Abstract
Leaf colour variation is observed in several plants. We obtained two types of branches with yellow and variegated leaves from Camellia sinensis. To reveal the mechanisms that underlie the leaf colour variations, combined morphological, histological, ionomic and proteomic analyses were performed using leaves from abnormal branches (variants) and normal branches (CKs). The measurement of the CIE-Lab coordinates showed that the brightness and yellowness of the variants were more intense than the CKs. When chloroplast profiles were analysed, HY1 (branch with yellow leaves) and HY2 (branch with variegated leaves) displayed abnormal chloroplast structures and a reduced number and size compared with the CKs, indicating that the abnormal chloroplast development might be tightly linked to the leaf colour variations. Moreover, the concentration of elemental minerals was different between the variants and the CKs. Furthermore, DEPs (differentially expressed proteins) were identified in the variants and the CKs by a quantitative proteomics analysis using the label-free approach. The DEPs were significantly involved in photosynthesis and included PSI, PSII, cytochrome b6/f complex, photosynthetic electron transport, LHC and F-type ATPase. Our results suggested that a decrease in the abundance of photosynthetic proteins might be associated with the changes of leaf colours in tea plants.
The leaves of plants are the major photosynthetic organs that provide energy for plant development. The leaf colour, size, and shape directly affect photosynthesis, yield and quality. Generally, the normal leaf colour is green, which depends on stabilised chloroplast development, chlorophyll and the biosynthesis of other pigments. However, the leaf-colour variations, including chlorina, albino, and striata, are observed in many higher plant species and are applied in breeding, such as rice1,2, wheat3,4, oilseed rapa5 and Camellia sinensis6,7,8. These mutants mentioned above serve as a perfect material to reveal the underlying mechanism involved in chlorophyll biosynthesis9, chloroplast structure and function1, the regulation of chloroplast development, and photosynthesis10. Dissecting the underlying mechanism of the leaf colour variations is of great importance for theoretical significances and for broad application prospects.
The occurrence of the variations in leaf colour is mainly determined by genetic and environmental factors. These variants mainly confer a change from green to white and yellow colours according to their phenotypes. The formation of leaf colour depends on several processes, including chloroplast development, the number and size of chloroplasts, and chlorophyll biosynthesis. Thus, any defect in these processes can result in the loss of the green colour in the leaf. For the internal factors, a gene mutation, including nuclear genes and cytoplasmic genes and the restraining protein transport, can result in variations in leaf colour. To date, many genes involved in chloroplast development and chlorophyll biosynthesis have been identified through the leaf colour mutants11,12,13. Those genes directly or indirectly regulate the structure of chloroplasts, chlorophyll biosynthesis and several metabolic processes that affect the depth of leaf colour14,15. Additionally, changes in environmental factors, including temperature6,16, light17, and elemental minerals18, can also lead to the variations in leaf colour. Therefore, the variations in leaf colour are caused by one or/and more of the factors mentioned above, which leads to difficulty in studying the underlying mechanism of leaf colour.
The tea plant, Camellia sinensis, is an economically important genus cultivated in China, Japan, and Korea19. Among the diverse cultivars, many materials showing variations in leaf colour have been obtained to expand the germplasms. At present, two main types of variations in leaf colour were identified in tea plants, including albino and chlorina6,7. They exhibit highly improved economic value depending on changes in their biochemical composition7,20. Therefore, the elucidation of the molecular mechanism underlying colour formation is important for tea plants breeding with variable leaf colours. However, to date, only a few studies have reported the molecular mechanisms involved in the changes of leaf colour in tea plants6,7,8. These studies found that the differentially expressed genes and proteins involved in the metabolism of amino acids, nitrogen and sulfur, photosynthesis, flavonoid biosynthesis, and chlorophyll biosynthesis are the major driving forces for the leaf colour changes. Although some mechanisms related to leaf colour (chlorina) in tea plants have been reported, the materials used in the previous studies did not share the same genetic background7,8, which indicates that these materials were not optimum for studying the molecular mechanism. Thus, to gain a true mechanistic view into this issue, it is essential to obtain leaf colour variants with the same genetic background compared with the normal plants.
Proteomics has emerged as a powerful tool that facilitates the study of global protein expression and is widely used in plants to address specific biological responses21,22,23. Additionally, proteomic analyses were used in studies of the leaf colour of tea plants through two different approaches (2D gel electrophoresis and iTRAQ)4,8. Thus, large-scale proteomic data derived from the leaf colour variants in tea plants are the basic information for the underlying mechanism of variations in leaf colour. In this study, we adopted a label-free MS-based approach.
We obtained two types of branches showing yellow and variegated colours in leaves from a population of “Yinghongjiuhao”. “Yinghongjiuhao” is a tea cultivar that was selected from Yunnan Big leaf tea. Investigating these variants is helpful to explore the molecular mechanisms underpinning leaf colour formation. Here, the growth performances were observed, and the leaf colour was identified through the CIE-Lab model, which provides digitalisation and visualisation results. Then, the number and ultra-structure of the chloroplasts were analysed to find the relationship between leaf colour and the characteristics of the chloroplasts. Furthermore, the ionomics and proteomics were performed to explore the mechanism of the leaf colour variations. Our findings reveal the complex process of leaf colour formation, involving phenotype, ultra-structure, mineral ions and protein, which will improve our understanding of phenotype in the leaf colour variants.
Results
Characterisation of leaf colour variants and their corresponding CKs
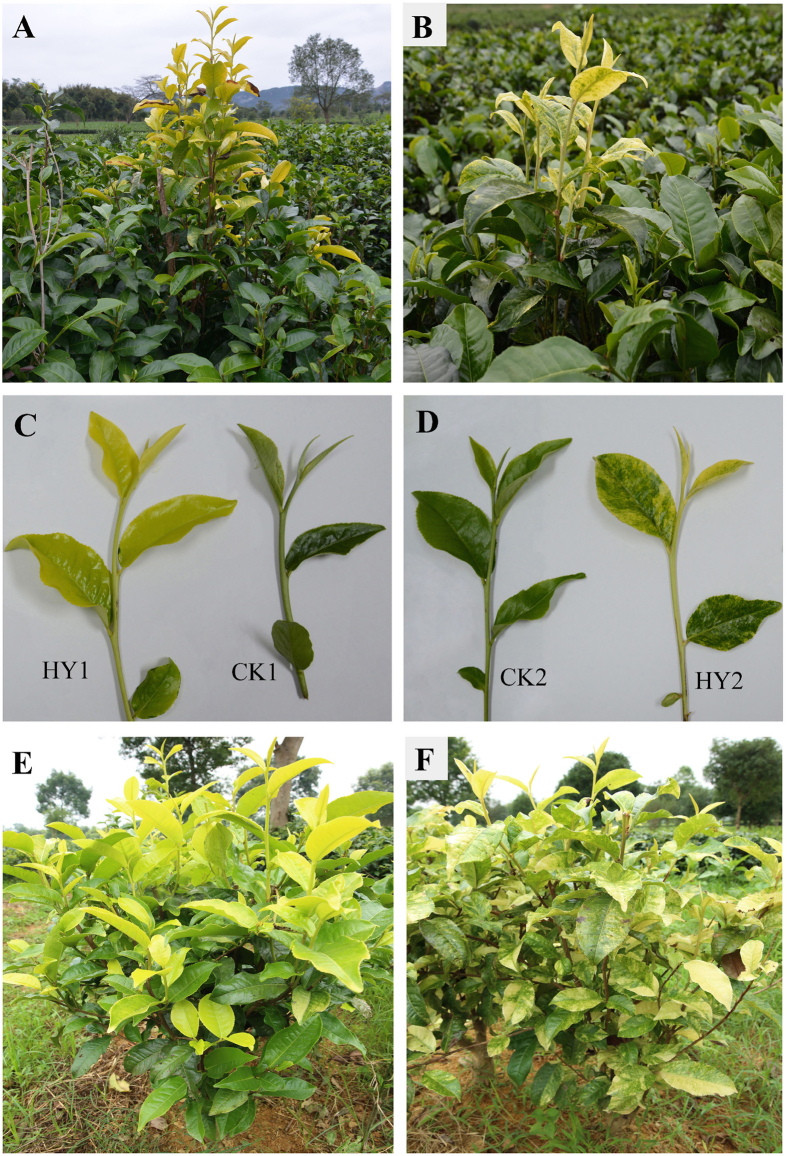
Compared with CK1 and CK2, the leaves from HY1 (Fig. 1A,C) and HY2 (Fig. 1B,D) exhibited yellow and variegated colour at an early stage of leaf development, respectively, and then the leaves tended to revert to green along with increasing maturity. Additionally, the variations in leaf colour can be stably maintained through grafting propagation of variants (Fig. 1E,F).
Figure 1. Phenotypes of leaves from the variants and the CKs of Camellia Sinensis.
(A) Performance of HY1 (showing yellow leaf) and CK1 (showing green leaf) in the field; (B) Performance of HY2 (showing variegated leaf) and CK2 (showing green leaf) in the field; (C) a comparison of the characteristics of HY1 and its corresponding CK1; (D) a comparison of HY2 and its corresponding CK2; (E) the stable variation of HY1 in leaf colour through the grafting propagation; (F) the stable phenotype in leaf colour of HY2 through the grafting propagation.
To characterise the colour changes between the variants and the CKs during leaf development, the measurement of the CIE-Lab coordinates was carried out, and the average and standard deviation of the L*, a* and b* parameters were calculated (Table 1). Due to the variegated colour at the different scanned points, the standard deviation of L*, a* and b* in HY2 was high. The L* and b* colour parameters showed that the brightness and yellowness, respectively, of the variants were more intense than the CKs. Moreover, the lower a* during leaf development indicated that the leaves from HY1 and HY2 were gradually greening, and the greenness of the four-leaf from HY2 (−8.59) approximated CK2 (−8.96), and at the same time, HY1’s (−3.39) did not achieve the level of CK1 (−8.82).
Table 1. Average data and standard deviation of the L*, a* and b* parameters of leaves from different leaf positions in the variants and the CKs.
| Sample | L* | a* | b* | |
|---|---|---|---|---|
| HY1 | 1st leaf | 66.04 ± 0.76a | 1.67 ± 1.14a | 56.78 ± 0.79b |
| 2nd leaf | 66.83 ± 1.43aE | 3.33 ± 2.56aE | 58.34 ± 1.83abE | |
| 3rd leaf | 66.08 ± 1.43a | 1.17 ± 3.19a | 59.87 ± 2.15a | |
| 4th leaf | 62.95 ± 0.96b | −3.39 ± 2.53b | 53.69 ± 3.11c | |
| CK1 | 1st leaf | 46.63 ± 1.23a | −7.67 ± 1.50ab | 27.90 ± 2.98a |
| 2nd leaf | 41.15 ± 1.60bF | −7.13 ± 0.58 aF | 24.26 ± 2.09bF | |
| 3rd leaf | 39.87 ± 2.18bc | −8.00 ± 0.26bc | 22.60 ± 2.73bc | |
| 4th leaf | 39.26 ± 1.47c | −8.82 ± 0.61c | 21.63 ± 1.39c | |
| HY2 | 1st leaf | 67.49 ± 0.64a | −1.39 ± 0.74a | 45.72 ± 4.39a |
| 2nd leaf | 64.31 ± 4.58aE | −3.53 ± 2.54bE | 48.17 ± 5.06aE | |
| 3rd leaf | 53.8 ± 4.00b | −6.17 ± 1.34c | 35.70 ± 2.93b | |
| 4th leaf | 52.18 ± 4.15b | −8.59 ± 0.41d | 34.51 ± 6.14b | |
| CK2 | 1st leaf | 41.49 ± 1.23a | −9.48 ± 0.42c | 24.97 ± 2.11a |
| 2nd leaf | 40.75 ± 0.83abF | −8.01 ± 0.89 aF | 24.19 ± 1.43 aF | |
| 3rd leaf | 39.56 ± 1.39b | −8.61 ± 0.61ab | 22.08 ± 1.69b | |
| 4th leaf | 40.66 ± 1.97ab | −8.96 ± 0.73bc | 23.91 ± 2.43ab |
L*: brightness, 0% (no reflection) for black-coloured objects and 100% for white-coloured objects; a*: redness, with negative values for green and positive values for red; and b*: yellowness, with negative values for blue and positive values for yellow. Within the same columns, a, b, c, d refer to significant difference (P < 0.05) among the different developmental stages of the same sample. E, F in the same columns between HY1 and CK1, HY2 and CK2, respectively, indicate significant difference (P < 0.01) in the 2nd leaf stage.
Profiles of chloroplasts in the variants and their CKs
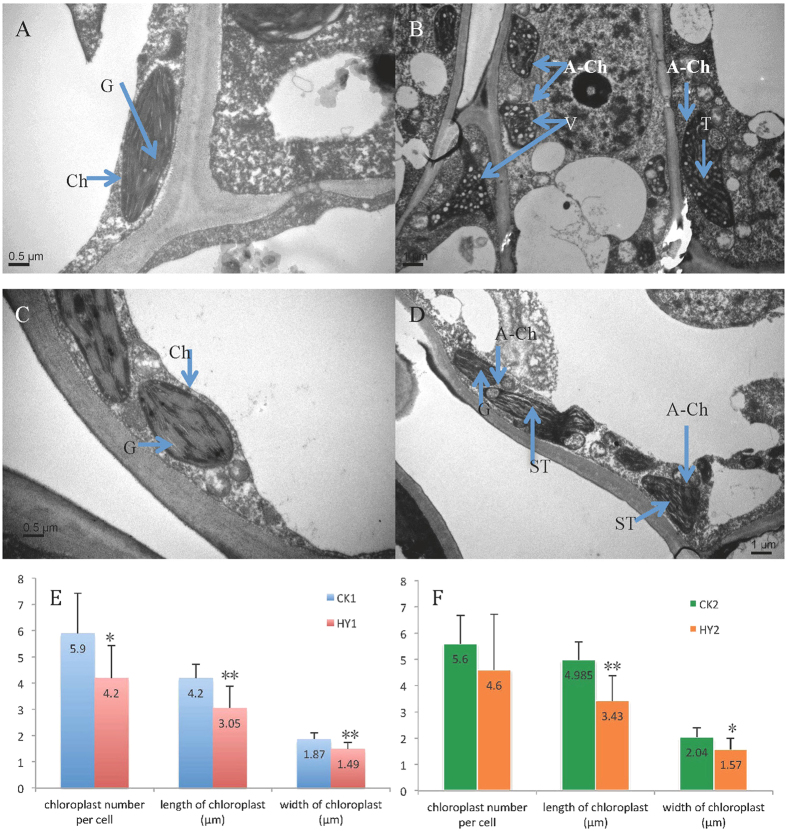
To further identify the relationship between the chloroplasts and leaf colour variations in our study, the number, size and ultrastructure of the chloroplasts were investigated using the second leaves from the one bud and four leaves stage. The chloroplasts showed well-developed membrane systems composed of grana connected by stroma lamellae in CK1 and CK2 (Fig. 2A,C). However, in HY1, the chloroplasts lacked well-structured thylakoid membranes, and some of the chloroplasts contained irregularly arranged vesicles, which led to a decrease of the number of thylakoids and the disappearance of the grana (Fig. 2B). In particular, a few chloroplasts in HY1 were almost completely filled with vesicles and almost no inner member structures (Fig. 2B). In the HY2, normal, or close normal chloroplasts, were observed; although, abnormal chloroplasts with swelling thylakoid and the disappearance of stacks of the thylakoid also existed (Fig. 2D). Moreover, the shape of the chloroplasts in CK1 and CK2 mainly displayed an ellipse, while those in HY1 and HY2 appeared as abnormal shapes (Fig. 2B,D). Additionally, there were significant differences in the number, length and width of the chloroplasts between HY1 and CK1, while in HY2 and CK2 insignificant differences in number were found (Fig. 2E,F).
Figure 2. Chloroplast profiles of the variants and the CKs.
(A–D) Chloroplast ultrastructure of the variants and the CKs; (A,C) chloroplast ultrastructure of CK1 and CK2, respectively (Bar = 0.5 μm); (B,D) Chloroplast ultrastructure of HY1 and HY2, respectively (Bar = 1 μm). (E,F) the difference of the number, length and width of the chloroplasts in the variants and the CKs; (E) a comparison of the number, length and width of the chloroplasts between HY1 and CK1; (F) a comparison of the number, length and width of the chloroplasts between HY2 and CK2. In these pictures, Ch refers to the chloroplast; G refers to the grana; A-Ch refers to an abnormal chloroplast; T refers to the thylakoid; ST refers to a swelling thylakoid; V refers to a vesicle; **indicates a significant difference (P < 0.01); *refers to a significant difference at the 0.05 level.
As described above, we found that the leaves of HY1 and HY2 gradually turned green in colour with the maturity of the leaves. As shown in Fig. S1, the dysfunctional structures of the chloroplasts gradually improved in the leaves with different maturity in HY1 and HY2, containing an increased number of lamellar structured and a well-structured thylakoid, which is consistent with the change in leaf colour. For example, in the first leaf of HY1, the chloroplast displayed a swelling thylakoid, while stacks of well-structured thylakoids were observed in the fourth leaf (Fig. S1A and S1D).
Ionomics on the leaves of the variants and the CKs
To identify the relationship between ion accumulation in leaves and leaf colour variation, ionome profiling was performed on the leaves of the variants and the CKs (Table 2). The concentrations of various macro- and microelements were observed in two comparisons. The Mn concentrations in two types of CKs were higher than in their corresponding variants. The concentrations of Na, K, Ca, Fe, As, Mo and Pb increased in two variants. The changes in the concentrations of Mg, Cr, Ni, Cu, Zn and Cd in the two comparisons showed a different tendency.
Table 2. Concentration of fourteen types of elemental minerals in the second leaves of CK1, HY1, CK2 and HY2.
| CK1 | HY1 | CK2 | HY2 | |
|---|---|---|---|---|
| Na (g/kg) | 0.09 ± 0.001B | 0.13 ± 0.004A | 0.03 ± 0.01 | 0.03 ± 0.006 |
| Mg (g/kg) | 1.85 ± 0.039B | 2.2 ± 0.035A | 2.37 ± 0.058 | 1.89 ± 0.227 |
| K (g/kg) | 18.33 ± 0.294B | 21.36 ± 0.331A | 20.6 ± 3.589 | 21.81 ± 2.95 |
| Ca (g/kg) | 2.37 ± 0.037B | 2.81 ± 0.08A | 3.63 ± 0.684 | 3.71 ± 0.509 |
| Cr (mg/kg) | 0.21 ± 0.066 | 0.28 ± 0.064 | 0.26 ± 0.023 | 0.2 ± 0.055 |
| Mn (mg/kg) | 678.61 ± 8.522 | 661.45 ± 7.215 | 464.4 ± 13.013 | 443.76 ± 39.674 |
| Fe (mg/kg) | 46.69 ± 0.947b | 49.21 ± 0.177a | 0.06 ± 0.012 | 0.07 ± 0.007 |
| Ni (mg/kg) | 3.83 ± 0.071 | 2.94 ± 0.036 | 3 ± 0.091 | 3.81 ± 0.514 |
| Cu (mg/kg) | 10.37 ± 0.248B | 11.21 ± 0.147A | 13.76 ± 0.427 | 13.63 ± 1.629 |
| Zn (mg/kg) | 22.89 ± 0.389A | 20.93 ± 0.106B | 14.84 ± 0.144B | 36.78 ± 0.808A |
| As (mg/kg) | 0.03 ± 0.004 | 0.04 ± 0.003 | 0.05 ± 0.015 | 0.06 ± 0.009 |
| Mo (mg/kg) | 0.02 ± 0.015B | 0.08 ± 0.099A | 0.02 ± 0.004 | 0.04 ± 0.02 |
| Cd (mg/kg) | 0.03 ± 0.006B | 0.06 ± 0.004A | 0.09 ± 0.016a | 0.06 ± 0.009b |
| Pb (mg/kg) | 0.37 ± 0.035b | 0.48 ± 0.029a | 0.31 ± 0.165 | 0.6 ± 0.114 |
The values, expressed as g/kg or mg/kg dry weight, are the mean ± SD of three independent experimental replicates. The different lowercase letters in the same row within the comparison (CK1 and HY1, CK2 and HY2, respectively) indicate a significant difference (P < 0.05), and the uppercase letters refer to a significant difference at the 0.01 level.
Quantitative identification of proteins from leaves of the variants and the CKs
To gain a global view of the molecular responses to leaf colour variations, total proteins in leaves were extracted from the variants and the corresponding CK branches in three independent biological experiments, and the protein expression profiles were explored using the label-free MS-based technique. We have deposited the LC-MS/MS data to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD004750. Overall, 1,185, 1,006, 2,280 and 1,836 proteins were identified in HY1, CK1, HY2 and CH2, respectively (Tables S2–5). The numbers of peptides, the mass, the sequence coverage and the description of the proteins are also provided. Additionally, among identified proteins, the number of proteins identified by one unique peptide with only one spectrum was 97, 255, 142 and 361 in CK1, CK2, HY1 and HY2, respectively, and the annotated spectra corresponding to proteins were shown in Fig. S2-5.
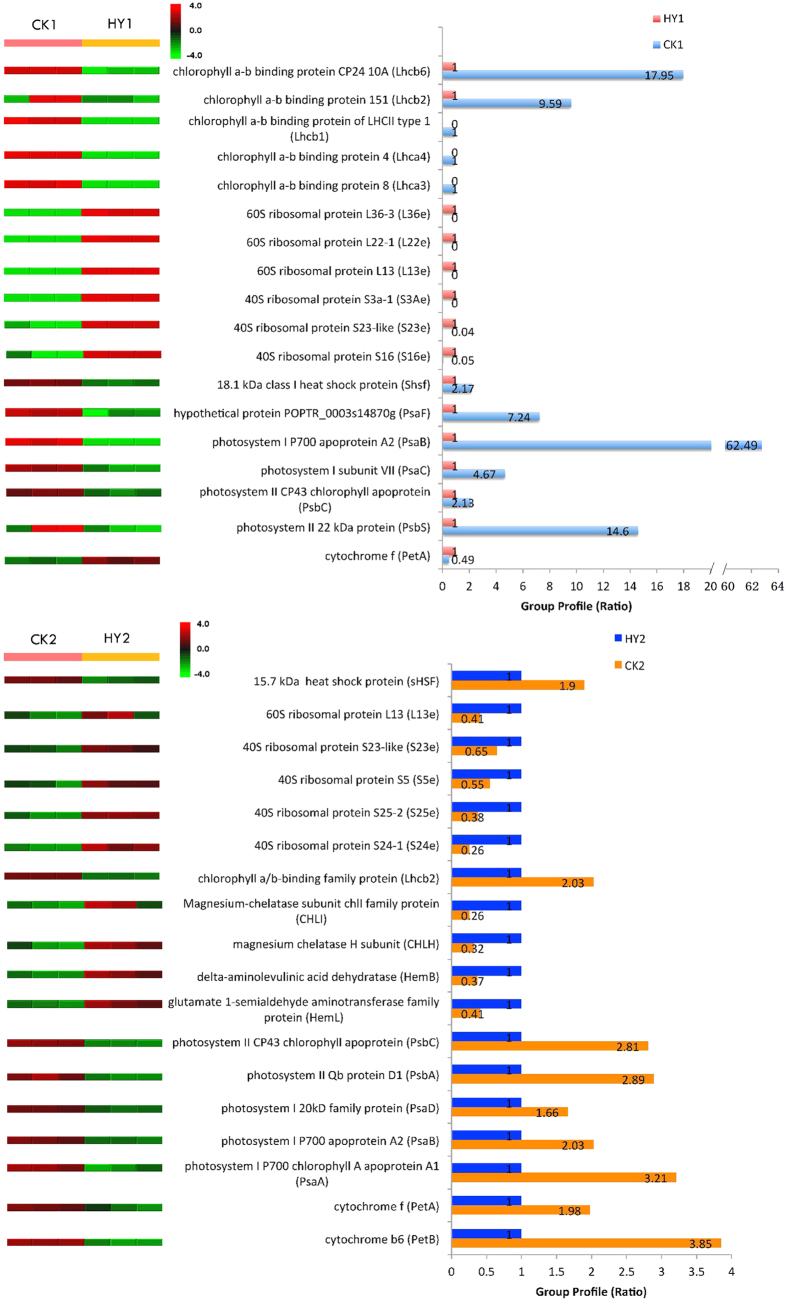
Changes in protein abundance in response to HY1 and HY2 variations were analysed, and 93 and 202 proteins were significantly different (Tables S6 and S7). Among the proteins identified, 45 and 32 proteins decreased their expression level to less than 0.67-fold with the yellow and variegation variations, respectively. However, the expression level of 48 and 170 proteins increased in HY1 and HY2, respectively (Tables S6 and S7). For example, as shown in Fig. 3, the expression of chlorophyll a-b binding proteins, heat shock proteins and proteins related to photosystem all decreased in HY1 and HY2, whereas the expression of ribosomal proteins in the two types of variants was significantly increased. However, cytochrome f protein showed a different expression pattern in the two comparisons, and four proteins related to chlorophyll synthesis, including CHLI, CHLH, HemB and HemL, were only found in the comparison between HY2 and CK2 (Fig. 3; Figs S6 and S7; Tables S6 and S7).
Figure 3. Quantitative comparison of several differentially expressed proteins between the variants and the CKs.
The right panels represent the fold change of the individual protein, and the left panels represent the reproducibility in three independent biological experiments. The up- or down-regulated proteins are indicated by the red and green colour code, respectively. The colour intensity changes with the protein expressional level.
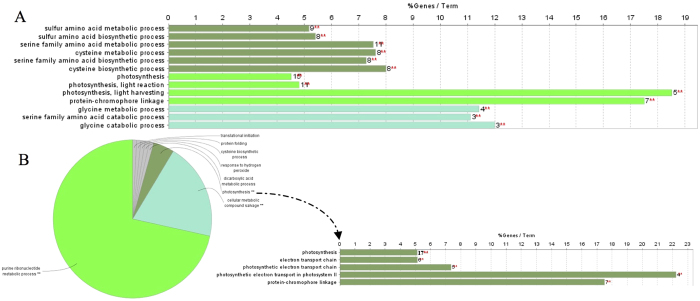
To elucidate the possible different biological events behind the proteomic data, all the DEPs (differentially expressed proteins) in the two comparisons were translated into Vitis vinifera orthologues (Tables S8 and S9) and were analysed using ClueGO with the Vitis vinifera database to detect the significantly enriched GO terms. In the comparison of HY1 and CK1, serine family amino acid metabolic process, photosynthesis and glycine metabolic process were significantly enriched (Fig. 4A). In another comparison, the proteins were enriched in eight major functional groups, and among these, photosynthesis was also significantly enriched (Fig. 4B; Fig. S8).
Figure 4. ClueGO analysis of differentially expressed proteins.
Single (*) or double (**) asterisk indicate significant enriched GO terms at the p < 0.05 and p < 0.01 statistical levels, respectively. The numbers of corresponding genes associated with a specific term are indicated. The percentage of genes associated with a specific term is listed on the bars. (A) Enriched GO terms of differentially proteins identified from the comparison of HY1 and CK1. (B) The left pie chart indicates overview specific cluster of differentially expressed proteins between HY2 and CK2. The right represents specific Go terms related to photosynthesis.
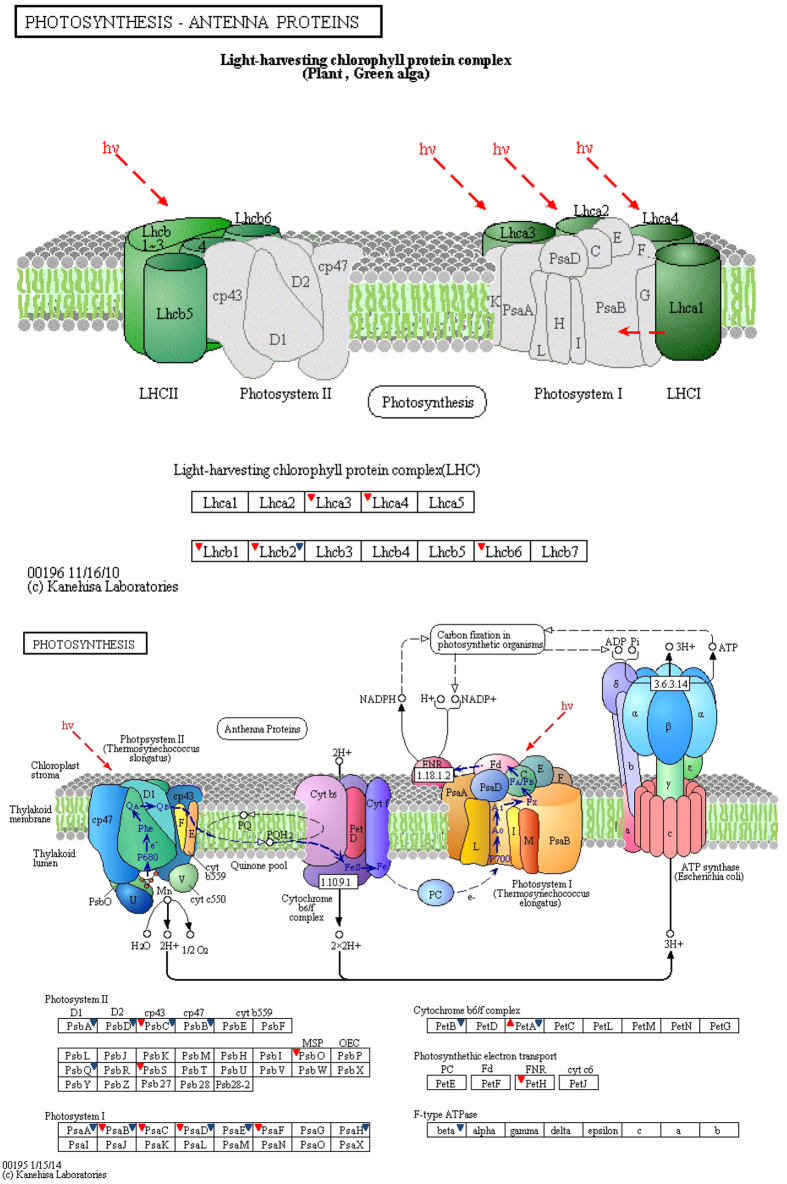
For a better understanding of the biological process of the DEPs, these proteins were further investigated using the KEGG database. The DEPs in the two comparisons were mapped to 50 and 63 KEGG pathways, respectively (Tables S10 and S11). The biological pathways involved in ribosome (16), photosynthesis (9) and photosynthesis – antenna proteins (5) were significantly enriched between HY1 and CK1 (Table 3). Moreover, four processes, including carbon metabolism (22), carbon fixation in photosynthetic organisms (11), photosynthesis (13) and glycolysis/gluconeogenesis (12), were significantly enriched between HY2 and CK2 (Table 3). Additionally, one protein derived from the DEPs between HY2 and CK2 was also annotated in photosynthesis – antenna proteins (Table S11). Moreover, almost all the proteins mapped in the photosynthesis pathway showed decreased expression in the two variants compared to their CKs, except for cytochrome f between CK1 and HY1 (Table 4). To survey the differences in photosynthesis between the variants and the CKs, we overlaid each protein profile onto a photosynthesis pathway (Fig. 5). The results showed that 14 differentially expressed proteins were associated with photosynthesis between HY1 and CK1, including 4 proteins in PSI, 3 proteins in PSII, 1 protein in the cytochrome b6/f complex, 1 protein in the photosynthetic electron transport, and 5 proteins in LHC; whereas between HY2 and CK2, 14 proteins, including 5 proteins in PSI, 5 proteins in PSII, 2 proteins in the cytochrome b6/f complex, 1 protein in the F-type ATPase, and 1 protein in LHC, were observed. These proteins may therefore be associated with the leaf colour variations.
Table 3. Significantly enriched pathways among differentially expressed proteins.
| Pathway | Number of the mapped DEPs | Number of reference proteins | Corrected P value | |
|---|---|---|---|---|
| HY1 VS. CK1 | Ribosome | 16 | 256 | 0.001225464 |
| Photosynthesis | 9 | 84 | 0.001225464 | |
| Photosynthesis- antenna proteins | 5 | 19 | 0.001225464 | |
| HY2 VS. CK2 | Carbon metabolism | 22 | 156 | 0.001632539 |
| Carbon fixation in photosynthetic organisms | 11 | 52 | 0.004375366 | |
| Photosynthesis | 13 | 84 | 0.009634559 | |
| Glycolysis/Gluconeogenesis | 12 | 74 | 0.009634559 |
Table 4. Identified differentially expressed proteins involved in photosynthesis.
| Accession | Protein | Gene | CK1/HY1 (Ratio) | CK2/HY22 (Ratio) |
|---|---|---|---|---|
| gi|671743315 | photosystem II CP43 chlorophyll apoprotein (chloroplast) | PsbC | 2.13:1.00 | 2.81:1.00 |
| gi|225459564 | PREDICTED: photosystem II 22 kDa protein chloroplastic | PsbS | 14.60:1.00 | / |
| gi|224084209 | O2 evolving complex 33kD family protein | PsbO | 2.99:1.00 | / |
| gi|552540956 | photosystem II Qb protein D1 (chloroplast) | PsbA | / | 2.89:1.00 |
| gi|568244554 | photosystem II protein D2 (plastid) | PsbD | / | 2.02:1.00 |
| gi|542688125 | photosystem II p680 chlorophyll A apoprotein CP-47 (chloroplast) | PsbB | / | 2.49:1.00 |
| gi|224078826 | Oxygen-evolving enhancer protein 3-1 | PsbQ | / | 1.50:1.00 |
| gi|671743230 | photosystem I P700 apoprotein A2 (chloroplast) | PsaB | 62.49:1.00 | 2.03:1.00 |
| gi|671743459 | photosystem I subunit VII (chloroplast) | PsaC | 4.67:1.00 | / |
| gi|566184073 | photosystem I 20kD family protein | PsaD | 2.49:1.00 | 1.66:1.00 |
| gi|566162704 | hypothetical protein POPTR_0003s14870g | PsaF | 7.24:1.00 | / |
| gi|552541026 | photosystem I P700 chlorophyll A apoprotein A1 (chloroplast) | PsaA | / | 3.21:1.00 |
| gi|225437898 | PREDICTED: photosystem I reaction center subunit IV B chloroplastic-like | PsaE | / | 2.08:1.00 |
| gi|224073967 | photosystem I 11 K family protein | PsaH | / | 1.76:1.00 |
| gi|552540866 | Cytochrome f (chloroplast) [Camellia danzaiensis] | PetA | 0.49:1.00 | 1.98:1.00 |
| gi|359492254 | PREDICTED: ferredoxin–NADP reductase leaf-type isozyme chloroplastic | PetH | 1.84:1.00 | / |
| gi|671743260 | cytochrome b6 (chloroplast) [Camellia petelotii] | PetB | / | 3.85:1.00 |
| gi|225436257 | PREDICTED: chlorophyll a-b binding protein 8 chloroplastic | Lhca3 | 1.00:0 | / |
| gi|225457389 | PREDICTED: chlorophyll a-b binding protein 4 chloroplastic | Lhca4 | 1.00:0 | / |
| gi|359483839 | PREDICTED: chlorophyll a-b binding protein of LHCII type 1 | Lhcb1 | 1.00:0 | / |
| gi|225447576 | PREDICTED: chlorophyll a-b binding protein 151 chloroplastic | Lhcb2 | 9.59:1.00 | 2.03:1.00 |
| gi|225447745 | PREDICTED: chlorophyll a-b binding protein CP24 10A chloroplastic | Lhcb6 | 17.95:1.00 | / |
| gi|542688129 | ATP synthase CF1 beta subunit (chloroplast) | beta | / | 1.61:1.00 |
“/” indicates not detect in the database.
Figure 5. Mapping differentially expressed proteins with known Photosynthesis pathway.
The images of known photosynthesis pathway were obtained from freely available KEGG database (www.kegg.jp). Regular triangle with red colour refers to higher expression in HY1 compared with CK1, while inverted triangles with red colour indicate the higher expression in CK1. Blue inverted triangles refer to lower expression of proteins in HY2 compared to CK2.
Transcriptional expression analysis of the differentially expressed proteins
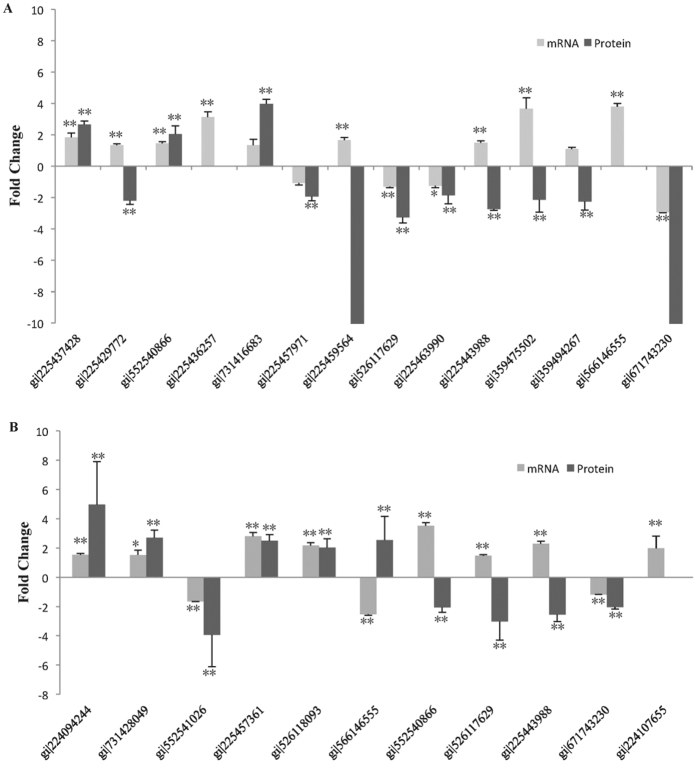
In order to assess the correlation of the expression levels between mRNA and protein, fourteen and eleven proteins were randomly selected, respectively, in the two comparisons and were analysed by quantitative RT-PCR (Fig. 6). Between HY1 and CK1, the expression of seven genes (gi|225437428, gi|552540866, gi|731416683, gi|225457971, gi|526117629, gi|225463990 and gi|671743230) is consistent with the corresponding proteins, while six genes (gi|224094244, gi|731428049, gi|552541026, gi|225457361, gi|526118093 and gi|671743230) showed similar protein and mRNA expression patterns in the comparison of HY2 and CK2 (Fig. 6; Tables S6 and S7). Additionally, the proteins (gi|225436257 and gi|566146555) were not detected in HY1 and CK1, respectively, while the protein (gi|224107655) was not detected in CK2. And the fold change in proteins expression of gi|225459564 and gi|671743230 more than ten. The QRT-PCR demonstrated that these genes displayed similar protein and mRNA expression patterns with divergent quantitative values. Additionally, other genes showed different expression patterns between the mRNAs and proteins, which might be a result of posttranscriptional and posttranslational regulatory processes.
Figure 6. Quantitative real time PCR analyses of fourteen and eleven differentially expressed proteins at mRNA level, respectively.
(A) The black and the gray bars represent fold changes of protein and mRNA between HY1 and CK1, respectively. The positive values indicate higher expression in the HY1 and negative values denote higher expression in CK1. (B) The black and the gray bars represent fold changes of protein and mRNA between HY2 and CK2, respectively. The positive values indicate higher expression in the HY2 and negative values denote higher expression in CK2. Error bar is standard deviation. *indicates significant difference (P < 0.05) at mRNA level, **refers to significant difference at 0.01 level at mRNA and protein level.
Discussion
Leaves with a green colour are the primary sites of photosynthesis and contribute to the biosynthesis of plant biomass and energy24,25. Currently, two types of abnormal branches showing yellow and variegated colour in leaves are observed from the tea cultivar “Yinghongjiuhao,” which leads to more uniform fermenting and a better presentation of tea in production; however, the mechanism of these variations is not fully understood. Accordingly, to reveal the mechanism, combined morphological, histological, ionomic and proteomic analyses were performed using leaves from the variants and their corresponding CKs. We found that the leaf colour in HY1 and HY2 both exhibited gradual greening along with increasing maturity. Moreover, abnormal chloroplast structures and a reduced number and size of chloroplasts were observed in the two variants. Finally, the difference in the concentration of elemental minerals and the protein expression between the variants and the CKs might be associated with the leaf colour changes.
The present variants are suitable materials for analysing the mechanism of leaf colour variations
The mechanism of leaf colour variations is a complicated biological process. As an excellent model in such previous studies, leaf colour mutants have gained more and more attention. In the present study, two types of variants, showing yellow leaf and variegated leaves, were selected as the materials. They were suitable for mechanism exploration because they are of the same genetic background compared with the CK branches. Furthermore, the leaf colour variations in HY1 and HY2 might be caused by some unknown mutation(s) and might belong to bud mutation, because of the very low frequency, the random and the stability of leaf colour variations. Thus, in our opinion HY1 and HY2 might be mutants derived from the CKs. Moreover, leaf colour was traditionally identified by subjective judgements in previous studies of leaf colour variations. In this study, the leaf colour variations were quantified from colour parameters (CIE Lab) using a chromameter, which facilitated objective results and helped us to accurately distinguish the differences of leaf colour among the different samples, especially the samples from the different developmental stages. The above results indicate that suitable samples were prepared for mechanism exploration of leaf colour variations in tea plants.
Leaf colour reflected the developmental characteristics of the chloroplast
Chloroplasts, composed of chloroplast membrane, thylakoid and matrix, is essential for carbon assimilation and amino acid synthesis26. The chloroplast profiles, including the structure, number and size are relatively stable, but they are apt to be influenced by genetic and environmental conditions27. Most affected plants with dysfunctional chloroplasts usually have leaves that lose their green colour. Thus, changes in leaf colour might reflect the abnormal development and function of the plastid. In our study, abnormal chloroplast structures and the improvement of abnormal chloroplasts structure were observed through lateral and vertical comparative assessments, respectively (Fig. 2; Fig. S1), which strongly suggests a relationship between the chloroplast and leaf colour. A previous study reported that abnormal chloroplasts might only be located on the variational positions, and our data showed that both normal and abnormal chloroplasts existed in the leaf of HY2, which could explain the phenotype of the variegated leaf. In addition, more types of abnormal chloroplasts were observed in the present variants than in those reported by Wang and Li6,7,8, and the number and size of the chloroplasts was firstly analysed in the leaf colour variants of the tea plants (Fig. 2). In HY1, a significantly decreased size of the chloroplast and a decreased number were observed, which indicates that the size and number of the chloroplasts are also related to leaf colour variations. These multiple comparisons provide more convincing results than in previous studies6,7,8. These findings demonstrate that the interruption of chloroplast development might be tightly linked to occurrence of the leaf colour variations. Additionally, the metabolism of ion might be influenced in leaves of variants, and several kinds of ions were associated with the chloroplast development. Based on the present data, the differences in the concentration of the elemental minerals in leaves were observed. Furthermore, the changes in concentration of Mn and Zn may be associated with the chloroplast development. For example, Zn plays a role in the formation of chloroplasts, and Mn is essential in the formation and maintenance of the normal structure of chloroplasts28.
Proteins expression patterns and leaf colour variations
The present variants showed abnormal leaf colour and chloroplast profiles, while also displaying changes in protein expression patterns. Proteomic profiling provides information regarding quantitative changes in protein expression, which will advance our understanding of the role of proteins involved in leaf colour variations. For tea plants, there have been only two reports regarding a protein analysis for the changes in leaf colour. One of the studies identified twenty-six differentially expressed proteins in three developmental stages of the albino tea cultivar using a comparative proteomic approach based on two-dimensional electrophoresis and mass spectrometry. However, the disadvantage of the 2D gel technique limited its application to a comprehensive analysis of proteome changes29,30,31,32. Fortunately, MS-based high-resolution proteomic approaches are a powerful tool for large-scale protein identification and quantitation and are successfully utilized for the comprehensive characterisation of the proteome. Currently, two main types of relative quantification strategies for MS-based proteomics analysis exist, including label-based and label-free MS-based approaches33. In another study of leaf colour from tea plants, iTRAQ (label-based) was used to analyse the differentially expressed proteins, which overcame the disadvantages of the 2D gel technique and identified more proteins8. It is reported that MS-based label-free quantitative proteomics studies are reliable, versatile, and a cost-effective alternative to label-based quantitation22, and the label-free method allows for a qualitative analysis based on the number of identified proteins. Conversely, the iTRAQ methodology does not allow for the identification of unique proteins because the protein ratio is only calculated when the protein is present in both of the tested conditions34. Additionally, Latosinska et al.35 reported that the label-free strategy provided a higher protein sequence coverage and ultimately detected a higher number of significant changes compared to the iTRAQ experiment35. Therefore, in this study, we adopted a quantitative proteomic method using the label-free MS based system, and a total of 6307 proteins were identified in the leaves of the variants and the corresponding CKs (Tables S2–S5). In agreement with a previous study, several unique proteins were only detected in one sample, such as chlorophyll a-b binding protein of LHCII type 1 (gi|359483839) and 3-oxoacyl-[acyl-carrier-protein]reductase (gi|224107655), which were identified in HY1 and CK1 comparison and in HY2 and CK2 comparison, respectively (Tables S6 and S7). Thus, the greater number of proteins identified in our study will provide an effective information pool to identify the related proteins involved in leaf colour variations. In our study, two types of variants sharing the same genetic background were selected to analyse the differences in the proteins, which facilitated the identification of the proteins involved in leaf colour variations because of the common property between the yellow and variegated leaf.
The chloroplast profile assays demonstrated that an abnormal structure was observed in HY1 and HY2 compared with the CKs (Fig. 2). Thus, we speculate that changes in protein expression might be related to the chloroplast development in the two variants. The biogenesis of chloroplasts is a complex process that is tightly regulated by the coordinated expression of both nuclear and plastid genes and requires communication among the nucleus, cytoplasm, chloroplast, and mitochondrion36,37. To date, many genes involved in chloroplast development have been identified, such as the plastid-encoded genes Lhcb, rbcL, rbcS, psaA and psbA38,39. Several lines of evidence demonstrate that the chlorotic phenotype is closely associated with a reduced level of plastid genes40,41,42. For example, impaired grana stacking and a reduced number of chloroplasts in the mutants are a result of the significantly decreased level of LHC proteins43,44. In our study, the expression levels of the proteins corresponding to the above five plastid-encoded genes, chlorophyll a-b binding protein, ribulose bisphosphate carboxylase, photosystem I P700 chlorophyll A apoprotein A1, and photosystem II Qb protein D1, were remarkably repressed in the HY2 (Table 4). Additionally, heat shock protein also affects the chloroplast development by regulating protein folding and protein transport45,46. Consistent with the previous findings, reduced expression of the heat shock proteins in the variants was also observed in our study (Fig. 3; Tables S6 and S7). Thus, we speculated that the abnormal chloroplast profiles in the present variants might be associated with the downregulation of the above-mentioned proteins.
Photosynthesis takes place in the chloroplast. Thus, abnormal chloroplast profiles might be linked to a dramatic down-regulation of proteins related to the photosystem. Consistent with this theory, the DEPs were both significantly enriched in the photosynthesis pathway in the two comparisons. The expression of these DEPs, including PSI subunits, PSII subunits, antenna proteins, cytochrome b6/f complex, and the beta F-type ATPase, was decreased in HY1 and/or HY2 (Fig. 5). The relationship between the leaf colour variations and the expression patterns of proteins was revealed through the proteomic analysis, which facilitated the exploration of the mechanism of leaf colour formation.
Conclusions
In conclusion, two types of leaf colour variations (yellow and variegated leaf colour) are observed from the tea cultivar “Yinghongjiuhao” in our study. To reveal the mechanisms, combined morphological, histological, ionomic and proteomic analyses were performed between variants and their corresponding CKs. The measurement of the CIE-Lab coordinates showed that HY1 (yellow colour in leaves) and HY2 (variegated colour in leaves) exhibited more brightness and yellowness according to the L* and b* colour parameters, and gradual greening was observed with increasing maturity. Moreover, the results of the chloroplast analyses indicated that abnormal chloroplast development was related to leaf colour variations. In addition, the changes in the concentration of elemental minerals may be associated with the leaf colour variations. Finally, a quantification of proteins with leaf colour variations was performed. A total of 93 and 202 DEPs in HY1 and HY2, respectively, were identified. Many pathways potentially associated with chloroplast development were revealed, such as photosynthesis. Further studies should mainly focus on the origin of the different observations showed in our present results between variants and CKs. We speculated that the ultimate cause of phenotype variations in our study must be a mutation (or mutations), which led to a series of changes including organelle and molecular levels and might ultimately regulate the formation of leaf colour in tea plants.
Materials and Methods
Plant Materials
The two leaf-colour variation branches of tea (Camellia sinensis) were derived from the cultivar “Yinghongjiuhao,” which was grown in the field of the Tea Research Institute at the Guangdong Academy of Agricultural Sciences in Yingde, China. The branches with yellow leaves and variegated leaves were named HY1 and HY2, respectively, and the remaining normal corresponding branches of the same plant were named CK1 and CK2 (See the details in Fig. 1). Additionally, the very low frequency of variations (approximately 0.001%) was observed in our tea plantation (about 800000 plants) based on three years statistics. All the leaves samples were harvested with three independent biological replicates, and the specific samples used for the different analyses were derived from the same sample pool.
Leaf colour determination of the variants and the CKs
To better illustrate the leaf colour differences between the variants and the CKs, the leaves at the developmental stage of one bud and four leaves were harvested as samples. The colour coordinates (CIE Lab) of the sampled leaves were quantified by a portable spectrophotometer (Datacolor CHECK II) (Datacolor, US). The leaf colour variations were also detected using leaves from four leaf positions of the variants and the CKs, which illustrate the changes in the leaf colour corresponding with the developmental stages. To determine leaf colour exactly, the samples were scanned in triplicate at three different locations (sections) away from the main leaf vein. Following the measurement, the mean of nine values was calculated as the averaged colour data for each sample. The statistical analyses were performed by one-way ANOVA (SPSS version 19.0, SPSS, Inc.) using Duncan’s multiple-range test.
Transmission electron microscopic (TEM) analysis of the chloroplasts
The ultrastructure of the chloroplasts from the leaves at the different developmental stages in the variants and the CKs was investigated via TEM, which clarified the basic explanation for the occurrence of varied characters. The samples were collected from the four leaf positions as described above. The differences between the variants and the CKs were elucidated using the second leaf. In addition, the profiles of the chloroplasts in the variants at different developmental stages were also analysed. The fresh samples were cut into 2 mm-wide pieces and were fixed with 2.5% glutaraldehyde overnight at 4 °C. Following a wash with phosphate buffer (0.1 M, pH 7.0, three times) for 15 min each time, the samples were incubated in 1% OsO4 for 2 h and washed again with 0.1 M phosphate buffer three times. Subsequently, the samples were subjected to a dehydration series in ascending concentrations of ethanol (30%, 50%, 70%, 80%, 90% and 95%), with each concentration step lasting 15 min, and were then soaked in 100% ethanol for 20 min. The dehydrated samples were drenched in acetone for 20 min followed by a drenching in epoxy resin and acetone (v/v = 1/1) for 1 h and epoxy resin and acetone (v/v = 3/1) for 3 h. The samples were finally embedded in a pure epoxy resin at 70 °C overnight. The thick sections (90 nm) were cut and stained with lead citrate and uranyl acetate in 50% ethanol for 10 min, respectively, and were visualised with a Hitachi H-7650 transmission electron microscope (Hitachi, Tokyo, Japan).
Multi-element analysis
To identify the relationship between elemental minerals and leaf colour variation, ionomics were performed using one bud and two leaves from the variants and the CKs. The freeze-dried samples were digested with HNO3-H2O2, and the final volume was 50.0 ml. Then, the metal concentrations of the digests were measured using an Agilent 7700x ICP-MS (Agilent Technologies, Palo Alto, CA). The ICP-MS operating parameters for the analysis were as follows: RF power 1500 W; RF matching 1.8 V; carrier gas 1.08 L/min; plasma gas 14.97 L/min; nebulizer pump 0.1 rps; and sample depth 8 mm. All the samples and the blank solutions were measured in triplicate. The statistical significance of differences of concentration of ions within the comparison were analysed using one-way ANOVA (SPSS version 19.0, SPSS, Inc.).
Protein Sample Preparation
Fresh leaves from the four materials (two variants and two CKs) in three independent biological replicates at the development stage of one bud and two leaves were collected for the proteomic analysis. Approximately 3 g of leaves from each biological replicate with 0.4 g of PVP (polyvinylpyrrolidone) and 0.1 g of DTT (dithiothreitol) were ground into a fine powder in liquid nitrogen and were suspended in 8 volumes of ice-cold acetone containing 10% (v/v) TCA (trichloroacetic acid) and 0.07% (v/v) β-Me (β-mercaptoethanol). After an intermittent ultrasonic treatment that was repeated 50 times (time for each treatment was 5 s, with a 15 s interval) on ice, the mixture was kept at −20 °C for 1 h and centrifuged at 15,000 x g for 15 min at 4 °C. Then, the supernatant was discarded, and the pellets were washed twice with 6 volumes of ice-cold acetone containing 0.07% β-Me, vacuum-dried and stored at −80 °C. The dried protein powders (50 mg of each sample) were re-suspended in 750 μL of lysis buffer (8 M urea, 30 mM DTT, 4% CHAPS, 2 M thiourea, and 20 mM Tris-base), vortexed, incubated at 37 °C for 2 h and centrifuged at 15,000 x g for 15 min at 4 °C. Four volumes of ice-cold acetone (containing 5 mM β-Me) were added to the above collected supernatant, and the mixture was kept at 4 °C for 2 h for protein precipitation and desalting. Subsequently, the mixture was centrifuged at 15,000 x g and 4 °C for 15 min. The supernatant was discarded, and the pellets were vacuum-dried. The pellets were dissolved in the lysis buffer described above and were incubated at 37 °C for 2 h. After centrifugation at 15,000 x g and 4 °C for 15 min, an aliquot of each supernatant was used to determine the protein concentration using the Bradford assay. A total of 1.5 mg of the dried pellets was dissolved thoroughly in 100 μL of 5 M urea buffer, and then 400 μL of a 40 mM NH4HCO3 buffer was added to the mixture. The proteins were reduced with 50 μL of 100 mM DTT and were then alkylated with 100 mM iodoacetamide. The proteins were digested using trypsin at a mass ratio of 1:50 (enzyme/protein) at 37 °C for 14 h. The enzymatic digestion was stopped by adding 1 μL of formic acid to the solution, and the samples were then vacuum-dried using a SpeedVac system (RVC 2-18, Marin Christ, Osterod, Germany). The samples were redissolved in 0.1% formic acid. The sample solution was centrifuged, and the supernatant was collected for the MS/MS analysis.
Nano-LC−MS/MS Analysis
The analysis was performed according to a method described by Han, et al.47. Briefly, each of the biological replicates was analysed using a Q-Exactive mass spectrometer coupled to a two-column EASY-nLC 1000 nanoflow system (Thermo Fisher Scientific). The samples were loaded onto a 2-cm long, 100-μm inner diameter fused silica trap column containing 5.0 μm Aqua C18 beads (Thermo Fisher Scientific) for 2 min in buffer A (0.1% acetic acid) at a flow rate of 5 μL/min prior to the analytical separation. A 15-cm fused silica trap column (75 μm i.d.) filled with 3.0 μm Aqua C18 beads (Thermo Fisher Scientific) was selected as the stationary phase, while 0.1% formic acid (A) and 0.1% formic acid in 80% acetonitrile (B) was chosen as the binary mobile phase. The peptides were separated by the following gradient elution: from 0 to 8% solvent B for 5 min; from 8 to 20% solvent B for 55 min; from 20 to 30% solvent B for 10 min; from 30 to 100% solvent B for 10 min; and 100% solvent B for 15 min. The eluate from the column was analysed with a Q-Exactive mass spectrometer (Thermo Fisher Scientific) using an interface with an ESI ionisation source. The MS and MS/MS information was collected in data-dependent mode using the following settings: one full scan (resolution 70 000 at m/z 400; m/z300−1800) followed by the top 20 MS/MS scans using a high-energy collision dissociation in the linear ion trap mass spectrometer (resolution: 17 500, isolation window: 2 m/z, normalized collision energy: 27) using dynamic exclusion (charge exclusion: unassigned 1, > 8; peptide match: preferred; exclude isotopes: on; dynamic exclusion: 10 s).
Protein Identification and Label-Free Abundance Quantitation
The raw peptide data were retrieved using Xcalibur software (version 2.2, Thermo Fisher Scientific) and were analysed and processed using the in-house PEAKS software (version 7.0, Bioinformatics Solutions). A composite database was downloaded with a total of 85,307 entities, containing 38,134 protein sequences of Vitis vinifera, 45,942 protein sequences of Populus trichocarpa and 1,231 sequences from Camellia. The search criteria were as follows: parent ion mass tolerance, 20.0 ppm; fragment ion mass tolerance, 0.05 Da; enzyme, trypsin; allowing a nonspecific cleavage at neither end of the peptide; maximum missed cleavages per peptide, 2; and maximum allowed variable post translational modification (PTM) per peptide, 3. A fusion target-decoy approach was used for the estimation of the false discovery rate (FDR) and was controlled at ≤1.0% (−10 log P ≥ 20.0) at both the protein and peptide level.
The relative quantification of the peptide abundance level was performed by the label-free approach in the PEAKS Q module. Feature detection was performed separately on each sample by using the expectation-maximization algorithm48. The features of the same peptide from different samples were aligned together using a high-performance retention time alignment algorithm49. The identification results were chosen so that they would attach as the last step of the label-free quantification. The peptide features and proteins were considered to be significantly changed between different samples using statistical p value < 0.01 and a fold change ≥ 1.5.
Bioinformatics Analysis
To create an expressional profile of the DEPs, unsupervised hierarchical clustering was analysed (gene cluster 3.0) using an uncentered Pearson correlation and an average linkage and was visualised by Java Treeview software.
To better understand the biological implications of the DEPs, we assigned the Vitis vinifera orthologues of the tea DEPs a UniProt identifier by searching the UniProt database, due to the high homology of the amino acid sequences between the tea plant and Vitis vinifera, and the more effective database of Vitis vinifera. Subsequently, the unique identifiers of the Vitis vinifera orthologues of the tea DEPs were used as an input for functional analyses using CluGO (version 2.1.6), a Cytoscape plug-in50. The significantly enriched, functional gene ontology (GO) categories in biological processes were reported using a right-sided hypergeometric test. This test compares the background set of the GO annotations in the entire Vitis vinifera database and then assesses the FDR by a Bonferroni stepdown test to correct the probability value. The results are represented visually in graphical form.
Additionally, the DEPs were analysed by the KEGG Orthology - Based Annotation System (KOBAS 2.0, http://kobas.cbi.pku.edu.cn) to significantly enrich the identified proteins in biological pathways. The DEPs were firstly translated into the Vitis vinifera orthologues and were blasted against the Vitis vinifera database, and the pathway enrichment was then conducted by a hypergeometric statistical test. Only a corrected p value < 0.05 was considered a statistically significant enriched biological pathway.
Quantitative Real-Time RT-PCR
Total RNA was extracted from one bud and two leaves of HY1, CK1, HY2 and CK2 using the RNAprep Pure Plant Kit (TIANGEN) and was converted to cDNA for qRT-PCR using the PrimeScript RT Reagent Kit (Takara). All the amplification reactions were performed with specific primers (Table S1) using 20-μl volumes of SYBR Green (Takara) and the ABI StepOne Real-Time PCR system. The 18 S ribosomal RNA gene was selected as a reference gene for normalisation7,8. Three biological replicates were included for each sample, and three technical replicates per each replicate were performed. And finally the fold change was calculated for each gene.
Additional Information
How to cite this article: Ma, C. et al. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 6, 33369; doi: 10.1038/srep33369 (2016).
Supplementary Material
Acknowledgments
This work was supported by the construction of industrial technology innovation platform for black tea of Guangdong big leaves species, and National Natural Science Foundation of China (grant no. 31500563).
Footnotes
Author Contributions A.M. and J.C. designed the experiments; C.M. and A.M. analyzed the characteristics and chloroplast profiles; C.M. and J.L. performed protein experiment; B.Z. and J.T. performed ionomics experiment; C.M. wrote the paper.
References
- Zhang F., Luo X., Hu B., Wan Y. & Xie J. YGL138(t), encoding a putative signal recognition particle 54kDa protein, is involved in chloroplast development of rice. Rice 6, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant physiology 145, 29–40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel T. G., Meehl J. B. & Staehelin L. A. Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant physiology 112, 821–832 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Jia J., Xia C., Liu X. & Kong X. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breeding science 63, 169–175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang M., Zhang Y., Du L. & Pan T. A chlorophyllreduced seedling mutant in oilseed rape, Brassica napus, for utilization in F1 hybrid production. Plant Breeding 119, 131–135 (2000). [Google Scholar]
- Li Q. et al. Proteomic analysis of young leaves at three developmental stages in an albino tea cultivar. Proteome science 9, 44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC plant biology 14, 352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. Journal of proteomics 130, 160–169 (2016). [DOI] [PubMed] [Google Scholar]
- Sakuraba Y. et al. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. The Plant journal: for cell and molecular biology 74, 122–133 (2013). [DOI] [PubMed] [Google Scholar]
- Deng X. J. et al. Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PloS one 9, e99564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C. D., Coe E. H. Jr. & Martienssen R. A. Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. The EMBO journal 11, 4037–4046 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt U., Grimm B. & Hortensteiner S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant molecular biology 56, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- Kato Y., Miura E., Matsushima R. & Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant physiology 144, 952–960 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. H. et al. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant & cell physiology 44, 463–472 (2003). [DOI] [PubMed] [Google Scholar]
- Chen G., Bi Y. R. & Li N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. The Plant journal: for cell and molecular biology 41, 364–375 (2005). [DOI] [PubMed] [Google Scholar]
- Du Y. et al. Effect of temperature on accumulation of chlorophylls and leaf ultrastructure of low temperature induced albino tea plant. African Journal of Biotechnology 7, 1881–1885 (2008). [Google Scholar]
- Sharma R. R., Patel V. B. & Krishna H. Relationship between light, fruit and leaf mineral content with albinism incidence in strawberry (Fragaria x ananassa Duch.). Scientia Horticulturae 109, 66–70 (2006). [Google Scholar]
- Kumar S., Asif M. H., Chakrabarty D., Tripathi R. D. & Trivedi P. K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Functional & integrative genomics 11, 259–273 (2011). [DOI] [PubMed] [Google Scholar]
- Chen L., Zhou Z.-X. & Yang Y.-J. Genetic improvement and breeding of tea plant (Camellia sinensis) in China: from individual selection to hybridization and molecular breeding. Euphytica 154, 239–248 (2006). [Google Scholar]
- Du Y. et al. A study on the chemical composition of albino tea cultivars. Journal of Horticultural Science and Biotechnology 81, 809–812 (2006). [Google Scholar]
- Lee J. & Koh H.-J. A label-free quantitative shotgun proteomics analysis of rice grain development. Proteome science 9, 1–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson K. A. et al. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553 (2011). [DOI] [PubMed] [Google Scholar]
- Mohayeji M. et al. Heterosis profile of sunflower leaves: a label free proteomics approach. Journal of proteomics 99, 101–110 (2014). [DOI] [PubMed] [Google Scholar]
- Evans J. R. & von Caemmerer S. Enhancing photosynthesis. Plant physiology 155, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant physiology 155, 125–129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. et al. Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta 236, 1165–1176 (2012). [DOI] [PubMed] [Google Scholar]
- Mullet J. E. Dynamic regulation of chloroplast transcription. Plant physiology 103, 309–313 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U. M., Sareen P., Sengar R. S. & Kumar A. Plant ionomics: a newer approach to study mineral transport and its regulation. Acta Physiologiae Plantarum 35, 2641–2653 (2013). [Google Scholar]
- Ciambella C., Roepstorff P., Aro E. M. & Zolla L. A proteomic approach for investigation of photosynthetic apparatus in plants. Proteomics 5, 746–757 (2005). [DOI] [PubMed] [Google Scholar]
- Wittmann-Liebold B., Graack H. R. & Pohl T. Two-dimensional gel electrophoresis as tool for proteomics studies in combination with protein identification by mass spectrometry. Proteomics 6, 4688–4703 (2006). [DOI] [PubMed] [Google Scholar]
- Chandramouli K. & Qian P. Y. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Human genomics and proteomics: HGP 2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owiti J. et al. iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. The Plant journal: for cell and molecular biology 67, 145–156 (2011). [DOI] [PubMed] [Google Scholar]
- Bantscheff M., Schirle M., Sweetman G., Rick J. & Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical and bioanalytical chemistry 389, 1017–1031 (2007). [DOI] [PubMed] [Google Scholar]
- Carvalhais V., Cerca N., Vilanova M. & Vitorino R. Proteomic profile of dormancy within Staphylococcus epidermidis biofilms using iTRAQ and label-free strategies. Applied microbiology and biotechnology 99, 2751–2762 (2015). [DOI] [PubMed] [Google Scholar]
- Latosinska A. et al. Comparative Analysis of Label-Free and 8-Plex iTRAQ Approach for Quantitative Tissue Proteomic Analysis. PloS one 10, e0137048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu F. & Rodermel S. Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. Journal of integrative plant biology 52, 750–761 (2010). [DOI] [PubMed] [Google Scholar]
- Cao Z. L. et al. A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development. Journal of integrative plant biology 53, 258–269 (2011). [DOI] [PubMed] [Google Scholar]
- Hirai A. et al. Rice chloroplast DNA: a physical map and the location of the genes for the large subunit of ribulose 1,5-bisphosphate carboxylase and the 32 KD photosystem II reaction center protein. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 70, 117–122 (1985). [DOI] [PubMed] [Google Scholar]
- Matsuoka M. Classification and characterization of cDNA that encodes the light-harvesting chlorophyll a/b binding protein of photosystem II from rice. Plant and Cell Physiology 31, 519–526 (1990). [Google Scholar]
- Loschelder H., Schweer J., Link B. & Link G. Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant physiology 142, 642–650 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A. L. et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. The Plant journal: for cell and molecular biology 56, 590–602 (2008). [DOI] [PubMed] [Google Scholar]
- Yue R. et al. A rice stromal processing peptidase regulates chloroplast and root development. Plant & cell physiology 51, 475–485 (2010). [DOI] [PubMed] [Google Scholar]
- Kim E. H. et al. The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: a study using two chlorophyll b-less mutants. Biochimica et biophysica acta 1787, 973–984 (2009). [DOI] [PubMed] [Google Scholar]
- Chu P. et al. iTRAQ-based quantitative proteomics analysis of Brassica napus leaves reveals pathways associated with chlorophyll deficiency. Journal of proteomics 113, 244–259 (2015). [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., Xu X. M. & Moller S. G. Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 232, 567–578 (2010). [DOI] [PubMed] [Google Scholar]
- Zhong L. et al. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. The Plant cell 25, 2925–2943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Zhang L., Feng M., Fang Y. & Li J. An integrated proteomics reveals pathological mechanism of honeybee (Apis cerena) sacbrood disease. Journal of proteome research 12, 1881–1897 (2013). [DOI] [PubMed] [Google Scholar]
- Han B. et al. Quantitative Neuropeptidome Analysis Reveals Neuropeptides Are Correlated with Social Behavior Regulation of the Honeybee Workers. Journal of proteome research 14, 4382–4393 (2015). [DOI] [PubMed] [Google Scholar]
- Lin H., He L. & Ma B. A combinatorial approach to the peptide feature matching problem for label-free quantification. Bioinformatics 29, 1768–1775 (2013). [DOI] [PubMed] [Google Scholar]
- Bindea G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.