Abstract
Polyphosphate is a polymer of phosphate residues linked by high energy phosphoanhydride bonds. Despite being highly conserved throughout nature, its function is poorly understood. Here we show that Dictyostelium cells accumulate extracellular polyphosphate, and this acts to inhibit proliferation at high cell densities. In shaking culture, extracellular polyphosphate concentrations increase as cell density increases, and if the concentration of polyphosphate observed at the stationary phase is added to cells at mid-log, proliferation is halted. Adding an exopolyphosphatase to cell cultures or stationary phase conditioned medium decreases polyphosphate levels and abrogates the anti-proliferative effect. The cells show saturable binding of polyphosphate, suggesting the presence of a cell surface polyphosphate receptor. Extracellular polyphosphate accumulation is potentiated by decreased nutrient levels, potentially as a means to anticipate starvation. Loss of the Dictyostelium polyphosphate kinase DdPpk1 causes intracellular polyphosphate levels to become undetectable and negatively affects fitness, cytokinesis, and germination. However, cells lacking DdPpk1 accumulate ∼50% normal levels of extracellular polyphosphate, suggesting an additional means of synthesis. We found that cells lacking inositol hexakisphosphate kinase, which is responsible for the synthesis of the inositol pyrophosphates IP7 and IP8, reach abnormally high cell densities and show decreased extracellular polyphosphate levels. Two different enzymes thus appear to mediate the synthesis of Dictyostelium extracellular polyphosphate, which is used as a signal in an autocrine negative feedback loop to regulate cell proliferation.
Keywords: cell biology, cell growth, cell proliferation, Dictyostelium, stress, inositol hexakisphosphate kinase, polyphosphate, polyphosphate kinase
Introduction
Polyphosphate is a linear polymer of five to hundreds of orthophosphates linked by high energy phosphoanhydride bonds and is found in all kingdoms of life (1). In bacteria, polyphosphate has a role in survival at stationary phase, stress responses, activation of Lon protease, virulence, quorum sensing, and biofilm formation (2–8). In higher eukaryotes, polyphosphate is present in the liver, lungs, kidneys, heart, and brain in subcellular compartments including vacuoles, mitochondria, and the nucleus (9). In platelets, polyphosphate is present in dense granules and upon platelet activation is released and modulates coagulation and fibrinolysis (10–13). Inositol hexakisphosphate kinase (I6kA), which is involved in inositol pyrophosphate synthesis, has been linked to polyphosphate regulation in yeast and mice (14–16).
The eukaryotic social amoeba Dictyostelium discoideum can be grown in shaking culture and at densities of ∼2 × 107 cells/ml reaches a stationary phase where the cells stop proliferating (17, 18). Adding nutrients to stationary phase cells does not cause any further increase in proliferation, indicating that a lack of nutrients is not the cause of the proliferation arrest (18). However, adding conditioned medium (CM)2 from stationary phase cells to cells at mid-log phase causes the mid-log cells to stop proliferating, indicating that stationary phase cells accumulate an extracellular inhibitor of proliferation (18). When resuspended in fresh medium, the inhibited cells resume proliferation, indicating that the extracellular inhibitor does not kill the cells (18).
Vegetative Dictyostelium cells accumulate intracellular polyphosphate, with the highest amounts observed at stationary phase (19). Dictyostelium is the only eukaryote known to have a homolog of polyphosphate kinase, an enzyme responsible for polyphosphate synthesis in bacteria, likely through horizontal gene transfer (20). Cells lacking the Dictyostelium polyphosphate kinase (DdPpk1) lack detectable levels of intracellular polyphosphate (21). In this report we show that the Dictyostelium stationary phase proliferation inhibitor is ∼9-mer polyphosphate and that both I6kA and polyphosphate kinase contribute to the extracellular accumulation of polyphosphate.
Results
Polyphosphate Has the Properties of the Dictyostelium Stationary Phase Factor
Previous work indicated that the Dictyostelium stationary phase factor is a <10-kDa heat-resistant molecule (9, 10). We observed that this factor is resistant to proteinase K, DNase, and RNase treatment; does not partition into hydrophobic organic solvents; binds anion but not cation exchange resins; passes through a 3-kDa filter; and is retained by a 2-kDa filter (Table 1). This suggests that the stationary phase factor is a negatively charged ∼2–3-kDa molecule.
TABLE 1.
Effect of materials added to stationary phase CM on the ability of the medium to inhibit proliferation
For proteinase K, RNase, and DNase, enzymes were added to CM for 18 h and were then removed with a 10-kDa cutoff spin filter. Organic extractions were done with an equal volume of solvent, vortexing for 20 min, and then centrifugation to separate phases. The separated phases were dried by lyophilization. Organic phases were resuspended in PBM, and the aqueous phase were resuspended in water. Approximately 30% (v/v) beads were added to CM, mixed gently for 1 h, and then removed by centrifugation. Treated CM was then incubated with wild type cells, and cell density was measured
| Material added to CM | CM activity |
|---|---|
| Proteinase K | + |
| RNase | + |
| DNase | + |
| Hexane extraction | |
| Organic | − |
| Aqueous | + |
| Ethyl acetate extraction | |
| Organic | − |
| Aqueous | + |
| Dichloromethane extraction | |
| Organic | − |
| Aqueous | + |
| Amberlite XAD-4 hydrophobic material: binding beads | − |
| 50-X8 cation exchange beads | − |
| Bio-Rex 70 cation exchange beads | − |
| 50W-X8 cation exchange beads | − |
| 1-X8 anion exchange beads | + |
| 1-X2 anion exchange beads | + |
| 3-kDa cutoff spin filter | |
| Retentate | − |
| Flow-through | + |
| 2-kDa cutoff spin filter | |
| Retentate | + |
| Flow-through | − |
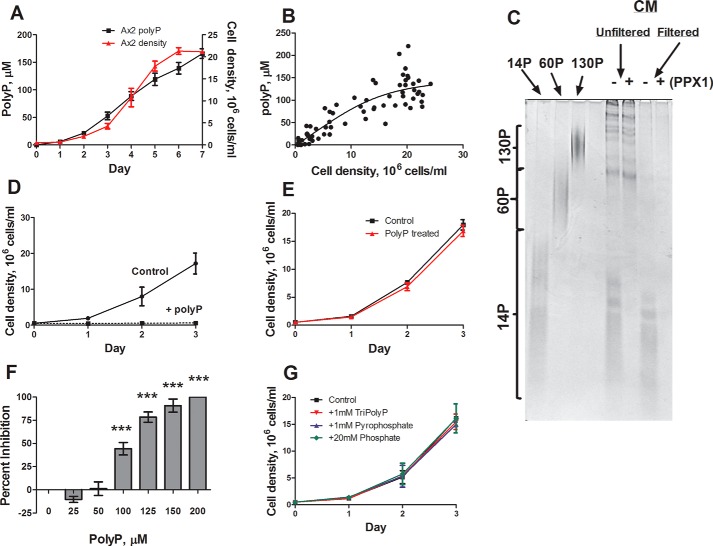
Small polyphosphate molecules share the observed properties of the stationary phase factor. To determine whether polyphosphate could be acting as the stationary phase factor in Dictyostelium, we first tested whether cells accumulate extracellular polyphosphate. Conditioned medium samples were taken every 24 h from wild type cells, and polyphosphate was identified and quantified using DAPI (22). Wild type cells accumulate extracellular polyphosphate, and the levels increase as cell density increases, with the highest amounts observed during stationary phase (Fig. 1, A and B). Although DAPI analysis suggests the presence of polyphosphate, currently the only way to definitively show the presence of polyphosphate in a sample is through PAGE analysis, in which polyphosphate appears as a smear, similar to RNA. Analysis of stationary phase conditioned medium using PAGE confirmed the presence of extracellular polyphosphate polymers of 5–13 phosphate residues with an average of 9 phosphate residues (Fig. 1C). Enzymatic treatment of stationary phase conditioned medium with the exopolyphosphatase ScPPX1 (23) removed the polyphosphate band, indicating that polyphosphate is responsible for the band. The accumulation of ∼150 μm of extracellular polyphosphate (following convention, the concentration is in units of phosphate monomers) coincided with the inhibition of proliferation at stationary phase. To determine whether polyphosphate affects proliferation, 150 μm polyphosphate was added to mid-log cells. This concentration of polyphosphate strongly inhibited proliferation, and upon resuspension in fresh medium, the cells resumed proliferation, suggesting that the inhibition was not due to cell death (Fig. 1, D and E). Polyphosphate concentrations as low as 100 μm inhibited proliferation, whereas lower concentrations showed no inhibition or slightly potentiated proliferation (Fig. 1F). Triphosphate and pyrophosphate at concentrations up to 1 mm and monophosphate up to 20 mm had no effect on proliferation (Fig. 1G).
FIGURE 1.
Wild type cells accumulate increasing amounts of extracellular polyphosphate as cell density increases. A and B, cells were cultured, and the polyphosphate (polyP) concentration in the conditioned medium was measured. C, polyP from stationary phase conditioned medium was resolved on a 25% polyacrylamide gel and stained with toluidine blue (representative image of three gels). D and F, cells were cultured with 150 μm polyP (D) or the indicated concentration of polyP (F) and density (D), and the percentage of inhibition after 24 h (F) was determined. E, wild type cells were cultured in the presence or absence of 150 μm polyP for 18 h, washed twice in HL5, resuspended in fresh HL5, and counted daily. G, wild type cells were cultured in the presence or absence of triphosphate, pyrophosphate, or sodium phosphate and counted daily. All values are means ± S.E., n ≥ 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (one-way analysis of variance).
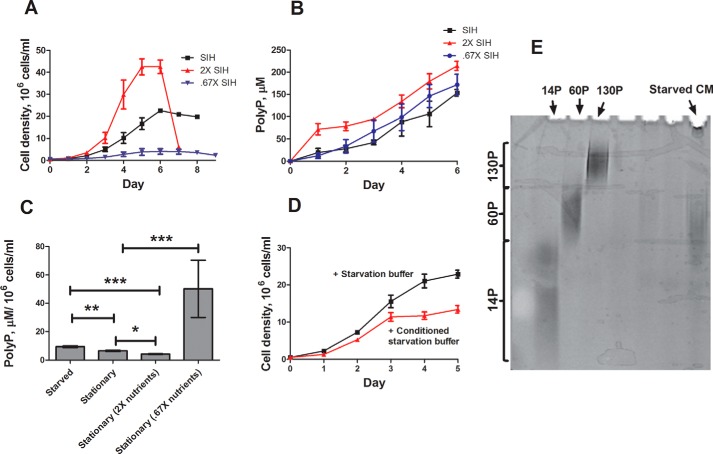
To determine whether nutrient levels altered polyphosphate accumulation, cells were cultured in concentrated SIH growth medium, a defined minimal medium, which caused increased proliferation and decreased polyphosphate accumulation per cell, whereas cells cultured in diluted growth medium or starvation buffer showed decreased proliferation and increased polyphosphate accumulation per cell (Fig. 2, A–C). Conditioned medium from starved cells also inhibited proliferation and contained ∼40-mer polyphosphate (Fig. 2, D and E). Together, these results suggest that extracellular polyphosphate is acting as an inhibitor of proliferation to regulate population density in Dictyostelium, and its accumulation is potentiated by decreased nutrient availability.
FIGURE 2.
Effect of nutrient levels on extracellular polyphosphate accumulation. A and B, SIH was generated at concentrations of 1×, 2×, and 0.67×; wild type cells were cultured and counted daily (A); and CM samples were harvested to determine polyP content (B). C, cells were cultured in the indicated conditions, and the polyP concentration in the conditioned medium was measured. D, wild type cells were starved for 5 h in PBM, and conditioned medium was harvested and concentrated using a 2-kDa size exclusion filter. Wild type cells were then cultured in the presence or absence of 50% of the resulting concentrated, starved CM and counted daily. E, polyP from starved CM was resolved using a 25% polyacrylamide gel, stained with toluidine blue, and destained with 20% methanol. All values are means ± S.E., n ≥ 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (one-way analysis of variance).
The Proliferation-inhibiting Activity Is Reduced by Polyphosphatase
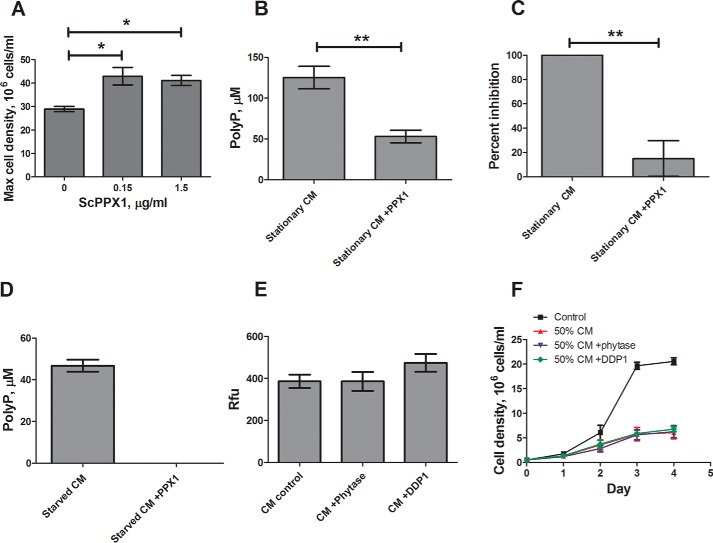
To test the hypothesis that Dictyostelium cells secrete polyphosphate to inhibit their proliferation, we treated cell cultures and conditioned medium with ScPPX1. Daily additions of ScPPX1 to cultures caused cells to proliferate to higher densities compared with untreated cells (Fig. 3A). ScPPX1 treatment of conditioned media reduced extracellular polyphosphate to ∼50 μm (Fig. 3B), a concentration where we observed little effect on proliferation (Fig. 1F), and this dramatically reduced the activity of the stationary phase factor in the conditioned medium (Fig. 3C). ScPPX1 treatment of conditioned starvation buffer also decreased polyphosphate levels (Fig. 3D).
FIGURE 3.
The proliferation inhibiting activity is reduced by exopolyphosphatase. A, wild type cells were treated with ScPPX1 or buffer (0) daily, and cell density was determined. B and C, stationary phase conditioned medium was treated with 0.15 μg/ml ScPPX1 for 3 h. The samples were then assayed for polyP content (B) and the ability to inhibit cell proliferation (C). D, starved CM was treated with ScPPX1 for 3 h, and polyP content was determined using DAPI. E and F, stationary phase CM was treated with phytase or DDP1, polyP fluorescence was determined using DAPI (E), and inhibitory activity was determined by culturing cells with 50% CM (treated or untreated) with daily counts (F). The values are means ± S.E., n ≥ 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with the no-PPX1 control (unpaired two-tailed t tests).
Because phytic acid can also interact and fluoresce with DAPI at the same wavelengths as polyphosphate (24), conditioned medium was treated with the phytic acid-degrading enzymes phytase and DDP1 (25, 26). Phytase or DDP1 treatments had no effect on DAPI fluorescence of conditioned medium or on the activity of the stationary phase factor (Fig. 3, E and F). These results indicate that the polyphosphate in Dictyostelium stationary phase conditioned medium inhibits proliferation.
Polyphosphate Inhibits Cytokinesis to Cause an Increase in Cell Mass
Proliferation (an increase in cell number with time) and growth (an increase in cell mass with time) can be regulated separately (27). The stationary phase factor inhibits proliferation, yet compared with mid-log cells, stationary phase cells are larger (17). To determine whether polyphosphate inhibits proliferation but not growth, the cells were treated with polyphosphate or conditioned medium. Compared with controls, the cells treated with polyphosphate or conditioned medium showed increased cell mass and forward scatter in a flow cytometer (Table 2). Cells in stationary phase showed an increase in multinucleate cells compared with mid-log cells (Table 3). To determine the effect of polyphosphate on cytokinesis, the cells were treated with polyphosphate for 18 h and then stained with DAPI. Polyphosphate caused a dramatic increase in multinucleate cells (Table 3). AprA, a secreted protein present in stationary conditioned medium, causes a decrease in the number of nuclei per cell (27), which may explain the increased number of nuclei per cell caused by polyphosphate compared with the number observed in stationary phase. These results show that the effects of polyphosphate on cell size match the observed properties of the stationary phase factor, appear to be due to polyphosphate inhibiting cytokinesis and suggest that polyphosphate has more of an inhibitory effect on cell proliferation than it does on cell growth or karyokinesis.
TABLE 2.
Effect of polyphosphate on mass and size of cells
Polyphosphate inhibits proliferation but not growth. Wild type cells were treated with polyP or stationary phase CM for 18 h, and cell mass was determined. Forward scatter of cells (arbitrary units) was measured with a flow cytometer. The values are means ± S.E., minimum n ≥ 4.
| Condition | Cell density after 18 h (106 cells/ml) | Cell mass (mg/106 cells/ml) | Forward scatter |
|---|---|---|---|
| HL5 | 1.51 ± .23 | 1.11 ± .17 | 37.6 ± 0.2 |
| Stationary CM | 0.62 ± .06a | 1.58 ± .16a | 51.1 ± 0.6b |
| 150 μm polyP | 0.60 ± .04b | 1.61 ± .18c | 63.5 ± 1.6b |
a p < 0.05 compared with the HL5 control (paired two-tailed t tests).
b p < 0.01 compared with the HL5 control (paired two-tailed t tests).
c p < 0.001 compared with the HL5 control (paired two-tailed t tests).
TABLE 3.
Effect of polyphosphate on number of nuclei
Polyphosphate inhibits proliferation but not growth. The cells were stained with DAPI, and the nuclei were counted by microscopy. The values are means ± S.E., minimum n ≥ 4.
| Condition | Percentage of cells |
Nuclei/100 cells | ||
|---|---|---|---|---|
| With one nucleus | With two nuclei | With three or more nuclei | ||
| Ax2 mid-log | 75 ± 2 | 23 ± 2 | 2 ± 1 | 125 ± 2 |
| Ax2 stationary | 62 ± 4a | 32 ± 4b | 6 ± 1b | 147 ± 4b |
| Ax2 + 150 μm polyP | 41 ± 4a | 35 ± 1a | 24 ± 4c | 216 ± 5a |
a p < 0.01 compared with mid-log cells (paired two-tailed t tests).
b p < 0.05 compared with mid-log cells (paired two-tailed t tests).
c p < 0.001 compared with mid-log cells (paired two-tailed t tests).
Dictyostelium Binds Polyphosphate
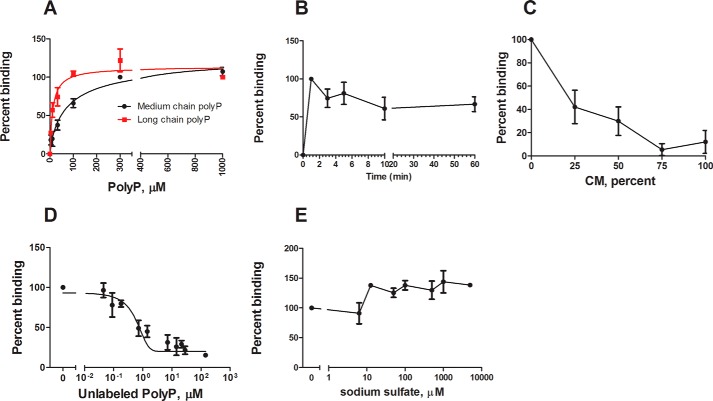
The presence of a cell surface polyphosphate receptor would require that cells show a class of saturable binding sites for polyphosphate. To determine whether polyphosphate binds to cells, the binding of biotinylated polyphosphate to wild type cells was measured. Long chain biotinylated polyphosphate showed saturatable binding to wild type cells with a KD of 12 ± 3 μm, whereas medium chain biotinylated polyphosphate had a KD of 78 ± 32 μm (mean ± S.E., n = 4), suggesting that longer polymers have a higher binding affinity (Fig. 4A). The Hill coefficient was 1.0 ± 0.3 for both chain lengths, suggesting that there is no binding cooperativity, and F tests comparing models with multiple binding sites indicated a single class of binding site. The Bmax of medium chain polyphosphate was 1.6 ± 1.1 × 106 binding sites/cell. Although this number is high compared with known Dictyostelium cell surface receptors (28–30), there are many examples of numbers of cell surface binding sites in this range and higher (31–35). The binding appeared to equilibrate within 1 min (Fig. 4B). The binding of medium chain biotinylated polyphosphate could be competed with both conditioned medium and unlabeled polyphosphate (Fig. 4, C and D). Using the equation of Cheng and Prusoff (36), unlabeled polyphosphate showed a Ki of 87.5 ± 7.2 μm. To check that the binding of polyphosphate to cells was not simply due to anionic interactions, binding was competed using sodium sulfate, another anionic molecule (Fig. 4E). Sodium sulfate was not able to compete with polyphosphate for binding, suggesting that polyphosphate binding is not simply an anionic interaction with the cell membrane. Together, these results indicate that polyphosphate shows saturatable binding to cells, and the binding can be competed with unlabeled polyphosphate and stationary phase conditioned media.
FIGURE 4.
Polyphosphate binds wild type cells. A, mid-log wild type cells were incubated with the indicated amounts of biotinylated polyP and a streptavidin-conjugated fluorophore. The cells were washed, and fluorescence was measured using a flow cytometer. B, cells were incubated with 200 μm medium chain biotinylated polyP for the indicated times, and binding was measured as in A. C, D, and E, competition assays were performed by incubating wild type cells with 200 μm medium chain biotinylated polyP, streptavidin-conjugated fluorophore, and either stationary phase conditioned medium (C), unlabeled polyP (D), or sodium sulfate (E) binding was measured as in A. Concentrations of unlabeled polyP (D) are shown as whole polyphosphate molecules as opposed to phosphate monomers. The values are means ± S.E., n ≥ 4.
Inositol Hexakisphosphate Kinase Regulate Extracellular Polyphosphate Accumulation
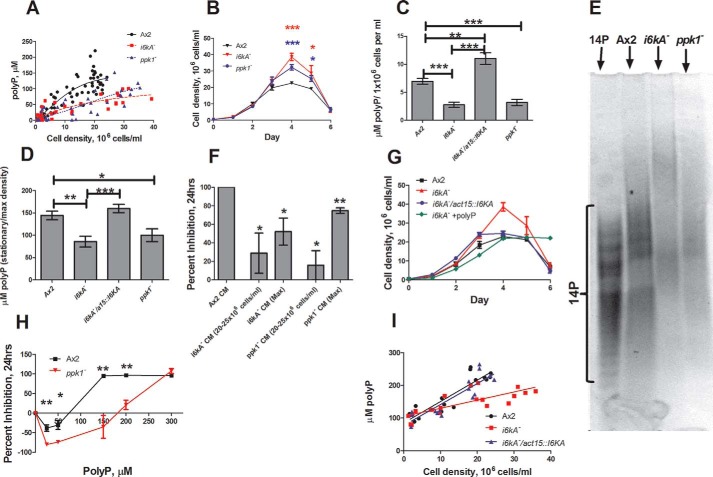
Inositol hexakisphosphate kinase (IP6K) uses IP6 as a substrate to generate the inositol pyrophosphates IP7 and IP8 (37, 38). Yeast lacking the IP6K Kcs1 have no observable polyphosphate accumulation (14, 16). Mice lacking IP6K have reduced polyphosphate levels in platelets (15). To test the hypothesis that IP6K also regulates polyphosphate levels in Dictyostelium, we measured extracellular polyphosphate in cells lacking the IP6K I6kA, as well as cells lacking DdPpk1 (21, 39). The cells lacking either I6kA or DdPpk1 proliferated to abnormally high cell densities and had reduced extracellular polyphosphate accumulation (Fig. 5, A–D). PAGE indicated that both mutants accumulate less extracellular polyphosphate than wild type (Fig. 5E). In agreement with the abnormally low levels of extracellular polyphosphate in cultures of i6kA− and ppk1− cells, conditioned medium from i6kA− and ppk1− cells harvested at densities in which wild type cells enter stationary (20–25 × 106 cells/ml) or at maximum densities (30–40 × 106 cells/ml) contained abnormally low levels of inhibitory activity (Fig. 5F). The abnormally high maximum density and low polyphosphate accumulation phenotypes exhibited by i6kA− cells were rescued by expressing the I6kA cDNA under the control of the actin 15 promoter (Fig. 5, C, D, and G). Polyphosphate supplementation effectively rescued the i6kA− proliferation defect, suggesting that this defect is due to reduced stationary phase factor production as opposed to reduced sensitivity to the factor itself (Fig. 5G). The proliferation of ppk1− cells could also be inhibited by exogenous polyphosphate, although these cells showed a roughly 2-fold reduction in sensitivity (Fig. 5H). Consistent with these observations, both I6kA and DdPpk1 are up-regulated upon transitioning from vegetative growth to starvation as reported by the DictyExpress. Together, these results show roles for both I6kA and DdPpk1 in regulating extracellular polyphosphate accumulation in Dictyostelium, as well as a conserved role for inositol hexakisphosphate kinase in polyphosphate accumulation in multiple eukaryotic models.
FIGURE 5.
Inositol hexakisphosphate kinase regulates extracellular polyphosphate accumulation. A and B, the indicated strains were cultured in SIH, and extracellular polyP accumulation was measured. C and D, conditioned medium from cells at stationary (WT) or maximum density (i6kA−, ppk1−) was assayed for polyP. E, polyP from conditioned medium was resolved on a 25% polyacrylamide gel and stained with toluidine blue. 14P indicates 14-mer polyP size standard. F, conditioned medium from cells at the indicated densities was assayed for their ability to inhibit the proliferation of wild type cells. G, proliferation of the indicated strains or of i6kA− cells with 30 μm polyP added to the culture at day 1, 20 μm added at days 2 and 3, and 10 μm added at days 4 and 5. H, wild type or cells lacking DdPpk1 were cultured in SIH in the presence or absence of polyP. Cell density was measured daily. I, the indicated strains were cultured in HL5, and intracellular polyP accumulation was measured. The values are means ± S.E., n ≥ 4. *, p < 0.05, compared with the Ax2 wild type control (unpaired two-tailed t tests in most panels; unpaired one-tailed t test in F).
To determine whether intracellular polyphosphate is also regulated by both DdPpk1 and I6kA, intracellular polyphosphate was measured in wild type cells and cells lacking I6kA. As previously reported, intracellular polyphosphate levels are highest as cells enter the stationary phase (19), showing a roughly 2-fold increase from mid-log to stationary (Fig. 5I). Cells lacking I6kA had normal amounts of intracellular polyphosphate at low cell densities but did not show the robust increase in intracellular polyphosphate production at high cell densities. Although DdPpk1 is essential for intracellular polyphosphate production at all cell densities, I6kA appears to have a role in regulating intracellular polyphosphate levels at high cell densities.
Mutant Proliferation Phenotypes Correlate with Extracellular Polyphosphate Accumulation
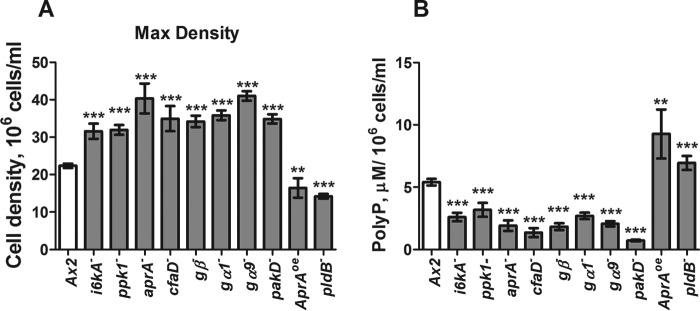
We previously identified transformants that proliferate to abnormally high or low stationary phase or maximum cell densities (27, 40–42). In addition to cells lacking I6kA and DdPpk1, cells lacking the G proteins Gβ, Gα1, and Gα9, the secreted proteins AprA and CfaD and the kinase PakD all proliferate to abnormally high cell densities (Fig. 6A). For instance, AprA and CfaD slow but do not stop cell proliferation and thus have some stationary phase factor-like properties (27, 40). All of these strains had abnormally low levels of extracellular polyphosphate/1 × 106 cells at their respective maximum densities (Fig. 6B). The cells lacking phospholipase D, a homolog of human PLD1, and the cells overexpressing AprA proliferate to abnormally low stationary cells densities (Fig. 6A) and had abnormally high levels of extracellular polyphosphate (Fig. 6B). These results suggest that for some transformants, maximal cell density shows an inverse correlation with extracellular polyphosphate level.
FIGURE 6.
Mutant proliferation phenotypes correlate with extracellular polyphosphate accumulation. A, wild type cells or transformants were cultured in SIH and counted daily, and the maximal density was recorded. B, the extracellular polyP concentration was measured at the maximal density and normalized to the cell density. The values are means ± S.E., n ≥ 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with the wild type control (unpaired two-tailed t tests).
Discussion
In this report, we found that the extracellular proliferation inhibitor originally identified by Yarger, Soll, and co-workers (17, 18), which causes Dictyostelium cells to stop proliferating at stationary phase (17, 18), is ∼9-mer polyphosphate. When extracellular polyphosphate is depleted by treatment with exopolyphosphatase, the cells grow to a higher density. At first glance, having polyphosphate inhibit proliferation would thus seem to be disadvantageous to the cells. However, we found that polyphosphate has more of an inhibitory effect on proliferation than on growth, resulting in high density cells stopping proliferation when they have large amounts of stored nutrients. This then suggests that when a population of cells starves, having somewhat fewer cells with more stored nutrients per cell is more advantageous than having more cells with less stored nutrients per cell. We envision that polyphosphate thus allows proliferating cells to anticipate the inevitable situation where a high density of cells overgrows its food supply and starves. Decreased nutrient levels potentiate extracellular polyphosphate accumulation, potentially allowing the cells to more accurately predict when they will starve.
When starved, Dictyostelium cells lacking DdPpk1, and therefore intracellular polyphosphate, form smaller fruiting bodies and have defective spore germination (21). We found that extracellular polyphosphate may help cells anticipate and prepare for starvation by stopping proliferation when they have relatively high levels of stored nutrients. Polyphosphate accumulation in Dictyostelium thus potentiates survival in stressful conditions as it does in many prokaryotes (1, 5).
Although polyphosphate present in starved conditioned medium can be degraded to undetectable levels, some of the polyphosphate present in stationary phase conditioned medium appears to be resistant to yeast exopolyphosphatase. Kornberg and co-workers (1) observed that preparations of polyphosphate from biological sources also show resistance to exopolyphosphatase activity, presumably through a terminal end modification, which can confer resistance to exopolyphosphatase (43). Some of the stationary phase extracellular polyphosphate may thus also have covalent modifications.
Although the saturable binding of polyphosphate to cells suggests the existence of a cell surface polyphosphate receptor, it does not prove the existence. If the observed binding is to a receptor that causes cells to stop proliferating, with the observed first order binding kinetics, using percentage bound = [ligand]/([ligand] + Ki), at 150 μm polyphosphate and the observed Ki of unlabeled polyphosphate (87.5 μm), the receptor stops proliferation when ∼63% of the receptors are occupied.
Cells lacking DdPpk1 have undetectable levels of intracellular polyphosphate (21), whereas we observed that these same cells have reduced but detectable levels of extracellular polyphosphate. Polyphosphate accumulation in yeast and mice involves inositol hexakisphosphate kinase, and we observed that this enzyme also contributes to the accumulation of extracellular and intracellular polyphosphate at high cell densities in Dictyostelium. Thus, for unknown reasons, Dictyostelium cells appear to use at least two different enzymes to generate polyphosphate.
The secreted proteins AprA and CfaD; the G protein subunits Gα1, Gα9 (which mediates AprA signaling (41)), and Gβ; and the putative kinases PakD and PldB all modulate extracellular polyphosphate accumulation and maximal cell density. A reasonable assumption is that these signals and pathways are involved in sensing cell density, nutrient status, or other conditions that to some extent help cells accumulate enough polyphosphate to anticipate starvation and stop proliferating.
Little is known about how the size or composition of a tissue is regulated. One possibility is that cells of a specific type secrete a factor called a chalone. Chalones are factors that inhibit the proliferation of the cells that secrete them (44, 45). As the number of cells secreting the chalone increases, the extracellular concentration of the chalone increases, and at some combination of cell number and chalone concentration, cell proliferation stops (45). Despite considerable evidence for their existence in mammalian tissues such as epidermis, intestine, spleen, liver, and kidney, for many chalones, attempts to identify them as either organic molecules or proteins have failed (45–51). Extracellular polyphosphate appears to be acting as a chalone in Dictyostelium. Together, these results suggest a new role for polyphosphate, as well as the intriguing possibility that chalones in higher eukaryotes might be compounds that are neither conventional organic molecules nor polypeptides.
Experimental Procedures
Reagents and Materials
HL5 and SIH growth media were from Formedium (Norfolk, UK). Polyphosphate was from both Spectrum (New Brunswick, NJ) and Merck. Phytic acid and high density nickel beads were from GoldBio (St. Louis, MO). Phytase, hexane, ethyl acetate, dichloromethane, sodium tripolyphosphate, sodium pyrophosphate, acidified phenol:chloroform, and Amberlite XAD-4 were from Sigma. DDP1 from MyBioSource (San Diego, CA). Biotinylated polyphosphate was from KeraFast Inc. (Boston, MA). Streptavidin-conjugated Alexa Fluor 647 was from Invitrogen. Blasticidin and G418 were from CalBioChem (Boston, MA). Vectors and mutant cell lines were obtained from dictyBase. Size exclusion spin filters were from Sartorius (Bohemia, NY) (2 kDa) and Aviva Biosciences (San Diego, CA) (10 and 3 kDa). Proteinase K, RNase, DNase, and restriction enzymes were from New England BioLabs (Ipswich, MA). All cation and anion exchange beads were from Bio-Rad.
Cell Culture and Proliferation Curves
The cells used were Dictyostelium wild type Ax2, aprA− (DB60T3–8), cfaD− (DB27C-1), gβ− (DBS0236531), gα1− (DBS02306088), gα9− (DBS0236109), i6kA− (DBS0236426), ppk1− (DBS0350686), pakD− (DBS0350281), pldB− (DBS0236796), and AprAOE (DBS0235510). Mutant genotypes were verified by PCR and kept under constant selection. The cells were grown in shaking culture using HL5 nutrient rich growth medium (10–12-h doubling time) or the synthetic medium SIH (∼22–26-h doubling time). Proliferation curves used mid-log cells diluted to 5 × 105 cells/ml, and density was measured every 24 h using a hemocytometer. Conditioned medium was harvested by centrifugation of cells at 800 × g for 10 min at 4 °C, and the supernatant was saved. The supernatant was clarified by centrifugation at 9000 × g for 10 min at 4 °C, and the resultant supernatant was filter sterilized using 0.22-μm filters (Genesee Scientific, San Diego, CA) and stored at 4 °C. The cells were starved in PBM (27) for 5 h at 5 × 106 cells/ml, and conditioned medium was collected. Proliferation inhibition by conditioned medium was assessed by culturing cells in 50% conditioned medium and 50% growth medium. The percentage of inhibition was determined by treating control cells as 100% proliferation, 0% inhibition and wild type conditioned medium as 100% inhibition, 0% proliferation. The cell mass and nuclei content were measured as previously described (27). Forward scatter (in arbitrary units) was measured on a BD Accuri C6 (BD Biosciences, San Jose, CA) flow cytometer.
Treatments of Conditioned Media
Proteinase K, RNase, and DNase were added to SIH conditioned medium to 0.1 mg/ml at 37 °C for 18 h, and the enzyme was then removed using a 10-kDa cutoff spin filter. Organic extractions were done by adding an equal volume of solvent, vortexing for 10 s every 2–3 min for 20 min, phase separation by centrifugation at 200 × g for 5 min, and then lyophilization of the organic and aqueous phases. The organic phase was resuspended in SIH, and the aqueous phase was resuspended in water. Bead treatments involved washing beads following the manufacturer's protocol, adding a 30% volume of bead slurry to conditioned medium, rotating gently for 1 h, letting the beads settle out, and then clarification of the supernatant at 200 × g for 5 min.
Polyphosphate Measurements
Polyphosphate levels in conditioned medium were assessed by incubating samples with DAPI and measuring fluorescence at 550 nm when excited at 415 nm as previously described (22) with the following modifications. Conditioned medium samples were generated in the synthetic media SIH to reduce the amount of background fluorescence from HL5 medium and were clarified using a 10-kDa spin filter (except for starved CM), and the filtrate was saved. Conditioned medium was then incubated with 25 μg/ml DAPI for 5 min, and the fluorescence was measured. Polyphosphate concentrations, in terms of phosphate monomers, were determined using polyphosphate standards generously provided by Dr. Toshikazu Shiba (RegeneTiss Inc.).
Intracellular polyphosphate levels were assessed by resuspending cell pellets in LETS buffer (100 mm LiCl, 10 mm EDTA, 10 mm Tris-HCl, pH 8, 0.5% SDS). One volume of acidic phenol:chloroform (pH 4) was added, and samples were vortexed for 5 min at 4 °C. The samples were then spun at 17,000 × g for 5 min at 4 °C. Two volumes of chloroform were added to the resulting aqueous phase, vortexed for 5 min at 4 °C, and then spun at 2000 × g for 5 min at 4 °C. The resulting aqueous phase was incubated with 2.5 volumes of 100% ethanol for 5 min at room temperature and then spun at 15,000 × g for 10 min at 4 °C. The pellet was resuspended in 100 μl of TE (10 mm Tris-HCL, pH 7.4, 1 mm EDTA, pH 8), pH 7.4, containing 1 μg/ml of RNase A and incubated for 15 min at room temperature. 2.5 volumes of 100% ethanol were then added, and the mixture was incubated at −80 °C for 1–2 h and then spun at 15,000 × g for 10 min at 4 °C. The pellet was resuspended in TE. The samples were then assayed for polyphosphate content as described above using DAPI.
PAGE Analysis
Polyphosphate was resolved on polyacrylamide gels as previously described (38). Stationary phase and starved conditioned media were concentrated 15-fold using 2-kDa size exclusion filters. Polymer lengths were calculated by plotting the log of polymer lengths against the position of the band on a gel.
Enzyme Treatments
Plasmids for purifying yeast PPX1 were kindly provided by Dr. Michael Gray (52). Recombinant protein purification was performed as previously described (27). Treatments of conditioned media using 0.15 μg/ml recombinant ScPPX1 were done for 4 h at 37 °C, supplementing the medium with 5 mm MgCl2. Phytase was added to 3 μg/ml (crude extract) for 4 h at pH 5.5 at 37 °C. pH was adjusted using 1 n HCl or 1 n NaOH followed by filter sterilization using 0.22-μm filters. DDP1 treatments at 0.15 μg/ml were done for 4 h at 37 °C in the presence of 5 mm MgCl2. The reactions were passed through 10-kDa cutoff spin filters to remove enzyme and then assayed for polyphosphate content and chalone activity. For ScPPX1 treatment of cell cultures, mid-log cells were inoculated at 5 × 106 cells/ml, and ScPPX1 or an equal volume of buffer was added daily.
Binding Assay with Biotinylated Polyphosphate
Binding of polyphosphate to wild type cells was determined using biotinylated polyphosphate. The cells at mid-log were washed twice with ice-cold PBM and then resuspended in ice-cold PBM. The samples were incubated with biotinylated compound (100 μm for competition assays) and streptavidin-conjugated Alexa Fluor 647 (1:200) on ice for 3 min, centrifuged at 200 × g for 3 min at 4 °C, and resuspended in ice-cold PBM, and fluorescence was measured using a flow cytometer. Conditioned medium was 10-kDa filtered to remove any large molecules or proteins.
The total number of binding sites/cell was calculated by incubating 107 cells for 3 min on ice with 150 μm biotinylated polyphosphate in 200 μl. The samples were then centrifuged at 200 × g for 3 min at 4 °C, and the supernatant was harvested without disruption of the cell pellet. The supernatant was then incubated with mid-log cells as described above. Comparison of the supernatant binding and the binding of different concentrations of biotinylated polyphosphate to cells was used to determine the amount of biotinylated polyphosphate bound to the 107 cells, and this was used to calculate the Bmax.
Expression of I6kA in i6kA− Cells
To generate the i6kA−/a15::I6kA cell line, the i6kA open reading frame was amplified by PCR from wild type vegetative cDNA with the primers 5′-CGCGAAGCTTATGCACATATTTTATTTAGTA and 5′-CCGCGGATCCTTAGTTTGTATTTATGACTGTAT with terminal HindIII and BamHI sites, respectively. This PCR product was cloned into pGEM-T vector (Promega). The i6kA open reading frame was isolated by HindIII and BamH1 digestion and gel purification and then ligated into the HindIII and BamH1 sites of PDXa-3C. Correct orientation of the open reading frame within the vector was confirmed by restriction mapping and sequencing. This vector was transformed into Dictyostelium cells lacking I6kA following (27), using 5 μg/ml G418 to select for cells containing the vector.
Author Contributions
P. M. S. designed, performed, and analyzed the experiments and wrote the paper. R. H. G. conceived and coordinated the study, acquired funding, and revised the final paper.
Acknowledgments
We thank Bethany Sump and Jacob Watson for assistance, Dr. Toshikazu Shiba of RegeneTiss Inc. for polyphosphate standards, Dr. Michael Gray for plasmids, and Thomas Livermore and the Saiardi Lab at University College London for advice on PAGE analysis of polyphosphate.
This work was supported by National Institutes of Health R01 GM102280. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CM
- conditioned medium
- polyP
- polyphosphate.
References
- 1. Rao N. N., Gómez-García M. R., and Kornberg A. (2009) Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605–647 [DOI] [PubMed] [Google Scholar]
- 2. Achbergerová L., and Nahálka J. (2011) Polyphosphate: an ancient energy source and active metabolic regulator. Microb. Cell Fact. 10, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuroda A., Nomura K., Ohtomo R., Kato J., Ikeda T., Takiguchi N., Ohtake H., and Kornberg A. (2001) Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708 [DOI] [PubMed] [Google Scholar]
- 4. Nikel P. I., Chavarría M., Martínez-García E., Taylor A. C., and de Lorenzo V. (2013) Accumulation of inorganic polyphosphate enables stress endurance and catalytic vigour in Pseudomonas putida KT2440. Microb. Cell Fact. 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rao N. N., and Kornberg A. (1996) Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178, 1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rao N. N., and Kornberg A. (1999) Inorganic polyphosphate regulates responses of Escherichia coli to nutritional stringencies, environmental stresses and survival in the stationary phase. Prog. Mol. Subcell. Biol. 23, 183–195 [DOI] [PubMed] [Google Scholar]
- 7. Rashid M. H., Rumbaugh K., Passador L., Davies D. G., Hamood A. N., Iglewski B. H., and Kornberg A. (2000) Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sureka K., Dey S., Datta P., Singh A. K., Dasgupta A., Rodrigue S., Basu J., and Kundu M. (2007) Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 65, 261–276 [DOI] [PubMed] [Google Scholar]
- 9. Kumble K. D., and Kornberg A. (1995) Inorganic polyphosphate in mammalian cells and tissues. J. Biol. Chem. 270, 5818–5822 [DOI] [PubMed] [Google Scholar]
- 10. Morrissey J. H., Choi S. H., and Smith S. A. (2012) Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood 119, 5972–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Müller F., Mutch N. J., Schenk W. A., Smith S. A., Esterl L., Spronk H. M., Schmidbauer S., Gahl W. A., Morrissey J. H., and Renné T. (2009) Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 139, 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruiz F. A., Lea C. R., Oldfield E., and Docampo R. (2004) Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 279, 44250–44257 [DOI] [PubMed] [Google Scholar]
- 13. Smith S. A., Mutch N. J., Baskar D., Rohloff P., Docampo R., and Morrissey J. H. (2006) Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. U.S.A. 103, 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Auesukaree C., Tochio H., Shirakawa M., Kaneko Y., and Harashima S. (2005) Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 280, 25127–25133 [DOI] [PubMed] [Google Scholar]
- 15. Ghosh S., Shukla D., Suman K., Lakshmi B. J., Manorama R., Kumar S., and Bhandari R. (2013) Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood 122, 1478–1486 [DOI] [PubMed] [Google Scholar]
- 16. Saiardi A. (2012) How inositol pyrophosphates control cellular phosphate homeostasis? Adv. Biol. Regul. 52, 351–359 [DOI] [PubMed] [Google Scholar]
- 17. Soll D. R., Yarger J., and Mirick M. (1976) Stationary phase and the cell cycle of Dictyostelium discoideum in liquid nutrient medium. J. Cell Sci. 20, 513–523 [DOI] [PubMed] [Google Scholar]
- 18. Yarger J., Stults K., and Soll D. R. (1974) Observations on the growth of Dictyostelium discoideum in axenic medium: evidence for an extracellular growth inhibitor synthesized by stationary phase cells. J. Cell Sci. 14, 681–690 [DOI] [PubMed] [Google Scholar]
- 19. Zhang H., Gómez-García M. R., Brown M. R., and Kornberg A. (2005) Inorganic polyphosphate in Dictyostelium discoideum: influence on development, sporulation, and predation. Proc. Natl. Acad. Sci. U.S.A. 102, 2731–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H., Gómez-García M. R., Shi X., Rao N. N., and Kornberg A. (2007) Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum, with a role in cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 104, 16486–16491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Livermore T. M., Chubb J. R., and Saiardi A. (2016) Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 113, 996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aschar-Sobbi R., Abramov A. Y., Diao C., Kargacin M. E., Kargacin G. J., French R. J., and Pavlov E. (2008) High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 18, 859–866 [DOI] [PubMed] [Google Scholar]
- 23. Wurst H., and Kornberg A. (1994) A soluble exopolyphosphatase of Saccharomyces cerevisiae: Purification and characterization. J. Biol. Chem. 269, 10996–11001 [PubMed] [Google Scholar]
- 24. Kolozsvari B., Parisi F., and Saiardi A. (2014) Inositol phosphates induce DAPI fluorescence shift. Biochem. J. 460, 377–385 [DOI] [PubMed] [Google Scholar]
- 25. Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C., and Saiardi A. (2011) Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peers F. G. (1953) The phytase of wheat. Biochem. J. 53, 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brock D. A., and Gomer R. H. (2005) A secreted factor represses cell proliferation in Dictyostelium. Development 132, 4553–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choe J. M., Bakthavatsalam D., Phillips J. E., and Gomer R. H. (2009) Dictyostelium cells bind a secreted autocrine factor that represses cell proliferation. BMC Biochem. 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caterina M. J., Hereld D., and Devreotes P. N. (1995) Occupancy of the Dictyostelium cAMP receptor, cAR1, induces a reduction in affinity which depends upon COOH-terminal serine residues. J. Biol. Chem. 270, 4418–4423 [DOI] [PubMed] [Google Scholar]
- 30. Wurster B., and Butz U. (1980) Reversible binding of the chemoattractant folic acid to cells of Dictyostelium discoideum. Eur. J. Biochem. 109, 613–618 [DOI] [PubMed] [Google Scholar]
- 31. Low J. E., and Jardine I. (1986) A novel cyclosporine binding assay. J. Pharmacol. Exp. Ther. 238, 39–45 [PubMed] [Google Scholar]
- 32. Müller A., Rice P. J., Ensley H. E., Coogan P. S., Kalbfleish J. H., Kelley J. L., Love E. J., Portera C. A., Ha T., Browder I. W., and Williams D. L. (1996) Receptor binding and internalization of a water-soluble (1[arrrow]3)-β-d-glucan biologic response modifier in two monocyte/macrophage cell lines. J. Immunol. 156, 3418–3425 [PubMed] [Google Scholar]
- 33. Yamanaka Y., Fowlkes J. L., Wilson E. M., Rosenfeld R. G., and Oh Y. (1999) Characterization of insulin-like growth factor binding protein-3 (IGFBP-3) binding to human breast cancer cells: kinetics of IGFBP-3 binding and identification of receptor binding domain on the IGFBP-3 molecule. Endocrinology 140, 1319–1328 [DOI] [PubMed] [Google Scholar]
- 34. Britigan B. E., Serody J. S., and Cohen M. S. (1994) The role of lactoferrin as an anti-inflammatory molecule. Adv. Exp. Med. Biol. 357, 143–156 [DOI] [PubMed] [Google Scholar]
- 35. Soda R., and Tavassoli M. (1984) Liver endothelium and not hepatocytes or Kupffer cells have transferrin receptors. Blood 63, 270–276 [PubMed] [Google Scholar]
- 36. Cheng Y., and Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 37. Morrissey J. H. (2013) One inositol ring to rule thrombosis. Blood 122, 1331–1332 [DOI] [PubMed] [Google Scholar]
- 38. Pisani F., Livermore T., Rose G., Chubb J. R., Gaspari M., and Saiardi A. (2014) Analysis of Dictyostelium discoideum inositol pyrophosphate metabolism by gel electrophoresis. PLoS One 9, e85533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luo H. R., Huang Y. E., Chen J. C., Saiardi A., Iijima M., Ye K., Huang Y., Nagata E., Devreotes P., and Snyder S. H. (2003) Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell 114, 559–572 [DOI] [PubMed] [Google Scholar]
- 40. Bakthavatsalam D., Brock D. A., Nikravan N. N., Houston K. D., Hatton R. D., and Gomer R. H. (2008) The secreted Dictyostelium protein CfaD is a chalone. J. Cell Sci. 121, 2473–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bakthavatsalam D., Choe J. M., Hanson N. E., and Gomer R. H. (2009) A Dictyostelium chalone uses G proteins to regulate proliferation. BMC Biol. 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Phillips J. E., and Gomer R. H. (2014) The p21-activated kinase (PAK) family member PakD is required for chemorepulsion and proliferation inhibition by autocrine signals in Dictyostelium discoideum. PLoS One 9, e96633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choi S. H., Collins J. N., Smith S. A., Davis-Harrison R. L., Rienstra C. M., and Morrissey J. H. (2010) Phosphoramidate end labeling of inorganic polyphosphates: facile manipulation of polyphosphate for investigating and modulating its biological activities. Biochemistry 49, 9935–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bullough W. S. (1965) Mitotic and functional homeostasis: a speculative review. Cancer Res. 25, 1683–1727 [PubMed] [Google Scholar]
- 45. Gomer R. H. (2001) Not being the wrong size. Nat. Rev. Mol. Cell Biol. 2, 48–54 [DOI] [PubMed] [Google Scholar]
- 46. Boldingh W. H., and Laurence E. B. (1968) Extraction, purification and preliminary characterisation of the epidermal chalone: a tissue specific mitotic inhibitor obtained from vertebrate skin. Eur. J. Biochem. 5, 191–198 [DOI] [PubMed] [Google Scholar]
- 47. Bullough W. S., Hewett C. L., and Laurence E. B. (1964) The epidermal chalone: a preliminary attempt at isolation. Exp. Cell Res. 36, 192–200 [DOI] [PubMed] [Google Scholar]
- 48. Metcalf D. (1964) Restricted growth capacity of multiple spleen grafts. Transplantation 2, 387–392 [DOI] [PubMed] [Google Scholar]
- 49. Michalopoulos G. K., and DeFrances M. (2005) Liver regeneration. Adv. Biochem. Eng. Biotechnol. 93, 101–134 [DOI] [PubMed] [Google Scholar]
- 50. Saetren H. (1956) A principle of auto-regulation of growth; production of organ specific mitose-inhibitors in kidney and liver. Exp. Cell Res. 11, 229–232 [DOI] [PubMed] [Google Scholar]
- 51. Sassier P., and Bergeron M. (1977) Specific inhibition of cell proliferation in the mouse intestine by an aqueous extract of rabbit small intestine. Cell Tissue Kinetics 10, 223–231 [DOI] [PubMed] [Google Scholar]
- 52. Gray M. J., Wholey W. Y., Wagner N. O., Cremers C. M., Mueller-Schickert A., Hock N. T., Krieger A. G., Smith E. M., Bender R. A., Bardwell J. C., and Jakob U. (2014) Polyphosphate is a primordial chaperone. Mol. Cell 53, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]