Abstract
Pseudomonas aeruginosa is a leading cause of hospital-acquired infections and is resistant to many antibiotics. Type IV pili (T4P) are among the key virulence factors used by P. aeruginosa for host cell attachment, biofilm formation, and twitching motility, making this system a promising target for novel therapeutics. Point mutations in the conserved PilMNOP alignment subcomplex were previously shown to have distinct effects on assembly and disassembly of T4P, suggesting that it may function in a dynamic manner. We introduced mutations encoding Cys substitutions into pilN and/or pilO on the chromosome to maintain normal stoichiometry and expression levels and captured covalent PilNO heterodimers, as well as PilN and PilO homodimers, in vivo. Most covalent PilN or PilO homodimers had minimal functional impact in P. aeruginosa, suggesting that homodimers are a physiologically relevant state. However, certain covalent homo- or heterodimers eliminated twitching motility, suggesting that specific PilNO configurations are essential for T4P function. These data were verified using soluble N-terminal truncated fragments of PilN and PilO Cys mutants, which purified as a mixture of homo- and heterodimers at volumes consistent with a tetramer. Deletion of genes encoding alignment subcomplex components, PilM or PilP, but not other T4P components, including the motor ATPases PilB or PilT, blocked in vivo formation of disulfide-bonded PilNO heterodimers, suggesting that both PilM and PilP influence the heterodimer interface. Combined, our data suggest that T4P function depends on rearrangements at PilN and PilO interfaces.
Keywords: cysteine-mediated cross-linking, protein-protein interaction, Pseudomonas aeruginosa (P. aeruginosa), type II secretion system (T2SS), type IV pili, twitching motility
Introduction
Many bacteria, including Pseudomonas aeruginosa, use type IV pili (T4P)3 for surface attachment/adhesion, biofilm formation, and twitching motility (1–6). T4P are divided into T4aP and T4bP subgroups, based on differences in pilin size and organization of the assembly machinery (6, 7). Here we focus on T4aP, henceforth referred to as T4P. T4P are long, thin, fibrous appendages, which extend from the bacteria, attach to a surface, and are retracted back into the cell, winching the cell forward. This flagellum-independent form of locomotion is called twitching motility. Four subcomplexes make up a T4P machine in P. aeruginosa as follows: the outer membrane (OM) secretin (PilQ) and its pilotin (PilF) (8–12); the inner membrane (IM) platform protein (PilC) and cytoplasmic motor proteins (ATPases PilBTU) (13, 14); the helical pilus fiber, composed mainly of the major pilin subunit (PilA) plus minor pilins (PilVWXE and FimU) and adhesin PilY1 (5, 15, 16); and the alignment subcomplex proteins (PilMNOP) that span from the cytoplasm to the OM secretin (17–20). PilN and PilO are bitopic IM proteins with similar predicted secondary structures. These proteins are essential for T4P function and connect the cytoplasmic alignment subcomplex component, PilM, with the IM-associated lipoprotein PilP that interacts with the N0 domain of the secretin monomer PilQ (19, 21). Proposed functions of the PilMNOP subcomplex include aligning the IM motor with the OM secretin, concentrating pilin subunits at the site of extension and retraction, and possibly transducing signals via conformational changes from the cytoplasmic ATPases to generate “open” or “closed” states of the gated secretin (19, 20).
The T4P and type II secretion (T2S) systems have analogous components and similar architecture (22, 23). Whereas the T4P system extends and retracts an elongated pilus, the T2S system specializes in secreting protein substrates from the periplasm into the environment, using a short piston-like “pseudopilus” (23–25). Although the systems have different outputs, their underlying functional mechanisms, which remain to be deciphered, are thought to be comparable. Orthologous components of the alignment subcomplexes in the two systems (PilMN/GspL, PilO/GspM, and PilP/GspC) have limited sequence identity, but atomic resolution structures revealed a high degree of structural similarity, and all three components form equimolar complexes with a ratio of 1:1:1 (18, 20, 26–30). PilN- and PilO-like proteins in the T2S system (GspL and GspM, respectively) form both homo- and heterodimers, although it is not yet clear whether the T4P system is similar (26, 27, 31–34). Previous work in P. aeruginosa showed that PilP fails to interact with a PilO homodimer and requires both PilN and PilO for stability (17, 20). Although PilO crystallized as a homodimer (18), complementation of a pilO mutant with a plasmid-encoded copy of the gene led to formation of predominantly PilO homodimers at the cost of PilN stability, and thus T4P function (17). In Thermus thermophilus, purified T4P alignment subcomplex components were estimated by negative stain cryoelectron microscopy to be in a ratio of 2:2:2 for PilM/PilN/PilO, where initial PilN homodimers in complex with PilM were disrupted by the addition of PilO, ultimately forming PilNO heterodimers (29). These observations, coupled with data from the P. aeruginosa system, suggested that the PilNO heterodimer was the relevant functional state in vivo, whereas PilN and PilO homodimers might represent intermediates during formation of the assembly system or in complementation experiments, artifacts of overexpression (20).
Cysteine disulfide-bonding analysis is useful for clarifying protein tertiary and quaternary structure and for investigating dynamic interactions among components of cellular machineries (35, 36). The ability of two Cys residues to form a disulfide bond depends on the proximity of their side chains, with bond formation occurring in an oxidizing environment at a Cβ-Cβ distance of 4–8 Å (37). Disulfide bonding imposes structural constraints, and analysis of its effects on function can reveal the importance of conformational changes. Using Cys substitution mutants, in vivo formation of both homo- and heterodimers of GspL and GspM was reported for the T2S system of Dickeya dadantii (34). The authors suggested that alternative interactions between homo- and heterodimers of GspL and GspM, i.e. GspLL and GspMM versus two of GspLM, occurred in vivo. Formation of disulfide bonds between residues that were predicted to be outwardly facing suggested that these interactions might be controlled through rotation of the proteins, including their transmembrane segments (TMSs) (34). This mechanism was proposed to propagate movement from the cytoplasmic to periplasmic regions of the T2S system machinery, when the ATPase GspE, equivalent to T4P extension ATPase PilB, interacted with the cytoplasmic domain of GspL, equivalent to PilM (34). The interaction itself, or the hydrolysis of ATP and resulting conformational changes in GspE, could induce a corresponding change in GspL, transduced to GspM through their interaction. Previous work in the T2S system and, more recently, the T4P system provided evidence for interactions between the equivalents of PilM and the cytoplasmic ATPase, PilB, either directly or in conjunction with the platform protein PilC (38–41). These interactions might similarly transduce conformational changes that contribute to pilus assembly or disassembly.
We hypothesized that dynamic movement of the T4P alignment subcomplex proteins PilN and PilO may occur during (or because of) extension and retraction of pili and that PilO's ability to interact with both itself and PilN may reflect a functionally relevant switch between homo- and heterodimers. To test this idea, we introduced Cys substitutions at predicted interfaces between PilNO and PilOO, and we used non-reducing SDS-PAGE to observe dimer formation. Covalent PilN and PilO homodimers, as well as PilNO heterodimers, were captured in vivo under physiological conditions. Depending on the location, disulfide bonding disrupted the normal function of the T4P system, inhibiting motility. Deletion of the motor ATPases had no effect on the dimerization state of PilN and PilO, arguing against a link between motor function and conformational switching, but loss of either PilM or PilP specifically blocked PilNO heterodimer formation. These data provide new insights into the architecture and dynamics of the T4P system.
Results
Selection of Residues for Cys Substitution
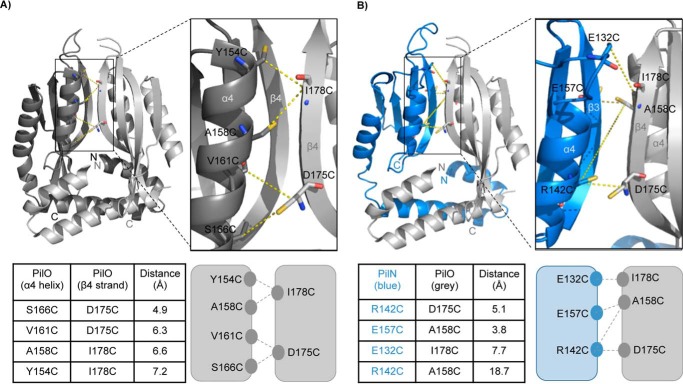
Our PilO homodimer structure (Protein Data Bank code 2RJZ) (18) was used to identify candidate residues predicted to be within appropriate distances for disulfide bond formation (Fig. 1A, top). In the PilO model, two relevant interfaces were identified as follows: the core, comprising mainly contacts between the α4-helix and β4-strand; and the coiled coils, between the α1- and α2-helices (18). In the PilOΔ68 model, the coiled-coils are truncated and folded back, because they are not anchored in the IM. Because of the potentially artificial nature of that particular interaction interface, we focused on residues in the core region previously shown to be important for function (42). Residues in the α4-helix and β4-strand of the PilO homodimer model with an estimated distance between β-carbons of less than 8 Å were selected for mutagenesis (Fig. 1A, bottom) (37, 43, 44).
FIGURE 1.
Locations of residues chosen for Cys substitution mutagenesis in PilN and PilO. A schematic diagram representation mapping the N and C termini, secondary structure elements, and structural domains of a PilOΔ68 dimer (2RJZ (18)) (A) or a PilNΔ57 (blue) and PilOΔ68 (gray) heterodimer using a Phyre2 model (45) (B) of PilN generated using the PilOΔ68 homodimer as template. The core-core domains consist of PilN α-helices 3–4 and β-strands 1–4 and PilO α-helices 3–4 and β-strands 1–5. Inset shows zoomed view of interaction interface with residue substitutions labeled accordingly. Bottom, tables indicate the distance between the Cβ of the substituted Cys residues (in angstroms) and schematic depiction of residue locations.
All attempts to purify soluble P. aeruginosa PilN for structural studies, or to solve an x-ray crystal structure of the PilNO heterodimer, have so far been unsuccessful. The structure of a periplasmic portion of PilN from T. thermophilus is available (29); however, due to the low primary sequence identity with PilN from P. aeruginosa, as well as a different arrangement of secondary structure elements (Thermus PilN has an extra α-helix inserted in the first αββ motif), we were unable to generate a high confidence model using that structural template. P. aeruginosa PilN is predicted to have a structural organization similar to PilO, and thus we used the truncated P. aeruginosa PilO homodimer structure (18) to generate a high confidence Phyre2 (45) model of a PilNΔ57 monomer, and we used the PilO homodimer interface to model a PilNO heterodimer using the same core-core interface (Fig. 1B, top) (42). This model was used to design a series of single Cys substitutions in the α4-helix and β4-strand of either PilN or PilO (Fig. 1B, bottom). The PilNO model has been partially validated in previous work (42), but for this study, we included a negative control Cys pair (PilNR142C/PilOA158C) at a Cβ distance of over 12 Å, considered too far for disulfide bond formation (37, 44).
To avoid issues arising from non-stoichiometric expression, the mutations encoding the Cys substitutions of interest were introduced at the native pilMNOPQ locus of P. aeruginosa PAK by homologous recombination. Screening of the sequence-verified mutants for effects on expression and stability of each of the inner membrane alignment subcomplex proteins revealed no effects of the Cys substitutions on PilMNOP levels relative to wild type (Fig. 2A).
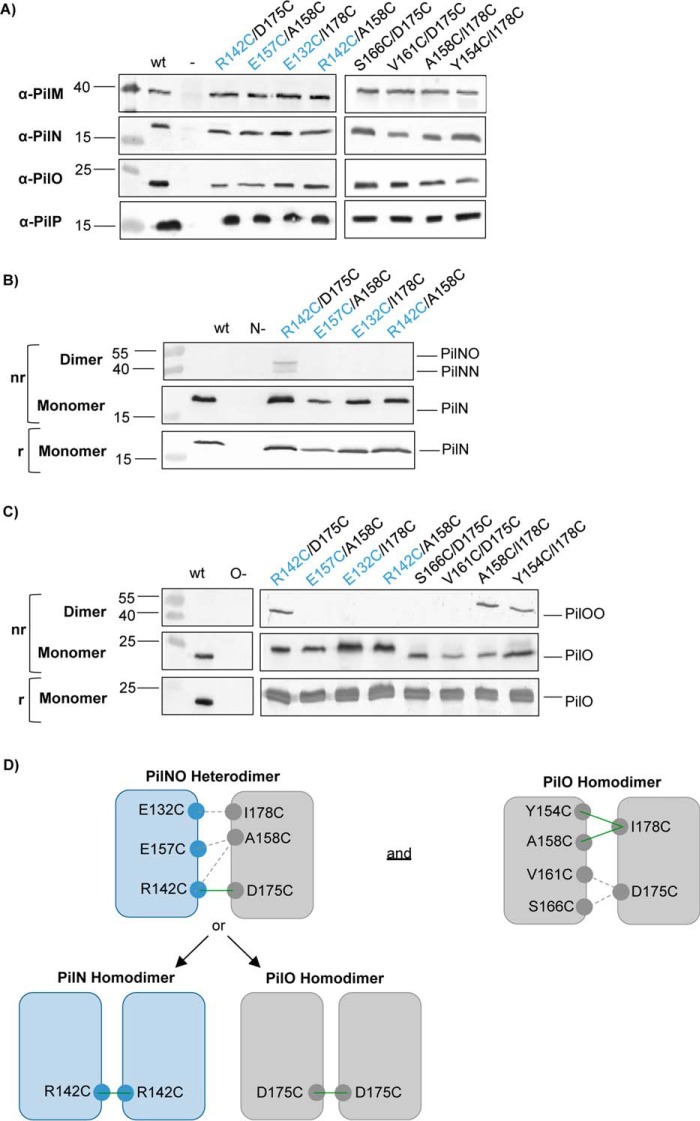
FIGURE 2.
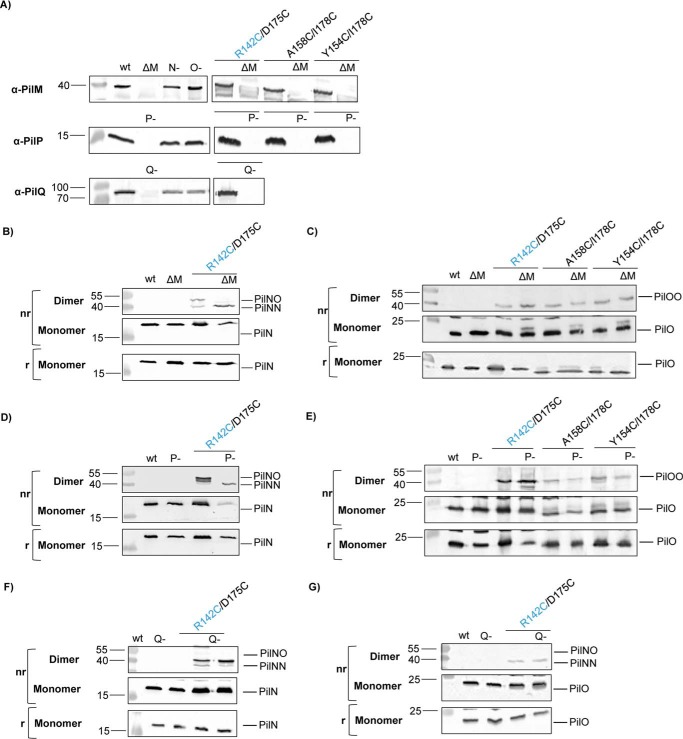
Formation of disulfide-bonded homo- and heterodimers in vivo. Lysates for the wild-type (wt), negative controls (−), and the Cys mutants were subject to SDS-PAGE under non-reducing (nr) and reducing (r) conditions and then blotted with specific antibodies to each alignment subcomplex component (α-PilM, α-PilN, α-PilO, and α-PilP) to confirm stability of each protein in the Cys mutants (A), PilN (B), and PilO (C). PilNR142C/PilOD175C was able to form PilNO heterodimers (44.7 kDa) and PilN homodimers (43.8 kDa), whereas PilOA158C/I178C and PilOY154C/I178C mutants were able to form disulfide bonds and form PilO homodimers (45.6 kDa) in vivo. Images are representative of at least three independent experiments. D, schematic of the residues that were able to form disulfide bonds (green lines) in the cross-linking experiments and their relative locations. Dashed lines indicate no dimer formation was observed. The PilNR142C/PilOD175C Cys mutants formed PilNO heterodimers but also PilNR142C and PilOD175C homodimers. PilN residues are indicated in blue text.
Formation of Cys Cross-linked PilNO and PilOO Dimers in Vivo
Whole cell lysates of all mutants were analyzed in parallel in reducing and non-reducing conditions, followed by immunoblotting with anti-PilN or PilO antibodies. Of the PilNO Cys substitutions, only the PilNR142C/PilOD175C variant formed significant levels of cross-linked dimers in vivo (Fig. 2, B and C). Two bands appeared on the anti-PilN blot (Fig. 2B), at the sizes consistent with in vivo formation of a PilNO heterodimer (44.7 kDa) and, unexpectedly, a PilN homodimer (43.8 kDa). Of the PilO double Cys mutants, both PilOA158C/I178C and PilOY154C/I178C variants formed cross-linked dimers in vivo (Fig. 2C) at a size consistent with that of PilO homodimers (45.6 kDa). Thus, at least one of each set of selected residue pairs formed the predicted linkage (Fig. 2D). This is the first evidence in any T4P system for PilN homodimerization under physiological conditions.
To further investigate PilN homodimerization, we leveraged a previously described PilNΔ44/PilOΔ51_His co-expression construct (20) that encodes truncated forms of both proteins. These proteins lack their short cytoplasmic N termini and transmembrane segments and encompass a larger portion of the periplasmic domains than the construct used to obtain the aforementioned PilOΔ68 homodimer structure. A PilN Cys mutant (PilNΔ44R142C/PilOΔ51), a PilO Cys mutant (PilNΔ44/PilOΔ51D175C), or a double PilNO Cys pair (PilNΔ44R142C/PilOΔ51D175C) were expressed in E. coli Origami 2 DE3, which has a non-reducing cytoplasm to allow for the formation of disulfide bonds, and purified using nickel-nitrilotriacetic acid affinity chromatography. Purified proteins were separated under reducing or non-reducing conditions and probed with anti-PilN and PilO antibodies. The PilNΔ44R142C/PilOΔ51 pair yielded disulfide-bonded PilN homodimers (∼34.0 kDa) (Fig. 3A). Interestingly, the same proteins also formed apparent PilNO heterodimers, as PilN monomers were still visible in the non-reducing lane. Because PilN is unstable/insoluble in the absence of PilO (20), this result suggests that a proportion of PilN monomers did not participate in covalent self-interactions but rather in non-covalent interactions with PilO. For PilNΔ44/PilOΔ51D175C, PilO homodimers (∼36.4 kDa) were clearly visible in the anti-PilO blot, showing that PilO occupies both homo- and heterodimer states (Fig. 3B). When both PilN and PilO contained Cys substitutions (PilNΔ44R142C/PilOΔ51D175C), three subpopulations were identified as follows: PilN homodimers, PilO homodimers, and PilNO heterodimers (∼35.2 kDa; Fig. 3, A and B). In this case, nearly all PilN monomers were involved in covalent interactions in non-reducing conditions (Fig. 3, A and B) when compared with PilO. This suggests that PilO homodimers might occupy multiple orientations, some disulfide-bonded and others not, implying in the latter case that the PilOD175C mutation was too far or incorrectly oriented for disulfide bond formation. These data confirm the observation that PilN and PilO form homodimers and heterodimers under physiological conditions in P. aeruginosa.
FIGURE 3.
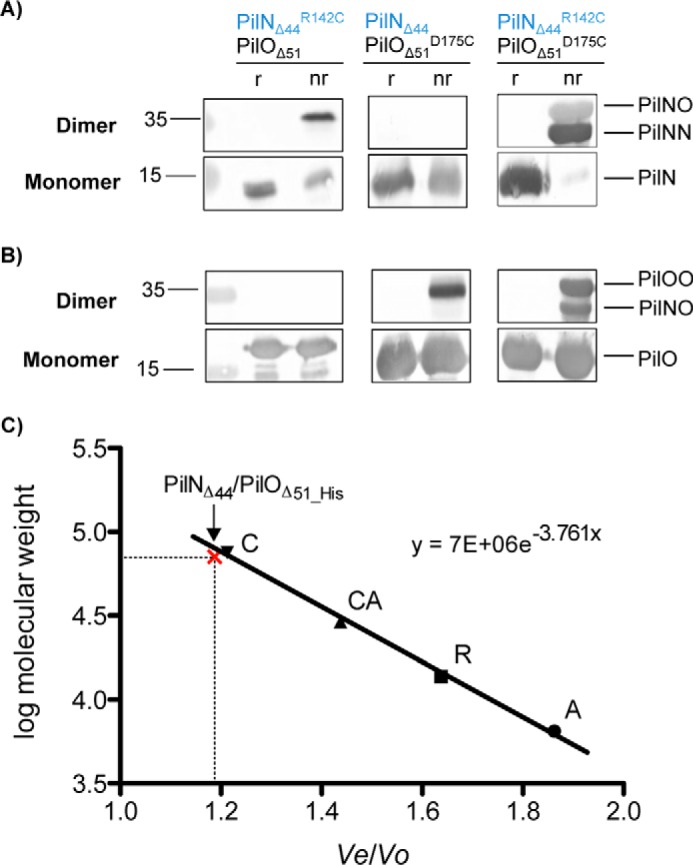
Purification of PilN and PilO tetramers. PilNΔ44 and PilOΔ51 were co-expressed with either PilNR142C, PilOD175C, or both the PilNR142C/PilOD175C Cys mutations. After nickel affinity purification, the proteins were subject to SDS-PAGE under either non-reducing (nr) or reducing (r) conditions and blotted with antisera specific for PilN (A) or PilO (B). We observed the following three disulfide-bonded subpopulations: PilN homodimers (34.0 kDa) in both the PilNR142C and PilNR142C/PilOD175C samples (left and right panels); PilO homodimers (36.4 kDa) in both the PilOD175C and PilNR142C/PilOD175C samples (center and right panels); and PilNO heterodimers (35.2 kDa) in the double Cys mutant as well (right). C, gel filtration calibration curve for the protein standards on a Superdex 75 10/300 GL column; aprotinin (A; 6.5 kDa), RNase A (R; 13.7 kDa), carbonic anhydrase (CA; 29.0 kDa), and conalbumin (C; 75.0 kDa). PilNΔ44 and PilOΔ51 co-eluted at a volume similar to conalbumin as indicated on graph with the red x The elution volumes (Ve) of the standards are divided by the void volume of the column (V0) and plotted against the log of the standards' molecular weights to give the slope of the line that was used to solve for the molecular weight of the eluted PilNΔ44/PilOΔ51_His complex (70.4 kDa). PilN residues indicated in blue text.
Because the untagged covalently linked PilNR142C homodimers purified with C-terminally His-tagged PilO, we realized that PilN homodimers must interact with PilO homodimers via an interface other than the one shown in Fig. 1B; the most likely candidate for a second interface was the coiled coils, below the core domains. Using size exclusion chromatography, we confirmed that PilNΔ44R142C/PilOΔ51_His (as well as unmodified PilNΔ44/PilOΔ51_His and other Cys mutants) eluted from an S75 size exclusion column at a volume similar to the conalbumin standard (∼75 kDa) (Fig. 3C), consistent with a PilNO tetramer (70.4 kDa). Thus, PilNO can simultaneously form homo- and heterodimers via two distinct interfaces.
Disulfide Bond Formation between PilN and/or PilO Can Disrupt T4P Function
To assess the functional consequences arising from formation of covalently linked dimers in vivo, the ability of the Cys mutant strains to assemble surface-exposed T4P was assessed (Fig. 4A). The PilNR142C/PilOD175C and PilOA158C/I178C mutants had very low levels of surface pili, and piliation of the PilOY154C/I178C mutant was comparable with negative controls. Both the PilOS166C/D175C and PilOV161C/D175C mutants had a moderate reduction in surface pili, possibly related to their shared D175C mutation. These data suggest that pilus biogenesis requires specific PilN and PilO configurations that are perturbed by these mutations.
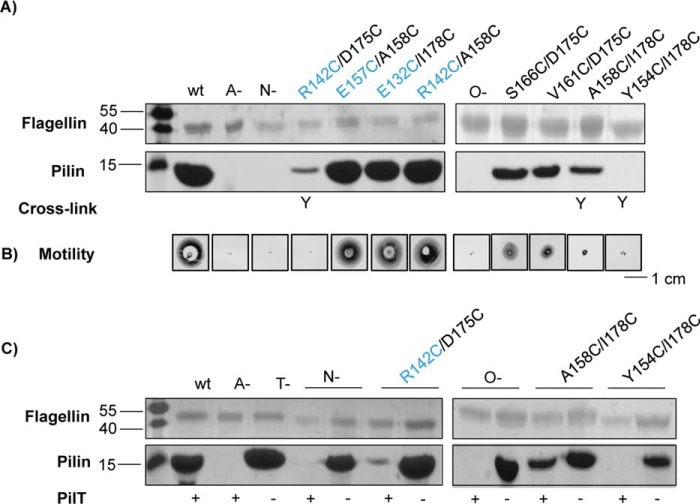
FIGURE 4.
Functional consequences of covalent dimer formation in wild type and retraction-deficient backgrounds. PilN and PilO Cys mutants, plus wild type (wt), non-piliated (A−), hyperpiliated (T−), and negative controls (N- and O-) were tested for sheared surface pili (A), and twitching motility (B). Sheared surface preparations revealed that the PilNR142C/PilOD175C and PilOA158C/I178C mutants exhibited severely reduced levels of surface pili, although the PilOY154C/I178C mutant exhibited no surface-exposed pili, and all were devoid of twitching motility similar to negative controls. Large disruptions in motility and piliation in the remaining PilNO Cys pairs were not observed. The ability of PilNO or PilOO Cys pairs to form Cys cross-links is indicated below (Y). C, select Cys mutants were introduced into a retraction-deficient background, which revealed that all three double Cys mutants assemble more pili without the retraction ATPase, PilT. PilT presence (+) or absence (−) in each strain is indicated below. Images are representative of at least three independent experiments.
Because T4P function relies on the dynamic assembly and disassembly of pili, it was important to assess motility as well as piliation levels (Fig. 4B). Strains forming covalently cross-linked dimers (PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C) were incapable of twitching. PilOS166C/D175C and PilOV161C/D175C strains had approximately half of wild-type motility, consistent with their decrease in piliation levels, although no in vivo cross-linked dimers were observed for these mutants. The remaining Cys mutants twitched similarly to the wild type.
Lack of Piliation Is Caused by Inefficient Assembly
Any pili assembled in strains lacking the retraction ATPase, PilT, become trapped on the cell surface (46). Loss of pilus retraction in alignment subcomplex mutants was shown previously (14) to allow for capture of a few pili on the surface of the cells. Because three of the double Cys mutants had reductions in the levels of surface pili (PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C), we hypothesized that they would assemble more pili in a retraction-deficient background, similar to pilN or pilO mutants. We disrupted pilT in these backgrounds and tested the resulting levels of surface piliation. As predicted, all mutants assembled more pili when retraction was disabled (Fig. 4C). This result suggests that all mutants are capable of extending pili but that assembly in those backgrounds is inefficient, especially in the PilOY154C/I178C mutant, resulting in net pilus retraction when PilT is active.
Specific Single Cys Mutations Affect Function of the T4P System
Because moderate reductions in twitching and piliation were observed in two PilO double Cys mutant strains that share a common substitution (PilOS166C/D175C and PilOV161C/D175C), we next investigated whether individual Cys substitutions disrupted T4P production and/or function. The stability of alignment subcomplex components in strains with single Cys substitutions was tested using specific antibodies, and the levels of PilMNOP were found to be comparable with wild type (Fig. 5A). Each single Cys mutant produced pili at levels similar to wild type, with the exception of PilNR142C that had a modest reduction in piliation (Fig. 5B). Interestingly, neither PilNR142C nor PilOD175C could twitch (Fig. 5C), even though both produced pili; this phenotype is a hallmark of retraction defects. When the retraction ATPase PilT was deleted from PilNR142C and PilOD175C, their piliation levels were similar to the pilT control, showing they are capable of wild-type pilus assembly (Fig. 5D). Motility, and therefore pilus retraction, of all other single Cys mutants was unaffected.
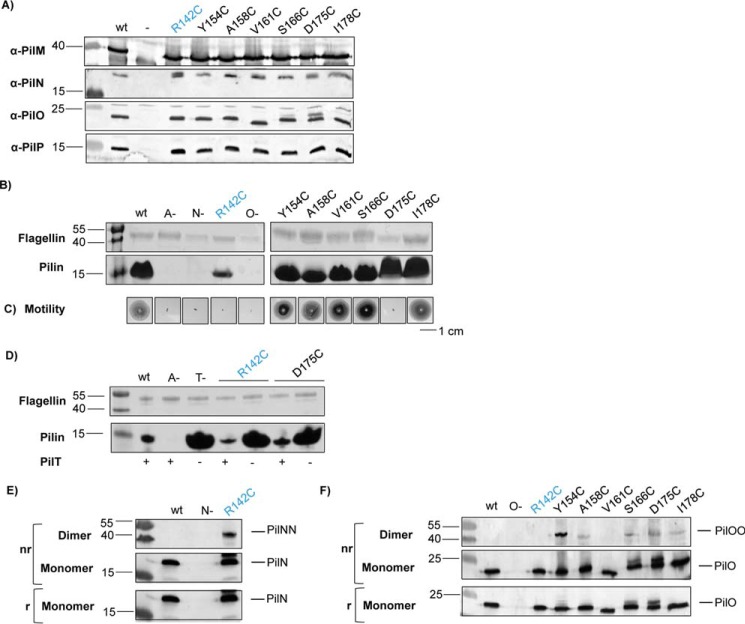
FIGURE 5.
PilNR142C and PilOD175C single Cys substitutions cause T4P dysfunction in wild-type and retraction-deficient backgrounds. Lysates for the wild type (wt), negative controls (−), and the Cys mutants were subject to SDS-PAGE and then blotted with specific antibodies to each alignment subcomplex component (α-PilM, α-PilN, α-PilO, and α-PilP) to confirm stability of each protein in the Cys mutants (A). Each mutant along with wild type, non-piliated (A−), hyperpiliated (T−), and negative controls (N- and O-) were tested for sheared surface pili (B), and twitching motility (C). PilNR142C had less pili and motility than WT, whereas PilOD175C had similar level of pili but no motility. D, PilNR142C and PilOD175C were introduced into a retraction-deficient background, which revealed that both Cys mutants can assemble more pili without the retraction ATPase, PilT. PilT presence (+) or absence (−) in each strain is indicated below. Whole cell lysates were then subjected to SDS-PAGE under non-reducing (nr) and reducing (r) conditions and then blotted with specific antibodies to PilN (E), and PilO (F). All single Cys mutants formed homodimers to varying extents. Images are representative of at least three independent experiments.
Single Cys Substitutions Allow for in Vivo Homodimer Formation, but Only a Subset Has Functional Consequences
To better understand why PilNR142C and PilOD175C single mutants could extend pili but not twitch, we hypothesized that these substitutions might trap PilN or PilO in specific homodimer configurations, impairing function. Whole cell lysates of all single Cys mutants were analyzed in reducing and non-reducing conditions followed by immunoblotting with PilN and PilO antibodies. We were surprised to see that all single Cys mutants formed covalent homodimers in vivo to varying degrees, with PilNR142C and PilOY154C forming the most, and fewer dimers captured for other Cys mutants such as PilOD175C and PilOV161C, where dimer formation is harder to detect (Fig. 5, E and F). These data suggest that with specific exceptions (PilNR142C and PilOD175C), covalent in vivo homodimerization of PilN and PilO can occur without functional consequences.
Heterodimer Formation of PilNR142C and PilOD175C Is Abolished in a pilM Background
As proposed for the T2S system (34), partner switching, or reorientation of PilN and PilO interfaces, might explain how PilN and PilO can form both homo- and heterodimers in vivo. We hypothesized that PilN might respond to conformational changes in its cytoplasmic partner PilM, repositioning it relative to PilO. To test whether formation of covalent dimers requires PilM, the pilM gene was deleted in each of the PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C strains. The absence of PilM was confirmed by Western blot (Fig. 6A), and whole cell lysates of Cys mutant strains lacking PilM were analyzed in reducing and non-reducing conditions, followed by immunoblot analysis with PilN or PilO antibodies. Although both PilOA158C/I178Cand PilOY154C/I178C formed covalent homodimers in the absence of PilM, PilNR142C/PilOD175C no longer formed covalent heterodimers (Fig. 6, B and C).
FIGURE 6.
Heterodimer formation is abolished in pilM or pilP backgrounds. Lysates of PilN and PilO Cys mutants, wild type (wt), PilM deletion (ΔM), PilP::FRT (P-), and the negative controls (N- and O-) were resolved using SDS-PAGE under non-reducing (nr) and reducing (r) conditions and subjected to Western blot detection using specific antibodies to PilM, PilP, and PilQ to show the loss of the proteins (A), PilN (B, D, and F), and PilO (C, E, and G). Images are representative of at least three independent experiments. In the absence of PilM or PilP, Cys cross-linked homodimers (PilOA158C/I178C and PilOY154C/I178C) still form, whereas the PilNR142C/PilOD175C heterodimer cannot. The absence of PilQ does not alter the dimerization profile of the PilNR142C/PilOD175C Cys pair.
Heterodimer Formation by PilNR142C and PilOD175C Is Also Abolished in a pilP Background
To determine whether the remaining member of the alignment subcomplex, PilP, had effects on PilNO dimer formation, we created a pilP strain expressing the Cys pairs of interest (PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C), confirmed loss of PilP by Western blot (Fig. 6A), and analyzed whole cell lysates as described above. The PilNR142C/PilOD175C strain failed to form covalent PilNO heterodimers in the absence of PilP; however, PilN and PilO formed homodimers (Fig. 6, D and E), as in the pilM mutant. To investigate whether the absence of PilP alone affected the PilNO interface, or whether the change in dimerization state was due to the lack of PilNO tethering via PilP to the N0 domain of PilQ (19), we created a pilQ strain expressing the PilNR142C/PilOD175C Cys pair, confirmed loss of PilQ by Western blot (Fig. 6A), and tested lysates as described above. Absence of PilQ had no effect on homo- or heterodimer formation (Fig. 6, F and G), suggesting that PilP specifically contributes to the formation of an interface conducive to the formation of a covalent bond between PilNR142C/PilOD175C.
Other T4P Components Do Not Modulate the PilNO Interface
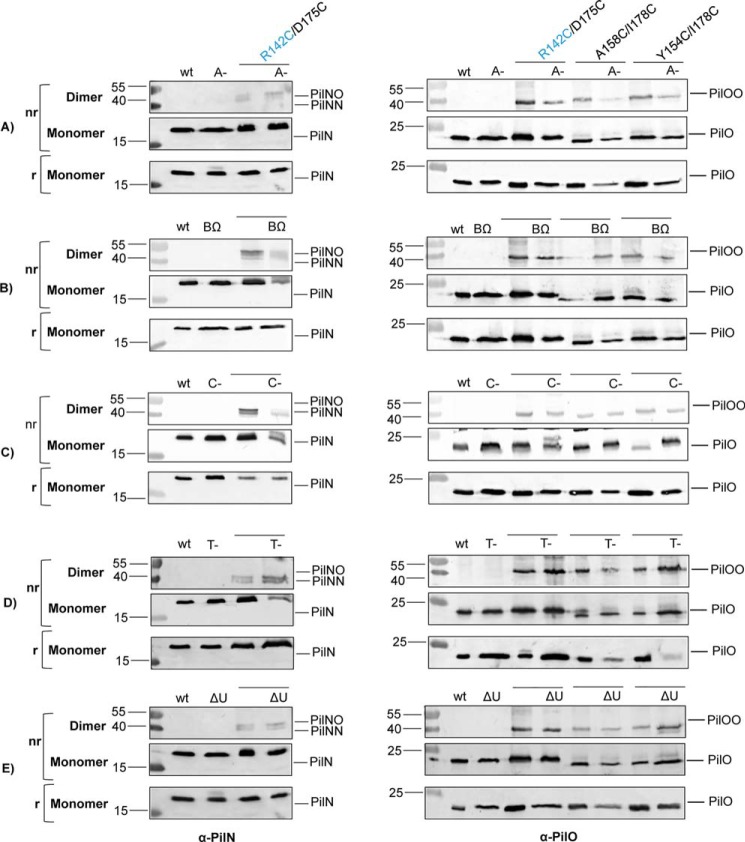
Recently, interactions between PilM and motor ATPases of the T4P system were identified for both P. aeruginosa and M. xanthus (40, 41). Because PilM was important for formation of covalent PilNR142C/PilOD175C heterodimers, we hypothesized that conformational changes in PilM, driven by its interaction with other cytoplasmic components of the T4P motor while bound to the N terminus of PilN, might be propagated to and affect the orientation of periplasmic PilNO interfaces. Three AAA+-ATPases modulate extension (PilB) and retraction (PilTU) of the pilus fiber (5, 47, 48). We created pilB, pilT, and pilU mutants containing the Cys pairs of interest (PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C) and confirmed the absence of each ATPase by Western blotting (Fig. 7). Unexpectedly, all strains formed covalent homo- and heterodimers (Fig. 8, B, D, and E), suggesting that the ATPases do not influence the PilNO interface.
FIGURE 7.
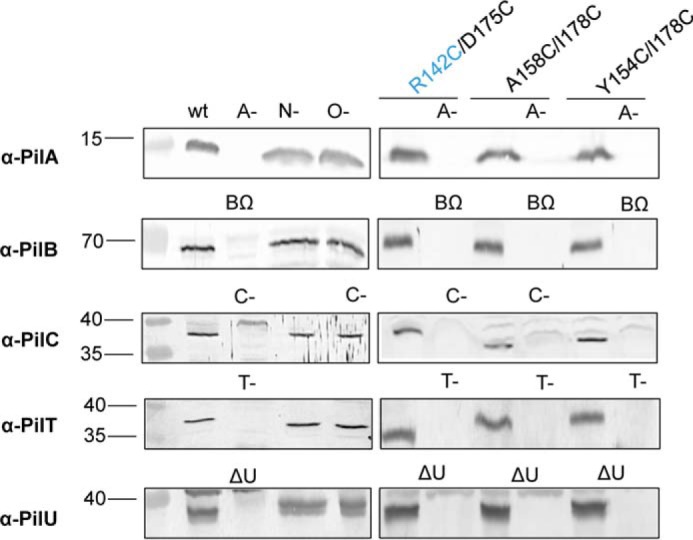
Disruption of pilA, pilB, pilC, pilT, and pilU in PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C backgrounds. Lysates of wild type (wt), negative control strains (A-, BΩ, C-, N-, O-, T-, and ΔU), and Cys mutants were tested for expression and stability of each protein using protein-specific antisera. Minus signs indicate the gene was disrupted via an FRT insertion; Δ indicates the gene was produced by a deletion of the gene; and Ω indicates the gene was disrupted by an Ω cassette insertion.
FIGURE 8.
Dimerization of PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C is unaffected by loss of non-alignment subcomplex components. Wild type (wt), negative controls (A-, BΩ, C-, N-, O-, T-, and ΔU) and Cys mutant lysates were tested for the ability of PilN and PilO to form homo- or heterodimers on non-reducing SDS-PAGE in various T4P mutant backgrounds: (A) pilA, (B) pilB, (C) pilC, (D) pilT, and (E) pilU. PilN (left) and PilO (right) were blotted for using protein-specific antisera. Minus signs indicate the gene was disrupted via an FRT insertion; Δ indicates the gene was produced by a deletion of the gene, and Ω indicates the gene was disrupted by an Ω cassette insertion. Images represent at least three independent experiments.
We further investigated whether other T4P components associated with PilM, PilN, or PilO, including PilA, the major component of the pilus fiber and PilC, the IM platform protein, influenced the dimerization state of PilN and PilO. Strains with disruptions in pilA or pilC, plus the above Cys substitutions (Fig. 7), formed covalent homo- and heterodimers similar to the wild type (Fig. 8, A and C). These data suggest that only interactions among members of the alignment subcomplex modulate the interface between PilN and PilO.
Discussion
The PilMNOP alignment subcomplex is highly conserved among T4aP-expressing bacteria (5). Previous studies demonstrated that PilN and PilO form stable heterodimers in a 1:1 stoichiometry (18); however, the precise interface(s) between PilN and PilO has not yet been elucidated. We used disulfide-bonding analysis to study the interactions between these components and to assess the functional consequences of covalently cross-linking PilN and/or PilO. Our results revealed that PilN and PilO form both homo- and heterodimers in vivo under physiological conditions and that conformational flexibility of these states is necessary for T4P function.
PilN and PilO May Form Functional Dimers of Dimers in Vivo
Studies in the T2S system showed that PilN- and PilO-like proteins (GspL and GspM, respectively) form homo- and heterodimers, possibly via a process called “partner switching,” implying large rotations in the TMS and core interfaces (34). Here, we captured PilO homodimers, previously observed only upon overexpression in trans (17, 18), when expressed at native levels and stoichiometry from the Pseudomonas chromosome, confirming that the PilO homodimer interface revealed by structural studies is physiological (Fig. 2C). Unexpectedly, while trying to understand whether PilNO heterodimers share the same interface as PilO homodimers, we identified multiple species on an anti-PilN Western blot (Fig. 2B), with masses corresponding to PilNO heterodimers and PilN homodimers. Although a PilN homodimer (bound to PilM) from the T4P system of T. thermophilus was observed in vitro (29), to our knowledge this is the first report of PilN and PilO homodimers in their native context.
When Cys-substituted soluble periplasmic fragments of PilN or PilO (PilNR142C and PilOD175C, Fig. 3, A and B) were expressed in E. coli, they formed covalent homodimers. Co-expression of PilOD175C with PilNR142C led to capture of three subpopulations: PilNR142C homodimers, PilOD175C homodimers, and PilNR142C/PilOD175C heterodimers (Fig. 3, A and B), suggesting that there are (at least) two possible interfaces for each protein. Interestingly, a large amount of PilO monomers remained in the non-reducing lanes, indicating that a substantial proportion of PilO was either present as a monomer or participating in non-covalent dimers, possibly as a result of non-permissive positioning of a PilO interaction interface for disulfide bond formation.
P. aeruginosa PilN expressed in vitro without PilO is mostly insoluble (20), which has precluded studies of PilN alone. We hypothesized that formation of covalent PilN homodimers might stabilize it in the absence of PilO, and we attempted to purify PilNΔ44_HisR142C under non-reducing conditions, but the protein remained mostly insoluble. However, untagged PilNR142C homodimers co-purified with tagged PilO, and the molecular mass of the complex was ∼75 kDa (Fig. 3C), likely corresponding to a 2:2 PilNΔ44/PilOΔ51_His complex. This stoichiometry is consistent with our previous data, where PilNOP were shown by gel filtration to elute as either a trimer or a dimer of trimers (20). A dimeric complex of PilMNO (2:2:2) was reported for T. thermophilus (29), fitting with the idea that P. aeruginosa PilN and PilO form a dimer of dimers (17, 19, 20, 29).
Only two of four predicted PilO Cys mutant pairs, and one of three PilNO pairs, formed disulfide bonds, suggesting that the truncated PilOΔ68 structure (Fig. 1A) (18) and the PilNΔ57/PilOΔ68 heterodimer homology model derived from it (Fig. 1B) do not exactly mirror the in vivo conformation of the proteins. However, the ability to cross-link both homo- and heterodimers of PilN and PilO in vivo confirms that select residues are close enough and in the appropriate orientation to allow for disulfide bond formation. The recent cryoelectron tomography study of the M. xanthus T4P system (49), with resolutions averaging 4 nm, now offers the exciting possibility that we could use similar approaches to image the T4P systems of our P. aeruginosa Cys mutants in situ, to visualize how cross-linking of PilNO affects the configuration of the machinery.
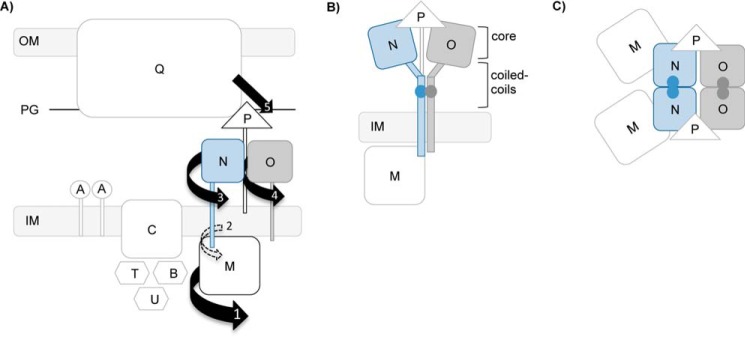
Interestingly, the relative position of the Cys residue pairs able to cross-link PilO homodimers in vivo were at the “top” of the predicted interface in most cases, although they were at the “bottom” for the PilNO heterodimer (Fig. 2D), suggesting that the homo- and heterodimer interfaces might be angled differently. Models of the core interaction interface between PilN and PilO in T. thermophilus (29) and M. xanthus (49) show them as tilted, possibly because of longer predicted coiled coils in PilO (18). The persistence of PilN and PilO monomers (Figs. 2, B and C, and 3, A and B) not involved in covalent interactions suggests the co-existence of multiple PilN and PilO interaction species. This scenario was reported for type II secretion system PilP- and PilQ-like proteins (GspC and GspD, respectively), where contacts were established at three distinct interfaces in vivo (50). These observations, combined with our results, suggest that homo- and heterodimers may occur simultaneously. Based on these new data, we propose that the oppositely charged coiled coils of PilNO (18) could serve as the primary interaction interface between heterodimers (Fig. 9B), whereas the core regions could mediate homodimer interactions (Fig. 9C). This revised model better explains our previous findings, where up to six combined point mutations along the PilN core (now proposed to be the homodimer interface) failed to disrupt PilNO heterodimers in a bacterial two-hybrid assay (42). Furthermore, this new model could explain why PilNR142C and PilOD75C residues at the bottom of the original model (Fig. 1) adjacent to the coiled coils participate in both homo- and heterodimers (Figs. 2, B and C, and 3, A and B). A tilted PilNO core interface could position these residues in close enough proximity to form a disulfide bond with either partner. Importantly, our data do not support the proposal of large rotations in the core-core interfaces, as suggested previously for the T2S system (34).
FIGURE 9.
Model of the interaction of PilN and PilO with other T4P components. A, schematic of the T4P system in P. aeruginosa with the Pil nomenclature. Major cell compartments are labeled as follows: inner membrane (IM), peptidoglycan (PG), and outer membrane (OM). Specific T4P components are highlighted as follows: PilQ, the secretin; PilC, the IM platform protein; PilBTU, the three cytoplasmic ATPases; PilA, the major pilin subunit; and PilMNOP, the alignment subcomplex components. Numbered arrows indicate order of hypothesized movement generated from the cytoplasm outwards. B, T4P system schematic model showing PilN and PilO heterodimer formation mediated through the coiled coils. C, top-down view of PilN and PilO homodimers mediated through their core domains. Domain movement in PilM could be transduced through PilN and PilO, ultimately leading to a movement of PilP, creating a signal transduction network between the inner and outer T4P components.
PilN and PilO Homodimers May Represent a Functional Rather Than Assembly Intermediate State of the T4P System
Our data show that covalent PilN or PilO homodimers are not necessarily detrimental to function. With the exception of PilNR142C and PilOD175C, both of which assembled pili but had retraction defects that abrogated motility, none of the single Cys mutants had obvious piliation or motility defects (Fig. 5, B and C), despite their capacity to form covalent homodimers in vivo (Fig. 5, E and F). The relative abundance of covalent homodimers formed by each mutant did not correlate with T4P dysfunction, as PilOY154C formed the most dimers, but without functional consequences. This result suggests that cross-linking of only specific residues affects function and supports the idea that PilN and PilO homodimers represent a normal functional state of the T4P system.
Three covalent pairs (PilNR142C/PilOD175C, PilOA158C/I178C, and PilOY154C/I178C) caused significant loss of surface piliation and complete loss of motility (Fig. 4, A and B). This result suggests that some configurations of PilNO can affect pilus function. In a retraction-deficient background, where assembled pili are trapped on the surface (46), all three double Cys mutants had more pili (Fig. 4C), confirming that covalently linked dimers permit pilus assembly. Thus, lack of motility in these strains more likely arises from extension-retraction imbalances. We showed previously (42) that substitution of specific residues at the coiled-coil interface of PilN and PilO also confers an assembly-competent yet motility-deficient phenotype. Together, these data emphasize that PilNO contribute to both pilus extension and retraction and that altering their interfaces could affect the ability of the T4P machinery to switch between those states.
Previous work in T. thermophilus suggested that a dimeric complex of PilMN (two PilMs and two PilNs) forms an intermediate transmembrane platform onto which other alignment subcomplex components dock (29). The addition of PilO to the PilMN subassembly was proposed to dissociate PilN homodimers, sequestering each monomer into a PilNO heterodimer. Our data support an alternative model, in which PilN and PilO might homodimerize via their core domains (Fig. 9C), and simultaneously heterodimerize via their coiled coils and potentially their TMS (Fig. 9B) (42). This scenario could explain how untagged covalent PilN homodimers were co-purified with tagged PilO homodimers, as both constructs retain their coiled coils (Fig. 3). Alternatively, tetrameric arrangement of PilN and PilO homodimers could suggest an alternative PilNO interface on the “side” of the core homodimer interface (Fig. 9B); however, we have no evidence to support that idea. The PilNR142C and PilOD175C residues are located near the base of the modeled core interfaces and could be oriented more laterally (Fig. 9B) than depicted by our static models (Fig. 1), explaining why these particular residues were the only ones that successfully formed cross-linked homo- and heterodimers.
Modulation of the PilNO Core Interface by the Other Alignment Subcomplex Components
Cross-linking of Cys residues at select locations within the PilNO equivalents (GspLM) in the T2S system suggested rotation of the proteins through their TMS and periplasmic cores (34). In the T2S system, PilM orthologs interact with the single cytoplasmic motor ATPase (51–54), and more recently, interactions between PilM and the T4P motor ATPases in P. aeruginosa and M. xanthus were confirmed (40, 41). Thus, we tested whether interactions with the cytoplasmic component of the alignment subcomplex, PilM, which is bound to the N terminus of PilN, modulated PilNO interfaces. PilNR142C/PilOD175C heterodimers, but not homodimers, were lost in the absence of PilM (Fig. 6, B and C), suggesting that the PilNO interface no longer adopts an orientation permissive for disulfide bond formation. New crystal structures of a P. aeruginosa PilM homodimer and of monomeric PilM fused to part of its binding partner, PilN(1–12), revealed that in the absence of PilN, the seven N-terminal residues of PilM (PilM(1–7)) bind in the PilN binding cleft of a second PilM monomer (41). Thus, PilM dimerizes in the absence of PilN. In this scenario, the lack of PilM-PilN interaction may favor PilN and PilO homodimer formation, whereas when PilM becomes monomeric upon binding PilN's N terminus, a corresponding shift in the position of the PilNO interfaces could induce heterodimer formation. PilM's interactions with its cytoplasmic partners, PilB, PilT, and/or PilC (41), differ depending on whether it is bound to PilN, potentially revealing a mechanism for switching between active and resting systems or between pilus assembly and disassembly. These differential interactions might similarly modulate PilNO homo- versus heterodimer states in the periplasm. Disruption of pilB, pilT, or pilU had no effect on PilN and PilO dimerization (Fig. 8, B, D, and E). We considered that in the absence of one ATPase (e.g. PilB), one of the other two (PilT or PilU) may interact with PilM, stabilizing a configuration permissive for the formation of PilNO homo- and heterodimers. However, removal of PilC, the proposed interaction partner for all three ATPases, had no effect on dimerization states (Fig. 8C).
The major pilin interacts with PilNOPQ in P. aeruginosa (19), but its specific interaction partner(s) are unknown. Transmission electron microscopy studies of T. thermophilus revealed a stable complex composed of soluble fragments of PilMNO-PilA (29) with PilN interacting directly with PilA. One could envision that PilN and PilO might help extract PilA from the IM during assembly, priming the base of the pilus fiber for incorporation of the next PilA subunit. However, loss of pilA had no effect on PilNO dimerization states (Fig. 8A).
In M. xanthus, PilQ recruits PilP, which binds PilN and PilO, and PilN binds PilM, creating a transenvelope complex (55). In P. aeruginosa, a similar pathway has been proposed, where PilP binds PilQ as well as PilNO but does not bind PilO homodimers (20). Here, loss of PilP prevented formation of PilNR142C/PilOD175C heterodimers (Fig. 6, D and E), although loss of PilQ had no effect (Fig. 6, F and G). The core regions of PilN or PilO may homodimerize (Fig. 9C), and the large electrostatic difference predicted for the coiled coils of PilN and PilO (18) could promote heterodimerization (Fig. 9B). The unstructured N terminus of PilP could interact with the core or coiled-coil regions of PilNO heterodimers, stabilizing the interface in an orientation that allowed for disulfide bond formation.
PilN and PilO Rearrangement during “Active” and “Resting” States of the T4P System
Elegant cryoelectron tomography studies of the M. xanthus T4P system allowed for derivation of a “pseudo-atomic” model that implied differences between active and resting states of the machinery (49). The active state occurs during assembly or disassembly of the fiber, when a PilB or PilT ATPase is engaged at the base of the system, whereas the resting state was ATPase-free. PilM was proposed to form a ring surrounding a dimer of PilC, and expansion of the ring upon binding of PilB or PilT to PilC during the active state, accompanied by rearrangement of the density associated with PilN and PilO's coiled coils, was observed (49). Without PilM, density corresponding to PilC and the ATPases was missing (49). In our work, only PilM or PilP could modulate the PilNO core interface. We speculate that subtle reorientation of PilN and PilO may occur during the transition between resting and active or “extension” versus “retraction” states of the T4P system, consistent with the hypothesis that PilM may disengage from PilN (41). Although a ratcheting-type mechanism, in which PilNO conformational dynamics might contribute to the stepwise addition or removal of pilin subunits cannot yet be ruled out, our data do not support this idea.
Experimental Procedures
Strains, Media, and Growth Conditions
Bacterial strains used in this study are listed in Table 1. E. coli and P. aeruginosa were grown at 37 °C in Luria-Bertani (LB) media supplemented with antibiotics at the following final concentrations when necessary (μg·ml−1): ampicillin, 100; carbapenem (Cb), 200; kanamycin, 50; gentamicin (Gm), 15 for E. coli and 30 for P. aeruginosa, unless otherwise specified. Bacterial strains were stored at −80 °C in LB with 20% glycerol (v/v). Plasmids were transformed by heat shock into chemically competent cells. All constructs were verified by DNA sequencing (MOBIX, McMaster University).
TABLE 1.
Bacterial strains
| Strain | Description | Source/Ref. |
|---|---|---|
| E. coli strains | ||
| DH5α | F−, ϕ80lacZ, M15,Δ (lacZYA-argF), U169, recA1, endA1, hsdR17 (rk−,mk+), phoAsupE44, thi-1, gyrA96, relA1, λ- | Invitrogen |
| Origami2 (DE3)pLysS | Δ(ara-leu)7697 ΔlacX74 ΔphoA PvuII phoR araD139 ahpC galE galK rpsL F`[lac+ lacIq pro] (DE3) gor522::Tn10 trxB pLysS (CamR, StrR, TetR) | Novagen |
| SM10 | thi-1, thr, leu, tonA, lacy, supE, recA, RP4-2-Tcr::Mu, Kmr; mobilizes plasmids into P. aeruginosa via conjugation | 62 |
| P. aeruginosa strains | ||
| PAK | Wild type | J. Boyd |
| ΔpilM | Deletion of pilM | 17 |
| pilN::FRT | FRT scar at position 124 within pilN | 17 |
| pilO::FRT | FRT scar at position 328 within pilO | 17 |
| pilP::FRT | FRT scar at position 86 within pilP | 17 |
| pilQ::FRT | FRT scar at position at 571 within pilQ | 60 |
| pilT::FRT | FRT scar at position 540 within pilT | 47 |
| pilA::FRT | FRT scar at SphI site within pilA | 59 |
| pilBΩ | pilB with Ω cassette insertion | 61 |
| ΔpilU | Deletion of pilU | Dr. M. Wolfgang |
| pilC::FRT | FRT scar at position 365 within pilC | 14 |
| PilNR142C/PilOD175C | pilN R142C and pilO D175C | This study |
| PilNE157C/PilOA158C | pilN E157C and pilO A158C | This study |
| PilNE132C/PilOI178C | pilN E132C and pilO I178C | This study |
| PilNR142C/PilOA158C | pilN R142C and pilO A158C | This study |
| PilOS166C/D175C | pilO S166C, D175C | This study |
| PilOV161C/D175C | pilO V161C, D175C | This study |
| PilOA158C/I178C | pilO A158C, I178C | This study |
| PilOY154C/I178C | pilO Y154C, I178C | This study |
| PilNR142C | pilN R142C | This study |
| PilOY154C | pilO Y154C | This study |
| PilOA158C | pilO A158C | This study |
| PilOV161C | pilO V161C | This study |
| PilOS166C | pilO S166C | This study |
| PilOD175C | pilO D175C | This study |
| PilOI178C | pilO I178C | This study |
| PilNR142C/PilOD175C ΔpilM | pilN R142C and pilO D175C with deletion of pilM | This study |
| PilOA158C/I178C ΔpilM | pilO A158C, I178C with deletion of pilM | This study |
| PilOY154C/I178C ΔpilM | pilO V154C, I178C with deletion of pilM | This study |
| PilNR142C/PilOD175C pilA::FRT | pilN R142C and pilO D175C with FRT insertion at SphI site within pilA | This study |
| PilOA158C/I178C pilA::FRT | pilO A158C, I178C with FRT insertion at SphI site within pilA | This study |
| PilOY154C/I178C pilA::FRT | pilO Y154C, I178C with FRT insertion at SphI site within pilA | This study |
| PilNR142C/PilOD175C pilBΩ | pilN R142C and pilO D175C with Ω cassette insertion pilB | This study |
| PilOA158C/I178C pilBΩ | pilO A158C, I178C with Ω cassette insertion pilB | This study |
| PilOY154C/I178C pilBΩ | pilO V154C, I178C with Ω cassette insertion pilB | This study |
| PilNR142C/PilOD175C pilC::FRT | pilN R142C and pilO D175C with FRT insertion at position 365 within pilC | This study |
| PilOA158C/I178C pilC::FRT | pilO A158C, I178C with FRT insertion at position 365 within pilC | This study |
| PilOY154C/I178C pilC::FRT | pilO V154C, I178C with FRT insertion at position 365 within pilC | This study |
| PilNR142C/PilOD175C pilP::FRT | pilN R142C and pilO D175C with FRT insertion at position 86 within pilP | This study |
| PilOA158C/I178C pilP::FRT | pilO A158C, I178C with FRT insertion at position 86 within pilP | This study |
| PilOY154C/I178C pilP::FRT | pilO V154C, I178C with FRT insertion at position 86 within pilP | This study |
| PilNR142C/PilOD175C pilQ::FRT | pilN R142C and pilO D175C with FRT insertion at position 571 within pilQ | This study |
| PilNR142C/PilOD175C pilT::FRT | pilN R142C and pilO D175C with FRT insertion at position 540 within pilT | This study |
| PilOA158C/I178C pilT::FRT | pilO A158C, D178C with FRT insertion at position 540 within pilT | This study |
| PilOY154C/I178C pilT::FRT | pilO V154C, D178C with FRT insertion at position 540 within pilT | This study |
| PilNR142C/PilOD175C ΔpilU | pilN R142C and pilO D175C with deletion of pilU | This study |
| PilOA158C/I178C ΔpilU | pilO A158C, I178C with deletion of pilU | This study |
| PilOY154C/I178C ΔpilU | pilO Y154C, I178C with deletion of pilU | This study |
Generation of Cys Mutants
Sets of double Cys mutations in PilO, as well as single Cys mutations in PilN and PilO, were chosen with reference to the PilOΔ68 homodimer crystal structure Protein Data Bank code 2RJZ (18) and a PilNΔ57/PilOΔ68 heterodimer homology model, created by substituting the Phyre2 generated PilNΔ57 monomer in place of a PilO subunit in the PilO homodimer structure (18, 45). Distances were measured in PyMOL, and residues were selected to ensure that the distance between Cβ atoms was less than 8 Å (Fig. 1). Codons for selected Cys substitutions were introduced into either a pEX18Gm::pilMNOP construct (for PilNO Cys pairs) or a pEX18Gm::pilNOP construct (for double PilO Cys mutants) using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. Mutations were verified by DNA sequencing (MOBIX).
Generation of pilN and pilO Cys Mutations onto the Chromosome of P. aeruginosa PAK
Cys-encoding chromosomal mutants of P. aeruginosa were generated by biparental mating with E. coli SM10 using a Flp-FRT (FLP recombination target) system as described previously (18). Constructs containing the desired Cys substitutions, pEX18Gm::pilMNOP for PilNO Cys mutants and pEX18Gm::pilNOP for PilO Cys mutants, were introduced into E. coli SM10 cells for biparental mating with PAK pilN::FRT and PAK pilO::FRT, respectively (12). The E. coli SM10 donor was counter-selected by plating on Pseudomonas isolation agar (PIA; Difco) containing Gm (100 μg·ml−1). Gm-resistant P. aeruginosa isolates were streaked on LB-no salt sucrose plates (1% w/v bacto-tryptone, 0.5% w/v bacto-yeast extract, 5% w/v sucrose) and then incubated for 16 h at 30 °C. Select colonies were restreaked in parallel on LB and LB plates supplemented with Gm. Gm-sensitive colonies were screened by PCR using pilN or pilO primer pairs (Table 2) to confirm replacement of the original FRT-disrupted gene, and PCR products of the expected size were DNA-sequenced (MOBIX) to confirm incorporation of the desired mutations.
TABLE 2.
Oligonucleotide primer sequences
Fwd means forward, and Rev means reverse.
| Primer name | Oligonucleotide Sequence (5`–3`) |
|---|---|
| PilA Fwd | TATATAACTCTAGAGATGAAAGCTCAAAAAGGCTTT |
| PilA Rev | TATTATAAGAATTCGTTACTTAGAGCAACC |
| PilB Fwd | TATATAACTCTAGAGATGAACGACAGCAT |
| PilB Rev | TATTATAAGAATTCGTTAATCCTTGGTCAC |
| PilC Fwd | TATATAACTCTAGAGATGGCGGACAAAGCGTTAAAAACCAG |
| PilC Rev | TATATATGGAATTCGTTATCCGACGACGTTGCCGAG |
| PilM Fwd | TATATATATCTAGAGGTGCTAGGGCTCATAAAGAAGAAAG |
| PilM Rev | TATTATAAGAATTCGTCAGTCGAAACTCCTCAACGCC |
| PilN Fwd | TATATAACTCTAGAGATGGCACGGATCAACCTTCTACCCTGG |
| PilN Rev | TATATATGGAATTCGTCATTTCTTGGCTCCTTGCGCAACCCC |
| PilO Fwd | TATATATATCTAGAGATGAGTCTGGCCAGTTCCCTG |
| PilO Rev | TATATATAGAATTCGTCATTTCTTCAGCCCCTTGTCG |
| PilP Fwd | TATATATGGAATTCGATGAGAGCCCGC |
| PilP Rev | TATATATAGAATTCGTCAGGAGCGTTCCTTGAGAGTC |
| PilT Fwd | TATATAACTCTAGAGATGGATATTACCGAGCTG |
| PilT Rev | TATATATAGAATTCGTCAGAAGTTTTCCGG |
| PilU Fwd | TATATAACTCTAGAGATGGAATTCGAAAAGCTG |
| PilU Rev | TATATATAGAATTCGTCAGCGGAAGCGCCG |
| PilN E132C Fwd | CGCCGGCGCGGCCTGCTCCAACAACCGCG |
| PilN E132C Rev | CGCGGTTGTTGGAGCAGGCCGCGCCGGCG |
| PilN R142C Fwd | CGTTTCCAATCTCATGTGCAACATGGACGCGTC |
| PilN R142C Rev | GACGCGTCCATGTTGCACATGAGATTGGAAACG |
| PilN E157C Fwd | GCCCCGACCCTGAACTGCGTCAAGGCGGTGACC |
| PilN E157C Rev | GGTCACCGCCTTGACGCAGTTCAGGGTCGGGGC |
| PilO Y154C Fwd | GTGGTCGGCGGCTGCCACGACTTGG |
| PilO Y154C Rev | CCAAGTCGTGGCAGCCGCCGACCAC |
| PilO A158C Fwd | GGCTACCACGACTTGTGCACCTTCGTCAGCGGC |
| PilO A158C Rev | GCCGCTGACGAAGGTGCACAAGTCGTGGTAGCC |
| PilO V161C Fwd | GACTTGGCGACCTTCTGCAGCGGCGTGTCCAG |
| PilO V161C Rev | CTGGACACGCCGCTGCAGAAGGTCGCCAAGTC |
| PilO S166C Fwd | GCGGCGTGTCCTGCCTGCCGCGG |
| PilO S166C Rev | CCGCGGCAGGCAGGACACGCCGC |
| PilO D175C Fwd | GGATCGTCACCCTGCATTGCTTCGAGATCAAGCCGG |
| PilO D175C Rev | CCGGCTTGATCTCGAAGCAATGCAGGGTGACGATCC |
| PilO I178C Fwd | CTGCATGACTTCGAGTGCAAGCCGGTCGCGCC |
| PilO I178C Rev | GGCGCGACCGGCTTGCACTCGAAGTCATGCAG |
Preparation of Whole Cell Lysates
P. aeruginosa strains were grown on LB agar plates overnight at 37 °C. Cells were scraped from the surface and resuspended in 1× PBS (pH 7.4) to an A600 of 0.6. A 1-ml aliquot of cells was collected by centrifugation at 2292 × g for 3 min in a microcentrifuge. The cell pellet was resuspended in 100 μl of either 1× SDS-PAGE loading buffer (125 mm Tris, pH 6.8, 2% (v/v) β-ME, 20% (v/v) glycerol, 4% (w/v) SDS, and 0.001% (w/v) bromphenol blue) or 1× non-reducing SDS-PAGE loading buffer (no β-ME) and subjected to Western blotting analysis. Biological and technical replicates of all strains were performed n ≥ 3.
Western Blotting Analysis
Samples were separated on 15% SDS-polyacrylamide gels for 1.5 h at 160 V and transferred to nitrocellulose membranes for 1 h at 225 mA. Membranes were blocked using a 5% (w/v) low fat skim milk powder in 1× PBS for 1 h at room temperature on a shaking platform, followed by incubation with protein-specific antibodies at the following dilutions: α-PilM, α-PilN, α-PilO, and α-PilP at dilutions of 1:1000 (17); α-PilB, α-PilT, and α-PilU at 1:2000, 1:500, and 1:5000, respectively (56); α-PilA at 1:5000 (57), and α-PilC at 1:1000 (14). Blots were incubated in primary antibody ∼16 h at 4 °C on a rocking platform. The membranes were washed twice in 1× PBS for 5 min and then incubated in goat anti-rabbit IgG-alkaline phosphatase-conjugated secondary antibody (Bio-Rad) at a dilution of 1:3000 for 1 h at room temperature. The membranes were washed twice in 1× PBS for 5 min and visualized with alkaline phosphatase developing reagent (Bio-Rad) following the manufacturer's protocol. Western blottings were performed n ≥ 3.
Expression and Purification of PilNΔ44 and PilOΔ51
N-terminally truncated versions of PilN and PilO were previously cloned into the EcoRI/HindIII and NdeI/XhoI cloning sites, respectively, of a pET28a vector, creating a vector that overexpresses untagged PilNΔ44 and C-terminal His6-tagged PilOΔ51_His (18, 20). Cys mutations were introduced into this construct using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions to create pET28a::pilNΔ44/pilOΔ51_His containing pilNΔ44R142C, pilOΔ51D175C, or a double pilNΔ44R142C/pilOΔ51D175C construct. Constructs were transformed into E. coli Origami2 (DE3) pLysS cells and plated on LB agar plates supplemented with 50 μg·ml−1 kanamycin. A single colony was used to inoculate 20 ml of LB containing 50 mg·ml−1 kanamycin and incubated overnight at 37 °C with shaking. The overnight culture was used to inoculate 1 liter of fresh LB (1:100 dilution) containing antibiotic, and the cells were grown at 37 °C, with shaking, to an A600 of 0.6. Protein expression was induced by adding isopropyl β-d-1-thiogalactopyranoside (Sigma) to the culture at a final concentration of 1 mm. The cells were allowed to grow for 4 h at 37 °C, prior to being harvested by centrifugation (3993 × g, 15 min, 4 °C) in an Avanti J-26 XPI centrifuge (Beckman Coulter). Bacterial pellets were frozen at −80 °C until further use.
Bacterial pellets were thawed and resuspended in 10 ml nickel A buffer (20 mm Tris-HCl, pH 7.2, 500 mm KCl, 10 mm imidazole, 10% (v/v) glycerol) and then combined in a 50-ml screw cap tube with one complete EDTA-free protease inhibitor tablet (Roche Applied Science). Cells were lysed via sonication on ice, on setting 4, for 2 min with cycles of 10 s on and 10 s off (Sonicator 3000; Misonix). The lysates were centrifuged (11,000 × g, 30 min, 4 °C) in an Avanti J-26 XPI centrifuge (Beckman Coulter) to remove intact cells and other cellular debris. Pelleted material was retained for analysis by SDS-PAGE, and supernatants were filtered through 0.22-μm Acrodisc syringe filter (Pall Corp.). The lysate was purified using nickel-nitrilotriacetic acid affinity chromatography on an AKTA start FPLC (VWR Scientific). Protein lysate was flowed through a 1-ml His-TrapTM FF column (GE Healthcare) pre-equilibrated with buffer and washed with nickel A buffer and then increasing amounts of nickel B buffer (20 mm Tris-HCl, pH 7.2, 500 mm KCl, 300 mm imidazole, 10% (v/v) glycerol) in a linear gradient. The bound protein was eluted from the column in pure nickel B buffer and collected in 1-ml fractions. An aliquot of each fraction was mixed 1:1 with 2× reducing or non-reducing SDS-PAGE loading and electrophoresed on a 15% SDS-polyacrylamide gel. The gel was stained using Coomassie Blue Staining solution (0.1% Coomassie Brilliant Blue R-250, 50% (v/v) methanol, and 10% (v/v) glacial acetic acid) or developed by Western blotting using protein-specific antibodies as described above.
Twitching Motility Assays
Twitching assays were performed as described previously (5, 47, 48, 58). Briefly, single colonies were stab inoculated to the bottom of a 1% LB agar plate. The plates were incubated for 36 h at 37 °C. Postincubation, the agar was carefully removed and the adherent bacteria stained with 1% (w/v) crystal violet dye, followed by washing with tap water to remove unbound dye. Twitching zone areas were measured using ImageJ software (National Institutes of Health). Statistical significance of differences compared with wild type was determined using Student's t test. All experiments were performed in triplicate with at least three independent replicates.
Sheared Surface Protein Preparation
Surface pili and flagella were analyzed as described previously (58) with the following modifications. P. aeruginosa strains of interest were streaked in a grid-like pattern on large LB 1.5% agar plates and incubated at 37 °C for ∼16 h. The cells were scraped from the plates with glass coverslips and resuspended in 4.5 ml of 1× PBS. Centrifugation of the cells was performed in 1.5-ml Eppendorf tubes at 11,688 × g. Cell pellets were resuspended in 150 μl of 1× SDS sample buffer (125 mm Tris, pH 6.8, 2% (v/v) β-ME, 20% (v/v) glycerol, 4% (w/v) SDS, and 0.001% (w/v) bromphenol blue) and visualized on a 15% SDS-polyacrylamide gel by staining with Coomassie Blue staining solution.
In Vivo Disulfide Cross-linking Analysis
Spontaneous disulfide bond formation between PilN and PilO Cys mutants was assessed in bacteria grown on solid media. Whole cell lysates of the various strains resuspended in reducing or non-reducing SDS-PAGE buffer were boiled for 10 min to lyse cells, electrophoresed on a 15% SDS-polyacrylamide gel, and subjected to Western blotting with protein-specific antibodies for PilN and PilO as described previously.
Generation of PilN and PilO Cys Pairs in Various T4P Component Mutant Backgrounds
To test the contributions of other T4P proteins to dimer formation, select Cys mutants were generated in previously generated T4P mutant backgrounds (Table 1). To create strains with Cys substitutions and devoid of PilM, a construct composed of 700 bp up- and downstream of the pilM gene (ΔpilM) was subcloned into pEX18Gm using XbaI/EcoRI. The resulting suicide construct was transformed into E. coli SM10 and transferred to P. aeruginosa PAK strains already containing pilNR142C/pilOD175C, pilOA158C/I178C, or pilOY154C/I178C by biparental mating, and mutants were selected as described above.
To make Cys mutant strains lacking PilA, PilC, PilQ, or PilT, we used previously engineered suicide constructs for pilA::FRT (59), pilC::FRT (14), pilQ::FRT (60), and pilT::FRT (47) mutants. These constructs were introduced into the P. aeruginosa PAK PilNR142C/PilOD175C, PilOA158C/I178C, or PilOY154C/I178C backgrounds, with the exception of pilQ::FRT, which was only introduced into the PilNR142C/PilOD175C background. Briefly, a suicide vector containing the target gene (pilA, pilC, pilQ, or pilT) disrupted by a gentamicin resistance cassette flanked by FRT sites (pEX18Ap pilA::GmFRT, pEX18Ap pilC::GmFRT, pEX18Ap pilQ::GmFRT, or pEX18Ap pilT::GmFRT, respectively) (22) were introduced into chemically competent E. coli SM10 cells. The constructs were transferred to P. aeruginosa PAK strains containing pilNR142C/pilOD175C, pilOA158C/I178C, or pilOY154C/I178Cby biparental mating as described above. After overnight incubation on sucrose plates, select colonies were plated in parallel on LB plates supplemented with either Cb or Gm, and Gm-resistant, Cb-sensitive colonies were selected. The integrated Gm cassette was removed by conjugally transferring the Flp-expressing pFLP2 from E. coli SM10 into P. aeruginosa as described previously (18). E. coli SM10 was counter-selected by plating on Pseudomonas isolation agar (Difco) containing Cb. pFLP2 was cured by streaking Cb-resistant isolates on LB-no salt plates containing 5% (w/v) sucrose and incubating for 16 h at 30 °C. Select colonies were streaked in parallel on LB plates, LB plus Cb, and LB plus Gm plates. Cb- and Gm-sensitive colonies were selected, and the pilA::FRT, pilC::FRT, pilQ::FRT, and pilT::FRT mutations were confirmed by PCR and DNA sequencing using the primers listed in Table 2.
In some cases, we started with various P. aeruginosa PAK mutant backgrounds, pilBΩ (61), pilP::FRT (17), and ΔpilU (gift of Dr. Matthew Wolfgang), and introduced suicide vectors containing the desired PilN and PilO Cys pairs (as described above). After selection for Gm-sensitive colonies, we screened for correct incorporation of PilN and PilO Cys pairs through PCR using pilN or pilO primer pairs (Table 2) and mutations were confirmed by DNA sequencing (MOBIX).
Author Contributions
T. L. L., D. H. Y., L. L. B., and P. L. H. designed the study and developed the methodology. T. L. L. and D. H. Y. created all Cys mutant strain variants. Experiments were performed by T. L. L. T. L. L., L. L. B., and P. L. H. wrote and edited the manuscript.
This work was supported by Operating Grant MOP-93585 from the Canadian Institutes of Health Research (to L. L. B. and P. L. H.). The authors declare that they have no conflicts of interest with the contents of this article.
- T4P
- type IV pili
- OM
- outer membrane
- IM
- inner membrane
- β-ME
- β-mercaptoethanol
- T2S
- type II secretion
- TMS
- transmembrane segment
- FRT
- FLP recombination target
- Cb
- carbenicillin
- Gm
- gentamicin.
References
- 1. Bradley D. E. (1980) A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26, 146–154 [DOI] [PubMed] [Google Scholar]
- 2. Burrows L. L. (2005) Weapons of mass retraction. Mol. Microbiol. 57, 878–888 [DOI] [PubMed] [Google Scholar]
- 3. Burrows L. L. (2012) Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 [DOI] [PubMed] [Google Scholar]
- 4. Leighton T. L., Buensuceso R., Howell P. L., and Burrows L. L. (2015) Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ. Microbiol. 17, 4148–4163 [DOI] [PubMed] [Google Scholar]
- 5. Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 [DOI] [PubMed] [Google Scholar]
- 6. Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 [DOI] [PubMed] [Google Scholar]
- 7. Strom M. S., and Lory S. (1993) Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47, 565–596 [DOI] [PubMed] [Google Scholar]
- 8. Burkhardt J., Vonck J., and Averhoff B. (2011) Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286, 9977–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins R. F., Davidsen L., Derrick J. P., Ford R. C., and Tønjum T. (2001) Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins R. F., Ford R. C., Kitmitto A., Olsen R. O., Tønjum T., and Derrick J. P. (2003) Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 185, 2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koo J., Burrows L. L., and Howell P. L. (2012) Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 328, 1–12 [DOI] [PubMed] [Google Scholar]
- 12. Koo J., Tammam S., Ku S. Y., Sampaleanu L. M., Burrows L. L., and Howell P. L. (2008) PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 190, 6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiang P., Habash M., and Burrows L. L. (2005) Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takhar H. K., Kemp K., Kim M., Howell P. L., and Burrows L. L. (2013) The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288, 9721–9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen Y., Jackson S. G., Aidoo F., Junop M., and Burrows L. L. (2010) Structural characterization of novel Pseudomonas aeruginosa type IV pilins. J. Mol. Biol. 395, 491–503 [DOI] [PubMed] [Google Scholar]
- 16. Nguyen Y., Sugiman-Marangos S., Harvey H., Bell S. D., Charlton C. L., Junop M. S., and Burrows L. L. (2015) Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 290, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayers M., Sampaleanu L. M., Tammam S., Koo J., Harvey H., Howell P. L., and Burrows L. L. (2009) PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 394, 128–142 [DOI] [PubMed] [Google Scholar]
- 18. Sampaleanu L. M., Bonanno J. B., Ayers M., Koo J., Tammam S., Burley S. K., Almo S. C., Burrows L. L., and Howell P. L. (2009) Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. J. Mol. Biol. 394, 143–159 [DOI] [PubMed] [Google Scholar]
- 19. Tammam S., Sampaleanu L. M., Koo J., Manoharan K., Daubaras M., Burrows L. L., and Howell P. L. (2013) PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J. Bacteriol. 195, 2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tammam S., Sampaleanu L. M., Koo J., Sundaram P., Ayers M., Chong P. A., Forman-Kay J. D., Burrows L. L., and Howell P. L. (2011) Characterization of the PilN, PilO and PilP type IVa pilus subcomplex. Mol. Microbiol. 82, 1496–1514 [DOI] [PubMed] [Google Scholar]
- 21. Balasingham S. V., Collins R. F., Assalkhou R., Homberset H., Frye S. A., Derrick J. P., and Tønjum T. (2007) Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J. Bacteriol. 189, 5716–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bose N., Payne S. M., and Taylor R. K. (2002) Type 4 pilus biogenesis and type II-mediated protein secretion by Vibrio cholerae occur independently of the TonB-facilitated proton motive force. J. Bacteriol. 184, 2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ayers M., Howell P. L., and Burrows L. L. (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5, 1203–1218 [DOI] [PubMed] [Google Scholar]
- 24. Douzi B., Filloux A., and Voulhoux R. (2012) On the path to uncover the bacterial type II secretion system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Durand E., Bernadac A., Ball G., Lazdunski A., Sturgis J. N., and Filloux A. (2003) Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185, 2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abendroth J., Bagdasarian M., Sandkvist M., and Hol W. G. (2004) The structure of the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae: an unusual member of the actin-like ATPase superfamily. J. Mol. Biol. 344, 619–633 [DOI] [PubMed] [Google Scholar]
- 27. Abendroth J., Kreger A. C., and Hol W. G. (2009) The dimer formed by the periplasmic domain of EpsL from the type 2 secretion system of Vibrio parahaemolyticus. J. Struct. Biol. 168, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abendroth J., Rice A. E., McLuskey K., Bagdasarian M., and Hol W. G. (2004) The crystal structure of the periplasmic domain of the type II secretion system protein EpsM from Vibrio cholerae: the simplest version of the ferredoxin fold. J. Mol. Biol. 338, 585–596 [DOI] [PubMed] [Google Scholar]
- 29. Karuppiah V., Collins R. F., Thistlethwaite A., Gao Y., and Derrick J. P. (2013) Structure and assembly of an inner membrane platform for initiation of type IV pilus biogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E4638–E4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karuppiah V., and Derrick J. P. (2011) Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J. Biol. Chem. 286, 24434–24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Py B., Loiseau L., and Barras F. (2001) An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandkvist M., Hough L. P., Bagdasarian M. M., and Bagdasarian M. (1999) Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J. Bacteriol. 181, 3129–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robert V., Hayes F., Lazdunski A., and Michel G. P. (2002) Identification of XcpZ domains required for assembly of the secreton of Pseudomonas aeruginosa. J. Bacteriol. 184, 1779–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lallemand M., Login F. H., Guschinskaya N., Pineau C., Effantin G., Robert X., and Shevchik V. E. (2013) Dynamic interplay between the periplasmic and transmembrane domains of GspL and GspM in the type II secretion system. PLoS One 8, e79562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winston S. E., Mehan R., and Falke J. J. (2005) Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry 44, 12655–12666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seeger M. A., von Ballmoos C., Eicher T., Brandstätter L., Verrey F., Diederichs K., and Pos K. M. (2008) Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 15, 199–205 [DOI] [PubMed] [Google Scholar]
- 37. Bass R. B., Butler S. L., Chervitz S. A., Gloor S. L., and Falke J. J. (2007) Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 423, 25–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arts J., de Groot A., Ball G., Durand E., El Khattabi M., Filloux A., Tommassen J., and Koster M. (2007) Interaction domains in the Pseudomonas aeruginosa type II secretory apparatus component XcpS (GspF). Microbiology 153, 1582–1592 [DOI] [PubMed] [Google Scholar]
- 39. Robert V., Filloux A., and Michel G. P. (2005) Subcomplexes from the Xcp secretion system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 252, 43–50 [DOI] [PubMed] [Google Scholar]
- 40. Bischof L. F., Friedrich C., Harms A., Søgaard-Andersen L., and van der Does C. (2016) The type IV pilus assembly ATPase PilB of Myxococcus xanthus interacts with the inner membrane platform protein PilC and the nucleotide binding protein PilM. J. Biol. Chem. 291, 6946–6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCallum M., Tammam S., Little D. J., Robinson H., Koo J., Shah M., Calmettes C., Moraes T. F., Burrows L. L., and Howell P. L. (2016) PilN binding modulates the structure and binding partners of the Pseudomonas aeruginosa type IVa pilus protein PilM. J. Biol. Chem. 291, 11003–11015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leighton T. L., Dayalani N., Sampaleanu L. M., Howell P. L., and Burrows L. L. (2015) A novel role for PilNO in type IV pilus retraction revealed by alignment subcomplex mutations. J. Bacteriol. 197, 2229–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Careaga C. L., and Falke J. J. (1992) Thermal motions of surface α-helices in the d-galactose chemosensory receptor. Detection by disulfide trapping. J. Mol. Biol. 226, 1219–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Careaga C. L., and Falke J. J. (1992) Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophys. J. 62, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kelley L. A., and Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 46. Wolfgang M., Lauer P., Park H. S., Brossay L., Hébert J., and Koomey M. (1998) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29, 321–330 [DOI] [PubMed] [Google Scholar]
- 47. Whitchurch C. B., Hobbs M., Livingston S. P., Krishnapillai V., and Mattick J. S. (1991) Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101, 33–44 [DOI] [PubMed] [Google Scholar]
- 48. Whitchurch C. B., and Mattick J. S. (1994) Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13, 1079–1091 [DOI] [PubMed] [Google Scholar]
- 49. Chang Y. W., Rettberg L. A., Treuner-Lange A., Iwasa J., Søgaard-Andersen L., and Jensen G. J. (2016) Architecture of the type IVa pilus machine. Science 351, aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X., Pineau C., Gu S., Guschinskaya N., Pickersgill R. W., and Shevchik V. E. (2012) Cysteine scanning mutagenesis and disulfide mapping analysis of arrangement of GspC and GspD protomers within the type 2 secretion system. J. Biol. Chem. 287, 19082–19093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Py B., Loiseau L., and Barras F. (1999) Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol. 289, 659–670 [DOI] [PubMed] [Google Scholar]
- 52. Sandkvist M., Bagdasarian M., Howard S. P., and DiRita V. J. (1995) Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14, 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gray M. D., Bagdasarian M., Hol W. G., and Sandkvist M. (2011) In vivo cross-linking of EpsG to EpsL suggests a role for EpsL as an ATPase-pseudopilin coupling protein in the type II secretion system of Vibrio cholerae. Mol. Microbiol. 79, 786–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamagata A., Milgotina E., Scanlon K., Craig L., Tainer J. A., and Donnenberg M. S. (2012) Structure of an essential type IV pilus biogenesis protein provides insights into pilus and type II secretion systems. J. Mol. Biol. 419, 110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Friedrich C., Bulyha I., and Søgaard-Andersen L. (2014) Outside-in assembly pathway of the type IV pilus system in Myxococcus xanthus. J. Bacteriol. 196, 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chiang P., Sampaleanu L. M., Ayers M., Pahuta M., Howell P. L., and Burrows L. L. (2008) Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154, 114–126 [DOI] [PubMed] [Google Scholar]
- 57. Harvey H., Habash M., Aidoo F., and Burrows L. L. (2009) Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J. Bacteriol. 191, 6513–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kus J. V., Tullis E., Cvitkovitch D. G., and Burrows L. L. (2004) Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150, 1315–1326 [DOI] [PubMed] [Google Scholar]
- 59. Kus J. V., Kelly J., Tessier L., Harvey H., Cvitkovitch D. G., and Burrows L. L. (2008) Modification of Pseudomonas aeruginosa Pa5196 type IV pilins at multiple sites with d-Araf by a novel GT-C family arabinosyltransferase, TfpW. J. Bacteriol. 190, 7464–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koo J., Tang T., Harvey H., Tammam S., Sampaleanu L., Burrows L. L., and Howell P. L. (2013) Functional mapping of PilF and PilQ in the Pseudomonas aeruginosa type IV pilus system. Biochemistry 52, 2914–2923 [DOI] [PubMed] [Google Scholar]
- 61. Nunn D., Bergman S., and Lory S. (1990) Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172, 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Simon V., and Schumann W. (1987) In vivo formation of gene fusions in Pseudomonas putida and construction of versatile broad-host-range vectors for direct subcloning of Mu d1 and Mu d2 fusions. Appl. Environ. Microbiol. 53, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]