Abstract
The ubiquitin (Ub) ligase anaphase promoting complex/cyclosome (APC/C) and the tumour suppressor retinoblastoma protein (pRB) play key roles in cell cycle regulation. APC/C is a critical regulator of mitosis and G1-phase of the cell cycle whereas pRB keeps a check on proliferation by inhibiting transition to the S-phase. APC/C and pRB interact with each other via the co-activator of APC/C, FZR1, providing an alternative pathway of regulation of G1 to S transition by pRB using a post-translational mechanism. Both pRB and FZR1 have complex roles and are implicated not only in regulation of cell proliferation but also in differentiation, quiescence, apoptosis, maintenance of chromosomal integrity and metabolism. Both are also targeted by transforming viruses. We discuss recent advances in our understanding of the involvement of APC/C and pRB in cell cycle based decisions and how these insights will be useful for development of anti-cancer and anti-viral drugs.
Keywords: anaphase promoting complex/cyclosome, cell cycle, FZR1, human papilloma virus, LxCxE, retinoblastoma
INTRODUCTION
Somatic cell cycle has alternating DNA synthetic (S) and mitotic (M) phases, separated by gap phases (G1 and G2). The correct sequence of events, a robust feature in cell cycle, is maintained by timely degradation of cell-cycle regulators by ubiquitin proteasome pathway (UPP). The UPP consists of the ubiquitin (Ub)-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub ligases (E3) that covalently link Ub on to target proteins either singly or in chains that are formed using various internal lysines of Ub. Although chains linked through Lys-48 and Lys-11 lead to destruction of substrate proteins by 26S proteasome, monoubiquitination and chains other than Lys-48 and Lys-11 linkages have non-proteolytic functions. Two related multi-subunit E3s, the anaphase promoting complex/cyclosome (APC/C) and the Skp1/Cul1/F-box (SCF) complex are crucial for timely proteolysis of cell cycle proteins (Figure 1A). Although SCF performs throughout the cell cycle, APC/C remains active from M to late G1 [1]. The APC/C has emerged as a critical regulator of mitosis not only due to its role in spindle assembly checkpoint (SAC) but also for being crucial for degradation of mitotic cyclins and securin that paves the way for completion of mitosis. On the other hand, it is equally important for post-mitotic decisions of the cell about proliferation, differentiation and quiescence. Other than cell cycle, the APC/C also regulates neuronal development and metabolism [2–4]. With 15 subunits in vertebrates, APC/C is one of the largest and the most complex E3s known to date and has been a subject of intense investigation due to its wide-ranging roles. Combined use of cryo-electron microscopy, mass-spectroscopy and docking of crystal structures and homology models allowed reconstruction of the pseudo atomic model and later with more advanced cryo-EM technology, of the atomic scale structure of its co-activator in complex with the E2s, UbcH10 and Ube2S and with one of its inhibitory protein, Emi1 [5–7]. These studies have given an insight into the mechanism of initiation of ubiquitination, inhibition of the complex and regulation by co-activators. However, we are still far from a complete mechanistic understanding of its various functions, its complete interactome and substrates and its regulation by phosphorylation. This review will focus on G1-S regulation by APC/C and the readers are directed to other excellent recent reviews on its other functions [1,8–10]. We review the recent advances in our understanding of how APC/C regulates G0/G1 stage and controls S-phase entry and discuss the implications of its interaction with the tumour suppressor protein retinoblastoma (pRB) for cell cycle regulation and development of anti-viral and anti-cancer drugs.
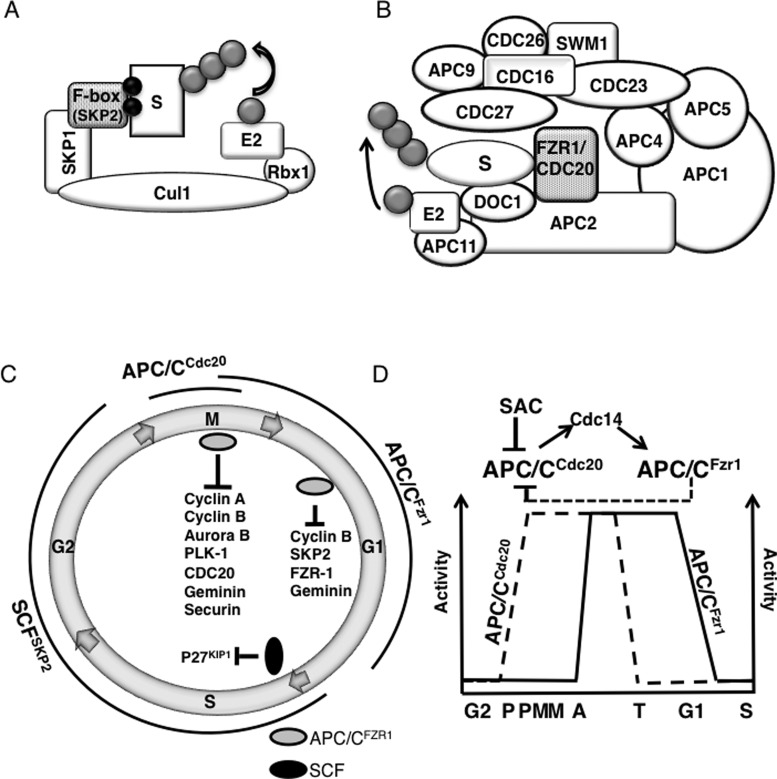
Figure 1. Regulation of cell cycle by the SCF and APC/C E3 ligases.
Schematic diagrams of the modular structure of the SCF (A) and APC/C (B) showing the relative positions of various subunits. The stages of the cell cycle where activities of these complexes regulates key events are shown in (C) along with some of the key substrates. The activities of two APC/C assemblies (APC/CCDC20 and APC/CFZR1) are regulated in opposite manner by phosphorylation resulting their manifestation at different stages (D). APC/CCDC20 is activated upon phosphorylation by the mitotic cyclin/CDK complex but its activity is kept in check by the SAC. Once SAC is satisfied, APC/CCDC20 targets mitotic cyclins resulting in decrease in kinase activity thus inactivating APC/CCDC20. At the same time, activation of the phosphatase Cdc14 dephosphorylate FZR1 resulting in activation of APC/CFZR1.
G1-S REGULATION BY APC/C
CDC20 and FZR1 are two related co-activators that recruit substrates to APC/C in two distinct assemblies, APC/CCDC20 and APC/CFZR1 [11]. Both CDC20 and FZR1 are structurally related and consist primarily of a seven-bladed WD40-repeat-propeller that facilitates protein–protein interactions. The CDC20 and FZR1 bound forms of APC/C demonstrate both different and overlapping substrate specificities [12,13]. The most well characterized degrons in APC/C substrates are the destruction box (D-box) [14] and the KEN-box [15], but other targeting motifs have also been identified [16–18]. CDC20 and FZR1 recognize specific target proteins depending on the recognition sequences present, such as the destruction box (D-box) for CDC20, and the D-box, KEN box and CRY box for FZR1 [19,20].
APC/CCDC20 is activated upon phosphorylation by mitotic cyclin-CDK but its activity is kept in check till the SAC is satisfied with CDC20 as a component of the mitotic checkpoint complex [21]. Once SAC is satisfied APC/CCDC20 activity is unleashed resulting in ubiquitination and destruction of many substrates. Critical among these are the mitotic cyclins and securin (Figure 1B). This leads to low kinase activity and inactivation of APC/CCDC20 during metaphase. Low kinase activity also results in dephosphorylation of FZR1 and activation of APC/CFZR1 that has a much broader substrate range. It also targets CDC20 and any remaining mitotic cyclin and some other mitotic substrates for degradation [22]. APC/CFZR1 continues working till late G1 and negatively regulates DNA replication and cell proliferation through degradation of multiple proteins that helps in maintenance of G1 state (Table 1) [10,23,24]. Some of these critical targets include pro-proliferative proteins like Polo-like kinase 1 (PLK1), Aurora kinase A and CDC25A, the activator of CDK1 and proteins required for DNA replication (e.g. geminin, Cdc6 and TK1). SKP2, a subunit of the SCF E3 is an important target of FZR1 that results in stabilization of CKI protein p27KIP1, further stabilizing the G1 state. Another critical target of APC/CFZR1 in maintaining G1 is the transcription factor E2F1 that promotes the S-phase genes once it is released by phosphorylation of pRB [25]. E2F1 is targeted for degradation by APC/CFZR1 until the G1/S transition. Hence APC/CFZR1 on one hand maintains low levels of E2F1 to inhibit S-phase; on the other it engages with pRB in mediating APC/CFZR1 dependent degradation of SKP2 and allowing the build-up of CKI proteins to inhibit S-phase, thus utilizing a two pronged strategy involving transcriptional and post-translational mechanisms to prevent cells from entering the S-phase.
Table 1. Substrates of APC/CFZR1.
| G1 | Mitosis | Reference |
|---|---|---|
| FZR1 | [27] | |
| SKP2 | [132] | |
| Cyclin B1 | Cyclin B1 | [133,134] |
| FoxM1 | [135] | |
| CDCA3 | [136] | |
| Anillin | [137] | |
| Nek2 | [15] | |
| B99 | [15] | |
| E2F1 | [25] | |
| TOME1 | [138] | |
| Geminin | Geminin | [139] |
| Aurora B | [140] | |
| CDC6 | [141] | |
| CKAP2 | [142] | |
| CDC20 | [143] | |
| Cyclin A | [144] | |
| ETS2 | [145] | |
| Claspin | [46] | |
| Id2 | [146] | |
| PLK1 | [147] | |
| Rcs1 | [148] | |
| Securin | [149] | |
| Sgo1 | [150] | |
| SnoN | [151] | |
| Tpx2 | [152] | |
| Xkid | [153] | |
| CLB2 | [154] | |
| CDC5 | [155] | |
| HSL1 | [155] | |
| CDC25A | [138] | |
| NDD1 | [156] | |
| CtIP | [157] |
Regulation of FZR1
For cells to enter the S-phase, the activity of APC/CFZR1 has to be brought down in a regulated manner. There are several mechanisms that regulate FZR1 levels in cells that allow APC/C to be shut-off in late G1. Although FZR1 RNA levels remain constant throughout the cell cycle, its protein levels fluctuate. FZR1 levels are high in mitosis, but lowered in late G1- and S-phases [26]. FZR1 mediates its own degradation in late G1. This process of self-destruction requires the two D-boxes of FZR1 [27]. Another mechanism of APC/CFZR1 inactivation is the ubiquitination of APC/C-specific E2 UBCH10 by APC/CFZR1 itself, thereby providing a negative feedback mechanism [28,29]. Phosphorylation also regulates FZR1. The binding of FZR1 to APC/C depends on FZR1 phosphorylation status. CDC28 mediated phosphorylation of FZR1 excludes it from the nucleus and dissociates it from the core APC/C resulting in FZR1 inactivation [30,31]. E2F mediated accumulation of cyclin A at the G1/S transition also results in phosphorylation of FZR1 [32]. Phosphorylated FZR1 is targeted by SCF E3 ligase, further limiting the activity of APC/CFZR1 [33]. In Caenorhabditis elegans, cyclin D1/CDK4 phosphorylates N terminus of FZR1 and linker domain of LIN-35, the pRB homologue thereby counteracting the cell cycle inhibitory functions of both the proteins in G1 [34]. In Drosophila endocycle in which cells undergo repeated rounds of DNA replication with no intervening mitosis, cyclin E/CDK2 mediated phosphorylation of FZR1 drive the periodicity of APC/CFZR1 activity [35].
In addition, inhibitory protein Emi1 (also known as FBXO5), not only inhibits APC/CCDC20 activity in S- and G2-phases but also inhibits APC/CFZR1 in interphase by binding like a pseudo-substrate to the APC/C and also by antagonizing the two E2s that function with APC/CFZR1 [7,36]. Similarly, the meiotic function of APC/CFZR1 is blocked by Emi2, a homologue of Emi1. The levels of Emi1 start increasing at the start of G1 and are brought down by SCFβTRCP1 in early mitosis to allow activation of APC/CCDC20 [37]. Interestingly, Emi1 expression is under the control of E2F which promotes G1-S transition when released by phosphorylation of pRB by cyclin/CDK4, 6.
Consequences of loss of FZR1
A number of studies have investigated the consequences of aberrant FZR1 expression and its loss on cell cycle and tumorigenicity (Table 2). In budding yeast, FZR1 is required for destruction of mitotic cyclin during mitotic exit [38] but in Drosophila and frog embryos it is not required for mitotic exit [39–41]. Major outcomes of FZR1 knockdown in different mammalian cells due to stabilization of several FZR1 substrates are shortening of G1-phase, a premature and prolonged S-phase, delayed entry into mitosis and aberrant chromosomal separation and cytokinesis [42]. Conditional knockout of FZR1 is lethal in mouse and embryos die at around E10 due to inability of placental trophoblast cells to endoreduplicate. This lethality is prevented when FZR1 is re-expressed in placenta [43]. Recent findings that FZR1 is required for regulation of G2/M transition during differentiation of placental trophoblast cells in mice [44], provide an explanation for the previous findings of Garcia-Higuera et al. [43]. Cells derived from FZR1 knockout mice develop both numeric and structural chromosomal defects indicating that FZR1 is needed for genomic stability [43]. FZR1 heterozygous mice develop tumours of the mammary gland, lung, kidney, testis, sebaceous glands and B-cell lymphomas [43]. More recent studies with oocyte specific deletion of FZR1 show that it is not required for completion of meiosis and viable pups could be obtained when FZR1 negative females were mated with normal males. However, absence of both female and male FZR1 led to major genomic instability with embryos arrested at first mitotic division [45]. All these studies suggest that FZR1 is essential for maintenance of genomic integrity and its deficiency leads to tumorigenesis. Therefore, FZR1 has been proposed to be a putative haploinsufficient tumour suppressor [24,42,43].
Table 2. Consequences of FZR1 depletion in cells and model animals.
| Cells/model | Effect of FZR1 depletion | Reference |
|---|---|---|
| Yeast | ||
| Fission yeast FZR1Δ gene disruption mutants | Sterility, defective in cell cycle arrest in the G1-phase upon starvation | [158] |
| Fission yeast FZR1Δ gene disruption mutants | Meiotic mutant with aberrant asci having one or two mature spores | [159] |
| Budding yeast gene replacement FZR1 mutants | Premature exit from meiotic prophase I | [160,161] |
| Budding yeast FZR1Δ mutants | Inhibition of mitotic cyclin degradation and inappropriately induced DNA replication | [162] |
| Mammalian cell lines | ||
| FZR1 shRNA treated rat cortical neurons and SH-SY5Y human neuroblastoma cells | Increased proportion of cells in S-phase, apoptosis | [3] |
| FZR1 siRNA treated Saos2 | Loss of cell cycle arrest, increased generation time | [64] |
| Lentiviral RNAi mediated KO of FZR1 in HeLa | Early onset of DNA replication | [42] |
| Human fibroblast cells | Premature senescence | [46] |
| MEFs from FZR1-KO mice | Poor proliferation, premature senescence | [163] |
| FZR1 shRNA treated HeLa | Increase in half-life of SKP2 | [164] |
| FZR1 shRNA treated HCT 116 | Sub-G1 DNA content | [164] |
| Arabidopsis | ||
| Arabidopsis xcm9 mutant with loss of function allele of FZR1 | Premature termination of floral shoots, disruption of cell cycle progression, defects in cyclin B1 expression, defects in endoreduplication | [165,166] |
| Drosophila | ||
| Loss of function Drosophila mutants of FZR1 | Reentry into the cell cycle following embryonic cycle 16 thereby bypassing the normal G1 arrest | [39] |
| RAP/FZR1 loss-of-function mutants of Drosophila | Changes in size and morphology of synapses, locomotion defects | [167] |
| C. elegans | ||
| RNAi mediated inactivation of FZR-1 in C. elegans | Sterility, aberrant germ cell proliferation | [168] |
| Mouse | ||
| Conditional knockout mouse | Embryonic lethality at E9.5–E10.5 | [43] |
| FZR1−/+ mouse | Increased susceptibility to spontaneous tumours | [43] |
| FZR1 KO mouse embryos | Embryonic lethality, lack of endoreduplication, placentation defects | [45] |
| Male FZR1 germline knockout (KO) mice | Abnormal proliferation of spermatogonia, infertility, failure of early meiotic prophase I in male germ cells | [169] |
| Female FZR1 germline KO mice | Premature onset of ovarian failure, subfertile females, defects in early meiotic prophase I | [169] |
FZR1, SKP2 and p27KIP1 in human cancers
Normal human fibroblasts undergo premature senescence after acute loss of FZR1, hinting at a built-in fail-safe mechanism against cancer development and the possible underlying molecular mechanism for the less frequently observed FZR1 loss in tumour cells. Thus, it is possible that loss of FZR1 occurs late in tumour development [46]. Nevertheless, SKP2, an FZR1 target, is up-regulated in many cancers [47–49]. SKP2 recruits the cyclin-dependent kinase inhibitory protein (CKI) p27KIP1 to the SCF complex for degradation. A variety of carcinoma show a low level of p27KIP1 [48,50,51]. Decreased p27KIP1 levels are correlated with high grade of malignancy, low survival rate, greater tumour size and histological differentiation suggesting possible role of p27KIP1 as a promising prognostic marker for cancer. A number of solid tumours including lung, breast, ovarian, prostate, colon and squamous cell carcinoma manifest conditions of high SKP2 accompanied by low p27KIP1considered to be associated with highly aggressive tumours [48,49,51–55]. Human colorectal tumour arrays show higher percentage of SKP2 positive samples and lower percentage of FZR1 and p27KIP1 positive samples and high FZR1 expression was associated with tumours showing lower grade histology [56]. These data demonstrate a pathological correlation between FZR1, SKP2 and p27KIP1 and suggested that FZR1 levels could be used as a prognostic marker in colorectal samples. Similar investigations in other types of cancers would indicate whether it is applicable to other cancers as well.
Reduced expression of FZR1 is observed in several other tumours other than colon, including brain, liver, ovary, breast and prostate [57] but it is also overexpressed in certain malignant tumours concomitantly with Emi1 [58]. This may represent a compensatory mechanism as overexpression of Emi1 can overcome the cell cycle block due to FZR1 overexpression [59]. It is noteworthy that in these studies levels of APC/C substrate SKP2, securin, aurora A, PLK1, FZR1 and Emi1 correlate positively with malignancy.
G1-S REGULATION BY RETINOBLASTOMA
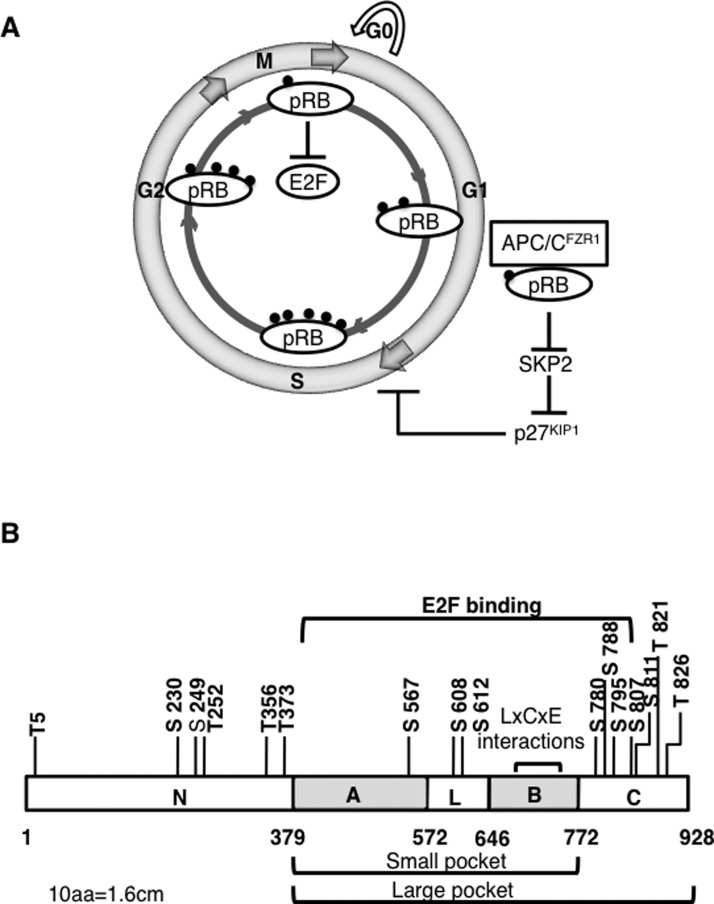
The tumour suppressor pRB plays a crucial role in not only regulating G1 to S transition, but also in quiescence, differentiation and senescence [60]. Its role in inhibiting G1-S transition by transcriptional regulation is the best understood among its many different functions. There are two other family members, p107 and p130, that also act as suppressors of cell proliferation by modulating transcription of genes required for cell cycle progression [61]. The canonical model for G1-S regulation by pRB is the sequestration of E2F family of transcription factors by hypophosphorylated pRB and release of E2F upon hyperphosphorylation of pRB that promotes the transcription of S-phase genes (Figure 2A) [62]. Although simple and elegant, this model does not explain the retention of tumour suppressor activity of pRB mutants that cannot bind E2F [63].
Figure 2. Different mechanisms of G1-S regulation by pRB.
Transcriptional and post-translational regulation of G1-S transition by pRB-E2F and pRB-APC/CFZR1 complexes. The phosphorylation status of pRB is indicated by black solid balls, with numbers of balls reflecting hypo- or hyper phosphorylation of pRB (not actual number of phosphorylation sites) (A). Domain organization of pRB showing phosphorylation sites and regions of interactions with various cellular proteins. Numbers show amino acid positions (B).
The search for an alternative mechanism of G1 regulation by pRB led to the discovery that APC/CFZR1 and SKP2 simultaneously bind to pRB resulting in SKP2 degradation and accumulation of p27KIP1 [64]. Wild-type pRB expressing cells can arrest in G1 before E2F mediated transcriptional repression, and a pRB mutant R661W, defective for binding to E2F in vitro, retains the ability to interfere with SCF mediated degradation of p27KIP1 [65]. In the absence of growth-promoting signals, pRB interacts with the N terminus of SKP2 and inhibits SKP2 mediated p27KIP1 degradation [65]. The findings of Binne et al. [64], provided a satisfactory explanation of these earlier studies and demonstrated the existence of a post-translational mechanism of pRB with APC/CFZR1 that may precede and is distinct from the pRB–E2F mediated transcriptional control for G1 arrest (Figure 2A). Apart from SKP2, PLK1 also showed significant accumulation when pRB levels were down-regulated by shRNA whereas other known cell cycle proteins like CDC20, aurora A or geminin were not affected [64]. Although pRB–APCFZR1 interaction was not detected in nontransformed, primary human fibroblasts growing asynchronously, it could be detected when these cells were contact inhibited suggesting additional factors controlling the interaction of pRB and FZR1. pRB depletion from contact inhibited U2OS osteosarcoma cell line caused an accumulation of SKP2 suggesting crucial role of this interaction for cellular differentiation compared with proliferation situations [64,66]. Accumulation of other FZR1 substrates like PLK1, but not those of CDC20 substrates, cyclin B1 and securin, in these cells also suggests that there may be other substrates whose levels may be regulated by pRB–FZR1 interaction. Identification of these substrates and what pathways they function in will allow a better understanding of mechanistic aspects of pRB mutations in various cancers.
Genetic investigations reveal that unlike p107 or p130, only pRB mutations are commonly found in human cancers. Studies on mice lacking different combinations of pocket pRB/p107/p130 genes suggest that pRB has significantly stronger tumour suppressor properties than p107 or p130 [61,67]. Retinoblastoma contains three functional domains: The N-terminal domain (RB-N), followed by the AB and the C pockets (Figure 2B). The pRB-AB pocket provides a conserved structural motif called the double cyclin fold found in cyclins which function as modules for protein recognition [68]. There is structural similarity between pRB N-terminal domain and the AB cyclin-like folds suggesting domain duplication [69]. Domains A and B interact with each other along an extended interdomain interface to form the central ‘pocket’ [70,71] which is essential for the tumour-suppressor activity of pRB [72]. Most mutations in pRB are associated with pocket domains. The AB pocket domain is not only crucial for interaction of pRB with E2F but also a target of several transforming viruses as detailed below.
Inhibition of pRB function by viral oncoproteins
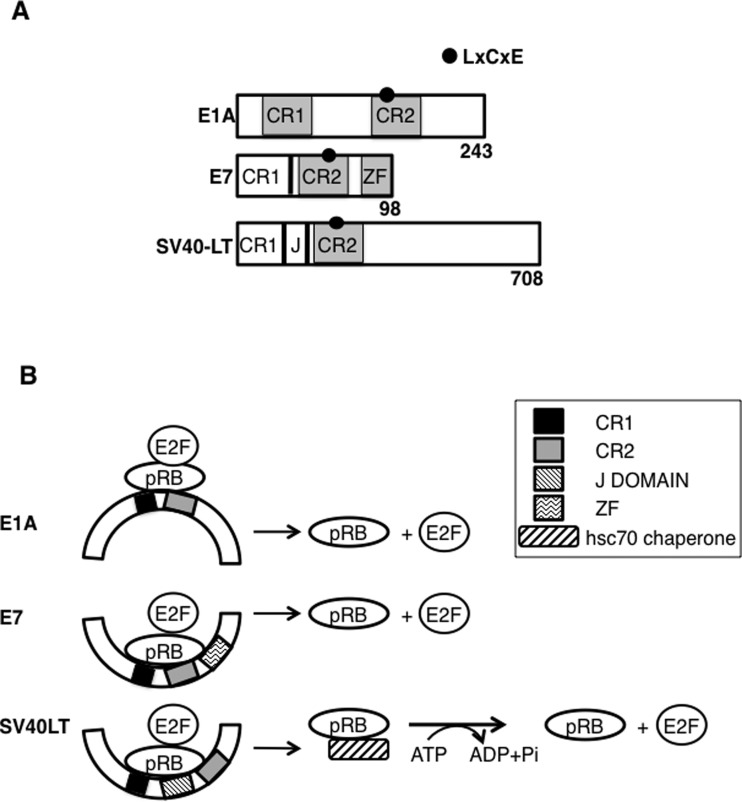
Transforming viruses like human papilloma virus (HPV), adenovirus and SV40 attack pRB via proteins containing an LxCxE motif (Figure 3A). HPV E7, SV40 large T-antigen (LT) and Adenovirus 5 E1A proteins bind to pocket proteins and displace E2F transcription factors. Like pRB, p130 and p107 are also predicted to contain LxCxE-binding clefts, which bind viral proteins. High risk HPV16 E7 and 48E7 target other pocket proteins like p130 in addition to pRB to overcome cell cycle block. LxCxE motif is a ligand short linear motif (SLiM) that often acts as a simple interface that recruits proteins to multi-protein complexes. This SLiM of viral and cellular proteins binds within a cleft located in the B-pocket of pRB (Figure 2B). The hydrophobic groove in pRB pocket domain B which forms the LxCxE motif binding site consists of four conserved amino acids Tyr-709, Lys-713, Tyr-756 and Asn-757 which are involved in contacting the backbone of the LxCxE peptide. Mutation of these contact amino acids inhibited binding of pRB to LxCxE motif carrying proteins [71,73–75]. Crystal structure of pRB reveals that the region of pRB where the LxCxE peptide binds consists of a patch of positively charged amino acids [71]. A series of E7- and HDAC-1-derived peptides with single or double amino acid substitutions in LxCxE motif was used to show that any positively charged residue in and around the LxCxE peptides had a significant effect on weakening the binding [76].
Figure 3. Domain organization of viral proteins.
LxCxE motif is indicated with solid black ball. Numbers indicate amino acids (A). Mechanisms of displacement of E2F from pRB by LxCxE containing viral proteins (B).
Although several oncogenic viral proteins display a high affinity binding to pRB via the LxCxE motif, the molecular mechanism of displacement of E2F from pRB is different in each case (Figure 3B). Small DNA viruses such as adenovirus, HPV and polyomavirus carry conserved regions (CRs). The LxCxE motif is found in the CR2 region in these DNA viruses (Figure 3A) [77]. In spite of the fact that these viral oncoproteins share the LxCxE motif, its location is in considerably dissimilar molecular context explaining the differential mechanisms of displacement of E2F by viral LxCxE proteins (Figure 3B). Although CRI of E1A is sufficient to compete with E2F for binding to pRB, the equivalent region of E7 is neither necessary nor sufficient [78–80]. HPV E7 uses high affinity LxCxE mediated binding followed by engagement of E7-C terminal region with pRB C domain which is required to displace pRB–E2F [81] and also destabilizes pRB through proteasomal degradation via reprogramming of the cullin 2 ubiquitin ligase complex [82–84]. CR1 region of Adenoviral-E1A and the transactivation domain (TA) of E2F compete for the same binding site on pRB and thus E1A uses a competitive binding to pRB (Figure 3B) [78]. SV40 LT antigen possess N-terminal J domain that recruits hsc70, a DnaK homologue and uses a chaperone like mechanism to displace pRB–E2F complex. The pRB–E2F complex is then transferred from SV40LT to the ATPase domain of hsc70 resulting in ATP hydrolysis and conformational changes in hsc70 thereby releasing pRB and E2F as separate molecules (Figure 3B) [85]. These results suggest that pRB associates with viral oncoproteins and E2F through overlapping but distinct domains and viral LxCxE is necessary for its binding to pRB, though not sufficient to displace E2F.
The binding kinetics of LxCxE to pRB is modulated by a casein kinase II and PEST sequence (CK2-PEST) present in all three viral proteins HPVE7, SV40LT and AdE1A. It is believed that the complex between LxCxE and pRB is stabilized by CK2-PEST by increasing electrostatic interactions that can aid in a faster kinetics of association [81]. Although the PEST sequence is supposed to contribute to long-range interactions, the phosphate groups contribute to both long and short-range interactions. The long range interactions have fewer conformational restrictions, the short range interactions will be conformationally restricted and therefore are likely to form later during association reaction [81].
Mutations in the LxCxE binding cleft of pRB do not prevent pRB from binding and inactivating E2Fs [86]. Full-length E7 protein carrying mutations outside the LxCxE motif inhibits RB-E2F binding, but retains cell cycle arrest [87], demonstrating that LxCxE dependent interactions of pRB to be distinct from pRB pool binding to E2Fs. These studies also indicate that E2F independent pRB pathways guard proliferative pathways. Biochemical, biophysical and molecular dynamics studies show that the binding of E7 peptide with pRB–E2F complex alone is not sufficient for the dissociation of E2F [88–90] leading to the idea that pharmacological molecules can be designed that can inhibit the binding of E7 without displacing E2F.
pRB mutations affecting LxCxE based interactions
Many mutations within the pRB LxCxE binding cleft disrupt the interaction of pRB with the viral oncoproteins [73]. An M704V variant of pRB retains its ability to interact with E2F3/DP1, but its ability to interact with LxCxE of SV40LTis greatly compromised [91]. A C706F variant found in small cell lung cancer is unable to interact with LxCxE motif of SV40-LT and adenovirus E1A, whereas retaining its interaction with E2F transcription factors [92,93].
A number of naturally occurring point mutations of pRB found in cancer cells result in disruptions of the integrity of the AB pocket [94,95]. V654L mutation results in reduced penetrance, but substitution of glutamic acid for valine at the same position yield a highly penetrant phenotype [96,97]. Structural studies reveal that this valine residue lie 90–100% buried within the pRB pocket domain. Hence substitution of valine to a charged residue may possibly alter the protein structure [71]. However, effect of this mutation in its binding to E2F and LxCxE has not been determined. Some pRB mutations have extremely low penetrance caused by pRB promoter mutations or splice mutations resulting in truncation of translation of unstable mRNA resulting in reduced pRB expressivity [98]. These weak alleles suppress tumorigenesis in the biallelic state but not in the monoallelic state. These include a deletion involving exons 24 and 25 [99], a splicing mutation C712R at the final base of exon 21 [100], a deletion of Asn-480 and a point mutation R661W [96,101]. Surprisingly, all these pRB mutations reside in the B box of pRB and result in reduced binding to LxCxE proteins and minimal binding to E2F [98,102]. C712R mutation is in proximity to Lys-713, which is a key residue in binding to LxCxE-containing proteins such as HDAC [71,86]. These low penetrant mutants defective in binding to E2Fs and LxCxE, retain partial tumour suppressor activity suggesting that E2F and LxCxE binding are not the only mechanisms through which pRB inhibits cell growth [103,104].
Many cellular proteins that interact with pRB also contain LxCxE motif and many of these proteins are chromatin modifiers [105]. It can be expected that mutations in LxCxE binding region in pRB potentially affect many cellular interactions and defining the molecular basis of any observed phenotype is difficult. Therefore, deletion or point mutation of LxCxE motif of individual pRB interacting proteins in isolation is required to understand the functional importance of this motif in different cellular partners of pRB.
INTERACTION OF LxCxE CONTAINING CELLULAR PROTEINS WITH pRB
Approximately 30 cellular proteins, including RBP1, RBP-2, CtIP, EID-1, MAP3K5 (ASK-1), gankyrin, cyclin D1, D2, D3 and HDAC-1 and -2, that bind to pRB have a conserved LxCxE motif. However, mutations in the LxCxE motif of cellular proteins have varied effects on interaction and function with pRB (Table 3). Mutations in LxCxE motif of cyclin D1, D2 and ASK-1 profoundly diminish their binding to pRB. The LxCxE motif of cyclin D1 is not necessary for its function, but the motif is required by cyclin D2 [106–108]. Although mutation in LxCxE motif of RBP-1 replacing ‘E’ to ‘K’ retains its binding to pRB [109], mutation at similar position replacing ‘E’ to ‘A’ in oncoprotein gankyrin abrogates its binding to pRB. Gankyrin binding to pRB accelerates phosphorylation and degradation of pRB, releasing E2F transcription factor to activate DNA synthesis genes [110]. Surprisingly, mutation within the LxCxE motif of RBP2 resulted in loss of ability to precipitate p107, whereas retaining its binding to pRB [109]. Replacement of LxCxE of BRCA1 to RxRxH retained its binding to pRB. However, RxRxH mutation altered the tumour suppressor activity of BRCA1 [111]. These studies suggest that the mere presence of the motif is not predictive of its importance for interaction and function of pRB.
Table 3. Effects of mutations in the LxCxE motif of cellular proteins on their function and interaction with pRB.
| pRB binding protein | LxCxE mutation | Effect on binding to pRB | Effect on function | References |
|---|---|---|---|---|
| RBP-1 | E to K | Retains binding to pRB | ND | [109] |
| HBP-1 | C to G in both LxCxE and LxCxE | No binding to pRB | Loss of transcriptional repression by HBPl | [170] |
| CtIP | Deletion | No binding to pRB | ND | [171] |
| HDAC-1 | Deletion | No binding to pRB | Inhibition of pRB repressive function | [172,173] |
| BRG-1 | Mutation in pRB LxCxE cleft | Retains binding to pRB | ND | [86] |
| EID-1 | Deletion | No binding to pRB | No effect | [174] |
| Gankyrin | E to A | No binding to pRB | Inhibition of phosphorylation and degradation of pRB | [110] |
| RFC-1 | C to G, E to K | No binding to pRB | Loss of RFC function in promoting cell survival after DNA damage | [175] |
| BRCA-1 | RXRXH | Retains binding to pRB | Altered tumour suppressor activity of BRCA1 | [111,172] |
| ASK-1 | LKCFE to VRCFD | No binding to pRB | Altered ASK-1 activity in induction of apoptosis | [106] |
| Cyclin D1, D2 | C to G, E to K | Profoundly diminished binding to pRB | Partial abrogation of pRB-induced growth arrest and senescence | [107,176] |
| RBBP-9 | L to Q | No binding to pRB | ND | [177] |
| NuMA | C to G, E to K | No binding to pRB | Abnormal organization of spindle microtubules | [178] |
Unlike HPVE7, SV40 LT and AdE1A, that all have the CK2-PEST sequences, only some cellular LxCxE containing proteins have an acidic region or phosphorylation site whereas others do not. No systematic study has been carried out to correlate the presence or absence of such regions on modulation of binding of LxCxE containing cellular proteins with pRB. Viral oncoproteins carrying LxCxE motif can displace cellular proteins like cyclin D, HDAC-1 and BRG1 from pRB [112]. Cyclin D and BRG1 do not have an acidic region whereas HDAC-1 has an acidic region. Thus, presence or absence of acidic region does not seem to determine whether viral proteins can displace cellular proteins from pRB. Rather, binding affinities and phosphorylation status may be more important.
pRB is a conformationally plastic protein in response to phosphorylation [113]. Among the 16 phosphorylation sites identified, no phosphorylation site is found in the B-pocket suggesting that modifications in this structure are not tolerated [114]. Phosphorylation at Thr-821 and Thr-826 inhibits binding of pRB B-box to LxCxE motif containing proteins without affecting pRB binding to E2Fs [113]. The AB domain is reported to be metastable whose native state can be destabilized even by mild perturbations [115]. RB-AB unfolding presents a three-state transition involving cooperative secondary and tertiary structure changes and a partially folded intermediate that can oligomerize. This property of pRB-AB may allow the formation of multi protein complexes, constituting a robust mechanism to retain its cell cycle regulatory role [115]. Equilibrium unfolding studies suggest that the equilibrium between native and non-native states of the AB domain is strongly influenced by LxCxE and other ligands, with degree of stabilization correlating with ligand binding free energy. Molecular dynamics studies suggest that the stability of pRB-AB in the apo- and in E7 bound forms is similar but it increases in the E2F bound form. The binding of E7 peptide through its LxCxE motif with the B box of pRB induces the conformational changes around A–B interface where E2F binds to pRB [115]. These studies suggest that pRB native structure is stabilized in vivo by interactions with its numerous ligands and the native state may be very sensitive to destabilization by mutations. There are many mutations reported for pRB in various databases of cancers but the mechanism by which they affect pRB function is not understood in each case. Although some of the missense mutations may have no effect, others may affect the stability and conformational dynamics of pRB and its ability to interact with LxCxE containing proteins that can compromise its function.
pRB–APC/CFZR1 interaction
Although there is lack of structural details of interactions between APC/C and pRB, studies on RB mutagenesis suggest that FZR1 binding involves the pRB LxCxE-binding cleft in the AB pocket [64]. Unlike E2Fs and viral oncoproteins, the possibility that the same RB molecule can regulate E2F and APC–SKP2–p27KIP1 activities simultaneously does not seem plausible. It is also unclear whether pRB–SKP2–p27KIP1 pathway is equivalent in importance with E2F transcriptional repression in cell cycle regulation. Interestingly, neither p107 nor p130 display any affinity for APC/C subunits [64]. Further, pRB associates exclusively with FZR1 and not CDC20 [64]. The C-terminus WD40 domain of FZR1 contains a motif similar to the LxCxE motif. Loss of the C-terminal 100 amino acids of FZR1 impairs pRB binding [64]. The presence of LxCxE motif within this deleted region hints at a possible role of this motif in binding of FZR1 to pRB but the importance of the motif for interaction with pRB is not known. HPV E7 protein bound to pRB establish a dual-contact mode with 90% of the binding energy determined by the E7 LxCxE motif, with an additional binding determinant located in the C-terminal domain of E7 [90]. Whether FZR1 has a single or multiple contact sites for pRB is not known. The modular structure of pRB and the fact that both pRB and FZR1 interact with multiple partners, binding between FZR1 and pRB is likely to be weaker compared with HPV E7 peptide to allow the periodic changes in the downstream targets like SKP2 that are required for normal cell cycle.
In vitro studies show that FZR1 can regulate E2F activity in G1 that involves its interaction with hypophosphorylated pRB but independent from its interaction with APC/C [46]. Another example of APC/C independent function of FZR1 is promotion of Smurf1 E3 activity in vitro by a C-box deletion mutant of FZR1 that cannot bind to the APC/C core implicating it in osteoblast differentiation [116], although pRB interaction was not checked in this study. If confirmed in vivo, these results show that there may be APC/C independent functions of FZR1–pRB other than cell cycle regulation and should be taken into consideration while designing any inhibitor or drug against these target proteins.
APC/C and pRB as drug targets
It is becoming clear in recent years that the role of proteins defined as tumour suppressors is not as clear cut as previously thought and the cellular and genetic context defines the functional outcome of mutations, inactivation and dosage effects. Both APC/C and pRB regulate many, and often contradictory functions in a cell and decide the fate of many proteins. Both are also targets of various transforming viruses. During cancer progression, changes in developmental phenotype are thought to involve pRB [117,118]. Due to their many interacting partners and the fact that APC/C is a multi-subunit assembly where the functions of each subunit are still not defined, both protein interface drug discovery and high-throughput screening approaches may need to be explored. The latter has been used to find promising lead compounds inhibiting APC/C and SKP2/CKS1 interaction [119,120].
Various components of the UPP are considered attractive drug targets and the success of Bortezomib, a proteasome inhibitor, in treatment of multiple myeloma has pushed UPP at the forefront as drug target. Of the three classes of enzymes in the pathway, E3s are considered to be better targets compared with E1 and E2 because they determine the substrate specificity thus potentially more targeted therapies can be developed. Conversely, multiplicity of substrates can be a challenge for targeted therapies. The other concerns are the therapeutic window and the selectivity between normal and cancer cells. Despite of these concerns, the attractive feature of APC/C as a potent drug target is that outcome of APC/C activity can be controlled by modulating either of the two adaptor proteins–FZR1 or CDC20. Anti-mitotic reagents like Taxol and Nocodazole, used as anti-cancer therapies, activate the SAC presumably through inhibiting APCCDC20 [121]. Therefore, development of inhibitors that will specifically target CDC20 could reduce the off-target and side effects. CDC20 is a preferred target compared with FZR1 as its function and substrates are mostly restricted to mitosis whereas FZR1 has a much broader substrate range and functions. However, due to its role in maintenance of G0/G1 state and inhibition of G1-S transition along with pRB, agonists can perhaps be designed that can promote these functions of FZR1. For example small molecules that may stabilize FZR1–pRB interaction or inhibit the release of FZR1 from pRB may prove useful to prevent proliferation. Interdependence between FZR1, SCF, CDC20 and Emi1 can also be exploited by combining antagonists and agonists [122]. Centrosome amplification, a common feature of many cancer cells has been proposed to drive genomic instability. Degradation of the motor protein kinesin-5 (Eg5) by APC/CFZR1 is required for the clustering of these extra centrosomes. Accumulation of Eg5 in cells expressing FZR1 carrying mutations in certain D- or KEN-box motifs causes cancer cell death [123]. These studies hint future prospects of developing drugs targeting FZR1 at specific motifs.
The strategies for drug development with pRB as a target may include pRB-mediated promotion of cell cycle inhibition, senescence, apoptosis or differentiation of cancer cells or exploiting its absence for targeted killing. Each of these strategies will have to take into account the status of pRB in different cancers. Structurally defining individual protein interaction surfaces within or outside the pocket domain of pRB that mediate some of the pRB-specific tumour suppressor functions and that are not conserved in p107 and p130 represent attractive drug targets for pRB. Cells deficient in pRB are more susceptible to apoptosis induced by DNA damage and this capability is linked to its growth suppression activity. This property is useful for cancers that are pRB negative. Indeed, pRB negative breast cancer cells are more sensitive to chemotherapy compared with pRB positive breast cancer cells [124]. Since cyclin D/CDK4, 6 mediated phosphorylation of pRB results in release of growth promoting E2F, CDK inhibition has been explored as a strategy to prevent pRB inactivation and reestablish cell cycle control. Among these inhibitors flavopiridol and roscovitine have a very broad range of targets but can inhibit cell cycle and cause cell death. Despite this flavopiridol is not effective in many cancers [125]. More specific inhibitors have been developed against CDK4/6 among which palbociclib, the most successful, has been evaluated in mono- as well as combination therapy and is in the Phase III trial [126]. However, a range of sensitivities to palbociclib was noted in breast cancer [127].
Since the binding of E7 LxCxE peptide with pRB–E2F complex alone is not sufficient for the dissociation of E2F, pharmacological molecules can be designed that can inhibit the binding of LxCxE dependent interactions of viral proteins with pRB without disturbing pRB/E2F interactions [88]. However, such an approach will need to consider the effect on cellular LxCxE containing proteins. Perhaps a better strategy will be to exploit phosphorylation mediated plasticity of pRB to prevent its inactivation. This approach has been used to investigate the efficacy of an LxCxE peptide as an inhibitor of phosphorylated T373 mediated conformational change that weakens the interaction of E2F with pRB [128]. The LxCxE peptide from HPV E7 was found to have a modest effect and full length HPV E7 had better effect on activation of phosphorylated pRB in vitro in this study. Significantly, LxCxE peptides derived from cyclin D could not activate pRB.
FUTURE DIRECTIONS AND PROSPECTS
Both APC/C and pRB are critical determinants of important cellular decisions regarding proliferation, differentiation and quiescence. Both are also targeted by several viruses for proliferative advantages. The interaction of these two important players adds another layer to the G1-S regulation which is an important phase of the cell cycle in which cells can choose between different fates. This interaction, although providing an explanation for the retention of growth suppressor activity of pRB mutants defective in E2F binding, opens many important new questions. Further research is required to understand the mechanisms that regulate the FZR1–pRB interaction, the number and type of APC/CFZR1 substrates that are affected by this interaction, whether pRB makes contacts with other APC/C subunits and the molecular mechanism by which pRB stimulates SKP2 ubiquitination. What other substrates, other than PLK1 and SKP2 are directly affected by this interaction is also an open question. Answers to these questions will provide further insight into the molecular mechanism of APC/C function and provide clues that can be exploited to develop better inhibitors.
It is interesting that among cellular proteins carrying LxCxE motif that bind to pRB, several are ubiquitin ligases. Apart from APC/CFZR1, BRCA-1 and NRBE3 are other E3 ligases that bind to pRB. NRBE3 is a novel pRB ligase that promotes pRB degradation and is transcriptionally regulated by E2F transcription factors [129]. All the three E3 ligases contain LxCxE motif (Ramanujan and Tiwari, unpublished observations). Future work is required to functionally and structurally dissect the role of ubiquitin ligases in association with pRB and to identify E3 ubiquitin ligases responsible for proteasome mediated degradation of pRB. There are increasing evidences of viruses inactivating key cell cycle players, the most recent being that of Epstein–Barr virus (EBV) protein kinase BGLF4 directly binding and phosphorylating CDC20, possibly allowing CDC20 to be targeted by SCF for degradation [130]. EBV nuclear antigen 3C (EBNA3C) is also linked to regulation of the SCF complex thereby mediating degradation of pRB through the SCFSKP2 complex [131]. We speculate that transforming viruses might disable APC/C throughout the cell cycle, by inactivating its activity mediated by both its adaptor proteins, FZR1 and CDC20. It is important to understand the mechanistic basis of these interactions and understand their functional implications to be able to develop better anti-viral and anti-cancer drugs.
Acknowledgments
We thank Devinder Sehgal for critical reading of the manuscript.
Abbreviations
- APC/C
anaphase promoting complex/cyclosome
- CK2-PEST
casein kinase II and PEST sequence
- CKI
cyclin-dependent kinase inhibitory protein
- CR
conserved region
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin ligase
- EBV
Epstein–Barr virus
- HPV
human papilloma virus
- LT
large T-antigen
- PLK1
Polo-like kinase-1
- pRB
retinoblastoma protein
- SAC
spindle assembly checkpoint
- SCF
Skp1/Cul1/F-box
- SLiM
short linear motif
- Ub
ubiquitin
- UPP
ubiquitin proteasome pathway
FUNDING
This work was supported by the Department of Biotechnology (DBT) [grant number BT/PR4444/BRB/10/1023/2012 (to S.T.)]; the DBT BUILDER [grant number BT/PR5006/INF/22/153/2012 (to S.T.)]; the DST-PURSE [grant number PAC-JNU-DST-PURSE-462 (Phase-II) (to S.T.)]; the University Grants Commission (UGC)-UPEII [grant number 43 (to S.T.)]; and fellowships to A.R. from DBT BUILDER and UGC.
References
- 1.Li M., Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 2009;4:2. doi: 10.1186/1747-1028-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konishi Y., Stegmuller J., Matsuda T., Bonni S., Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 3.Almeida A., Bolanos J.P., Moreno S. Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. J. Neurosci. 2005;25:8115–8121. doi: 10.1523/JNEUROSCI.1143-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almeida A., Bolanos J.P., Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber A., Stengel F., Zhang Z., Enchev R.I., Kong E.H., Morris E.P., Robinson C.V., da Fonseca P.C., Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 6.Chang L., Zhang Z., Yang J., McLaughlin S.H., Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang L., Zhang Z., Yang J., McLaughlin S.H., Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manchado E., Eguren M., Malumbres M. The anaphase-promoting complex/cyclosome (APC/C): cell-cycle-dependent and -independent functions. Biochem. Soc. Trans. 2010;38:65–71. doi: 10.1042/BST0380065. [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Bonni A. A decade of the anaphase-promoting complex in the nervous system. Genes Dev. 2016;30:622–638. doi: 10.1101/gad.274324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Wan L., Dai X., Sun Y., Wei W. Functional characterization of anaphase promoting complex/cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis. Biochim. Biophys. Acta. 2014;1845:277–293. doi: 10.1016/j.bbcan.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters J.M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 12.Fang G., Yu H., Kirschner M.W. Control of mitotic transitions by the anaphase-promoting complex. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999;354:1583–1590. doi: 10.1098/rstb.1999.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfleger C.M., Lee E., Kirschner M.W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glotzer M., Murray A.W., Kirschner M.W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 15.Pfleger C.M., Kirschner M.W. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 16.Castro A., Vigneron S., Bernis C., Labbe J.C., Prigent C., Lorca T. The D-Box-activating domain (DAD) is a new proteolysis signal that stimulates the silent D-Box sequence of Aurora-A. EMBO Rep. 2002;3:1209–1214. doi: 10.1093/embo-reports/kvf241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro A., Vigneron S., Bernis C., Labbe J.C., Lorca T. Xkid is degraded in a D-box, KEN-box, and A-box-independent pathway. Mol. Cell. Biol. 2003;23:4126–4138. doi: 10.1128/MCB.23.12.4126-4138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Araki M., Wharton R.P., Tang Z., Yu H., Asano M. Degradation of origin recognition complex large subunit by the anaphase-promoting complex in Drosophila. EMBO J. 2003;22:6115–6126. doi: 10.1093/emboj/cdg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis A., Levasseur M., Chang H.Y., Elliott D.J., Jones K.T. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO Rep. 2006;7:1040–1045. doi: 10.1038/sj.embor.7400772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zur A., Brandeis M. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 2002;21:4500–4510. doi: 10.1093/emboj/cdf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell. Biol. 2002;14:706–714. doi: 10.1016/S0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- 22.Kramer E.R., Scheuringer N., Podtelejnikov A.V., Mann M., Peters J.M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper J.W., Burton J.L., Solomon M.J. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 24.Bochis O.V., Fetica B., Vlad C., Achimas-Cadariu P., Irimie A. The importance of ubiquitin E3 ligases, SCF and APC/C, in human cancers. Clujul Med. 2015;88:9–14. doi: 10.15386/cjmed-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budhavarapu V.N., White E.D., Mahanic C.S., Chen L., Lin F.T., Lin W.C. Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle. 2012;11:2030–2038. doi: 10.4161/cc.20643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prinz S., Hwang E.S., Visintin R., Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 1998;8:750–760. doi: 10.1016/S0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- 27.Listovsky T., Oren Y.S., Yudkovsky Y., Mahbubani H.M., Weiss A.M., Lebendiker M., Brandeis M. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rape M., Kirschner M.W. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 29.Rape M., Reddy S.K., Kirschner M.W. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 30.Jaquenoud M., van Drogen F., Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1) EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaspersen S.L., Charles J.F., Morgan D.O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 1999;9:227–236. doi: 10.1016/S0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 32.Lukas C., Sorensen C.S., Kramer E., Santoni-Rugiu E., Lindeneg C., Peters J.M., Bartek J., Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- 33.Benmaamar R., Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- 34.The I., Ruijtenberg S., Bouchet B.P., Cristobal A., Prinsen M.B., van Mourik T., Koreth J., Xu H., Heck A.J., Akhmanova A., et al. Rb and FZR1/Cdh1 determine CDK4/6-cyclin D requirement in C. elegans and human cancer cells. Nat. Commun. 2015;6:5906. doi: 10.1038/ncomms6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narbonne-Reveau K., Senger S., Pal M., Herr A., Richardson H.E., Asano M., Deak P., Lilly M.A. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development. 2008;135:1451–1461. doi: 10.1242/dev.016295. [DOI] [PubMed] [Google Scholar]
- 36.Reimann J.D., Gardner B.E., Margottin-Goguet F., Jackson P.K. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 2001;15:3278–3285. doi: 10.1101/gad.945701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guardavaccaro D., Kudo Y., Boulaire J., Barchi M., Busino L., Donzelli M., Margottin-Goguet F., Jackson P.K., Yamasaki L., Pagano M. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev. Cell. 2003;4:799–812. doi: 10.1016/S1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 38.Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W., Jang J., Shevchenko A., Charbonneau H., Deshaies R.J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/S0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 39.Sigrist S.J., Lehner C.F. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/S0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 40.Lorca T., Castro A., Martinez A.M., Vigneron S., Morin N., Sigrist S., Lehner C., Dorée M., Labbé J.C. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs H., Richter D., Venkatesh T., Lehner C. Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr. Biol. 2002;12:1435–1441. doi: 10.1016/S0960-9822(02)01074-6. [DOI] [PubMed] [Google Scholar]
- 42.Engelbert D., Schnerch D., Baumgarten A., Wasch R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene. 2008;27:907–917. doi: 10.1038/sj.onc.1210703. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Higuera I., Manchado E., Dubus P., Canamero M., Mendez J., Moreno S., Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 44.Naoe H., Chiyoda T., Ishizawa J., Masuda K., Saya H., Kuninaka S. The APC/C activator Cdh1 regulates the G2/M transition during differentiation of placental trophoblast stem cells. Biochem. Biophys. Res. Commun. 2013;430:757–762. doi: 10.1016/j.bbrc.2012.11.075. [DOI] [PubMed] [Google Scholar]
- 45.Seah M.K., Holt J.E., Garcia-Higuera I., Moreno S., Jones K.T. The APC activator fizzy-related-1 (FZR1) is needed for preimplantation mouse embryo development. J. Cell Sci. 2012;125:6030–6037. doi: 10.1242/jcs.110155. [DOI] [PubMed] [Google Scholar]
- 46.Gao D., Inuzuka H., Korenjak M., Tseng A., Wu T., Wan L., Kirschner M., Dyson N., Wei W. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol. Biol. Cell. 2009;20:3305–3316. doi: 10.1091/mbc.E09-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan C.H., Morrow J.K., Li C.F., Gao Y., Jin G., Moten A., Stagg L.J., Ladbury J.E., Cai Z., Xu D., et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gstaiger M., Jordan R., Lim M., Catzavelos C., Mestan J., Slingerland J., Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Signoretti S., Di Marcotullio L., Richardson A., Ramaswamy S., Isaac B., Rue M., Monti F., Loda M., Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J. Clin. Invest. 2002;110:633–641. doi: 10.1172/JCI0215795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Catzavelos C., Tsao M.S., DeBoer G., Bhattacharya N., Shepherd F.A., Slingerland J.M. Reduced expression of the cell cycle inhibitor p27Kip1 in non-small cell lung carcinoma: a prognostic factor independent of Ras. Cancer Res. 1999;59:684–688. [PubMed] [Google Scholar]
- 51.Chu I.M., Hengst L., Slingerland J.M. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 52.Hafez M.M., Alhoshani A.R., Al-Hosaini K.A., Alsharari S.D., Al Rejaie S.S., Sayed-Ahmed M.M., Al-Shabanah O.A. SKP2/P27Kip1 pathway is associated with Advanced Ovarian Cancer in Saudi Patients. Asian Pac. J. Cancer Prev. 2015;16:5807–5815. doi: 10.7314/APJCP.2015.16.14.5807. [DOI] [PubMed] [Google Scholar]
- 53.Yang G., Ayala G., De Marzo A., Tian W., Frolov A., Wheeler T.M., Thompson T.C., Harper J.W. Elevated Skp2 protein expression in human prostate cancer: association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin. Cancer Res. 2002;8:3419–3426. [PubMed] [Google Scholar]
- 54.Chen H., Mo X., Yu J., Huang S., Huang Z., Gao L. Interference of Skp2 effectively inhibits the development and metastasis of colon carcinoma. Mol. Med. Rep. 2014;10:1129–1135. doi: 10.3892/mmr.2014.2308. [DOI] [PubMed] [Google Scholar]
- 55.Qiu L., Lv J., Chen Y., Wang J., Wu R. Expression of Skp2 and p27 proteins in hypopharyngeal squamous cell carcinoma and its clinical significance. Oncol Lett. 2015;10:3756–3760. doi: 10.3892/ol.2015.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita T., Liu W., Doihara H., Wan Y. Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am. J. Pathol. 2008;173:217–228. doi: 10.2353/ajpath.2008.070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujita T., Liu W., Doihara H., Date H., Wan Y. Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clin. Cancer Res. 2008;14:1966–1975. doi: 10.1158/1078-0432.CCR-07-1585. [DOI] [PubMed] [Google Scholar]
- 58.Lehman N.L., Tibshirani R., Hsu J.Y., Natkunam Y., Harris B.T., West R.B., Masek M.A., Montgomery K., van de Rijn M., Jackson P.K. Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. Am. J. Pathol. 2007;170:1793–1805. doi: 10.2353/ajpath.2007.060767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu J.Y., Reimann J.D., Sorensen C.S., Lukas J., Jackson P.K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat. Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- 60.Du W., Searle J.S. The rb pathway and cancer therapeutics. Curr. Drug Targets. 2009;10:581–589. doi: 10.2174/138945009788680392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cobrinik D., Lee M.H., Hannon G., Mulligan G., Bronson R.T., Dyson N., Harlow E., Beach D., Weinberg R.A., Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 62.Chellappan S.P., Hiebert S., Mudryj M., Horowitz J.M., Nevins J.R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-F. [DOI] [PubMed] [Google Scholar]
- 63.Sellers W.R., Novitch B.G., Miyake S., Heith A., Otterson G.A., Kaye F.J., Lassar A.B., Kaelin W.G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Binne U.K., Classon M.K., Dick F.A., Wei W., Rape M., Kaelin W.G., Jr, Näär A.M., Dyson N.J. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- 65.Ji P., Jiang H., Rekhtman K., Bloom J., Ichetovkin M., Pagano M., Zhu L. An Rb-Skp2-p27 pathway mediates acute cell cycle inhi6bition by Rb and is retained in a partial-penetrance Rb mutant. Mol. Cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 66.Ruijtenberg S., van den Heuvel S. Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15:196–212. doi: 10.1080/15384101.2015.1120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee M.H., Williams B.O., Mulligan G., Mukai S., Bronson R.T., Dyson N., Harlow E., Jacks T. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 68.Noble M.E., Endicott J.A., Brown N.R., Johnson L.N. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem. Sci. 1997;22:482–487. doi: 10.1016/S0968-0004(97)01144-4. [DOI] [PubMed] [Google Scholar]
- 69.Hassler M., Singh S., Yue W.W., Luczynski M., Lakbir R., Sanchez-Sanchez F., Bader T., Pearl L.H., Mittnacht S. Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture. Mol. Cell. 2007;28:371–385. doi: 10.1016/j.molcel.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chow K.N., Dean D.C. Domains A and B in the Rb pocket interact to form a transcriptional repressor motif. Mol. Cell. Biol. 1996;16:4862–4868. doi: 10.1128/MCB.16.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J.O., Russo A.A., Pavletich N.P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 72.Qin X.Q., Chittenden T., Livingston D.M., Kaelin W.G., Jr Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 73.Dick F.A., Dyson N.J. Three regions of the pRB pocket domain affect its inactivation by human papillomavirus E7 proteins. J. Virol. 2002;76:6224–6234. doi: 10.1128/JVI.76.12.6224-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dick F.A., Sailhamer E., Dyson N.J. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 2000;20:3715–3727. doi: 10.1128/MCB.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Talluri S., Francis S.M., Dick F.A. Mutation of the LXCXE binding cleft of pRb facilitates transformation by ras in vitro but does not promote tumorigenesis in vivo. PLoS One. 2013;8:e72236. doi: 10.1371/journal.pone.0072236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singh M., Krajewski M., Mikolajka A., Holak T.A. Molecular determinants for the complex formation between the retinoblastoma protein and LXCXE sequences. J. Biol. Chem. 2005;280:37868–37876. doi: 10.1074/jbc.M504877200. [DOI] [PubMed] [Google Scholar]
- 77.Felsani A., Mileo A.M., Paggi M.G. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene. 2006;25:5277–5285. doi: 10.1038/sj.onc.1209621. [DOI] [PubMed] [Google Scholar]
- 78.Fattaey A.R., Harlow E., Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol. Cell. Biol. 1993;13:7267–7277. doi: 10.1128/MCB.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu X., Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007;21:2711–2716. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang P.S., Patrick D.R., Edwards G., Goodhart P.J., Huber H.E., Miles L., Garsky V.M., Oliff A., Heimbrook D.C. Protein domains governing interactions between E2F, the retinoblastoma gene product, and human papillomavirus type 16 E7 protein. Mol. Cell. Biol. 1993;13:953–960. doi: 10.1128/MCB.13.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chemes L.B., Sanchez I.E., de Prat-Gay G. Kinetic recognition of the retinoblastoma tumor suppressor by a specific protein target. J. Mol. Biol. 2011;412:267–284. doi: 10.1016/j.jmb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 82.Huh K., Zhou X., Hayakawa H., Cho J.Y., Libermann T.A., Jin J., Harper J.W., Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boyer S.N., Wazer D.E., Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 84.Jones D.L., Thompson D.A., Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 85.Kim H.Y., Ahn B.Y., Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 2001;20:295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dahiya A., Gavin M.R., Luo R.X., Dean D.C. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 2000;20:6799–6805. doi: 10.1128/MCB.20.18.6799-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Helt A.M., Galloway D.A. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 2001;75:6737–6747. doi: 10.1128/JVI.75.15.6737-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramakrishnan C., Subramanian V., Balamurugan K., Velmurugan D. Molecular dynamics simulations of retinoblastoma protein. J. Biomol. Struct. Dyn. 2013;31:1277–1292. doi: 10.1080/07391102.2012.732345. [DOI] [PubMed] [Google Scholar]
- 89.Patrick D.R., Oliff A., Heimbrook D.C. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J. Biol. Chem. 1994;269:6842–6850. [PubMed] [Google Scholar]
- 90.Chemes L.B., Sanchez I.E., Smal C., de Prat-Gay G. Targeting mechanism of the retinoblastoma tumor suppressor by a prototypical viral oncoprotein. Structural modularity, intrinsic disorder and phosphorylation of human papillomavirus E7. FEBS J. 2010;277:973–988. doi: 10.1111/j.1742-4658.2009.07540.x. [DOI] [PubMed] [Google Scholar]
- 91.Henley S.A., Francis S.M., Demone J., Ainsworth P., Dick F.A. A cancer derived mutation in the retinoblastoma gene with a distinct defect for LXCXE dependent interactions. Cancer Cell. Int. 2010;10:8. doi: 10.1186/1475-2867-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kratzke R.A., Otterson G.A., Lin A.Y., Shimizu E., Alexandrova N., Zajac-Kaye M., Horowitz J.M., Kaye F.J. Functional analysis at the Cys706 residue of the retinoblastoma protein. J. Biol. Chem. 1992;267:25998–26003. [PubMed] [Google Scholar]
- 93.Kaye F.J., Kratzke R.A., Gerster J.L., Horowitz J.M. A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6922–6926. doi: 10.1073/pnas.87.17.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zacksenhaus E., Bremner R., Phillips R.A., Gallie B.L. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 1993;13:4588–4599. doi: 10.1128/MCB.13.8.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamel P.A., Phillips R.A., Muncaster M., Gallie B.L. Speculations on the roles of RB1 in tissue-specific differentiation, tumor initiation, and tumor progression. FASEB J. 1993;7:846–854. doi: 10.1096/fasebj.7.10.8344484. [DOI] [PubMed] [Google Scholar]
- 96.Lohmann D.R., Brandt B., Hopping W., Passarge E., Horsthemke B. Distinct RB1 gene mutations with low penetrance in hereditary retinoblastoma. Hum. Genet. 1994;94:349–354. doi: 10.1007/BF00201591. [DOI] [PubMed] [Google Scholar]
- 97.Hung C.C., Lin S.Y., Lee C.N., Chen C.P., Lin S.P., Chao M.C., Chiou S.S., Su Y.N. Low penetrance of retinoblastoma for p.V654L mutation of the RB1 gene. BMC Med. Genet. 2011;12:76. doi: 10.1186/1471-2350-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harbour J.W. Molecular basis of low-penetrance retinoblastoma. Arch. Ophthalmol. 2001;119:1699–1704. doi: 10.1001/archopht.119.11.1699. [DOI] [PubMed] [Google Scholar]
- 99.Bremner R., Du D.C., Connolly-Wilson M.J., Bridge P., Ahmad K.F., Mostachfi H., Rushlow D., Dunn J.M., Gallie B.L. Deletion of RB exons 24 and 25 causes low-penetrance retinoblastoma. Am. J. Hum. Genet. 1997;61:556–570. doi: 10.1086/515499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richter S., Vandezande K., Chen N., Zhang K., Sutherland J., Anderson J., Han L., Panton R., Branco P., Gallie B. Sensitive and efficient detection of RB1 gene mutations enhances care for families with retinoblastoma. Am. J. Hum. Genet. 2003;72:253–269. doi: 10.1086/345651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Onadim Z., Hogg A., Baird P.N., Cowell J.K. Oncogenic point mutations in exon 20 of the RB1 gene in families showing incomplete penetrance and mild expression of the retinoblastoma phenotype. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6177–6181. doi: 10.1073/pnas.89.13.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Otterson G.A., Chen W., Coxon A.B., Khleif S.N., Kaye F.J. Incomplete penetrance of familial retinoblastoma linked to germ-line mutations that result in partial loss of RB function. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12036–12040. doi: 10.1073/pnas.94.22.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park Y., Kubo A., Komiya T., Coxon A., Beebe K., Neckers L., Meltzer P.S., Kaye F.J. Low-penetrant RB allele in small-cell cancer shows geldanamycin instability and discordant expression with mutant ras. Cell Cycle. 2008;7:2384–2391. doi: 10.4161/cc.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whitaker L.L., Su H., Baskaran R., Knudsen E.S., Wang J.Y. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol. Cell. Biol. 1998;18:4032–4042. doi: 10.1128/MCB.18.7.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takahashi C., Sasaki N., Kitajima S. Twists in views on RB functions in cellular signaling, metabolism and stem cells. Cancer Sci. 2012;103:1182–1188. doi: 10.1111/j.1349-7006.2012.02284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dasgupta P., Betts V., Rastogi S., Joshi B., Morris M., Brennan B., Ordonez-Ercan D., Chellappan S. Direct binding of apoptosis signal-regulating kinase 1 to retinoblastoma protein: novel links between apoptotic signaling and cell cycle machinery. J. Biol. Chem. 2004;279:38762–38769. doi: 10.1074/jbc.M312273200. [DOI] [PubMed] [Google Scholar]
- 107.Landis M.W., Brown N.E., Baker G.L., Shifrin A., Das M., Geng Y., Sicinski P., Hinds P.W. The LxCxE pRb interaction domain of cyclin D1 is dispensable for murine development. Cancer Res. 2007;67:7613–7620. doi: 10.1158/0008-5472.CAN-07-1207. [DOI] [PubMed] [Google Scholar]
- 108.Baker G.L., Landis M.W., Hinds P.W. Multiple functions of D-type cyclins can antagonize pRb-mediated suppression of proliferation. Cell Cycle. 2005;4:330–338. doi: 10.4161/cc.4.2.1485. [DOI] [PubMed] [Google Scholar]
- 109.Kim Y.W., Otterson G.A., Kratzke R.A., Coxon A.B., Kaye F.J. Differential specificity for binding of retinoblastoma binding protein 2 to RB, p107, and TATA-binding protein. Mol. Cell. Biol. 1994;14:7256–7264. doi: 10.1128/MCB.14.11.7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Higashitsuji H., Itoh K., Nagao T., Dawson S., Nonoguchi K., Kido T., Mayer R.J., Arii S., Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 111.Fan S., Yuan R., Ma Y.X., Xiong J., Meng Q., Erdos M., Zhao J.N., Goldberg I.D., Pestell R.G., Rosen E.M. Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding. Oncogene. 2001;20:4827–4841. doi: 10.1038/sj.onc.1204666. [DOI] [PubMed] [Google Scholar]
- 112.Morris E.J., Dyson N.J. Retinoblastoma protein partners. Adv. Cancer. Res. 2001;82:1–54. doi: 10.1016/S0065-230X(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 113.Knudsen E.S., Wang J.Y. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J. Biol. Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 114.Chen P.L., Scully P., Shew J.Y., Wang J.Y., Lee W.H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 115.Chemes L.B., Noval M.G., Sanchez I.E., de Prat-Gay G. Folding of a cyclin box: linking multitarget binding to marginal stability, oligomerization, and aggregation of the retinoblastoma tumor suppressor AB pocket domain. J. Biol. Chem. 2013;288:18923–18938. doi: 10.1074/jbc.M113.467316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wan L., Zou W., Gao D., Inuzuka H., Fukushima H., Berg A.H., Drapp R., Shaik S., Hu D., Lester C., et al. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol. Cell. 2011;44:721–733. doi: 10.1016/j.molcel.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y., Clem B., Zuba-Surma E.K., El-Naggar S., Telang S., Jenson A.B., Wang Y., Shao H., Ratajczak M.Z., Chesney J., Dean D.C. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell. 2009;4:336–347. doi: 10.1016/j.stem.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kitajima S., Kohno S., Kondoh A., Sasaki N., Nishimoto Y., Li F., Abdallah Mohammed M.S., Muranaka H., Nagatani N., Suzuki M., et al. Undifferentiated state induced by Rb-p53 double inactivation in mouse thyroid neuroendocrine cells and embryonic fibroblasts. Stem Cells. 2015;33:1657–1669. doi: 10.1002/stem.1971. [DOI] [PubMed] [Google Scholar]
- 119.Huang J., Sheung J., Dong G., Coquilla C., Daniel-Issakani S., Payan D.G. High-throughput screening for inhibitors of the e3 ubiquitin ligase APC. Methods Enzymol. 2005;399:740–754. doi: 10.1016/S0076-6879(05)99049-6. [DOI] [PubMed] [Google Scholar]
- 120.Huang K.S., Vassilev L.T. High-throughput screening for inhibitors of the Cks1-Skp2 interaction. Methods Enzymol. 2005;399:717–728. doi: 10.1016/S0076-6879(05)99047-2. [DOI] [PubMed] [Google Scholar]
- 121.Huang H.C., Shi J., Orth J.D., Mitchison T.J. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16:347–358. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cardozo T., Pagano M. Wrenches in the works: drug discovery targeting the SCF ubiquitin ligase and APC/C complexes. BMC Biochem. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2091-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Drosopoulos K., Tang C., Chao W.C., Linardopoulos S. APC/C is an essential regulator of centrosome clustering. Nat. Commun. 2014;5:3686. doi: 10.1038/ncomms4686. [DOI] [PubMed] [Google Scholar]
- 124.Witkiewicz A.K., Ertel A., McFalls J., Valsecchi M.E., Schwartz G., Knudsen E.S. RB-pathway disruption is associated with improved response to neoadjuvant chemotherapy in breast cancer. Clin. Cancer Res. 2012;18:5110–5122. doi: 10.1158/1078-0432.CCR-12-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bose P., Simmons G.L., Grant S. Cyclin-dependent kinase inhibitor therapy for hematologic malignancies. Expert Opin. Investig. Drugs. 2013;22:723–738. doi: 10.1517/13543784.2013.789859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hamilton E., Infante J.R. Targeting CDK4/6 in patients with cancer. Cancer Treat. Rev. 2016;45:129–138. doi: 10.1016/j.ctrv.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 127.Finn R.S., Dering J., Conklin D., Kalous O., Cohen D.J., Desai A.J., Ginther C., Atefi M., Chen I., Fowst C., et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pye C.R., Bray W.M., Brown E.R., Burke J.R., Lokey R.S., Rubin S.M. A strategy for direct chemical activation of the retinoblastoma protein. ACS Chem. Biol. 2016;11:1192–1197. doi: 10.1021/acschembio.6b00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y., Zheng Z., Zhang J., Kong R., Liu J., Zhang Y., Deng H., Du X., Ke Y. A novel retinoblastoma protein (RB) E3 ubiquitin ligase (NRBE3) promotes RB degradation and is transcriptionally regulated by E2F1 transcription factor. J. Biol. Chem. 2015;290:28200–28213. doi: 10.1074/jbc.M115.655597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li R., Liao G., Nirujogi R.S., Pinto S.M., Shaw P.G., Huang T.C., Wan J., Qian J., Gowda H., Wu X., et al. Phosphoproteomic profiling reveals Epstein–Barr virus protein kinase integration of DNA damage response and mitotic signaling. PLoS Pathog. 2015;11:e1005346. doi: 10.1371/journal.ppat.1005346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Knight J.S., Sharma N., Robertson E.S. Epstein–Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wei W., Ayad N.G., Wan Y., Zhang G.J., Kirschner M.W., Kaelin W.G., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 133.Hershko A., Ganoth D., Pehrson J., Palazzo R.E., Cohen L.H. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J. Biol. Chem. 1991;266:16376–16379. [PubMed] [Google Scholar]
- 134.Homer H.A., McDougall A., Levasseur M., Murdoch A.P., Herbert M. Mad2 is required for inhibiting securin and cyclin B degradation following spindle depolymerisation in meiosis I mouse oocytes. Reproduction. 2005;130:829–843. doi: 10.1530/rep.1.00856. [DOI] [PubMed] [Google Scholar]
- 135.Park H.J., Costa R.H., Lau L.F., Tyner A.L., Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol. Cell. Biol. 2008;28:5162–5171. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Uchida F., Uzawa K., Kasamatsu A., Takatori H., Sakamoto Y., Ogawara K., Shiiba M., Tanzawa H., Bukawa H. Overexpression of cell cycle regulator CDCA3 promotes oral cancer progression by enhancing cell proliferation with prevention of G1 phase arrest. BMC Cancer. 2012;12:321. doi: 10.1186/1471-2407-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao W.M., Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J. Biol. Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 138.Donzelli M., Squatrito M., Ganoth D., Hershko A., Pagano M., Draetta G.F. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 2002;21:4875–4884. doi: 10.1093/emboj/cdf491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McGarry T.J., Kirschner M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/S0092-8674(00)81209-X. [DOI] [PubMed] [Google Scholar]
- 140.Stewart S., Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- 141.Duursma A.M., Agami R. CDK-dependent stabilization of Cdc6: linking growth and stress signals to activation of DNA replication. Cell Cycle. 2005;4:1725–1728. doi: 10.4161/cc.4.12.2193. [DOI] [PubMed] [Google Scholar]
- 142.Seki A., Fang G. CKAP2 is a spindle-associated protein degraded by APC/C-Cdh1 during mitotic exit. J. Biol. Chem. 2007;282:15103–15113. doi: 10.1074/jbc.M701688200. [DOI] [PubMed] [Google Scholar]
- 143.Visintin R., Prinz S., Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 144.Geley S., Kramer E., Gieffers C., Gannon J., Peters J.M., Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li M., Shin Y.H., Hou L., Huang X., Wei Z., Klann E., Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat. Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lasorella A., Stegmuller J., Guardavaccaro D., Liu G., Carro M.S., Rothschild G., de la Torre-Ubieta L., Pagano M., Bonni A., Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 147.Lindon C., Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell. Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhao W.M., Coppinger J.A., Seki A., Cheng X.L., Yates J.R., 3rd, Fang G. RCS1, a substrate of APC/C, controls the metaphase to anaphase transition. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13415–13420. doi: 10.1073/pnas.0709227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zur A., Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20:792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Karamysheva Z., Diaz-Martinez L.A., Crow S.E., Li B., Yu H. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J. Biol. Chem. 2009;284:1772–1780. doi: 10.1074/jbc.M807083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stroschein S.L., Bonni S., Wrana J.L., Luo K. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 2001;15:2822–2836. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stewart S., Fang G. Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol. Cell. Biol. 2005;25:10516–10527. doi: 10.1128/MCB.25.23.10516-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Funabiki H., Murray A.W. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/S0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 154.Carlile T.M., Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Simpson-Lavy K.J., Brandeis M. Clb2 and the APC/C(Cdh1) regulate Swe1 stability. Cell Cycle. 2010;9:3046–3053. doi: 10.4161/cc.9.15.12457. [DOI] [PubMed] [Google Scholar]
- 156.Sajman J., Zenvirth D., Nitzan M., Margalit H., Simpson-Lavy K.J., Reiss Y., Cohen I., Ravid T., Brandeis M. Degradation of Ndd1 by APC/C(Cdh1) generates a feed forward loop that times mitotic protein accumulation. Nat. Commun. 2015;6:7075. doi: 10.1038/ncomms8075. [DOI] [PubMed] [Google Scholar]
- 157.Lafranchi L., de Boer H.R., de Vries E.G., Ong S.E., Sartori A.A., van Vugt M.A. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J. 2014;33:2860–2879. doi: 10.15252/embj.201489017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kitamura K., Maekawa H., Shimoda C. Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol. Biol. Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Asakawa H., Kitamura K., Shimoda C. A novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol. Genet. Genomics. 2001;265:424–435. doi: 10.1007/s004380000429. [DOI] [PubMed] [Google Scholar]
- 160.Okaz E., Arguello-Miranda O., Bogdanova A., Vinod P.K., Lipp J.J., Markova Z., Zagoriy I., Novak B., Zachariae W. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell. 2012;151:603–618. doi: 10.1016/j.cell.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 161.Cooper K.F., Mallory M.J., Egeland D.B., Jarnik M., Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schwab M., Lutum A.S., Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/S0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 163.Sigl R., Wandke C., Rauch V., Kirk J., Hunt T., Geley S. Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J. Cell Sci. 2009;122:4208–4217. doi: 10.1242/jcs.054197. [DOI] [PubMed] [Google Scholar]
- 164.Wiebusch L., Hagemeier C. p53- and p21-dependent premature APC/C-Cdh1 activation in G2 is part of the long-term response to genotoxic stress. Oncogene. 2010;29:3477–3489. doi: 10.1038/onc.2010.99. [DOI] [PubMed] [Google Scholar]
- 165.Liu Y., Ye W., Li B., Zhou X., Cui Y., Running M.P., Liu K. CCS52A2/FZR1, a cell cycle regulator, is an essential factor for shoot apical meristem maintenance in Arabidopsis thaliana. BMC Plant Biol. 2012;12:135. doi: 10.1186/1471-2229-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lammens T., Boudolf V., Kheibarshekan L., Zalmas L.P., Gaamouche T., Maes S., Vanstraelen M., Kondorosi E., La Thangue N.B., Govaerts W., et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wise A., Schatoff E., Flores J., Hua S.Y., Ueda A., Wu C.F., Venkatesh T. Drosophila-Cdh1 (Rap/Fzr) a regulatory subunit of APC/C is required for synaptic morphology, synaptic transmission and locomotion. Int. J. Dev. Neurosci. 2013;31:624–633. doi: 10.1016/j.ijdevneu.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fay D.S., Keenan S., Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–517. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Holt J.E., Pye V., Boon E., Stewart J.L., Garcia-Higuera I., Moreno S., Rodríguez R., Jones K.T., McLaughlin E.A. The APC/C activator FZR1 is essential for meiotic prophase I in mice. Development. 2014;141:1354–1365. doi: 10.1242/dev.104828. [DOI] [PubMed] [Google Scholar]
- 170.Tevosian S.G., Shih H.H., Mendelson K.G., Sheppard K.A., Paulson K.E., Yee A.S. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 171.Meloni A.R., Smith E.J., Nevins J.R. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 173.Magnaghi-Jaulin L., Groisman R., Naguibneva I., Robin P., Lorain S., Le Villain J.P., Troalen F., Trouche D., Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 174.Miyake S., Sellers W.R., Safran M., Li X., Zhao W., Grossman S.R., Gan J., DeCaprio J.A., Adams P.D., Kaelin W.G., Jr Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol. 2000;20:8889–8902. doi: 10.1128/MCB.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pennaneach V., Salles-Passador I., Munshi A., Brickner H., Regazzoni K., Dick F., Dyson N., Chen T.T., Wang J.Y., Fotedar R., Fotedar A. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell. 2001;7:715–727. doi: 10.1016/S1097-2765(01)00217-9. [DOI] [PubMed] [Google Scholar]
- 176.Baker J.E., Krementsova E.B., Kennedy G.G., Armstrong A., Trybus K.M., Warshaw D.M. Myosin V processivity: multiple kinetic pathways for head-to-head coordination. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5542–5546. doi: 10.1073/pnas.0307247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vorobiev S.M., Su M., Seetharaman J., Huang Y.J., Chen C.X., Maglaqui M., Janjua H., Proudfoot M., Yakunin A., Xiao R., et al. Crystal structure of human retinoblastoma binding protein 9. Proteins. 2009;74:526–529. doi: 10.1002/prot.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Uchida C., Hattori T., Takahashi H., Yamamoto N., Kitagawa M., Taya Y. Interaction between RB protein and NuMA is required for proper alignment of spindle microtubules. Genes Cells. 2014;19:89–96. doi: 10.1111/gtc.12119. [DOI] [PubMed] [Google Scholar]