Abstract
Intimin is an essential adhesin of attaching and effacing organisms such as entropathogenic Escherichia coli. It is also the prototype of type Ve secretion or inverse autotransport, where the extracellular C-terminal region or passenger is exported with the help of an N-terminal transmembrane β-barrel domain. We recently reported a stalled secretion intermediate of intimin, where the passenger is located in the periplasm but the β-barrel is already inserted into the membrane. Stalling of this mutant is due to the insertion of an epitope tag at the very N terminus of the passenger. Here, we examined how this insertion disrupts autotransport and found that it causes misfolding of the N-terminal immunoglobulin (Ig)-like domain D00. We could also stall the secretion by making an internal deletion in D00, and introducing the epitope tag into the second Ig-like domain, D0, also resulted in reduced passenger secretion. In contrast to many classical autotransporters, where a proximal folding core in the passenger is required for secretion, the D00 domain is dispensable, as the passenger of an intimin mutant lacking D00 entirely is efficiently exported. Furthermore, the D00 domain is slightly less stable than the D0 and D1 domains, unfolding at ∼200 piconewtons (pN) compared with ∼250 pN for D0 and D1 domains as measured by atomic force microscopy. Our results support a model where the secretion of the passenger is driven by sequential folding of the extracellular Ig-like domains, leading to vectorial transport of the passenger domain across the outer membrane in an N to C direction.
Keywords: atomic force microscopy (AFM), bacterial pathogenesis, immunoglobulin-like domain, protein folding, protein secretion, type V secretion system
Introduction
Intimin is a well known virulence factor of attaching and effacing pathogens, such as enterohemorrhagic and enteropathogenic Escherichia coli (EPEC),3 Citrobacter rodentium, and Hafnia alvei (1). Intimin is an adhesin that mediates tight attachment to epithelial cells, which ultimately leads to the effacement of microvilli and formation of actin pedestals on the host cell. Remarkably, the main receptor for intimin is not a host cell receptor but a bacterial protein called Tir for translocated intimin receptor. This protein is secreted by the pathogens directly into the host cells via a type III secretion system (2).
Intimin consists of three functional components: an extracellular domain, a transmembrane domain, and a small periplasmically located domain. The adhesive, extracellular region (referred to as the passenger) of intimin is an extended, rod-like structure (Fig. 1A). The crystal structure of the intimin C terminus shows three domains: two immunoglobulin (Ig)-like domains (D1–2) capped by a C-type lectin-like domain (D3) (3). The Ig-like domain D2 and the lectin-like domain D3 together form the Tir binding region (4). In addition, a third Ig-like domain (D0) was predicted at the N terminus of the passenger. Later, an additional domain at the very N terminus of the extracellular domain has been identified, referred to as the D00 domain, although its fold was not conclusively predicted (5). Intimin is anchored in the outer membrane by a 12-stranded β-barrel domain (5). In addition, intimin contains a short N-terminal periplasmic region containing a lysin motif, which mediates dimerization and binding to peptidoglycan at low pH (6).
FIGURE 1.
Intimin structure and location of double HA tag. A, schematic structure of an intimin dimer. Where experimental structural information is available, the structures are depicted in schematic representation. The periplasmic domain, which mediates dimerization, is colored yellow. The β-barrel domain is blue, the periplasmically located α-helical turn is salmon, and the linker is orange. The extracellular domains, D00-D3, are colored green, red, gray, light blue, and violet, respectively. Protein database identifiers for the structures used in the figure are 2mpw (periplasmic LysM domain), 4e1s (β-barrel domain), and 1f00 (extracellular domains D1–3). B, schematic of the stalled intimin variant, Int HA453. In this mutant, the double HA tag inserted into the N terminus of the passenger domain is surface-exposed, but the C terminus of the protein remains in the periplasm (11). The double HA tag is colored in red, the other features are colored as in panel A. C, insertion site of the double HA tag in Int HA453. A stretch of EPEC intimin sequence is shown spanning the C terminus of the β-barrel domain, the linker sequence, and the beginning of the D00 domain in the passenger. Secondary structure elements (from the intimin β-barrel crystal structure or predicted for the D00 domain) are shown as cylinders for α-helices or arrows for β-strands. The insertion site of the double HA tag in Int HA453 is in the first predicted β-strand of the D00 domain. The coloring corresponds to panels A and B.
As a surface-exposed protein, the intimin passenger is secreted across the outer membrane to the cell surface. This is accomplished by an autotransport process, termed type Ve secretion (7, 8). Intimin is the prototype of type Ve secretion, also known as inverse autotransport (9). In the current model for type Ve secretion, the β-barrel domain acts as a secretion pore through which the passenger domain is transported via a hairpin intermediate, such that the N terminus of the passenger is the first to reach the extracellular space (5, 8). Secretion of the entire passenger then proceeds N to C, with the C terminus the last to reach the surface. This model is supported by the crystal structure of the intimin β-barrel domain, where the linker region connecting the passenger and the β-barrel domain is located within the pore of the β-barrel (5). However, outer membrane insertion of intimin is dependent on the β-barrel assembly machinery or BAM complex (10), and the BAM complex may also be involved in passenger secretion (11). Additionally, the periplasmic chaperone SurA is required for inverse autotransporter biogenesis (8, 10, 11). Thus, the term “autotransporter” is somewhat misleading but may still be valid for the export of the passenger.
This study was prompted by our recent observation that introduction of a double hemagglutinin (HA) tag (sequence GSGYPYDVPDYAGSGYPYDVPDYAGSG) at the N terminus of the passenger domain prevents secretion of the passenger and traps intimin in what is most probably the hairpin intermediate (11). In this mutant the β-barrel domain is fully formed and inserted into the membrane, but the C terminus of the passenger is located in the periplasm (Fig. 1B). The double HA tag of this mutant is in fact located in the beginning of the D00 domain sequence, where it disrupts a predicted β-strand (Fig. 1C).
In this study we investigated the effect of this insertion on the D00 domain and its implications for passenger secretion. The C-terminal, membrane-proximal region of several classical autotransporter passengers functions as a highly stable folding core that is necessary for efficient secretion (12–15). This region has earlier been termed an “autochaperone” domain, although this name is no longer recommended (16). By analogy, the N-terminal region of inverse autotransporters has been suggested to play a similar role in the secretion process by promoting protein folding (11, 17). We, therefore, set out to test whether the D00 domain acts as a folding core in inverse autotransport. Our results show that disruption of this domain leads to misfolding of D00 and stalling of passenger secretion; however, the D00 domain is not required for passenger secretion, and it does not form a stable folding core. Rather, our results support a model where sequential folding of individual Ig-like domains at the cell surface provides most of the free energy for inverse autotransporter passenger secretion.
Results
The D00 Domain of Intimin Is an Ig-like Domain
When investigating the borders of the β-barrel domain of Intimin, Fairman et al. (5) identified a protease-resistant extracellular fragment that they termed domain D00. This domain was not recognized as either a bacterial Ig-like or a C-type lectin domain by the Pfam database; therefore, Fairman et al. (5) suggested it might adopt a different fold. However, when we subjected the amino acid sequence of the D00 domain to sensitive homology-based structure prediction using HHPred (18), all the best hits were to Ig-like domains (Fig. 2A). In addition, secondary structure prediction for an alignment of D00 domains from various inverse autotransporters shows seven predicted β-strands, consistent with an Ig-like topology (Fig. 2B). Furthermore, we made a homology model of the D00-D0 domain pair, which clearly shows both adopting an Ig-like fold (Fig. 2C and supplemental File 1). To test our prediction, we produced and purified the D00 domain using recombinant techniques. The circular dichroism (CD) spectrum of the protein shows a broad minimum centered around 212 nm that is typical of β-structured proteins, consistent with our predictions of an Ig-like domain (Fig. 2D).
FIGURE 2.
D00 is an Ig-like domain. A, homology-based structure prediction for the intimin β-barrel domain and D00 domain using HHPred (18). The best hits (in red) for the D00 domain both have a fibronectin module III-like fold. Fibronectin module III belongs to the immunoglobulin superfamily (47). PDB IDs displayed for the β-barrel domain are 4t1s (EHEC intimin β-barrel; probability = 100%, E (expect)-value = 7.1E-83), 4e1t (Yersinia pseudotuberculosis Invasin β-barrel, probability = 100%, E-value = 3E-82), 3qra (Yersinia pestis attachment and invasion locus protein Ail, probability = 96.7%, E-value = 0.032), 2jmm (E. coli OmpA, probability = 96.2%, E-value = 0.49), 2k0l (Klebsiella pneumoniae OmpA, probability = 96, E-value = 0.51), and 2lhf (Pseudomonas aeruginosa H1 outer membrane protein, probability = 95.9%, E-value = 0.16). PDB IDs displayed for the D00 domain are 3pe9 (fibronectin module III-like module from cellobiohydrolase A, probability = 97.4%, E-value = 0.0018), 3pdd (fibronectin module III-like module from cellobiohydrolase A, probability = 96.6%, E-value = 0.026), 2yn5 (Salmonella SiiE Ig-domain adhesin, probability = 85%, E-value = 1.2), and 1gl4 (Mus musculus heparin sulfate proteoglycan core protein Ig-like domain, probability = 78.9, E-value = 9.9). Probability refers to the probability of the hit being a true positive, E-value denotes the average number of false positives with a score better than the one for the template when scanning the database. B, secondary structure prediction for an alignment of selected D00 domains from various inverse autotransporters. The intimin sequence is on the top. Consensus secondary structure elements are shown above the alignment: blue arrows denote β-strands and pink cylinders α-helices. The strength of the secondary structure prediction is demonstrated by the intensity of the colors (blue for β-strands and pink for α-helices). C, homology model of the intimin D00-D0 region. In the model both domains adopt an Ig-like fold. D00 is in green, and D0 is in red. The PDB file of the model is available as supplemental File 1. D, CD spectrum of purified intimin D00 domain.
The Introduction of the Double HA Tag Leads to Misfolding of the D00 Domain
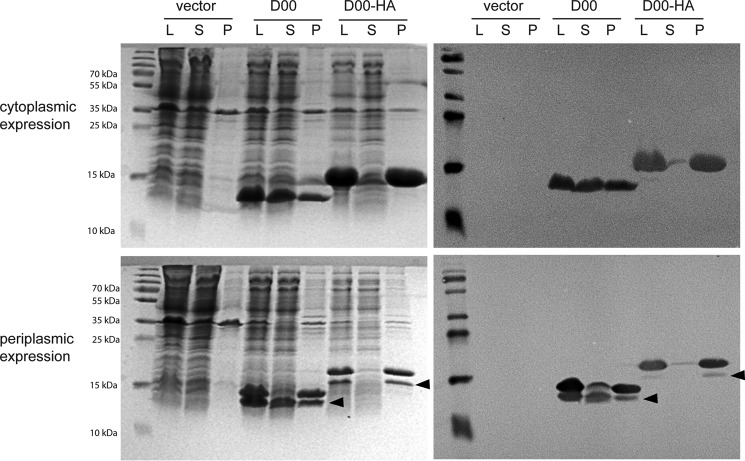
Position 453 lies within the first predicted β-strand of the intimin D00 (Fig. 1C). Thus, the introduced double HA tag presumably leads to misfolding of the D00 domain. To test this hypothesis, we produced both the wild-type (WT) D00 domain and the HA-tagged version, referred to as D00-HA, recombinantly in E. coli. Cytoplasmically produced D00 was soluble to a large extent, whereas D00-HA remained almost entirely in the pellet even after extraction with Triton X-100, showing that the protein was deposited in inclusion bodies (Fig. 3, top). When targeted to the periplasm by the addition of an OmpA signal peptide, most of the processed D00 domain (i.e. with cleaved signal peptide, the lower bands denoted by the arrowheads in Fig. 3, bottom) is soluble. In contrast, D00-HA is mostly preprotein, and only a small amount of it is processed. However, even the protein with cleaved signal peptide is insoluble. This demonstrates that insertion of the HA tag into the D00 domain at position 453 leads to misfolding of the domain and its deposition in inclusion bodies. Unfortunately, we were unable to purify the tiny amount of D00-HA in the supernatant and could, therefore, not measure a CD spectrum to show that the protein is unfolded.
FIGURE 3.
HA tag insertion disrupts folding of D00. Intimin D00 and D00-HA domains were produced in E. coli either in the cytoplasm (top) or with a periplasmic export signal (bottom). The cells were lysed, and samples were taken from the lysate (L), the supernatant after centrifugation of the lysate (S), or the pellet after extracting twice with detergent (P). The panels on the left are Coomassie-stained gels after SDS-PAGE; the panels on the right are Western blots using an anti-StrepII antibody to detect the C-terminal StrepII tag of the constructs. Arrowheads show the presumed processed form of the protein with the signal peptide removed.
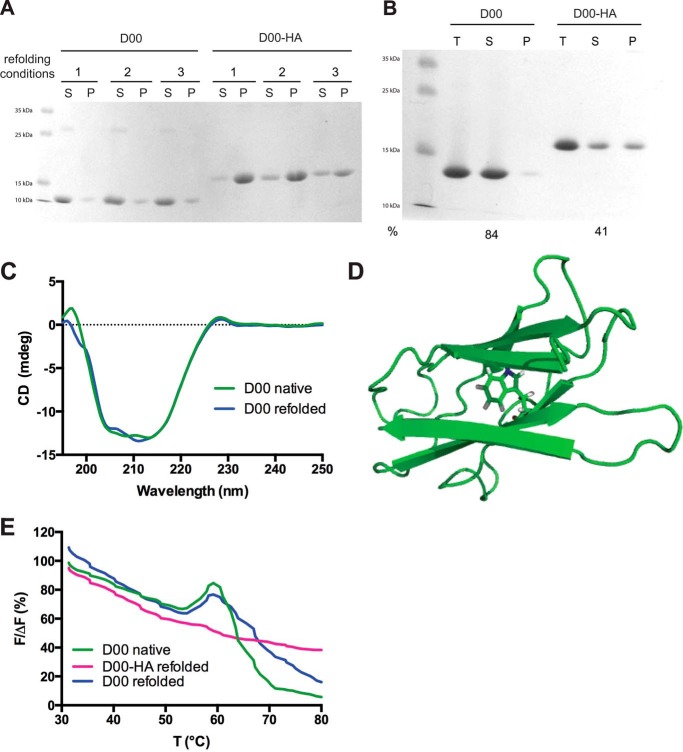
Therefore, we decided to test whether the introduction of the double HA tag would interfere with refolding of the protein. The WT D00 domain refolded robustly in a matrix of 9 conditions when diluted from 6 m guanidine hydrochloride. In all the conditions, almost all the protein was found in the supernatant after refolding (Fig. 4A). In contrast, D00-HA did not refold efficiently, as shown by the large amount of protein that precipitated after dilution into the refolding buffers (Fig. 4A). For a larger scale refolding experiment we chose conditions where we saw an appreciable amount of D00-HA in the supernatant (condition 3 in Fig. 4A; this was also the condition where the deletion mutant D00Δ472–477 was most soluble; see below). In this experiment, ∼60% of D00-HA was lost after ultracentrifugation, whereas >80% of D00 remained soluble (Fig. 4B). The CD spectrum of the refolded D00 showed that it had adopted a similar fold to purified D00 before denaturing in guanidine (Fig. 4B). For D00-HA, more protein was lost during dialysis and subsequent concentration; therefore, we were unable to concentrate the small amount of D00-HA from the supernatant to measure a CD spectrum, suggesting that even this soluble fraction was not correctly folded. However, we did have enough protein to perform differential scanning fluorometry. The D00 domain contains a single tryptophan residue (Trp-487), which is predicted to be buried in our homology model (Fig. 4D). In addition, the C-terminal StrepII tag contains a tryptophan residue that we assume is fully solvent-exposed. Upon unfolding, the buried tryptophan should become more accessible to the solvent, with a concomitant increase of emission at 350 nm. In the assay, native D00 displayed a clear increase in fluorescence at 350 nm centered at 58 °C, presumably corresponding to the melting point of the Ig-like fold (Fig. 4E). Refolded D00 displayed a similar transition (Fig. 4E). In contrast, refolded D00-HA did not have a similar transition, suggesting that the protein does not have a defined globular fold (Fig. 4E). Taken together, the fact that D00-HA ends up in inclusion bodies and does not refold in conditions where WT D00 is largely soluble indicates that the insertion of the double HA tag greatly reduces the propensity of D00-HA to fold correctly.
FIGURE 4.
HA tag insertion reduces the efficiency of D00 refolding. A, refolding of guanidine-solubilized D00 and D00-HA in three representative conditions: 1 = 50 mm Tris, pH 8.0; 2 = 50 mm MOPS, pH 7.0, 500 mm NaCl; 3 = 50 mm MES, pH 6.0, 150 mm NaCl. S = supernatant; P = pellet. B, large-scale refolding of D00 and D00-HA in condition 3. T = total protein after refolding, S = supernatant after ultracentrifugation, P = resuspended pellet, and % = percentage of soluble protein after ultracentrifugation compared with total protein based on absorbance at 280 nm. C, CD spectra of native (green) and refolded (blue) D00. D, homology model of D00 (in schematic representation) showing the position of the buried Trp-487. E, differential scanning fluorometry of native D00 (green), refolded D00 (blue), and refolded D00-HA (magenta).
Misfolding of the D00 Domain by Deleting a β-Strand Also Prevents Passenger Secretion
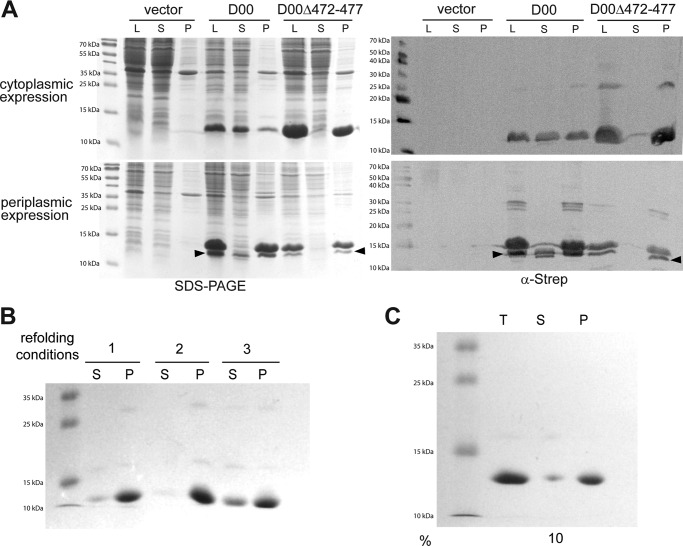
To test whether disrupting the Ig fold of the D00 domain by a different method would lead to a similar stalled secretion phenotype as in the D00-HA mutant, we deleted residues 472–477. These correspond to the second predicted β-strand of the D00 domain (sequence IQLIVK, Fig. 1C). When we produced this protein (D00Δ472–477) recombinantly, both with and without a signal peptide, it was again found almost entirely in the pellet, similar to D00-HA (Fig. 5A). Note that the amount of soluble, processed WT D00 found in the supernatant varies to some degree, but soluble protein was always present for the WT (compare Fig. 3, bottom panel and Fig. 5A). Furthermore, D00Δ472–477 also failed to refold in the conditions where WT D00 refolds efficiently (Fig. 5B). Some protein was soluble in condition 3 (Fig. 5B); therefore, we performed a larger scale refolding experiment in these conditions. After ultracentrifugation, only 10% was soluble (Fig. 5C). So little protein remained in solution after refolding, dialysis, and concentration that we were unable to either obtain a CD spectrum or perform differential scanning fluorometry. This is consistent with the interpretation that deleting the β-strand results in misfolding and subsequent aggregation of D00.
FIGURE 5.
Deletion of an internal β-strand in D00 causes the domain to misfold. A, intimin D00 and D00Δ472–477 domains were produced in E. coli either in the cytoplasm (top) or with a periplasmic export signal (bottom). The cells were lysed, and samples were taken from the lysate (L), the supernatant after centrifugation of the lysate (S), or the pellet after extracting twice with detergent (P). The panels on the left are Coomassie-stained gels after SDS-PAGE; the panels on the right are Western blots using an anti-StrepII antibody to detect the C-terminal StrepII tag of the constructs. Arrowheads show the processed protein lacking a signal peptide. B, refolding of guanidine-solubilized D00Δ472–477 in three representative conditions. The conditions are the same as in Fig. 4A. S = supernatant, P = pellet. C, large scale refolding of D00Δ472–477 in condition 3. T = total protein after refolding, S = supernatant after ultracentrifugation, P = resuspended pellet, % = percentage of soluble protein after ultracentrifugation compared with total protein based on absorbance at 280 nm.
We then tested whether this mutant would have an effect on intimin passenger secretion. To this end, we introduced the deletion of residues 472–477 into full-length intimin (IntΔ472–477). For detection, we included a C-terminal StrepII tag, because periplasmically localized intimin passenger is coated with SurA, which prevents immunological detection using an antibody against the C terminus of the passenger (11). The StrepII tag does not affect passenger export or adhesion to Tir-expressing cells (11).
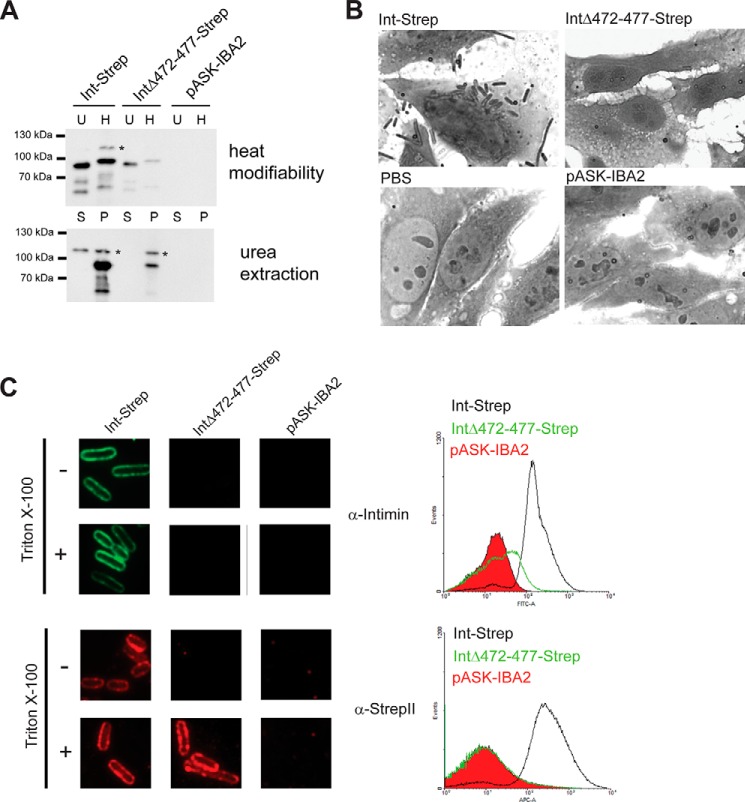
Folded β-barrel proteins are intrinsically resistant to SDS denaturation and remain folded in sample buffer unless they are boiled. The unheated β-barrel thus usually migrates at lower apparent molecular weight than the boiled sample. Properly integrated β-barrels are also resistant to extraction from outer membrane vesicles in 6 m urea. Int-Strep shows two bands: the heat-modifiable band which, when denatured, migrates at the expected size of intimin (99 kDa). We often observe an additional band in the intimin samples, migrating at ∼115 kDa (indicated by an asterisk in Figs. 6A, 8A, and 9A). This band is not heat modifiable and is partially urea-extractable. The cause of this band is not entirely clear, but it has been observed before (11). IntΔ472–477-Strep was produced in E. coli, albeit at lower levels than WT Intimin; however, IntΔ472–477-Strep was heat-modifiable and resistant to urea extraction, showing that the β-barrel domain of this lower amount of protein is correctly folded and inserted into the membrane (Fig. 6B). It is not clear why the antibody does not stain the denatured form of IntΔ472–477-Strep as strongly the native form (the heated and unheated samples derive from the same OM preparation).
FIGURE 6.
Deletion of a β-strand in intimin D00 leads stalling of passenger secretion. A, membrane insertion and folding of intimin Δ472–477. Western blots using an anti-intimin antibody showing heat modifiability (above) and urea extraction (below) of WT Int-Strep and IntΔ472–477-Strep. Bacteria with the empty vector (pASK-IBA2) were the negative control. For heat modifiability assays, the outer membrane sample was split in half, and one duplicate (H) was heated for 10 min at 95 °C before loading. The other, unheated duplicate (U) was kept at room temperature until loaded. In urea extraction experiments, outer membrane samples were treated with 6 m urea for 1 h and then pelleted at 150,000 × g. Samples were loaded from the supernatant (S) and resuspended pellet (P). Asterisks denote the occasionally observed intimin band at ∼115 kDa of uncertain provenance. B, adhesion of bacteria expressing Int-Strep and IntΔ472–477-Strep to Tir-primed HeLa cells. Bacteria containing the empty vector (pASK-IBA2) and a sample with no bacteria (PBS) served as negative controls. C, surface exposure of IntΔ472–477. Surface exposure of the intimin passenger was assayed both by immunofluorescence microscopy (left) and flow cytometry (right). In both, the protein was detected with both an anti-intimin antibody (upper panels) and an anti-StrepII tag antibody (lower panels). In immunofluorescence microscopy, surface-exposed epitopes are detected both with and without treatment with the detergent Triton X-100, but periplasmically located epitopes can only be detected when the cells are permeabilized with the detergent. In flow cytometry, the histogram for Int-Strep is given in black, the histogram for IntΔ472–477-Strep is in green, and the histogram for the vector control (pASK-IBA2) filled in with red.
FIGURE 8.
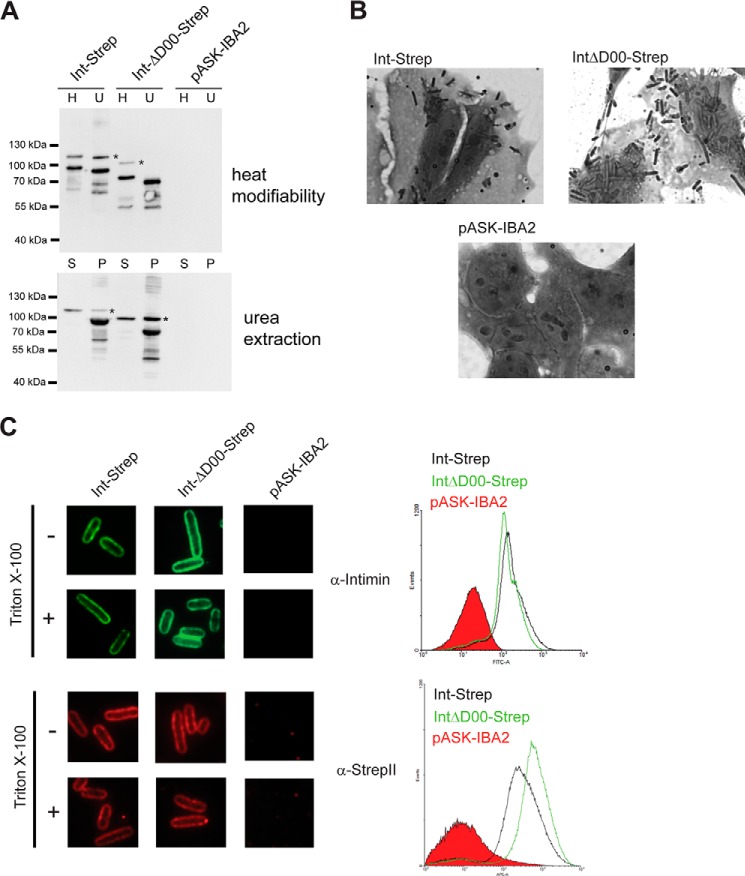
Deletion of the D00 domain does not impair intimin passenger secretion. A, membrane insertion and folding of intimin ΔD00. Western blots using an anti-intimin antibody showing heat modifiability (above) and urea extraction (below) of Int-Strep and IntΔD00-Strep. Bacteria with the empty vector (pASK-IBA2) were the negative control. For heat modifiability assays, the outer membrane sample was split in half and one duplicate (H) was heated for 10 min at 95 °C before loading. The other, unheated duplicate (U) was kept at room temperature until loaded. Note that the samples were loaded differently compared with Fig. 6A (H before U). In urea extraction experiments, outer membrane samples were treated with 6 m urea for 1 h and then pelleted at 150 000 × g. Samples were loaded from the supernatant (S) and resuspended pellet (P). Asterisks denote the occasionally observed intimin band of uncertain provenance. B, adhesion of bacteria expressing Int-Strep and IntΔD00-Strep to Tir-primed HeLa cells. Bacteria containing the empty vector (pASK-IBA2) served as the negative control. C, surface exposure of IntΔD00. Surface exposure of the intimin passenger was assayed both by immunofluorescence microscopy (left) and flow cytometry (right). In both, the protein was detected with both an anti-intimin antibody (upper panels) and an anti-StrepII tag antibody (lower panels). In immunofluorescence microscopy, surface-exposed epitopes are detected both with and without treatment with the detergent Triton X-100, but periplasmically located epitopes can only be detected when the cells are permeabilized with the detergent. In flow cytometry, the histogram for Int-Strep is given in black, the histogram for IntΔD00-Strep is in green, and the histogram for the vector control (pASK-IBA2) filled in with red.
FIGURE 9.
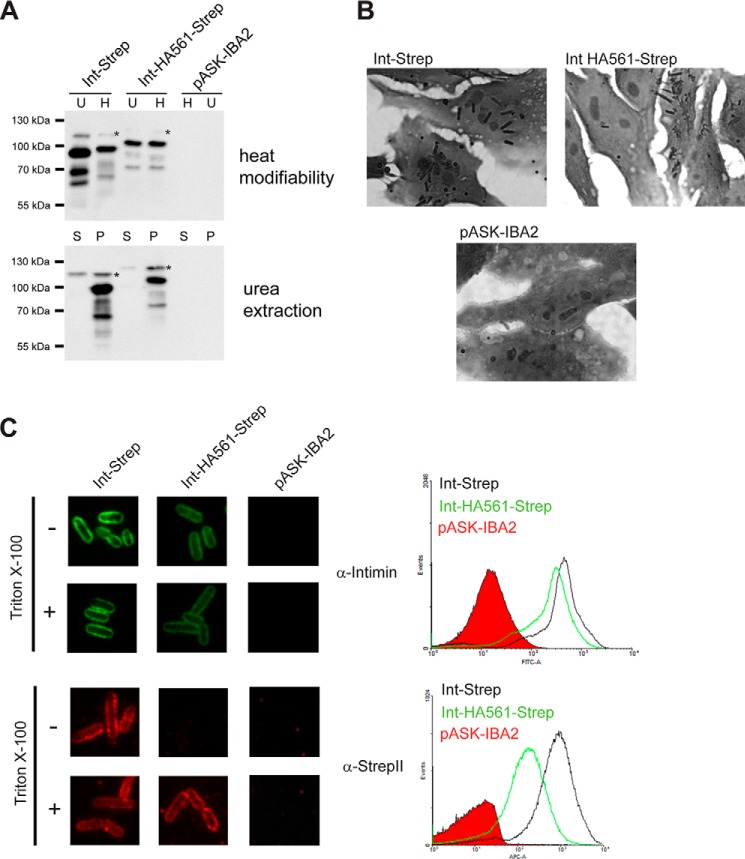
Insertion of a double HA tag into the intimin D0 domain leads to impaired passenger secretion. A, membrane insertion and folding of Int HA561; Western blots using an anti-intimin antibody showing heat modifiability (above) and urea extraction (below) of Int-Strep and Int HA561-Strep. Bacteria with the empty vector (pASK-IBA2) were the negative control. For heat modifiability assays, the outer membrane sample was split in half, and one duplicate (H) was heated for 10 min at 95 °C before loading. The other, unheated duplicate (U) was kept at room temperature until loaded. In urea extraction experiments, outer membrane samples were treated with 6 m urea for 1 h and then pelleted at 150,000 × g. Samples were loaded from the supernatant (S) and resuspended pellet (P). Asterisks denote the occasionally observed intimin band of uncertain provenance. B, adhesion of bacteria expressing Int-Strep and Int HA561-Strep to Tir-primed HeLa cells. Bacteria containing the empty vector (pASK-IBA2) served as the negative control. C, surface exposure of Int HA561. Surface exposure of the intimin passenger was assayed both by immunofluorescence microscopy (left) and flow cytometry (right). In both, the protein was detected with both an anti-intimin antibody (upper panels) and an anti-StrepII tag antibody (lower panels). In immunofluorescence microscopy, surface-exposed epitopes are detected both with and without treatment with the detergent Triton X-100, but periplasmically located epitopes can only be detected when the cells are permeabilized with the detergent. In flow cytometry, the histogram for Int-Strep is given in black, the histogram for Int HA561-Strep is in green, and the histogram for the vector control (pASK-IBA2) filled in with red.
IntΔ472–477-Strep failed to adhere to Tir-primed HeLa cells, suggesting that the passenger is not surface-exposed in this mutant (Fig. 6C). Indeed, when we probed the cells expressing IntΔ472–477-Strep with an anti-intimin or an anti-StrepII antibody, we could not detect any surface-exposed intimin in immunofluorescence microscopy or flow cytometry (Fig. 6D). Only when the cells were permeabilized with detergent did we see a signal with the anti-StrepII antibody in immunofluorescence microscopy, suggesting that the C terminus of IntΔ472–477-Strep is located in the periplasm (Fig. 6D). As expected, we could not detect the intimin passenger with the anti-intimin antibody, even after membrane permeabilization, because SurA covers the epitopes in the passenger when in the periplasm (11). Thus, the phenotype of IntΔ472–477-Strep, although less abundant, is otherwise similar to that observed in Int HA453, which shows that disrupting the folding of D00 leads to inefficient passenger secretion.
Disrupting the D00 Domain Does Not Influence the Topology of the β-Barrel
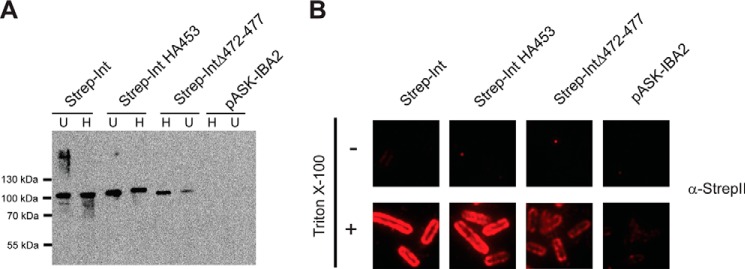
Our results with Int HA453 and IntΔ472–477 show that the C terminus of intimin is located in periplasm, whereas the β-barrel domain is correctly folded. We have interpreted this to mean that we have caught an intimin in a translocation intermediate with both the N and C termini in the periplasm (11). However, another possible, albeit unlikely, reason for these observations is that disrupting the D00 domain leads to an inverted topology, i.e. with the N terminus of intimin outside the cell. To rule out this possibility, we introduced a StrepII tag into WT intimin, Int HA453, and IntΔ472–477 at the N terminus, i.e. on the periplasmic domain, to produce the constructs Strep-Int, Strep-Int HA453, and Strep-IntΔ472–477, respectively. We then produced these proteins in E. coli. As expected, all three proteins were found in the outer membrane fraction and displayed heat modifiability (Fig. 7A). As for the C-terminally tagged IntΔ472–477-Strep, Strep-IntΔ472–477 was produced at lower levels than the other two proteins. We then investigated the topology of the constructs by immunofluorescence microscopy using an antibody against the StrepII tag (Fig. 7B). For all three constructs, the StrepII tag could only be detected when the cells were permeabilized with detergent, demonstrating that the N terminus is in the periplasm and that disruption of the D00 domain does not affect the topology of the β-barrel.
FIGURE 7.
Disrupting the D00 domain does not affect the topology of Intimin. A, Western blot of N-terminally (periplasmically) StrepII-tagged intimin variants with anti-intimin antibody. U = unheated; H = heated at 95 °C for 10 min. B, surface exposure of Strep-Int and derivatives. Surface exposure of the intimin passenger was assayed by immunofluorescence microscopy. Proteins were detected with an anti-StrepII tag antibody. Surface-exposed epitopes are detected both with and without treatment with the detergent TritonX-100, but periplasmically located epitopes can only be detected when the cells are permeabilized with the detergent.
The D00 Domain Is Not Required for Efficient Passenger Secretion
Our original hypothesis was that the D00 domain acts as a membrane-proximal folding core, similar to that found in many classical autotransporters. To test this hypothesis, we deleted the entire D00 domain and observed its effect on passenger secretion. The IntΔD00-Strep protein was produced at similar amounts compared with Int-Strep, and it was heat-modifiable and urea extraction-resistant (Fig. 8A). In a HeLa cell adhesion assay, IntΔD00-Strep adhered at similar levels to the WT, demonstrating that the passenger domain was correctly exported and folded (Fig. 8B). Deleting the D00 domain did not have a significant effect on passenger surface display, and the amount exported was similar for both WT Intimin-Strep and IntΔD00-Strep (Fig. 8C). Our results demonstrate that the D00 domain is not required for efficient passenger secretion, and its deletion has no deleterious effects on passenger surface display. This is in contrast to many classical autotransporters, where the folding core is required for passenger export and/or folding (12–14).
Introduction of the Double HA Tag into the D0 Domain Impairs Passenger Secretion
To determine whether disruption of the second Ig-like domain, D0, would have an effect similar to disrupting D00, we introduced the double HA tag into the D0 domain at position 561, which is the equivalent position to the HA453 insertion in D00. Surprisingly, this protein did not display heat modifiability, probably because the insertion destabilizes the β-barrel, which then unfolds in SDS (Fig. 9A). However, Int HA561-Strep was resistant to urea extraction, showing that the β-barrel is fully integrated in the outer membrane (Fig. 9A). In the HeLa cell adhesion assay, some Int HA561-Strep-expressing cells adhered to Tir-primed HeLa cells but at a lower level than WT Int-Strep (Fig. 9B). This shows that at least some of the passenger is secreted to the surface. Consistent with this, we observed reduced surface exposure of the passenger for Int HA561 in immunofluorescence microscopy and flow cytometry (Fig. 9C). Although the reduction of surface exposure was not as dramatic as for HA453, it was clear when using both the anti-intimin and especially anti-Strep antibodies in flow cytometry. In immunofluorescence microscopy, staining with the anti-intimin antibody resulted in less intense staining than the WT in both the intact and permeabilized cells. However, we only saw a signal with the anti-Strep antibody with permeabilized cells. We interpret these results to mean that some of the passenger domain is exported but a significant proportion remains in the periplasm. As for IntΔ472–477-Strep, periplasmically located intimin passenger will not stain with the anti-intimin antibody; thus, the anti-intimin antibody only binds to surface-exposed Int HA561-Strep, explaining why the staining is equal for the intact and permeabilized cells.
The D00 Domain Is No More Stable Than the Other Ig Domains of the Intimin Passenger
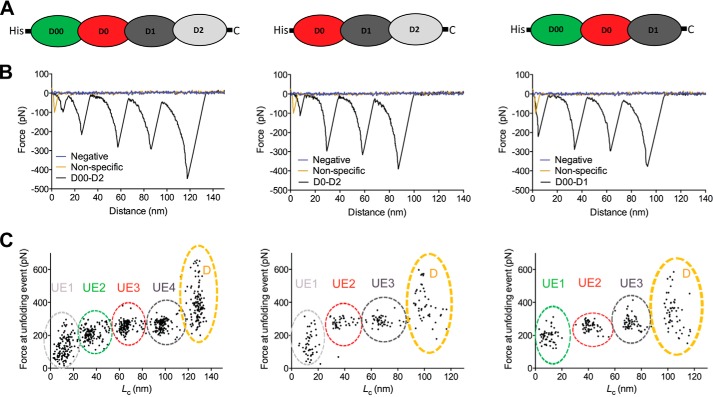
The membrane-proximal folding cores of classical autotransporters form particularly stable structures (15, 19). We wished to test whether the D00 domain is more stable than the other Ig domains of the intimin passenger. We, therefore, produced a construct containing the four Ig domains (D00, D0, D1, and D2). We then subjected the purified D00-D2cys protein to atomic force microscopy (AFM). The C terminus of the protein was coupled to a gold surface via the engineered C-terminal cysteine (after reduction of the disulfides), and the N terminus containing the His tag was bound to the AFM tip. The force extension curves generated in the AFM experiments show four unfolding events, with a contour length increment (ΔLc) of ∼30 nm (Fig. 10 and Table 1). 30 nm is close to the ΔLc of the extended Ig domains previously measured with AFM (20). Two of these unfolding events have a force peak of ∼260 pN, and a third domain unfolds at ∼210 pN, whereas the fourth has a lower force maximum at ∼130 pN (Fig. 10 and Table 1). All these values are consistent with values observed for the unfolding of Ig domains in titin (150–300 pN) (21).
FIGURE 10.
AFM measurements on intimin passenger constructs. A, schematic depiction of the three constructs used in AFM. The construct consist of three to four Ig-like domains (D00, D0, D1, and D2) of the intimin passenger, with an N-terminal His tag and C-terminal cysteine for coupling to the gold substrate. The coloring of the schematic matches the coloring of the domains in Fig. 1. B, representative force extension curves (in black) corresponding to the construct depicted above. For comparison, a negative force extension curve (no protein attached to the substrate, in blue) and an unspecific force extension curve (in yellow) are depicted. C, the force peaks from each unfolding event from multiple experiments plotted against contour length (Lc). The Lc values were calculated for each force-distance curve by fitting to a wormlike chain model. The clusters of unfolding events (UE) are indicated, and the colors match the schematic depictions in panel A. The unfolding events for D00 and D2 can be unambiguously assigned based on the differences between the constructs, but the unfolding events for D0 and D1 are arbitrarily assigned. D stands for detachment of the sample from the cantilever tip. See Table 1 for statistics.
TABLE 1.
Contour length increments and force peaks derived from AFM experiments
Values for contour length increments (ΔLc) and force peaks are given as the mean of all experiments ± S.D. ΔLc is defined as the distance from the force peak of an unfolding event to force peak of the following one. N/A = not applicable.
| D00-D2 | D0-D2 | D00-D1 | |
|---|---|---|---|
| Number of measurements | 125 | 45 | 55 |
| Unfolding event 1 | |||
| ΔLc (nm) | 25.1 ± 6.2 | 24.9 ± 6.0 | 31.3 ± 3.3 |
| Max. force (pN) | 130.9 ± 64.2 | 148.0 ± 71.4 | 196.4 ± 40.1 |
| Unfolding event 2 | |||
| ΔLc (nm) | 28.2 + 5.0 | 31.0 ± 3.5 | 31.5 ± 1.8 |
| Max. force (pN) | 210.3 ± 41.6 | 276.1 ± 38.2 | 255.28 |
| Unfolding event 3 | |||
| ΔLc (nm) | 30.6 ± 2.7 | 31.3 ± 1.5 | 31.5 ± 2.8 |
| Max. force (pN) | 255.5 ± 31.3 | 286.6 ± 29.4 | 279.1 ± 38.9 |
| Unfolding event 4 | |||
| ΔLc (nm) | 30.5 ± 4.0 | N/A | N/A |
| Max. force (pN) | 264.0 ± 15.0 | N/A | N/A |
Based on these results, we could not determine which unfolding event corresponds to which Ig domain. Therefore, we produced two other constructs, D00-D1cys and D0-D2cys, that lack one of the two outermost Ig domains. When we performed unfolding experiments on these, the D00-D1cys protein gave two peaks of ∼260 pN and one at ∼200 pN (Fig. 10 and Table 1). In contrast, the D0-D2cys showed 2 peaks at ∼280 pN and 1 peak at ∼150 pN (Fig. 10 and Table 1). As the Ig domain with this lowest mechanical stability is present in both D00-D2 and D0-D2, this must the distal D2 domain. In the native protein, this domain forms a superdomain with the lectin-like D3 domain. Thus, outside its native context, this domain is probably less stable than the other intimin Ig domains. The D00 domain appears to be the next one to unfold under an applied force of ∼200 pN. The other two Ig domains (D0 and D1) are of similar mechanical stability. Our experiments demonstrate that D00 is not more stable than the two Ig domains following it and in fact is slightly less mechanically stable.
Discussion
Protein secretion in Gram-negative bacteria poses special challenges; not only must the secreted polypeptides cross two membranes, but the outer membrane is also absent of obvious energy sources. The periplasm is devoid of ATP, and the permeability of the outer membrane to low molecular weight compounds prevents maintaining electrochemical gradients across it. Thus, the energy for outer membrane translocation in two-step secretion systems (such as type V secretion systems and pili assembled by the chaperone-usher pathway) must come from alternative sources. In pilus biogenesis, the energy for subunit translocation was long thought to derive from the formation of a helical quaternary structure by the rod outside the cell. However, recently this assumption was shown to be incorrect, and other factors than coiling of the rod must prevent backsliding during pilus assembly (22). Protein folding has been suggested to provide the free energy to drive secretion in two-partner (type Vb) secretion systems and for passenger domain secretion in classical (type Va) autotransporters (23, 24). The exported domain of these proteins typically comprises a stable β-helical structure. In classical autotransporters, folding of the passenger starts at the C terminus at a particularly stable region earlier referred to as the autochaperone domain. Folding then proceeds in a vectorial manner to the N terminus (15, 25). This sequential folding would function as a Brownian ratchet that prevents backsliding of the polypeptide chain into the periplasm (24).
Recently, this model has also been questioned. Based on calculations of the free energy required for secretion, Bernstein and co-worker (26) have argued that protein folding alone cannot provide sufficient free energy to drive passenger translocation. Introducing mutations that locally destabilize the β-helix into the medial and N-terminal regions of the passenger of the classical autotransporter EspP only had a moderate effect on passenger secretion (26). The translocator of EspP can secrete an intrinsically unfolded polypeptide efficiently, and this secretion is dependent on the negative charge of the polypeptide rather than protein folding (27). Similarly, the passengers of autotransporters are usually acidic, which might promote secretion through electrostatic repulsion (27). However, very recent work on the classical autotransporter pertactin showed that protein folding was the dominant driving force for passenger secretion (16). In addition, many classical autotransporters contain passenger-associated transport repeat motifs, which aid in passenger folding and secretion (28).
Here, we provide evidence to show that in the case of inverse (type Ve) autotransporters, protein folding appears to be the main driving force for secretion. Disrupting the folding of the D00 domain by insertion of a double HA tag or by deleting a predicted β-strand leads to stalling of passenger secretion. Passenger secretion can also be impaired by introducing the double HA tag into the second Ig-like domain D0.
Our results lead us to two conclusions about the mechanism of type Ve secretion. First, in contrast to many classical autotransporters, there is no folding core or autochaperone region in intimin (or by extension other inverse autotransporters). The D00 domain is not more stable than the other Ig-like domains; in fact, our AFM measurements show it is somewhat less stable than the two subsequent Ig-like domains (D0-D1). Furthermore, the D00 domain can be removed in its entirety without any defects in secretion. The D0 domain can thus substitute the function of the D00 domain in the secretion process. Although we did not test this, it seems reasonable to assume that also the D1 domain could act as a secretion initiator in the absence of both the D00 and D0 domains. D1, like D00 and D0, is an Ig-like domain, and, similar to D0, it is more stable than D00. These repeated Ig-like domains act as independent folding modules; thus, one domain can fold at the cell surface regardless of whether it is preceded by other domains. It is possible that the marginal instability of D00 plays a subtle role in the initiation of the autotransport process that is not detected with the assays we used here. However, we prefer the interpretation that there is nothing intrinsically exceptional about the D00 domain regarding initiation of folding, and it can be functionally replaced by the D0 domain for efficient passenger secretion.
Second, our results suggest that protein folding is the main driving force for passenger domain secretion in type Ve autotransporters. Our refolding experiments show that the D00 domain folds robustly in a number of conditions, in contrast to D00-HA and D00Δ472–477, which both failed to refold properly under the same conditions. Robust folding is to be expected for extracellular autotransporter domains to provide the free energy for transport.
Although we cannot fully exclude the possibility that other factors, such as charge distribution in the polypeptide sequence or molecular crowding in the periplasm, do play a role in inverse autotransport, it appears that folding of the individual Ig-like domains is the main mechanism for providing the free energy to pull the polypeptide chain to the cell surface. The addition of the HA tag changes the calculated pI of the D00 from 9.1 to 5.9, thus making it significantly more acidic. However, the deletion of the second β-strand (Δ472–477) results in a modest increase, giving a calculated pI of 9.6. Thus, the deletion of residues 472–477 demonstrates that the stalled phenotype in both IntΔ472–477 and Int HA453 is not due to a large change in the pI of the D00 domain but because these changes prevent the domain from folding correctly, which in turn prevents secretion of the rest of the passenger.
In contrast to mutations that impair folding proximal to the membrane, mutations in the distal passenger (C-terminal in inverse and N-terminal in classical autotransporters) might not have an equally large effect on secretion efficiency. Due the extended shape of autotransporter passengers, a large portion of the polypeptide will already be outside the cell by the time distal mutations are exported, and correct folding at the distal end is, therefore, not needed for secretion. This is supported by our results with the HA561 mutant, where the D0 domain is disrupted. Here, we observed partial export of the passenger and not complete stalling as for the HA453 mutant. The D00 domain is intact in the HA561 mutant, and therefore, the stalling at the D0 domain only occurs once an appreciable amount of the polypeptide is already outside the cell. This might be enough to allow diffusion of the passenger to the outside of the cell, albeit with low efficiency. In the classical autotransporter EspP, distal destabilizing mutations did not significantly impair secretion, and even combinations of such mutations had only moderate effects (26). These results are similar to what we observe with Int HA561.
As a final note, passenger secretion may not be an autonomous process of the autotransporter polypeptide chain. BamA, the central component of the BAM complex, has been implicated not only in membrane insertion of classical autotransporters but also in the secretion of passengers (13, 29). Furthermore, a subset of classical autotransporters including antigen 43, is dependent on a BamA homologue, TamA, for efficient secretion (30). TamA interacts with an inner membrane protein, TamB, which together form the translocation and assembly (TAM) complex (30). The TAM complex has recently been shown to promote membrane insertion of antigen 43 in a reconstituted system (31). Our work on the stalled mutant Int HA453 showed that BamA is associated with the mutant protein despite the β-barrel apparently being fully folded and inserted into the membrane (11). This suggests that the intimin passenger domain, still located in the periplasm, is connected to BamA. Thus, BamA might also be involved in passenger secretion, e.g. by shuttling the unfolded passenger domain along its POTRA (polypeptide transport-associated) domains toward the intimin β-barrel or by acting as the secretion pore itself. The BamA POTRA domains interact with amphiphilic motifs present in C-terminal β-strands of transmembrane β-barrels, but these motifs are not very sequence-specific (32, 33). Autotransporter passengers are rich in β-structures; classical autotransporter passengers and TpsA proteins of two-partner secretion systems mostly consist of β-helices, and many inverse autotransporters have Ig-like (all-β) domains. These β-structures also have amphipathic character; thus, it could be that BamA interacts with these kinds of proteins in a nonspecific manner, recognizing amphiphilic β-motifs also in the passengers. However, the exact role of BamA and its homologue TamA in autotransporter secretion remains enigmatic, and elucidating their contribution to passenger translocation and the energetics of secretion will require further investigation.
Experimental Procedures
Bioinformatics and Protein Structure Prediction
Sequence analysis was performed using the programs of the online bioinformatics toolkit of the Max Planck Institute for Developmental Biology (34). For retrieval of D00 domain sequences, we submitted the intimin β-barrel and D00 domain sequence to a position-specific iterated BLAST (35) search against the nr_bac70 database with five iterations. The resulting sequences were then aligned using Kalign (36), and the β-barrel sequences were removed based on this alignment. Secondary structure prediction for the D00 domains was performed with Ali2D, and the alignment was manually edited. The three-dimensional atomic model for D00-D0 domain was modeled from multiple threading alignments (37) and iterative structural assembly simulations followed by structure refinement using the Discovery Studio software suite version 4.5 as described recently (38, 39).
Cloning and Mutagenesis
All plasmids used in this study are summarized in Table 2. To clone the D00 domain, the DNA sequence coding for the D00 domain was amplified from wt EPEC E2348/69 intimin (8) using PCR with Q5 polymerase (New England BioLabs). D00-HA was amplified from pIBA2-Int HA453 (11). The PCR products were digested with the enzyme BsaI (enzymes were from New England BioLabs) and ligated into the expression vector pASK-IBA3 (for cytoplasmic production, from IBA GmbH) or pASK-IBA2 (for periplasmic production). The ligation mixture was transformed into chemically competent E. coli TOP10 (Invitrogen), and transformants were selected on lysogeny broth (LB) supplemented with 100 μg/ml ampicillin (40). Positive clones were screened for by colony PCR and the correctness of the constructs was verified by sequencing. The resulting constructs included an N-terminal hexahistidine tag for purification, a short stretch of sequence preceding the D00 domain (starting at residue 443, so that we could use the same primers for cloning both the WT and D00-HA), the D00, and a C-terminal StrepII tag.
TABLE 2.
Plasmids used in this study
| Plasmid | Insert | Notes | Source |
|---|---|---|---|
| pIBA2-D00 | Intimin residues 443–550 | For periplasmic production of wild-type intimin D00, includes OmpA signal peptide for periplasmic export, an N-terminal His tag after the signal peptide cleavage site and a C-terminal StrepII tag. | This study |
| pIBA3-D00 | Intimin residues 443–550 | For cytoplasmic production of WT intimin D00, includes an N-terminal His tag and a C-terminal StrepII tag. | This study |
| pASK-IBA2 | None | Expression vector for periplasmic protein targeting | IBA GmbH |
| pASK-IBA3 | None | Expression vector for cytoplasmic targeting | IBA GmbH |
| pIBA2-D00-HA | Intimin residues 443–550, with double HA tag | For periplasmic production of intimin D00-HA. Identical to pIBA2-D00,but includes double HA tag at position 453. | This study |
| pIBA3-D00-HA | Intimin residues 443–550, with double HA tag | For cytoplasmic production of intimin D00-HA. Identical to pIBA3-D00 but includes double HA tag at position 453. | This study |
| pIBA3-D00 Δ472–477 | Intimin residues 443–550, with residues 474–7 deleted | For cytoplasmic production of intimin D00. Identical to pIBA3-D00, but second predicted β-strand of D00 is deleted | This study |
| pIBA2-D00 Δ472–477 | Intimin residues 443–550, with residues 472–477 deleted | For periplasmic production of intimin D00. Identical to pIBA3-D00, but second predicted β-strand of D00 is deleted. | This study |
| pIBA3-IntD00-D2cys | Intimin residues 450–841 | For production of all Ig-like domains of intimin passenger. Includes N-terminal His tag for purification, and C-terminal cysteine for coupling to a gold surface for AFM experiments. | This study |
| pIBA3-IntD0-D2cys | Intimin residues 550–841 | For production of 3 Ig-like domains of intimin passenger. Includes N-terminal His tag for purification, and C-terminal cysteine for coupling to a gold surface for AFM experiments. | This study |
| pIBA3-IntD00-D1cys | Intimin residues 450–753 | For production of 3 Ig-like domains of intimin passenger. Includes N-terminal His tag for purification, and C-terminal cysteine for coupling to a gold surface for AFM experiments. | This study |
| pIBA2-Int-Strep | Full-length intimin | For production of full-length intimin, with OmpA signal peptide for periplasmic export. Includes C-terminal StrepII tag for detection. | This study |
| pIBA2-IntΔ472–477-Strep | Full-length intimin, with residues 472–477 deleted | For production of full-length intimin, but second predicted β-strand of D00 is deleted. Includes C-terminal StrepII tag for detection. | This study |
| pIBA2-Int HA561-Strep | Full-length intimin, with double HA tag at position 561 | For production of full-length intimin, but a double HA tag has been introduced into the D0 domain. Includes C-terminal StrepII tag for detection. | This study |
| pIBA2-Strep-Int | Full-length intimin | For production of full-length intimin, with OmpA signal peptide for periplasmic export. Includes N-terminal StrepII tag for detection. | This study |
| pIBA2-Strep IntΔ472–477 | Full-length intimin with residues 472–477 deleted | For production of full-length intimin, but second predicted β-strand of D00 is deleted. Includes N-terminal StrepII tag for detection. | |
| pIBA2-Strep-Int HA453 | Full-length intimin with double HA tag inserted at position 453 | For production of full-length intimin with a double HA tag introduced into the D00 domain. Includes N-terminal StrepII tag for detection. | This study |
The full-length intimin construct was produced by amplifying the DNA coding for the mature protein (without signal peptide) by PCR from EPEC E2348/69 genomic DNA. This was then digested with BsaI and ligated into pASK-IBA2 such as to include a C- or N-terminal StrepII tag. The ligation mixture was transformed into E. coli TOP10, and the correct clones were screened for and verified as above.
For mutagenesis to produce the HA561, Δ472–477, and ΔD00 constructs, we employed an inverse PCR protocol to amplify the entire plasmid (41). Briefly, after the PCR reaction, the amplified PCR products were phosphorylated with polynucleotide kinase, and the template plasmid was digested with DpnI. The PCR products where then circularized using T4 DNA ligase and transformed into E. coli TOP10. Transformants were selected on LB plus ampicillin. Correct clones were identified by sequencing. Primer sequences are available on demand. For HA561, we introduced the same double HA tag (GSGYPYDVPDYAGSGYPYDVPDYAGSG) as used in the HA453 mutant. The double HA tag was originally used to boost the fluorescence signal in immunofluorescence microscopy (8).
To produce the D00-D2cys construct, the part of the intimin gene coding for the Ig-like domains D00, D0, D1, and D2 (but not the lectin-like D3 domain) was amplified by PCR. The D00-D1cys and D0-D2cys fragments were produced similarly, but each lacked one of the outer Ig domains. The primers were designed so that a hexahistidine tag was included at the N terminus, and a free cysteine residue was added at the very C terminus (no other cysteines are coded for in the resulting fragment). This fragment was then cloned into pASK-IBA3 by Gibson assembly (42). The assembly mix was cloned into TOP10, and positive colonies were screened for as above.
Protein Production and Detection
For production of the D00 domain and its derivatives, plasmids were transformed into the expression strain BL21Gold(DE3) (Novagen). To produce the proteins and determine their solubility, overnight cultures were diluted 1:200 into 50 ml of ZYP medium (43) supplemented with ampicillin at 100 μg/ml. The cultures were grown at 37 °C till mid-log phase (A600 ∼0.5), at which time protein production was induced with anhydrotetracycline (IBA GmbH) at 0.2 μg/ml. The cultures were grown for a further 2 h. The cells were collected by centrifugation (10 min at 4000 × g) and resuspended in 1 ml of PBS (20 mm sodium phosphate, 150 mm NaCl, pH 7.4). We then added MgCl2 and MnCl2, both to 1 mm, and a pinch of DNase I. The cells were lysed using a French pressure cell (2 passes at 18 000 p.s.i). Cellular debris was pelleted by centrifugation (10 min at 16,000 × g). The pellet was extracted twice with PBS and 1.5% Triton X-100 and finally resuspended in 1 ml of PBS. For detection, samples from the lysate (before centrifugation), the first supernatant after centrifugation, and the resuspended pellet after detergent extraction were subjected to SDS-PAGE. For Western blotting, the separated proteins were transferred to a nitrocellulose (Protran BA 85, GE Healthcare) or a polyvinylidene fluoride membrane using a semi-dry apparatus. The membrane was blocked with 2% fat-free milk powder dissolved in TBS (20 mm Tris, pH 7.4, and 150 mm NaCl), after which an anti-StrepII antibody (monoclonal from IBA GmbH or polyclonal from Abnova), diluted 1:200 (monoclonal) or 1:3000 (polyclonal) in blocking buffer, was applied to the membrane. After 1 h at room temperature, the membrane was washed 3 times with TBS and 0.05% Tween 20, and a secondary antibody (goat anti-mouse-alkaline phosphatase from Dianova or goat anti-rabbit-HRP from Santa Cruz), diluted 1:10 000 in blocking buffer, was added. After 3 washes with TBS and 0.05% Tween 20, chromogenic detection was performed using the alkaline phosphatase substrate nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate, diluted to 33 and 17 ng/ml, respectively, in 100 mm Tris, pH 9.5, 100 mm NaCl, 5 mm MgCl2. For HRP detection, an enhanced chemiluminescent substrate was added (Pierce), and imaging was performed using a Kodak Image Station 4000R.
Purification of Intimin D00 and Passenger Constructs for Atomic Force Microscopy
For purification of intimin D00, BL21Gold(pIBA3-D00) was grown in 2 liters of ZYP medium with ampicillin (100 μg/ml) till mid-log, when protein production as induced with anhydrotetracycline as above. After 3 h of induction, the cells were pelleted as above and resuspended in 40 mm sodium phosphate, pH 8.0, 400 mm NaCl. After lysis by French pressing, as above, the lysate was centrifuged for 1 h at 100,000 × g. The clarified supernatant was applied to a 5-ml HisTrap column (GE Healthcare), and bound protein was eluted with a concentration gradient of imidazole. The eluted protein was dialyzed overnight against 10 mm HEPES, pH 7.3. The following day the protein was applied to a cation exchange column (1 ml of Mono S from GE Healthcare) equilibrated with 10 mm HEPES, pH 7.3, and eluted with a NaCl gradient in the same buffer. The resulting D00 protein was pure.
D00-D2cys, D00-D1cys, and D0-D2cys were produced as above. After cell lysis using a French pressure cell as above, the proteins were purified first by applying to HisTrap column as above. The eluate from this column was dialyzed against 10 mm Tris, pH 7.5, and then applied to a HiTrap SP cation exchange column (GE Healthcare) from which is was eluted with a sodium chloride gradient (up to 1 m) in the same buffer. The peak containing the D00-D2cys protein was concentrated (Vivaspin centrifugal concentrator, 30-kDa molecular weight cut off) and applied to a 26/60 Sephacryl200 column (GE Healthcare) equilibrated with 20 mm Tris, pH 7.5, 200 mm NaCl, and 0.02% sodium azide. The peak containing the intimin fragment was collected and concentrated as above. The protein was pure. However, we noticed that the purified proteins tended to migrate at the expected size of a dimer (∼80 kDa) in SDS-PAGE, due to disulfide formation by the C-terminal cysteines. Therefore, the disulfides were reduced with tris(2-carboxyethyl)phosphine before AFM measurements (see below).
Refolding of D00 and Its Variants
For refolding experiments, D00 was produced and purified as above. The purified protein was then concentrated to 10 mg/ml and diluted to 1:4 in denaturing buffer (8 m guanidine hydrochloride, 10 mm HEPES, pH 7.4) to give a concentration of 2.5 mg/ml D00. The protein was then refolded in a matrix of 9 conditions by diluting it 1:20 into the various buffers. The buffer matrix was prepared by combining three different NaCl concentrations (0, 150 and 500 mm, final concentrations after dilution) with 3 different buffers (Tris, pH 8.0; MOPS, pH 7.0; or MES, pH 6.0, each at 50 mm final concentration). After incubating for 1 h at room temperature, the samples were centrifuged (15 min at 16,000 × g), and the supernatant was removed. The pellet was dissolved in 1 × SDS-PAGE sample buffer. To analyze the protein content of the supernatant, the proteins were pelleted by ethanol precipitation: 9 volumes of cold (−20 °C) ethanol was added, the samples were incubated for 20 min at −20 °C and then centrifuged as above. The supernatant was discarded, and the pellet was dissolved in 1× SDS-PAGE sample buffer. The samples were then subjected to SDS-PAGE to determine the amount of soluble protein.
The D00 variants D00-HA and D00Δ472–477 were produced as inclusion bodies under the same conditions as the WT. The cell pellets were resuspended in PBS and lysed using a French pressure cell as above. The inclusion bodies were pelleted by centrifugation (10 min at 3000 × g), extracted twice with PBS plus 1% Triton X-100, and finally washed once with PBS. The inclusion body pellet was then resuspended in PBS plus 6 m guanidine hydrochloride, and the inclusion bodies were solubilized overnight at room temperature. The supernatant was centrifuged (15 min at 4000 × g) and applied to a Nickel-Sepharose Excel column (GE Healthcare). The protein was then eluted under denaturing conditions with an imidazole gradient. The resulting protein was pure. We measured the concentration based on absorbance at 280 nm, and the concentration of the protein was adjusted to 2.5 mg/ml. Refolding was then assayed in the same conditions as for the WT followed by analysis by SDS-PAGE as described above.
Circular Dichroism Spectroscopy
For CD spectrum measurement, intimin D00 was diluted to ∼0.2 mg/ml in 10 mm HEPES, pH 7.3, and spectra were measured using a 1-mm quartz cuvette and a Jasco J-810 machine. The presented spectrum is the average of 5 individual measurements, accumulated at a scan speed of 200 nm/min (response time 1 s) with a data pitch of 0.1 nm (0.5 for refolding experiments) and a bandwidth of 1 nm.
Differential Scanning Fluorometry
For differential scanning fluorometry, D00 and refolded D00-HA were diluted to ∼10 μm in 10 mm HEPES, pH 7.4, 10 mm NaCl. Emission intensities were measured with a Jasco-815 machine at 350 nm (bandwidth 10 nm). Tryptophan fluorescence was excited at 295 nm (bandwidth 5 nm). Intensities were measured for each degree between 25 °C and 85 °C. To see transitions clearly, the fluorescence was normalized by dividing the fluorescence intensity by the difference between the maximum and minimum fluorescence (ΔF = Fmax − Fmin) obtained for the sample and expressed as a percentage (F × 100%/ΔF).
Adhesion Assay
HeLa cells were seeded onto coverslips and grown overnight in RPMI 1640 (Biochrom, Berlin, Germany). The next day cells were washed and incubated in medium without antibiotics for 1 h. Afterward HeLa cells were infected at a multiplicity of infection value of 100 with the EPEC strain E2348/69 ΔeaeA for 2 h. After 4 washing steps, the remaining adherent bacteria were killed by incubation with gentamicin (100 μg/ml) for 1 h. Finally, cells were washed with medium without antibiotics. For infection with E. coli strains expressing WT intimin or intimin mutants, bacteria were centrifuged onto preinfected HeLa cells and incubated for 2 h. After three washing steps with PBS, the cells were fixed overnight with 4% paraformaldehyde in PBS and stained with fuchsine for 60 s before mounting the coverslips in Entellan (Merck). Samples were analyzed with a light microscope with a 100-fold magnification. For detailed protocol information, see Oberhettinger et al. (11).
Assessing Outer Membrane Insertion and Folding
For heat modifiability assays, outer membrane fractions were prepared essentially as described in Leo et al. (44). Briefly, an amount of BL21omp2 cells (45) transformed with the appropriate plasmid and grown in ZYP medium and induced with anhydrotetracycline (50 ng/ml), corresponding to 50 ml at an A600 value of 1.0, were pelleted. The cells were resuspended in 1 ml of lysis buffer (10 mm HEPES, pH 7.4, 1 mm MgCl2, 1 mm MnCl2, 0.1 mg/ml lysozyme, and a pinch of DNase I) and disrupted using a bead beater (Thermo FastPrep FP120). After a short centrifugation to remove cell debris and unlysed cells, membranes were pelleted by centrifuging at 16,000 × g for 30 min. The pelleted membranes were resuspended, and the inner membrane was solubilized in 1% N-lauroyl sarcosine for 30 min at room temperature. The outer membrane vesicles were pelleted as above, and after 1 wash with 10 mm HEPES, pH 7.4, the outer membrane pellet was resuspended in 45 μl of 10 mm HEPES, pH 7.4, and 15 μl of 4 × SDS-PAGE sample buffer was added. The sample was then split into two: one-half was heated for 10 min at 95 °C, and the other half was incubated at room temperature. 5 μl of the samples were then applied to a 10% polyacrylamide gel. After electrophoresis, the samples were transferred to a polyvinylidene fluoride membrane (Thermo) using a semidry apparatus. After transfer, the membrane was blocked with 2% skimmed milk powder in PBS (w/v) either 1 h at room temperature or overnight. The primary antibody, a rabbit polyclonal antibody against the C terminus of intimin (11), was diluted 1:5000 in blocking buffer, and the membrane was incubated with the antibody for 1 h at room temperature. After 2 washes with PBS plus 0.05% Tween 20, the membrane was incubated with the secondary antibody (goat-anti-rabbit-HRP, from Santa Cruz, diluted 1:10,000 in blocking buffer) for 1 h at room temperature. For detection, we used enhanced chemiluminescence (WesternBright substrate from Advansta), and imaging was performed using a Kodak Image Station 4000R.
For urea extraction experiments cells were grown and induced as above, and an amount corresponding to 500 ml at an A600 value of 1.0 was pelleted. The cells were resuspended in lysis buffer as above and disrupted using a French pressure cell (3 passes at 18 000 p.s.i). The outer membrane fraction was then isolated essentially as above, although centrifugations were performed at 22,000 × g. The purified outer membrane pellet was resuspended in 1 ml of urea buffer (15 mm HEPES, pH 7.4, 100 mm glycine, 6 m urea) and incubated with shaking for 1 h at 37 °C. The insoluble membranes were then pelleted for 1 h at 150,000 × g. A sample was taken from the supernatant for SDS-PAGE, and the rest of the supernatant was discarded. The pellet was thoroughly resuspended in 1 ml of 10 mm HEPES, pH 7.4, and a sample was taken for SDS-PAGE. The samples were heated at 95 °C for 10 min and then applied to a 10% polyacrylamide gel. Detection was done by Western blot as above.
Immunofluorescence Microscopy and Flow Cytometry
Immunofluorescence staining was performed as described in Oberhettinger et al. (11). Briefly, bacteria on coverslips were fixed with 4% paraformaldehyde in PBS (w/v) and subsequently blocked with 1% BSA in PBS (w/v). Staining of periplasmically localized StrepTagII was performed after permeabilization of bacterial cells with 0.5% Triton X-100/PBS (v/v); the C terminus of the intimin passenger was detected using antibodies directed against EaeA. Subsequently, secondary antibodies were incubated for 2 h in a dark chamber before mounting the coverslips with Mowiol. Fluorescence images were obtained using an upright DMRE fluorescence microscope (Leica, Wetzlar, Germany).
For quantitation of surface-exposed intimin by cytometry, bacteria expressing intimin were harvested by centrifugation. Cells were washed with PBS, fixed with 4% paraformaldehyde, and finally, blocked with 1% BSA in PBS. Afterward, cells were stained with rabbit anti-intimin (1:200) or mouse anti-Strep tagII, respectively, overnight at 4 °C followed by incubation with anti-rabbit Cy2- (1:100; Dianova) or anti-mouse DyLight649 (1:100; Dianova) secondary antibody for 2 h at room temperature. Surface localization of the intimin C terminus was measured by flow cytometry using an LSRFortessa cell analyzer (BD Biosciences). Data were analyzed with WinMDI (J. Trotter) software.
Atomic Force Microscopy Measurements
To prepare nickel-nitrilotriacetic acid-functionalized probes, silicon nitrite gold-coated cantilevers (BL-RC150VB-C1: Olympus, Japan) were cleaned in piranha solution (H2SO4:30% H2O2 = 3:1) for 5 min and then washed with pure water. The nickel-nitrilotriacetic acid-functionalized probes (spring constant = 20–35 pN/nm) were calibrated by the thermal noise method (46) before the measurement. The cantilevers were incubated in 0.1 mm NTA-SAM formation reagent (Dojindo Laboratories, Tokyo, Japan) dissolved in ethanol for 2 h and then washed with ethanol. After this, the probes were incubated in 40 mm NiSO4 for 1 h and washed with pure water. 1 mg/ml purified protein (Intimin D00-D2, D00-D1 or D0-D2) was reduced in 5 mm tris(2-carboxyethyl)phosphine in PBS for 1 h and placed for 30 min on a gold substrate (ARGOLD-15 mm: Asylum Research) cleaned with piranha solution and then washed with PBS. Force spectroscopy measurements were performed with an MFP-3D-BIO atomic force microscope (Asylum Research) in PBS. Using contact mode, the probe was brought to the gold substrate and pushed at a force of 100 pN for 1 s then pulled up at a velocity of 1 μm/s. All AFM experiments were performed at room temperature.
Author Contributions
J.C.L., P. O., M. S, K. H., and D. L. conceived and coordinated the study. J. C. L., P. O., S. Y., and D. L. wrote the paper. P. O. performed the immunofluorescence microscopy, flow cytometry, and adhesion assays. S. Y. performed and analyzed the AFM measurements. D. B. R. K. G. U. produced the D0-D00 homology model. J. P. M. performed the differential scanning fluorometry measurements. J. C. L. performed the cloning and protein purification and designed and performed all other experiments.
Supplementary Material
Acknowledgments
We are grateful to Prof. Per Eugen Kristiansen (University of Oslo) for help with CD spectroscopy and to Ina Meuskens and Hawzeen Salah Khalil for technical assistance. We thank Ingo B. Autenrieth for continuing support and fruitful scientific discussions.
This work was supported by the Fortüne Program F1433253 (to P. O.), a grant from VISTA, a basic research program funded by Statoil in collaboration with the Norwegian Academy of Science and Letters (to J. C. L.), grants from the German Research Council (DFG SFB766) and the German Center for Infection Research (DZIF) (to M. S.), and by a Molecular Life Science (MLS) initiative grant from the University of Oslo and by the Research Council of Norway, Grant 230576-BACTAD (to D. L.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental File 1.
- EPEC
- enteropathogenic E. coli
- AFM
- atomic force microscopy
- BAM
- β-barrel assembly machinery
- HA
- hemagglutinin
- Ig
- immunoglobulin
- LB
- lysogeny broth
- TAM
- translocation and assembly machinery
- pN
- piconewtons.
References
- 1. Schmidt M. A. (2010) LEEways: tales of EPEC, ATEC, and EHEC. Cell. Microbiol. 12, 1544–1552 [DOI] [PubMed] [Google Scholar]
- 2. Kenny B., DeVinney R., Stein M., Reinscheid D. J., Frey E. A., and Finlay B. B. (1997) Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520 [DOI] [PubMed] [Google Scholar]
- 3. Luo Y., Frey E. A., Pfuetzner R. A., Creagh A. L., Knoechel D. G., Haynes C. A., Finlay B. B., and Strynadka N. C. (2000) Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 4. Kelly G., Prasannan S., Daniell S., Fleming K., Frankel G., Dougan G., Connerton I., and Matthews S. (1999) Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Biol. 6, 313–318 [DOI] [PubMed] [Google Scholar]
- 5. Fairman J. W., Dautin N., Wojtowicz D., Liu W., Noinaj N., Barnard T. J., Udho E., Przytycka T. M., Cherezov V., and Buchanan S. K. (2012) Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 20, 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leo J. C., Oberhettinger P., Chaubey M., Schütz M., Kühner D., Bertsche U., Schwarz H., Götz F., Autenrieth I. B., Coles M., and Linke D. (2015) The intimin periplasmic domain mediates dimerisation and binding to peptidoglycan. Mol. Microbiol. 95, 80–100 [DOI] [PubMed] [Google Scholar]
- 7. Leo J. C., Grin I., and Linke D. (2012) Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos. Trans. R Soc. Lond. B. Biol. Sci. 367, 1088–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oberhettinger P., Schütz M., Leo J. C., Heinz N., Berger J., Autenrieth I. B., and Linke D. (2012) intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS ONE 7, e47069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leo J. C., Oberhettinger P., Schütz M., and Linke D. (2015) The inverse autotransporter family: intimin, invasin, and related proteins. Int. J. Med. Microbiol. 305, 276–282 [DOI] [PubMed] [Google Scholar]
- 10. Bodelón G., Marín E., and Fernández L. A. (2009) Role of periplasmic chaperones and BamA (YaeT/Omp85) in folding and secretion of intimin from enteropathogenic Escherichia coli strains. J. Bacteriol. 191, 5169–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oberhettinger P., Leo J. C., Linke D., Autenrieth I. B., and Schütz M. (2015) The inverse autotransporter intimin exports its passenger domain via a hairpin intermediate. J. Biol. Chem. 290, 1837–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soprova Z., Sauri A., van Ulsen P., Tame J. R., den Blaauwen T., Jong W. S., and Luirink J. (2010) A conserved aromatic residue in the autochaperone domain of the autotransporter Hbp is critical for initiation of outer membrane translocation. J. Biol. Chem. 285, 38224–38233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peterson J. H., Tian P., Ieva R., Dautin N., and Bernstein H. D. (2010) Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc. Natl. Acad. Sci. U.S.A. 107, 17739–17744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oliver D. C., Huang G., Nodel E., Pleasance S., and Fernandez R. C. (2003) A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol. Microbiol. 47, 1367–1383 [DOI] [PubMed] [Google Scholar]
- 15. Junker M., Schuster C. C., McDonnell A. V., Sorg K. A., Finn M. C., Berger B., and Clark P. L. (2006) Pertactin β-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drobnak I., Braselmann E., Chaney J. L., Leyton D. L., Bernstein H. D., Lithgow T., Luirink J., Nataro J. P., and Clark P. L. (2015) Of linkers and autochaperones: An unambiguous nomenclature to identify common and uncommon themes for autotransporter secretion. Mol. Microbiol. 95, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai J. C., Yen M.-R, Castillo R., Leyton D. L., Henderson I. R., and Saier M. H. Jr., (2010) The bacterial intimins and invasins: a large and novel family of secreted proteins. PLoS ONE 5, e14403–e14414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Söding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Renn J. P., and Clark P. L. (2008) A conserved stable core structure in the passenger domain β-helix of autotransporter virulence proteins. Biopolymers 89, 420–427 [DOI] [PubMed] [Google Scholar]
- 20. Rief M., Gautel M., Schemmel A., and Gaub H. E. (1998) The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys. J. 75, 3008–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rief M., Gautel M., Oesterhelt F., Fernandez J. M., and Gaub H. E. (1997) Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 22. Hospenthal M. K., Redzej A., Dodson K., Ukleja M., Frenz B., Rodrigues C., Hultgren S. J., DiMaio F., Egelman E. H., and Waksman G. (2016) Structure of a chaperone-usher pilus reveals the molecular basis of rod uncoiling. Cell 164, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jacob-Dubuisson F., Guérin J., Baelen S., and Clantin B. (2013) Two-partner secretion: as simple as it sounds? Res. Microbiol. 164, 583–595 [DOI] [PubMed] [Google Scholar]
- 24. Leyton D. L., Rossiter A. E., and Henderson I. R. (2012) From self-sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat. Rev. Microbiol. 10, 213–225 [DOI] [PubMed] [Google Scholar]
- 25. Renn J. P., Junker M., Besingi R. N., Braselmann E., and Clark P. L. (2012) ATP-independent control of autotransporter virulence protein transport via the folding properties of the secreted protein. Chem. Biol. 19, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang'ethe W., and Bernstein H. D. (2013) Stepwise folding of an autotransporter passenger domain is not essential for its secretion. J. Biol. Chem. 288, 35028–35038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang'ethe W., and Bernstein H. D. (2013) Charge-dependent secretion of an intrinsically disordered protein via the autotransporter pathway. Proc. Natl. Acad. Sci. U.S.A. 110, E4246–E4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doyle M. T., Tran E. N., and Morona R. (2015) The passenger-associated transport repeat promotes virulence factor secretion efficiency and delineates a distinct autotransporter subtype. Mol. Microbiol. 97, 315–329 [DOI] [PubMed] [Google Scholar]
- 29. Ieva R., and Bernstein H. (2009) Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Selkrig J., Mosbahi K., Webb C. T., Belousoff M. J., Perry A. J., Wells T. J., Morris F., Leyton D. L., Totsika M., Phan M.-D., Celik N., Kelly M., Oates C., Hartland E. L., Robins-Browne R. M., Ramarathinam S. H., et al. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol. 19, 506–510 [DOI] [PubMed] [Google Scholar]
- 31. Shen H.-H., Leyton D. L., Shiota T., Belousoff M. J., Noinaj N., Lu J., Holt S. A., Tan K., Selkrig J., Webb C. T., Buchanan S. K., Martin L. L., and Lithgow T. (2014) Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nat. Commun. 5, 5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paramasivam N., Habeck M., and Linke D. (2012) Is the C-terminal insertional signal in Gram-negative bacterial outer membrane proteins species-specific or not? BMC Genomics 13, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robert V., Volokhina E. B., Senf F., Bos M. P., Van Gelder P., and Tommassen J. (2006) Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLos Biol. 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biegert A., Mayer C., Remmert M., Söding J., and Lupas A. N. (2006) The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34, W335–W3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul S. F., and Koonin E. V. (1998) Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem. Sci. 23, 444–447 [DOI] [PubMed] [Google Scholar]
- 36. Lassmann T., and Sonnhammer E. L. (2005) Kalign, an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu S., and Zhang Y. (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Udatha D. B., Topakas E., Salazar M., Olsson L., Andersen M. R., and Panagiotou G. (2015) Deciphering the signaling mechanisms of the plant cell wall degradation machinery in Aspergillus oryzae. BMC Syst. Biol. 9, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Udatha D. B., Madsen K. M., Panagiotou G., and Olsson L. (2015) Multiple nucleophilic elbows leading to multiple active sites in a single module esterase from Sorangium cellulosum. J. Struct. Biol. 190, 314–327 [DOI] [PubMed] [Google Scholar]
- 40. Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Byrappa S., Gavin D. K., and Gupta K. C. (1995) A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5, 404–407 [DOI] [PubMed] [Google Scholar]
- 42. Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 43. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 44. Leo J. C., Oberhettinger P., and Linke D. (2015) Assessing the outer membrane insertion and folding of multimeric transmembrane β-barrel proteins. Methods Mol. Biol. 1329, 157–167 [DOI] [PubMed] [Google Scholar]
- 45. Prilipov A., Phale P. S., Van Gelder P., Rosenbusch J. P., and Koebnik R. (1998) Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 163, 65–72 [DOI] [PubMed] [Google Scholar]
- 46. Hutter J. L., and Bechhoefer J. (1993) Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64, 1868–1873 [Google Scholar]
- 47. Bodelón G., Palomino C., and Fernández L. (2013) Á. Immunoglobulin domains in Escherichia coli and other enterobacteria: from pathogenesis to applications in antibody technologies. FEMS Microbiol. Rev. 37, 204–250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.