Key Points
CM impairs erythroid lineage specification and differentiation from Pre-Meg/E progenitors.
CM creates abnormal preleukemic Pre-Meg/E progenitors predisposed to leukemia initiation.
Abstract
Acute myeloid leukemia (AML) arises through multistep clonal evolution characterized by stepwise accumulation of successive alterations affecting the homeostasis of differentiation, proliferation, self-renewal, and survival programs. The persistence and dynamic clonal evolution of leukemia-initiating cells and preleukemic stem cells during disease progression and treatment are thought to contribute to disease relapse and poor outcome. Inv(16)(p13q22) or t(16;16)(p13.1;q22), one of the most common cytogenetic abnormalities in AML, leads to expression of a fusion protein CBFβ-SMMHC (CM) known to disrupt myeloid and lymphoid differentiation. Anemia is often observed in AML but is presumed to be a secondary consequence of leukemic clonal expansion. Here, we show that CM expression induces marked deficiencies in erythroid lineage differentiation and early preleukemic expansion of a phenotypic pre-megakaryocyte/erythrocyte (Pre-Meg/E) progenitor population. Using dual-fluorescence reporter mice in lineage tracking and repopulation assays, we show that CM expression cell autonomously causes expansion of abnormal Pre-Meg/E progenitors with compromised erythroid specification and differentiation capacity. The preleukemic Pre-Meg/Es display dysregulated erythroid and megakaryocytic fate-determining factors including increased Spi-1, Gata2, and Gfi1b and reduced Zfpm1, Pf4, Vwf, and Mpl expression. Furthermore, these abnormal preleukemic Pre-Meg/Es have enhanced stress resistance and are prone to leukemia initiation upon acquiring cooperative signals. This study reveals that the leukemogenic CM fusion protein disrupts adult erythropoiesis and creates stress-resistant preleukemic Pre-Meg/E progenitors predisposed to malignant transformation. Abnormality in Meg/E or erythroid progenitors could potentially be considered an early predictive risk factor for leukemia evolution.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematopoietic malignancy with an overall survival rate of ∼20% in patients treated with current standard chemotherapy.1,2 Refractory and relapse are thought to be due to persistence of leukemia-initiating cells or leukemia stem cells, which display resistance to chemotherapy and unlimited self-renewal capacity.3,4 AML transformation requires multiple cooperating mutations that coordinately deregulate cell proliferation and impair normal differentiation.5 Multiple lines of evidence support the model that AML arises through clonal evolution characterized by stepwise accumulation of successive mutations. The majority of the initiating mutations are thought to occur in the long-lived, self-renewing hematopoietic stem cells (HSCs); however, more differentiated cell types could also acquire certain mutations conferring self-renewal properties.6,7 Recent genomic sequencing studies in AML patients revealed the presence of preleukemic stem cells, harboring only a few founding mutations and can persist during remission, that thus serve as potential reservoir for subsequent leukemia relapse.8-10 These findings highlight the complex dynamic clonal evolution and the need to eradicate preleukemic clones predisposed for additional mutations driving leukemia relapse. Therefore, it is pertinent to define early preleukemic alterations and cellular subsets predisposed to malignant transformation.
The core-binding factor (CBF) complex, consisting of RUNX and CBFβ, is a master hematopoietic transcriptional regulator frequently disrupted in leukemias.11 Inv(16)(p13q22) or t(16;16)(p13.1;q22) [henceforth inv(16)] is a recurrent chromosomal rearrangement found in ∼5% to 12% of human AML cases and is associated with the M4Eo subtype.12 Inv(16) results in a fusion of CBFB with the MYH11 gene, which encodes a smooth muscle myosin heavy chain, creating a fusion protein CBFβ-SMMHC (CM).13,14 Knocking in 1 allele of Cbfb-MYH11 resulted in lethal defects in definitive hematopoiesis, identical to phenotypes of Runx1- or Cbfb-null mice,15-17 thus establishing CM as a dominant inhibitor of CBF function. Expression of CM predisposes for AML development; however, additional genetic events are required for full leukemia transformation.18 Our previous studies using a conditional Cbfb-MYH11 knock-in mouse model further recapitulated somatic acquisition of inv(16) and the stepwise progression of inv(16) leukemogenesis.19 We showed that CM expression leads to impaired differentiation of both myeloid and lymphoid lineages19-21 and produces preleukemic stem cells and abnormal myeloid progenitors (AMP) that are at risk for acquiring additional mutations.19 Although erythrocytes derived from CM knock-in embryonic stem cells were detected in the circulating blood in chimeric mice,18 the consequences of CM in the development and maturation of adult erythroid lineage have not been investigated. In this study, we further defined the early alterations preceding leukemia progression and identified a preleukemic pre-megakaryocyte/erythrocyte (Pre-Meg/E) progenitor subset that are impaired in erythroid differentiation and predisposed for leukemia initiation.
Materials and methods
Mice
Cbfb+/56M/Mx1-Cre (129SvEv) mice19 were backcrossed to C57BL/6 strain for >10 generations. ROSAmT/mG mice (C57BL/6) were purchased from the Jackson Laboratory. To induce CM, 4- to 8-week-old Cbfb+/56M/Mx1-Cre mice were injected with 250 μg poly (I:C) (InvivoGen) every other day for 7 doses. Similarly treated Cbfb+/56M or Mx1-Cre littermates were used as control. Lineage-tracking repopulation experiments were performed via intravenous injection of Cbfb+/56M/Mx1-Cre/mTmG+ or Mx1-Cre/mTmG+ cells together with 2 × 105 mTmG+ supporter cells into lethally irradiated (11 Gy; 2 split doses) 6- to 8-week-old CD45.1+ congenic C57BL/6 mice. Secondary or tertiary AML transplantation was performed using 1 × 106 bone marrow (BM) cells from moribund mice and injected intravenously into sublethally irradiated (6.5 Gy) 6- to 8-week-old CD45.1+ recipient mice. All mice were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited animal facility, and all experimental procedures were performed in accordance with federal and state government guidelines and established institutional guidelines and protocols approved by the Institutional Animal Care and Use Committee at the Beckman Research Institute of City of Hope.
Cell isolation and flow cytometry
BM mononuclear cells were isolated from femurs, tibias, and pelvis as previously described.19 For fluorescence activated cell-sorting (FACS) analyses, cells were stained in phosphate-buffered saline (PBS) with 0.5% bovine serum albumin for 15 minutes on ice with fluorescently labeled antibodies for cell surface markers, cKit, Sca1, CD16/32, CD105, CD150, CD48, CD41, CD71, and Ter119. Lineage cocktail includes biotin-conjugated CD3, CD4, CD8, B220, CD19, IgM, NK1.1, CD11b, CD11c, and IL7Rα. For cell sorting, lineage-negative cells were enriched using EasySep selection reagents (StemCell Technologies). For apoptotic analysis, cells were stained for surface antigens followed by Annexin V staining according to the manufacturer’s protocol (eBioscience). For cell cycle analysis, cells were stained for surface markers followed by fixation, permeabilization, and staining with Ki67 antibody and 4′,6-diamidino-2-phenylindole (DAPI). All antibodies were purchased from BioLegend, BD Biosciences, or eBiosciences. Flow cytometry was performed using a 5-laser, 15-detector BD LSRII, and sorting was performed using BD ARIA-III. Acquired data were analyzed by Flowjo software (Tree Star Inc). Phenotypic hematopoietic stem and progenitor cell (HSPC) populations were defined as LSK (Lin−ckit+Sca1+); myeloid progenitors (MPs; Lin−ckit+Sca1−); pre–granulocyte-macrophage (Pre-GM; Lin−ckit+Sca1−CD41−CD16/32−/loCD105−CD150−); granulocyte-macrophage progenitors (GMPs; Lin−ckit+Sca1−CD41−CD16/32+CD150−); Pre-Meg/E (Lin−ckit+Sca1−CD41−CD16/32−/loCD105−CD150+); megakaryocyte progenitors (MkPs; Lin−ckit+Sca1−CD41+CD150+); pre–colony-forming unit–erythroid (Pre-CFU-E; Lin−ckit+Sca1−CD41−CD16/32−/loCD105+CD150+); colony-forming unit–erythroid (CFU-E; Lin−ckit+Sca1−CD41−CD16/32−/loCD105+CD150−Ter119−); pro-erythrocytes (ProEry; Lin−ckit+Sca1−CD41−CD16/32−/loCD105+CD150−Ter119+); erythroid progenitors (EPs; Lin−ckit+Sca1−CD41−CD16/32−/loCD105+).
In vitro erythroid differentiation culture
Erythroid differentiation liquid culture was performed as previously described.22 In brief, sorted Pre-Meg/E and EP cells (2 × 104) were seeded in 96-well flat-bottom plates and cultured in Iscove’s Modified Dulbecco’s Medium containing 15% fetal bovine serum (StemCell Technologies), 1% detoxified bovine serum albumin (StemCell Technologies), 500 μg/mL holo-transferrin (Sigma), 10 μg/mL recombinant human insulin (Sigma), 2 mM l-glutamine, and 0.5 U/mL erythropoietin (Epo) (R&D Systems). After 3 days, cells were counted and stained for CD71 and Ter119 to determine the stages of maturation as previously described.22
BM transduction and transplantation
For murine stem cell virus (MSCV)–ires-green fluorescent protein (GFP) (MIG) retrovirus transduction, BM HSPCs were sorted and cultured in Iscove’s Modified Dulbecco’s Medium with 20% fetal bovine serum, stem cell factor (SCF) (20 μg/mL), interleukin 3 (IL3 ;10 μg/mL), IL6 (6 μg/mL), and thrombopoietin (20 μg/mL) for 4 hours, followed by spinoculation in culture media supplemented with a final concentration of 5 μg/mL polybrene (American Bioanalytical) and 7.5 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer.
Transduced cells were harvested and transplanted into lethally irradiated (11 Gy) congenic CD45.1+ C57BL/6 mice together with 2 × 105 CD45.1+ wild-type BM supporter cells. Mice were monitored for signs of leukemia by peripheral blood analysis every month up to 6 months and analyzed when moribund.
Colony-forming assays
Erythroid-specific colony-forming cell (CFC) assays were conducted using Epo containing MethoCult (M3334; StemCell Technologies) according to the manufacturer’s protocol. CFU-E and mature burst-forming unit-erythroid (BFU-E) were counted at day 3, and BFU-E (containing ≥250 clusters of erythroblasts) was counted at day 10. Myeloid/erythroid CFC assays were performed using MethoCult containing SCF, IL3, IL6, and Epo (M3434; StemCell Technologies), and colonies were counted at day 7 according to the manufacturer’s protocol. For replating assays, cells from each plate were harvested and replated at 2 × 104 cells per dish in duplicate.
Quantitative reverse transcriptase–polymerase chain reaction
RNA was isolated using the RNeasy micro Kit (Qiagen) following the manufacturer’s protocol. First-strand cDNA was generated using SuperScript III reverse transcriptase (Life Technologies). Quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) was performed using Taqman gene expression assays (supplemental Table 3, available on the Blood Web site) and the Biomark PCR system (Fluidigm). Assay results were first normalized to Hprt levels by the Ct method, and relative expression was calculated as 2−ΔCt.
Statistics
Statistical analyses were performed with the Student t test or analysis of variance for normal distributions. Mann-Whitney U tests were used when the criteria for a normal distribution were not satisfied. P < .05 was considered statistically significant.
Results
CM expression induces expansion of phenotypic Pre-Meg/E progenitors
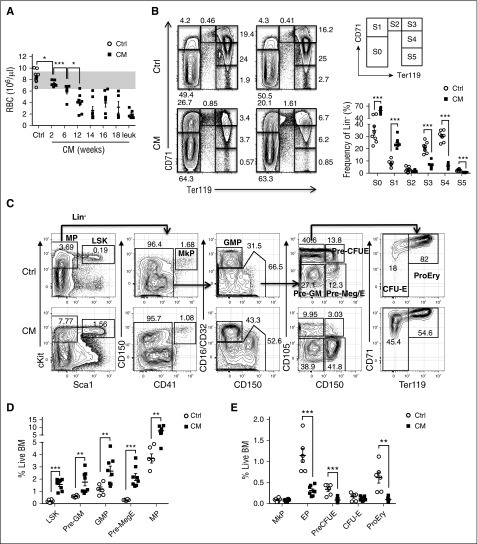
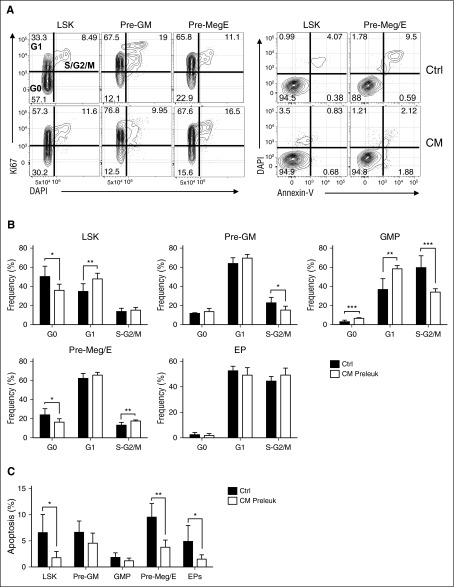
To take advantage of the wealth of phenotypic markers well defined in the C57BL/6 strain and further delineate the preleukemic progenitors and leukemia-initiating populations, we backcrossed Cbfb+/56M/Mx1-Cre (129SvEv) into C57BL/6 for >10 generations. Similar to previous studies in the 129SvEv strain, CM expression in adult C57BL/6 mice leads to induction dose–dependent and cell number–dependent development of AML (supplemental Figure 1A-B). We noted that red blood cell (RBC) counts were significantly reduced as early as 2 weeks after CM induction and progressively decreased over time through leukemia progression (Figure 1A). To examine erythroid maturation, we analyzed cell surface markers CD71 and Ter119 commonly used to define the maturation stages of erythroid progenitors (Figure 1B).23,24 At 2 weeks after CM induction, we observed significantly more immature S0 (CD71−/Ter119−; 59.08% vs 34.75%) and S1 (CD71hi/Ter119−; 25.11% vs 7.69%) populations and correspondingly reduced S3 (CD71hi/Ter119hi; 6.03% vs 20.86%), S4 (CD71loTer119hi; 6.01% vs 30.25%), and S5 (CD71−Ter119hi; 0.63% vs 2.32%) populations (Figure 1B), indicating a severe block in erythroid maturation. We further examined myeloid-erythroid progenitor populations including Pre-GM (Lin−cKit+Sca1−CD16/32-/loCD150−CD105−), GMPs (Lin−cKit+Sca1−CD16/32hiCD150−), Pre-Meg/E (Lin−cKit+Sca1−CD16/32−/loCD150+CD105−), and EPs (Lin−cKit+Sca1−CD16/32−/loCD105hi; including Pre-CFU-E, CFU-E, ProEry) as previously defined (Figure 1C).25 Analysis of preleukemic BM 2 weeks after induction showed significant increase of LSK (Lin−/cKit+/Sca1+) and MPs (Lin−/cKit+/Sca1−), consistent with previous studies.19 Further separation of phenotypic myeloid-erythroid progenitor subsets showed ∼2-fold increased frequency and numbers of Pre-GM, GMP, and a 5.7-fold expansion of Pre-Meg/E progenitors compared with similarly treated control mice (Figure 1C-D; supplemental Figure 1C). Notably, the increase in Pre-Meg/E was accompanied by significant decrease of EP subsets including Pre-CFU-E, CFU-E, and pro-erythrocytes, whereas MkPs seemed unaffected (Figure 1E; supplemental Figure 1D). Cell cycle analysis by Ki67 and DAPI staining in various phenotypic subsets revealed compartment selective effects induced by CM expression. Increased cell cycle progression was seen in LSK and Pre-Meg/E with significantly less cells in G0 phase and more in G1 and S-G2/M phases, respectively (Figure 2A-B). On the contrary, a reduced fraction in S-G2/M phase was observed in Pre-GM and GMP populations. No significant change in cell cycle was seen in EP population (Figure 2B), suggesting that the decrease in EPs is not due to alterations in cell proliferation. In addition, enhanced survival of LSK, Pre-Meg/E, and EP populations was evident with reduced Annexin V+ apoptotic cells in CM-expressing mice (Figure 2A,C). Altogether, we show that CM induces cellular compartment-dependent alterations in the preleukemic BM.
Figure 1.
CM expression leads to preleukemic expansion of a phenotypic Pre-Meg/E progenitor population. (A) RBC counts in peripheral blood of CM mice (n = 5-8) over time (2-18 weeks) and when moribund with leukemia (n = 6) compared with similarly treated control mice (n = 13). The gray area indicates the typical normal range. (B) Representative FACS plots showing various erythroid progenitor subsets defined by the expression of Ter119 and CD71 in CM-expressing preleukemic BM 2 weeks after poly (I:C) compared with control (Ctrl) BM analyzed by flow cytometry. Numbers indicate the frequency of each gate in Lin− BM cells. Right panel shows the gating strategy (top) and the frequency of each phenotypic erythroid subsets (S0-S5) in Lin− BM as defined above (bottom). Mean ± standard error of the mean (SEM; Ctrl, n = 8: CM, n = 8) are shown. (C) Representative FACS plots showing the gating strategy of various phenotypic progenitor populations in CM preleukemic BM compared with Ctrl BM analyzed by flow cytometry. The frequency within total BM is shown for LSK and MP (first plot from left), and the frequency in the respective parent gate is shown for the other populations. (D-E) Frequency of various CM (n = 8) or Ctrl (n = 6) phenotypic progenitor populations in total BM as defined above. Mean ± SEM is shown. *P < .05; **P < .01; ***P < .001.
Figure 2.
CM expression alters the proliferation and apoptosis of preleukemic Pre-Meg/E progenitor population. (A) Representative FACS plots of Ki67/DAPI (left) or Annexin-V/DAPI (right) staining and gating strategy in CM or control BM HSPCs. (B) Frequency (%) of CM preleukemic (n = 6) or control (n = 6) LSK, Pre-GM, GMP, Pre-Meg/E, and EP in G0, G1, or S/G2/M phases of cell cycle as defined above. Bar graphs and lines indicate mean + standard deviation (SD). (C) The frequency (%) of apoptotic cells defined by Annexin-V+ in CM (n = 6) or Ctrl (n = 5) LSK, Pre-GM, GMP, Pre-Meg/E, and EP. Bars and lines indicate mean + SD; *P < .05; **P < .01; ***P < .001.
CM impairs erythroid lineage specification and differentiation from Pre-Meg/E progenitors
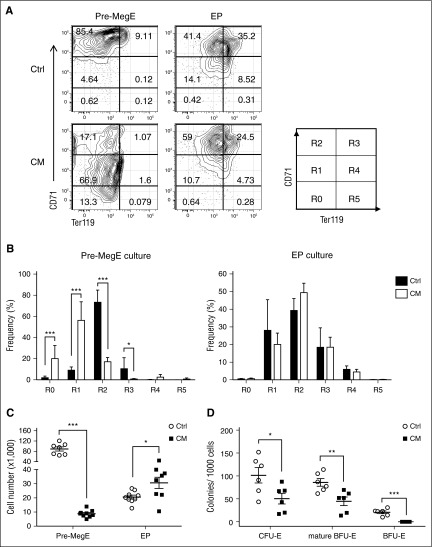
The expansion of phenotypic Pre-Meg/Es in CM preleukemic BM was accompanied by a marked reduction of EPs with seemingly normal proliferation. Therefore, we hypothesized that CM may impair erythroid lineage specification and differentiation from Pre-Meg/E progenitors. To track CM-expressing cells, we generated Cbfb+/56M/Mx1-Cre mice with a double fluorescent Rosa26mT/mG Cre reporter allele, which allows expression of a membrane targeted dTomato fluorescent protein before Cre induction and GFP expression after Cre-mediated deletion. We sorted GFP+ Pre-Meg/Es and EPs from induced Cbfb+/56M/Mx1-Cre/mTmG+ or Mx1-Cre/mTmG+ control mice and performed an in vitro differentiation assay using a liquid culture supplemented with Epo and transferrin.22 The differentiating cells were counted at day 3 and stained for erythroid differentiation markers CD71 and Ter119 as a measure of their maturation status (Figure 3A). Although control Pre-Meg/E cultures had matured to R2 (CD71hiTer119−) and transitioning to R3 (CD71hiTer119+), CM Pre-Meg/E mostly remained in R1 (CD71loTer119−) (Figure 3A-B), indicating a block in erythroid differentiation. Similar analysis of EP cultures did not show significant changes in maturation, suggesting that the main deficiency was at the transition from Pre-Meg/E progenitors. Consistent with these results, CM-expressing Pre-Meg/E cultures generated >10-fold less cells compared with the control, whereas increased output was observed for EP cultures (Figure 3C). We also used a methylcellulose-based CFC assay optimized for erythroid progenitors to evaluate the differentiation capacity of CM Pre-Meg/E progenitors. We found a significant decrease in CFU-E, mature BFU-E, and BFU-E numbers in CM Pre-Meg/E cultures compared with controls (Figure 3D). Taken together, these results demonstrate that CM expression impairs erythroid differentiation capacity of Pre-Meg/E progenitors.
Figure 3.
CM-expressing Pre-Meg/Es are impaired in erythroid differentiation potential in vitro. (A) Representative FACS plots and gating strategy (R0-R5) of CD71 and Ter119 expression of sorted CM or control Pre-Meg/E and EPs after 3-day erythroid differentiation culture. (B) The frequency of differentiating erythroid cells at various maturation stage (R0-R5) from Pre-Meg/E (left) or EPs (right) at day 3. Bars and lines represent mean + SD (n = 7-9) from 3 independent experiments. (C) Total cell number output from CM or control Pre-Meg/E and EPs after 3 days of differentiation culture. Results include 3 independent experiments performed in triplicate. Mean ± SEM (×1000) is shown. (D) Number of CFU-E, mature BFU-E, and BFU-E colonies from CM or control Pre-Meg/Es (per 1000 cells). Results include 3 independent experiments performed in duplicate. CFU-E and mature BFU-E colonies were counted at day 3, and BFU-E colonies were counted at day 10. Mean ± SEM is shown. *P < .05; **P < .01; ***P < .001.
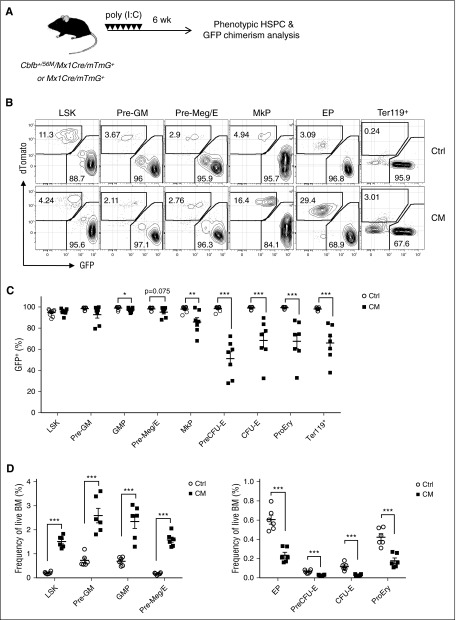
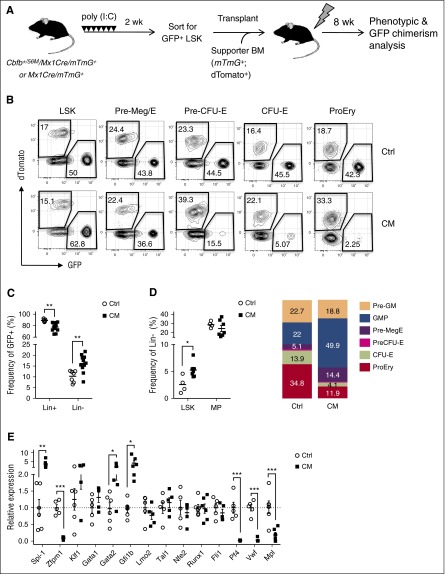
To further assess the effects of CM on homeostatic erythroid differentiation in vivo, we tracked GFP+ chimerism indicative of CM+ cells in Cbfb+/56M/Mx1-Cre/mTmG+ mice (Figure 4A). Phenotypic analysis 6 weeks after poly (I:C) induction showed that the vast majority (>90%) of total live cells were GFP+ in both CM/mTmG+ and control (Mx1-Cre/mTmG+) mice (supplemental Figure 2A). Significantly reduced GFP+ chimerism was observed in B, T, and myeloid lineages (supplemental Figure 2A-B), consistent with impaired lymphoid and myeloid differentiation previously described. Consistent with phenotypes in CM mice, significant increase of phenotypic HSPCs including LSK, Pre-GM, and Pre-Meg/E was observed in CM/mTmG+ (supplemental Figure 2C). Similar GFP+ chimerism was detected in LSK, Pre-GM, and Pre-Meg/E progenitors in CM/mTmG+ and control mice (Figure 4B-C). In contrast, GFP+ chimerism was significantly reduced in EP subsets including Pre-CFU-E, CFU-E, ProEry, and Ter119+ erythroid cells in CM/mTmG+ mice. Concurrently, the frequency and absolute numbers of GFP+ EP populations were significantly reduced (Figure 4D; supplemental Figure 2D). These results confirmed that CM expression impairs erythroid differentiation at the Pre-Meg/E stage in vivo. We also observed reduced GFP+ chimerism in MkPs (Lin−cKit+Sca1−CD41+CD150+; Figure 4B-C), consistent with previous findings that CM impairs megakaryocytic differentiation.19
Figure 4.
CM expression results in reduced homeostatic erythroid differentiation in vivo. (A) Schematic of experimental design. Cbfb+/56M/Mx1-Cre/Rosa26mT/mG (CM) or control Mx1-Cre/Rosa26mT/mG (Ctrl) dual fluorescent reporter mice were injected with 7 doses of poly (I:C) to induce the expression of Cre. Contribution of CM-expressing cells (GFP+) in HSPC subsets were assessed 6 weeks after induction. (B) Representative FACS plots of GFP+ (CM-expressing) and dTomato+ (non-CM cells) in various phenotypic HSPC subsets. (C) Frequency (%) of GFP+ cells in each phenotypic compartment of CM (n = 7) or Ctrl mice (n = 9). Mean ± SEM is shown. (D) Frequencies of GFP+ LSK and myeloid/erythroid progenitor subsets within total CM (n = 6) or Ctrl (n = 6) BM. Mean ± SEM is shown. *P < .05; **P < .01; ***P < .001.
We further determined whether this differentiation block is cell autonomous by transplanting sorted GFP+ CM or control LSK cells for repopulation into irradiated (11 Gy) wild-type C57BL/6 mice (Figure 5A). Eight weeks after transplantation, we examined GFP+ chimerism in various phenotypic HSPC subsets by flow cytometry. Similar GFP+ contribution was seen among various phenotypic subsets in control recipients, whereas significantly reduced GFP+ chimerism was selectively detected in erythroid-restricted progenitors including Pre-CFU-E, CFU-E, and ProEry in CM recipients (Figure 5B). There were significantly more lineage-negative progenitors composed of increased LSK, GMP, and Pre-Meg/E and reduced Pre-CFU-E, CFU-E, and ProEry derived from CM GFP+ cells (Figure 5C-D). These results indicate that CM causes erythroid differentiation block at the Pre-Meg/E stage in a cell autonomous fashion in vivo.
Figure 5.
CM cell autonomously impairs erythroid lineage commitment and deregulates expression of key erythroid/Mk lineage-determining regulators. (A) Schematic of experimental design. Cbfb+/56M/Mx1-Cre/Rosa26mT/mG (CM) or control Mx1-Cre/Rosa26mT/mG (Ctrl) mice (CD45.2+) were induced as above. Sorted CM-expressing or Ctrl GFP+ LSKs (500 cells) were cotransplanted with 2 × 105 mTmG+ (CD45.2+; dTomato+) supporter BM cells into lethally irradiated (11 Gy) CD45.1+ congenic recipients. Phenotypic HSPCs and GFP chimerism in the BM were analyzed 8 weeks after transplantation by flow cytometry. (B) Representative FACS plots of GFP+ and dTomato+ chimerism within each HSPC compartment. (C) The frequency of lineage-committed (Lin+) and lineage-negative (Lin−) cells within Ctrl (n = 7) or CM (n = 11) GFP+ donor-derived population. (D) The frequency of LSK or MP (left), and phenotypic composition (right) of GFP+/Lin− cells derived from Ctrl (n = 4) or CM (n = 7) donors. Mean ± SEM is shown. (E) Relative expression levels of Spi-1, Zfpm1, Klf1, Gata1, Gata2, Gfi1b, Lmo2, Tal1, Nfe2, Runx1, Fli1, Pf4, Vwf, and Mpl mRNA in sorted Ctrl or CM Pre-Meg/Es (n = 4-6) assessed by Taqman-based qRT-PCR assays. Shown is the relative expression level (mean ± SEM) for each gene normalized to the mean expression levels in Ctrl Pre-Meg/Es (indicated by dashed line). *P < .05; **P < .01; ***P < .001.
Hematopoietic lineage differentiation is determined by lineage-specific gene expression profile tightly regulated by networks of signaling and transcriptional regulators. We examined whether the erythroid and megakaryocytic (Mk) fate determining program is disrupted in CM preleukemic Pre-Meg/Es. We sorted GFP+ Pre-Meg/E cells from CM/mTmG+ preleukemic and control mTmG+ mice for qRT-PCR analysis. Significantly altered expression of a number of erythroid and megakaryocytic regulators, including reduced expression of Zfpm1 (encodes Fog1), Pf4, Vwf, and Mpl and increased expression of Spi-1 (encodes Pu.1), Gata2, and Gfi1b was seen in CM Pre-Meg/Es (Figure 5E). These alterations in erythroid/Mk gene expression program are consistent with defects in erythroid/Mk lineage specification.
CM-expressing preleukemic Pre-Meg/E progenitors are prone to leukemia initiation
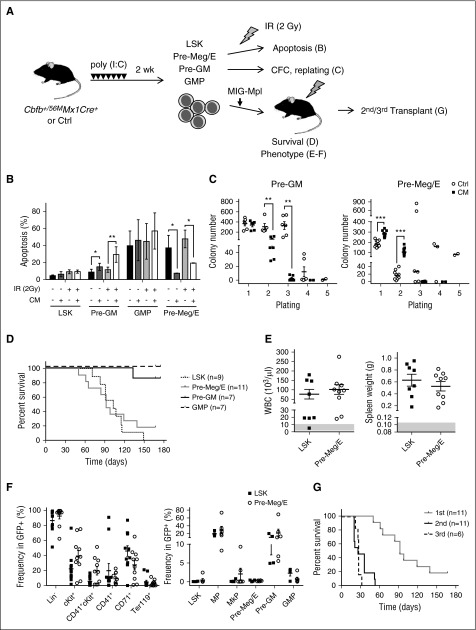
Given our finding that CM differentially enhanced homeostatic proliferation and survival in the Pre-Meg/E subset among MPs, we reasoned that CM Pre-Meg/E might be more stress resistant and are prone to leukemic transformation. First, we tested whether CM expression affects the sensitivity to genotoxic stress in various HSPC subsets. We sorted live LSK, Pre-GM, GMP, and Pre-Meg/E cells and subjected them to irradiation (IR; 2 Gy) followed by Annexin V/DAPI staining as a measure for apoptosis 24 hours later (Figure 6A, top). Notably, apoptosis rates in response to IR and in vitro culturing varied greatly among HSPC subsets. Consistent with reduced spontaneous apoptosis seen in Pre-Meg/E cells ex vivo (Figure 2C), CM-expressing Pre-Meg/Es were significantly protected from apoptosis in culture (unirradiated) and upon IR (Figure 6B). In contrast, significantly increased apoptosis was observed in CM Pre-GM, whereas no significant change in apoptosis was found in LSK and GMP populations. Next, we sought to examine the clonogenic activity and replating capacity of Pre-Meg/Es using methylcellulose-based CFC assays with SCF, IL3, IL6, and Epo (Figure 6A, middle). After 7 days of culture, we observed significantly more CFU-Cs in CM Pre-Meg/E compared with control but not in CM Pre-GM subset (Figure 6C). Flow cytometry analysis of these colonies revealed that CM Pre-Meg/E generated predominantly CFU-Meg/Es (supplemental Figure 3A). Increased CD41+cKit+ cells were seen in CM Pre-Meg/E cultures (supplemental Figure 3B), consistent with retaining a more primitive progenitor phenotype.26 We further assessed the proliferative capacity by serially replating the CFU-Cs every 7 days. CM Pre-Meg/E showed enhanced replating capacity, whereas reduced replating was seen for CM Pre-GM in the second plating (Figure 6C). However, the enhanced progenitor activity seemed to be transient because few CFU-Cs were detected in further replating assays.
Figure 6.
Expanded CM Pre-Meg/E progenitors are capable of leukemia-initiation. (A) Schematic of leukemia initiation experiment design. Cbfb+/56M/Mx1-Cre mice were induced by poly (I:C). LSKs, Pre-Meg/Es, Pre-GMs, and GMPs were sorted 2 weeks later and subjected to (B) apoptosis analysis, (C) CFC and replating, or (D) transduced with MIG-Mpl retrovirus followed by transplantation and monitored for survival, (E-F) phenotype, and (G) engraftment in serial transplants. (B) The frequency (%) of apoptotic cells defined by Annexin-V+ in CM or Ctrl LSK, Pre-GM, GMP, or Pre-Meg/E cells 24 hours after IR (2 Gy). Bars and lines indicate mean ± SD. (C) Number of CFCs derived from Pre-GM (left) or Pre-Meg/Es (right) in first plating (1000 cells) and in serial replating (2 × 104 cells) using MethoCult M3434. Shown is mean ± SEM of ≥3 independent experiments performed in duplicate. (D) Kaplan-Meier survival curves of recipients transplanted with CM/Mpl LSKs (7-8 × 103 cells), Pre-Meg/Es, Pre-GMs, or GMPs (2.5 × 104). (E) Peripheral white blood cell counts (103/μL; left) and spleen weights (g; right) in CM-Mpl LSK– or Pre-Meg/E–derived leukemic mice. The gray area indicates the normal range detected in normal Ctrl mice (n = 8). (F) The frequency (%) of various immunophenotypic populations in GFP+ BM of leukemic mice transplanted with CM-Mpl LSK (n = 4) or CM-Mpl Pre-Meg/E (n = 6). (G) Kaplan-Meier survival curves of secondary (n = 11) and tertiary (n = 6) recipients transplanted with 1 × 106 BM cells from moribund mice transplanted with CM-Mpl Pre-Meg/E. *P < .05; **P < .01; ***P < .001.
It is known that Mpl overexpresion cooperates with CM to induce leukemogenesis in mice.27 To directly test whether CM preleukemic Pre-Meg/Es are susceptible to leukemic transformation, we introduced Mpl overexpression and assessed leukemia onset and survival in transplant recipients (Figure 6A, bottom). Sorted CM LSK, Pre-Meg/E, Pre-GM, and GMP cells were transduced with MIG-Mpl retrovirus. We transplanted 7 to 8 × 103 CM/Mpl (GFP+) LSK or 2.5 × 104 CM/Mpl Pre-MegE/Pre-GM/GMP based on the approximate number of each population detected in 0.5 to 1 × 106 BM cells (supplemental Table 1), which we determined not sufficient to induce spontaneous AML development (supplemental Figure 1B). All CM/Mpl LSK recipients (9 of 9) developed leukemia with a median survival of 105 days, similar to previous studies.19 Most of the CM/Mpl Pre-Meg/E recipients (9 of 11) also developed lethal leukemia, with a median survival of 92 days (Figure 6D). In contrast, only 14% (1 of 7) of Pre-GM recipients and none (0 of 7) of GMP recipients developed leukemia. The frequency of leukemia-initiating cells was significantly higher in LSK (1 in 2419) compared with Pre-Meg/E (1 in 19 528), as estimated by limiting-dilution transplantation (supplemental Table 2). All diseased mice presented pathologic hallmarks of leukemia including high white blood cell counts (Figure 6E, left), enlarged spleens (Figure 6E, right), and massive expansion of Lin− (CD3, CD4, CD8, B220, CD19, IgM, NK1.1, CD11c, CD11b, Gr1) and cKit+ blasts (Figure 6F; supplemental Figure 4B-D). Variable amounts of CD41+ or CD71+ cells could be seen in both LSK- and Pre-Meg/E–derived leukemias (Figure 6F, left; supplemental Figure 4A). The frequency of phenotypic LSK (LSK group: 0.074 ± 0.027%; Pre-Meg/E group: 0.0467 ± 0.0399%; n = 4-6) and Pre-Meg/E (LSK group: 0.269 ± 0.110%; Pre-Meg/E group: 0.228 ± 0.054%; n = 4-6) was relatively low in the leukemic BM, irrespective of the immunophenotype of the initiating cell population (Figure 6F, right). We confirmed that these Pre-Meg/E–derived leukemias are readily transplantable, as aggressive leukemia was seen in all secondary and tertiary transplantation recipients with a median survival of 26 to 27 days (Figure 6G). These results indicate that, although leukemia-initiating activity is enriched in the LSK population, CM preleukemic Pre-Meg/Es are susceptible to transformation and are capable of initiating leukemia in mice upon acquisition of cooperating alterations.
Discussion
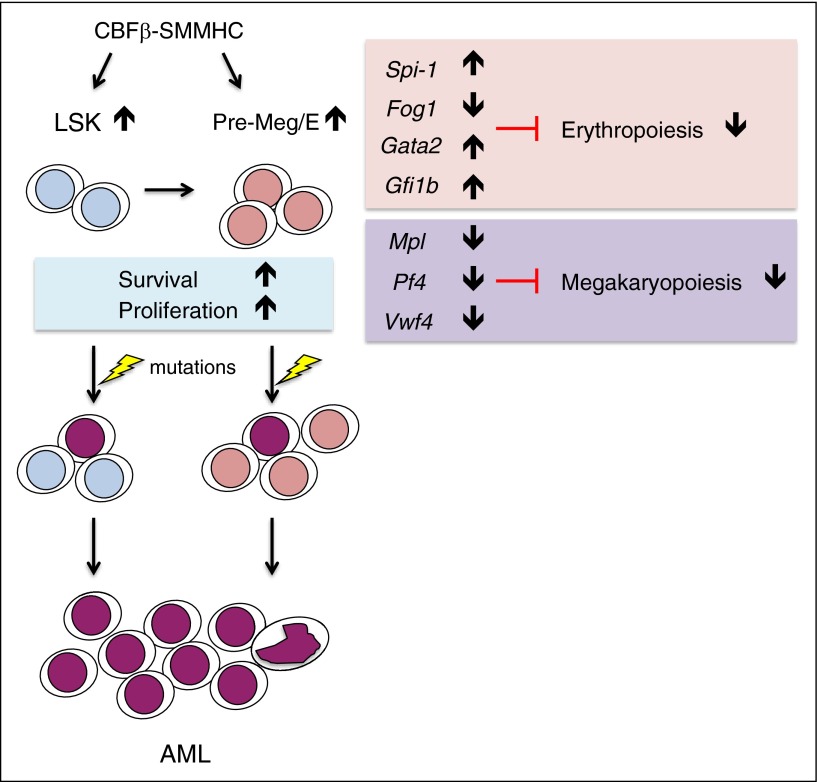
We demonstrate here that the leukemogenic CM fusion protein induces the expansion of preleukemic Pre-Meg/E progenitors, which feature a dysregulated erythroid/Mk transcriptional network and impaired erythroid differentiation. These poorly differentiating progenitors display enhanced survival and serve as a cellular reservoir prone to malignant transformation (Figure 7). Our studies reveal that CM profoundly impairs erythroid lineage commitment at the early Pre-Meg/E stage, a previously unrecognized function of the inv(16) fusion protein. Previous studies demonstrated that CM expression impaired multiple hematopoietic lineages including lymphocytes, myeloid cells, and megakaryocytes.15,19-21 These defects are presumably due to Runx1 inhibition because similar deficiencies were seen in Runx1-deficient mice.28,29 Previous studies also noted that CM expression affected primitive erythropoiesis; however, CM knocked-in cells were detected in the c-kit+/Ter119+ BM population in CM knock-in chimeras.18 It is therefore thought that CM does not affect adult erythropoiesis, although this has not been formally tested. Our in vitro differentiation assays and in vivo lineage tracking studies provide evidence that CM expression in adult BM causes impaired erythroid differentiation at the stage of early Pre-Meg/E progenitors, leading to a drastic decrease of committed erythroid progenitors. Conditional deletion of Cbfb resulted in severe reduction of CD105+ erythroid progenitors followed by anemia.30 In a recent study, an aberrant Meg progenitor population and impaired erythroid differentiation were also observed in Runx1-deficient mice.31 Therefore, the defects in erythroid differentiation observed in CM mice could be caused by inhibition of CBF/Runx1 function.
Figure 7.
Model for CM-induced preleukemic HSPC alterations and predisposition for leukemia evolution. Somatic expression of CM induces the expansion of preleukemic HSPCs including LSK and Pre-Meg/E progenitors with dysregulated erythroid (Spi-1, Fog1, Gata2, and Gfi1b) and megakaryocytic (Mpl, Pf4, and Vwf4) gene expression program, leading to impaired erythroid and megakaryocytic differentiation. With increased survival and proliferation rates, poorly differentiating preleukemic Pre-Meg/Es accumulate and serve as a cellular reservoir predisposed for additional mutations and AML transformation.
Consistent with the requirement of FOG1 (encoded by ZFPM1) for both erythroid and Mk lineage commitment,32 we found that Fog1 expression is greatly reduced in CM Pre-Meg/E cells. Given that FOG1 physically interacts with GATA1/GATA2 and GATA1 requires FOG1 for chromatin loop formation,33-35 the observed deficiency in FOG1 might be the predominant factor contributing to impaired erythroid commitment. The expression of GATA1 and GATA2 are dynamically and reciprocally regulated by multiple transcription factors including themselves, with high levels of GATA2 in primitive stem cells switching to high levels of GATA1 during erythropoiesis, referred to as the GATA switch.36 Although Gata1 levels appear to be unchanged in CM Pre-Meg/Es, much higher Gata2 expression was observed possibly because Gata2 could not be switched off in the absence of Fog1.33 In addition, Spi-1 (PU.1) and Gfi1b levels were increased approximately four- to fivefold in CM Pre-Meg/Es. PU.1 is known to inhibit erythroid differentiation by functioning as an integral repressor of a core erythroid network.37 Gfi1b is required for erythroid development beyond the bipotential Pre-Meg/E progenitors38 and enforced expression of GFI-1B reportedly induced expansion of immature erythroblast and resulted in increased GATA2 expression.39 The combined effect of Fog1 deficiency and sustained expression of Spi-1 and Gata2 together with Gfi1b could possibly form a regulatory feedback network underlying the erythroid differentiation block in CM Pre-Meg/Es. Mk-lineage transcription factors including Pf4, Vwf, and Mpl were greatly reduced in CM Pre-Meg/Es, consistent with our previous finding that CM expression impairs Mk differentiation.19
We previously reported an AMP population able to initiate leukemic transformation in the 129SvEv strain.19 However, these AMPs lack cell surface markers, CD34 and CD16/32, used to define classical MP subsets.40 Our current study use the well-characterized C57BL/6 background and additional myeloid-erythroid phenotypic markers25 to further define the cellular states. Although all phenotypically defined MP subsets seem to be expanded in CM preleukemic BM, we found that CM Pre-Meg/Es are selectively more resistant to spontaneous apoptosis in culture or the apoptosis induced by genotoxic stress. The sensitivity to stress, introduced by IR and/or in vitro culturing, varies greatly among phenotypically defined HSPC subsets. LSK cells are relatively more resistant to apoptosis induced by IR, consistent with a previous report that primitive stem/progenitor cells predominantly survive and undergo growth arrest after IR.41 Although CM expression protects LSK cells from spontaneous apoptosis in freshly isolated BM (Figure 2C), there is no apparent change in survival after culturing in vitro for 24 hours with or without IR (Figure 6B). This might be due to the low intrinsic apoptosis observed in LSK and/or the need of additional extrinsic factors to act in concert with CM to protect from apoptosis in vitro. In addition, we show that CM Pre-Meg/E is more susceptible to leukemic transformation compared with Pre-GM or GMP populations. Similar results were obtained when another cooperating oncogene FLT3-ITD42 were introduced (supplemental Figure 5A-B), suggesting that the susceptibility to transformation is not specific to Mpl overexpression. The immunophenotypes of the resulting leukemia are predominately lineage negative regardless of whether the leukemia originated from LSK or Pre-Meg/E. Variable fractions of both LSK- or Pre-Meg/E–derived leukemia cells express CD71 or CD41 and cKit (Figure 6F; supplemental Figure 4A). Irrespective of the initial cell of origin, the majority of MPs showed a Pre-GM–like immunophenotype in the leukemia BM (Figure 6F), and the frequency of phenotypic Pre-Meg/E cells in these leukemias was very low (Figure 6F). The leukemia-initiating activity appears to be enriched in the LSK and the side population, defined by their ability to efflux Hoechst 33342 dye43 (supplemental Figure 6A). Similar numbers (15 000-20 000) of phenotypic Pre-Meg/Es isolated from leukemia BM were unable to initiate leukemia when transplanted into secondary recipients (supplemental Figure 6B). These results suggest that there exist high degrees of phenotypic plasticity during leukemic transformation and pathogenesis. This could reflect the molecular and cellular reprograming caused by cell intrinsic oncogenic alterations, as well as the interplay of compensatory or microenvironmental signals in the leukemic state.
The complex coevolving clonal heterogeneity and phenotypic plasticity collectively control the dynamic homeostasis in leukemic hematopoiesis. We show that CM fusion protein impairs erythroid lineage specification, in addition to blocking differentiation of myeloid and lymphoid lineages. Consequently, CM causes context-selective alterations creating highly stress resistant premalignant progenitors that are predisposed for leukemia transformation. Abnormal erythroid progenitor proliferation and differentiation has been reported in human preleukemia cases, some of which was noted to subsequently succumb to chronic or acute leukemia.44 Thus, defects in erythroid progenitors could possibly be considered a predictive risk factor for leukemia evolution. Runx1 deficiency is recently shown to reduce ribosome biogenesis, lower p53 levels, and confer stress resistance to hematopoietic stem and progenitor cells.45 In addition, we recently showed that CM inhibits p53 activity through HDAC8-mediated deacetylation.46 Further understanding of how stress resistance and survival programs are differentially wired in distinct cellular compartments may inform potential approaches to eradicate chemoresistant preleukemic populations and prevent leukemia evolution.
Acknowledgments
This work was supported in part by the Gehr Family Center for Leukemia Research, American Cancer Society Research Scholar grant 123278-RSG-12-140-01-CSM (Y.-H.K.), and National Institutes of Health National Cancer Institute award R01CA178387 (Y.-H.K.). Research reported in this publication included work performed in the Analytical Cytometry Core and Animal Resource Center supported by National Cancer Institute award P30CA33572.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Q.C. designed and performed research, analyzed data, and wrote the manuscript; R.J., W.-K.H., G.J.C., B.Z., J.Q., H.L., and L.L. performed research, analyzed data, and reviewed the manuscript; C.-C.C. and G.M. designed research and reviewed manuscript; and Y.-H.K. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ya-Huei Kuo, Division of Hematopoietic Stem Cell and Leukemia Research, Gehr Family Center for Leukemia Research, Hematologic Malignancies and Stem Cell Transplantation Institute, Beckman Research Institute, City of Hope Medical Center, 1500 E. Duarte Rd, Duarte, CA 91010; e-mail: ykuo@coh.org.
References
- 1.Bloomfield CD, Marcucci G, Döhner K, Döhner H. Introduction: Acute myeloid leukemia. Semin Oncol. 2008;35(4):324–325. doi: 10.1053/j.seminoncol.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 3.Chan W-I, Huntly BJP. Leukemia stem cells in acute myeloid leukemia. Semin Oncol. 2008;35(4):326–335. doi: 10.1053/j.seminoncol.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Reinisch A, Chan SM, Thomas D, Majeti R. Biology and clinical relevance of acute myeloid leukemia stem cells. Semin Hematol. 2015;52(3):150–164. doi: 10.1053/j.seminhematol.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dash A, Gilliland DG. Molecular genetics of acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14(1):49–64. doi: 10.1053/beha.2000.0115. [DOI] [PubMed] [Google Scholar]
- 6.Passegué E, Jamieson CHM, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA. 2003;100(Suppl 1):11842–11849. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huntly BJP, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5(4):311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 8.Corces-Zimmerman MR, Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 2014;28(12):2276–2282. doi: 10.1038/leu.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shlush LI, Zandi S, Mitchell A, et al. HALT Pan-Leukemia Gene Panel Consortium. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corces-Zimmerman MR, Hong W-J, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 12.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol. 2009;37(6):649–658. doi: 10.1016/j.exphem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Liu P, Tarlé SA, Hajra A, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 14.Liu PP, Wijmenga C, Hajra A, et al. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chromosomes Cancer. 1996;16(2):77–87. doi: 10.1002/(SICI)1098-2264(199606)16:2<77::AID-GCC1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Castilla LH, Wijmenga C, Wang Q, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87(4):687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Stacy T, Miller JD, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 17.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 18.Castilla LH, Garrett L, Adya N, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23(2):144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 19.Kuo Y-H, Landrette SF, Heilman SA, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9(1):57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Kuo Y-H, Gerstein RM, Castilla LH. Cbfbeta-SMMHC impairs differentiation of common lymphoid progenitors and reveals an essential role for RUNX in early B-cell development. Blood. 2008;111(3):1543–1551. doi: 10.1182/blood-2007-07-104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao L, Cannons JL, Anderson S, et al. CBFB-MYH11 hinders early T-cell development and induces massive cell death in the thymus. Blood. 2007;109(8):3432–3440. doi: 10.1182/blood-2006-10-051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102(12):3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 23.Pop R, Shearstone JR, Shen Q, et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010;8(9):e1000484. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koulnis M, Pop R, Porpiglia E, Shearstone JR, Hidalgo D, Socolovsky M. Identification and analysis of mouse erythroid progenitors using the CD71/TER119 flow-cytometric assay. J Vis Exp. 2011;(54):2809. [DOI] [PMC free article] [PubMed]
- 25.Pronk CJH, Rossi DJ, Månsson R, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1(4):428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Miyawaki K, Arinobu Y, Iwasaki H, et al. CD41 marks the initial myelo-erythroid lineage specification in adult mouse hematopoiesis: redefinition of murine common myeloid progenitor. Stem Cells. 2015;33(3):976–987. doi: 10.1002/stem.1906. [DOI] [PubMed] [Google Scholar]
- 27.Landrette SF, Madera D, He F, Castilla LH. The transcription factor PlagL2 activates Mpl transcription and signaling in hematopoietic progenitor and leukemia cells. Leukemia. 2011;25(4):655–662. doi: 10.1038/leu.2010.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 30.Satpathy AT, Briseño CG, Cai X, et al. Runx1 and Cbfβ regulate the development of Flt3+ dendritic cell progenitors and restrict myeloproliferative disorder. Blood. 2014;123(19):2968–2977. doi: 10.1182/blood-2013-11-539643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behrens K, Triviai I, Schwieger M, et al. Runx1 downregulates stem cell and megakaryocytic transcription programs that support niche interactions. Blood. 2016;127(26):3369–3381. doi: 10.1182/blood-2015-09-668129. [DOI] [PubMed] [Google Scholar]
- 32.Mancini E, Sanjuan-Pla A, Luciani L, et al. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J. 2012;31(2):351–365. doi: 10.1038/emboj.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal S, Cantor AB, Johnson KD, et al. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA. 2004;101(4):980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vakoc CR, Letting DL, Gheldof N, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17(3):453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Jing H, Vakoc CR, Ying L, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29(2):232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko H, Shimizu R, Yamamoto M. GATA factor switching during erythroid differentiation. Curr Opin Hematol. 2010;17(3):163–168. doi: 10.1097/MOH.0b013e32833800b8. [DOI] [PubMed] [Google Scholar]
- 37.Wontakal SN, Guo X, Smith C, et al. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci USA. 2012;109(10):3832–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foudi A, Kramer DJ, Qin J, et al. Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J Exp Med. 2014;211(5):909–927. doi: 10.1084/jem.20131065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osawa M, Yamaguchi T, Nakamura Y, et al. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 2002;100(8):2769–2777. doi: 10.1182/blood-2002-01-0182. [DOI] [PubMed] [Google Scholar]
- 40.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 41.Mohrin M, Bourke E, Alexander D, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H-G, Kojima K, Swindle CS, et al. FLT3-ITD cooperates with inv(16) to promote progression to acute myeloid leukemia. Blood. 2008;111(3):1567–1574. doi: 10.1182/blood-2006-06-030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chui DH, Clarke BJ. Abnormal erythroid progenitor cells in human preleukemia. Blood. 1982;60(2):362–367. [PubMed] [Google Scholar]
- 45.Cai X, Gao L, Teng L, et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell. 2015;17(2):165–177. doi: 10.1016/j.stem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi J, Singh S, Hua W-K, et al. HDAC8 inhibition specifically targets Inv(16) acute myeloid leukemic stem cells by restoring p53 acetylation. Cell Stem Cell. 2015;17(5):597–610. doi: 10.1016/j.stem.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]