Abstract
Copper is an essential transition metal for living organisms. In the plant model Arabidopsis thaliana, half of the copper content is localized in the chloroplast, and as a cofactor of plastocyanin, copper is essential for photosynthesis. Within the chloroplast, copper delivery to plastocyanin involves two transporters of the PIB-1-ATPases subfamily: HMA6 at the chloroplast envelope and HMA8 in the thylakoid membranes. Both proteins are high affinity copper transporters but share distinct enzymatic properties. In the present work, the comparison of 140 sequences of PIB-1-ATPases revealed a conserved region unusually rich in histidine and cysteine residues in the TMA-L1 region of eukaryotic chloroplast copper ATPases. To evaluate the role of these residues, we mutated them in HMA6 and HMA8. Mutants of interest were selected from phenotypic tests in yeast and produced in Lactococcus lactis for further biochemical characterizations using phosphorylation assays from ATP and Pi. Combining functional and structural data, we highlight the importance of the cysteine and the first histidine of the CX3HX2H motif in the process of copper release from HMA6 and HMA8 and propose a copper pathway through the membrane domain of these transporters. Finally, our work suggests a more general role of the histidine residue in the transport of copper by PIB-1-ATPases.
Keywords: Arabidopsis, ATPase, chloroplast, copper transport, plant biochemistry, copper release site, PIB-1-ATPases
Introduction
Copper is an essential micronutrient involved in many cellular processes. However, because of its redox properties, copper is also highly toxic when present in excess, and thus its homeostasis in the cell is finely tuned thanks to a complex network of processes involving acquisition, storage, and transport. This tuning is especially crucial in the chloroplast where, under high light condition, electrons can escape from the photosynthetic machinery and directly reduce molecular oxygen to superoxide and hydrogen peroxide. The presence of free copper would convert these molecules into the highly toxic hydroxyl radical (OH•) through the Haber-Weiss cycle (1). In plant, copper is the cofactor of proteins involved in key cellular functions like the cytochrome c oxidase for respiration in mitochondria and plastocyanin for photosynthesis in chloroplasts. Plastocyanin, one of the most abundant plant cuproproteins, allows the transfer of photosynthetic electrons between cytochrome b6f and photosystem I. It is located in the thylakoid lumen, where it acquires copper (2). Copper delivery to apoplastocyanin involves two transporters of the P-ATPases family, HMA6 and HMA8, located at the chloroplast envelope (3, 4) and the thylakoid membranes (5, 6), respectively. Both transporters are high affinity transporters for monovalent copper (7–9). HMA6 and HMA8 loss of function mutations strongly reduce chloroplast copper content, in the stroma for the hma6 mutant and in thylakoids for the hma8 mutant. Consequently, the activity of the copper/zinc superoxide dismutase in the stroma is reduced in the hma6 mutant, and plastocyanin-dependent photosynthetic electron transfer in the thylakoids is reduced in both mutants (4, 5). Several steps of copper transport from the cytosol to the lumen of thylakoids have been identified. The copper chaperone PCH1, located in the intermembrane space of the chloroplast envelope, could bring copper to HMA6 (7). HMA6 would then transfer copper in the stroma to the chaperone CCS for the activation of the copper/zinc superoxide dismutase (4, 10, 11). However, this interaction has never been experimentally validated. Although CCS can interact and transfer copper to HMA8 in vitro (7), in planta studies of a ccs mutant suggest that another chaperone might be involved in the transport of copper from HMA6 to HMA8 (12). So far, the mechanism of copper transfer from HMA8 to plastocyanin is not known. However, combining enzymatic and structural approaches, we proposed that the two proteins could have favorable electrostatic interactions allowing a direct copper transfer without requirement of a metallochaperone (9). As for HMA6 and CCS, the interaction between HMA8 and plastocyanin needs to be experimentally validated.
P-ATPases form a large family of transmembrane proteins that translocate ions or lipids across membranes thanks to the hydrolysis of ATP (13). P-ATPases are divided into five subfamilies of different transport specificities. HMA6 and HMA8 belong to the PIB-ATPases subfamily, which gathers heavy metal ions transporting P-ATPases. In this subfamily, they are classified as PIB-1-ATPases corresponding to Cu+/Ag+-ATPases (14, 15). PIB-1-ATPases display the same core topology as other P-ATPases with six transmembrane segments (TM1–TM6)4 and three cytoplasmic domains constituting the catalytic moiety of the transporter (A, N, and P). However, PIB-1-ATPases have unique features like one to six cytoplasmic metal-binding domains and two supplementary transmembrane helices at their N-terminal part (TMA and TMB; Fig. 1A). TMB is a kinked helix displaying an amphipathic platform that would serve as a docking interface for the copper chaperone (16, 17).
FIGURE 1.
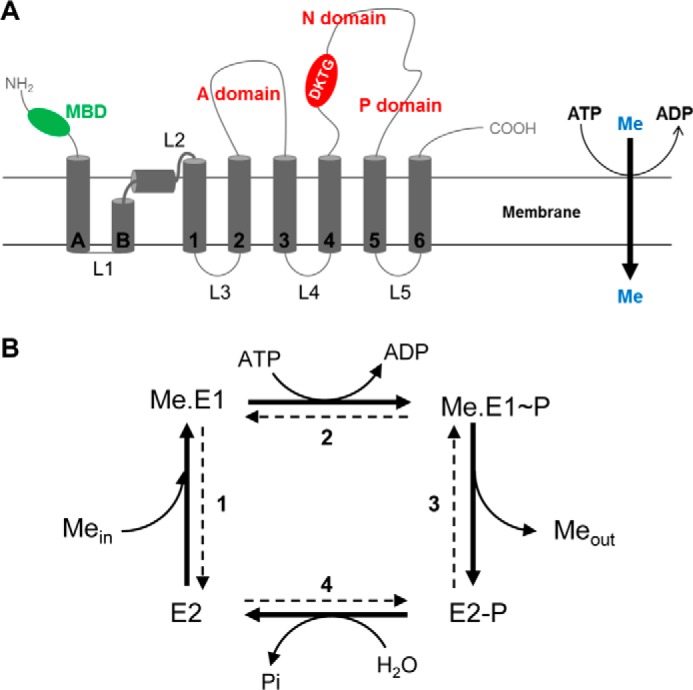
A, scheme representing the membrane topology of a typical PIB-1-ATPase. Transmembrane segments are represented in dark gray. With other P-ATPases, PIB-1-ATPases share a core topology with six transmembrane segments (TM1–TM6) and three cytoplasmic domains (actuator domain (A), nucleotide-binding domain (N), and phosphorylation domain (P) shown in red) constituting the catalytic moiety of the transporter. PIB-1-ATPases have specific features like several cytosolic metal-binding domains (MBD in green) and two supplementary transmembrane helices at their N-terminal part (TMA and TMB). TMB forms a kinked helix. B, catalytic cycle of PIB-ATPases. The bold arrows correspond to the forward cycle of PIB-ATPases. E2, Me.E1, Me.E1∼P, and E2-P represent the major catalytic intermediates of the enzyme. Me represents the transported metal. Mein and Meout represent the cytosolic and extracellular/luminal metal, respectively.
The catalytic cycle of a PIB-ATPase is a four-step process (Fig. 1B) characterized by the formation of a transient phosphorylated intermediate on a conserved aspartate residue of the DKTGT motif (13, 18). In its free state (E2), the ATPase binds metal (Me.E1) from the cytoplasm (or from the stroma for HMA8) at its high affinity transport site in the membrane domain (step 1). ATP bound to the large cytoplasmic domain of the transporter is then hydrolyzed, leading to the formation of a phosphorylated enzyme (step 2). The metal-bound phosphorylated state of the enzyme (Me.E1∼P) undergoes conformation changes leading to metal release at the other side of the membrane (step 3). In the metal-free phosphorylated enzyme (E2-P), the aspartyl phosphate bond is then hydrolyzed to return the enzyme to its free state (E2; step 4). As we demonstrated for HMA6 (8), P-ATPases can be phosphorylated from inorganic phosphate (Pi) in the reverse order (step 4). Although phosphorylation from ATP requires metal binding to the transporter, phosphorylation from Pi only occurs on the metal-free state of the ATPase. Involving a simple equilibrium reaction (and not the whole catalytic cycle), phosphorylation from Pi affords greater precision in calculating the affinity of the ATPase for the transported ion.
In the last few years, biochemical, biophysical, and structural studies have provided significant insights into the transport mechanism of Cu-ATPases. It is now widely believed that the copper chaperone would transfer copper to the ATPase by docking with the amphipathic platform created by the TMB kink, through the formation of transient complexes involving amino acids from both proteins (19, 20). In the crystal structures of the Cu-ATPase LpCopA from Legionella pneumophila (16, 21, 22), this entry site involves the methionine residue Met148 in TM1 and the glutamate residue Glu205 and the aspartate residue Asp337 in TM2 and TM3, respectively. Relocation of the copper ion to the high affinity transport site would involve copper coordination exchanges between amino acids constituting the entry site and cysteine residues of the conserved CPC motif in TM4 and the methionine residue of the conserved MX2SS motif in TM6. In LpCopA, the high affinity transport site comprises the cysteine residues Cys382 and Cys384 and the methionine residue Met717. On the other hand, the mechanism of copper release from the PIB-1-ATPases and its transfer to final acceptors is less documented. Studies of CopA from Escherichia coli (23) and of the human ATPase ATP7A (24, 25) have pointed out the importance of a region comprising the end of TMA and the first luminal loop, as well as invariant methionine and glutamate residues in copper release and transfer. These results also suggested that the functional difference between the highly homologous ATP7A and ATP7B proteins could be explained by the nature of the amino acids present in the L1 (the L1 loop of ATP7A is particularly enriched in histidine and methionine residues as compared with ATP7B). In LpCopA, the release site would be composed of the glutamate residue Glu89 in TM2 and the two methionine residues Met100 and Met711 in TMA and TM6, respectively.
In Arabidopsis chloroplasts, the two Cu-ATPases HMA6 and HMA8 act sequentially to deliver copper in the thylakoid lumen for the maturation of plastocyanin. However, because of their distinct localization patterns in chloroplast membranes, the two ATPases are thought to interact with different partners and to have distinct functions. Indeed, HMA6 is thought to interact with different chaperones to provide copper to the copper/zinc superoxide dismutase and to HMA8 (5, 26). The latter could interact directly with plastocyanin (9). In both case, the interaction between the ATPase and its recipient protein has not been experimentally confirmed. To identify residues of HMA6 and HMA8 involved in copper release, we first compared the TMA-L1-TMB region of Cu-ATPases from prokaryotes and eukaryotes. This allowed us to point out the presence of a unique cysteine- and histidine-rich sequence containing the conserved CX3HX2H motif, exclusively found in Cu-ATPases localized in chloroplasts of eukaryotic organisms (plant and algae). Through a structure/function study using expression in yeast and in vitro biochemical characterization of mutated proteins after expression in Lactococcus lactis, we identified two critical residues, a cysteine and a histidine, involved in copper release. From this work, we propose a metal pathway through the membrane domain of HMA6 and HMA8, and highlight some structural differences that exist between Cu-ATPases from plant and bacteria. In addition, our work suggests a more general, and so far unknown, role of the histidine residue in copper transport by PIB-1-ATPases.
Results
The TMA-L1 Region of Chloroplast Cu-ATPases Contains Conserved Cysteine and Histidine Residues
To identify residues specifically involved in copper release during the catalytic cycle of chloroplast Cu-ATPases, we compared the primary sequence corresponding to the TMA-L1-TMB region of 140 PIB-1-ATPases from prokaryotes and eukaryotes. This comparison allowed the identification of 66 unique sequences containing relatively well conserved proline, cysteine, histidine, or methionine residues in the TMA-L1-TMB region. From this analysis, it was not possible to find a unique motif shared by all the Cu-ATPases but rather three homology groups. Group A comprises sequences with a conserved proline residue in TMA, a predominance of methionine residues at the TMA-L1 junction, and histidine residues exclusively found in the L1 loop (Fig. 2A). In this group, only very few sequences contain cysteine residues in TMA. Within this group are the well characterized Cu-ATPases CopA from bacteria, Ccc2p from yeast, and the human ATP7A and ATP7B. Group B and C sequences differ from group A sequences by the absence of a proline residue in TMA. Group B sequences, exclusively found in bacteria, are characterized by an enrichment in methionine residues in TMA and a conserved histidine residue in the L1 loop (Fig. 2B). Group C sequences are enriched in cysteine and histidine residues and display a strongly conserved CX3HX2H motif (Fig. 2C). This group comprises Cu-ATPases like HMA6 and HMA8, localized in the chloroplast membranes of photosynthetic eukaryotes (algae and plant). Note that the two other plant Cu-ATPases, HMA5 and HMA7, localized at the plasma and ER membranes, respectively, are found in the group A in agreement with a recent phylogenetic study on the classification on Cu-ATPases (27).
FIGURE 2.
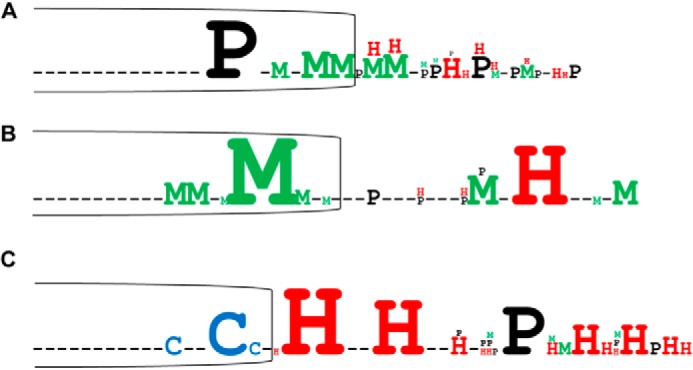
Conserved residues found in the TMA-L1-TMB region of PIB-1-ATPases. 66 unique sequences were deduced from sequence alignments of the TMA-L1-TMB region of 140 PIB-1-ATPases from prokaryote and eukaryote organisms. These sequences were classified into three homology groups according to the presence of proline (black), methionine (green), cysteine (blue), and histidine (red) residues (corresponding to panels A, B and C in the figure). The size of the letter is proportional to the conservation of the residue in each group. The limit of the TMA domain (shown by the open box) was defined in each group either from the crystal structure of LpCopA (group A) or from structural models (constructed from LpCopA structure) for the two other groups. Group A (panel A) contains Cu-ATPases from all kingdoms like CopA from L. pneumophila and E. coli, the human ATP7A and ATP7B ATPases, the yeast Ccc2p, PacS from cyanobacteria but also the plant ATPases HMA5 and HMA7. Group B (panel B) only comprises so far uncharacterized bacterial Cu-ATPases. Group C (panel C) is specific to chloroplast Cu-ATPases from photosynthetic eukaryotes (including chloroplast HMA6 and HMA8) and plasma membrane Cu-ATPases from cyanobacteria (CtaA).
Functional Expression of Mutated Forms of HMA6 and HMA8 in Yeast
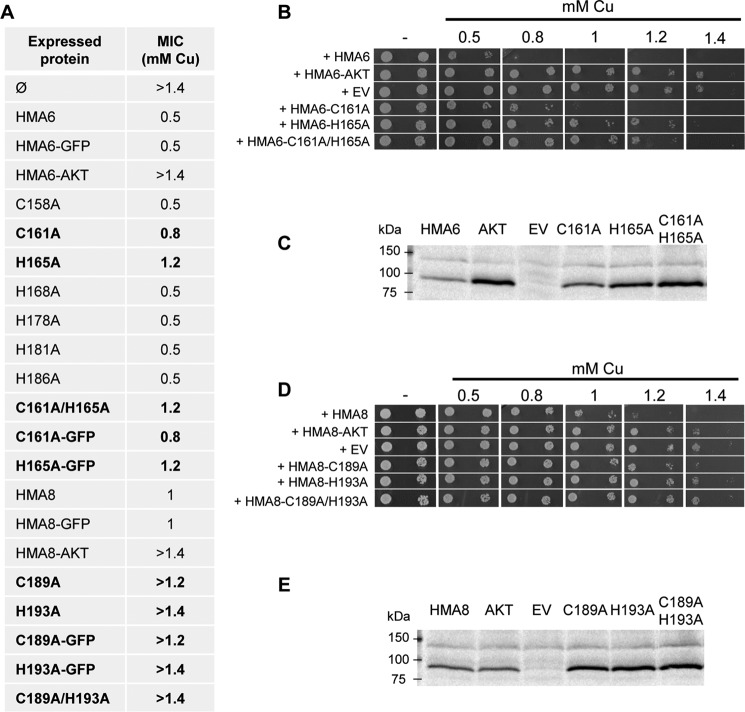
To more accurately model the positioning of the conserved histidine and cysteine residues in the TMA-L1-TMB region, we generated three-dimensional models of HMA6 and HMA8 from the crystal structure of the L. pneumophila Cu-ATPase LpCopA as described in Ref. 9 (Fig. 3). This in silico analysis allowed us to identify seven putative residues in HMA6 (two cysteines and five histidines, some of them belonging to the CX3HX2H motif), which may participate in copper release. In our previous studies, we have shown that yeast is a useful tool for the functional characterization of plant PIB-ATPases, among them the chloroplast PIB-ATPases HMA1, HMA6, and HMA8 (8, 9, 28). Indeed, expression of an active form of these ATPases induces growth arrest provided that the transported metal is present in the culture medium at the adequate concentration. We have shown that, for HMA6 and HMA8, the phenotype is linked to the localization of the transporters at the ER membrane and toxic accumulation of metal in this compartment (9). The case of HMA6 is particularly illustrative: expression of the active form of the transporter induces yeast growth arrest at 0.5 mm copper, whereas a strain expressing HMA6-AKT (an inactive form of HMA6 mutated in the phosphorylation site) tolerates up to 1.4 mm copper (Fig. 4 and Ref. 8). This range of copper concentration allows us to accurately assess the copper sensitivity of strains expressing targeted mutants of HMA6 in the TMA-L1-TMB region and establish a minimum inhibitory concentration (MIC).
FIGURE 3.
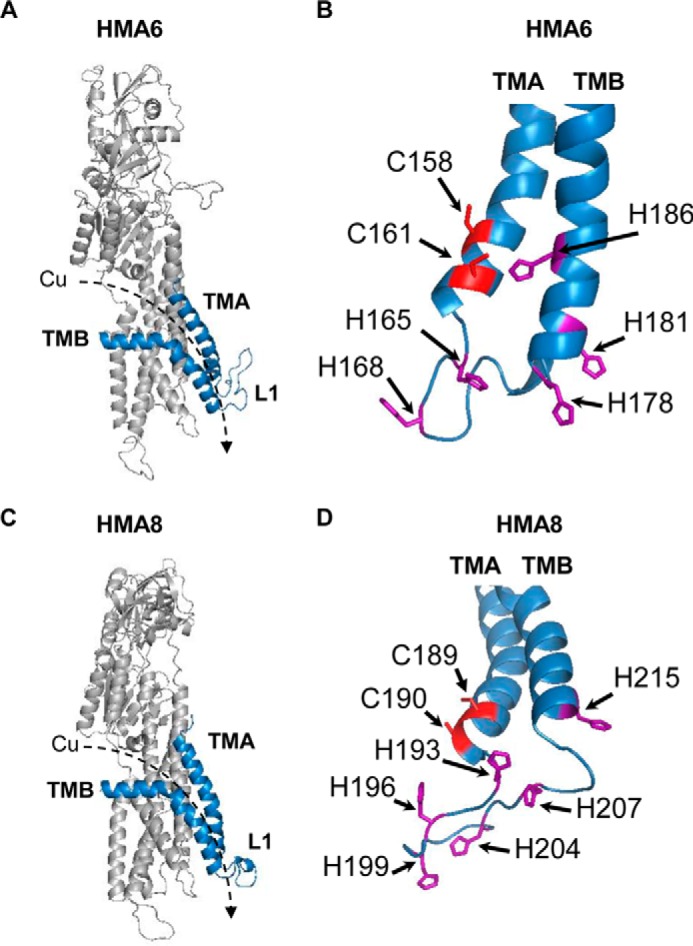
Structural models of HMA6 and HMA8 from A. thaliana. A and C, structural models were obtained from the structure of LpCopA (Protein Data Bank code 4BBJ) (21) with the MODELLER program (40) as previously described (9). The TMA and TMB helices are shown in blue. The dashed arrows indicate the orientation of copper transport. B and D, focus on the cysteine and histidine residues found in the TMA-L1-TMB region of HMA6 (B) and HMA8 (D).
FIGURE 4.
Functional expression of HMA6 and HMA8 mutants in yeast. A, MIC of copper observed for each yeast strain expressing the different forms of HMA6 and HMA8. Mutants affecting yeast sensitivity to copper as compared with the native protein are indicated in bold. B, yeast strains expressing HMA6 and HMA6 mutants (AKT, C161A, H165A, and C161A/H165A) were grown on selective media supplemented with copper at the indicated concentrations. For each expression condition, two independent transformants were spotted at dilutions 1 and 1/10, as 2-μl drops (only one transformant is shown). Dilution 1 corresponds to an optical density of 1.5 at 600 nm. EV, empty vector. C, expression of HMA6 and HMA6 mutants (AKT, C161A, H165A, and C161A/H165A) in yeast membranes was revealed by Western blotting using a Strep-tactin HRP conjugate. D, phenotypic tests as described in B for yeast strains expressing HMA8 and HMA8 mutants (AKT, C189A, H193A, and C189A/H193A). E, expression of HMA8 and HMA8 mutants (AKT, C189A, H193A, and C189A/H193A) in yeast membranes was revealed by Western blotting using a Strep-tactin HRP conjugate.
Each of the seven candidate residues in HMA6 were mutated and expressed in their mature forms (i.e. without the chloroplast transit peptide) and with a Strep-tag II fused at their C terminus. As shown in Fig. 4A, yeast strains expressing the C158A, H168A, H178A, H181A, and H186A mutants behave like a strain expressing the native form of HMA6, with a MIC of 0.5 mm copper, suggesting that these mutations have no impact on the activity of the transporter. This is not the case for the mutations of the His165 and Cys161 residues. Indeed, a strain expressing the H165A mutant displays a MIC of 1.2 mm copper close to that of strains containing the empty vector or expressing the inactive HMA6-AKT protein (Fig. 4, A and B). A strain expressing the C161A mutant displays a lower sensitivity to copper as compared with the strain expressing the native form of HMA6 (MIC of 0.8 and 0.5 mm copper, respectively). Importantly, the copper sensitivity of a strain expressing the double mutant C161A/H165A is similar to that of a strain expressing H165A, suggesting that the inhibitory effect of single mutations are not cumulative and that the H165A mutation is phenotypically “dominant.”
Because the copper sensitivity of the strain is a result of the concentration of copper accumulated in the organism, we assessed whether the wild type and mutants showed differences in protein abundance. Applying the Michaelis-Menten concept, a lower amount of the transporter might require more copper to produce the same inhibitory effect. If the Cys161 and His165 mutants had the same enzymatic properties of HMA6, a lower amount of these proteins in yeast would displace their corresponding MIC toward higher concentrations. To check this, we have assessed the amount of the different HMA6 forms in yeast by Western blotting. As shown in Fig. 4C, the C161A, H165A, and C161A/H165A mutants are more abundant than HMA6. Hence, one can exclude the possibility that the lower sensitivity to copper induced by these mutants is due to a lower expression level in yeast. Note that the other mutants were all expressed at the same level as HMA6 (data not shown).
Phenotypes might also be linked to intracellular localization of the expressed proteins. We thus localized the different forms of HMA6 in yeast using GFP fusions and checked the expression level, as well as the phenotype induced by these new forms of HMA6. As shown in Fig. 5A, all mutants are expressed in intracellular structures similar to the ones described for HMA6 (9). Critically, the addition of GFP has no impact on the phenotype because copper MICs of strains expressing either the mutant or mutant-GFP fusion are identical (Fig. 4A). In conclusion, these phenotypic tests allowed us to identify two residues, Cys161 and His165, which could have an important role on the enzymatic properties of HMA6.
FIGURE 5.
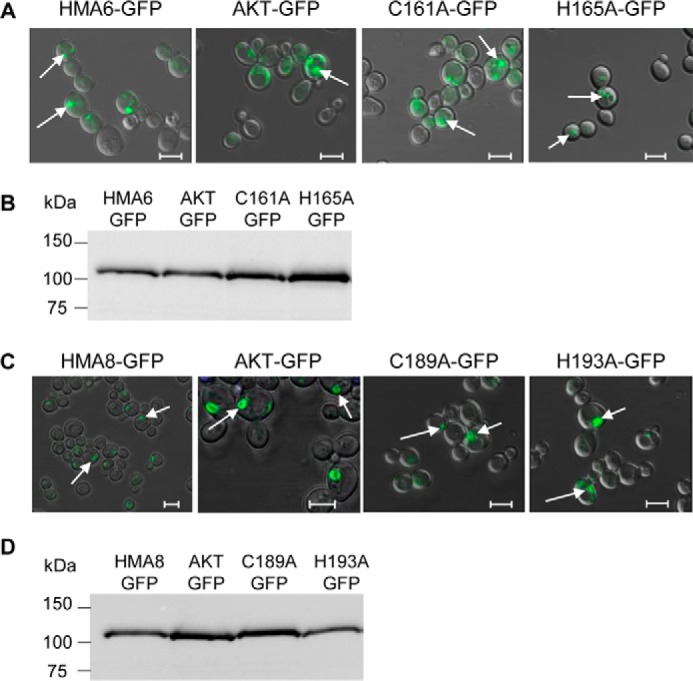
Functional expression of HMA6-GFP and HMA8-GFP mutants in yeast. A and B, the intracellular localization in yeast of HMA6 and HMA6 mutants fused to GFP (AKT-GFP, C161A-GFP, and H165A-GFP) was determined by confocal fluorescence microscopy. The GFP signal (green fluorescence) is indicated by the arrows (A). The expression of each protein in yeast membranes was analyzed by Western blotting using a Strep-tactin HRP conjugate (B). C and D, same analyses as described for A and B for HMA8 and HMA8 mutants (AKT, C189A, and H193A) fused to the GFP protein. The scale bars correspond to 5 μm.
HMA8 displays similar sequence features as HMA6: a CX3HX2H motif at the TMA-L1 junction (Fig. 2C) and several histidine residues in the L1 loop and in TMB. Remarkably, HMA8 possesses Cys189 and His193 equivalent to Cys161 and His165 of HMA6 (Fig. 3, B and D). As with HMA6, we mutated these two residues into alanine (we also performed the double mutation C190A/H193A) and used the yeast expression system to test the impact of the mutations on the activity of the transporter. We have already shown that the expression of HMA8 increases yeast sensitivity to copper but to a lower extent than HMA6 (MIC of 1 mm copper for HMA8 versus 0.5 mm copper for HMA6 (Fig. 4A and Refs. 8 and 9). As shown in Fig. 4D, the strain expressing the C189A mutant displays a lower sensitivity to copper than a strain expressing HMA8, with a MIC of 1.2 mm copper. Strains expressing the H193A and C189A/H193A mutants behave like a strain expressing the nonfunctional protein HMA8-AKT. As shown in the case of HMA6, the phenotype of the HMA8 mutants is not due to a lower expression level (Fig. 4E) nor to a different intracellular localization, as compared with the native protein (Fig. 5, C and D).
Finally, the screening in yeast allowed us to identify in chloroplast PIB-1-ATPases two residues of the CX3HX2H motif, the cysteine and the first histidine, whose mutations affect the activity of the transporters. Note that the histidine residue, whose mutation has the strongest impact on the activity of the transporter, is also the most highly conserved amino acid in the CX3HX2H motif (Fig. 2C).
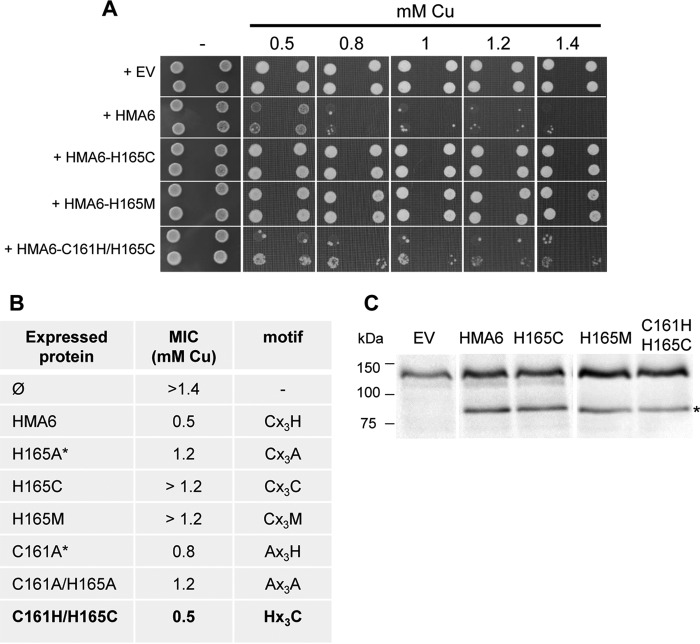
To further assess the importance of the specificity of the residue histidine at position 165 in HMA6, we have mutated this residue into methionine or cysteine, other residues known to bind copper. As shown in Fig. 6, yeast strains expressing the CX3CX2H or CX3MX 2H mutants (named H165C and H165M in Fig. 6) display the same phenotype as a yeast strain expressing the CX3AX2H mutant (named H165A in Fig. 4), with a MIC of 1.2 mm copper. This result suggests that the first histidine residue is strictly required in the CX3HX2H motif, i.e. it cannot be replaced by any other residues, even metal-coordinating ones. Interestingly, the double substitution HX3CX2H (HMA6-C161H/H165C in Fig. 6), which inverts the positions of the cysteine and first histidine residues compared with the native CX3HX2H motif, does not impair the function of HMA6 as shown by the conserved copper sensitivity of a yeast strain expressing this mutant (MIC of 0.5 mm copper).
FIGURE 6.
Functional expression of H165C, H165M, and C161H/H165C HMA6 mutants in yeast. A, yeast strains expressing HMA6 and HMA6 mutants were grown on selective media supplemented with copper at the indicated concentrations. For each expression condition, two independent transformants were spotted at dilutions 1 and 1/10, as 2-μl drops. Dilution 1 corresponds to an optical density of 1.8 at 600 nm. EV, empty vector. B, MIC of copper observed for each yeast strain expressing the different forms of HMA6 (the asterisks correspond to values obtained from Fig. 4). Mutants leading to the same yeast sensitivity to copper as HMA6 are indicated in bold. C, expression of HMA6 and HMA6 mutants (H165C, H165M, and C161H/H165C) in yeast membranes was revealed by Western blotting using a Strep-tactin HRP conjugate. The asterisk indicates HMA6, HMA6-H165C, HMA6-H165M, and HMA6-C161H/H165C.
At least two hypotheses could explain the observed phenotypes induced by HMA6 and HMA8 mutants. On one hand, mutations might have lowered the affinity of the transporter for copper, thus explaining that higher copper concentrations are required to inhibit yeast growth. On the other hand, mutations might have slowed down the catalytic cycle of the transporters. To test these hypotheses, we first produced the different forms of HMA6 and HMA8 in L. lactis, a bacterium that was found well suited for the production of plant PIB-ATPases (29). As previously shown, HMA6 is produced in larger quantities than HMA8 (∼10 times more) in Lactococcus (9). This is also the case for the mutated forms of the two transporters (Fig. 7). To determine some of the enzymatic properties of HMA6 and HMA8 mutants, we carried out experiments of phosphorylation from [γ-32P]ATP and 32Pi. In our previous studies, we have shown that in Lactococcus membranes HMA6 could be phosphorylated from ATP and Pi, allowing an easy determination of equilibrium and kinetic constants (8). In contrast, HMA8 was more difficult to study. We were unable to precisely determine an apparent affinity for copper by any of the two types of phosphorylation but could conclude from dephosphorylation kinetics that in vitro HMA8 had a lower catalytic activity than HMA6 (9).
FIGURE 7.
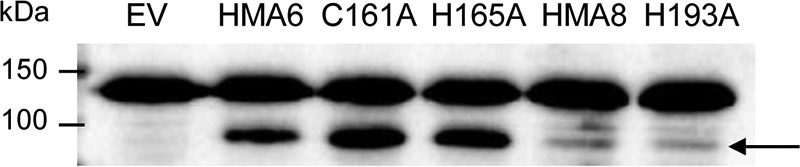
Production of native and mutated forms of HMA6 and HMA8 in Lactococcus membranes. Total membrane proteins (20 μg) were separated by SDS-PAGE. The different forms of HMA6 and HMA8 were detected using a Strep-tactin HRP conjugate. The arrow indicates HMA6, HMA6-C161A, HMA6-H165A, HMA8, and HMA8-H193A. EV, membrane extract derived from bacteria containing the empty pNZ8148 vector.
In HMA6, Mutations of Cys161 and His165 Residues of the CX3HX2H Motif Do Not Affect the Affinity for Copper
To determine whether mutations of His165 and Cys161 modified HMA6 affinity for copper, we carried out two types of phosphorylation, both depending on the binding of the metal to the transport site located in the membrane domain of the transporter. In a phosphorylation assay using ATP and at the lowest metal concentrations, the amount of phosphorylated protein is correlated to the amount of the enzyme in the Me.E1 state (8). This increasing phosphorylation phase can be used to determine the apparent affinity for the transported metal. At higher metal concentrations (beyond the apparent Kd value), the phosphorylation level can be relatively stable or may decrease as shown for some Cu-ATPases like HMA6, HMA8, and ATP7A (8, 30, 31). In the phosphorylation assay using Pi, the amount of phosphorylated protein is inversely correlated to the amount of the enzyme in the Me.E1 state because Pi only binds to the metal-free state (E) of the transporter (8).
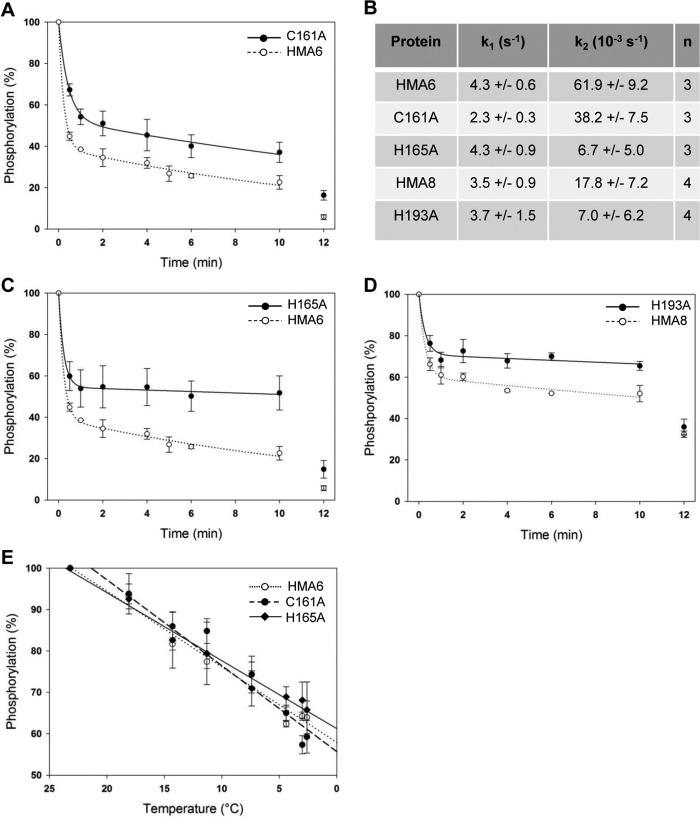
Phosphorylation reactions from Pi were performed at copper concentrations ranging from 0.05 to 300 μm on Lactococcus membrane extracts containing HMA6 or the HMA6 mutants (C161A or H165A). The reaction was carried out for 10 min at 30 °C in the presence of 20% DMSO at pH 6 and in the absence of potassium. As shown in Fig. 8A, the phosphorylation profiles of HMA6 and the C161A mutant are very similar with half-inhibition of the phosphorylation signal at 1.7 ± 0.7 and 1.5 ± 0.3 μm, respectively. For the H165A mutant, this inhibition is observed at a slightly higher copper concentration (3.1 ± 0.9 μm), but the difference with HMA6 is not significant. The phosphorylation assays using ATP were performed at copper concentrations ranging from 0.05 to 50 μm, for 30 s at room temperature and at pH 7. As shown in Fig. 8B, the phosphorylation profiles are biphasic. As explained above, the first increasing phase is likely to represent the normal activation process of a P-ATPase, with the progressive filling of the transport site and the concomitant phosphorylation of the transporter. The inhibition phase is currently not explained but as mentioned above is a characteristic of some eukaryotic Cu-ATPases. The apparent affinity of HMA6 for copper estimated from the increasing phase is 0.4 ± 0.03 μm, a value close to that measured in the previous biochemical characterization of this transporter (8). The apparent affinity values of the C161A and H165A mutants (0.74 ± 0.1 and 0.82 ± 0.2 μm, respectively) are slightly above that of HMA6.
FIGURE 8.
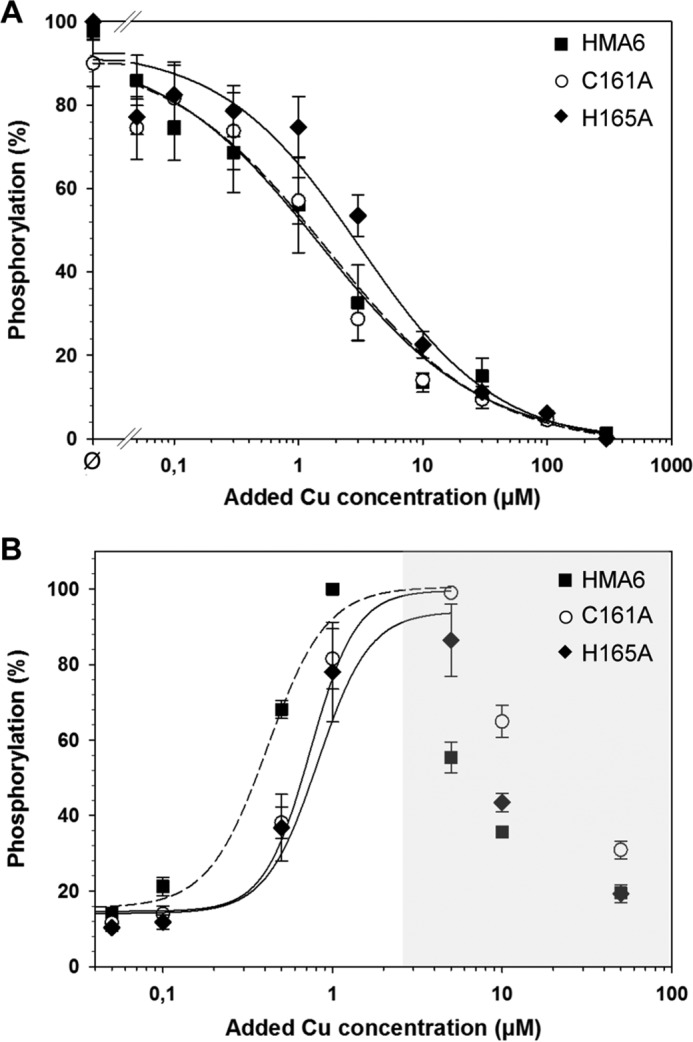
Copper dependence of HMA6 mutants phosphorylations from Pi and ATP. A, membrane fractions (100 μg) containing HMA6 or the HMA6 mutants (C161A and H165A) were incubated with various concentrations of CuSO4 under reducing condition (500 μm Na2SO3). Phosphorylation was initiated by the addition of 100 μm 32Pi and stopped after 10 min at 30 °C by addition of 1 mm KH2PO4, 7% (v/v) TCA. Samples were submitted to an acidic SDS-PAGE, and phosphorylation was detected using a phosphorimaging device. Phosphorylation intensities were quantified using the Optiquant Software. 100% represents the phosphorylation level of HMA6 or HMA6 mutants in the absence of added metal (Ø). Fittings were performed using a Hill equation. The values correspond to the means of five independent experiments. B, membrane preparations (50 μg) containing HMA6 or HMA6 mutants (C161A and H165A) were phosphorylated from ATP under reducing conditions (500 μm Na2SO3) with various concentration of CuSO4. Samples were treated as described above. 100% corresponds to the maximum phosphorylation level of HMA6 or HMA6 mutants. The values correspond to the means of four independent experiments. The error bars indicate standard deviation.
Overall, the results obtained from the two types of phosphorylation show no significant difference of apparent affinity for copper between the native and the mutant forms of HMA6. This means that mutations in the TMA-L1-TMB region do not affect copper binding to the transporter. The quantitative difference between the apparent Kd values estimated by the phosphorylation from Pi (comprised between 1 and 2.5 μm copper) and those estimated by the phosphorylation from ATP (∼0.5 μm) can be explained by the fact that the two assays imply different enzymatic mechanisms: phosphorylation from ATP involves all the steps of the catalytic cycle and will tend to overestimate the apparent Kd, whereas phosphorylation from Pi involving simple equilibrium reactions will provide a more realistic estimation of the affinity for copper.
Mutations of the Cysteine and the First Histidine Residues of the CX3HX2H Motif Impact the Dephosphorylation Step
One way to estimate the overall catalytic activity of an ATPase is to measure its dephosphorylation kinetics. In such a measurement, the use of ADP will discriminate between the two transient phosphorylated states of the transporter: the metal-bound Me.E1∼P state sensitive to ADP and the metal-free E2-P state insensitive to ADP (Fig. 1B). Hence, a quantification of these two states will indirectly inform on the efficiency of ion release. To measure the dephosphorylation kinetics of the different forms of HMA6 and HMA8, we first determined the reaction time required to reach the maximum phosphorylation level. We then added a mix of the copper chelators BCA/BCS to prevent the initiation of any new catalytic cycle and followed the dephosphorylation over a time range of 10 min. The reaction was performed at 4 °C to slow down the catalytic cycle of the ATPases, and at the end of the kinetics, ADP was added to quantify the proportion of the remaining Me.E1∼P and E2-P states.
As shown in Fig. 9 (A–D), the dephosphorylation process is biphasic for the different forms of HMA6 and HMA8, with a rapid phase and a slow phase of dephosphorylation. The rapid phase could correspond to the hydrolysis of the aspartyl-phosphate bond and then to the E2-P to E2 transition (note that potassium present in the assay is likely to favor this transition). The slow phase could correspond to dephosphorylation from the Me.E1∼P state, whose velocity is limited by the Me.E1∼P to E2-P transition. It is not unexpected that this transition is slower than the hydrolysis of the aspartyl-phosphate bond because it involves important conformation changes both in the catalytic and membrane domains required for metal release.
FIGURE 9.
Dephosphorylation kinetics. A–D, dephosphorylation kinetics in presence of copper chelators after phosphorylation from ATP. Experiments were performed at 4 °C to slow down the reaction. Lactococcus membranes (50 μg) containing HMA6 or HMA6 mutants (C161A, A, and H165A, C) were incubated under reducing conditions (500 μm Na2SO3) with 5 μm CuSO4 and 1 μm [γ-32P]ATP. After 2 min, an aliquot was taken and acid-quenched. On the remaining sample, a mix of BCA/BCS was added, and the reaction was stopped at the indicated times. After 10 min of incubation with BCA/BCS, 1 mm ADP was added to the reaction for 2 min more. In the case of HMA8 and the mutated form HMA8-H193A (D), 150 μg of Lactococcus membrane were incubated under reducing conditions (500 μm Na2SO3) with 1 μm CuSO4 and 1 μm [γ-32P]ATP. After 4 min, an aliquot was taken and acid-quenched. The experiment then proceeded as for HMA6. Quantifications were made using the Optiquant software. 100% corresponds to the maximum phosphorylation level of the different proteins in the presence of copper before the addition of chelators. Values correspond to means of at least three independent experiments (three for HMA6, C161A, and H165A and four for HMA8 and H193A). The error bars indicate the standard deviation. Fittings were made using the SigmaPlot software and were used to determine the different dephosphorylation rates listed in B. E, temperature-dependent dephosphorylation after phosphorylation from Pi. Membrane fractions (50 μg) containing HMA6 or the HMA6 mutants (C161A and H165A) were phosphorylated in the same conditions as described in Fig. 8A in the presence of BCA/BCS (100 μm/10 μm). After 10 min, samples were put on ice, and the reaction was stopped at different times (between 20 and 280 s) by addition of 1 mm KH2PO4, 7% (v/v) TCA. Samples were submitted to an acidic SDS-PAGE, and phosphorylation was detected using a phosphorimaging device. Phosphorylation intensities were quantified using the Optiquant Software. In parallel, a time-dependent temperature decrease was measured in the same experimental conditions (without radioactive Pi). The values correspond to the mean of four independent experiments. 100% corresponds to the phosphorylation level after 20 s on ice and correspond to a temperature of 23 °C. The error bars indicate the standard deviation. Fittings were made using the SigmaPlot software with slopes of −1.8 ± 0.12, −2.1 ± 0.2, and −1.6 ± 0.1 for HMA6, C161A, and H165A, respectively.
In the case of HMA6, the rapid phase of dephosphorylation leads to the diminution of 60% of the total phosphorylation signal with a rate constant k1 of 4.3 s−1. The remaining phosphorylation signal slowly disappears with a rate constant k2 of 62 × 10−3 s−1, to reach ∼20% after 10 min. At that time, addition of ADP almost completely dephosphorylates the protein (∼6% of residual signal), thus confirming that the remaining phosphorylated state is Me.E1∼P (Fig. 9, A–C).
The dephosphorylation kinetics of the H165A and C161A mutants are obviously different from that of HMA6. First, the amplitude of the rapid phase of dephosphorylation is reduced in the two mutants as compared with the native protein (47% for the two mutants versus 60% for HMA6). Note that the rate constant k1 of the H165A mutant is identical to that of HMA6 and approximately two times lower for the C161A mutant (Fig. 9, A–C). Second, the velocity of the slow phase is reduced in the two mutants as compared with HMA6. Although this reduction is weak for the C161A mutant (of ∼40%), it is much more important for the H165A mutant. Indeed, the rate constant k2 of the H165A mutant is nearly 10 times lower than that of HMA6 (6.6 × 10−3 s−1 versus 62 × 10−3 s−1). Third, the phosphorylation signal after the addition of ADP at the end of the kinetics is three times higher for the two mutants than for HMA6.
We have previously shown that HMA8 has an intrinsically low rate of dephosphorylation (9). As shown in Fig. 9D, the rapid phase of the dephosphorylation kinetic of HMA8 is characterized by a k1 constant similar to that of HMA6. This was expected because this phase would correspond to the hydrolysis of the aspartyl-phosphate bond, a process supposed to be identical in the two transporters. However, the amplitude of this phase is much reduced compared with that of HMA6. This is also the case for the amplitude and the rate constant of the slow phase (Fig. 9, B and D). The incomplete dephosphorylation of HMA8 (Fig. 9D) could be due to the existence of a particularly stable E1-P.[Moccluded] ADP-insensitive state (9) not observed for HMA6. In that particular context, study of the H193A mutant was more complicated. However, as shown in Fig. 9D, the behavior of this mutant qualitatively mimics that of the H165A mutant, with a significant reduction of the amplitude of the rapid phase (without modifying the rate constant k1) and a strong reduction of the velocity of the slow phase.
To know whether mutations affect the E2-P to E2 transition, we measured the temperature-dependent dephosphorylation of the E2-P state of HMA6 and its mutated forms, obtained after phosphorylation from Pi (note that HMA8 cannot be phosphorylated from Pi). Because the amount of E2P depends on the absolute values and the ratio between the kinetics constants of the E2 to E2P and E2P to E2 reactions, this experiment will indirectly provide information about the kinetics properties of the E2-P to E2 transition. As shown in Fig. 8E, there are no significant differences between HMA6, H165A, and C161A, showing that the mutations do not impact this catalytic step.
In summary, the mutations of the Cys161 and His165 residues in HMA6, as well as the His193 residue in HMA8, qualitatively produce the same effects: a reduction of the amplitude of the rapid phase of dephosphorylation and a reduction of the velocity of the second phase. In the case of HMA6, these reductions are more pronounced for the histidine mutant. These results suggest that the mutants accumulate in a “non E2-P” phosphorylated state. In the case of the HMA6 mutants, the partial effect of ADP indicates that this “non E2-P” phosphorylated intermediate still present at the end of the kinetics comprises the Me.E1∼P ADP sensitive state and another so far undefined ADP-insensitive phosphorylated state. In the catalytic cycle, this state should logically be localized between the Me.E1∼P and E2-P states. Note that the C161A mutant has a lower rate constant k1 than HMA6 in dephosphorylation assays after phosphorylation from ATP, suggesting that the E2-P to E2 transition might be somehow affected by the mutation (Fig. 9, A and B). However, this is not confirmed by the dephosphorylation assay from E2P (Fig. 8E).
In conclusion, our results reveal an essential and conserved role of the first histidine of the CX3HX2H motif in the activity of HMA6 and HMA8. Mutations of this histidine residue in the two chloroplast PIB-1-ATPases, as well as the Cys161 in HMA6, slow down the Me.E1∼P to E2 transition, suggesting that these residues are essential for metal release.
Discussion
Evolution of the Copper Release Site in Chloroplast Cu-ATPases
Chloroplast Cu-ATPases, as exemplified by HMA6 and HMA8, have different roles and localizations within the chloroplast. HMA6, at the chloroplast envelope, brings copper to the stroma where it is required for the copper/zinc superoxide dismutase, whereas HMA8, at the thylakoid membranes, brings copper in the lumen for the electron carrier, plastocyanin. Despite these differences, the chloroplast Cu-ATPases would have evolved from the same ancestor, CtaA, one of the two Cu-ATPases from cyanobacteria (32). In these photosynthetic bacteria, from which chloroplasts originated, CtaA is found at the plasma membrane, whereas PacS is located at the thylakoids respectively. In plants, HMA6 and HMA8 are functional homologs of CtaA and PacS but exclusively co-orthologs of CtaA (7, 27). In agreement, our focus on the sole TMA-L1-TMB region shows that with a short HX2H motif in TMA, CtaA belongs to group C, whereas PacS, lacking this motif, is found in the group A. Furthermore, this suggests that the TMA-L1 region of chloroplast Cu-ATPases, which seems essential in copper release, could have evolved by complexification of the initial HX2H motif to the CX3HX2H motif found in algae (e.g. CTP2 and CTP4 from Chlamydomonas reinhardtii) and plants (e.g. HMA6 and HMA8 from Arabidopsis thaliana).
A Model of Copper Transport through the Chloroplast Cu-ATPases
So far, LpCopA from L. pneumophila is the only Cu-ATPase whose crystal structure has been solved. It is important to note that none of the LpCopA structures have been obtained in presence of copper. Nevertheless, a combination of structural and functional data leads to a mechanistic model of copper transport through the membrane domain of this transporter (16, 21, 22, 33). This model proposes that during its motion from the cytoplasmic to the extra-cytoplasmic face of the transporter, the metal successively occupies three sites: an entry site, a high affinity membrane site, and a release site.
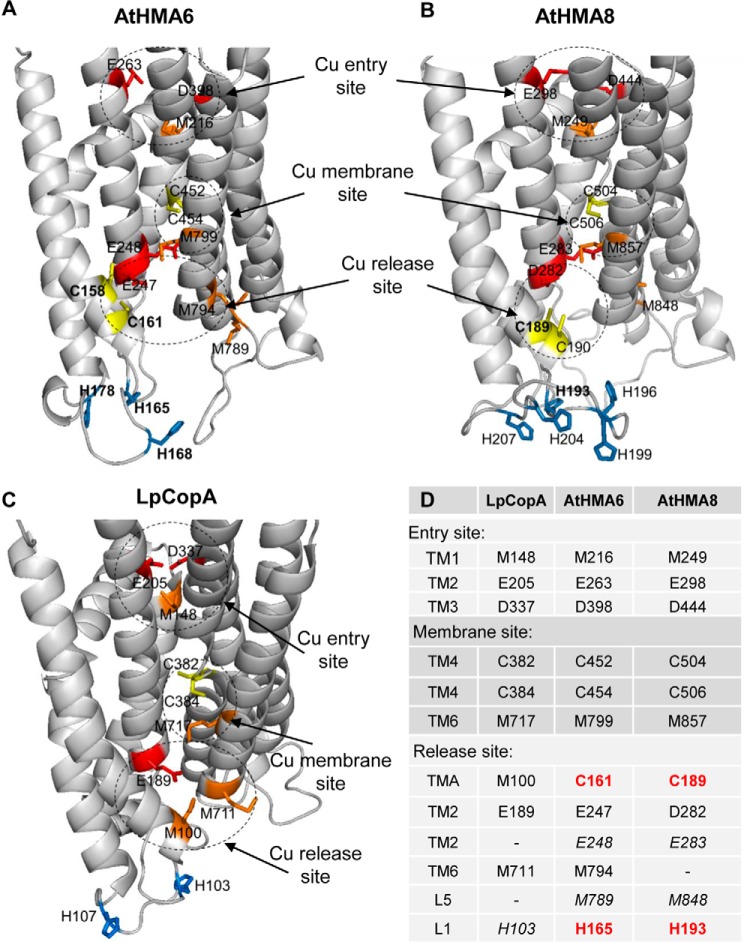
HMA6 and HMA8 possess ∼55% similarity with LpCopA. Focusing specifically on the membrane domain reveals that amino acids that could constitute the copper entry site in LpCopA (Met148, Glu205, and Asp337) are also conserved in HMA6 (Met216, Glu263, and Asp398) and HMA8 (Met249, Glu298, and Asp444) (Fig. 10). The high affinity membrane site in LpCopA is composed of residues Cys382 and Cys384 of the conserved CPC motif in TM4 and Met717 of the conserved MX2SS motif in TM6. As signature sequences of PIB-1-ATPases, these residues are also conserved in HMA6 (Cys452 and Cys454 in TM4 and Met799 in TM6) and HMA8 (Cys504 and Cys506 in TM4 and Met857 in TM6). In LpCopA, copper transfer from the entry to the membrane site would imply a coordination exchange between Met148 and Met717. By analogy, this exchange would involve Met216 and Met799 in HMA6 and Met249 and Met857 in HMA8. Once copper is bound at the membrane site, the ATPase can be phosphorylated from ATP. This phosphorylation triggers major rearrangements of the transmembrane helices, allowing copper transfer to the release site within the Me.E1∼P to E2-P transition.
FIGURE 10.
Models of copper transport through HMA6 and HMA8. The copper pathway through AtHMA6 and AtHMA8 (A and B) is deduced from that proposed for LpCopA (C) (16, 21, 22) and from results obtained in this study. The figure points out the high conservation of residues involved in the coordination of copper at the entry and the high affinity membrane sites. Cysteine residues are yellow, histidine residues are blue, methionine residues are orange, and glutamate and aspartate residues are red. Bold type for HMA6 and HMA8 indicates residues that have been mutated in the present study. D, list of the amino acids involved in the entry, membrane, and release sites in LpCopA, AtHMA6, and AtHMA8. HMA6 and HMA8 residues involved in copper release and identified in this work are shown in red. Additional residues that could participate in the release site are noted in italics.
In LpCopA, the predicted copper release site would involve residue Glu189 in TM2, as well as residues Met100 and Met711 in TMA and TM6, respectively (21, 22). In the beginning of TM2, HMA6 and HMA8 have two acidic residues (Glu247 and Glu248 in HMA6 and Asp282 and Glu283 in HMA8), instead of the unique glutamate of LpCopA. From our 3D models, Glu247 in HMA6 and Asp282 in HMA8 could be the equivalent of Glu189 in LpCopA. Met100 in LpCopA is not conserved in the chloroplast ATPases but is instead replaced by the cysteine residues Cys161 in HMA6 and Cys189 in HMA8 whose mutation was shown in the present study to reduce the activity of the transporters. Interestingly, it was shown in CopA from E. coli that mutation of residue Met204 (the equivalent of Met100 in LpCopA) into a cysteine alters neither the enzymatic properties of the Cu-ATPase nor the copper transfer to its copper-target protein (23). Methionine and cysteine residues are among the main Cu+ ligands in proteins (34). In chloroplast Cu-ATPases, the presence of specific cysteine residues, known to bind copper with higher affinity than methionine (35), could be linked to a specific regulation of copper transfer to its target(s) (chaperones or copper-binding proteins). Note that another cysteine residue (Cys158 in HMA6 and Cys190 in HMA8), which does not correspond to the cysteine of the CX3HX2H motif, is also present in this region. We have shown that, in HMA6, the mutation of this cysteine residue has no effect on the activity of the transporter. However, we cannot exclude that the presence of these residues somehow attenuate the effect of the mutation of the cysteine residue present in the CX3HX2H motif (Cys161 in HMA6 and Cys189 in HMA8). Met711, the other methionine residue of the release site of LpCopA, has its counterpart (Met794) at the beginning of TM6 of HMA6 but not HMA8. In the same region, the two chloroplast ATPases, but not LpCopA, possess another methionine residue (Met789 in HMA6 and Met848 in HMA8) in the loop connecting TM5 and TM6 (L5 loop; Fig. 1A) that could participate in the release site. Together, these observations show that the release site of the chloroplast Cu-ATPases is more complex than that of LpCopA, suggesting that it could accommodate more than one copper ion.
A Particular Role of Histidine Residues in Copper Release from Cu-ATPases
Our present work points out the important role in copper transport of the cysteine residue belonging to the conserved CX3HX2H motif and located in TMA of chloroplast Cu-ATPases from eukaryotes. Thereby, it complements the model of metal transport by PIB-1-ATPases previously proposed from LpCopA structure (Fig. 10D). Importantly, it highlights the so far unknown and critical role of the first histidine of the CX3HX2H motif on the activity of HMA6 and HMA8. Note that other histidine residues are found in the L1 loop of HMA6 and HMA8 (Figs. 3 and 10). In HMA6, single mutations of these histidine have no effect on the activity of the transporter as deduced from the phenotypes (Fig. 4A). However, we cannot totally exclude the possibility that the flexibility of the L1 loop allows compensatory effects by the remaining histidine residues. From the three-dimensional models, we could hypothesize that, in addition to the first histidine of the CX3HX2H motif, histidine residues clustered in the L1 loop could be a relay in copper transfer from the ATPase to its acceptor. This hypothesis will need to be tested when recipient proteins are identified and their interaction with the chloroplast ATPases is validated. Hence, there might be two distinct classes of histidine residues: one involved in copper motion in the membrane domain of the ATPase (like His165 in HMA6 and His193 in HMA8) and the other involved in copper transfer to a recipient protein (other His residues located in the L1 loop). The essential role of a histidine residue located in the L1 loop might be extended to most of the PIB-1-ATPases. Indeed, one to five histidine residues are found in the L1 loop of most of the Cu-ATPases (Fig. 2). As an example, LpCopA has two histidine residues in the L1 loop (Fig. 10C). His103 would be the equivalent of His165 in HMA6 and His193 in HMA8, but its role in the activity of the bacterial transporter has not been yet analyzed. CopA from E. coli is one of the few Cu-ATPases lacking histidine residue in the L1 loop. Instead, this region contains methionine residues that have been shown to be important for copper release and transfer to CusF, its periplasmic acceptor (23).
Our results also suggest that coordination properties of the cysteine and the first histidine residues of the CX3HX2H motif, more than their location in the release cavity, are essential for copper motion into the ATPase. Indeed, the double reciprocal mutation leading to a HX2CX3H motif has no impact on the activity of the transporter (C161H/H165C mutant in Fig. 6). Such a double mutant behaves like the wild type protein. A modification of this metal binding motif by single mutations (even by other metal coordinating residues) strongly affects the activity of the transporter. Given that histidine, cysteine, and methionine residues have different copper affinity (Cys > His > Met at pH 7.4) (35), it may be that copper motion from the copper release site is strongly impaired in the CX3AX2H and CX3MX2H mutants. On the other hand, in the case of the CX3CX2H mutant and because of the strong affinity of the two cysteine residues, copper would be bound too tightly, preventing its release from the ATPase. These results highlight the importance of the cysteine and the first histidine residues of the CX3HX2H motif and point out that these specific residues are necessary to fine-tune the binding of copper allowing an optimal functioning of the transporter.
Cysteine and histidine residues are known to have higher affinity for copper than methionine residues. The enrichment in these residues of the TMA-L1 region in HMA6 and HMA8 (CX3HX2H motif) suggests that copper release from chloroplast Cu-ATPases could be more controlled than in other Cu-ATPases. Copper would be more strongly coordinated in the release cavity of the chloroplast Cu-ATPases, and its transfer would then strictly depend on specific interactions between the transporter and a specific acceptor.
Experimental Procedures
Cloning of HMA6 and HMA8 Mutants
HMA6 (At4g33520, Q9SZC9) and HMA8 sequences (At5g21930, B9DFX7) used in the present study code for the precursor forms of the proteins, i.e. containing the 102 and 65 first amino acid residues, respectively corresponding to the predicted chloroplast transit sequence (ChloroP (36)). Directed mutagenesis was performed on the pKS-HMA6 (8) and pKS-HMA8 (9) plasmids using the QuikChange® kit (Stratagene), allowing the production of cDNAs coding the mature and mutated forms HMA6-C161A, HMA6-H165A, HMA6-H165C, HMA6-H165M HMA6-H168A, HMA6-H178A, HMA6-H181A HMA6-H186A, HMA6-C161A/H165A, HMA6-C161H/H165C, HMA8-C189A, HMA8-H193A, and HMA8-C189A/H193A. These plasmids were then used for cloning the mutated cDNAs into the pCEN yeast expression vector and the pNZ8148 Lactococcus expression vector. For confocal imaging in yeast, HMA6, HMA8, and their mutated sequences have been fused to the green fluorescent protein (GFP) encoding sequence in the pCEN expressing vector. All the proteins expressed contain a Strep-Tag II (IBA, Goettingen, Germany) fused at their C-terminal end.
Expression and Phenotypic Tests in Saccharomyces cerevisiae
Mature forms of HMA6, HMA8, and their mutated forms were expressed as already described (8). For maintenance, the S. cerevisiae strain BY4741 was grown at 30 °C in rich YD medium (1% (w/v) yeast extract KAT, 2% (w/v) glucose). The cells were transformed as described in Ref. 37. Plasmid selection was performed on solid synthetic minimal medium containing 2% (w/v) glucose, 0.17% (w/v) yeast nitrogen base, 0.5% (w/v) ammonium sulfate, 0.7 g/liter dropout powder without leucine, and 2% (w/v) Agar-Y (Bio 101® Systems). Drop test experiments were performed using CuSO4 at various concentrations.
Yeast Protein Extracts
Total yeast proteins were extracted according to the method described by Volland et al. (38) with the following modifications: after 24 h of culture at 30 °C, cells expressing the chloroplast HMAs (HMA6, HMA8, and their mutated forms) were spun down (5 min at 5,000 × g at 4 °C) and resuspended in 500 μl of distilled water. The cells were lysed with 0.185 n NaOH for 10 min at 4 °C, and proteins were precipitated in 5% (v/v) trichloroacetic acid for 10 min at 4 °C. After centrifugation (10 min at 15,000 × g at 4 °C), the pellet was washed with 500 μl of 1 m Tris-Base (pH 7.5) and then resuspended in 50 μl of 50 mm Tris (pH 6.8), 2 mm EDTA, 10% (w/v) glycerol, 2% (v/v) β-mercaptoethanol. The total protein extract was boiled at 95 °C for 5 min, and 10 μl was typically loaded onto SDS-PAGE gel.
Microscopy
Yeast cells were analyzed by confocal laser scanning microscopy (Leica TCS-SP2 operating system, Deerfield, IL). Observations were done with a 40× oil immersion objective. GFP was excited at 490, and the fluorescence was collected from 500 to 535 nm. Transmission images where recorded using differential interference contrast optics.
Expression in L. lactis
The expression of mature HMA6, HMA8, and the mutated forms of HMA6 and HMA8 from the pNZ8148 vector, as well as the preparation of bacterial membranes, were performed as previously described (8, 9, 29).
SDS-PAGE and Chemoluminiscence Detection of Strep-tag Proteins
Protein content was estimated using the Bio-Rad protein assay. SDS-PAGE analyses were performed as described by Chua (39). Detection of the Strep-Tag II was performed using the Strep-Tactin HRP conjugate (IBA, Goettingen, Germany) at a 1/10,000 dilution, followed by ECL detection.
Phosphorylation Assays
Phosphorylation assays from ATP and Pi were performed as previously described (8). Phosphorylation from ATP was performed in 100 μl of 20 mm HEPES (pH 6.0), 100 mm KCl, 10 mm MgCl2, 300 mm sucrose, 500 μm of the reducing agent Na2SO3, and either 50 μg of membranes containing HMA6 (or mutated forms of HMA6) or 150 μg of membranes containing HMA8 (or mutated forms of HMA8). Phosphorylations were performed at room temperature or at 4 °C in the presence of various concentrations of copper (as indicated) or chelators (1 mm BCA, 0.1 mm BCS) either alone or in combination. The reaction was started by addition of 1 μm [γ-32P]-ATP (50–500 μCi nmol−1) and stopped, 30 s later, by the addition of 1 ml of ice-cold 1 mm KH2PO4 in 7% (v/v) trichloroacetic acid. Phosphorylation from Pi was performed at 30 °C in a medium containing 1 mg ml−1 of L. lactis membrane preparations, 20 mm HEPES (pH 6.0), 10 mm MgCl2, and 20% (v/v) Me2SO (dimethyl sulfoxide), in the presence of various concentrations of copper. After a 5-min incubation at 30 °C, the reaction was started by the addition of 100 μm 32Pi (10–100 μCi nmol− 1) and stopped, 10 min later, by the addition of 1 ml of ice-cold 1 mm KH2PO4 in 7% (v/v) trichloroacetic acid. After a 30-min incubation on ice, samples were treated as described in Ref. 8. In the dephosphorylation kinetics from E2P, samples were first phosphorylated from Pi as described above during 10 min at 30 °C. They were then put on ice and stopped at different times by addition of 1 ml of ice-cold 1 mm KH2PO4 in 7% (w/v) trichloroacetic acid. The phosphorylation signal was revealed using a phosphorimaging device (Cyclone, PerkinElmer Life Science) and analyzed using the Optiquant Software (PerkinElmer Life Sciences). The amount of loaded proteins was checked after Coomassie Blue staining.
Author Contributions
E. S., P. C., and D. S.-B. conceived and designed the experiments. E. S., C. G., T.-V. D., L. M., D. S., and S. C. performed the experiments. E. S., C. G., T.-V. D., L. M., D. S., S. C., N. R., P. C., and D. S.-B. analyzed the data. P. C. and D. S.-B. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Simon Conn for critical reading of the manuscript.
This work was supported by the Commissariat à l'Energie Atomique et aux Energies Alternatives, the Centre National de la Recherche Scientifique, the French National Institute for Agricultural Research, and the University of Grenoble Alpes. The authors declare that they have no conflicts of interest with the contents of this article.
- TM
- transmembrane segment
- BCA
- bicinchoninic acid
- BCS
- bathocuproine disulfonate
- MIC
- minimum inhibitory concentration.
References
- 1. Ravet K., and Pilon M. (2013) Copper and iron homeostasis in plants: the challenges of oxidative stress. Antioxid. Redox Signal. 19, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li H. M., Theg S. M., Bauerle C. M., and Keegstra K. (1990) Metal-ion-center assembly of ferredoxin and plastocyanin in isolated chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 87, 6748–6752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., and Rolland N. (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2, 325–345 [DOI] [PubMed] [Google Scholar]
- 4. Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K. K., and Pilon M. (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15, 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdel-Ghany S. E., Müller-Moulé P., Niyogi K. K., Pilon M., and Shikanai T. (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17, 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomizioli M., Lazar C., Brugière S., Burger T., Salvi D., Gatto L., Moyet L., Breckels L. M., Hesse A. M., Lilley K. S., Seigneurin-Berny D., Finazzi G., Rolland N., and Ferro M. (2014) Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol. Cell. Proteomics 13, 2147–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blaby-Haas C. E., Padilla-Benavides T., Stübe R., Argüello J. M., and Merchant S. S. (2014) Evolution of a plant-specific copper chaperone family for chloroplast copper homeostasis. Proc. Natl. Acad. Sci. U.S.A. 111, E5480–E5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Catty P., Boutigny S., Miras R., Joyard J., Rolland N., and Seigneurin-Berny D. (2011) Biochemical characterization of AtHMA6/PAA1, a chloroplast envelope Cu(I)-ATPase. J. Biol. Chem. 286, 36188–36197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sautron E., Mayerhofer H., Giustini C., Pro D., Crouzy S., Ravaud S., Pebay-Peyroula E., Rolland N., Catty P., and Seigneurin-Berny D. (2015) HMA6 and HMA8 are two chloroplast Cu+-ATPases with different enzymatic properties. Biosci. Rep. 35, e00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu C. C., Lee W. C., Guo W. Y., Pan S. M., Chen L. J., Li H. M., and Jinn T. L. (2005) A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 139, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang C. H., Kuo W. Y., Weiss C., and Jinn T. L. (2012) Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 158, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohu C. M., Abdel-Ghany S. E., Gogolin Reynolds K. A., Onofrio A. M., Bodecker J. R., Kimbrel J. A., Niyogi K. K., and Pilon M. (2009) Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an Arabidopsis mutant. Mol. Plant 2, 1336–1350 [DOI] [PubMed] [Google Scholar]
- 13. Kühlbrandt W. (2004) Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 [DOI] [PubMed] [Google Scholar]
- 14. Argüello J. M. (2003) Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J. Membr. Biol. 195, 93–108 [DOI] [PubMed] [Google Scholar]
- 15. Smith A. T., Smith K. P., and Rosenzweig A. C. (2014) Diversity of the metal-transporting P1B-type ATPases. J. Biol. Inorg. Chem. 19, 947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gourdon P., Liu X. Y., Skjørringe T., Morth J. P., Møller L. B., Pedersen B. P., and Nissen P. (2011) Crystal structure of a copper-transporting PIB-type ATPase. Nature 475, 59–64 [DOI] [PubMed] [Google Scholar]
- 17. Gourdon P., Sitsel O., Lykkegaard Karlsen J., Birk Møller L., and Nissen P. (2012) Structural models of the human copper P-type ATPases ATP7A and ATP7B. Biol. Chem. 393, 205–216 [DOI] [PubMed] [Google Scholar]
- 18. Palmgren M. G., and Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 19. González-Guerrero M., and Argüello J. M. (2008) Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. U.S.A. 105, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Padilla-Benavides T., McCann C. J., and Argüello J. M. (2013) The mechanism of Cu+ transport ATPases: interaction with Cu+ chaperones and the role of transient metal-binding sites. J. Biol. Chem. 288, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andersson M., Mattle D., Sitsel O., Klymchuk T., Nielsen A. M., Møller L. B., White S. H., Nissen P., and Gourdon P. (2014) Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 21, 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mattle D., Zhang L., Sitsel O., Pedersen L. T., Moncelli M. R., Tadini-Buoninsegni F., Gourdon P., Rees D. C., Nissen P., and Meloni G. (2015) A sulfur-based transport pathway in Cu+-ATPases. EMBO Reports 16, 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Padilla-Benavides T., George Thompson A. M., McEvoy M. M., and Argüello J. M. (2014) Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 289, 20492–20501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barry A. N., Otoikhian A., Bhatt S., Shinde U., Tsivkovskii R., Blackburn N. J., and Lutsenko S. (2011) The lumenal loop Met672–Pro707 of copper-transporting ATPase ATP7A binds metals and facilitates copper release from the intramembrane sites. J. Biol. Chem. 286, 26585–26594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otoikhian A., Barry A. N., Mayfield M., Nilges M., Huang Y., Lutsenko S., and Blackburn N. J. (2012) Lumenal loop M672-P707 of the Menkes protein (ATP7A) transfers copper to peptidylglycine monooxygenase. J. Am. Chem. Soc. 134, 10458–10468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burkhead J. L., Reynolds K. A., Abdel-Ghany S. E., Cohu C. M., and Pilon M. (2009) Copper homeostasis. New Phytol. 182, 799–816 [DOI] [PubMed] [Google Scholar]
- 27. Hanikenne M., and Baurain D. (2013) Origin and evolution of metal P-type ATPases in Plantae (Archaeplastida). Front. Plant Sci. 4, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seigneurin-Berny D., Gravot A., Auroy P., Mazard C., Kraut A., Finazzi G., Grunwald D., Rappaport F., Vavasseur A., Joyard J., Richaud P., and Rolland N. (2006) HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 281, 2882–2892 [DOI] [PubMed] [Google Scholar]
- 29. Frelet-Barrand A., Boutigny S., Moyet L., Deniaud A., Seigneurin-Berny D., Salvi D., Bernaudat F., Richaud P., Pebay-Peyroula E., Joyard J., and Rolland N. (2010) Lactococcus lactis, an alternative system for functional expression of peripheral and intrinsic Arabidopsis membrane proteins. PLoS One 5, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hung Y. H., Layton M. J., Voskoboinik I., Mercer J. F., and Camakaris J. (2007) Purification and membrane reconstitution of catalytically active Menkes copper-transporting P-type ATPase (MNK; ATP7A). Biochem. J. 401, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Voskoboinik I., Mar J., Strausak D., and Camakaris J. (2001) The regulation of catalytic activity of the menkes copper-translocating P-type ATPase: role of high affinity copper-binding sites. J. Biol. Chem. 276, 28620–28627 [DOI] [PubMed] [Google Scholar]
- 32. Blaby-Haas C. E., and Merchant S. S. (2012) The ins and outs of algal metal transport. Biochim. Biophys. Acta 1823, 1531–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sitsel O., Grønberg C., Autzen H. E., Wang K., Meloni G., Nissen P., and Gourdon P. (2015) Structure and function of Cu(I)- and Zn(II)-ATPases. Biochemistry 54, 5673–5683 [DOI] [PubMed] [Google Scholar]
- 34. Holm R. H., Kennepohl P., and Solomon E. I. (1996) Structural and functional aspects of metal sites in biology. Chem. Rev. 96, 2239–2314 [DOI] [PubMed] [Google Scholar]
- 35. Rubino J. T., Chenkin M. P., Keller M., Riggs-Gelasco P., and Franz K. J. (2011) A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics 3, 61–73 [PubMed] [Google Scholar]
- 36. Emanuelsson O., Nielsen H., and von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuo C. L., and Campbell J. L. (1983) Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8–1 mutation. Mol. Cell. Biol. 3, 1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volland C., Galan J. M., Urban-Grimal D., Devilliers G., and Haguenauer-Tsapis R. (1994) Endocytose and degradation of the uracil permease of S. cerevisiae under stress conditions: possible role of ubiquitin. Folia Microbiol. (Praha) 39, 554–557 [DOI] [PubMed] [Google Scholar]
- 39. Chua N. H. (1980) Electrophoretic analysis of chloroplast proteins. Methods Enzymol. 69, 434–436 [Google Scholar]
- 40. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]