Abstract
Downstream of receptor tyrosine kinase and G protein-coupled receptor (GPCR) stimulation, the phosphatidylinositol 3,4,5-trisphosphate (PIP3)-dependent Rac exchange factor (PREX) family of guanine nucleotide exchange factors (GEFs) activates Rho GTPases, leading to important roles for PREX proteins in numerous cellular processes and diseases, including cancer. PREX1 and PREX2 GEF activity is activated by the second messengers PIP3 and Gβγ, and further regulation of PREX GEF activity occurs by phosphorylation. Stimulation of receptor tyrosine kinases by neuregulin and insulin-like growth factor 1 (IGF1) leads to the phosphorylation of PREX1; however, the kinases that phosphorylate PREX1 downstream of these ligands are not known. We recently reported that the p21-activated kinases (PAKs), which are activated by GTP-bound Ras-related C3 botulinum toxin substrate 1 (Rac1), mediate the phosphorylation of PREX2 after insulin receptor activation. Here we show that certain phosphorylation events on PREX1 after insulin, neuregulin, and IGF1 treatment are PAK-dependent and lead to a reduction in PREX1 binding to PIP3. Like PREX2, PAK-mediated phosphorylation also negatively regulates PREX1 GEF activity. Furthermore, the onset of PREX1 phosphorylation was delayed compared with the phosphorylation of AKT, supporting a model of negative feedback downstream of PREX1 activation. We also found that the phosphorylation of PREX1 after isoproterenol and prostaglandin E2-mediated GPCR activation is partially PAK-dependent and likely also involves protein kinase A, which is known to reduce PREX1 function. Our data point to multiple mechanisms of PREX1 negative regulation by PAKs within receptor tyrosine kinase and GPCR-stimulated signaling pathways that have important roles in diseases such as diabetes and cancer.
Keywords: G protein-coupled receptor (GPCR); guanine nucleotide exchange factor (GEF); insulin; phosphatidylinositol 3-kinase (PI 3-kinase); Rac (Rac GTPase); receptor tyrosine kinase; signal transduction; phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 (PREX1); p-21-activated kinase (PAK)
Introduction
The Rho family of GTPases, part of the Ras superfamily, contains multiple subgroups, including the Ras-related C3 botulinum toxin substrate (Rac), cell division control protein 42 homolog (CDC42), and Rho-like GTPases (1). Rho GTPases exist in the cell either in an inactive, GDP-bound or an active, GTP-bound conformation. The cycling between these two conformational states can be regulated by guanine nucleotide exchange factors (GEFs),2 which promote the release of GDP and subsequent binding of GTP to activate the Rho GTPase (2). While in the GTP bound state, Rho GTPases can then bind to their downstream effectors, which activate many intracellular signaling pathways. For example, Rac1-GTP binds and activates the p21-activated kinases (PAKs), which are critical for cell motility, survival, and proliferation (3). Phosphatidylinositol 3,4,5-trisphosphate (PIP3)-dependent Rac exchanger (PREX1), along with its homolog PREX2, are Rho family GEFs (4–6). The ability of PREX proteins to activate Rho GTPases makes them important regulators of cell motility, proliferation, glucose uptake, and reactive oxygen species generation (7). PREX1 activates Rac1, Rac2, Rac3, and RhoG both in vitro and in cells, and also activates TC10 and CDC42 in vitro (4, 8–11).
The regulation of Rho GTPases occurs at the cell membrane, typically after the activation of a membrane-bound receptor (12). This is evident in the regulation of PREX1 and PREX2, as their GEF activity toward Rho GTPases is stimulated by PIP3 and by the Gβγ heterodimer, both of which are second messengers that are produced at the membrane after receptor activation (4–6). PIP3 can regulate the activity of other Rho GEFs at the membrane as well, including VAV and SOS1 (13, 14). PIP3 is the product of class I PI3K activation downstream of both receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCRs), and Gβγ is released following GPCR activation. Given that PREX1 and PREX2 are activated in the same manner, it is important to identify the similarities and differences in their physiological functions and regulatory mechanisms. Part of their physiological specificity likely arises from different protein expression patterns. PREX1 protein is strongly expressed in leukocytes and in the brain, and PREX2 protein is also strongly expressed in the brain in addition to other tissues but not in leukocytes (4, 5, 15, 16). In the brain, where both PREX2 and PREX1 are expressed, there does appear to be some redundancy in function. Prex2−/− mice show motor function deficiencies that are exacerbated in Prex1−/−Prex2−/− mice despite the fact that Prex1−/− mice do not show these defects (16). This suggests that PREX1 can partially compensate for the loss of PREX2 and that these two proteins can potentially work in similar pathways.
Given the importance of RTKs and GPCRs in mediating numerous signaling outputs in multiple cell types, PREX proteins are uniquely positioned to impact many cellular processes. Following the stimulation of certain RTKs, including the neuregulin-activated ErbB receptors, PREX1 regulates Rac activation, proliferation, adhesion, colony formation, and invasion of breast cancer cells (17–19). In breast cancer cell lines with HER2 amplification or PIK3CA-activating mutations, PREX1 is necessary for activation of ERK signaling through a Rac and PAK-dependent mechanism (20). PREX1 also has a role in Rac-mediated glucose uptake in mouse adipocytes downstream of the insulin receptor (21). Prex2−/− mice have deficient insulin signaling and decreased glucose uptake, likely because of the dual role of PREX2 as both a Rac GEF and as an inhibitor of PTEN phosphatase activity (15, 22). Interestingly, PREX1 does not bind PTEN and does not appear to affect PTEN function in cells (15). Furthermore, many of the known PREX1 physiological effects are due to its role in GPCR signaling pathways. For example, in neutrophils and macrophages, PREX1 is important for GPCR-dependent Rac and RhoG activation to regulate reactive oxygen species formation and cell migration (8–10).
PREX1 also has a role in tumorigenesis and cancer metastasis. PREX1 is overexpressed in a variety of human cancers, including melanoma and breast, prostate, and colon cancer (17, 23–26). PREX1 knockdown decreases tumor growth in xenograft mouse models of cancer using breast cancer cell lines (17, 18). Additionally, PREX1 appears to have a specific role in metastasis. The expression of PREX1, but not a GEF-dead mutant, induces spontaneous metastasis in a xenograft prostate cancer mouse model (24), and PREX1 inactivation in mice reduces metastasis in a melanoma transgenic mouse model (23). PREX2 is also overexpressed in numerous cancer types and is frequently mutated in cancer, with especially high mutation rates in melanoma (25–27). PREX2 mutations found in cancer activate its GEF activity and promote tumorigenesis in a melanocyte xenograft mouse model (27, 28). Furthermore, certain mutations can also evade PTEN-mediated inhibition of PREX2-driven breast cancer cell invasion (29).
Although PREX1 is clearly important for the physiological outputs of receptor-activated signaling and in cancer, important questions remain regarding how its GEF activity is regulated downstream of receptor activation, particularly by phosphorylation. It is known that PREX1 is phosphorylated in the cell, demonstrated by the fact that after gel electrophoresis, it is detected as multiple bands that are collapsed by both λ-phosphatase and protein phosphatase 1α (PP1α) (30, 31). λ-Phosphatase and PP1α dephosphorylation of PREX1 increase GEF activity, which is consistent with reports showing that PKA phosphorylation of PREX1 leads to a reduction in its in vitro GEF activity both in vitro and in cells (31, 32, 49). Importantly, PREX1 is phosphorylated after the activation of specific RTKs and GPCRs, indicating that the regulation of PREX1 by phosphorylation has a role in these signaling pathways. The stimulation of β-adrenergic GPCRs with isoproterenol results in the phosphorylation of PREX1, which is likely due to the activation of PKA downstream of the β-adrenergic receptor (31). Similarly, downstream of sphingosine 1-phosphate, PKA phosphorylates PREX1 to regulate an intramolecular interaction that reduces PREX1 GEF activity (49). Additionally, PREX1 is phosphorylated after the stimulation of RTKs with neuregulin, insulin-like growth factor 1 (IGF1), platelet-derived growth factor, and fibroblast growth factor, but the kinases that are responsible for these phosphorylation events are not known (18, 19).
We recently identified PAKs as mediators of the phosphorylation of PREX2 after insulin receptor activation. PAK-dependent phosphorylation of PREX2, which occurs downstream of Rac activation, reduces PIP3 and Gβγ binding as well as PIP3 and Gβγ-stimulated PREX2 GEF activity, suggesting that these phosphorylation events are part of a negative feedback circuit to reduce Rac activation after insulin stimulation (33). In this study, we found that PREX1 is phosphorylated after insulin treatment of breast cancer cells and that insulin, neuregulin, and IGF1-mediated phosphorylation of PREX1 are PI3K- and PAK-dependent. Importantly, phosphorylated PREX1 does not efficiently bind to PIP3, and the activation of PREX1 GEF activity by PIP3 is reduced when PREX1 is phosphorylated in cells that co-express PAK1. Last, we identified a GPCR ligand, prostaglandin E2 (PGE2), that can induce PREX1 phosphorylation, and we found that PGE2 and isoproterenol both stimulate PAK-dependent phosphorylation events on PREX1. Altogether, we show that PREX1 and PREX2 are both similarly regulated by phosphorylation downstream of RTKs through a PAK-dependent mechanism, demonstrating that negative feedback on these GEFs could have a significant impact on Rho GTPase function.
Results
PREX1 Is Phosphorylated through a PI3K-dependent Mechanism Downstream of Receptor Tyrosine Kinase Activation
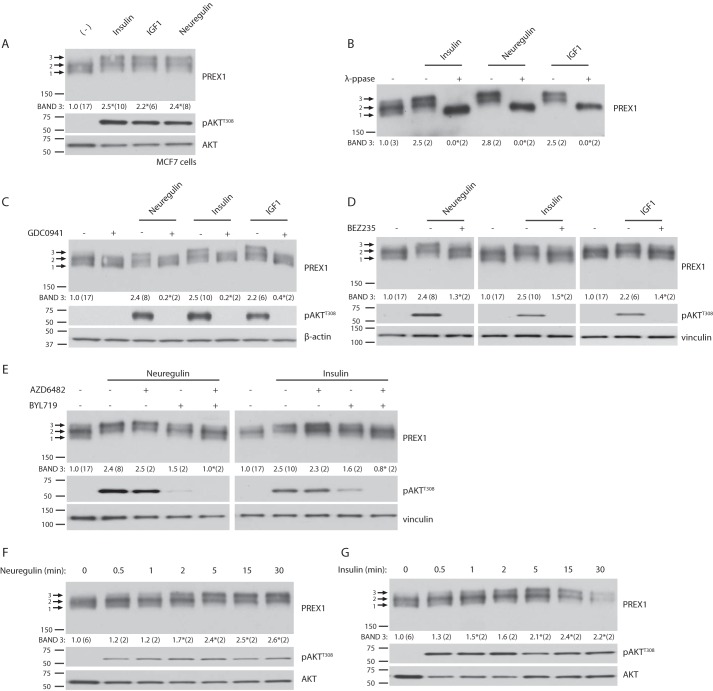
PREX1 is phosphorylated downstream of neuregulin and IGF1 (18, 19), and multiple studies have demonstrated that phosphorylation causes an electrophoretic mobility shift in PREX1 (18, 30, 31). In MCF7 breast cancer cells, we found that insulin also caused a PREX1 mobility shift (Fig. 1A). PREX1 can be comprised of three bands when gel electrophoresis is performed on MCF7 cell lysates with a 4% polyacrylamide gel, and, in the starved state, the bottom band (Fig. 1A, 1) and the middle band (Fig. 1A, 2) are enriched. However, treatment with insulin, IGF1, or neuregulin reduced the amount of the bottom, fastest migrating band (Fig. 1A, 1) and significantly enriched the upper, slowest migrating band (Fig. 1A, 3). Furthermore, treatment with λ-phosphatase ablated the RTK-induced electrophoretic mobility shift, demonstrating that these changes to PREX1 were the result of phosphorylation (Fig. 1B). λ-Phosphatase treatment also compressed band 2, which is seen in the starved state, indicating that steady-state phosphorylation is present on PREX1. However, from these data it is unclear whether the RTK-induced band shift (to band 3) is a result of a single or multiple phosphorylation events.
FIGURE 1.
Receptor tyrosine kinases regulate the phosphorylation of PREX1 through a PI3K-dependent mechanism. For all quantified Western blots, the ratio of the intensity of band 3 by densitometry to that of total PREX1 protein was calculated, and the number of independent samples used for each quantification is shown in parenthesis. A, Western blotting analysis of MCF7 cells that were starved and then treated with 5 μg/ml insulin, 50 ng/ml IGF1, or 10 nm neuregulin for 30 min. Arrows indicate the three different PREX1 bands in MCF7 cells. Band 3 was quantified as described above. *, p < 0.05 compared with the starved sample. B, Western blotting analysis of λ-phosphatase (λ-ppase)- or mock-treated cell lysates from MCF7 cells that were treated with insulin, neuregulin, or IGF1 for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding mock-treated sample for that treatment. C, Western blotting analysis of MCF7 cells that were starved and then treated with 500 nm GDC0941 (PI3K inhibitor) for 15 min, followed by treatment with neuregulin, insulin, or IGF1 for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding ligand treatment without GCD0941. D, Western blotting analysis of MCF7 cells that were starved and then treated with 500 nm BEZ235 (PI3K/mTOR inhibitor) for 15 min, followed by treatment with neuregulin, insulin, or IGF1 for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding ligand treatment without BEZ235. E, Western blotting analysis of MCF7 cells that were starved and then treated with 1 μm AZD6482 (PI3Kβ inhibitor) and/or 1 μm BYL719 (PI3Kα inhibitor) for 15 min, followed by treatment with neuregulin or insulin for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding ligand treatment without AZD6482 or BYL719. F, Western blotting analysis of MCF7 cells that were starved and then treated with neuregulin for the indicated time periods. Band 3 was quantified as described above. *, p < 0.05 compared with the starved sample. G, Western blotting analysis of MCF7 cells that were starved and then treated with insulin for the indicated time periods. Band 3 was quantified as described above. *, p < 0.05 compared with the starved sample. A phospho-specific antibody to AKT (Thr-308) was used to detect signaling alterations downstream of receptor tyrosine kinase ligands and small molecule inhibitors.
It is currently unknown which kinases are regulating PREX1 phosphorylation downstream of RTK activation. We found that neuregulin, insulin, and IGF1-dependent PREX1 phosphorylation was sensitive to PI3K inhibition after treatment with either GDC0941, a pan-class I PI3K inhibitor, or BEZ235, a class I PI3K/mTOR dual inhibitor (Fig. 1, C and D). This suggests that the kinase that is regulating PREX1 is downstream of PI3K-dependent PIP3 production. Interestingly, our experiments show that the phosphorylation of PREX1 is not as sensitive to BEZ235 as it is to GDC0941, as the induced upper band is still faintly present after BEZ235 treatment, whereas the upper band is completely gone after treatment with GDC0941. Similarly, we previously reported that the phosphorylation of PREX2 downstream of insulin is more sensitive to GDC0941 than to BEZ235 (33). The reduced sensitivity of PREX phosphorylation to BEZ235 could be because BEZ235 is a less potent inhibitor of PI3Kβ, and PI3Kβ can help to maintain higher PIP3 levels when PI3Kα is efficiently inhibited. This is supported by the fact that the phosphorylation of PREX1 was more sensitive to a combination of a PI3Kβ and a PI3Kα inhibitor than it was to either drug alone (Fig. 1E). Collectively, these data demonstrate that the RTK-mediated phosphorylation events that regulate PREX1 require PI3K activation.
Receptor Tyrosine Kinase-dependent Phosphorylation of PREX1 Is Regulated by PAKs
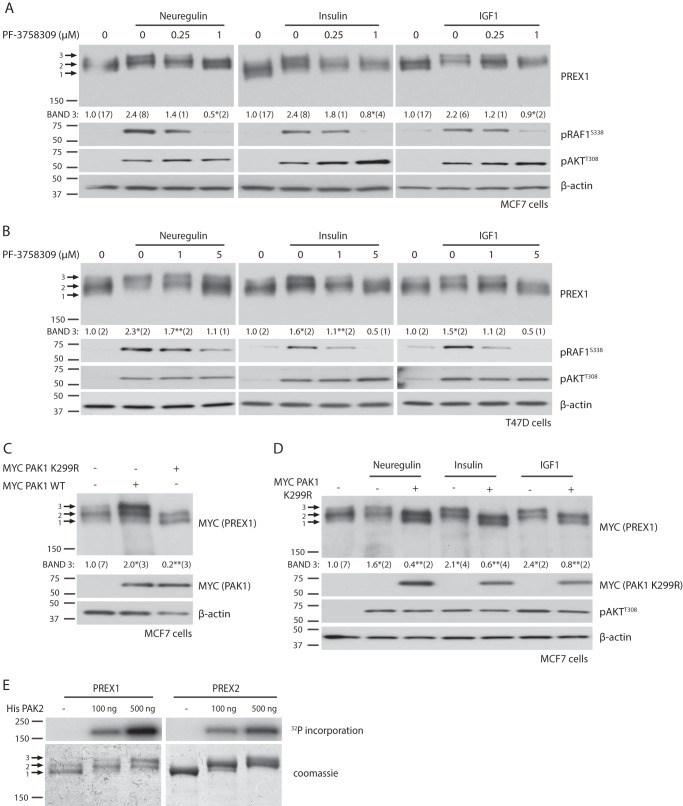
We then analyzed the kinetics of PREX1 phosphorylation in MCF7 cells after either neuregulin or insulin stimulation and found that the phosphorylation of PREX1 was very low for the first minute after stimulation and did not reach peak levels of phosphorylation until 5–15 min (Fig. 1, F and G). AKT, which is activated by PIP3 production, was significantly phosphorylated within the first 30 s. This time course of PREX1 phosphorylation is consistent with the time course of PAK-dependent PREX2 phosphorylation downstream of insulin receptor activation (33). Additionally, insulin-mediated phosphorylation of PREX2 also requires PI3K, and because of these similarities between PREX2 and PREX1, we tested whether PREX1 phosphorylation downstream of RTKs is also mediated by PAKs. Indeed, the pan-PAK inhibitor PF-3758309 reduced the neuregulin-, insulin-, and IGF1-dependent phosphorylation of PREX1 in MCF7 cells (Fig. 2A). Phosphorylation of the PAK substrate RAF1 at Ser-338 was used in this experiment to show that the PAKs were stimulated by RTK activation and that the inhibitor efficiently blocked PAK function. We also tested the effect of PAK inhibitors on PREX1 electrophoretic mobility in T47D breast cancer cells, a cell line where PREX1 is important for Rac activation in response to neuregulin, EGF, and TGFα (17). Similar to what we observed in MCF7 cells, the enrichment of a slower migrating band of PREX1 that was induced by neuregulin, insulin, and IGF1 was sensitive to the inhibition of PAKs with PF-3758309 (Fig. 2B). Co-expression of PREX1 and PAK1 in starved MCF7 cells caused the same type of electrophoretic band shift that was seen after treatment with RTK agonists, and co-expression of the kinase-dead, dominant negative K299R mutant of PAK1 caused an enrichment of the lowest, presumably dephosphorylated PREX1 band (Fig. 2C). The K299R PAK1 mutant also prevented neuregulin-, insulin-, and IGF1-mediated phosphorylation of PREX1 (Fig. 2D), providing further evidence that PAKs are regulating the phosphorylation of PREX1 downstream of RTK activation. After finding that PAKs could regulate PREX1 phosphorylation in cells, we tested whether PREX1 was a direct substrate of PAKs. Previous studies from our laboratory have demonstrated that PREX2 is phosphorylated in vitro by recombinant PAK2 (33), which is constitutively active when produced in bacteria (34). In an in vitro kinase assay, PREX1 was also phosphorylated by PAK2 in a dose-dependent manner, and the levels of phosphorylation were comparable with those seen with PREX2 (Fig. 2E). In addition, in vitro phosphorylation of PREX1 by PAK2 resulted in electrophoretic mobility shifts in PREX1 at both the lower and higher doses of PAK2, as shown by Coomassie staining. This indicates that PAKs contribute to the PREX1 mobility shifts that are seen in cells in response to RTK activation. Taken together, these data implicate PAKs as important mediators of PREX1 phosphorylation downstream of receptor tyrosine kinases.
FIGURE 2.
PAKs mediate receptor tyrosine kinase-dependent phosphorylation of PREX1. For all quantified Western blots, the ratio of the intensity of band 3 by densitometry to that of total PREX1 (or MYC-tagged PREX1) protein was calculated, and the number of independent samples used for each quantification is shown in parenthesis. A, Western blotting analysis of MCF7 cells that were starved and treated with the indicated concentrations of PF-3758309 (PAK inhibitor) for 15 min, followed by treatment with 10 nm neuregulin, 5 μg/ml insulin, or 50 ng/ml IGF1 for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding ligand treatment without PF-3758309. B, Western blotting analysis of T47D cells that were starved and treated with the indicated concentrations of PF-3758309 (PAKi) for 15 min, followed by treatment with neuregulin, insulin, or IGF1 for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the starved sample; **, p < 0.05 compared with the corresponding ligand treatment without PF-3758309. C, Western blotting analysis of MCF7 cells co-expressing MYC PREX1 and either MYC PAK1 WT or MYC PAK1 K299R. Cells were starved before being harvested. Band 3 was quantified as described above. *, p < 0.05 compared with the sample with no PAK expression; **, p < 0.05 compared with the WT PAK-expressing sample. D, Western blotting analysis of MCF7 cells expressing MYC PREX1 with or without co-expression of MYC PAK1 K299R. Cells were starved and treated with neuregulin, insulin, or IGF1 for 30 min. Band 3 was quantified as described above. *, p < 0.05 compared with the starved sample; **, p < 0.05 compared with the corresponding ligand treatment without MYC PAK1 K299R co-expression. E, in vitro kinase assay with purified His PAK2 and MYC PREX1 or V5 PREX2 isolated on either MYC or V5-agarose beads. Incorporation of [γ-32P]ATP was analyzed. A representative exposure is shown of two independent experiments. Phosphospecific antibodies to AKT (Thr-308) and RAF1 (Ser-338) were used to detect signaling alterations downstream of receptor tyrosine kinase ligands and small molecule inhibitors.
Phosphorylation of PREX1 Reduces PIP3 Binding and PIP3-mediated GEF Activation
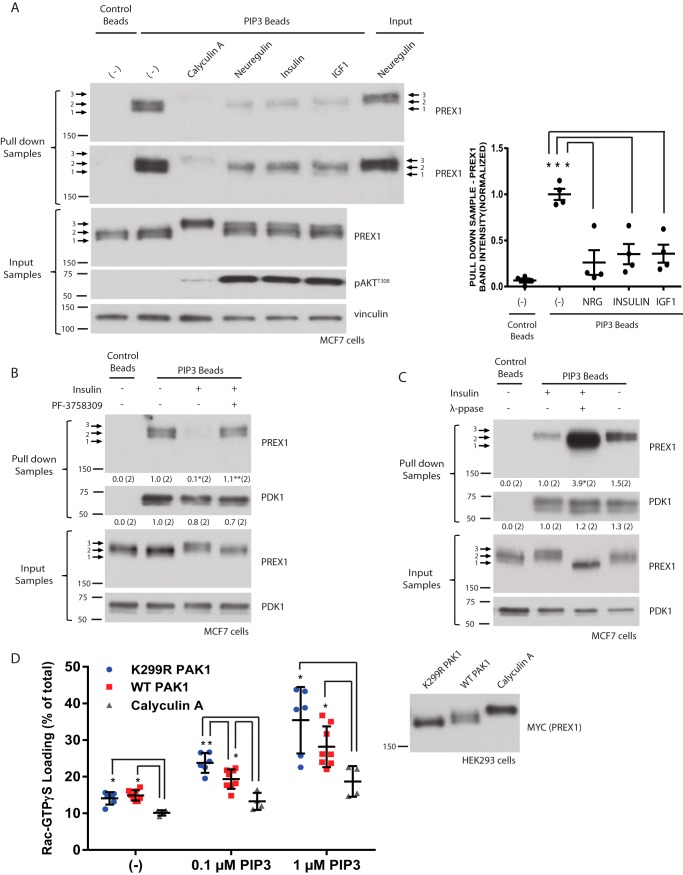
We then sought to determine the functional consequences of RTK-dependent phosphorylation of PREX1. We have shown previously that insulin-mediated phosphorylation of PREX2 reduces binding to PIP3 to prevent PIP3-dependent activation of GEF activity (33). Furthermore, multiple studies on PREX1 have shown that phosphorylation can affect GEF activity. Dephosphorylation of PREX1 with either λ-phosphatase or PP1α increases GEF activity, and PKA-mediated phosphorylation of PREX1 reduces both the binding of PREX1 to Gβγ and Gβγ-dependent activation of PREX1 GEF activity (30–32). By performing PIP3 pulldowns in MCF7 cells, we found that treatment with neuregulin, insulin, or IGF1 for 30 min reduced the total amount of PREX1 that bound to PIP3 (Fig. 3A). A longer exposure of the Western blot revealed that the upper or slowest migrating PREX1 band that is induced by RTK activation did not efficiently bind to PIP3 despite being enriched in the input sample, providing further evidence that RTK-dependent phosphorylation reduces PREX1 binding to PIP3. Complete enrichment of the upper PREX1 band after treatment with calyculin A, a PP1α and PP2A phosphatase inhibitor, resulted in a further reduction in PREX1 binding to PIP3. To test whether this decrease in PIP3 binding was PAK-dependent, we used the PAK inhibitor PF-3758309 and found that it reversed the insulin-induced reduction in binding to PIP3 (Fig. 3B). The PIP3 binding of phosphoinositide-dependent kinase-1 (PDK1), a well established PIP3-interacting protein, was unaffected by insulin treatment or PAK inhibition. Furthermore, abolishing the insulin-induced phosphorylation of endogenous PREX1 with in vitro λ-phosphatase treatment also rescued PREX1 PIP3 binding, whereas λ-phosphatase had no effect on the binding of PDK1 to PIP3 (Fig. 3C).
FIGURE 3.
Phosphorylation of PREX1 reduces PIP3 binding and in vitro GEF activity. A, Western blotting analysis of PIP3 bead pulldowns from MCF7 cells that were starved and treated with 10 nm neuregulin, 5 μg/ml insulin, 50 ng/ml IGF1, or 100 nm calyculin A for 30 min. A representative blot is shown of four independent experiments. Right panel, quantification of PREX1 bound to the PIP3 beads for each condition. The ratio of PREX1 band intensity for the pulldown sample over the PREX1 band intensity for the input was calculated for each sample. These values were then normalized to the average ratio for the starved/PIP3 bead samples. *, p < 0.05 compared to the starved/PIP3 bead sample. B, Western blotting analysis of PIP3 bead pulldowns from MCF7 cells that were starved and treated with insulin or 1 μm of PF-3758309 (PAKi) for 15 min, followed by treatment with insulin. A representative blot is shown of two independent experiments. The ratio of PREX1 or PDK1 band intensity for the pulldown sample over the band intensity for the input was calculated for each sample. These values were then normalized to the average ratio for the starved/PIP3 bead samples, and these values are shown below the corresponding pulldown blot. *, p < 0.05 compared with the starved/PIP3 bead sample; **, p < 0.05 compared with the insulin/PIP3 bead sample. C, Western blotting analysis of PIP3 bead pulldowns from λ-phosphatase- or mock-treated cell lysates from MCF7 cells that were starved and then treated with insulin. A representative blot is shown of two independent experiments. The ratio of PREX1 or PDK1 band intensity for the pulldown sample over the band intensity for the input was calculated for each sample. These values were then normalized to the average ratio for the insulin/mock-treated samples, and these values are shown below the corresponding pulldown blot. *, p < 0.05 compared with the insulin/mock-treated sample. D, MYC PREX1 was purified from HEK293 cells co-expressing MYC PAK1 WT or K299R or from HEK293 cells that were treated with 100 nm calyculin A for 30 min (a representative Western blot is shown in the right panel). The ability of increasing doses of PIP3 to stimulate PREX1 GEF activity toward GST Rac1 was assessed in an in vitro GEF assay. Data are mean ± S.D. for at least two experiments with samples done in duplicate at each PIP3 concentration. *, p < 0.05 by t test.
Given that PIP3 is critical for PREX1 activation during RTK signaling, we then used an in vitro Rac1 activation assay to test whether the phosphorylated form of PREX1 that is induced after RTK stimulation also shows reduced PIP3-mediated activation of GEF activity. Compared with dephosphorylated PREX1 that was purified from HEK293 cells co-expressing dominant negative PAK1, phosphorylated PREX1 that was purified from calyculin A-treated cells had significantly reduced PIP3-stimulated GEF activation (Fig. 3D). This is consistent with previous reports showing that dephosphorylation of PREX1 by protein phosphatases activates GEF activity (30, 31). In addition, PREX1 that was purified from cells co-expressing PAK1 showed reduced GEF activity at multiple PIP3 concentrations compared with dephosphorylated PREX1, with a significant difference in GEF activation in the presence of 0.1 μm PIP3. Taken together, it appears that the phosphorylation of PREX1 downstream of RTK activation and PAKs reduces binding to PIP3, and PAK-mediated phosphorylation decreases PIP3-dependent stimulation of GEF activity. When combined with the delayed time course of phosphorylation shown in Fig. 1, F and G, these data suggest that the phosphorylation of PREX1 is negatively regulating GEF activity at later time points after stimulation, potentially as part of feedback to turn off GTPase activation.
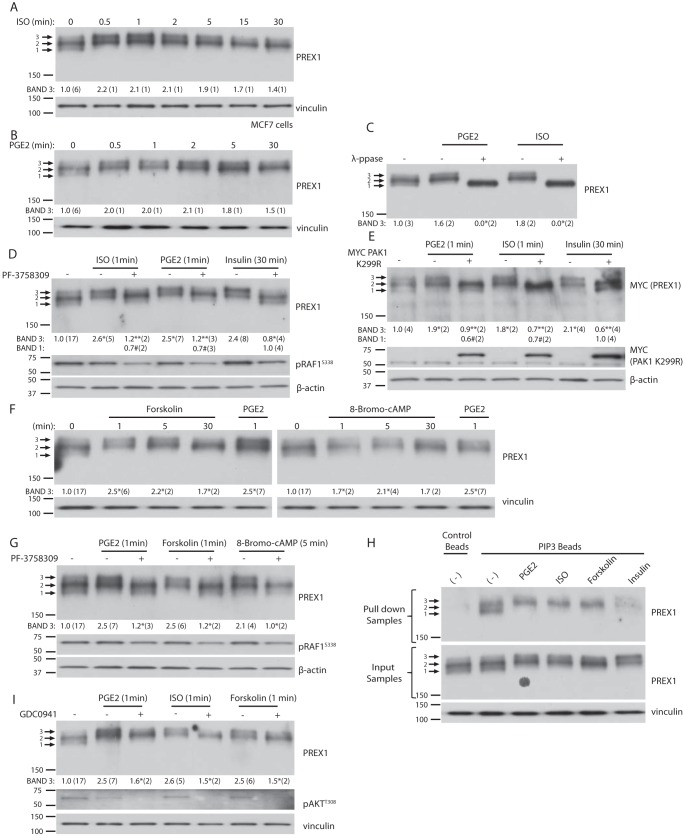
Isoproterenol and Prostaglandin E2 Stimulation Leads to the Phosphorylation of PREX1 through a Mechanism That Is Distinct from Phosphorylation Downstream of Receptor Tyrosine Kinases
PREX proteins are also activated by Gβγ, and PREX1 is both activated and phosphorylated after GPCR activation with the β-adrenergic receptor agonist isoproterenol (31, 35). Thus far, we have established that RTK-dependent phosphorylation of PREX1 is PAK-dependent, and next we sought to determine whether certain GPCR-mediated phosphorylation events on PREX1 also require PAKs. We analyzed the electrophoretic mobility of PREX1 after isoproterenol treatment in MCF7 cells to see whether it mimics the changes seen after RTK stimulation. Isoproterenol did induce an electrophoretic band shift similar to that seen after RTK activation; however, the upper band (band 3) was strongly enriched within the first 30 s of treatment (Fig. 4A). In contrast, after RTK activation, full enrichment of the upper band was not seen until the 5-min time point (Fig. 1, F and G). We then screened other GPCR agonists to see whether this type of regulation extends beyond β-adrenergic receptors, and we found that treatment of MCF7 cells with PGE2 resulted in the same electrophoretic mobility changes in PREX1 that were seen with isoproterenol (Fig. 4B). Furthermore, the mobility shifts induced by isoproterenol and PGE2 treatment were sensitive to λ-phosphatase, confirming that these changes were a result of the phosphorylation of PREX1 (Fig. 4C). As we established previously, RTK-mediated phosphorylation of PREX1 requires PAKs, and because isoproterenol and PGE2-dependent phosphorylation resulted in the same electrophoretic mobility changes in PREX1 that were seen after RTK activation, we tested whether PAKs regulate the phosphorylation of PREX1 downstream of these GPCR agonists. Treatment of MCF7 cells with the PAK inhibitor PF-3758309 or co-expression of the dominant negative PAK1 K299R mutant reduced enrichment of the upper PREX1 band (band 3) that was induced by isoproterenol or PGE2, suggesting that these agonists do cause PAK-dependent phosphorylation events (Fig. 4, D and E).
FIGURE 4.
Isoproterenol and prostaglandin E2 stimulation lead to a PAK-dependent mechanism of phosphorylation for PREX1 that is distinct from receptor tyrosine kinase activation. For all quantified Western blots, the ratio of the intensity of band 3 or band 1 (as indicated) by densitometry to that of total PREX1 (or MYC-tagged PREX1) protein was calculated, and the number of independent samples used for each quantification is shown in parenthesis. A, Western blotting analysis of MCF7 cells that were starved and then treated with 1 μm of isoproterenol (ISO) for the indicated times. Band 3 was quantified as described above. B, Western blotting analysis of MCF7 cells that were starved and then treated with 10 nm of PGE2 for the indicated times. Band 3 was quantified as described above. C, Western blotting analysis of λ-phosphatase (λ-ppase) or mock-treated cell lysates from MCF7 cells that were treated with PGE2 or isoproterenol for 1 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding mock-treated sample for that treatment. D, Western blotting analysis of MCF7 cells that were treated with 1 μm of PF-3758309 (PAKi) for 15 min, followed by treatment with isoproterenol or PGE2 for 1 min or 5 μg/ml insulin for 30 min. Either band 3 or band 1 was quantified as described above. *, p < 0.05 compared with the starved sample for band 3; **, p < 0.05 compared with the corresponding ligand treatment without PF-3758309 for band 3; #, p < 0.05 compared with the insulin- and PF-3758309-treated sample for band 1. E, Western blotting analysis of MCF7 cells expressing MYC PREX1 with or without co-expression of MYC PAK1 K299R. Cells were starved and treated with isoproterenol or PGE2 for 1 min or insulin for 30 min. Either band 3 or band 1 was quantified as described above. *, p < 0.05 compared with the starved sample for band 3. **, p < 0.05 compared with the corresponding ligand treatment without MYC PAK1 K299R co-expression for band 3; #, p < 0.05 compared with the insulin-treated sample with co-expression of MYC PAK1 K299R for band 1. F, Western blotting analysis of MCF7 cells that were starved and then treated with 100 nm forskolin, 500 μm 8-bromo-cAMP, or 10 nm PGE2 for the indicated times. Band 3 was quantified as described above. *, p < 0.05 compared with the starved sample. G, Western blotting analysis of MCF7 cells that were treated with 1 μm of PF-3758309 (PAKi) for 15 min, followed by treatment with PGE2 or forskolin for 1 min, or 8-bromo-cAMP for 5 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding ligand treatment without PF-3758309. H, Western blotting analysis of PIP3 bead pulldowns from MCF7 cells that were starved and treated with PGE2 or isoproterenol for 1 min, forskolin for 5 min, or insulin for 30 min. A representative blot is shown of two independent experiments. I, Western blotting analysis of MCF7 cells that were treated with 500 nm of GDC0941 (PI3Ki) for 15 min, followed by treatment with PGE2, isoproterenol, or forskolin for 1 min. Band 3 was quantified as described above. *, p < 0.05 compared with the corresponding ligand treatment without GCD0941. Phosphospecific antibodies to AKT (Thr-308) and RAF1 (Ser-338) were used to detect signaling alterations.
Despite PAKs having a role in the phosphorylation of PREX1 downstream of both RTKs and GPCRs, our data show that the mechanism of phosphorylation downstream of PGE2 and isoproterenol is different from the mechanism of phosphorylation that occurs downstream of RTKs. First, the time course of phosphorylation is much faster downstream of PGE2 and isoproterenol, and second, as shown in Fig. 4, D and E, PAK inhibition did not enrich the lower PREX1 band (band 1) to the extent that it did in the context of insulin treatment. One possible explanation for this observation is that another kinase in addition to PAKs is involved in the phosphorylation of PREX1 after isoproterenol and PGE2 treatment. We hypothesized that PKA could have a role because it is known to phosphorylate PREX1 and is activated downstream of both PGE2 and isoproterenol (31). We found that activation of PKA with forskolin or 8-bromo-cAMP resulted in a PREX1 band shift that was identical to the PGE2- and isoproterenol-stimulated shift, suggesting that PKA might also have a role in phosphorylating PREX1 downstream of these receptors (Fig. 4F). Interestingly, pharmacological inhibition of PAKs blocked the forskolin and 8-bromo-cAMP-dependent enrichment of the PREX1 upper band (band 3), demonstrating that PAKs act downstream of PKA in the same pathway (Fig. 4G).
Because PAK-mediated phosphorylation reduced the binding of PREX1 to PIP3 after RTK stimulation, we tested whether the phosphorylation of PREX1 after GPCR activation had the same effect. We found that the PAK-dependent upper band that was induced by PGE2, isoproterenol, and forskolin did not efficiently bind to PIP3, which was similar to the effect of PAK-mediated phosphorylation downstream of RTKs (Fig. 4H). However, compared with the level of PIP3 binding after RTK activation, the reduction in overall binding of PREX1 was not as striking after PGE2, isoproterenol, and forskolin treatment, further suggesting that the regulation of PREX1 downstream of these particular GPCRs differs from the mode of regulation downstream of RTKs. The fact that GPCR-dependent phosphorylation of PREX1 affects PIP3 binding suggests cross-talk between RTKs and GPCRs in the regulation of PREX1. This idea is supported by the fact that PI3K inhibition moderately reduced the enrichment of the PGE2-, isoproterenol-, and forskolin-dependent upper band of PREX1 (Fig. 4I). MCF7 cells contain an activating mutation on the catalytic subunit of PI3Kα, and these data indicate that the basal levels of PIP3 that likely result from this mutation are important for the stimulation of PAK that occurs downstream of GPCRs and PKA activation. Taken together, these data demonstrate that PAKs have a role in the phosphorylation of PREX1 downstream of isoproterenol and PGE2; however, the mechanism and consequences of phosphorylation, perhaps to the differential role of PKA, appear to be different from the phosphorylation of PREX1 downstream of RTKs.
Discussion
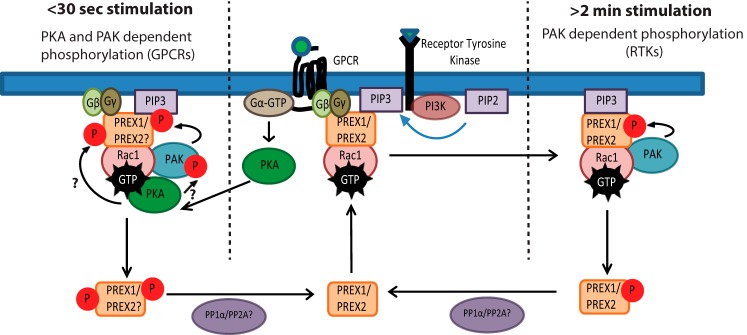
Although PREX1 and PREX2 are both activated by PIP3 and Gβγ downstream of RTKs and GPCRs, it was previously unclear whether they were similarly regulated by phosphorylation in these signaling pathways. We have shown previously that PREX2 is phosphorylated by PAKs after insulin receptor activation, and here we report that PREX1 is regulated in a similar manner by numerous RTKs. We show that these phosphorylation events on PREX1 after neuregulin, insulin, and IGF1 treatment cause a band shift and that these particular phosphorylation events that regulate the band shift require PAK activation. RTK- and PAK-dependent phosphorylation of PREX1 in MCF7 cells reduced the ability of PREX1 to bind to PIP3, and PAK1-mediated phosphorylation of PREX1 reduced PIP3-stimulated GEF activity. These data suggest that, like PREX2, PREX1 appears to be part of a negative feedback circuit where PAKs, which are activated by GTP-bound Rac, phosphorylate PREX1 after PREX1 has been activated to reduce its GEF activity and turn off the signaling pathway (Fig. 5). For illustrative purposes, our model shows that phosphorylated PREX1 is not at the membrane because it does not bind to PIP3; however, given that PREX1 has been shown previously to have PIP3-independent associations with the membrane, it is possible that phosphorylation has a significant impact on GEF activation by PIP3 while only weakening overall membrane binding (36).
FIGURE 5.
Model for PAK-dependent regulation of PREX1 phosphorylation and function by membrane receptors. PREX phosphorylation can be regulated by either RTKs or GPCRs. After RTK activation of PREX GEF activity, the Rac-dependent PAKs stimulate the phosphorylation of PREX proteins around 1–2 min after treatment with various RTK ligands. This causes a reduction in PIP3 binding and a reduction in PIP3-stimulated GEF activity and potentially weakens the membrane binding of PREX proteins. Downstream of certain GPCRs that are known to activate PKA, PAKs mediate the phosphorylation of PREX1 within 30 s of treatment with an agonist. PREX proteins are activated by Gβγ and PIP3 to promote GTP loading on Rac, which can then activate PAK-dependent phosphorylation of PREX1 or PREX2. Additionally, PKA is activated by Gα, and it then potentially binds to Rac-GTP to phosphorylate and activate PAKs and increase the PAK signal. This would allow for PAK-mediated phosphorylation of PREX proteins that relies on both PAK and PKA. While in this signaling complex, PKA may also phosphorylate PREX, and both PAK and PKA-mediated phosphorylation could provide negative feedback by reducing the binding of PREX to Gβγ and PIP3. PP1α and PP2A dephosphorylation may complete the circuit and allow for PREX proteins to become activated by future stimulation of the receptor.
PREX1 is critical for Rac activation downstream of many growth factors, such as neuregulin, IGF1, EGF, and TGFα. Furthermore, PREX1 knockdown reduces cell proliferation, adhesion, colony formation, and invasion after neuregulin stimulation, and also reduces adhesion and cell growth after IGF1 stimulation (17–19). Phosphorylation events on PREX1 have also previously been identified after both neuregulin and IGF1 treatment (18, 19). Our results further implicate PREX1 in these signaling pathways, demonstrating that it is targeted by downstream kinases, such as PAKs, to reduce its GEF activity. The importance of RTK signaling in cancer is well studied, and additionally, there is an emerging interest in targeting PAKs in cancer (37). We have now identified PAK-dependent negative feedback for both PREX1 and PREX2, and these signaling mechanisms may need to be considered when determining how to most effectively utilize PAK inhibitors in patients. Targeting PAKs in the context of PREX1 or PREX2 overexpression may reduce the efficacy of these inhibitors through prolongation of PREX protein GEF activation of Rho GTPases. Many other negative feedback mechanisms like the one described here have been identified, and importantly, some appear to have an impact on therapeutics. For example, mTORC1 is a key signaling protein in cancer that is downstream of RTKs and PI3K, and the reduced efficacy of mTORC1 inhibitors in the clinic appears to be the result of the shutting off of negative feedback for IGF1 signaling that is downstream of the mTORC1-dependent ribosomal S6 kinase (38). It will be interesting to see whether this type of paradigm is also true with PAK inhibitors, and, further, to potentially correlate the efficacy of PAK inhibitors in cancer with PREX expression.
This study also provides a further rationale to study PREX1 and PREX2 in the insulin signaling pathway and in insulin-related disease, such as insulin resistance and type 2 diabetes. We have now shown that both PREX1 and PREX2 are regulated by phosphorylation after insulin stimulation, which is in agreement with previous studies showing key roles for both proteins in insulin signaling and insulin-mediated glucose uptake (15, 21). Furthermore, multiple SNPs that are near the PREX1 gene locus and are associated with type 2 diabetes have been identified in independent populations, although it is unclear whether these SNPs are actually altering PREX1 expression or function (39). Downstream of PREX1, Rac1 and PAKs are also key mediators of insulin-mediated glucose uptake, and both have been implicated in insulin resistance (40, 41). Given that both PREX1 and PREX2 are negatively regulated by PAK-mediated phosphorylation in the insulin signaling pathway, it will be crucial to study the role of these phosphorylation events in insulin resistance and diabetes. It is possible that PREX proteins have high levels of phosphorylation in diabetic patients and that increasing PREX phosphorylation downstream of PAKs is a mechanism for insulin resistance, contributing to the onset of diabetes.
We also studied the phosphorylation of PREX1 after GPCR stimulation. It is known that PREX1 can be phosphorylated downstream of isoproterenol and sphingosine 1-phosphate (31, 49), and in this study we identified additional phosphorylation events on PREX1 after treatment with another GPCR agonist, PGE2. PGE2 is produced through the processing of the fatty acid arachidonic acid and has wide-ranging and diverse roles in physiology in addition to being implicated in cancer (42). Cyclooxygenase enzymes, a family of enzymes in the PGE2 synthesis pathway, are targets of non-steroidal anti-inflammatory drugs such as aspirin, which are widely used pain relievers and anti-inflammatory medications. These drugs have also been linked to reductions in colorectal cancer, a disease where PGE2 levels are sometimes increased (42). Our data showing that PREX1 is phosphorylated after PGE2 stimulation suggest that PREX1 is involved in this signaling pathway. Given the importance of PGE2 for basal cellular function and in disease, it will be important to determine whether PREX1 (or PREX2) has a significant role in PGE2 signaling, and, if so, whether this contributes to the role of PREX proteins in cancer. Interestingly, PGE2 has been shown previously to regulate Rac1, but the data are not clear, as PGE2 can both activate and inhibit Rac1 function (43, 44). It is possible that the effect of PGE2 on Rac1 is dependent on the timing after stimulation, the cell type, and the isoform of the PGE receptor that is expressed.
The phosphorylation events downstream of isoproterenol and PGE2 treatment were at least partially dependent on PAKs, demonstrating that PAKs can regulate the phosphorylation of PREX1 after GPCR activation. Our data also suggest that PKA, an established PREX1 kinase, likely has a role in the phosphorylation of PREX1 after isoproterenol and PGE2 treatment. PKA activation resulted in PAK-dependent phosphorylation of PREX1, suggesting that PKA is upstream of PAKs. A recent study showed that Rac binds to a PKA regulatory subunit and that PKA phosphorylates and activates PAK, setting up a model where PKA binds Rac-GTP to help in the stimulation of PAK and its downstream signals (45). It is also important to note that PREX1 is both activated by isoproterenol in cells and negatively regulated by PKA in vitro (10, 31, 32, 46). One possible model that incorporates all of these data is that, after GPCR stimulation, Gβγ signals to PREX1 to activate Rac and Gα signals to PKA to help in the activation of PAK by Rac-GTP, and then these two events lead to PAK-mediated negative feedback on PREX1 to turn off Rac (Fig. 5). It is also possible that PKA phosphorylates PREX1 in this pathway as an additional node of negative feedback. Additionally, very little is known about GPCR regulation of PREX2, so it will be important to study whether PKA can also regulate the phosphorylation of PREX2, particularly downstream of isoproterenol and PGE2.
Taken together, our data demonstrate that negative feedback mediated by PAKs extends beyond PREX2, as it is involved in the regulation of PREX1. These studies highlight the possibility that negative regulation of GEFs by downstream signals may play a significant role in shutting off GTPase activation. It will be important to study the contribution of these negative feedback circuits to the functional outputs of these signaling pathways in more depth and also to determine whether PAKs can regulate other GEFs. More generally, given how many cancer therapeutics target kinases such as PAK, it is imperative to continue to uncover these instances of negative feedback to fully understand and predict the contexts in which these kinase-targeting drugs will be effective.
Experimental Procedures
Plasmids and Constructs
Full-length PREX1 with an N-terminal MYC tag in the pCMV3 vector was a gift from Heidi Welch (4). MYC-pCMV6M-PAK1 WT and PAK1 K299R (Addgene plasmids 12209 and 12210) and His6-pET28-PAK2 WT were gifts from Jonathan Chernoff (47). PREX2 was cloned into the pcDNA3.1 V5/His vector (Life Technologies) as described previously (22).
Antibodies
The anti-PREX1 antibody (clone 6F12) was provided by Heidi Welch. The β-actin (A5316) antibody, anti-V5-agarose affinity gel (A7345), and anti-MYC agarose affinity gel (A7470) were purchased from Sigma-Aldrich. Rabbit total AKT (9272) and p308 AKT (4056) primary antibodies were purchased from Cell Signaling Technologies. The p338 RAF1/CRAF (05-538) primary antibody was purchased from Millipore. Anti-His HRP antibody (34460) was purchased from Qiagen. Secondary antibodies directed against rabbit and mouse IgG conjugated to HRP were purchased from Pierce.
Cell Lines, Transfection, and Drug Treatments
HEK293 and MCF7 cells were cultured in DMEM (Cellgro) supplemented with 10% (v/v) FBS, 100 IU penicillin, and 100 μg/ml streptomycin (Cellgro). T47D cells were cultured in RPMI (Cellgro) supplemented with 10% (v/v) FBS, 100 IU penicillin, and 100 μg/ml streptomycin. When indicated, cells were starved for 16 h in the appropriate medium without FBS before treatment. Transfections were performed with Lipofectamine 2000 (Life Technologies) following the protocol of the manufacturer. Unless otherwise indicated, cells were treated with 5 μg/ml insulin (Sigma), 10 nm neuregulin (Prospec), 50 ng/ml IGF1 (Sigma), and 100 nm calyculin A (Cell Signaling Technology) for 30 min. In addition, 1 μm isoproterenol (Sigma), 10 nm PGE2 (Sigma), 100 nm forskolin (Tocris), and 500 μm 8-bromo-cAMP (Tocris) were used for the indicated amounts of time. For treatment of cells with small molecule inhibitors, cells were incubated with the indicated concentrations for 15 min prior to addition of growth factor or GPCR agonist. The signaling experiments were performed in 6-well plates with the exception of the time course experiments with neuregulin, insulin, isoproterenol, and PGE2, which were performed in separate 35-mm plates.
In Vitro λ-Phosphatase Assay
MCF7 cells were plated in 6-well plates, starved overnight, and then treated as indicated. The cells were then harvested in 150 μl of 1× NEBuffer for Protein Metallophosphatases (New England Biolabs) that was supplemented with 1% Triton, 100 nm calyculin A, and 1× eukaryotic protease inhibitor mixture (Sigma). The lysates were then vortexed, sonicated, and centrifuged at 4 °C for 30 min. The 40-μl phosphatase reaction was then set up with 35 μl of the lysate, 4 μl of 10× MnCl2 (New England Biolabs), and 1 μl of either λ-phosphatase (New England Biolabs) or 1× NEBuffer for Protein Metallophosphatases. The reactions were incubated at 30 °C for 30 min. At the end of the incubation, 40 μl of 2× Laemmli sample buffer (125 mm Tris (pH 6.8), 4% SDS, 20% glycerol, 10% β-mercaptoethanol, and 0.05% bromphenol blue) was added to each mixture, and the samples were analyzed by Western blotting.
In Vitro PAK2 Kinase Assay
His6-PAK2 was purified as described previously (33). For the kinase assay, MYC PREX1 or V5 PREX2-expressing HEK293 cells were starved and then treated with 500 nm GDC0941 and 5 μm PF-3758309 for 30 min. One 15-cm plate was transfected for every three kinase reactions in a given experiment. Each plate was harvested into 1.5 ml of high-salt lysis buffer containing 25 mm Tris (pH 7.5), 0.1% Triton X-100, 1 m NaCl, and 1× eukaryotic protease inhibitor mixture, and the lysates were then vortexed, sonicated, and centrifuged for 30 min at 4 °C. The lysates were combined and then incubated with V5 or MYC-agarose beads for 16 h while rotating at 4 °C. The MYC PREX1 and V5 PREX2 beads were washed three times with lysis buffer, followed by three washes with 1× phosphobuffer containing 50 mm HEPES (pH 7.5), 25 mm NaCl, 1.25 mm MgCl2, and 1.25 mm MnCl2. The beads were divided equally into the appropriate number of tubes and used in a 30-μl reaction consisting of the indicated amount of His6-PAK2, 100 μm cold ATP, and 5 μCi [γ-32P]ATP (PerkinElmer Life Sciences), all diluted in 1× phosphobuffer. The reactions were incubated at 30 °C for 45 min. The beads were then washed two times with kinase buffer and resuspended in 2× Laemmli sample buffer. Incorporation of [γ-32P]ATP was detected by gel electrophoresis.
PIP3 Bead Pulldown of PREX1
10-cm plates of MCF7 cells were starved overnight and then treated as indicated before, being harvested in 1 ml of PIP3 binding/lysis buffer (20 mm HEPES (pH 7.4), 0.25% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 1 mm Na3VO4, 1 mm NaF, 100 nm calyculin A, and 1× eukaryotic protease inhibitor mixture). The lysates were then vortexed, sonicated, and centrifuged for 30 min at 4 °C. The supernatant was combined with 15 μl of PIP3 beads (Echelon), and these mixtures were rotated at 4 °C for 4 h. The beads were then washed five times in PIP3 binding/lysis buffer and resuspended in 30 μl of 2× Laemmli sample buffer to be analyzed by Western blotting.
PIP3 Bead Pulldown of PREX1 after in Vitro λ-Phosphatase Assay
15-cm plates of MCF7 cells were starved overnight and then were either left starved or treated with insulin before being harvested in 300 μl of 1× NEBuffer for Protein Metallophosphatases (New England Biolabs) that was supplemented with 1% Triton, 100 nm calyculin A, and 1× eukaryotic protease inhibitor mixture. The lysates were then vortexed, sonicated, and centrifuged at 4 °C for 30 min. The 190-μl phosphatase reaction was then set up with 168 μl of the lysate (the starved and insulin-treated lysates were each divided into two reactions), 19 μl of 10× MnCl2 (New England Biolabs), and 3 μl of either λ-phosphatase (New England Biolabs) or 1× NEBuffer for Protein Metallophosphatases. The reactions were incubated at 30 °C for 45 min. Next, 800 μl of PIP3 binding/lysis buffer (20 mm HEPES (pH 7.4), 0.25% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 1 mm Na3VO4, 1 mm NaF, 100 nm calyculin A, and 1× eukaryotic protease inhibitor mixture), and 15 μl of PIP3 beads (Echelon) was added to each reaction; these mixtures were rotated at 4 °C for 4 h. The beads were then washed five times in PIP3 binding/lysis buffer and resuspended in 30 μl of 2× Laemmli sample buffer to be analyzed by Western blotting.
In Vitro Rac-GEF Assay
MYC PREX1 was purified from HEK293 cells using the same method as described previously for V5 PREX2 purification, except MYC-agarose and MYC peptide (Sigma) were used (33). In vitro analysis of PIP3-stimulated MYC PREX1 GEF activity toward GDP-loaded GST Rac1 was performed as described previously, except glutathione-Sepharose beads (GE Healthcare Life Sciences) were used to isolate the GST Rac1 after incubation with MYC PREX1 (4, 48). GST Rac1 was purified, eluted, and loaded with GDP as described previously (33). For the in vitro GEF assay, PIP3 diC16 was purchased from Echelon Biosciences and was incorporated into liposomes. The final concentrations of GST Rac1 and MYC PREX1 in the reaction were 100 nm and 5 nm, respectively. Purified PREX1 and GST Rac1 were incubated in a final reaction volume of 10 μl with PIP3, 5 μm cold GTPγS (Sigma), and 1 μCi [35S]GTPγS (PerkinElmer Life Sciences) for 10 min at 30 °C. GST Rac1 was isolated on glutathione-Sepharose beads, and the loading of [35S]GTPγS by GST Rac1 was measured by scintillation counting.
Statistical Analysis
Student's t test was used to determine statistical significance, and p < 0.05 was considered significant. Scatter plots show the average value of all the samples in that dataset ± S.D.
Author Contributions
D. B. designed experiments, performed experiments, analyzed data, and wrote the paper. J. Z. H. performed experiments. R. P. designed the studies, analyzed data, and wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank all members of the Parsons laboratory for reading the manuscript and providing feedback.
This work was partially supported by National Institutes of Health Grants R01CA155117 (to R. P.) and R01CA184016 (to R. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GEF
- guanine nucleotide exchange factor
- PAK
- p21-activated kinase
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- RTK
- receptor tyrosine kinase
- GPCR
- G protein-coupled receptor
- PGE2
- prostaglandin E2
- mTOR
- mechanistic target of rapamycin
- PREX
- PIP3-dependent Rac exchange factor
- PTEN
- phosphatase and tensin homolog.
References
- 1. Rojas A. M., Fuentes G., Rausell A., and Valencia A. (2012) The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J. Cell Biol. 196, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cherfils J., and Zeghouf M. (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 [DOI] [PubMed] [Google Scholar]
- 3. King H., Nicholas N. S., and Wells C. M. (2014) Role of p-21-activated kinases in cancer progression. Int. Rev. Cell Mol. Biol. 309, 347–387 [DOI] [PubMed] [Google Scholar]
- 4. Welch H. C., Coadwell W. J., Ellson C. D., Ferguson G. J., Andrews S. R., Erdjument-Bromage H., Tempst P., Hawkins P. T., and Stephens L. R. (2002) P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108, 809–821 [DOI] [PubMed] [Google Scholar]
- 5. Donald S., Hill K., Lecureuil C., Barnouin R., Krugmann S., John Coadwell W., Andrews S. R., Walker S. A., Hawkins P. T., Stephens L. R., and Welch H. C. (2004) P-Rex2, a new guanine-nucleotide exchange factor for Rac. FEBS Lett. 572, 172–176 [DOI] [PubMed] [Google Scholar]
- 6. Rosenfeldt H., Vázquez-Prado J., and Gutkind J. S. (2004) P-REX2, a novel PI-3-kinase sensitive Rac exchange factor. FEBS Lett. 572, 167–171 [DOI] [PubMed] [Google Scholar]
- 7. Welch H. C. (2015) Regulation and function of P-Rex family Rac-GEFs. Small GTPases 6, 49–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Welch H. C., Condliffe A. M., Milne L. J., Ferguson G. J., Hill K., Webb L. M., Okkenhaug K., Coadwell W. J., Andrews S. R., Thelen M., Jones G. E., Hawkins P. T., and Stephens L. R. (2005) P-Rex1 regulates neutrophil function. Curr. Biol. 15, 1867–1873 [DOI] [PubMed] [Google Scholar]
- 9. Dong X., Mo Z., Bokoch G., Guo C., Li Z., and Wu D. (2005) P-Rex1 is a primary Rac2 guanine nucleotide exchange factor in mouse neutrophils. Curr. Biol. 15, 1874–1879 [DOI] [PubMed] [Google Scholar]
- 10. Damoulakis G., Gambardella L., Rossman K. L., Lawson C. D., Anderson K. E., Fukui Y., Welch H. C., Der C. J., Stephens L. R., and Hawkins P. T. (2014) P-Rex1 directly activates RhoG to regulate GPCR-driven Rac signalling and actin polarity in neutrophils. J. Cell Sci. 127, 2589–2600 [DOI] [PubMed] [Google Scholar]
- 11. Waters J. E., Astle M. V., Ooms L. M., Balamatsias D., Gurung R., and Mitchell C. A. (2008) P-Rex1: a multidomain protein that regulates neurite differentiation. J. Cell Sci. 121, 2892–2903 [DOI] [PubMed] [Google Scholar]
- 12. Bishop A. L., and Hall A. (2000) Rho GTPases and their effector proteins. Biochem. J. 348, 241–255 [PMC free article] [PubMed] [Google Scholar]
- 13. Das B., Shu X., Day G. J., Han J., Krishna U. M., Falck J. R., and Broek D. (2000) Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J. Biol. Chem. 275, 15074–15081 [DOI] [PubMed] [Google Scholar]
- 14. Han J., Luby-Phelps K., Das B., Shu X., Xia Y., Mosteller R. D., Krishna U. M., Falck J. R., White M. A., and Broek D. (1998) Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279, 558–560 [DOI] [PubMed] [Google Scholar]
- 15. Hodakoski C., Hopkins B. D., Barrows D., Mense S. M., Keniry M., Anderson K. E., Kern P. A., Hawkins P. T., Stephens L. R., and Parsons R. (2014) Regulation of PTEN inhibition by the pleckstrin homology domain of P-REX2 during insulin signaling and glucose homeostasis. Proc. Natl. Acad. Sci. U.S.A. 111, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donald S., Humby T., Fyfe I., Segonds-Pichon A., Walker S. A., Andrews S. R., Coadwell W. J., Emson P., Wilkinson L. S., and Welch H. C. (2008) P-Rex2 regulates Purkinje cell dendrite morphology and motor coordination. Proc. Natl. Acad. Sci. U.S.A. 105, 4483–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sosa M. S., Lopez-Haber C., Yang C., Wang H., Lemmon M. A., Busillo J. M., Luo J., Benovic J. L., Klein-Szanto A., Yagi H., Gutkind J. S., Parsons R. E., and Kazanietz M. G. (2010) Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol. Cell 40, 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montero J. C., Seoane S., Ocaña A., and Pandiella A. (2011) P-Rex1 participates in Neuregulin-ErbB signal transduction and its expression correlates with patient outcome in breast cancer. Oncogene 30, 1059–1071 [DOI] [PubMed] [Google Scholar]
- 19. Montero J. C., Seoane S., and Pandiella A. (2013) Phosphorylation of P-Rex1 at serine 1169 participates in IGF-1R signaling in breast cancer cells. Cell Signal. 25, 2281–2289 [DOI] [PubMed] [Google Scholar]
- 20. Ebi H., Costa C., Faber A. C., Nishtala M., Kotani H., Juric D., Della Pelle P., Song Y., Yano S., Mino-Kenudson M., Benes C. H., and Engelman J. A. (2013) PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. U.S.A. 110, 21124–21129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balamatsias D., Kong A. M., Waters J. E., Sriratana A., Gurung R., Bailey C. G., Rasko J. E., Tiganis T., Macaulay S. L., and Mitchell C. A. (2011) Identification of P-Rex1 as a novel Rac1-guanine nucleotide exchange factor (GEF) that promotes actin remodeling and GLUT4 protein trafficking in adipocytes. J. Biol. Chem. 286, 43229–43240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fine B., Hodakoski C., Koujak S., Su T., Saal L. H., Maurer M., Hopkins B., Keniry M., Sulis M. L., Mense S., Hibshoosh H., and Parsons R. (2009) Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science 325, 1261–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindsay C. R., Lawn S., Campbell A. D., Faller W. J., Rambow F., Mort R. L., Timpson P., Li A., Cammareri P., Ridgway R. A., Morton J. P., Doyle B., Hegarty S., Rafferty M., Murphy I. G., McDermott E. W., et al. (2011) P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat. Commun. 2, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin J., Xie Y., Wang B., Hoshino M., Wolff D. W., Zhao J., Scofield M. A., Dowd F. J., Lin M. F., and Tu Y. (2009) Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene 28, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berger M. F., Hodis E., Heffernan T. P., Deribe Y. L., Lawrence M. S., Protopopov A., Ivanova E., Watson I. R., Nickerson E., Ghosh P., Zhang H., Zeid R., Ren X., Cibulskis K., Sivachenko A. Y., et al. (2012) Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lissanu Deribe Y., Shi Y., Rai K., Nezi L., Amin S. B., Wu C. C., Akdemir K. C., Mahdavi M., Peng Q., Chang Q. E., Hornigold K., Arold S. T., Welch H. C., Garraway L. A., and Chin L. (2016) Truncating PREX2 mutations activate its GEF activity and alter gene expression regulation in NRAS-mutant melanoma. Proc. Natl. Acad. Sci. U.S.A. 113, E1296–E1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mense S. M., Barrows D., Hodakoski C., Steinbach N., Schoenfeld D., Su W., Hopkins B. D., Su T., Fine B., Hibshoosh H., and Parsons R. (2015) PTEN inhibits PREX2-catalyzed activation of RAC1 to restrain tumor cell invasion. Sci. Signal. 8, ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barber M. A., Hendrickx A., Beullens M., Ceulemans H., Oxley D., Thelen S., Thelen M., Bollen M., and Welch H. C. (2012) The guanine-nucleotide-exchange factor P-Rex1 is activated by protein phosphatase 1α. Biochem. J. 443, 173–183 [DOI] [PubMed] [Google Scholar]
- 31. Mayeenuddin L. H., and Garrison J. C. (2006) Phosphorylation of P-Rex1 by the cyclic AMP-dependent protein kinase inhibits the phosphatidylinositol (3,4,5)-trisphosphate and Gβγ-mediated regulation of its activity. J. Biol. Chem. 281, 1921–1928 [DOI] [PubMed] [Google Scholar]
- 32. Urano D., Nakata A., Mizuno N., Tago K., and Itoh H. (2008) Domain-domain interaction of P-Rex1 is essential for the activation and inhibition by G protein βγ subunits and PKA. Cell Signal. 20, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 33. Barrows D., Schoenfeld S. M., Hodakoski C., Silkov A., Honig B., Couvillon A., Shymanets A., Nürnberg B., Asara J. M., and Parsons R. (2015) p21-activated kinases (PAKs) mediate the phosphorylation of PREX2 protein to initiate feedback inhibition of Rac1 GTPase. J. Biol. Chem. 290, 28915–28931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De la Mota-Peynado A., Chernoff J., and Beeser A. (2011) Identification of the atypical MAPK Erk3 as a novel substrate for p21-activated kinase (Pak) activity. J. Biol. Chem. 286, 13603–13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lucato C. M., Halls M. L., Ooms L. M., Liu H. J., Mitchell C. A., Whisstock J. C., and Ellisdon A. M. (2015) The phosphatidylinositol (3,4,5)-trisphosphate-dependent Rac exchanger 1.Ras-related C3 botulinum toxin substrate 1 (P-Rex1.Rac1) complex reveals the basis of Rac1 activation in breast cancer cells. J. Biol. Chem. 290, 20827–20840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cash J. N., Davis E. M., and Tesmer J. J. (2016) Structural and biochemical characterization of the catalytic core of the metastatic factor P-Rex1 and its regulation by PtdIns(3,4,5)P3. Structure 24, 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baker N. M., Yee Chow H., Chernoff J., and Der C. J. (2014) Molecular pathways: targeting RAC-p21-activated serine-threonine kinase signaling in RAS-driven cancers. Clin. Cancer Res. 20, 4740–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., and Rosen N. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewis J. P., Palmer N. D., Ellington J. B., Divers J., Ng M. C., Lu L., Langefeld C. D., Freedman B. I., and Bowden D. W. (2010) Analysis of candidate genes on chromosome 20q12–13.1 reveals evidence for BMI mediated association of PREX1 with type 2 diabetes in European Americans. Genomics 96, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sylow L., Jensen T. E., Kleinert M., Højlund K., Kiens B., Wojtaszewski J., Prats C., Schjerling P., and Richter E. A. (2013) Rac1 signaling is required for insulin-stimulated glucose uptake and is dysregulated in insulin-resistant murine and human skeletal muscle. Diabetes 62, 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sylow L., Kleinert M., Pehmøller C., Prats C., Chiu T. T., Klip A., Richter E. A., and Jensen T. E. (2014) Akt and Rac1 signaling are jointly required for insulin-stimulated glucose uptake in skeletal muscle and downregulated in insulin resistance. Cell Signal. 26, 323–331 [DOI] [PubMed] [Google Scholar]
- 42. Greenhough A., Smartt H. J., Moore A. E., Roberts H. R., Williams A. C., Paraskeva C., and Kaidi A. (2009) The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 30, 377–386 [DOI] [PubMed] [Google Scholar]
- 43. Nagasawa S. Y., Takuwa N., Sugimoto N., Mabuchi H., and Takuwa Y. (2005) Inhibition of Rac activation as a mechanism for negative regulation of actin cytoskeletal reorganization and cell motility by cAMP. Biochem. J. 385, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Birukova A. A., Zagranichnaya T., Fu P., Alekseeva E., Chen W., Jacobson J. R., and Birukov K. G. (2007) Prostaglandins PGE(2) and PGI(2) promote endothelial barrier enhancement via PKA- and Epac1/Rap1-dependent Rac activation. Exp. Cell Res. 313, 2504–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bachmann V. A., Riml A., Huber R. G., Baillie G. S., Liedl K. R., Valovka T., and Stefan E. (2013) Reciprocal regulation of PKA and Rac signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 8531–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao T., Nalbant P., Hoshino M., Dong X., Wu D., and Bokoch G. M. (2007) Signaling requirements for translocation of P-Rex1, a key Rac2 exchange factor involved in chemoattractant-stimulated human neutrophil function. J. Leukocyte Biol. 81, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 47. Sells M. A., Knaus U. G., Bagrodia S., Ambrose D. M., Bokoch G. M., and Chernoff J. (1997) Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 7, 202–210 [DOI] [PubMed] [Google Scholar]
- 48. Hill K., and Welch H. C. (2006) Purification of P-Rex1 from neutrophils and nucleotide exchange assay. Methods Enzymol. 406, 26–41 [DOI] [PubMed] [Google Scholar]
- 49. Chávez-Vargas L., Adame-García S. R., Cervantes-Villagrana R. D., Castillo-Kauil A., Bruystens J. G., Fukuhara S., Taylor S. S., Mochizuki N., Reyes-Cruz G., and Vázquez-Prado J. (2016) Protein kinase A (PKA) type I interacts with P-Rex1, a Rac guanine nucleotide exchange factor: effect on PKA localization and P-Rex1 signaling. J. Biol. Chem. 291, 6182–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]