Abstract
Starch synthesis in cereal grain endosperm is dependent on the concerted actions of many enzymes. The starch plastidial phosphorylase (Pho1) plays an important role in the initiation of starch synthesis and in the maturation of starch granule in developing rice seeds. Prior evidence has suggested that the rice enzyme, OsPho1, may have a physical/functional interaction with other starch biosynthetic enzymes. Pulldown experiments showed that OsPho1 as well as OsPho1 devoid of its L80 region, a peptide unique to higher plant phosphorylases, captures disproportionating enzyme (OsDpe1). Interaction of the latter enzyme form with OsDpe1 indicates that the putative regulatory L80 is not responsible for multienzyme assembly. This heterotypic enzyme complex, determined at a molar ratio of 1:1, was validated by reciprocal co-immunoprecipitation studies of native seed proteins and by co-elution chromatographic and co-migration electrophoretic patterns of these enzymes in rice seed extracts. The OsPho1-OsDpe1 complex utilized a broader range of substrates for enhanced synthesis of larger maltooligosaccharides than each individual enzyme and significantly elevated the substrate affinities of OsPho1 at 30 °C. Moreover, the assembly with OsDpe1 enables OsPho1 to utilize products of transglycosylation reactions involving G1 and G3, sugars that it cannot catalyze directly.
Keywords: glycosyltransferase, metabolism, phosphorylase, protein complex, protein-protein interaction, D-enzyme, disproportionating enzyme, rice endosperm, starch phosphorylase, starch synthesis
Introduction
In higher plants starch phosphorylase (Pho1, EC 2.4.1.1) consists of two isoforms, Pho1 and Pho2, located in separate intracellular compartments. Pho1 is located in the plastid, whereas Pho2 is distributed to the cytoplasm (1–4). Although both isoforms catalyze the same reversible reaction (Glc1P2 + α-glucan (n) ⇔ α-glucan (n + 1) + orthophosphate), they have distinct roles in starch metabolism. The cytoplasmic Pho2 is involved in the metabolism of maltose, the amylolytic degraded starch product transported from the chloroplast to the cytoplasm. A role of Pho1 in starch biosynthesis was first reported for the plastidial PhoB enzyme from Chlamydomonas reinhardtii (5) where strains containing mutations in the structural gene STA4 exhibit reduced levels of starch and a modified amylopectin structure and elevated amylose content. The role of starch phosphorylase in higher plants remained enigmatic until the identification and characterization of pho1 mutants in rice, which exhibited pronounced alterations in seed starch. When grown at normal temperature, pho1 plants produced seeds ranging from shrunken in size to pseudo-normal-appearing seeds, the latter having reduced weight compared with normal seeds produced by wild type plants (6). At lower temperature, pho1 plants bore mainly shrunken seeds, indicating that Pho1 plays a significant role for starch biosynthesis at low temperature, whereas its activity at higher temperature is partially compensated by one or more other factors yet to be identified. The biosynthetic role of the rice Pho1 (OsPho1) is substantiated by its catalytic properties. The rice Pho1 is a homodimer (210 kDa) that exhibits considerably higher catalytic activity in the synthesis direction, having equilibrium constants ranging from 13 to 45 (7) depending on the acceptor substrate. The biosynthetic role is conspicuously displayed when OsPho1 is incubated with Pi alone using maltohexaose as a substrate. Although degradative products are produced, near equal amounts of synthetic products are also generated. Moreover, the enzyme showed significantly increased substrate affinities and catalytic activities at subsaturating substrate concentrations when the reaction temperature was lowered from 36 °C to 18 °C (4), supporting the enzyme's physiological role at low growth temperature.
The higher plant Pho1 is structurally homologous to Pho2 and microbial and animal glycogen phosphorylases with one significant exception. The enzyme from higher plants contains an additional 78–80-amino acid peptide (L78/L80) positioned near the middle of the primary sequence, which is not found in Pho2, maltodextrin phosphorylase, or glycogen phosphorylases (4, 8). This region, which has a number of charged amino acids, forms an undefined loop structure (supplemental Fig. S1) on the surface of the enzyme when modeled based on the crystal structure of Pho2 from Arabidopsis thaliana (9). Removal of L80 from OsPho1 did not affect the catalytic properties of OsPho1 for Glc1P, amylopectin, and malto-oligosaccharide but significantly increased substrate affinity when glycogen was used as the substrate (4), suggesting the L80 domain per se is not directly involved in catalysis in rice. It has also been suggested that L80 is not important for regulation of the enzyme but, instead, protects the OsPho1 protein under heat stress conditions (4). In sweet potato roots, high molecular weight complexes showing Pho1 activity contained the 20S proteasome that interacts with the L78 region of Pho1 and degrades the Pho1 protein under heat stress (10). In addition, like the L80 of OsPho1 the L78 of sweet potato, Pho1 has a number of charged amino acids that have been suggested to be potential targets for protein kinase activity (11). It was proposed that L78 serves as a switch controlling the catalytic direction of the enzyme reaction as proteolytic removal of the L78 region induced by the phosphorylation event resulted in alteration of the enzyme's affinities toward substrates (8).
A number of starch-metabolizing enzymes form multiprotein complexes in rice (12), wheat (13, 14), maize (15–19), and barley (20). In rice endosperm, protein-protein interactions occur between or among starch synthases (SSs), branching enzymes (BEs), pullulanase, and OsPho1 (21). In wheat, it has been reported that Pho1 interacts with BEI and BEIIb in an ATP-dependent manner in soluble extracts of developing endosperms harvested at mid-to-late developmental stage (14). Interestingly, the same interaction was not detected in developing endosperms harvested at the early stage possibly due to low BEI expression (13). In maize endosperm, a novel multienzyme complex consisting of SSIIa, SSIII, BEIIa, BEIIb, ADP-glucose pyrophosphorylase (AGPase) large and small subunits, and pyruvate phosphate dikinase (PPDK) was reported (15). Interestingly, the maize Pho1 was found associated with SSI, SSIIa, BEI, and BEIIa in the starch granule when expression of BEIIb was abolished in an amylose extender (ae−) mutant (17, 22). However, direct evidence for an interaction between the maize endosperm Pho1 and BEIIb/BEI has not been established. A very weak interaction between OsPho1 and BEIIa was observed in rice endosperm when co-immunoprecipitation (co-IP) was performed with Pho1 or BEIIa antibodies, a view supported by co-electrophoresis migration patterns as assessed by blue-native PAGE (12). Alternatively, the interaction between OsPho1 and BE may be indirect, as OsPho1 and BE may form an indirect physical association on glucans to synthesize branched α-glucans or maltodextrins (23).
To identify potential protein-interacting partners of the rice Pho1, we incubated developing rice seed extracts with immobilized OsPho1 or a L80-deleted form of OsPho1 (OsPho1ΔL80) as bait. Although starch synthases and branching enzyme proteins were expected, we instead identified disproportionating enzyme (d-enzyme or OsDpe1) as a prominent interacting partner of OsPho1. Results are presented that OsPho1 and OsDpe1 form a heteromeric multisubunit protein complex, and both enzymes are functional in the complex to cooperatively synthesize glucan substrates.
Results
Both OsPho1 and OsPho1ΔL80 Interact with OsDpe1
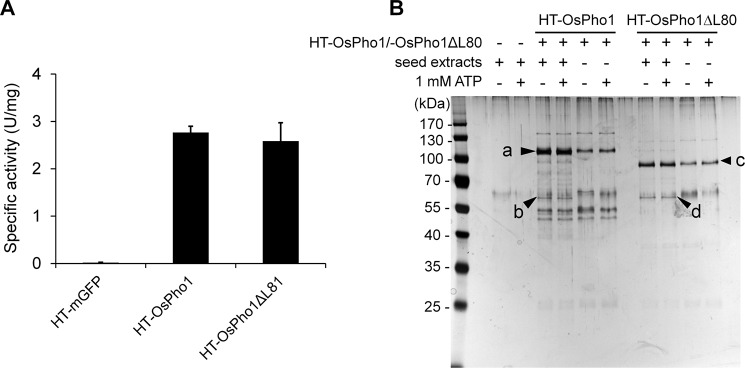
Our earlier study showed that OsPho1 and OsPho1ΔL80 are homodimers that showed similar kinetic properties when their recombinant forms were assayed (4). If the extended L80 region (supplemental Fig. S1) is responsible for the protein-protein interaction, the potential partner proteins would interact with OsPho1 but not with OsPho1ΔL80 (14). To identify proteins interacting with the OsPho1 proteins, we repeatedly attempted a pulldown assay using His6-tagged OsPho1 or OsPho1ΔL80 bound to Ni+-agarose resins but were unsuccessful due to instability of protein binding and background problems. Thus, we employed the HaloTag® pulldown technology as this method allows for stringent washing conditions to reduce undesirable background of nonspecific interacting proteins. Three fusion proteins, HaloTag®;green fluorescence protein (HT-GFP; control), HT-OsPho1, and HT-OsPho1ΔL80 (supplemental Fig. S2), were successfully overexpressed in Escherichia coli, and starch phosphorylase activities of the fusion proteins in the cleared cell lysates were measured (Fig. 1A). Both HT-OsPho1 and HT-OsPho1ΔL80 showed substantial levels of starch phosphorylase activities, whereas HT-GFP emitted strong fluorescence under 360-nm ultraviolet light, indicating that the recombinant-expressed proteins fused to HT maintained their active forms. The proteins were then purified to near homogeneity and covalently coupled to HaloLink resins. The resins were extensively washed with binding buffer and incubated with protein extracts from immature rice seeds (10 days after flowering (DAF)) in the absence or presence of 1 mm ATP to enable potential phosphorylation-dependent OsPho1 interaction with other proteins, as demonstrated by the phosphorylation-dependent interaction of SBEIIb and Pho1 in wheat amyloplast extracts (14). When the bound proteins were eluted and subjected to SDS-PAGE analysis, the silver-stained polyacrylamide gel showed unique bands at ∼63 kDa in both HT-OsPho1 (labeled b in Fig. 1B) and HT-OsPho1ΔL80 (labeled d) samples irrespective of the presence of 1 mm ATP. Unique polypeptide bands at 105 kDa (labeled a), 95 kDa (labeled c), and 63 kDa (labeled b and d) from both samples were analyzed by using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Mass spectrometry results revealed that the bands at 105 kDa and 95 kDa were OsPho1 and OsPho1ΔL80, respectively, whereas the 63-kDa bands were identified as 4-α-glucanotransferase (disproportionating enzyme, Dpe1 or D-enzyme; EC 2.4.1.25). The molecular mass of rice Dpe1 (OsDpe1), as estimated by mobility on SDS-polyacrylamide gels, corresponds well with the calculated molecular mass (62 kDa) of the matured monomeric form of OsDpe1 (GenBankTM accession number AK318541). No protein bands corresponding to the size of rice BEs (>80 kDa) (12) were identified in the presence or absence of 1 mm ATP, a result unexpected in light of the previous reports for the protein interacting properties of the wheat Pho1 (14) and maize Pho1 (22, 24).
FIGURE 1.
Pulldown assay of OsPho1. A, starch phosphorylase activities of HT-linked proteins. Cleared lysates from E. coli EA3457 cells expressing HT-GFP, HT-OsPho1, and HT-OsPho1ΔL80 were used to measure Pho1 activity with 2 mg/ml amylopectin and 20 mm Glc1P as described (4). The values were calculated from at least three replicates of independent reactions. Error bars, S.E. B, analysis of seed proteins eluted from a pulldown experiment with HT-OsPho1 and HT-OsPho1Δ80. Soluble extracts were prepared from immature (10 DAF) rice seeds. The arrows (a–d) indicate the protein bands, which were later excised for LC-MS/MS analysis.
Validation of Protein-Protein Interaction between OsDpe1 and OsPho1
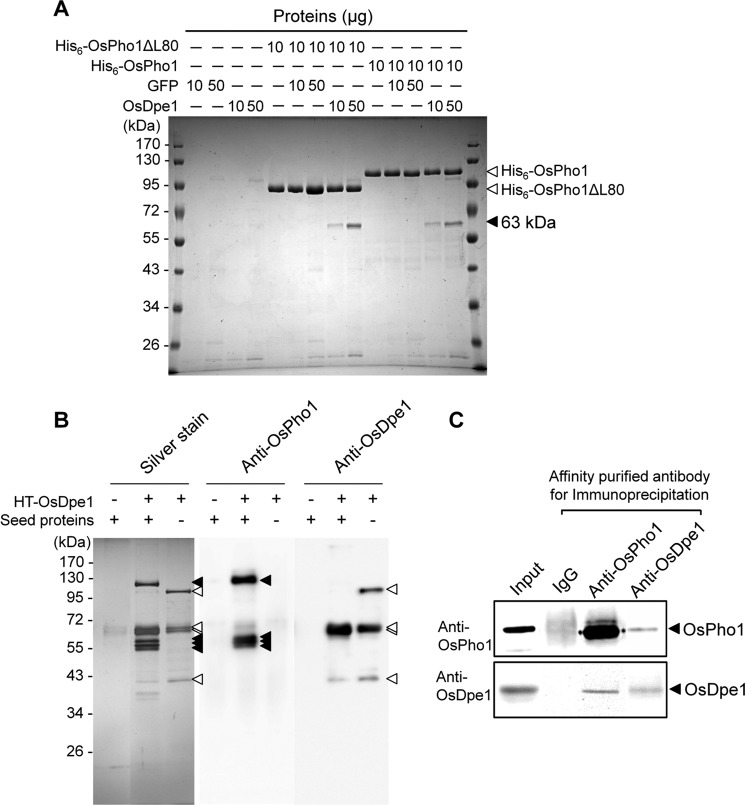
To confirm the interaction between OsPho1 and OsDpe1, the partially purified tag-less OsDpe1 and GFP proteins were mixed with His6-OsPho1 or with His6-OsPho1ΔL80 and then subjected to immobilized metal affinity chromatography (IMAC). After the IMAC resins were washed extensively, the proteins were eluted with 100 mm imidazole and analyzed by SDS-PAGE. No promiscuous bands were detected when GFP was incubated with OsPho1 or OsPho1ΔL80. By contrast, the 62-kDa OsDpe1 was captured by both OsPho1 forms at amounts relative to their input (Fig. 2A).
FIGURE 2.
Verification of OsPho1-OsDpe1 interaction. A, 10 μg each of His6-OsPho1 and His6-OsPho1ΔL80 were incubated 30 min at room temperature with 10 or 50 μg each of the partially purified tag-less GFP or OsDpe1, and the mixtures were applied to the TALON-IMAC column. Bound proteins were eluted in 1× SDS-PAGE sample buffer and then subjected to SDS-PAGE analysis. Arrowheads indicate, from top to bottom, His6-OsPho1, His6-OsPho1ΔL80, and OsDpe1 protein bands. B, pulldown experiment of OsDpe1-interacting proteins from immature rice seeds. Experiments were performed in the same manner as described in Fig. 1, except HT-OsDpe1 was used in place of HT-OsPho1. The eluted proteins were separated by SDS-PAGE and blotted onto nitrocellulose membrane. Antibodies raised against OsPho1 and OsDpe1 were used to detect OsPho1 and OsDpe1 proteins, respectively. Note that some bands detected at smaller molecular weights are the truncated forms of both proteins. Intact HT-OsDpe1 was not detected when incubated with seed protein extract, possibly due to increased degradation of the protein that was not covalently bound to the resin. Open triangle, OsDpe1; filled triangle, OsPho1. C, co-immunoprecipitation analysis of interaction between OsPho1 and OsDpe1. IPs were generated by incubating rice seed extracts with protein A beads conjugated with affinity-purified OsPho1 and OsDpe1 antibodies, which were then subjected to immunoblot analysis to detect the respective antigens. Rice seed extracts (Input) and immunoglobulin G (IgG) were used as positive and negative controls, respectively.
The physiological interaction of OsDpe1 with OsPho1 was confirmed using rice-developing seed extracts. HT-OsDpe1 fusion protein was constructed, expressed in E. coli, and captured on HaloLink resin. Soluble protein extracts from immature rice seeds were incubated with the HaloLink-OsDpe1 resin, and the bound proteins were resolved by SDS-PAGE and analyzed by silver staining of the polyacrylamide gel and immunoblotting (Fig. 2B). The silver-stained polyacrylamide gel displayed a complex distribution of polypeptide bands. When we performed immunoblot analysis with anti-OsPho1 (supplemental Fig. S3 A) and anti-OsDpe1 (supplemental Fig. S3B) antibodies, the major bands (denoted by triangles) in the silver-stained gel from the HT-OsDpe1 sample alone strongly reacted with anti-OsDpe1 antibody, whereas the bands seen at different sizes (denoted by filled triangles) from HT-OsDpe1 incubated with rice extracts reacted with OsPho1 antibody. This result indicates that intact and degraded forms of OsPho1 were captured by HaloLinked OsDpe1 and that OsPho1 is a major interacting partner of OsDpe1.
Co-IP assays were performed to verify the in situ interaction between OsPho1 and OsDpe1 in developing rice seeds. Affinity purified anti-OsPho1 and anti-OsDpe1 antibodies were immobilized on protein-A beads and incubated with soluble protein extracts from immature rice seeds. The bound proteins were eluted and analyzed by immunoblotting with anti-OsPho1 or anti-OsDpe1 antibodies (Fig. 2C). Immunoprecipitates (IPs) generated with the anti-OsPho1 antibody contained its respective antigen, OsPho1, as well as OsDpe1. Likewise, IPs formed using anti-OsDpe1 antibody contained its respective antigen, OsDpe1, as well as OsPho1. Overall, the capturing of recombinant and native OsDpe1 by recombinant OsPho1 and the reciprocal co-IP results indicate that OsPho1 and OsDpe1 form a stable protein complex.
OsDpe1 Is Less Abundant Than OsPho1
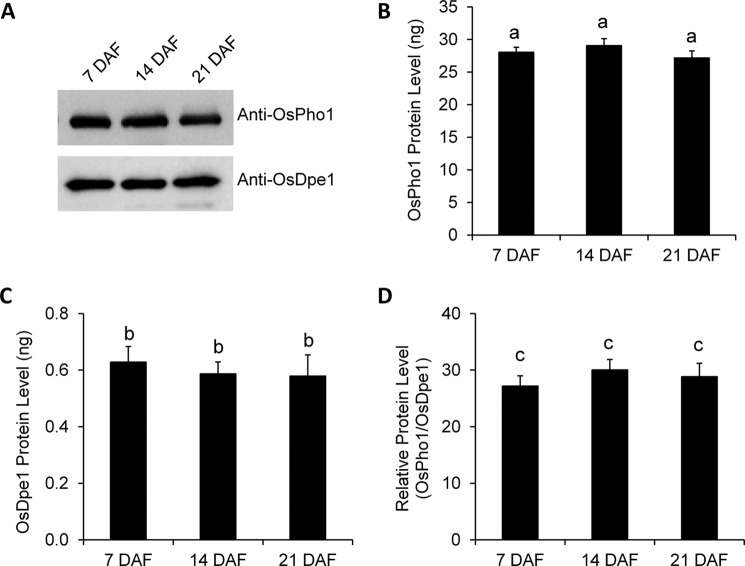
The expression levels of OsPho1 and OsDpe1 in immature seeds harvested at 7, 14, and 21 days after flowering were measured using the purified recombinant OsDpe1 and OsPho1 proteins as references (Fig. 3). Immunoblot analysis using anti-OsPho1 and anti-OsDpe1 antibodies (Fig. 3A) showed that the levels of OsDpe1 and OsPho1 protein were fairly constant throughout seed development (Fig. 3, A, B, and C). When the relative levels of the enzymes were calculated, OsPho1 is found to be much more abundant (>25-fold) than OsDpe1 during all development stages examined in this study (Fig. 3D).
FIGURE 3.
Protein levels of OsPho1 and OsDpe1 in developing rice seeds. A, proteins were extracted from immature seeds harvested at 7, 14, and 21 days after flowering and probed using anti-OsPho1 and anti-OsDpe1 antibodies. Protein amounts were measured by using standard curves from immunoblot analysis of varied amount (5–250 ng) of the purified recombinant OsPho1 and OsDpe1. Relative protein levels (D) were calculated by dividing molar quantity of OsPho1 (B) with molar quantity of OsDpe1 (C). Lowercase letters indicate the difference of means, by Tukey's critical range test, at α = 0.05.
OsPho1 and OsDpe1 Form a High molecular Weight Multiprotein Complex in Vitro and in Vivo
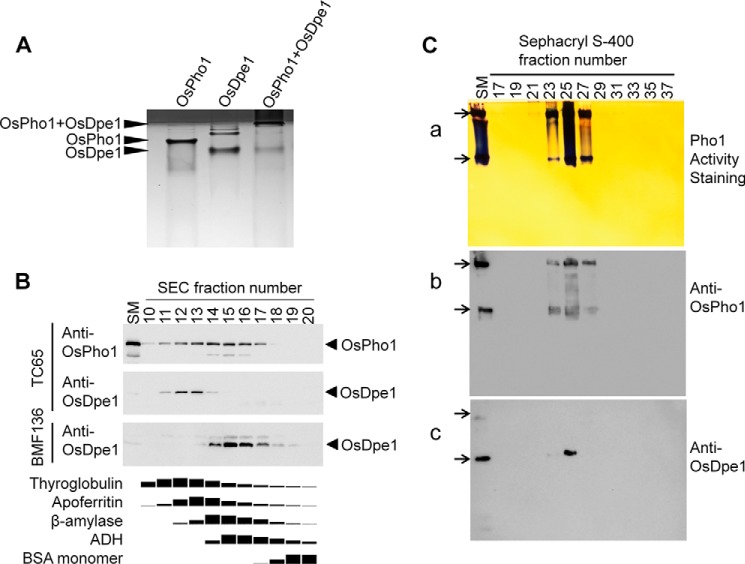
The recombinant OsDpe1 was subjected to size exclusion chromatography to measure its molecular weight. Our result showed that OsDpe1 has a molecular mass of 121 kDa (supplemental Fig. S4A), suggesting the rice Dpe1 forms a homodimer when taking into account that OsDpe1 monomer is 62 kDa as assessed by SDS-PAGE analysis (Fig. 2). An equal amount of the recombinant OsPho1 and OsDpe1 was mixed and incubated at room temperature for 1 h, and nondenaturing native polyacrylamide gel (10%) electrophoresis was performed in Tris-Tricine buffer. The protein bands were then visualized by Coomassie Brilliant Blue (CBB) staining. Fig. 4A shows that OsDpe1 protein migrated noticeably faster than OsPho1 protein, whereas a mixture of OsPho1 and OsDpe1 proteins barely migrated and was detected at the top part of the gel. This result indicates a formation of a high molecular weight complex between the two proteins.
FIGURE 4.
Interaction of OsPho1 and OsDpe1. A, mobility patterns of OsPho1, OsDpe1, and a mixture of OsPho1 and OsDpe1. Native polyacrylamide gel (10%) electrophoresis was performed in Tris/Tricine buffer, and the CBB staining result shows stacking (dark) and separation (bright) gels. B, immunoblot analysis of seed proteins in fractions (1 ml) resolved by Superdex-200 SEC chromatography. 10 μl of each fraction were subjected to 12% SDS-PAGE and subsequently analyzed by immunoblotting using anti-OsPho1 and anti-OsDpe1. The bottom panel depicts the elution profiles of the molecular mass standard proteins (669 kDa, thyroglobulin; 443 kDa, apoferritin; 200 kDa, β-amylase; 150 kDa, alcohol dehydrogenase; 66 kDa, BSA) under the same chromatography conditions. Amounts of the marker proteins in the fractions were determined by using the image analysis software Image J 1.48v after scanning the CBB-stained SDS-PAGE gel. C, co-migration of OsPho1 and OsDpe1. The Sephacryl S-400 fractions were separated on a 7.5% Tris/glycine non-denaturing polyacrylamide gel containing 0.8% glycogen. Activity staining and immunoblot analysis methods are described “Experimental Procedures.” Arrows indicate two OsPho1 activity bands (a) on the native gel and their corresponding locations on the immunoblot membranes of OsPho1 (b) and OsDpe1 (c). SM denotes starting material containing 15 μg of soluble proteins from immature rice seeds (see “Experimental Procedures”).
We further analyzed the protein complex formation by using Superdex-200 size exclusion chromatography (supplemental Fig. S5). The two dimeric forms of the recombinant enzymes were mixed at three different molar ratios (2:1, 1:1, and 1:2) and resolved by chromatography, and the amounts of each protein in each fraction estimated by the CBB staining of polypeptide bands was resolved by SDS-PAGE. At a OsPho1-OsDpe1 ratio of 1:2, two major peaks were observed on the chromatogram (result not shown), and SDS-PAGE analysis showed two proteins were at a similar molar ratio in the early fractions, but OsDpe1 bands were more prevalent in the later fractions, indicating a large amount of free OsDpe1 dimers were eluted in the later fractions. At 2:1, OsPho1 bands were prevalent in the early fractions, but OsDpe1 band declined drastically, and OsPho1 remained at higher levels thereafter, indicating a large amount of free OsPho1 dimers were eluted in the later fractions. However, at 1:1, two proteins were present at similar levels throughout the elution profile. These results suggest that OsPho1 and OsDpe1 interact with each other at the stoichiometric molar ratio of 1:1. The molecular size of the OsPho1-OsDpe1 complex was estimated to be 620 kDa (supplemental Fig. S4B), indicating the protein complex likely contains 4 subunits of OsPho1 and 4 subunits of OsDpe1.
Native OsDpe1 Is Eluted as a High Molecular Weight
To demonstrate the physiological existence of the OsPho1-OsDpe1 complex, we analyzed the elution profiles of OsDpe1 and OsPho1 present in soluble protein extract from immature rice seeds (10 DAF) by performing Superdex-200 chromatography and subsequent analysis of the fractions by immunoblot analysis with antibodies raised against OsPho1 and OsDpe1 (Fig. 4B). Although OsDpe1 co-eluted with OsPho1 in a small range of high molecular weight fractions (fractions 11–14), OsPho1 proteins were detected in a broader range of fractions (10–18) with the latter fractions overlapping with the elution profile of β-amylase (200 kDa) that was used as a molecular size marker. When a protein extract from immature seeds of BMF136 (pho1 null mutant) was analyzed, OsDpe1 proteins were no longer eluted in the high molecular weight fractions but, instead, were detected in lower molecular weight fractions (Fig. 4B). Because the molecular size of native OsDpe1 dimer is 124 kDa, most if not all of the OsDpe1 proteins are complexed with the 210-kDa OsPho1, whereas OsPho1 proteins unreacted to OsDpe1 elute in the later fractions as dimeric forms of OsPho1. This result is not surprising as the amounts of OsPho1 protein are substantially much higher than OsDpe1 in rice endosperm (Fig. 3), and thus a large portion of the OsPho1 proteins are not bound with OsDpe1 and elute as a dimer.
To obtain further evidence for physiological existence of the OsPho1-OsDpe1 complex, we analyzed soluble rice seed proteins fractionated by Sephacryl S-400 column chromatography by Pho1 enzyme assays in solution and by enzyme activity staining on native glycogen-containing polyacrylamide gels (Fig. 4C). The highest starch phosphorylase activity was measured in fractions 24 through 26 with lower amounts of enzyme activity in fractions 23 and 27 (result not shown). These fractions showed two Pho1 activity bands on native polyacrylamide gels (Fig. 4Ca) with the highest activity in fraction 25 coincident with the highest Pho1 enzyme activity measured in solution (result not shown). Despite repeated attempts, Dpe1 activity staining on native polyacrylamide gels was unsuccessful likely due to the low abundance of OsDpe1 in rice endosperm as evident in Fig. 3. Immunoblot analysis of the fractions using anti-OsPho1 and anti-OsDpe1 antibodies (Fig. 4Cb) showed that the strongest signals for OsPho1 proteins were detected in fraction 25 with two bands being recognized in fractions 23, 25, and 27, a result consistent with the activity staining result shown in Fig. 4Ca. OsDpe1 protein was also detected in the fraction 25 and weakly in the fraction 23 but not in the fraction 27 (Fig. 4Cc). The mobility of OsDpe1 was the same as that of the lower OsPho1 band. Thus, OsDpe1 not only co-eluted with OsPho1, but co-migrated with OsPho1 in the glycogen-containing polyacrylamide gel.
Analysis of Enzyme Action Patterns of OsDpe1 and OsPho1-OsDpe1 Complex
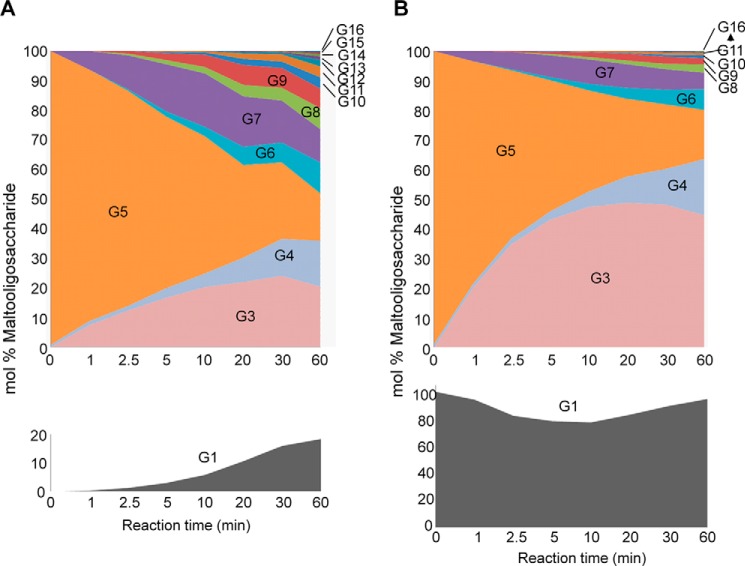
To characterize the enzymatic properties of the cloned OsDpe1, we purified the recombinant protein and performed enzyme reaction with various MOSs. As previously shown for Dpe1 from different plants, OsDpe1 is unable to metabolize maltose as a donor, whereas its action patterns with larger MOSs were very similar to that displayed by the sweet potato Dpe1 (25). Its disproportionating (or transglycosylase) activity hydrolyzes MOSs (Gn) into smaller MOSs (Gn−2) and larger ones (Gn+2) or synthesizes in the reverse way. It has been suggested that G1 serves as an acceptor for maltosyl groups as the OsDpe1 reaction with G1 and G7 produces G3 and G5 (26). To test this, we examined the time course of OsDpe1 activity using G5 alone and G5 with G1. Fluorophore-assisted carbohydrate electrophoresis (FACE) results of the OsDpe1 reaction with G5 alone and with equal molar amounts of G5 and G1 for 0–60 min are shown in supplemental Fig. S6 and Fig. 5. When G5 alone was used as substrate, the G1 level increased as the G5 level decreased, indicating that G5 served as both a donor and acceptor as the initial products from the reaction were G3 and G7. The reaction products were then reused as a donor and acceptor for OsDpe1 to produce a range of MOSs, and the amounts of the reaction products tended to be evenly distributed after 60 min (Fig. 5A). When G1 was added with G5 in the reaction, the action patterns of OsDpe1 showed a markedly different action pattern (Fig. 5B). G5 levels decreased more rapidly, and correspondingly, G3 levels were quickly accumulated as a major product maintained over the time. G7 levels were increased temporarily and subsequently decline, whereas the G1 level was slightly decreased after the reaction started but climbed back to its initial level. This result indicates that G5 + G1 ⇔ G3 reaction is preferred and that the presence of G1 promotes the accumulation of G3 from G5. The relative levels of large MOSs were significantly lowered by the addition of G1 compared with the reaction with G5 alone. This action pattern of OsDpe1 is very similar to that observed for Dpe1 from pea (27).
FIGURE 5.
Enzyme reaction profiles of OsDpe1 with maltopentaose (G5) in the absence (A) and presence (B) of glucose. The band densities of the glucans insupplemental Fig. S3 were analyzed using image analysis software from KODAK, and mol% of each glucan was calculated with respect to the total molar amount of glucans including G5 and G1. The initial mol% of the G5 was considered 100% in B, and the initial G1 and G5 mol% were 100% as an equal molar amount of both molecules were used for the reaction.
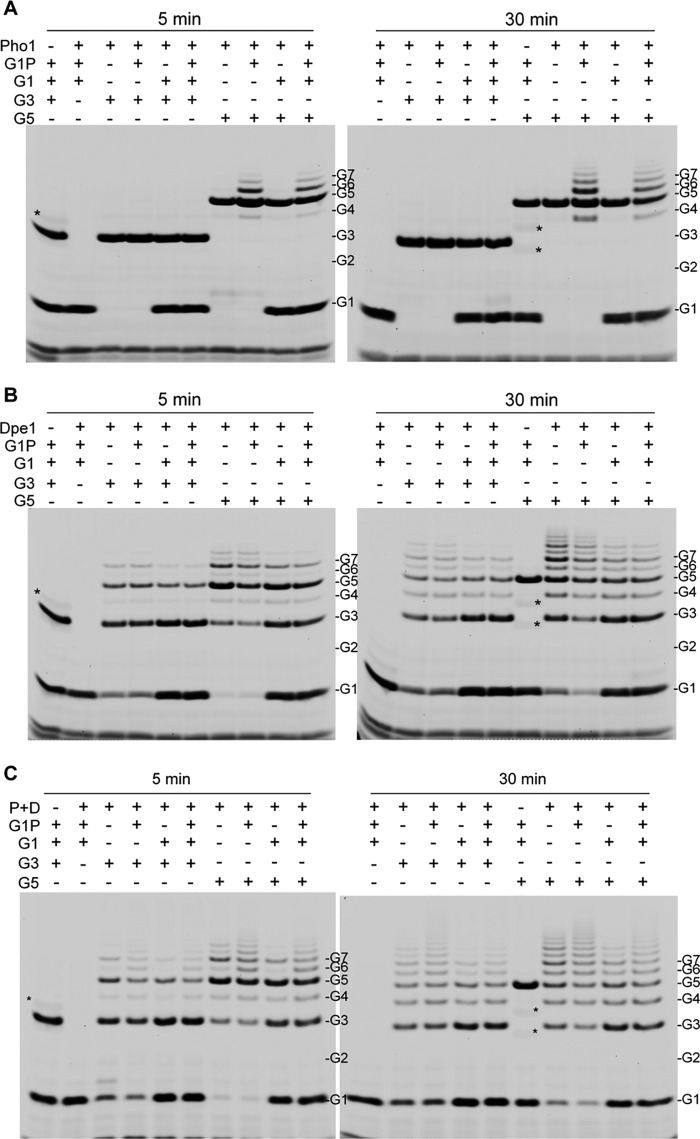
We further compared the enzymatic products of the gel filtration-purified recombinant OsPho1-OsDpe1 multiprotein complex to purified OsPho1 and OsDpe1. The reactions were performed with various combinations of substrates, G1, G3, G5, and/or Glc1P for 5 and 30 min at room temperature. As expected from our earlier study (7), OsPho1 alone did not show any enzyme activity with G3 in the presence of Glc1P or G1 but produced a narrow range of MOSs (G4 to G8) from G5 in the presence of Glc1P (Fig. 6A). Production of MOSs larger than G5 was increased after 30 min of reaction with the addition of G1 lowering the production of MOSs. OsDpe1 alone was unable to use Glc1P as substrate, but the enzyme catalyzed the disproportionation reaction of G3 into G1 and G5 as major products and into G4, G6, G7 and larger MOSs as minor products after 5 min of reaction (Fig. 6B). Longer incubation (30 min) produced a wide range of MOS (G1 to >G10). Transglycosylation activity of OsDpe1 also converted G5 into G3 and G7 as the major products and G4, G6, and other larger MOSs as minor products. Longer incubation with G5 increased the amounts of G3 and G7 and larger MOSs. The addition of G1 in the reactions maintained high levels of G3 but lowered the production of larger MOSs as evident in Fig. 5B.
FIGURE 6.
FACE analysis of the reaction products of OsPho1 (A), OsDpe1 (B), and OsPho1-OsDpe1 complex (C). G1, glucose; G2, maltose; G3, maltotriose; G4, maltaotetraose; G5, maltopentaose; G6, maltohexaose; G7, maltoheptaose; P + D, OsPho1-OsDpe1 complex. Asterisks (*) indicate the tracking dyes used in the sample buffer. Shown are the negative images converted from those taken with CCD camera. Note that Glc1P was not detected.
The initial reaction products of the OsPho1-OsDpe1 complex with G3 alone and G5 alone were G1+G5 and G3+G7, respectively (Fig. 6C). The OsPho1-OsDpe1 complex produced similar amounts of larger MOSs as seen for OsDpe1 (Fig. 6B). The addition of Glc1P to the reaction mixture profoundly accelerated the synthesis of larger MOSs, and the levels of the products were more evenly distributed than without Glc1P, indicating a phosphorylase reaction occurred with both G3 (via OsDpe1) and G5. G1, as an acceptor, again lowered the synthesis of larger MOSs from G5 and promoted the accumulation of G3. These results suggest that both OsPho1 and OsDpe1 are active in the complex and exert their respective catalytic activities. The two enzymes cooperatively work to produce more and evenly distributed levels of MOSs by sharing the intermediate MOS products larger than G3.
OsDpe1 Significantly Increases the Substrate Affinities of OsPho1 in a Multiprotein Complex
The substrate affinities of OsPho1 were significantly increased by lowering the reaction temperature of the enzyme from 36 °C to 18 °C (4). To determine whether low temperature affects the catalytic properties of the OsPho1-OsDpe1 complex, we measured the S0.5 values of the complex for Glc1P in the presence of 2 mg/ml amylopectin or 2 mm G5 and the S0.5 values for G5 in the presence of 20 mm Glc1P (Table 1). OsPho1 showed significantly increased substrate affinities toward Glc1P at the lower temperature: 8.2-fold in the presence of 2 mg/ml amylopectin and 2.2-fold in the presence of 2 mm G5. Substrate affinity for G5 was slightly increased by lowering the temperature. Although slightly different conditions were used for this study, these results were consistent with those reported in our earlier study (4). Interestingly, formation of a multiprotein complex with OsDpe1 increased the substrate affinity of OsPho1 for Glc1P at 30 °C by 8.1-fold to a value close to that measured at 20 °C in the presence of amylopectin. A similar effect was also observed for Glc1P in the presence of G5 but to lesser extent. Interestingly, this temperature effect on substrate affinity was not apparent for G5 as the substrate as no significant differences were evident at the two temperatures.
TABLE 1.
Effect of temperature on substrate affinities of OsPho1 and OsPho1:OsDpe1
S0.5 values (mm), substrate concentrations at 50% of maximum reaction velocities, were measured in the presence of 2 mg/ml amylopectin, 2 mm maltopentaose, or 20 mm Glc1P and are represented together with S.E. from at least two independent experiments. The values in parentheses are Hill numbers (nH).
| Enzyme | Reaction temperature | S0.5 |
|||||
|---|---|---|---|---|---|---|---|
| Glc1P (2 mg/ml amylopectin) | nH | Glc1P (2 mm maltopentaose) | nH | Maltopentaose (20 mm Glc1P) | nH | ||
| mm | mm | mm | |||||
| Pho1 | 20 °C | 1.10 ± 0.04 | 1.0 | 1.18 ± 0.03 | 0.9 | 0.48 ± 0.00 | 0.7 |
| 30 °C | 9.00 ± 0.42 | 0.6 | 2.53 ± 0.09 | 0.9 | 0.62 ± 0.10 | 0.7 | |
| Pho1 + Dpe1 | 20 °C | 0.89 ± 0.09 | 1.0 | 0.92 ± 0.01 | 0.9 | 0.31 ± 0.07 | 0.7 |
| 30 °C | 1.11 ± 0.08 | 0.9 | 1.18 ± 0.03 | 1.0 | 0.40 ± 0.05 | 0.7 | |
Discussion
Our earlier studies (6) revealed that the plastidial form of starch phosphorylase is important for normal starch synthesis in rice seeds and plays a significant role in starch initiation and maturation at low temperature. Kinetic studies of OsPho1 (4) supported its low temperature role as the substrate affinities were significantly increased by lowering the reaction temperature. Catalytic activity of the enzyme was also higher at low temperature when assayed under limiting substrate conditions. Thus, the kinetic properties of the enzyme are consistent with its requirement for starch initiation and maturation processes under low temperature growth conditions. Under normal growth conditions, OsPho1 is not required for starch initiation, indicating that another factor, yet to be identified, is necessary. It is likely that OsPho1 interacts with other amyloplast enzymes to initiate starch synthesis as well as aid in the maturation of the starch grain. Indeed, several starch biosynthetic enzymes are functionally associated with Pho1 in the higher plants. In rice, it has been shown that branched glucans mediate a functional interaction between OsPho1 and BEs (23). Analysis of size exclusion chromatography (SEC) elution patterns of soluble proteins extracted from rice endosperm showed that SSIIa, SSIIIa, SSIVb, OsPho1, BEIIa, and isoamylase (ISA1) co-elute in fractions containing high molecular weight proteins far in excess of their predicted molecular weights (12). Reciprocal co-IP studies demonstrated that OsPho1 interacts with BEIIa, whereas the wheat endosperm Pho1 interacts with BEIIb and BEI in the presence of ATP (14).
To identify rice protein(s) that interacts with OsPho1 or its L80 domain (supplemental Fig. S1), we employed a pulldown assay using HT-OsPho1 and HT-OsPho1ΔL80. These assays were performed in the presence or absence of 1 mm ATP to test whether binding of OsPho1 to other proteins is dependent on protein phosphorylation as shown by the phosphorylation-dependent interaction of wheat endosperm SBEIIb and starch phosphorylase (14). It is noteworthy that none of the purified recombinant proteins used in this study is phosphorylated (supplemental Fig. S7). We detected an ∼63-kDa protein in developing rice seeds (10 DAF) that interacted with both proteins regardless of the presence of ATP (Fig. 1B), indicating the interaction is neither mediated by the L80 region nor dependent on protein phosphorylation of the region. However, we still cannot rule out a possibility that phosphorylation at one or more site(s) other than the L80 region of OsPho1 is responsible for the protein-protein interaction. LCMS/MS analysis identified the 63-kDa protein as α-4-glucanotransferase, also known as disproportionating enzyme, D-enzyme, or Dpe1. Dpe1 is one of the two distinct types of α-4-glucanotransferases found in plants: a plastidial Dpe1 and cytosolic Dpe2. The two enzymes are functionally and structurally distinct. Phylogenetic analysis indicates that Dpe2 shows very little sequence identity (13.2%) and similarity (23.1%) relatedness to Dpe1 and is more closely related to the bacterial amylomaltase (MalQ) than to Dpe1 from plants (26, 28, 29). Moreover, the action pattern of products produced by the higher plant Dpe2 enzymes (26, 29, 30) is similar to that generated by MalQ. Lastly, the predicted size of mature OsDpe1 is 62 kDa (GenBankTM accession number BAC07076), whereas that of OsDpe2 is 106 kDa (BAC22431).
The interaction between OsPho1 and OsDpe1 was validated by several different approaches. Purified His6-tagged OsPho1 or OsPho1ΔL80 captured tag-free OsDpe1 in a dosage-dependent manner (Fig. 2A). A reciprocal pulldown assay using HT-OsDpe1 as bait showed that it was able to bind to native OsPho1 present in developing seed extracts (Fig. 2B). Finally, the two enzymes were found in immunoprecipitates generated with affinity-purified anti-OsPho1 or anti-OsDpe1 antibodies (Fig. 2C). Moreover, the OsPho1-OsDpe1 complex is stable as evident by the large molecular weight species seen on a native polyacrylamide gel (Fig. 4A) and by their co-elution off of gel filtration columns (Fig. 4B) and co-migration on native glycogen-containing polyacrylamide gels (Fig. 4C). Overall, the results provide compelling evidence that the two enzymes interact to form a stable protein complex in both the recombinant and native states.
During our immunoblot analysis of OsPho1 and OsDpe1 in extracts from immature seeds, we observed that the OsDpe1 signal was much weaker than that of OsPho1 (Fig. 2C) even if similar titer values of antibodies were used for generating IPs and the subsequent immunoblot analysis. Quantitative immunoblot analyses showed that OsDpe1 is present at significantly lower levels than OsPho1 in developing rice seeds (Fig. 3), a result consistent with an estimated 6∼7-fold lower mRNA expression of OsDPE1 compared with OsPHO1 (31). The expression profiles of both genes are very similar in being initiated at an early stage of seed development and attaining their highest levels at 3∼5 DAF and drastically declining thereafter. The early activation of OsPHO1 and OsDPE1 is consistent with their involvement in the initiation event of starch biosynthesis in endosperm of rice seeds (26, 31). However, despite their highest expression levels at 3∼5 DAF, the protein levels of OsPho1 and OsDpe1 remained relatively unchanged in developing rice seeds at 7 DAF through 21 DAF (Fig. 3), indicating that these enzymes are fairly stable and not subjected to significant proteolytic turnover during seed development.
Estimation of the molecular weight of OsDpe1 by SEC using a Superdex-200 column revealed that the recombinant OsDpe1 elutes at 121 kDa (supplemental Fig. S4A). As the OsDPE1 complementary DNA (cDNA) open reading frame codes for a polypeptide of 62 kDa, OsDpe1 exists as a dimer, a quaternary structure identical to that seen for the homodimer OsPho1 containing subunits of ∼105 kDa. Stoichiometry studies indicate that these two homodimers assemble to form a multiprotein complex at a 1:1 ratio (supplemental Fig. S5) with a molecular size >620 kDa as estimated by SEC (supplemental Fig. S4B) and by its mobility on native polyacrylamide gels (Fig. 4A). Based on elution properties from SEC, a pair(s) of OsPho1 dimers assembles with a pair(s) of OsDpe1 dimers to form a higher order protein structure (supplemental Fig. S4B). In this higher order conformation state, both proteins were highly susceptible to proteolysis as evident by the polypeptide pattern exhibited by OsPho1 and OsDpe1 polypeptide on SDS-polyacrylamide gel and immunoblots (Fig. 2B).
Evidence for the presence of the OsPho1-OsDpe1 complex in developing rice seed extracts was also obtained. Despite substantial differences in the molecular sizes of OsPho1 (210 kDa) and OsDpe1, these proteins co-elute in high molecular weight protein fractions (fractions 11–14) from the Superdex-200 column (Fig. 4B). Further analysis of the chromatographic fractions by native polyacrylamide gel electrophoresis in the presence of 0.8% glycogen indicates co-migration of OsPho1 and OsDpe1 (Fig. 4C, fractions 23–25). On native gels containing glycogen, OsPho1 migrates as two bands with the faster moving polypeptide band containing OsDpe1. As OsPho1 binds weakly to glycogen (4), the slower moving, more abundant OsPho1 band may be due to this enzyme interacting with other proteins, which collectively retards its mobility in the presence of glycogen.
Dpe1 was initially viewed as an enzyme activity involved in starch degradation as it was capable of using G3, a MOS resistant to further degradation by amylases or phosphorylase (32, 33). Dpe1 converts G3 into G1 and G5 and vice versa (9, 25, 26). When the reaction time was extended, a range of different MOSs such as G6 and G7 were produced by the enzyme, indicating the enzyme also used its reaction products as substrates. These larger MOSs can then be acted upon by amylases and phosphorylases. Such a degradative role for Dpe1 in starch metabolism has been suggested in Arabidopsis and potato leaves. In an Arabidopsis dpe1-1 mutant lacking D-enzyme activity in leaves, no alteration of chain length distribution was observed for amylopectin compared with wild type, whereas MOS levels, especially of G3, drastically increased during starch degradation (34). Similar effects were observed from potato leaves where DPE1 expression was repressed (35).
Other studies have suggested a biosynthetic role for Dpe1 in starch metabolism. The potato Dpe1 is capable of transferring long α-glucan chains where amylopectin and amylose can serve as both donor and acceptor molecules (36, 37), although repression of D-enzyme expression did not affect starch content and amylopectin structure in potato tuber (38). In contrast, removal or decreased level of Dpe1 in Chlamydomonas led to an increase in the amylose to amylopectin ratio and an increased number of very small chains on amylopectin molecules, suggesting D-enzyme is required for normal amylopectin synthesis (39). Repression or overexpression of the rice Dpe1 significantly altered the size, morphology, and crystallinity of starch granules in rice seeds (40), indicating that this enzyme OsDpe1 plays an important role in starch synthesis despite being at low amounts in developing rice seeds.
We demonstrated earlier that L80 peptide domain of OsPho1 is not important for catalysis and regulation of the enzyme (4). The L80 domain of OsPho1 is also not involved in protein-protein interaction for OsPho1 as both OsPho1 and OsPho1ΔL80 can efficiently bind OsDpe1 in rice seed extracts. It should be noted that our pulldown experiment failed to identify an interaction of OsPho1 with BEIIb and BEI even in the presence of 1 mm ATP, a result consistent with the Co-IP results for the maize Pho1 (ZmPho1). Co-IP experiments for wheat Pho1 showed that the enzyme interacts with BEIIb and BEI in endosperms harvested at 12–25 days after pollination (DAP) (14), whereas the interaction was not identified in endosperms at ∼15 DAP (13). A plausible explanation for the difference was that the protein expression of BEI, one of the Pho1interacting proteins potentially playing a bridge role for Pho1 and BEIIb interaction, does not occur until at least 20 DAP (13), and thus the interaction was not detected at the early-to-mid endosperm developmental stage. Likewise, the interaction of OsPho1 with BEI and BEIIb was not detected in young rice endosperms in this study. However, earlier studies reported that mRNA expression of rice BEI and BEIIb appears to be abundant from the early stage of endosperm development (31, 41). Thus, it is likely that BEI and BEIIb do not interact with OsPho1 despite abundant expression of all the aforementioned three enzymes in rice. Nevertheless, we cannot rule out a possibility that these two proteins may interact with OsPho1 in rice endosperms at the mid-to-late developmental stage as evident by the result for wheat endosperms (14).
We demonstrated earlier that the substrate affinity of OsPho1 for Glc1P, G5, and amylopectin was significantly increased by lowering the reaction temperature from 36 °C to 18 °C (4). Similar results were obtained for OsPho1 when the temperature was lowered from 30 °C to 20 °C (Table 1). Interestingly, the OsPho1-OsDpe1 complex showed enhanced substrate affinities at 30 °C similar to those measured at 20 °C, indicating that the interaction of OsPho1 with OsDpe1 eliminated the temperature effect on substrate binding properties. The enhanced binding to Glc1P at 30 °C by OsPho1 when complexed with OsDpe1 likely alters the structural conformation of OsPho1. Such structural changes in OsPho1 as well as in OsDpe1 are suggested by the increased sensitivities of these enzymes to proteolysis in the complex form (Fig. 2B).
Both enzymes in the OsPho1-OsDpe1 complex were found to cooperate to more efficiently synthesize longer MOSs as well as a larger distribution of MOSs (Fig. 6). The interaction with OsDpe1 also enables OsPho1 to potentially utilize MOSs smaller than G4, substrates that OsPho1 cannot directly extend. OsDpe1 can utilize G3 as a donor and acceptor of a maltosyl moiety where it will produce G5, which can be used by OsPho1, as well as glucose. OsPho1 effectively extends G5 to larger MOSs using Glc1P as substrate. Hence, the OsPho1-OsDpe1 complex can significantly enhance the synthesis of larger MOSs by utilizing more diverse substrates available in rice endosperm.
OsDpe1 plays an active role in starch synthesis and in starch granule formation in developing rice endosperm (40). We are currently undergoing a study on the physiological significance of OsPho1-OsDpe1 complex using transgenic OsDpe1 overexpressing and repressing lines generated in wild type and BMF136 lines (6). The study of such plant lines may provide insights on the roles of the OsPho1-OsDpe1 complex in initiating starch synthesis and how low growth temperature affects the plants when expression levels of OsPho1 and/or OsDpe1 are altered.
Experimental Procedures
Construction of Plasmid DNAs for Protein Expression
PCR was performed on pFN18A (Promega) with oligonucleotides Halo-Bm and Halo-NcSc (supplemental Table S1) using Pfu/Taq (1:1) DNA polymerases to amplify the HT insert. The PCR product was cut with NcoI and BamHI and cloned into pGEM58ZN (7). After the sequence was verified by DNA sequencing, the HaloTag® (HT) insert was re-cloned into the BamHI and NcoI sites of pSH481 (4) to make pSH582. The GFP sequence was removed by NcoI/HindIII from pSH280 (4) and cloned into pSH582 to make pSH583, which contains a HaloTag·GFP fusion (supplemental Fig. S2). Under UV light (365 nm) the E. coli cells harboring this plasmid emitted a strong green fluorescence, indicating that the HT-GFP fusion protein is abundantly produced and active.
A DNA fragment containing Pho1 sequence was PCR-amplified from SH407 (7) using Pho1-ATG and 274seq-r primers (supplemental Table S1). After digestion with NcoI and PstI (made blunt by Pfu DNA polymerase), the Pho1 DNA was cloned in-frame downstream of the HT sequence of pSH583 digested with NcoI and HindIII (made blunt by Pfu DNA polymerase) by replacing the GFP sequence to make pSH585. A plasmid DNA (pSH642) carrying HT-OsPho1ΔL80 was also constructed from pSH489 (4) in the same way. The cloned OsPho1 and OsPho1ΔL80 sequences were verified by DNA sequencing. These plasmids, pSH585 and pSH642, were introduced into E. coli EA3457 cells (7) to produce HT-OsPho1 and HT-OsPho1ΔL80, respectively. To test if the OsPho1 in the fusion proteins still retains catalytic activity, we measured Pho1 activity after protein expression after cell disruption using sonication. Both HT fusions retained catalytic activities at levels similar to that measured for the wild type enzyme (Fig. 1A).
Protein Purification
HT-tagged GFP, OsPho1, and OsPho1ΔL80 and His6-tagged OsPho1 and OsPho1ΔL80 (4, 7) proteins overexpressed in E. coli EA3457 cells were purified using TALON Superflow (GE Healthcare), a cobalt-based IMAC resin, and DEAE-Sepharose FF (GE Healthcare) to near homogeneity as described previously (4, 42, 43). Phosphorylation status of the purified recombinant proteins (2 μg each) were examined using Pro-Q Diamond phosphoprotein gel stain (Invitrogen) after being resolved by SDS-PAGE according to the manufacturer's instructions.
Preparation of Soluble Proteins from Immature Rice Seeds
Immature seeds from Oryza sativa L. cv. Taichung-65 (TC65) and mutant BMF136 (6) grown in an environment-controlled chamber at Washington State University were harvested at 10 days after flowering (DAF) and frozen at −80 °C before use. About 15 g of frozen seeds were placed in a chilled mortar, and 15 ml of protein extraction buffer containing 50 mm HEPES-NaOH, pH 7.4, 150 mm NaCl, 0.05% (v/v) Nonidet P-40, 1× proteinase inhibitors (Roche Applied Science) was added. The seeds were squeezed by using a pestle chilled in liquid nitrogen to avoid extraction of unnecessary proteins from the outer seed coats. Afterward, the seed extracts were cleared by successive centrifugations at 800 × g for 5 min at 4 °C and then at 17,500 × g for 10 min at 4 °C.
Pulldown Experiment of OsPho1 and OsPho1ΔL80
OsPho1 or OsPho1ΔL80 was coupled to HaloLink resin as follows. 100 μl of HaloLink resin (Promega) were washed twice with 1 ml of binding buffer containing 50 mm HEPES-NaOH, pH 7.4, 150 mm NaCl, and 0.05% v/v Nonidet P-40, and the wash solution was removed by centrifugation of the resin at 800 × g for 1 min. The Halolink resin was then placed in 100 μl of washing buffer containing 50 mm HEPES-NaOH, pH 7.4, 150 mm NaCl, 0.05% v/v Nonidet P-40, and 1 mg/ml bovine serum albumin (fraction V) followed by the addition of 150 μg each of the purified recombinant proteins, HT-GFP, HT-OsPho1, or HT-OsPho1ΔL80. After mixing for 30 min at room temperature, the resins/proteins were washed twice with 1 ml of washing buffer and then twice with 1 ml of binding buffer to remove uncoupled HT-tagged proteins.
Chromatographic pulldown assays were performed by incubating immobilized HaloLinked enzymes with rice developing seed extracts. 1 ml of developing rice seed extracts (∼3.5 mg of proteins) were incubated with the HaloLinked enzyme resins, and the samples were incubated on a rotating platform for 2 h in the presence or absence of 1 mm ATP to test if protein capture is dependent on protein phosphorylation by endogenous kinases present in the cell extract. The mixture was then washed twice with 1 ml of washing buffer and twice with 1 ml of binding buffer. Bound proteins were then eluted with 50 μl of 8 m urea in 50 mm Tris-HCl, pH 7.5, for 30 min at room temperature. After centrifugation at 800 g for 5 min, the resulting supernatant was mixed with equal volumes of deionized water and 3× SDS protein sample buffer (150 mm Tris-HCl, pH 6.8, 6% w/v SDS, 30% v/v glycerol, 6% v/v β-mercaptoethanol, 0.03% w/v bromphenol blue). After denaturation by boiling, the proteins were then resolved on 12% SDS-polyacrylamide gels and visualized by silver staining.
Purification of Multiprotein Complexes Using Immobilized Metal Affinity Chromatography Medium
750 μl of a cobalt-based TALON Superflow (GE Healthcare) medium was prepared by washing 4 times with PI buffer containing 100 mm HEPES-NaOH, pH 7.4, and 150 mm NaCl and then adjusting the volume to 1.5 ml with the same buffer. 100 μl each of the pre-equilibrated resin was added to 14 1.7-ml tubes containing PI buffer (4 tubes), His6-OsPho1 (5 tubes containing 10 μg each), or His6-OsPho1ΔL80 (5 tubes containing 10 μg each). A tube from each group was incubated with PI buffer, partially purified GFP (10 μg and 50 μg), or OsDpe1 (10 μg and 50 μg). After incubation for 5 min at room temperature with occasional mixing, the resin was washed three times with 0.9 ml of PI buffer. The supernatant was carefully discarded, and the sample volume was adjusted to 100 μl with PI buffer. 30 μl of 2 × SDS sample buffer was added to the tube and boiled for 3 min. After centrifugation at 800 × g for 1 min, 10 μl of the supernatant was recovered and subjected to SDS-PAGE.
Native PAGE, SDS-PAGE, and Silver Staining
SDS-PAGE analysis of proteins was performed with 12% SDS-polyacrylamide gels as described (44). Native PAGE for recombinant proteins was performed using 10% native polyacrylamide gels with Tris-Tricine buffer system (45) without SDS. Native PAGE for zymogram analysis was performed using 7.5% non-denaturing polyacrylamide gels containing 0.8% oyster glycogen (Sigma) with running buffer (25 mm Tris, 192 mm glycine). Silver staining of the gels was performed (46) with some modifications as described below. The SDS-PAGE gel was briefly washed in water and incubated for 30 min in ethanol:acetic acid:water (4:1:5) followed by 30 min in sensitizing solution (500 mm sodium acetate, 0.8 mm sodium thiosulfate, 30% ethanol). The gel was washed 3 times for 5 min in water, incubated 20 min in the dark in 0.6 mm silver nitrate, 0.04% formaldehyde (37%), and washed twice 1 min in water. Staining was done 2–15 min with 283 mm Na2CO3, 0.02% formaldehyde, and stopped with either 5% acetic acid or 50 mm EDTA.
LC-MS/MS
In-gel digestion of proteins was performed as described previously (47). The protein bands of interest were excised from the silver-stained gels and cut into cubes (∼1 × 1 mm) using a razor blade. The gel pieces were placed in a siliconized microcentrifuge tube, destained with 40 μl of 15 mm potassium ferricyanide and 50 mm sodium thiosulfate, washed 4 times with deionized water, and dehydrated with acetonitrile for 30 min. The samples were dried in centrifugal vacuum concentrator and incubated with 40 μl of 10 mm DTT in 100 mm NH4HCO3 for 45 min at 56 °C. The liquid was replaced by 40 μl of 55 mm iodoacetamide in 100 mm NH4HCO3, and the sample was kept for 30 min at room temperature in the dark. The sample was then washed once with 100 mm NH4HCO3 for 10 min and then dehydrated and dried as described above. 40 μl of the digestion solution containing 50 mm NH4HCO3, 5 mm CaCl2, and 15 ng/μl Trypsin Gold (Promega; Mass Spectrometry Grade) was added and incubated for 45 min on ice. The liquid phase was then removed and replaced with 10 μl of the same solution lacking the enzyme, and in-gel digestion was continued overnight at 37 °C. The samples were sent to the University of Idaho Mass Spectrometry Core Lab for protein identification by LC-MS/MS.
Cloning of OsDPE1 cDNA, Protein Expression, and Purification
OsDPE1 cDNA (AK318541, RGRC ID: J075088C22) was obtained from the Rice Genome Resource Center (RGRC, Tsukuba, Japan). ChloroP 1.1 (ChloroP 1.1 Server) was used to predict a transit peptide (37 amino acids) in OsDpe1, and PCR primers were designed to amplify the coding region of the mature form of OsDpe1. To amplify the 5′-untranslated region and coding sequence of OsDPE1 cDNA, the first round of PCR using Pfu DNA polymerase with the primers (DPE1-F and DPE1-Sal-R) (supplemental Table S1) was performed. The PCR products were then used as a template for the second round of PCR with PCR primers (DPE1-Bam-F and DPE1-Sal-R) to amplify the cDNA sequence encoding the mature OsDpe1 open reading frame. The PCR product was digested with BamHI and SalI and cloned into the expression vectors pSH280 and pSH659 (4) to construct pSH670 (for His6-OsDpe1) and pSH675 (for tag-less OsDpe1), respectively. An NcoI-HindIII fragment of pSH670 was cloned into pSH583 by replacing GFP sequence to construct pSH757 (HT-OsDpe1). DNA sequence was verified by sequencing with internal primers (supplemental Table S1). The tag-less GFP (pSH274A) (43), OsDpe1, and HT-OsDpe1 proteins were abundantly expressed in E. coli EA3457 cells harboring rare tRNA genes (4). A majority of soluble proteins was able to be extracted in the range of pH 7–7.5. GFP and OsDpe1 protein were partially purified at pH 7.5 using and were partially purified at pH 7.5 using DEAE-Sepharose chromatography (43), whereas HT-OsDpe1 was purified as described above for His6-tagged OsPho1 and OsPho1ΔL80 proteins (7, 43).
Antibody Production
The purified recombinant OsPho1 and OsDpe1 proteins were boiled in the presence of 0.1% SDS. The denatured proteins were then mixed with Sigma Adjuvant System® (Sigma), and a polyclonal antibody was generated after three periodic administrations of the antigen proteins (250 μg of protein each) into a rabbit housed at the Washington State University Office of the Campus Veterinarian. Cleared antiserum was obtained by centrifugation at 2000 × g for 5 min of the blood sample after being kept overnight at 4 °C. The antiserum was mixed slowly with increasing amounts of cold saturated (100%) ammonium sulfate solution (up to 60%) and centrifuged at 5000 × g for 10 min to precipitate the proteins. The pellet was resuspended in 0.5 original volumes of phosphate-buffered saline (PBS) supplemented with 50% glycerol, and aliquots were made and stored at −20 °C before use. The polyclonal antibodies against OsPho1 and OsDpe1 were specific to their respective antigens and displayed no cross-reactivity toward other proteins present in rice endosperm extracts (supplemental Fig. S3).
FACE
Enzyme reaction for OsDpe1 was performed at room temperature in a reaction mixture (100 μl) containing 50 mm HEPES-NaOH, pH 7, 4 mm maltooligosaccharide or glucose, and 0.5 μg of the purified OsDpe1. The enzyme reaction was stopped by mixing with 100 μl of 95% ethanol. 10 μl was taken from each sample and dried at 45 °C using the centrifugal vacuum concentrator (Eppendorf). The dried sample was resuspended in 10 μl of 200 mm 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS; Sigma) in 15% acetic acid and 10 μl of 1 m sodium cyanoborohydride in dimethyl sulfoxide. The mixture was incubated at 37 °C for 16 h in the dark and dried again at 45 °C for 2 h in the concentrator. The pellet was dissolved in 50 μl of FACE sample buffer containing 62.5 mm Tris-HCl, pH 6.8, and 15% glycerol. 10 μl of the resulting sample was used to resolve and measure ANTS-labeled oligosaccharides by resolved in a 31% polyacrylamide gel by electrophoresis at 30 mA and 4 °C. The gel contained 10% glycerol for better resolution, and two gel running buffers were used for better separation; the anode buffer contained 100 mm Tris base and 25 mm HCl, whereas the cathode buffer had 100 mm Tris base and 100 mm Tricine. The ANTS-labeled MOS standard mixture was also prepared in the same way with same molar amount of glucose (G1) through maltoheptaose (G7) without OsDpe1 enzyme. After electrophoresis, the gel was briefly washed with deionized water, and the image was taken at the wavelength of 306 nm using a CCD camera on Kodak Gel Logic 212 Imaging System. The image was converted to a negative, and the resolved oligosaccharides in the gel were quantified by using Kodak Molecular Imaging Software version 4.5.
Enzyme Assay
Unless otherwise indicated, all enzyme reactions were performed at room temperature. Pho1 activity was measured by the PAMb method as described previously (4, 7). One unit of enzyme activity is defined as the amount of enzyme that catalyzes the production of 1 μmol of product per minute. Kinetic parameters were calculated by fitting the data to the Hill equation (43) by using Kaleidagraph 4.5.
Purification of OsPho1-OsDpe1 Complex by SEC
The native molecular weights of proteins were estimated using a Superdex-200 column (1 × 30 cm; Amersham Biosciences) on DuoFlow FPLC system (Bio-Rad) with a SEC buffer containing 25 mm HEPES-NaOH, pH 7.4, 150 mm NaCl, and 5% glycerol at a flow rate of 1 ml/min. A high molecular weight gel filtration calibration kit (Sigma) containing thyroglobulin (669 kDa), apoferritin (450 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (45 kDa), and carbonic anhydrase (25 kDa) was used as the molecular mass standards. Purification of the OsPho1-OsDpe1 complex was accomplished by several rounds of SEC after mixing 2 proteins at a 1:1 molar ratio in the SEC buffer. The eluent was collected in a series of fractions on DuoFlow FPCL system. Size exclusion chromatography of soluble seed proteins was performed using Sephacryl S-400 column (1 × 50 cm) or Superdex-200 column (1 × 30 cm) using the same buffer system.
Immunoblot Analysis
Proteins separated by SDS-PAGE were electroblotted onto nitrocellulose membranes. Proteins resolved by native polyacrylamide gel (7.5%) electrophoresis were placed in 50 ml of SDS-PAGE running buffer (25 mm Tris base, 192 mm glycine, 0.1% SDS) and denatured briefly by microwaving until the solution just began to boil. After electroblotting, the nitrocellulose membranes were blocked in phosphate-buffered saline containing 5% skim milk, and the target proteins were incubated with anti-OsPho1 followed by goat anti-rabbit IgG (H+L) secondary antibody, HRP conjugate (Thermo Fisher Scientific). The antibody-antigen reaction was detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce) and Fujifilm Luminescent Image Analyzer LAS-3000.
Activity Staining of OsPho1
Two parts of the proteins samples were mixed with one part of the sample buffer (150 mm Tris-HCl, pH 6.8, 30% glycerol, 0.03% Bromophenol blue) and applied to 2 7.5% non-denaturing polyacrylamide gels containing 0.8% oyster glycogen. Gel electrophoresis was performed at 80 V for 2–3 h in the cold room. One gel was washed once with 20 ml of 100 mm MES-NaOH, pH 6.5, 0.05% soluble starch, and 20 mm Glc1P and incubated overnight in 20 ml of the same solution at room temperature with continuous mixing. The gel was briefly rinsed with water and placed in iodine solution containing 0.67% iodine and 3.33% potassium iodide until proper staining was achieved. The other gel was used for immunoblot analysis as described above.
Cross-linking of Antibodies to Protein A-agarose Beads
All steps were performed at 4 °C unless otherwise indicated. Antibody raised against OsPho1 was affinity-purified as described previously (48). In parallel, 25 μl of protein A-agarose slurry (Invitrogen) was centrifuged at 1000 × g for 1 min to remove the supernatant and was washed twice with 500 μl of PBS buffer. 300 μl (40 μg) of the purified antibody in PBS was incubated with the protein A-agarose beads overnight at 4 °C. The supernatant was discarded after centrifugation at 1000 × g for 1 min, and the unbound antibodies were removed by washing twice with 500 μl of PBS. Antibodies were cross-linked to the beads by incubating for 1 h with 100 μl of cross-linking solution containing 82 μl of PBS and 18 μl of 2.5 mm disuccinimidyl suberate in dimethyl sulfoxide at room temperature with constant mixing. After removal of the supernatant, non-cross-linked antibodies were removed by washing the resins twice with 50 μl of 100 mm glycine, pH 2.8, and twice with PBS buffer.
Co-immunoprecipitation
All steps were performed at 4 °C unless otherwise indicated. Protein A-agarose resins cross-linked with affinity-purified antibodies were washed twice with protein extraction buffer containing 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 2 mm ATP, 2 mm NaF, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor mixture. In parallel, 3 g of mid-stage developing seeds were ground in 3 ml of protein extraction buffer. Seed extracts were centrifuged sequentially at 1,500 × g and then 15,000 × g to remove starch and cell debris. 1 ml of the clarified seed extract was applied to the protein A resin cross-linked either Pho1 antibodies or IgG and incubated overnight at 4 °C with constant mixing. The mixture was then centrifuged at 1000 × g for 1 min to remove unbound proteins, and the resins were washed twice with 500 μl of wash buffer (protein extraction buffer minus Nonidet P-40) and once with 500 μl of PBS buffer to remove unbound proteins. Resins were pre-eluted with 20 μl of 100 mm glycine, pH 2.8, and eluted with 60 μl of 100 mm glycine, pH 2.8, for 10 min at room temperature. Samples were neutralized with 1 m Tris-HCl, pH 8.8, immediately after elution.
Author Contributions
S.-K. H. conducted most of the experiments, analyzed the results, and wrote most of the paper. K. K. conducted the co-immunoprecipitation experiment. S.-K. H., H. S., and T. W. O. conceived the idea for the project. S.-K. H., K. K., and H. S. wrote the paper with T. W. O.
Supplementary Material
Acknowledgments
We thank Lee Deobald for LC-MS/MS service and Salvinder Singh, Israel Santana, Colleen Kao, and Fabian Botero for technical assistance.
This work was supported in part by DE-FG02-12ER20216 from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy (to T. W. O, S.-K. H., and K. K.) and by project WNP00590, USDA-NIFA (United States Department of Agriculture National Institute of Food and Agriculture), and the Agricultural Research Center, College of Agricultural, Human, and Natural Resource Sciences, Washington State University. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1–S7.
- Glc1P
- glucose 1-phosphate
- SS
- starch synthase
- BE
- branching enzyme
- MOS
- malto-oligosaccharide
- IP
- immunoprecipitate/immunoprecipitation
- IMAC
- immobilized metal affinity chromatography
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CBB
- Coomassie Brilliant Blue
- FACE
- fluorophore-assisted carbohydrate electrophoresis
- SEC
- size exclusion chromatography
- DAP
- days after pollination
- HT
- HaloTag®
- DAF
- days after flowering
- cDNA
- complementary DNA
- ANTS
- 8-aminonaphthalene-1,3,6-trisulfonic acid.
References
- 1. Shimomura S., Nagai M., and Fukui T. (1982) Comparative glucan specificities of two types of spinach leaf phosphorylase. J. Biochem. 91, 703–717 [DOI] [PubMed] [Google Scholar]
- 2. Nakano K., and Fukui T. (1986) The complete amino acid sequence of potato α-glucan phosphorylase. J. Biol. Chem. 261, 8230–8236 [PubMed] [Google Scholar]
- 3. Mori H., Tanizawa K., and Fukui T. (1993) A chimeric α-glucan phosphorylase of plant type L and H isozymes: functional role of 78-residue insertion in type L isozyme. J. Biol. Chem. 268, 5574–5581 [PubMed] [Google Scholar]
- 4. Hwang S. K., Singh S., Cakir B., Satoh H., and Okita T. W. (2016) The plastidial starch phosphorylase from rice endosperm: catalytic properties at low temperature. Planta 243, 999–1009 [DOI] [PubMed] [Google Scholar]
- 5. Dauvillée D., Chochois V., Steup M., Haebel S., Eckermann N., Ritte G., Ral J. P., Colleoni C., Hicks G., Wattebled F., Deschamps P., d'Hulst C., Liénard L., Cournac L., Putaux J. L., Dupeyre D., and Ball S. G. (2006) Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48, 274–285 [DOI] [PubMed] [Google Scholar]
- 6. Satoh H., Shibahara K., Tokunaga T., Nishi A., Tasaki M., Hwang S. K., Okita T. W., Kaneko N., Fujita N., Yoshida M., Hosaka Y., Sato A., Utsumi Y., Ohdan T., and Nakamura Y. (2008) Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20, 1833–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hwang S. K., Nishi A., Satoh H., and Okita T. W. (2010) Rice endosperm-specific plastidial α-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Arch. Biochem. Biophys. 495, 82–92 [DOI] [PubMed] [Google Scholar]
- 8. Chen H. M., Chang S. C., Wu C. C., Cuo T. S., Wu J. S., and Juang R. H. (2002) Regulation of the catalytic behaviour of L-form starch phosphorylase from sweet potato roots by proteolysis. Physiol. Plant 114, 506–515 [DOI] [PubMed] [Google Scholar]
- 9. O'Neill E. C., Rashid A. M., Stevenson C. E. M., Hetru A. C., Gunning A. P., Rejzek M., Nepogodiev S. A., Bornemann S., Lawson D. M., and Field R. A. (2014) Sugar-coated sensor chip and nanoparticle surfaces for the in vitro enzymatic synthesis of starch-like materials. Chem. Sci. 5, 341–350 [Google Scholar]
- 10. Lin Y. C., Chen H. M., Chou I. M., Chen A. N., Chen C. P., Young G. H., Lin C. T., Cheng C. H., Chang S. C., and Juang R. H. (2012) Plastidial starch phosphorylase in sweet potato roots is proteolytically modified by protein-protein interaction with the 20S proteasome. PLoS ONE 7, e35336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Young G. H., Chen H. M., Lin C. T., Tseng K. C., Wu J. S., and Juang R. H. (2006) Site-specific phosphorylation of L-form starch phosphorylase by the protein kinase activity from sweet potato roots. Planta 223, 468–478 [DOI] [PubMed] [Google Scholar]
- 12. Crofts N., Abe N., Oitome N. F., Matsushima R., Hayashi M., Tetlow I. J., Emes M. J., Nakamura Y., and Fujita N. (2015) Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 66, 4469–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tetlow I. J., Beisel K. G., Cameron S., Makhmoudova A., Liu F., Bresolin N. S., Wait R., Morell M. K., and Emes M. J. (2008) Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol 146, 1878–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tetlow I. J., Wait R., Lu Z., Akkasaeng R., Bowsher C. G., Esposito S., Kosar-Hashemi B., Morell M. K., and Emes M. J. (2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16, 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hennen-Bierwagen T. A., Lin Q., Grimaud F., Planchot V., Keeling P. L., James M. G., and Myers A. M. (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149, 1541–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hennen-Bierwagen T. A., Liu F., Marsh R. S., Kim S., Gan Q., Tetlow I. J., Emes M. J., James M. G., and Myers A. M. (2008) Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 146, 1892–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu F., Makhmoudova A., Lee E. A., Wait R., Emes M. J., and Tetlow I. J. (2009) The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 60, 4423–4440 [DOI] [PubMed] [Google Scholar]
- 18. Liu F., Ahmed Z., Lee E. A., Donner E., Liu Q., Ahmed R., Morell M. K., Emes M. J., and Tetlow I. J. (2012) Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J. Exp. Bot. 63, 1167–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu F., Romanova N., Lee E. A., Ahmed R., Evans M., Gilbert E. P., Morell M. K., Emes M. J., and Tetlow I. J. (2012) Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. Biochem. J. 448, 373–387 [DOI] [PubMed] [Google Scholar]
- 20. Ahmed Z., Tetlow I. J., Ahmed R., Morell M. K., and Emes M. J. (2015) Protein-protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 233, 95–106 [DOI] [PubMed] [Google Scholar]
- 21. Abe N., Asai H., Yago H., Oitome N. F., Itoh R., Crofts N., Nakamura Y., and Fujita N. (2014) Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol. 14, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grimaud F., Rogniaux H., James M. G., Myers A. M., and Planchot V. (2008) Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 59, 3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura Y., Ono M., Utsumi C., and Steup M. (2012) Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins. Plant Cell Physiol. 53, 869–878 [DOI] [PubMed] [Google Scholar]
- 24. Subasinghe R. M., Liu F., Polack U. C., Lee E. A., Emes M. J., and Tetlow I. J. (2014) Multimeric states of starch phosphorylase determine protein-protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 83, 168–179 [DOI] [PubMed] [Google Scholar]
- 25. Suganuma T., Setoguchi S., Fujimoto S., and Nagahama T. (1991) Analysis of the characteristic action of D-enzyme from sweet-potato in terms of subsite theory. Carbohydr. Res. 212, 201–212 [Google Scholar]
- 26. Akdogan G., Kubota J., Kubo A., Takaha T., and Kitamura S. (2011) Expression and characterization of rice disproportionating enzymes. J. Appl. Glycosci. 58, 99–105 [Google Scholar]
- 27. Kakefuda G., and Duke S. H. (1989) Characterization of pea chloroplast D-enzyme (4-α-d-glucanotransferase). Plant Physiol. 91, 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu Y., Steichen J. M., Yao J., and Sharkey T. D. (2006) The role of cytosolic α-glucan phosphorylase in maltose metabolism and the comparison of amylomaltase in Arabidopsis and Escherichia coli. Plant Physiol. 142, 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu Y., and Sharkey T. D. (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218, 466–473 [DOI] [PubMed] [Google Scholar]
- 30. Lloyd J. R., Blennow A., Burhenne K., and Kossmann J. (2004) Repression of a novel isoform of disproportionating enzyme (stDPE2) in potato leads to inhibition of starch degradation in leaves but not tubers stored at low temperature. Plant Physiol. 134, 1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohdan T., Francisco P. B. Jr., Sawada T., Hirose T., Terao T., Satoh H., and Nakamura Y. (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56, 3229–3244 [DOI] [PubMed] [Google Scholar]
- 32. Okita T. W., and Preiss J. (1980) Starch degradation in spinach leaves: isolation and characterization of the amylases and R-enzymes of spinach leaves. Plant Physiol. 66, 870–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okita T. W., Greenberg E., Kuhn D. N., and Preiss J. (1979) Subcellular localization of the starch degradative and biosynthetic enzymes of spinach leaves. Plant Physiol. 64, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Critchley J. H., Zeeman S. C., Takaha T., Smith A. M., and Smith S. M. (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26, 89–100 [DOI] [PubMed] [Google Scholar]
- 35. Lütken H., Lloyd J. R., Glaring M. A., Baunsgaard L., Laursen K. H., Haldrup A., Kossmann J., and Blennow A. (2010) Repression of both isoforms of disproportionating enzyme leads to higher malto-oligosaccharide content and reduced growth in potato. Planta 232, 1127–1139 [DOI] [PubMed] [Google Scholar]
- 36. Takaha T., Yanase M., Takata H., Okada S., and Smith S. M. (1996) Potato D-enzyme catalyzes the cyclization of amylose to produce cycloamylose, a novel cyclic glucan. J. Biol. Chem. 271, 2902–2908 [DOI] [PubMed] [Google Scholar]
- 37. Takaha T., Yanase M., Takata H., Okada S., and Smith S. M. (1998) Cyclic glucans produced by the intramolecular transglycosylation activity of potato D-enzyme on amylopectin. Biochem. Biophys. Res. Commun. 247, 493–497 [DOI] [PubMed] [Google Scholar]
- 38. Takaha T., Critchley J., Okada S., and Smith S. M. (1998) Normal starch content and composition in tubers of antisense potato plants lacking D-enzyme (4-α-glucanotransferase). Planta 205, 445–451 [Google Scholar]
- 39. Colleoni C., Dauvill e D., Mouille G., Morell M., Samuel M., Slomiany M. C., Li nard L., Wattebled F., d'Hulst C., and Ball S. (1999) Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 120, 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dong X., Zhang D., Liu J., Liu Q. Q., Liu H., Tian L., Jiang L., and Qu le Q. (2015) Plastidial disproportionating enzyme participates in starch synthesis in rice endosperm by transferring maltooligosyl groups from amylose and amylopectin to amylopectin. Plant Physiol. 169, 2496–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang H., Dian W., and Wu P. (2003) Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 63, 53–59 [DOI] [PubMed] [Google Scholar]
- 42. Hwang S. K., Nagai Y., Kim D., and Okita T. W. (2008) Direct appraisal of the potato tuber ADP-glucose pyrophosphorylase large subunit in enzyme function by study of a novel mutant form. J. Biol. Chem. 283, 6640–6647 [DOI] [PubMed] [Google Scholar]
- 43. Hwang S.-K, Hamada S., and Okita T. W. (2007) Catalytic implications of the higher plant ADP-glucose pyrophosphorylase large subunit. Phytochemistry 68, 464–477 [DOI] [PubMed] [Google Scholar]
- 44. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 45. Schägger H. (2006) Tricine-SDS-PAGE. Nat. Protoc 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 46. Sørensen B. K., Højrup P., Østergård E., Jørgensen C. S., Enghild J., Ryder L. R., and Houen G. (2002) Silver staining of proteins on electroblotting membranes and intensification of silver staining of proteins separated by polyacrylamide gel electrophoresis. Anal. Biochem. 304, 33–41 [DOI] [PubMed] [Google Scholar]
- 47. Gharahdaghi F., Weinberg C. R., Meagher D. A., Imai B. S., and Mische S. M. (1999) Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605 [DOI] [PubMed] [Google Scholar]
- 48. Crofts A. J., Crofts N., Whitelegge J. P., and Okita T. W. (2010) Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 231, 1261–1276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.