.8 | 69 (25–113) | 0d | 0 | ||||
| 1.2 | 18/20 | 57.5 | 54 (0–113) | 4.7d | 10 | |||||
| D1H | 2.3 | 1:4.5 | 24K | 85K | ND | 0/20 | 0 | 0 (0–0) | 100d | 100 |
| 2.4 | 47K | 158K | ND | 0/20 | 0 | 0 (0–0) | 100d | 100 | ||
| 2.3 | 1:16 | 1/20 | 0.05 | 0 (0–1) | 99.9d | 95 | ||||
| 2.4 | 0/20 | 0 | 0 (0–0) | 100d | 100 | |||||
| D1M | 1 | 1:4.5 | ND | ND | 131K | 0/20 | 0 | 0 (0–0) | 100e | 100 |
| 2 | ND | ND | 220K | 0/20 | 0 | 0 (0–0) | 100e | 100 | ||
| 1 | 1:9 | 0/20 | 0 | 0 (0–0) | 100f | 100 | ||||
| 2 | 0/20 | 0 | 0 (0–0) | 100f | 100 | |||||
| 1 | 1:27 | 3/20 | 0.15 | 0 (0–1) | 99.7f | 85 | ||||
| 2 | 0/20 | 0 | 0 (0–0) | 100f | 100 | |||||
a ELISA titer reported as reciprocal dilution at an OD1.
b ND, not done.
c % Reduction of oocysts are derived from the denominator as average number of oocysts in adjuvant control (average 25.2, median 24, range minimum 0 and maximum 68).
d % Reduction of oocysts are derived from the denominator as average number of oocysts in adjuvant control (average 61.4, median 58.5, range minimum 0 and maximum 114).
e % Reduction of oocysts and prevalence are derived from a denominator as the average number of oocysts in adjuvant control (average 19.2, median 19, range minimum 9 and maximum 36).
f % reduction of oocysts and prevalence are derived from a denominator as the average number of oocysts in adjuvant control (average 53.4, median 51, range minimum 7 and maximum 150).
Transmission of Thai Isolates of P. falciparum Were Blocked by Rabbit Pfs230D1M-specific Antiserum
To further evaluate the blocking activity of Pfs230D1-specific antibodies, the rabbit Pfs230D1M antiserum was evaluated for blocking transmission of patient isolates of P. falciparum parasites. Two patient isolates of P. falciparum from Thailand were mixed with rabbit Pfs230D1M-specific antiserum or control serum and fed to mosquitoes and the TBA was determined using a membrane feeding assay. As shown in Table 3, rabbit Pfs230D1M-specific antiserum completely or nearly completely blocked transmission, even at a 1:16 dilution of whole serum. Using genomic DNA isolated from dried blood spots collected from the two patients, the Pfs230D1 gene sequence was amplified and sequenced. A single amino acid substitution, which is discussed below, was observed in both samples corresponding to a Gly to Ser substitution at position 605 of Pfs230 (accession number XP_001349600.1).
TABLE 3.
Membrane feeding assay results using human infected P. falciparum RBCs mixed with rabbit Pfs230D1M antiserum
| ID | Antigen | Rb ID | Serum dilution | No. oocysts |
Minimum-maximum | Reduction prevalance | |
|---|---|---|---|---|---|---|---|
| Total | Avg. | ||||||
| % | |||||||
| 1 | D1M | 5 | 1:8 | 0 | 0 | 0–0 | 100 |
| 6 | 2 | 0.05 | 1–1 | 95 | |||
| 5 | 1:16 | 0 | 0 | 0–0 | 100 | ||
| 6 | 0 | 0 | 0–0 | 100 | |||
| Adjjusted | 7 | 1:8 | 149 | 3.7 | 1–15 | 17.5 | |
| Concentration | 1:16 | 96 | 2.4 | 1–8 | 25 | ||
| Naive | 116 | 2.9 | 1–10 | 30 | |||
| 2 | D1M | 5 | 1:8 | 0 | 0 | 0–0 | 100 |
| 6 | 0 | 0 | 0–0 | 100 | |||
| 5 | 1:16 | 0 | 0 | 0–0 | 100 | ||
| 6 | 0 | 0 | 0–0 | 100 | |||
| Adjusted | 7 | 1:8 | 155 | 7.8 | 1–40 | 50 | |
| Concentration | 1:16 | 367 | 18.4 | 2–38 | 15 | ||
| Naive | 546 | 27.3 | 3–59 | 15 | |||
Analysis of Polymorphism within Pfs230 Domain 1
To gain a better understanding of the conformation of the protein, the disulfide bond arrangements of Pfs230D1M were determined using enzymatic digestion and subsequent analysis by LC/MS/MS. The disulfide bond structure was identified as C1 to C2 (Cys593-Cys611) and C3 to C4 (Cys626-Cys706). Each disulfide peptide was confirmed with elevated energy fragmentation (data not shown). Native Pfs230 is expressed in gametocytes and is known to contain polymorphic amino acids. We evaluated the minor allelic frequency (MAF) of amino acid substitutions in over 2000 primary sequences for Pfs230D1 reported in the MalariaGen P. falciparum database (28), which represents a large geographical distribution of P. falciparum malaria. Only two non-synonymous substitutions with a global MAF above 0.02 were observed, G605S and K661N with global MAFs of 0.052 and 0.189, respectively (accession number XP_001349600.1). In addition, the MAF varied in different geographical areas, in West Africa the MAF for G605S and K661N were 0.111 and 0.339, respectively, with the next highest non-synonymous MAF being 0.04.
To visualize the disulfides, polymorphisms, and overall structure of Pfs230D1M, a comparative model was built using the recent crystal structures of Pf12 and Pf41 as templates (Fig. 4). The predicted secondary structure content of the model (α-helix, 12%; β-sheet, 22%; β-turn, 27%; and unordered, 37%) is consistent with the secondary structure content observed by circular dichroism spectroscopy. The relative locations of the polymorphic amino acid substitutions are shown on a surface plot of Pfs230D1M (Fig. 4B). Interestingly, the two dominant locations for amino acid substitutions, G605S and K661N, in addition to a number of lower frequency substitutions are predicted to be primarily located on the face of the putative β-sheet of domain 1 made up of β-strands 2, 4, 7, and 8.
FIGURE 4.
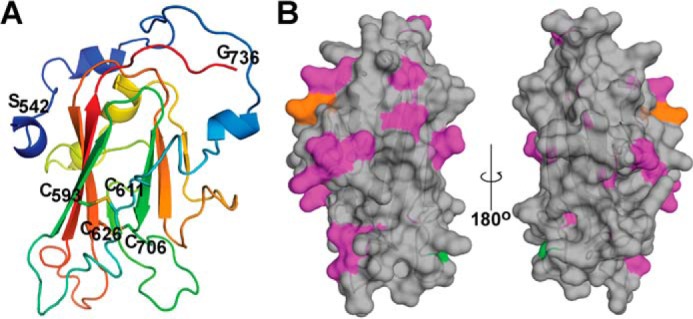
Ribbon diagram and amino acid substitutions corresponding to minor allelic variants identified in Pfs230D1. A, ribbon diagram of a Pfs230D1M comparative model. The cysteines participating in the two predicted disulfide bonds and the amino and carboxyl termini of the Pfs230D1M construct are labeled. B, the minor allelic variants are identified on a surface plot of Pfs230D1M: G605S, green; K661N, orange; and others, magenta.
Discussion
The evaluation of Pfs230 as a transmission blocking vaccine candidate for investigational human studies has not been possible until now. The limitation was due to an inability to produce a recombinant form of Pfs230 with the necessary purity, quantity, and quality characteristics required for human studies. Despite over a decade of experimental failures that targeted the production of functional 6-cysteine proteins (data not shown), here we report on the successful expression and characterization of Pfs230 subdomains. To achieve this aim, two different expression systems were evaluated based on earlier work reported by Williamson et al. (23) and more recently by Tachibana et al. (13). We first produced a refolded E. coli-expressed Pfs230 protein comprised of the non-structured domain through domain 2 (Pfs230D1–2) and then using the methylotrophic yeast P. pastoris a second recombinant protein was produced that included the non-structured domain through domain 1 (Pfs230D1H). Both proteins contained a His6 fusion tag to facilitate purification. The Pfs230D1H gene expression cassette included the non-structured domain, which was apparently unstable in P. pastoris as it was determined by Edman degradation that the N terminus was devoid of a segment of 98 amino acids. This deletion event was consistent with the secreted form of Pfs230D1H being smaller than expected by Coomassie Blue-stained SDS-PAGE (Fig. 1B) and by intact mass (Fig. 1C). However, the secreted form of Pfs230D1H appeared stable and was used for pre-clinical immunological studies. Pfs230-specific antibodies generated in rabbits were shown to block P. falciparum transmission in a titratable manner (Table 2). Most importantly, in a head to head comparative SMFA, rabbit Pfs230D1H-specific antibodies were observed to induce a more potent blocking activity than Pfs230D1–2 (p < 0.001). There was a marked difference (approximately 4-fold) in the domain 1-specific titer comparing the Pfs230D1–2 and Pfs230D1 antibody responses by ELISA (Table 2). Based on these observations, a quality by design approach was used to produce a new P. pastoris expression clone such that the synthetic gene product was in agreement with the observed translated, secreted protein, and was named Pfs230D1M. Due to the product-like nature of Pfs230D1M, no heterologous amino acids were included in the expressed protein. Pfs230D1M appeared comparable biochemically and biophysically to Pfs230D1H with the exception of the designed changes. In addition, the two disulfide bonds within Pfs230D1M were mapped and assigned as C1 to C2 and C3 to C4, which was in agreement with the NMR analysis of recombinant Pfs12, another malaria protein with a comparable 6-cysteine-rich motif (29). It is interesting to note that the latter disulfide is homologous to the C3-C6 disulfide pairing observed for Pf12 and Pf41 (29, 30).
During preclinical development, the P. pastoris-expressed Pfs230D1H protein appeared superior to the E. coli expressed and refolded Pfs230D1–2 even though the latter protein constituted a greater portion of native Pfs230. The basis for this increased potency is unclear. Recombinant Pfs230D1–2 protein was refolded and disulfide bond formation was confirmed by mobility shift following disulfide bond reduction. However, Western blotting analysis with mAb 4F12, which recognized a conformation-dependent epitope within native Pfs230 domain 1 demonstrated a stronger reactivity for Pfs230D1H/M than Pfs230D1–2 protein. Thus, the weaker reactivity of mAb 4F12 by Western blotting against the refolded Pfs230D1–2 may suggest that there is a large proportion of misfolded protein although this seems unlikely given the quality characteristics observed for the biophysical characterizations shown here. An alternative explanation may be that the non-structured domain and/or domain 2 interferes with mAb 4F12 binding to domain 1 such that these regions hinder the development of functional antibodies against domain 1. Previous reports demonstrated that blocking transmission by mAbs (31, 32) and polyclonal antibodies, including human antibodies targeting Pfs230 required complement (20). This is the first report showing that a Pfs230-specific mAb with an IgG1 isotype, which does not activate complement by the classical pathway (33) may block transmission in the presence or absence of human complement (Table 1). This result suggests that the biological function of Pfs230 may be disrupted directly by the formation of an antigen-antibody complex or possibly interfere with the formation of Pfs230 protein complexes (34).
Pfs230D1-specific human antibodies have been detected at low levels in endemic immune sera (35, 36). Native Pfs230 may provide T cell help because Pfs230 is naturally expressed during gametocytogenesis. This latter point may be a distinctive benefit for the inclusion of Pfs230 in a TBV to help maintain sufficient Pfs230D1-specific blocking antibodies. Pfs230 has been reported to contain a larger number of polymorphic residues than Pfs48/45 (37), another TBV candidate and Pfs25, which is not expressed in gametocytes. Even though Pfs230D1 appears to be under immune pressure, the observed changes in the MAFs, which are supported by our field observations for G605S, suggest that the polymorphisms are unlikely to interfere with our initial phase 1 trial in Mali. Looking ahead, a more detailed epidemiological analysis of patient-derived gametocyte isolates from Mali and elsewhere will be necessary in conjunction with ex vivo analysis by membrane feeding assays with a K661N parasite variant.
Two clinical trials have been conducted with recombinant Pfs25 to evaluate safety and immunogenicity, and this information can facilitate the design of Pfs230 clinical trials. In the first trial, recombinant Pfs25 (38) was formulated in Montanide ISA51 (3). Human Pfs25-specific transmission blocking antibodies were produced as shown by the ex vivo SMFA, however, the formulation was determined to be unsuitable for a public health vaccine due to its reactogenicity. A more recent human study evaluated a Pfs25-EPA chemical conjugate that was shown to have the characteristics of a nanoparticle (7). In the second study, the Pfs25-EPA formulated on Alhydrogel® was safe and immunogenic.3 Following four doses of the vaccine, significant biological activity was observed (11 of 12 vaccines providing greater than 50% TBA) as judged by the ex vivo SMFA. How Pfs230D1M chemically conjugated to EPA will perform alone, or in combination with a Pfs25-EPA conjugate, independently formulated on Alhydrogel® in humans is currently being evaluated in a United States/Mali phase 1 study (ClinicalTrials.gov Identifier: NCT02334462). Of note, after only two doses of Pfs230D1M-EPA alone in naive volunteers in the United States (n = 5), two volunteers with higher vaccine-specific antibody responses also had the highest functional activity in the SMFA (>80%), but this sample size was powered for safety, not immunogenicity.4 Taken all together, the development of Pfs230D1M increases the potential for achieving the overall goal of developing a vaccine to interrupt malaria transmission.
Experimental Procedures
Ethics Statement
The study protocol (number TMEC 11-033) for blood collection from Thai patients was approved by the Ethical Committees from the Faculty of Tropical Medicine, Mahidol University, and the Ministry of Public Health, Thailand. All animals used for this project were approved by the National Institute of Allergy and Infectious Diseases, Directorate of Intramural Research, Animal Care and Use Committee, protocol ASP LMIV 1E, at the National Institutes of Health, which is AAALAC accredited and OLAW assured. The NIAID DIR Animal Care and Use Program acknowledges and accepts responsibility for the care and use of animals involved in activities covered by NIH IRP's PHS Assurance number A4149-01.
Production Cell Lines
The amino acid sequence of the 3D7 Pfs230 protein (GenBankTM accession number XP_001349600.1) was used to design the Pichia codon-optimized Pfs230D1H and D1M genes (corresponding to Tyr444 to Gly736 and Ser542 to Gly736, respectively). Both genes had Asn585 mutated to Gln to remove the N-glycosylation site. The XhoI/XbaI genes were cloned into the XhoI/AvrII sites of a modified version of pPIC9K (Invitrogen) in which the XhoI site at position 5709 had been converted to an XbaI site. The synthetic genes contained a Lys-Arg Kex2 cleavage site immediately following the 5′ XhoI cloning site resulting in secretion of mature Pfs230D1M protein that contained no heterologous amino acids and mature Pfs230D1H protein that included heterologous GPHHHHHH amino acids at the carboxyl terminus. The expression plasmids were linearized with SacI and transformation into P. pastoris.
The E. coli expressed Pfs230D1–2 protein (XP_001349600.1 Glu443 to Asn915) was encoded by a gene codon optimized for expression in E. coli. The synthetic gene was cloned into the NdeI and XhoI sites of pET-24a(+) (Novagen) downstream of the T7 promoter. The expression plasmid was transformed into the E. coli BL21(DE3) expression line (Invitrogen) for recombinant expression. E. coli expressed Pfs230D1–2 protein contained heterologous LEHHHHHH amino acids at the carboxyl terminus.
Protein Production (Fermentation and Purification)
Pfs230D1–2 was refolded and purified following methodology previously reported (39). In brief, Pfs230D1–2 was refolded in 50 mm Tris, 9.6 mm NaCl, 0.4 mm KCl, 2 mm MgCl2, 2 mm CaCl2, 0.4 m sucrose, 0.05% polyethylene glycol 3350, 5% Triton X-100, 1 mm glutathione reduced, 0.1 mm glutathione oxidized, pH 8.0, and purified by sequential chromatographic separations using Ni-Sepharose HP, Q-Sepharose HP, and Superdex 75 chromatographic media (GE Healthcare). The Pfs230D1H and Pfs230D1M were fermented as previously described (38) with the exception that the methanol-induction conditions used were 25 °C, pH 5.0, and methanol feed rate of 375 ml/liter/h for 48 to 52 h.
Recombinant Pfs230D1H was purified from concentrated and dialyzed fermentation supernatant in 20 mm phosphate buffer with 300 mm NaCl, pH 7.4, prior to capture using a Ni-Sepharose HP (GE Healthcare) column equilibrated in 20 mm sodium phosphate, 300 mm NaCl, 25 mm imidazole, pH 7.4, and eluted with a 25–400 mm gradient of imidazole or a step elution with 300 mm imidazole. Fermentation supernatant containing Pfs230D1M was concentrated and dialyzed using a 3K NMWC hollow fiber cartridge with 20 mm sodium phosphate buffer with 300 mm NaCl, pH 7.8, and stored frozen at less than −70 °C. Prior to loading a MEP HyperCel (Pall BioSciences) mixed mode column equilibrated in 50 mm sodium phosphate, 100 mm arginine HCl, 500 mm ammonium sulfate, 500 mm sodium chloride, pH 7.25, the Pfs230D1M load was diluted with an equal volume of 150 mm sodium phosphate, 300 mm arginine HCl, 1.5 m ammonium sulfate, 1.2 m sodium chloride, pH 7.25. Pfs230D1M was loaded at 50 cm/h. The elution was a step with 50 mm sodium phosphate, 100 mm arginine HCl, 250 mm ammonium sulfate, 500 mm sodium chloride, pH 6.5. The eluate was concentrated and dialyzed into 50 mm glycine buffer, pH 9.5, in preparation for loading onto a Q-Sepharose FF (GE Healthcare) column equilibrated in the same buffer. The Pfs230D1M product eluted from the column in 50 mm glycine, 300 mm sodium chloride, pH 9.5, over 1 column volume. The Q-Sepharose FF elution pool was concentrated to a volume to allow for 5% loads (v/v) onto a Superdex 75 (GE Healthcare) size exclusion column. The size exclusion column was equilibrated in 23 mm sodium phosphate, monobasic, 75 mm sodium phosphate, dibasic, 154 mm sodium chloride, 5 mm EDTA, pH 7.3. The monomeric protein fractions were pooled and sterile filtered through a Millipak-60 (Millipore Corp). The pooled material was stored at less than −70 °C until further characterizations.
Protein Characterizations
Each recombinant protein was fully characterized using similar techniques to that reported (39, 40) for reversed-phase HPLC, and analytical SEC with in-line MALS and quasielastic light scatter detectors (Wyatt Technologies, Santa Barbara, CA). Protein separation was done on a TSKgel G2000SWxl column (Tosoh Biosciences). Amino-terminal sequencing and intact mass analysis were performed by the Research Technology Branch, NIAID, NIH. Edman degradation was done on a Sequenator model Procise 494 HT (Applied Biosystems) as previously described (39). Electrospray ionization mass spectrometry was done on an automated chip-based nanoelectrospray unit, TriVersa Nanomate (Advion BioSciences) as previously described (39). Circular dichroism (CD) spectroscopy was performed on a Jasco J-815 spectropolarimeter as previously described (41). Secondary structure content was calculated using the DICHROWEB web server (42).
Disulfide Bond Mapping
The disulfide bonding arrangement of PpPfs230D1M was identified using BiopharmLynx in association with a Waters XEVO G2 Q-TOF mass spectrometer. Recombinant PpPfs230D1M (90 μg) was diluted to 100 μl of buffer yielding a final concentration of 50 mm Tris, pH 7.8, and 0.06% RapiGestTM SF (Waters, p/n 186001861). Prior to digestion, the protein was denatured at 80 °C for 15 min. The sample was returned to room temperature, the free cysteine residues blocked by incubating with 10 mm iodoacetamide at room temperature for 30 min in the dark. To test for artificial disulfide-bond scrambling potentially introduced during the digestion, a digest prepared without alkylation was directly compared with a digest prepared in parallel with iodoacetamide alkylation before digestion. Samples were digested with sequencing-grade trypsin at 37 °C overnight. Trifluoroacetic acid (0.05% v/v) was used to quench the enzymatic reaction and degrade the RapiGestTM SF. The pH of the samples was adjusted to pH 10 by adding 10 μl of 1 n NH4OH for effective trapping on the 1st dimension column. Samples were spiked with alcohol dehydrogenase before loading the two-dimensional nanoACQUITY UPLC® for load normalization. Data acquisition was done on Xevo G2 Q-TOF mass spectrometer in positive ion mode using a nanoLockSprayTM source controlled by MassLynxTM 4.1 software. Digested samples were loaded on the first dimension trap column (300 μm × 50 mm XBridgeTM BEH130 C18 5 μm) at pH 10 and eluted to a second dimension trap column (180 μm × 20 mm Symmetry® C18 5 μm) with 50% acetonitrile and 50% 20 mm ammonium formate at pH 10 (mobile phases for first dimension pump) at a flow rate of 2 μl/min. Peptides eluted from the first dimension were then diluted and acidified with 0.1% formic acid in water at 20 μl/min flow prior to trapping onto the second dimension trap column. The peptides were separated on the second dimension analytical column (75 μm × 100 mm HSS T3, 1.8 μm) with a 60-min gradient from 3 to 60% mobile phase B (0.1% formic acid in acetonitrile). An alternating low collision energy (6 V) and elevated collision energy (ramping from 15 to 35 V) acquisition was used to acquire peptide precursor (MS) and fragmentation (MSE) data. Data were acquired at a scan rate of 1 spectrum/s. The capillary voltage was 3.6 kV, source temperature 100 °C, and cone voltage 45 V. Sampling of the lock spray channel was performed every 30 s. Spectra were recorded from m/z 50 to 1990. Samples were run in triplicates. LC/MSE data were processed and searched against Pfs230 protein sequences using BiopharmaLynxTM1.3. BiopharmaLynx identified the expected disulfide linkages with accurate mass. The mass accuracy of all identified disulfide-bonded peptides was within 5 ppm. All disulfide bond assignments were confirmed with elevated-energy fragmentation (MSE).
Immunogenicity and Blocking Activity of Pfs230 Antibodies
Paired New Zealand White rabbits were immunized three times (days: 0, 28, and 56) with 50 μg of purified Pfs230D1–2, Pfs230D1H, or Pfs230D1M formulated in ISA 720 VG (Seppic, Inc., Fairfield, NJ) administered subcutaneously. Rabbits were bled for sera on day 0 and 2 weeks following their third immunization for subsequent testing. Rabbit IgG purification used Protein G column chromatography as suggested by the manufacturer (GE Healthcare). All IgG samples were dialyzed extensively in PBS, pH 7.4. The SMFA assessing the blocking of parasite transmission of P. falciparum NF54 cultured in vitro and membrane feeding assay using isolates from Thai patients were performed as described (27).
ELISAs
The ELISA method used for Pfs230 followed the procedure previously reported for Pfs25 (43).
Production and Characterization of Hybridomas
Hybridomas were prepared by Precision Antibody (Columbia, MD) by immunizing mice with enriched preparations of P. falciparum female gametocytes. Hybridomas selected for development were screened by IFA against fixed gametocytes and by ELISA using the various recombinant Pfs230 proteins.
Surface and Modified Surface Immunofluorescence Assay
Surface immunofluorescence assay analysis was performed essentially as reported (22). In brief, gametocyte-enriched parasites were incubated in exflagellation media followed by incubations with appropriate primary antibody or mAbs for 30 min at 4 °C, washed, then incubated for 30 min at 4 °C with fluorescent labeled secondary antibodies, washed, and subsequently mounted in Vectashield for analysis. For the modified live surface IFA, the method has been previously described using sporozoites (40). In brief, the gametocytes or gametes/zygotes were washed following the primary antibody incubation and then spotted on glass slides, air dried, and stored at −80 °C. Slides were thawed, methanol-fixed, and reacted with primary and secondary antibodies diluted in blocking buffer (1× PBS, 3% BSA) at 4 °C. All primary and secondary antibody incubations were carried out at room temperature separated by extensive washing with PBS containing 0.05% Tween 20 (Bio-Rad). The samples were mounted under coverslips using Vectashield hard-set mounting medium and stored at 4 °C until images were acquired by using an Olympus fluorescence microscopy or confocal microscopy as described previously (39, 40).
Structural Modeling
The comparative model of Pfs230D1M was constructed with Rosetta (44) using the crystal structures of Pf12, PDB code 2YMO (29), and Pf41, PDB code 4YS4 (30), as templates. Secondary structure analysis was performed with STRIDE (45).
Genomic Sequencing of Pfs230
The genomic Pfs230D1 sequence of the two Thai patients whose P. falciparum isolates were blocked by rabbit Pfs230D1M-specific antisera (Table 3) was determined. The Pfs230D1 sequence was amplified and sequenced from genomic DNA isolated from dried blood spots using Qiagen's QIAamp DNA mini kit (Valencia, CA) and forward primer: 5′-GATGAGGATGAAGATTCTGTAGAAGC-3′ and reverse primer: 5′-GCATGCAATGTACATTCATGTGTATC-3′. Nucleic acid sequencing was performed by Eurofins Genomics.
Statistical Analyses
The statistical significance of the transmission blocking activity was assessed using a Mann-Whitney Rank Sum Test on each group of individual mosquitoes, which ignores potential variation within each container of mosquitoes (8).
Author Contributions
N. J. M., V. N., M. B., K. R., R. S., R. H., K. K., O. M., J. A., L. L., K. R., N. D., J. P., Y. W., J. S., X. A., and D. L.N. designed and performed research; N. J. M., V. N., M. B., K. R., R. S., R. H., K. K., O. M., J. A., J. P., P. E. D., Y. W., J. S., X. A., and D. L. N. analyzed data; N. J. M., K. R., K. K., X. A., and D. L. N. wrote the paper.
Acknowledgments
We thank Dr. Buddhadeb Mallik, Dr. Guan-Hong Song, Dr. Yanling Zhang, and Christopher Rowe for technical assistance within the Laboratory of Malaria Immunology and Vaccinology. Dr. Carl Hammer, Mark Garfield, from Research Technologies Branch, NIAID, for analytical support, and Dr. Michael Fay, from Biostatistics Research Branch, NIAID, for statistical support. Dr. Michael Nold from Waters Inc. for assistance with disulfide mapping studies, and Dr. Julian Rayner for the assistance with genotypic analysis of Pfs230 from the Wellcome Trust Sanger Institute. Finally we appreciate Dr. Louis Miller for his support and helpful discussion, This publication uses data from the MalariaGEN Pfalciparum Community Project as described in Ref. 28. Genome sequencing was performed by the Wellcome Trust Sanger Institute and the Community Projects is coordinated by the MalariaGENResource Centre with funding from the Wellcome Trust Grants 098051 and 090770.
This work was supported, in whole or in part, by the Intramural Research Program of NIAID, National Institutes of Health. The authors declare that they have no conflicts of interests with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
K. R. Talaat, R. D. Ellis, H. Hurd, A. Hentrich, E. Gabriel, N. A. Hynes, K. Rausch, D. Zhu, O. Muratova, C. Anderson, D. Jones, J. Aebig, S. Brockley, N. J. MacDonald, M. P. Fay, S. A. Healy, A. P. Durbin, D. L. Narum, Y. Wu, and P. E. Duffy, unpublished data.
S. A. Healy, A. Mwakingwe, C. Anderson, K. Rausch, D. L. Narum, D. L. Jones, N. J. MacDonald, D. Zhu, S. Brockley, J. Aebig, O. Muratova, J. C. Hume, S. Wong-Madden, E. Gabriel, Y. Wu, and P. E. Duffy, personal communication.
- Pfs25
- P. falciparum 25-kDa zygote and ookinete protein
- Pfs230
- P. falciparum 230-kDa sexual stage protein
- TBV
- transmission blocking vaccine
- EPA
- ExoProtein A
- TBA
- transmission blocking activity
- SMFA
- standard membrane feeding assay
- SEC-MALS
- size exclusion chromatography-multiangle light scattering.
References
- 1. Agnandji S. T., Lell B., Soulanoudjingar S. S., Fernandes J. F., Abossolo B. P., Conzelmann C., Methogo B. G., Doucka Y., Flamen A., Mordmuller B., Issifou S., Kremsner P. G., Sacarlal J., Aide P., Lanaspa M., et al. (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N. Engl. J. Med. 365, 1863–1875 [DOI] [PubMed] [Google Scholar]
- 2. RTS,S. Clinical Trials Partnership (2015) Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet 386, 31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu Y., Ellis R. D., Shaffer D., Fontes E., Malkin E. M., Mahanty S., Fay M. P., Narum D., Rausch K., Miles A. P., Aebig J., Orcutt A., Muratova O., Song G., Lambert L., et al. (2008) Phase 1 trial of malaria transmission blocking vaccine candidates Pfs25 and Pvs25 formulated with montanide ISA 51. PLoS ONE 3, e2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu Y., Przysiecki C., Flanagan E., Bello-Irizarry S. N., Ionescu R., Muratova O., Dobrescu G., Lambert L., Keister D., Rippeon Y., Long C. A., Shi L., Caulfield M., Shaw A., Saul A., Shiver J., and Miller L. H. (2006) Sustained high-titer antibody responses induced by conjugating a malarial vaccine candidate to outer-membrane protein complex. Proc. Natl. Acad. Sci. U.S.A. 103, 18243–18248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qian F., Wu Y., Muratova O., Zhou H., Dobrescu G., Duggan P., Lynn L., Song G., Zhang Y., Reiter K., MacDonald N., Narum D. L., Long C. A., Miller L. H., Saul A., and Mullen G. E. (2007) Conjugating recombinant proteins to Pseudomonas aeruginosa ExoProtein A: a strategy for enhancing immunogenicity of malaria vaccine candidates. Vaccine 25, 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qian F., Rausch K. M., Muratova O., Zhou H., Song G., Diouf A., Lambert L., Narum D. L., Wu Y., Saul A., Miller L. H., Long C. A., and Mullen G. E. (2008) Addition of CpG ODN to recombinant Pseudomonas aeruginosa ExoProtein A conjugates of AMA1 and Pfs25 greatly increases the number of responders. Vaccine 26, 2521–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shimp R. L. Jr, Rowe C., Reiter K., Chen B., Nguyen V., Aebig J., Rausch K. M., Kumar K., Wu Y., Jin A. J., Jones D. S., and Narum D. L. (2013) Development of a Pfs25-EPA malaria transmission blocking vaccine as a chemically conjugated nanoparticle. Vaccine 31, 2954–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miura K., Deng B., Tullo G., Diouf A., Moretz S. E., Locke E., Morin M., Fay M. P., and Long C. A. (2013) Qualification of standard membrane-feeding assay with Plasmodium falciparum malaria and potential improvements for future assays. PLoS ONE 8, e57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vincent A. A., Fanning S., Caira F. C., and Williamson K. C. (1999) Immunogenicity of malaria transmission-blocking vaccine candidate, y230.CA14 following crosslinking in the presence of tetanus toxoid. Parasite Immunol. 21, 573–581 [DOI] [PubMed] [Google Scholar]
- 10. Outchkourov N. S., Roeffen W., Kaan A., Jansen J., Luty A., Schuiffel D., van Gemert G. J., van de Vegte-Bolmer M., Sauerwein R. W., and Stunnenberg H. G. (2008) Correctly folded Pfs48/45 protein of Plasmodium falciparum elicits malaria transmission-blocking immunity in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 4301–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chowdhury D. R., Angov E., Kariuki T., and Kumar N. (2009) A potent malaria transmission blocking vaccine based on codon harmonized full length Pfs48/45 expressed in Escherichia coli. PLoS ONE 4, e6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farrance C. E., Rhee A., Jones R. M., Musiychuk K., Shamloul M., Sharma S., Mett V., Chichester J. A., Streatfield S. J., Roeffen W., van de Vegte-Bolmer M., Sauerwein R. W., Tsuboi T., Muratova O. V., Wu Y., and Yusibov V. (2011) A plant-produced Pfs230 vaccine candidate blocks transmission of Plasmodium falciparum. Clin. Vaccine Immunol. 18, 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tachibana M., Wu Y., Iriko H., Muratova O., MacDonald N. J., Sattabongkot J., Takeo S., Otsuki H., Torii M., and Tsuboi T. (2011) N-terminal prodomain of Pfs230 synthesized using a cell-free system is sufficient to induce complement-dependent malaria transmission-blocking activity. Clin. Vaccine Immunol. 18, 1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quakyi I. A., Carter R., Rener J., Kumar N., Good M. F., and Miller L. H. (1987) The 230-kDa gamete surface protein of Plasmodium falciparum is also a target for transmission-blocking antibodies. J. Immunol. 139, 4213–4217 [PubMed] [Google Scholar]
- 15. Templeton T. J., and Kaslow D. C. (1999) Identification of additional members define a Plasmodium falciparum gene superfamily which includes Pfs48/45 and Pfs230. Mol. Biochem. Parasitol. 101, 223–227 [DOI] [PubMed] [Google Scholar]
- 16. Kocken C. H., Jansen J., Kaan A. M., Beckers P. J., Ponnudurai T., Kaslow D. C., Konings R. N., and Schoenmakers J. G. (1993) Cloning and expression of the gene coding for the transmission blocking target antigen Pfs48/45 of Plasmodium falciparum. Mol. Biochem. Parasitol. 61, 59–68 [DOI] [PubMed] [Google Scholar]
- 17. Kumar N. (1987) Target antigens of malaria transmission blocking immunity exist as a stable membrane bound complex. Parasite Immunol. 9, 321–335 [DOI] [PubMed] [Google Scholar]
- 18. Brooks S. R., and Williamson K. C. (2000) Proteolysis of Plasmodium falciparum surface antigen, Pfs230, during gametogenesis. Mol. Biochem. Parasitol. 106, 77–82 [DOI] [PubMed] [Google Scholar]
- 19. Bustamante P. J., Woodruff D. C., Oh J., Keister D. B., Muratova O., and Williamson K. C. (2000) Differential ability of specific regions of Plasmodium falciparum sexual-stage antigen, Pfs230, to induce malaria transmission-blocking immunity. Parasite Immunol. 22, 373–380 [DOI] [PubMed] [Google Scholar]
- 20. Healer J., McGuinness D., Hopcroft P., Haley S., Carter R., and Riley E. (1997) Complement-mediated lysis of Plasmodium falciparum gametes by malaria-immune human sera is associated with antibodies to the gamete surface antigen Pfs230. Infect. Immun. 65, 3017–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roeffen W., Beckers P. J., Teelen K., Lensen T., Sauerwein R. W., Meuwissen J. H., and Eling W. (1995) Plasmodium falciparum: a comparison of the activity of Pfs230-specific antibodies in an assay of transmission-blocking immunity and specific competition ELISAs. Exp. Parasitol. 80, 15–26 [DOI] [PubMed] [Google Scholar]
- 22. Eksi S., Czesny B., van Gemert G. J., Sauerwein R. W., Eling W., and Williamson K. C. (2006) Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol. Microbiol. 61, 991–998 [DOI] [PubMed] [Google Scholar]
- 23. Williamson K. C., Keister D. B., Muratova O., and Kaslow D. C. (1995) Recombinant Pfs230, a Plasmodium falciparum gametocyte protein, induces antisera that reduce the infectivity of Plasmodium falciparum to mosquitoes. Mol. Biochem. Parasitol. 75, 33–42 [DOI] [PubMed] [Google Scholar]
- 24. Gerloff D. L., Creasey A., Maslau S., and Carter R. (2005) Structural models for the protein family characterized by gamete surface protein Pfs230 of Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 102, 13598–13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arredondo S. A., Cai M., Takayama Y., MacDonald N. J., Anderson D. E., Aravind L., Clore G. M., and Miller L. H. (2012) Structure of the Plasmodium 6-cysteine s48/45 domain. Proc. Natl. Acad. Sci. U.S.A. 109, 6692–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu D., Saul A. J., and Miles A. P. (2005) A quantitative slot blot assay for host cell protein impurities in recombinant proteins expressed in E. coli. J. Immunol. Methods 306, 40–50 [DOI] [PubMed] [Google Scholar]
- 27. Cheru L., Wu Y., Diouf A., Moretz S. E., Muratova O. V., Song G., Fay M. P., Miller L. H., Long C. A., and Miura K. (2010) The IC50 of anti-Pfs25 antibody in membrane-feeding assay varies among species. Vaccine 28, 4423–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manske M., Miotto O., Campino S., Auburn S., Almagro-Garcia J., Maslen G., O'Brien J., Djimde A., Doumbo O., Zongo I., Ouedraogo J. B., Michon P., Mueller I., Siba P., Nzila A., et al. (2012) Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tonkin M. L., Arredondo S. A., Loveless B. C., Serpa J. J., Makepeace K. A., Sundar N., Petrotchenko E. V., Miller L. H., Grigg M. E., and Boulanger M. J. (2013) Structural and biochemical characterization of Plasmodium falciparum 12 (Pf12) reveals a unique interdomain organization and the potential for an antiparallel arrangement with Pf41. J. Biol. Chem. 288, 12805–12817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parker M. L., Peng F., and Boulanger M. J. (2015) The Structure of Plasmodium falciparum Blood-Stage 6-Cys Protein Pf41 Reveals an Unexpected Intra-Domain Insertion Required for Pf12 Coordination. PLoS ONE 10, e0139407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roeffen W., Geeraedts F., Eling W., Beckers P., Wizel B., Kumar N., Lensen T., and Sauerwein R. (1995) Transmission blockade of Plasmodium falciparum malaria by anti-Pfs230-specific antibodies is isotype dependent. Infect. Immun. 63, 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Read D., Lensen A. H., Begarnie S., Haley S., Raza A., and Carter R. (1994) Transmission-blocking antibodies against multiple, non-variant target epitopes of the Plasmodium falciparum gamete surface antigen Pfs230 are all complement-fixing. Parasite Immunol. 16, 511–519 [DOI] [PubMed] [Google Scholar]
- 33. Strait R. T., Posgai M. T., Mahler A., Barasa N., Jacob C. O., Köhl J., Ehlers M., Stringer K., Shanmukhappa S. K., Witte D., Hossain M. M., Khodoun M., Herr A. B., and Finkelman F. D. (2015) IgG1 protects against renal disease in a mouse model of cryoglobulinaemia. Nature 517, 501–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simon N., Kuehn A., Williamson K. C., and Pradel G. (2016) Adhesion protein complexes of malaria gametocytes assemble following parasite transmission to the mosquito. Parasitol. Int. 65, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williamson K. C. (2003) Pfs230: from malaria transmission-blocking vaccine candidate toward function. Parasite Immunol. 25, 351–359 [DOI] [PubMed] [Google Scholar]
- 36. Jones S., Grignard L., Nebie I., Chilongola J., Dodoo D., Sauerwein R., Theisen M., Roeffen W., Singh S. K., Singh R. K., Singh S., Kyei-Baafour E., Tetteh K., Drakeley C., and Bousema T. (2015) Naturally acquired antibody responses to recombinant Pfs230 and Pfs48/45 transmission blocking vaccine candidates. J. Infect. 71, 117–127 [DOI] [PubMed] [Google Scholar]
- 37. Niederwieser I., Felger I., and Beck H. P. (2001) Limited polymorphism in Plasmodium falciparum sexual-stage antigens. Am. J. Trop Med. Hyg 64, 9–11 [DOI] [PubMed] [Google Scholar]
- 38. Tsai C. W., Duggan P. F., Shimp R. L. Jr., Miller L. H., and Narum D. L. (2006) Overproduction of Pichia pastoris or Plasmodium falciparum protein disulfide isomerase affects expression, folding and O-linked glycosylation of a malaria vaccine candidate expressed in P. pastoris. J. Biotechnol. 121, 458–470 [DOI] [PubMed] [Google Scholar]
- 39. Uchime O., Herrera R., Reiter K., Kotova S., Shimp R. L. Jr, Miura K., Jones D., Lebowitz J., Ambroggio X., Hurt D. E., Jin A. J., Long C., Miller L. H., and Narum D. L. (2012) Analysis of the conformation and function of the Plasmodium falciparum merozoite proteins MTRAP and PTRAMP. Eukaryot. Cell 11, 615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herrera R., Anderson C., Kumar K., Molina-Cruz A., Nguyen V., Burkhardt M., Reiter K., Shimp R. Jr., Howard R. F., Srinivasan P., Nold M. J., Ragheb D., Shi L., DeCotiis M., et al. (2015) Reversible conformational change in the Plasmodium falciparum circumsporozoite protein masks its adhesion domains. Infect. Immun. 83, 3771–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Plassmeyer M. L., Reiter K., Shimp R. L. Jr., Kotova S., Smith P. D., Hurt D. E., House B., Zou X., Zhang Y., Hickman M., Uchime O., Herrera R., Nguyen V., Glen J., Lebowitz J., et al. (2009) Structure of the Plasmodium falciparum circumsporozoite protein, a leading malaria vaccine candidate. J. Biol. Chem. 284, 26951–26963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whitmore L., and Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 43. Miura K., Keister D. B., Muratova O. V., Satta Bongkot J., Long C. A., and Saul A. (2007) Transmission-blocking activity induced by malaria vaccine candidates Pfs25/Pvs25 is a direct and predictable function of antibody titer. Malar J. 6, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song Y., DiMaio F., Wang R. Y., Kim D., Miles C., Brunette T., Thompson J., and Baker D. (2013) High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frishman D., and Argos P. (1995) Knowledge-based protein secondary structure assignment. Proteins 23, 566–579 [DOI] [PubMed] [Google Scholar]