Abstract
The recombinant ALVAC vaccine coupled with the monomeric gp120/alum protein have decreased the risk of HIV and SIV acquisition. Ab responses to the V1/V2 regions have correlated with a decreased risk of virus acquisition in both humans and macaques. We hypothesized that the breadth and functional profile of Abs induced by an ALVAC/envelope protein regimen could be improved by substituting the monomeric gp120 boost, with the full-length single-chain (FLSC) protein. FLSC is a CD4-gp120 fusion immunogen that exposes cryptic gp120 epitopes to the immune system. We compared the immunogenicity and relative efficiency of an ALVAC-SIV vaccine boosted either with bivalent FLSC proteins or with monomeric gp120 in alum. FLSC was superior to monomeric gp120 in directing Abs to the C3 α2 helix, the V5 loop, and the V3 region that contains the putative CCR5 binding site. In addition, FLSC boosting elicited significantly higher binding Abs to V2 and increased both the Ab-dependent cellular cytotoxicity activity and the breadth of neutralizing Abs. However, the FLSC vaccine regimen demonstrated only a trend in vaccine efficacy, whereas the monomeric gp120 regimen significantly decreased the risk of SIVmac251 acquisition. In both vaccine regimens, anti-V2 Abs correlated with a decreased risk of virus acquisition but differed with regard to systemic or mucosal origin. In the FLSC regimen, serum Abs to V2 correlated, whereas in the monomeric gp120 regimen, V2 Abs in rectal secretions, the site of viral challenge, were associated with efficacy.
Introduction
A protective role of vaccine-induced Abs has been suggested by the correlate analysis of the RV144 Thai trial, the first phase III clinical trial to show partial efficacy. Vaccination with the canarypox vector ALVAC expressing HIV gag-pro and gp120-TM genes and bivalent gp120 proteins (AIDSVAX B/E) formulated in alum significantly reduced the risk of HIV infection with an estimated efficacy rate of 31.2% (1). Reduced HIV risk was primarily associated with Abs directed to the V1/V2 region of gp120, whereas Abs that mediated other functions, including Ab-dependent cellular cytotoxicity (ADCC), were correlates in secondary analyses (2, 3). The results of the Thai trial have engendered optimism in the HIV vaccine field; however, the efficacy of the ALVAC/gp120 vaccine regimen needs to be augmented.
Neutralizing Abs are an effective antiviral response, and when passively administered systemically or mucosally, they can prevent simian HIV intravaginal infection (4–6). Nonneutralizing Abs are readily induced by many vaccination regimens and can coordinate with innate effector cells to opsonize virus or kill infected cells. Multifunctional Abs that mediate ADCC, phagocytosis, or Ab-dependent cell-mediated viral inhibition have been associated with vaccine-induced protection from infection, reduced virus transmission, and virus control post SIV/HIV infection (3, 7–14). Thus, strategies that alter the breadth and/or the functional capacity of the Ab response may provide increased clinical benefits.
HIV binding and entry into target cells is a two-step process: binding to CD4 induces a conformational change in gp120 that brings together several variable and constant regions forming the coreceptor binding site. Coreceptor binding then facilitates virus entry (15). The CD4-induced conformational change in gp120 reveals conserved intermediate structures that are presented transiently to the immune system before virus-target cell fusion (16). These conserved moieties are antigenic and can be divided into three epitope clusters: A, B, and C (17). Cluster A epitopes are occluded by gp41 in envelope trimers and become exposed during entry. They are functional targets for ADCC and include the A32-directed conformational epitope in the C1 region of gp120 (18, 19). Cluster B and C epitopes are proximal to or include the coreceptor binding site. They are absent, occluded, or only minimally displayed on monomeric or trimeric gp120. Cluster B epitopes are also ADCC targets. Cluster C epitopes induce Abs with a range of neutralization potency and breadth, and include broadly neutralizing Abs like 17b (20). Interestingly, CD4 induced epitopes (CD4i) can also be formed on the surface of cells when recently synthesized envelope proteins interact with CD4 (19). Because these structures represent a possible Achilles heel for HIV, the virus has evolved a strategy to minimize their exposure by downregulating CD4 from the surface of HIV-infected cells through the function of the Nef and Vpu proteins (19).
The full-length single-chain (FLSC) protein was generated by fusing gp120 with a flexible linker and CD4 to allow the gp120/CD4 interaction and exposure of cryptic epitopes (21). Furthermore, FLSC induced Abs to CD4i epitopes were associated with accelerated simian HIV control in nonhuman primate studies (22). Recently, a DNA prime-FLSC protein boost vaccination regimen reduced the rate of SIVmac251 mucosal acquisition (23). Thus, our goal was to test whether in macaques, the protective efficacy of an RV144-like regimen using an ALVAC-SIV prime and a protein boost could be improved by inducing Abs to CD4i epitopes. Thus, we directly compared the Ab and cell-mediated immune response in ALVAC-SIV primed animals, boosted with either monomeric gp120 (24) or FLSC proteins. We found that although the nature of the protein boost significantly altered Ab responses, it had no impact on the T cell response and surprisingly did not improve vaccine efficacy against repeated intrarectal low doses of SIVmac251. However, both vaccination regimens induced V2-directed Abs that were associated with delayed SIV acquisition.
Materials and Methods
Animals, vaccination, and SIV challenge
Seventy-seven rhesus macaques (Macaca mulatta) of Indian origin were used in this study. All animals were housed and cared for under the guidelines of the Association for the Assessment and Accreditation of Laboratory Animal Care. The study was conducted with the approval of the Institutional Animal Care and Use Committee at Advanced BioSciences Laboratories (Rockville, MD).
Fifty-three animals were vaccinated in the quadriceps at weeks 0, 4, 12, and 24 with 108 PFU of ALVAC (vCP2432) expressing SIV genes gag-pro and gp120 transmembrane (TM) domain manufactured by Sanofi Pasteur (Fig. 1). The sequence of the SIV genes was obtained from a mucosally transmitted founder variant of SIVmac251 designated M766 (25). The gag-pro gene was codon optimized and constructed using human codon bias leaving the stem loop and slippery sequences intact to allow for the −1 frameshift required for pol translation. The gag-pro encoded the entire gag followed by pol through the end of the protease. The gp120TM gene was codon optimized and constructed using human codon bias. The native signal peptide was replaced with a synthetic signal based on the human tissue plasminogen activator signal peptide (MDAMKRGLCCVLLLCGAVFVTTTEA). The SIVM766 gp120 included the stretch from isoleucine at position 20 to arginine at position 527 and the 22-aa residue of the gp41 TM domain from tyrosine 695 to leucine 716, following the numbering based on SIVmac251. Six residues of the HIV-1LAI cytoplasmic tail, asparagine 706 to glycine 711, within the TM domain (numbering based on HIV-1HXB2 numbering convention) were added.
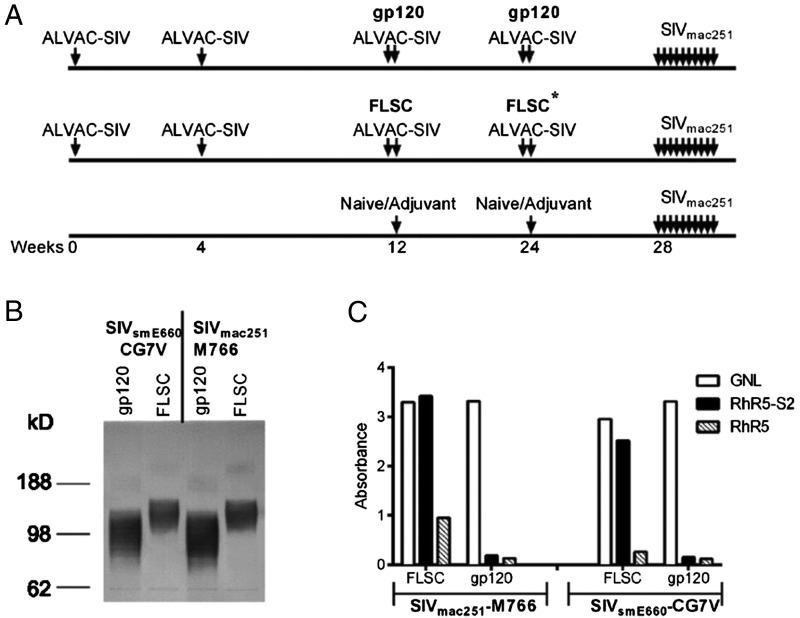
FIGURE 1.
Vaccination regimen and antigenicity of the FLSC proteins. (A) Schematic of the vaccination regimens used in this study. Vaccinated animals were all given ALVAC-SIV vaccines at weeks 0, 4, 12, and 24 and then boosted with either monomeric gp120 or FLSC proteins at weeks 12 and 24. Control animals were either naive or given an adjuvant at weeks 12 and 24. At week 28 all vaccinated animals and controls were given a low-dose SIVmac251 intrarectal challenge, which was repeated weekly for up to 10 wk based on SIV status. Asterisk refers to the fact that the boost at week 24 in the FLSC arm contained 50 mg of FLSC-M766 and 150 mg of FLSC SIV239 pw/eis. (B) Western blot showing the size of monomeric gp120 and FLSC proteins. 293 cells were transfected with plasmids expressing gp120-CG7V, FLSC-CG7V, gp120-M766, or FLSC-M766. Cells/media were harvested and immunoblotted with SIVmac251+ sera or SIVsmE660+ sera. (C) ELISA showing binding of gp120 and FLSC proteins from M766 and CG7V binding to GNL, a CCR5 peptide (RhR5), and a sulfonated CCR5 peptide (RhR5-S2). Both gp120 and FLSC bind to GNL, whereas only rhFLSC proteins bind to the sulfonated CCR5 peptide, indicating that an appropriately folded CD4-gp120 complex is expressed that reveals the coreceptor binding site.
At weeks 12 and 24, 27 animals were given a bivalent monomeric gp120 protein boost, formulated in alum, containing 200 μg of the SIVmac251-M766 protein and 200 μg of protein from a mucosally transmitted founder variant of SIVsmE660 designated CG7V, as previously described (Fig. 1) (24, 26). In brief, monomeric gp120 proteins were produced from Chinese Hamster Ovary cell lines stably transfected with plasmid DNA. Codon-optimized genes expressing M766 gp120, I20-R527, and CG7V gp120, V23-R527 (numbering based on SIVmac251) were inserted into the expression plasmid pSWTIPK3 following the 53 amino-terminal residues of the HSV-1 glycoprotein D (gD). Upon processing at the predicted cleavage site of the HSV-1 leader the gp120s are composed of the 28-residue gD N terminus, an alanine and serine linker, and the full-length gp120. Plasmids contain the CMV promoter for gp120 expression and an IRES puromycin-acetyltransferase cassette for selection of stable clones using the antibiotic puromycin. Monomeric gD-gp120 proteins were purified at Advanced BioScience Laboratories (Rockville, MD) by lectin affinity chromatography using Galanthus nivalis lectin agarose (Vector Laboratories, Burlingame, CA) and anion exchange chromatography using Q-Sepharose fast flow (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, U.K.) in flow-through mode.
Twenty-six ALVAC-SIV–vaccinated animals were boosted with rhesus FLSC (rhFLSC) proteins at weeks 12 and 24 (Fig. 1). The FLSC boost also consisted of M766 and CG7V gp120 proteins formulated in alum, but the gp120 proteins were linked to the D1D2 domains of rhesus CD4 by a 20-aa flexible linker that allowed self-association of gp120 and the CD4 molecule. The FLSC proteins were manufactured by Profectus Bioscience and are described later. The week 12 protein boosts consisted of 200 μg of FLSC-M766 and 200 μg of FLSC-CG7V. Similarly, at 24 wk, all animals were given 200 μg of FLSC-CG7V; however, because of manufacturing problems at week 24, a mixture of FLSC-SIVmac proteins were given: 50 μg of M766 along with 150 μg of SIVmac239 protein.
Vaccinated animals were compared with 47 controls including 23 historical controls that were exposed by the same operators to the same dose of the same virus stock in the same animal facility, and 24 contemporaneous controls that were either naive or given adjuvants (24). Four weeks after the last immunization, all vaccinated animals and controls were challenged with SIVmac251 via the rectal route at a dose of 120 TCID50 (Fig. 1A). Animals that tested negative for SIV RNA in plasma were rechallenged, and a maximum of 10 weekly challenges was administered.
rhFLSC protein production and formulation
Expi293F cells were transfected with pCDNA5 FRT TO plasmids expressing either codon-optimized M766, SIVmac239, CG7V gp120, or FLSC using the Expi293 expression system (Life Technologies). The HSV-1 gD was not added to these constructs. Media were collected and clarified by centrifugation at 4 d posttransfection. The FLSCs from the clarified supernatants were purified using lectin agarose from Galanthus nivalis (snow-drop; Vector Labs, Burlingame, CA). All steps were performed at 4°C. In brief, 16-mm columns containing 20 ml of lectin agarose were equilibrated with 60–80 ml of sterile filtered buffer A (0.65 M NaCl, 0.25% Empigen BB [Sigma] in 1× PBS) until the UV absorbance trace at 280 nm reached baseline. The clarified supernatants were adjusted to 0.65 M NaCl plus 0.25% Empigen BB, sterile filtered through a 0.22-μm filter, and loaded onto the lectin agarose columns overnight. After samples were loaded onto the columns, 20–30 ml of buffer A was added and the flow-through was collected until the UV absorbance trace decreased to baseline. The columns were washed four consecutive times with 40 ml each of sterile filtered buffer B (0.25% Empigen BB in 1× PBS), buffer C (1× PBS), buffer D (1 M of NaCl in 1× PBS), and then buffer C again. Buffer E (1 M of methyl α-d-mannopyranoside [MP Biomedicals, Sana Ana, CA] in 1× PBS) was added to the columns and allowed to flow through until the void column volume was reached and the UV absorbance increased (∼10–15 ml). The columns were allowed to sit for 30–60 min in the buffer E before elution of the FLSCs with an additional 20–40 ml of buffer E. The eluted proteins were then dialyzed 3× in 2 l for ≥4 h each time in 10,000 MWCO Slide-A-Lyzer dialysis cassettes (Thermo Fisher). The proteins were then concentrated to 1–3 mg/ml in Amicon Ultra-15 with Ultracel-10 centrifuge filters (10,000 MWCO; Thermo Fisher).
The purified FLSC proteins were formulated with 20 mg/ml aluminum phosphate (Alum; Catalent Pharma Solutions, Middleton, WI) at a protein/alum ratio of 1:8 in 5 mM acetate buffer (pH 5.1) with 40 mg/ml d-mannitol, pH 6.2.
SIV viral load
Plasma SIV RNA was quantified by nucleic acid sequence-based amplification as previously described (27). SIV DNA was quantified in mucosal tissues 3 wk post SIV infection by a real-time quantitative PCR assay with sensitivity of 10 copies/106 cells as previously described (28). In brief, genomic DNA was extracted from the rectal biopsies with the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s protocol as previously described (29). The quantity and quality of the DNA were assessed by OD260 measurements using an ND-1000 spectrophotometer (NanoDrop). The TaqMan probe and PCR primers for the real-time PCR were designed within the conserved gag gene of SIVmac239, and probe and primer sequences were also used for monkey albumin gene detection. The reaction conditions were as follows: the PCR mixture consisted of 500 ng of genomic DNA extracted from tissues; 200 nM of primers; 100 nM of probe; 2× TaqMan Universal PCR Mastermix (Applied Biosystems) consisting of 10 mM of Tris-HCl (pH 8.3); 50 mM of KCl; 5 mM of MgCl2; 300 μM each of deoxyadenosine triphosphate, deoxycytidine triphosphate, deoxyguanosine triphosphate; 600 μM of deoxyuridine triphosphate; 0.625 U of AmpliTaq Gold DNA polymerase; and 0.25 U of uracil N-glycosylase. Amplification was performed using 1 cycle at 50°C for 2 min and 1 cycle at 95°C for 10 min followed by a two-step PCR procedure consisting of 50 cycles of 15 s at 95°C and 1 min at 60°C. PCR amplification was performed using the ABI Prism 7500 Sequence Detector System (Applied Biosystems). The normalized value of the SIV proviral DNA load was calculated as SIV DNA copy number/Macaque albumin gene copy number × 2 × 106 and expressed as the number of SIV proviral DNA copies per 106 mononuclear cells.
Neutralization assays
Neutralization was measured as a reduction in luciferase reporter gene expression after a single round of infection in TZM-bl cells as described previously (30). TZM-bl cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Virus was incubated with serial 3-fold dilutions of samples in duplicate. Freshly trypsinized cells were added to each well. One set of control wells received cells and virus (virus control), and another set received cells only (background control). After the 48-h incubation, cells were transferred to 96-well black solid plates (Costar) for measurements of luminescence. Neutralization titers are the dilution at which relative luminescence units were reduced by 50% compared with that in virus control wells after subtraction of background relative luminescence units. Values are considered positive for neutralizing Ab activity based on the criterion of >3× the observed background against a negative control simian virus amphotropic murine leukemia pseudovirus (SVA-MLV) or signal detected in the sera from control unvaccinated animals, whichever is higher. Assay stocks of molecularly cloned Env-pseudotyped viruses SIVmac251.6, SIVmac251.30, SIVsmE660CR54-PK2A5, and SVA-MLV were prepared by transfection in 293T cells and were titrated in TZM-bl cells. The SIVmac251 challenge stock that had been expanded on rhesus PBMCs and titered in vivo was obtained from Nancy Miller, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
ADCC assay
ADCC was assessed in serum samples collected 2 wk after the final vaccination. The rapid and fluorometric–ADCC assay was performed as previously described using human PBMCs as effectors. SIVmac251 gp120-coated CEM.NKr cells were used as target cells, with an effector to target ratio of 50:1 (31). Serial dilutions of serum were evaluated and the ADCC titer defined as the reciprocal serum dilution at which the percent killing is greater than that of the mean of control samples plus 3 SDs. The ADCC activity as measured by this assay does include activity through trogocytosis as also described by others (32).
Western blot
Expi293F cells were transfected with pCDNA5 FRT TO plasmids expressing either M766 or CG7V gp120 or FLSC using the Expi293 expression system (Life Technologies). Media were collected and clarified by centrifugation at 2 d posttransfection. The clarified supernatants were subjected to SDS-PAGE under reducing conditions on a Bis-Tris 4–12% NuPAGE gel (Life Technologies) in 1× MOPS buffer. The proteins were transferred to a polyvinylidene difluoride membrane and detected using sheep anti–HIV-gp120 D7324 Ab (Aalto Bio Reagents, Dublin, Ireland) and an alkaline phosphatase–conjugated donkey anti-sheep secondary Ab. BCIP/NBT solution (Kirkegaard & Perry Laboratories, Gaithersburg, MD) was used to visualize the alkaline phosphatase–conjugated secondary Ab.
Binding to rhesus CCR5 peptide by ELISA
High-binding 96-well plates were coated overnight with 1 μg/ml streptavidin or Galanthus nivalis lectin (GNL). Plates were then blocked with blocking buffer (5% w/v BSA in 1× TBST) for 1 h. Saturating concentrations of biotinylated sulfonated (RhR5-S2) and unsulfonated (RhR5) peptide representing the N-terminal sequence of CCR5 were then added to the streptavidin plate. rhFLSC or gp120 proteins derived from SIVmac251 and SIVsmE543 transmitted/founder strains M744 and CG7V, respectively, were added at 1 μg/ml. Bound proteins were detected with antisera derived from macaques infected with SIVmac251 or SIVsmE543 followed by HRP-conjugated goat anti-monkey IgG. Between steps, the plates were washed 3× with 400 μl/well with 1× TBS with 0.1% Tween 20 (TBST) in an automated plate washer. Amount of bound protein was shown as absorbance at 450 nm.
Avidity index capture ELISAs
High-binding plates were coated with 1 mg/ml D7324 Ab (Aalto Bio Reagents) overnight at 4°C. The plates were washed 3× with 400 μl/well with 1× TBST in an automated plate washer, blocked with blotto buffer (5% w/v nonfat milk in 1× TBST) for 1 h, and washed again as before. SIVmac239 gp130 was added to the plates at 1 μg/ml, incubated for 1 h, and washed again. Test sera serially diluted in blotto buffer were added to the assay plates for 1 h. After washing, the wells were incubated for 30 min in 1.5 M sodium thiocyanate (NaSCN; Sigma Aldrich) or 1× PBS and washed again. HRP-conjugated goat anti-monkey Ab (KPL) was added at 1:1000 and incubated for 1 h. The plates were washed as before and incubated in SureBlue TMB Microwell Peroxidase Substrate (KPL) for 10 min. The reaction was stopped with 1 N H2SO4, and the plates were read on a spectrophotometer at 450 nm. The avidity index was calculated by taking the ratio of the NaSCN-treated plasma to the PBS-treated plasma.
Binding Abs to Fcγ receptors
High-binding plates were coated overnight at 4°C with 2 μg/ml of SIVmac239 gp130. The plates were washed 3× with 400 μl/well with 1× TBST in an automated plate washer, blocked with blotto buffer for 1 h, and washed again as before. Test sera serially diluted in blotto buffer were added to the assay plates for 1 h. Biotinylated Fcγ receptor (FcγR; Sino Biological, Beijing, China) was preincubated for 30 min with HRP-conjugated poly streptavidin (Life Technologies) at final concentrations of 0.3 and 0.8 μg/ml, respectively. After washing, the FcγR-poly streptavidin mixture was added to the plates for 1 h. The plates were washed as before and incubated in SureBlue TMB Microwell Peroxidase Substrate (KPL) for 10 min. The reaction was stopped with 1 N H2SO4 and the plates were read on a spectrophotometer at 450 nm.
Ab binding to FLSC, gp120, and SIVmac251 peptides
Capture ELISAs were performed as described earlier excluding incubation with NaSCN. In brief, plates were coated overnight with the D7324 capture Ab. After blocking, the plates were incubated with the indicated protein Ag (i.e., M766 and CG7V gp120 or FLSC). Serially diluted test sera were added to the coated/captured plates, and bound Ab was detected with HRP-conjugated goat anti-monkey Ab (KPL). HRP was detected using a colorimetric TMB reaction as described earlier.
Similarly, the Ab response to overlapping peptides spanning gp120 was determined by ELISA. A serial dilution of plasma was added to microtiter plates coated with individual peptides, and the titer was determined. The absorbance at OD450 nm is reported for peptide mapping.
Ab binding to scaffolded V1/V2 in blood
Binding response for gp70-SIV V1V2 scaffolds was evaluated using a custom SIV binding Ab multiplex assay performed as previously described (33, 34). In brief, gp70-SIV V1V2 scaffold proteins (produced by Dr. Abraham Pinter’s laboratory) were covalently coupled to carboxylated fluorescent beads (Luminex, Austin, TX). Ag-coupled beads were incubated with serially diluted serum samples, and after washes, subsequently incubated with biotinylated goat anti-monkey IgG (Rockland Immunochemicals, Limerick, PA). Beads were then washed and acquired on a Bio-Plex instrument (Bio-Rad, Hercules, CA), and mean fluorescence intensity (MFI) values were obtained for binding of each sample to each Ags coupled to the beads. MFI of binding to the gp70 backbone (MuLV gp70) was subtracted from the MFI of each gp70-SIV V1/V2 scaffold. Anti-SIV V1V2 levels are shown as MFI area under the curve computed over the dilutions series.
Ab binding to cyclic V2 peptides
Ninety-six-well Immuno 2U-bottom ELISA plates were coated overnight at 4°C with 100 μl of 2 μg/ml streptavidin (Sigma-Aldrich) in bicarbonate buffer, pH 9.6, followed by 100 μl of 1 μg/ml biotinylated cyclic V2 (cV2) peptide (synthesized by JPT Peptide Technologies) for 1 h at 37°C and then blocked with blocking buffer (0.5% milk in 1× PBS, 0.1% Tween 20, pH 7.4) overnight at 4°C. The contents were then dumped, and 100 μl of serum samples diluted in blocking buffer was added. Serum was initially diluted 1:100 in blocking buffer and then serial 2-fold dilutions were performed and added to wells for 1 h at room temperature. Wells were washed four times with wash buffer (PBS with 0.1% Tween 20, pH 7.4) using an automatic plate washer (BioStack washer, BioTek Instruments), and HRP-conjugated affinity-purified goat anti-monkey IgG (1:1000 in blocking buffer; The Binding Site) was added to wells for 1 h at room temperature. Plates were washed four times with wash buffer, 100 μl/well ABTS substrate was added, and color was allowed to develop at room temperature for 1 h in the dark. Plates were read at A405nm using an ELISA reader SpectraMax Plus (Molecular Devices). The data are expressed as end-point titers, with the titers being defined as the reciprocal of the highest dilution that yielded an absorbance value greater than twice the background value (wells that did not contain peptides).
SIV V2 peptide was synthesized by JPT Peptide Technologies GmbH, Berlin, Germany. The peptide was allowed to fold and cyclize under thermodynamic control at high dilution, and the purity was determined to be >90% by high-performance liquid chromatography and mass spectrometry. The amino acid sequence of the SIV V2 peptide is based on the SIVsmE543-3 V2 domain from GenBank, accession number U72748. The SIV V2 peptide sequences contain an N-terminal biotin tag. The sequences are as follows: GF SIVsmE543: CIKNNSCAGLEQEPMIGCKFNMTGLKRDKKIEYNETWYSRDLICEQPANGSESKCY; and GF SIVmac251 full: CIAQNNCTGLEQEQMISCKFNMTGLKRDKTKEYNETWYSTDLVCEQGNSTDNESRCY.
Reagents and surface plasmon resonance
CM5 chips and the BIAcore amine coupling kit were purchased from GE Healthcare (Piscataway, NJ). Streptavidin was purchased from Invitrogen (Grand Island, NY). Affinity-purified goat anti-monkey IgG (γ-chain) Ab was purchased from Rockland Immunochemicals (Gilbertsville, PA).
Surface plasmon resonance measurements were conducted with a BIAcore T200 using the CM5 chip. Streptavidin was immobilized onto the chip using the amine coupling kit as directed by the immobilization wizard packaged within the T200 control software. A total of 6700 response units (RU) of 1 μM of streptavidin in 20 mM of sodium formate, pH 4.2 (10-min contact time, 10 μl/min flow rate) was immobilized. The biotinylated peptide was prepared at a concentration of 1 μM in 20 mM of TRIS, pH 7.4, and allowed to flow (at 10 μl/min) over the streptavidin-coated surface of flow cell 4 until 3500 RU of SIV V2 peptide was captured.
The mucosal swabs were thawed on ice and centrifuged at 16,100 relative centrifugal force, 4°C, for 5 min. The supernatant was diluted 10-fold in TBS, pH 7.4, and then analyzed on the BIAcore. The diluted mucosal samples were passed over the chip surface at a flow rate of 30 μl/min for 3 min, followed by a 5-min dissociation period. At the end of the 5-min period, a 20 μg/ml solution of affinity-purified γ-chain–specific goat anti-monkey IgG Ab was passed over the peptide-coated Ig-bound surface for 2 min at a flow rate of 10 μl/min. After a 70-s dissociation period, the chip surface was regenerated and data analyzed as previously described using the BIAevaluation 4.1 software (35). The reported response units (RU) for the IgG-specific values are the difference between the average value of a 5-s window taken 60 s after the end of the anti-IgG injection and the average value of a 5-s window taken 10 s before the beginning of the anti-IgG injection. The data (RU) are presented for individual mucosal samples.
Chemstrips were used to determine the blood contamination in mucosal samples. Ten microliters of the mucosal supernatant sample was spotted onto a Chemstrip 5 OB Urine Test Strip. After 60 s, any change in color was recorded for comparison with the manufacturer’s color chart.
Phagocytosis
Phagocytosis was measured using methods based on those of Hartshorn et al. (36) and Huber et al. (37). SIVmac251 virus stock was concentrated with Lenti-X Concentrator (Clontech, Mountain View, CA) according to the manufacturer’s protocol and resuspended in complete medium (RPMI 1640 supplemented with 10% FBS and penicillin-streptomycin l-glutamine. FITC-labeled virus was prepared as described previously (36). In brief, concentrated virus stock was incubated with FITC at a 10:1 (virus/FITC) ratio by volume for 1 h and dialyzed overnight against PBS. FITC-labeled virus was stored in single-use aliquots in liquid nitrogen until use.
FITC-labeled virus (10 μl based on previous titration, data not shown) was incubated in screw cap Eppendorf tubes with heat-inactivated serum from preimmunized and week 26 samples (1:100 final dilution) in complete medium for 1 h at 37°C in 250 μl of total volume. After 1 h, tubes were spun at 21,920 × g at 4°C for 1 h. Without touching the bottom of the tube, the top 200 μl of supernatant from each tube was removed and discarded. Two hundred microliters of fresh complete medium was added to each tube and vigorously vortexed. One hundred microliters from each tube was dispensed into duplicate wells of a 96-well round-bottom microtiter plate. One hundred microliters of complete medium was added to two separate wells to serve as the cell only control. One hundred microliters of THP-1 cells (20,000 cells) was added to each well and the plate was incubated for 1 h on an orbital shaker at 37°C. After 1 h, cells were washed three times with PBS, trypsinized to remove bound but not internalized FITC-labeled virus, and resuspended in 150 μl of 4% paraformaldehyde. The microtiter plate was stored at 4°C in the dark overnight. The next day, the entire volume of each well was transferred to snap cap Eppendorf tubes and read on an Accuri C6 flow cytometer using CFlow Plus software. Data were collected and analyzed by setting the FL1 gate at <1% for the cell only control (Supplemental Fig. 1). A phagocytic index was obtained by multiplying the percent positive cells by the median MFI as previously described (38). The net phagocytic index is reported and was obtained by subtracting the mean of the indices from a pool of 16 preimmunized serum samples.
Intracellular cytokine assay
Mononuclear cells from blood were isolated and viably cryopreserved. Cells were thawed, rested overnight, and 2 × 106 cells were stimulated with 2 μg/ml cognate peptides for 6 h in RPMI 1640 containing 10% human serum in the presence of 5 μg/ml brefeldin A (Sigma-Aldrich). Nonstimulated cells, as well as cells stimulated with superantigen (staphylococcal enterotoxin A + staphylococcal enterotoxin B; Sigma-Aldrich), were used as controls. These cells were then stained with the following surface markers for 15 min at 4°C: CD4 (clone L200-PerCP-Cy5.5; BD Biosciences), CD8 (CD8–PE–Texas Red [ECD]; Cedarlane), CD95 (clone DX2-PE-Cy5; BD Biosciences), CD28 (clone CD28.2-Pacific Blue, custom-made; BD Biosciences), CCR7 (clone 3D12-PE-Cy7; BD Biosciences), and PD-1 (clone MIH4-FITC; BD Biosciences). Cells were fixed for 10 min in 100 μl of 2% paraformaldehyde at room temperature (25°C). To stain cells with Abs specific for intracellular cytokines (IFN-γ–Alexa-700, IL-2–allophycocyanin, TNF-α–PE; BD Biosciences), we incubated the cells with Abs in 0.25% saponin (Sigma-Aldrich) for 30 min at 25°C and analyzed them using the BD LSRII flow cytometer. Between 250,000 and 1 × 106 events were acquired for each condition. Data were then analyzed using DIVA software (BD Biosciences), and the frequency of cytokine-producing T cells is presented.
Statistical analysis
The Mann–Whitney–Wilcoxon test for continuous factors was used to compare groups. Correlation analyses were performed using the Spearman rank correlation method with exact permutation p values calculated. Significant differences between the FLSC group and the gp120 group in the binding of each of the 89 overlapping gp120 peptides were tested using the exact Mann–Whitney–Wilcoxon test, with the p values corrected for multiple comparisons by the Hochberg method. Because of the stringency of the correction, uncorrected p < 0.001 was taken as the cutoff for significance, corresponding to corrected p < 0.056. Peptide breadth was defined as the number of peptides in each group with an absorbance greater than background (0.058), and the difference was evaluated using McNemar test. Increased magnitude was defined as the number of peptides with a difference in median binding >0.25 and tested against the binomial distribution with a null proportion of 50% in each group. The number of challenges administered before SIV acquisition was assessed using the score test of the discrete-time proportional hazards model. Graphical analysis was performed using GraphPad Prism. Arithmetic or geometric means are shown on graphs and error bars represent the SE.
Ethics
All animals were housed and cared for under the guidelines of the Association for the Assessment and Accreditation of Laboratory Animal Care, the Office of Laboratory Animal Welfare, and the U.S. Department of Agriculture. The study was conducted with the approval of the Institutional Animal Care and Use Committee at Advanced BioSciences Laboratories (protocol no. 491). Animals were closely monitored by the staff veterinarian, and several steps were taken to prevent or alleviate suffering; these included pair housing during the vaccination phase of the study and close monitoring specifically following study-related procedures such as rectal or lymph node biopsies and after SIV infection.
The following steps were implemented should animals exhibit some pain or discomfort during or after biopsy procedures. Animals will be treated with buprenorphine (0.01–0.05 mg/kg body weight, intramuscular) for 12–96 h after surgery to ease pain and discomfort. Other analgesic agents (such as butorphanol 0.1–0.2 mg/kg, fentanyl 5–10 μg/kg) may be used at the discretion of the veterinarian. Tylenol (acetaminophen) (80 mg) may also be used for pain for minor problems such as trauma. Ketofen (ketoprofen) (5 mg/kg) or Banamine (flunixin meglumine) (1–2 mg/kg) is also used occasionally for pain associated with inflammation. Antibiotics such as ampicillin or amoxicillin (10 mg/kg) and cephalexin (25 mg/kg) are also given postsurgery. Appetite, drinking, and behavior are observed closely for at least 1 wk after surgery. In some instances, SIV-infected macaques may develop a marked drop in blood CD4+ T cells, severe weight loss, diarrhea, and so on. These animals may also acquire opportunistic infections caused by the drop of CD4+ T cells. An animal that is not responsive to treatment may constitute an end point as determined by the Advanced BioSciences Laboratories veterinarian.
Humane euthanasia criteria end points included diarrhea, inappetence, and/or weight loss (>20%, with reduced body condition), as well as tumor, infection, injury, neurologic signs, behavioral abnormality, or other condition that results in pain, distress, or a significant compromise to the animal’s well-being and cannot be alleviated by appropriate treatment. If any end point was reached, animals would be euthanized with an overdose given i.v. of a barbiturate euthanasia agent.
Results
Study design and antigenicity of the monomeric gp120 and the FLSC proteins
Rhesus macaques were vaccinated with ALVAC expressing SIV genes gag-pro and gp120TM at weeks 0, 4, 12, and 24 and given bivalent boosts with SIVmac251-M766 and SIVsmE660-CG7V gp120 or rhFLSC in alum at weeks 12 and 24 (Fig. 1A).
The expression of both the gp120 and the FLSC proteins was analyzed by Western blot, and as expected, the FLSC protein had a higher m.w. than the monomeric gp120 (Fig. 1B). The antigenicity of these proteins was evaluated by assessing binding to GNL and a CCR5 peptide (Fig. 1C). As expected, both gp120 and FLSC proteins bound to GNL; however, only the FLSC protein demonstrated robust binding to a sulfonated CCR5 peptide (Fig. 1C). Binding to the CCR5 peptide confirms that a correctly folded FLSC protein was expressed, and that the gp120/CD4 interaction exposes the coreceptor binding site more efficiently than monomeric gp120.
FLSC and monomeric gp120 induce Abs with qualitatively different binding profiles
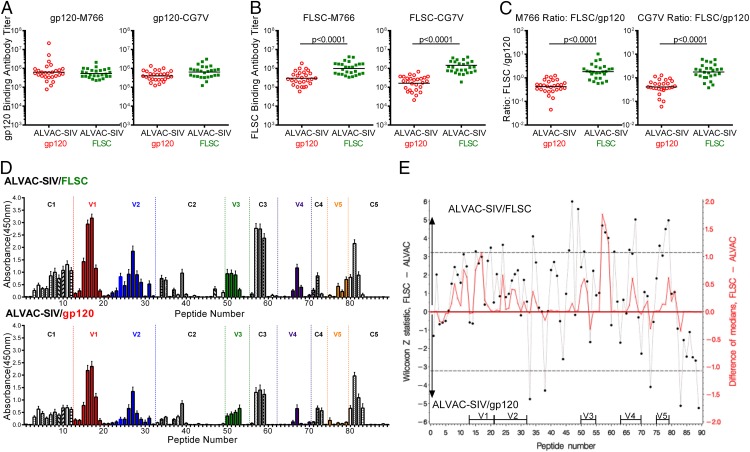
We compared the Ab response in ALVAC-SIV primed gp120 or FLSC-boosted animals by measuring binding Abs in serum 2 wk after the last vaccination (Fig. 2). Both groups developed equivalent high-titer binding Abs to monomeric gp120-M766 and gp120-CG7V (Fig. 2A). As expected, sera from the FLSC boosted animals had significantly higher titers of binding Abs that recognized the FLSC-M766 and the FLSC-CG7V immunogens compared with the monomeric gp120 boosted animals p < 0.0001 (Fig. 2B). The comparison of each animal’s binding titer to the FLSC-immunogen versus the gp120-immunogen gives an indication of the abundance of FLSC-specific Abs. Indeed, the FLSC/gp120 ratio was significantly higher in the FLSC group compared with the gp120 group: p < 0.0001 (Fig. 2C). This difference was not due to reactivity to the CD4 protein that is part of the FLSC immunogen because no seroreactivity was detected against CD4 alone (data not shown).
FIGURE 2.
ALVAC-SIV–primed FLSC boosted animals have increased recognition of gp120 peptides. (A) Binding Ab titers to monomeric gp120 proteins in ALVAC-SIV/gp120 and ALVAC-SIV/FLSC–vaccinated animals measured in serum 2 wk after the last vaccination. (B) Binding Ab titers to the FLSC immunogen in vaccinated animals measured in serum 2 wk after the last vaccination. Significantly higher binding was observed in the ALVAC-SIV/FLSC group compared with the ALVAC-SIV/gp120 group measured by the Mann–Whitney–Wilcoxon test with a p value <0.0001 for both proteins. (C) All data for the ALVAC-SIV/gp120 group have been previously reported (24). Ratio of Abs binding to the FLSC immunogen versus the monomeric gp120 immunogen (FLSC/gp120). This ratio demonstrates the abundance of FLSC-specific Abs. It was significantly higher in the ALVAC-SIV/FLSC group compared with the ALVAC-SIV/gp120 group evaluated by the Mann–Whitney–Wilcoxon test with p values <0.0001 for both proteins. (D) Peptide binding to 89 overlapping linear peptides that span SIVmac251 gp120. Shown is the average binding measured as absorbance to animals in the ALVAC-SIV/FLSC group on top and the ALVAC-SIV/gp120 group on the bottom. The constant regions of gp120 are shown in black and/or white bars, whereas the variable loops are in color (E). Comparative recognition of overlapping peptides presented as the difference between the median absorbance in the ALVAC-SIV/FLSC group, relative to the ALVAC-SIV/gp120 group, is shown in red on the right y-axis. The statistical significance of the differences is shown by the Z statistic of the Mann–Whitney–Wilcoxon test in black, on the left y-axis, with the dashed lines marking significance at the p < 0.056 level after a stringent correction for multiple comparisons by the Hochberg method. Increased recognition by the ALVAC-SIV/FLSC group is presented as positive (0–6) on the top half of the graph, whereas increased recognition in the ALVAC-SIV/gp120 group is presented as negative (−6 to 0) on the lower half of the graph.
Next, we assessed Ab recognition of overlapping linear peptides spanning the variable and constant regions of gp120 (Fig. 2D). ALVAC-SIV/gp120 vaccination of macaques, similar to ALVAC-HIV/gp120 vaccination in humans, induces recognition of C1, V2, V3, and C5 regions of gp120 (39). FLSC boosting significantly increased the magnitude of Ab recognition (p = 0.0005), assessed as peptides with differences in median absorbance >0.25. Similarly, the FLSC group demonstrated increased Ab breadth, with a greater number of peptides recognized relative to the gp120 group. The binding to each peptide in the two vaccination regimens was evaluated using the Z statistic of the Mann–Whitney–Wilcoxon test with the Hochberg method used to correct for multiple comparisons (Fig. 2E). Twenty-two peptides were differentially recognized; the FLSC group demonstrated better Ab binding to 17 peptides relative to the gp120 group (Table I). Of the 17 peptides, 6 mapped to the C3 and V3 domains. Abs to the α2 helix of the C3 domain correlated with vaccine-induced protection from SIVsmE660 (34). Overlapping peptides 57, 58, and 59 used in this study contain the analogous C3 α2 helix of SIVmac251 and were all significantly better recognized in the FLSC group compared with the gp120 group: p < 0.0001, p = 0.0004, and p = 0.0021, respectively (Table I).
Table I. Differential recognition of peptides by ALVAC-SIV/gp120 and ALVAC-SIV/FLSC vaccines.
| Peptide | Sequence | Region | Is Greater Than | p Value |
|---|---|---|---|---|
| 15 | PCVKLSPLCITMRCNKSETD | V1 | FLSC > gp120 | 0.053 |
| 20 | ITTAAPTSAPVSEKIDMVNE | V1 | FLSC > gp120 | 0.024 |
| 24 | IAQNNCTGLEQEQMISCKFT | V2 | FLSC > gp120 | 0.013 |
| 34 | NTSVIQESCDKHYWDTIRFR | C2 | FLSC > gp120 | 0.0013 |
| 46 | NRTYIYWHGRDNRTIISLNK | C2 | FLSC > gp120 | 0.046 |
| 47 | WHGRDNRTIISLNKYYNLTM | C2 | FLSC > gp120 | <0.0001 |
| 49 | NKYYNLTMKCRRPGNKTVLP | C2/V3 | FLSC > gp120 | <0.0001 |
| 51 | PGNKTVLPVTIMSGLVFHSQ | V3 | FLSC > gp120 | 0.0530 |
| 57 | WKDAIKEVKQTIVKHPRYTG | C3 | FLSC > gp120 | <0.0001 |
| 58 | EVKQTIVKHPRYTGTNNTDK | C3 | FLSC > gp120 | 0.0004 |
| 59 | VKHPRYTGTNNTDKINLTAP | C3 | FLSC > gp120 | 0.0021 |
| 61 | DKINLTAPGGGDPEVTFMWT | C3 | FLSC > gp120 | 0.0560 |
| 66 | MNWFLNWVEDRDVTTQRPKE | V4 | FLSC > gp120 | 0.039 |
| 68 | VTTQRPKERHRRNYVPCHIR | V4 | FLSC > gp120 | <0.0001 |
| 76 | TSLIANIDWTDGNQTSITMS | V5 | FLSC > gp120 | 0.015 |
| 78 | NQTSITMSAEVAELYRLELG | V5 | FLSC > gp120 | 0.0001 |
| 79 | MSAEVAELYRLELGDYKLVE | V5 | FLSC > gp120 | <0.0001 |
| 33 | CYMNHCNTSVIQESCDKHYW | C2 | gp120 > FLSC | <0.0001 |
| 38 | PGYALLRCNDTNYSGFMPKC | C2 | gp120 > FLSC | 0.0005 |
| 73 | KNVYLPPREGDLTCNSTVTS | C4 | gp120 > FLSC | 0.0013 |
| 83 | GLAPTDVKRYTTGGTSRNKR | C5 | gp120 > FLSC | <0.0001 |
| 89 | AMGAASLTLTAQSRTLLAGI | C5 | gp120 > FLSC | <0.0001 |
Boldface indicates the region of peptide overlap. The putative CCR5 binding sites in V3 are underlined: CRR…SGLV…QAWC.
The V3 loop of gp120 is essential for coreceptor binding and determines CCR5 usage in HIV and SIV (40–43). Analogy with the HIV gp120 CCR5 binding domain (16, 44, 45) indicates that the V3 region CRR…SGLVF…QAWC of SIVmac251 contributes to CCR5 binding. In addition, an L→W mutation in the SGLVF sequence confers CXCR4 usage to a SIVmac239 variant (45). The FLSC group had greater binding Abs to peptides 49 and 51 (p < 0.0001 and p = 0.053) that together contain the CRR…SGLVF sequence (Table I). Thus, FLSC boosting significantly increased Ab recognition of epitopes that may be important for virus control.
The FLSC protein boost induces Abs with improved breadth of neutralization and increased ADCC
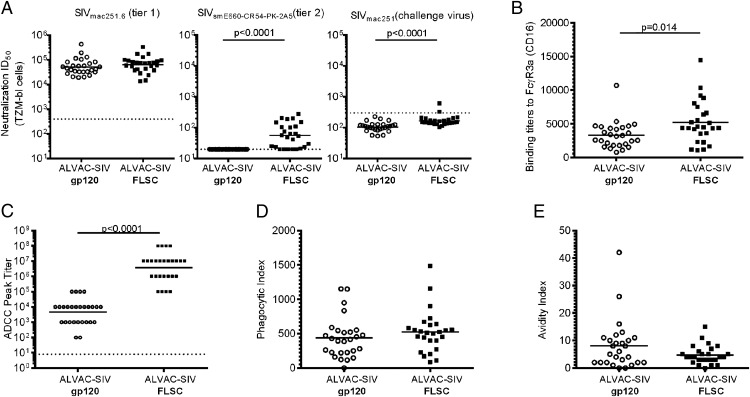
We next investigated whether the FLSC boosting affected Ab function and we used a panel of SIV strains that vary in their sensitivity to neutralization to assess the in vitro neutralization capacity of the sera from vaccinated macaques. We found that Abs induced by both the gp120 and the FLSC regimens similarly neutralized the tier-1–like (“easy to neutralize”) SIVmac251.6 viruses (Fig. 3A, left panel). In contrast, only the FLSC boosted animals also neutralized a tier-2–like (“difficult to neutralize”) SIVsmE660 virus (Fig. 3A, middle panel), with a significant difference in neutralization capacity (p < 0.0001) observed between the FLSC and gp120 groups. Animals boosted with FLSC demonstrated significantly higher neutralization ID50 values for the SIVmac251 challenge virus, p < 0.0001, although only 2 of the 53 vaccinated animals (both in the FLSC group) were above the cutoff level of the assay (Fig. 3A, right panel). None of the vaccinated animals neutralized a tier-2–like SIVmac251.30 isolate (data not shown).
FIGURE 3.
Vaccination with FLSC proteins altered select Ab functions. (A) Serum neutralization of SIVmac251.6 (left), SIVsmE660-CR54-PK-2A5 (middle), and the SIVmac251 challenge virus stock (right). Neutralization was assessed in TZMbl cells 2 wk postvaccination with gp120 boosted animals in open circles and the FLSC boosted animals in closed squares. Sera is considered positive if the sample value is >3× the observed background of a control murine leukemia pseudovirus or unvaccinated controls. Dashed horizontal lines on the graph indicate background levels. Both vaccine groups showed equivalent neutralization of the tier 1–like SIVmac251.6 isolate, whereas the ALVAC-SIV/FLSC group demonstrated significantly better neutralization of the tier 2–like SIVsmE660 pseudovirus; p < 0.0001 measured by the Mann–Whitney–Wilcoxon test. The ALVAC-SIV/FLSC group also showed significantly higher neutralization ID50 values for the SIVmac251 challenge virus, p < 0.0001, although only two animals had values that scored positive, that is, were greater than the cutoff level of the assay. (B) All data for the ALVAC-SIV/gp120 group in (A) have been previously reported (24). Serum binding Ab titers to FcγR3a (CD16), the ALVAC-SIV/FLSC group, had significantly higher binding titers relative to the ALVAC-SIV/gp120 group; p = 0.014 evaluated by a Mann–Whitney–Wilcoxon test. (C) ADCC was measured in serum from vaccinated animals 2 wk after the last vaccination. The ALVAC-SIV/FLSC group demonstrated significantly higher ADCC titers, p < 0.0001, by the Mann–Whitney–Wilcoxon test. (D) The data for the ALVAC/gp120 group have been previously reported (24). Equivalent opsonization of FITC-coated SIV virions presented as phagocytic index for the ALVAC-SIV/gp120 group and the ALVAC-SIV/FLSC group. (E) The avidity of Abs to gp120 presented as avidity index was similar in both groups.
High-avidity nonneutralizing Abs can prevent or control infection via Fcγ-mediated interactions with macrophages and NK cells. Thus, we compared binding to the FcγR (CD16), ADCC, phagocytosis, and avidity of Abs in the two vaccinated groups. FLSC significantly increased Ag-specific Abs that could bind to recombinant FcγR3a-CD16 (p = 0.014; Fig. 3B), whereas binding to recombinant FcγR2a and FcγR1 was similar in the FLSC and gp120 groups (data not shown). NK cells expressing FcγR3a can coordinate with Abs in the killing of infected cells. FLSC boosting also significantly increased the titer of Abs mediating ADCC (p < 0.0001; Fig. 3C) and increased maximum killing of gp120-coated target cells compared with the gp120 group (p < 0.0001, data not shown). Titers of Ag-specific Abs bound by recombinant FcγR3a were directly correlated with ADCC titer (r = 0.38, p = 0.005). Both vaccination regimens induced Abs that facilitated equivalent phagocytosis by the THP-1 cell line of FITC-labeled SIV virus and bound gp120 with similar avidity (Fig. 3D, 3E).
Serum Abs to the gp120 V2 region decrease the risk of SIVmac251 acquisition
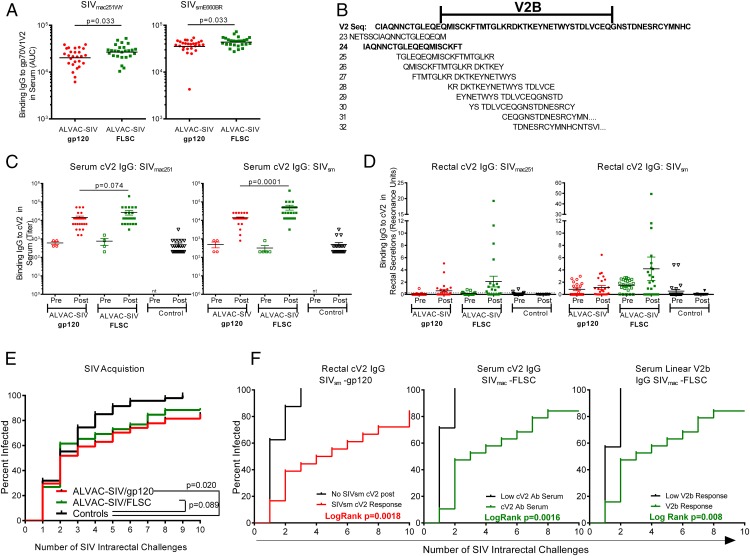
Reduced risk of HIV acquisition in the RV144 trial was correlated with serum vaccine-induced binding Abs to the V1/V2 region of gp120 (2, 46). V2 reactivity in the RV144 trial was tested using the V1/V2 loops fused to the murine leukemia virus gp70 (gp70-V1/V2), linear peptides spanning the V2 loop, as well as V2 peptides cyclized by disulfide bonds (39, 47). ALVAC-HIV/gp120 vaccines induced two potentially distinct V2 Ab responses: one directed to a conformational V1/V2 epitope measured by the gp70-V1/V2 scaffold, and the other to a linear epitope in V2 proximal to the putative α4β7 binding site (47). Using similar reagents constructed with SIVmac251 and SIVsmE543 variable loop sequences, we measured serum binding Ab levels to gp70-V1/V2 proteins, cV2, and linear V2 peptides at 2 wk after the last vaccination. ALVAC-SIV–primed FLSC-boosted animals had significantly higher levels of gp70-V1/V2–directed Abs (p = 0.033; Fig. 4A). Fig. 4B shows the V2 sequence used to measure serum Ab binding to 10 overlapping linear peptides that spanned the V2 region. The FLSC group demonstrated significantly better recognition of peptide 24 that spans the first third of the V2 loop, compared with the gp120 group (p = 0.013, Supplemental Fig. 2A). The second half of the V2 loop designated (V2b) spans peptides 26–28 and contains the region proximal to the putative α4β7 binding site. The majority of animals recognized the V2b region, and the magnitude of the response did not differ between the monomeric gp120 and the FLSC groups (Supplemental Fig. 2B).
FIGURE 4.
FLSC boosting increased Abs directed to the V2 loop of gp120. (A) Serum binding Abs to the gp-70 scaffolded V1/V2 proteins from SIVmac251WY (left) and SIVsmE660BR (right) measured 2 wk after the last vaccination. Significantly higher Abs are observed in the FLSC boosted animals compared with the gp120 boosted group evaluated by the Mann–Whitney–Wilcoxon test: p = 0.033 for both proteins. (B) Sequence of the V2 region of SIVmac251 and the 10 linear overlapping peptides that were used to map V2 responses. The sequence of peptide 24 is bolded. (C) Serum binding IgG titers to full-length cV2 peptides from SIVmac251 (left) and SIVsmE543 (right) measured 2 wk after the last vaccination. Significantly higher SIVsm cV2 Abs were seen in the FLSC group versus the gp120 group, p = 0.0001, whereas cV2 Abs to SIVmac251 approached, but did not attain, statistical significance: p = 0.074 measured by the Mann–Whitney–Wilcoxon test. (D) Binding Abs (IgG) in rectal secretions measured as resonance units to the full-length cV2 immunogens SIVmac251 (left) and SIVsmE543 (right) in vaccinated groups and controls. (E) Rate of SIV acquisition, shown as the percent infected after weekly intrarectal exposure to SIVmac251. ALVAC-SIV/gp120 vaccinated significantly decreased the rate of SIV acquisition compared with controls (p = 0.02) measured by the score test of the discrete time proportional hazards model. (F). SIV acquisition in the ALVAC-SIV/gp120 group dichotomized based on the presence or absence of cV2 Abs (IgG) to SIVsm in the rectal secretions (left panel). Animals with detectable rectal IgG to SIVsm had a slower rate of SIV infection, p = 0.0018, measured by the score test of the discrete time proportional hazards model. In the middle and right panels, animals in each group were dichotomized based on whether their serum cV2 or linear V2b peptide response fell above or below the 25th percentile. If the response was below the 25th percentile, the animals were considered low responders. ALVAC-SIV/FLSC–vaccinated animals with cV2 (middle panel) and V2b responses (right panel) above the 25th percentile had a slower rate of SIV infection (p = 0.0016 and p = 0.008), measured by the score test of the discrete time proportional hazards model. The data for the ALVAC-SIV/gp120 group (A and C–F) have been previously reported (24).
To confirm V2 reactivity, we compared binding Abs with cV2 peptides from SIVmac251 and SIVsmE543 in serum and rectal secretions. We found significantly higher SIVsm-cV2 responses in the serum of FLSC boosted animals compared with the gp120 group (p = 0.0001; Fig. 4C). A nonsignificant increase in serum SIVmac251 cV2 responses was also observed in the FLSC group (p = 0.074; Fig. 4C). Similarly, a nonsignificant increase in rectal cV2 Abs was observed in the FLSC group, directed to both SIVmac251 and SIVsmE543 Ags (Fig. 4D).
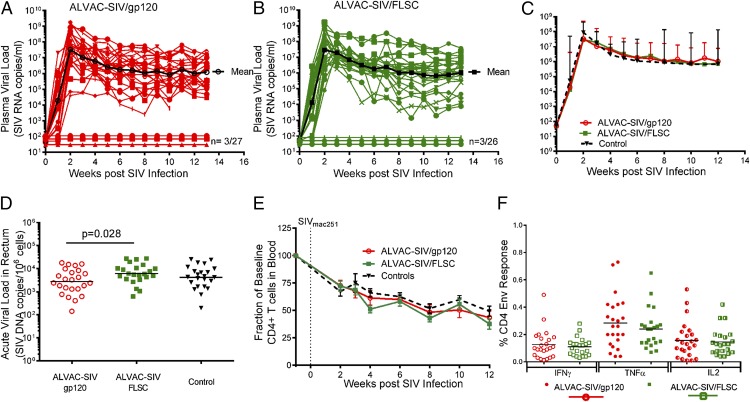
We next assessed vaccine efficacy by comparing the rate of intrarectal acquisition over the course of 10 weekly intrarectal exposures to SIVmac251 virus in vaccinated and unvaccinated animals. Animals were challenged with 120 TCID50 of SIVmac251, a dose that infects an average of 37% of the controls at each exposure. Seven days after SIV exposure, we measured SIV RNA in plasma by nucleic acid sequence-based amplification, and animals with <50 copies/ml at that time were considered SIV− and rechallenged. This study was powered to compare SIV acquisition between vaccinees and controls, but not to compare the rate of SIV acquisition between the two vaccine groups. As demonstrated previously, vaccination with ALVAC-SIV and monomeric gp120 significantly decreased the rate of SIV acquisition compared with controls (p = 0.020; Fig. 4E), with an estimated vaccine efficacy rate of 44% at each mucosal challenge (24). ALVAC-SIV–primed FLSC boosted animals demonstrated a decreased SIVmac251 acquisition rate compared with controls, which approached, but did not attain, statistical significance (p = 0.089; Fig. 4E), with an estimated vaccine efficacy rate of 34%. Notably, at the end of the challenge phase, that is, 2 wk after the 10th SIV challenge, three animals in the gp120 group and three in the FLSC group remained uninfected.
Next, we determined whether any immunologic responses measured were associated with delayed SIVmac251 acquisition. No correlation was found with T cell responses; however, delayed acquisition in both groups was associated with V2 responses. Animals in each group were dichotomized based on the presence or absence of a cV2 response in serum or rectal secretions. In the ALVAC-SIV/gp120 regimen, a decreased rate of SIV acquisition was observed in animals with mucosal cV2 responses directed to the SIVsmE543 Ag (p = 0.0018; Fig. 4F, left panel) (24). A similar association was not observed with the level of serum CV2 responses (data not shown). In contrast, in the ALVAC-SIV/FLSC regimen, serum, but not mucosal, Ab responses greater than the 25th percentile directed to cV2 or linear V2b peptides were associated with delayed SIV acquisition; p = 0.0016 and p = 0.008, respectively (Fig. 4F, middle and right panels, and data not shown). Importantly, the acquisition curves of animals that had Abs to cV2 or V2b differed significantly from that of control animals (Supplemental Fig. 3A, 3B).
FLSC boosting does not improve T cell responses or postinfection virus control
The infected vaccinated animals had peak viremia ranging from 106 to109 and set point viremia ranging from 104 to 108 (Fig. 5A, 5B). Next, we compared the geometric mean SIV plasma virus load among the gp120 group, FLSC group, and controls, and found no significant differences in peak or set-point viremia (Fig. 5C). Two to three weeks post SIV infection we obtained rectal pinch biopsies from all vaccinated animals and controls, and quantified the level of SIV DNA per 106 cells. ALVAC-SIV/gp120 vaccines had significantly lower SIV DNA levels in the rectum compared with the FLSC group (p = 0.028) but had a nonsignificant reduction in SIV DNA compared with controls. However, when the SIV DNA levels in all three groups are compared, there is no significant difference among the FLSC, gp120, and control groups (p = 0.074; Fig. 5D). One consequence of robust HIV/SIV virus replication is the loss of CD4+ T cells, a marker of disease progression. All vaccinated animals and controls had a progressive decline in CD4+ T cells and lost on average 50–65% of their CD4+ T cells within the first 12 wk of infection (Fig. 5E).
FIGURE 5.
Lack of virus control in ALVAC-SIV–primed gp120 or FLSC boosted infected macaques. (A) SIV plasma virus in ALVAC-SIV/gp120 animals over the course of the study; the geometric mean of all SIV-infected animals is shown in black. Three of 27 animals remained SIV− (SIV RNA < 50 copies/ml) over the 13 wk of follow-up. (B) SIV plasma virus in ALVAC-SIV/FLSC animals over the course of the study; the geometric mean of all SIV-infected animals is shown in black. Three of 26 vaccinated animals remained SIV− (SIV RNA < 50 copies/ml) over the 13 wk of follow-up. (C) Comparison of the geometric mean of plasma virus in SIV-infected animals from the gp120 group (red), FLSC group (green), and controls (black). No difference in peak or set point plasma virus is observed between vaccinated animals and controls. (D) Virus burden in the rectal mucosa measured as SIV DNA/106 mononuclear cells. Rectal biopsies were obtained 3 wk post SIV infection, and the ALVAC-SIV/gp120 animals are shown in red open circles, the ALVAC-SIV/FLSC animals in green squares, and the controls in black triangles. A significantly lower level of SIV DNA was observed in the gp120 group compared with the FLSC group (p = 0.028), but when both vaccine groups and controls are compared the differences are not significant (p = 0.074). (E) Percentage of baseline CD4 T cells in the blood over the course of the study. A similar progressive loss of CD4 T cells is observed post SIV infection over the 12 wk of follow-up in vaccinated animals (red lines, gp120 group; green lines, FLSC group) and controls (black line). (F) Envelope (Env)-specific CD4+ T cell responses measured in PBMCs 1 wk before SIV challenge. Shown is the frequency of IFN-γ+, TNF-α+, or IL2+ T cells after stimulation with overlapping SIVmac251-m766 Env peptides. ALVAC-SIV/gp120–vaccinated animals are shown by circles, whereas ALVAC-SIV/FLSC animals are shown by squares. A similar frequency of Env-specific CD4 T cells is observed in both groups after background subtraction of unstimulated cells. The data for the ALVAC-SIV/gp120 group (A and C–F) have been reported previously (24).
ALVAC/gp120 vaccination characteristically induces low T cell responses in humans and nonhuman primates (1, 35). One week before the first SIV challenge, we stimulated PBMCs with overlapping peptides that spanned SIVmac251 Gag or Env and evaluated the cytokine profile of SIV-specific CD4+ and CD8+ T cells. We observed negligible anti-Env and anti-Gag CD8+ T cell responses, no Gag-specific CD4 responses (data not shown), and comparable Env-specific CD4 T cell responses after the last immunization in both groups (Fig. 5F). The lack of virus control once infection occurred is likely due to the limited cell-mediated CD8+ T cell responses induced by these vaccine modalities.
Discussion
Vaccine-mediated protection from HIV infection appeared an unachievable goal until 2009, when the results of the RV144 Thai trial demonstrated that a recombinant ALVAC-HIV vaccine given with a bivalent gp120 Clade B/E boost could significantly reduce the risk of HIV acquisition (1). The protective efficacy rate of this regimen was estimated at 31.2% in a population primarily at low risk of HIV infection. To date, vaccine efficacy or a lack thereof, evaluated using mucosal exposure to SIV as a challenge model, has mirrored the results of all six HIV vaccine efficacy trials in humans (48–50). In this study, we evaluated relative vaccine efficacy by using low repeated doses of the pathogenic SIVmac251 propagated in macaque cells (35). This virus stock is highly genetically diverse, uses the CCR5 coreceptor, and similar to most primary HIV strains, is resistant to neutralization. One of the vaccine regimens reported in this study was designed to model RV144 in macaques and included an ALVAC vector expressing SIVmac251 Ags and two boosts with the two monomeric recombinant gp120 SIVmac251 and SIVsmE660 proteins (24). With this regimen, we observed an estimated vaccine efficacy of 44% (24) measured as a decreased risk of SIVmac251 acquisition at each mucosal challenge compared with controls (p = 0.020; Fig. 4E).
In this study, we aimed to improve the Ab breadth and functional profile induced by the ALVAC/gp120 vaccine by substituting the monomeric gp120 boosting of ALVAC-SIV–primed animals with an FLSC protein boost consisting of SIV-gp120 linked to rhFLSC CD4. Similar to previous studies using HIV-FLSC proteins (21), we show that SIV-FLSC proteins indeed adopt a conformation where the coreceptor binding site is exposed and can be bound by CCR5 peptides (Fig. 1).
ALVAC-SIV–primed FLSC-boosted macaques demonstrated increased Ab magnitude and breadth compared with animals boosted with monomeric gp120 (Fig. 2, Table I). Importantly, FLSC also induced Abs with increased recognition of peptides 49 and 51 that include the putative CCR5 binding domain in the V3 loop of SIV gp120 (Table I). The V3 loop of both HIV and SIV is critical for coreceptor binding and determines which coreceptor is used for entry (40–45). A CXCR4 using SIVmac239 variant was described with three mutations in the V3 loop sequence: NKTVLPVTIMSGLVF (positions shown in bold). These mutations either conferred or improved CXCR4 usage, demonstrating the importance of this V3 sequence for coreceptor usage (45). This identical sequence is present in peptide 51 that showed increased recognition by the FLSC group, further confirming that FLSC boosting induced Abs to the coreceptor binding site.
Surprisingly, FLSC-increased immunogenicity including the induction of Abs directed to the CCR5 binding site, high neutralizing Abs to a tier 2 virus, and increased ADCC (Fig. 3) but did not result in increased vaccine efficacy. Vaccine efficacy was significant in the ALVAC-SIV/gp120 group (44%) and not in the ALVAC-SIV/FLSC group. There was a trend for reduced SIV acquisition (p = 0.089), and a lower estimated vaccine efficacy rate of 34% at each mucosal challenge. At the end of the challenge phase, both vaccine groups had three animals that remained uninfected. Neither regimen decreased acute or set-point viremia or curtailed peripheral CD4+ T cell loss (Fig. 5). Similarly, a lack of virus control was observed in both macaque and human studies using ALVAC/gp120 regimens that induce limited CD8 responses (1, 35). FLSC boosting did not improve the T cell response relative to gp120-boosted animals (Fig. 5F).
In humans, the ALVAC-HIV/gp120 vaccine regimens induced V2-directed Abs measured by gp70-V1/V2 scaffolds and linear sequences that mapped to the midregion of V2 proximal to the α4β7 binding site, and were a correlate of reduced risk of acquisition of HIV (2, 39, 47). Furthermore, V2 Abs to residue 169 exerted immunologic pressure on transmitted viruses, producing a sieve effect in HIV-infected vaccinees (51). The FLSC boost increased binding Abs directed to the scaffolded gp70-V1/V2, a linear V2 peptide, and cV2 peptides (Fig. 4). In neither regimen was the decreased risk of SIV acquisition associated with serum Abs to the gp70 V1/V2 scaffold, as observed in the RV144 trial in humans. Rather, serum Abs to both cyclic and linear V2 peptides were associated with delayed SIV acquisition in the FLSC group (Fig. 4F). In the monomeric gp120 group it was the mucosal, and not the serum, Ab response to cV2 that correlated with decreased risk of SIV acquisition (Fig. 4F) (24). These data suggest that there may be differences in the Ab specificity and function that populate mucosal sites in animals immunized with these different Ags. Limitations in samples obtained from the mucosal secretions prevented the functional characterization of mucosal Abs. It is also possible that FLSC may induce different protective immune responses as compared with monomeric gp120 (23).
Apart from increasing binding Abs, boosting the Ab response with FLSC proteins also increased Ab function (Fig. 3). ADCC was a secondary correlate in RV144- (2) and ALVAC-HIV/gp120–induced Abs directed to the C1 region of gp120 that were potent mediators of ADCC in vitro (3). Similar ADCC epitopes have not been defined for SIV, but we were intrigued to find increased recognition of C1 peptides in the FLSC group and significantly increased ADCC titer (Fig. 3). However, none of these responses correlated with a decreased risk of SIVmac251 acquisition.
Neither vaccine regimen induced neutralizing Abs with sufficient breadth and potency to block SIVmac251 entry into target cells in vitro. It is notable that the FLSC-immunized animals, boosted with the FLSC-SIVsmE660-CG7V immunogen, derived from a virus that demonstrates intermediate neutralization resistance (52), neutralized the resistant pseudovirus SIVsmCR54-PK-2A5 (Fig. 3A). None of the animals in the gp120 group had Abs that could neutralize this tier-2–like SIVsm virus. FLSC immunization also significantly increased binding to the C3 α2 helix region of SIVmac251 (peptides 57–59; Table I). An Ab response to the C3 α2 helix region of gp120, AIQEVKETLVKHPRYTGT, was significantly associated with protection from SIVsmE660 (34). The analogous α2 helix is a neutralization target in subtype C HIV (53). Anti-C3 Abs can exert immunologic pressure to reduce HIV viremia, as evidenced by the escape mutations that emerge followed by virus rebound (53). In this study, however, increased C3 peptide recognition was not associated with delayed SIVmac251 acquisition. It is possible that the FLSC-induced anti-C3 Abs may prevent the transmission of neutralization-sensitive viruses as opposed to the neutralization-resistant challenge stock of SIVmac251 used in this study (52).
Our study demonstrates that the FLSC immunogen does improve Ab function and confirms the importance of Abs targeting the V2 region of gp120 in protection from SIVmac251 infection. However, a significant vaccine efficacy was observed only in the ALVAC-gp120 animals boosted with monomeric gp120. One difference between the groups is that the same gp120 immunogen is present in the prime and boost of the ALVAC-gp120 group, whereas the FLSC group contains the gp120 immunogen in the ALVAC-SIV prime and was given the FLSC immunogen as the boost. In contrast, a DNA prime-envelope boost regimen that included four vaccinations with matched FLSC immunogens significantly reduced the rate of transmission of the same stock of SIVmac251 (23). These results suggest that perhaps matching the structure of the envelope immunogens used to prime and boost the immune system may be critical to induce Abs with sufficient potency, specificity, and breadth to prevent SIVmac251 transmission.
Supplementary Material
Acknowledgments
We thank Drs. Deborah Weiss, Jim Treece, Maria Grazia Ferrari, Ranajit Pal, and Irene Kalisz at Advanced Bioscience Laboratories, Inc. for care of the animals. We also thank Nancy Miller, John Warren, Anthony DeVico, Robert Gallo, and George Lewis for helpful discussion, and David Abram and Jason Knight for editing the manuscript.
This work was supported by the National Institutes of Health Intramural Program.
The online version of this article contains supplemental material.
- ADCC
- Ab-dependent cellular cytotoxicity
- CD4i
- CD4 induced epitope
- cV2
- cyclic V2
- FcγR
- Fcγ receptor
- FLSC
- full-length single-chain
- gD
- glycoprotein D
- GNL
- Galanthus nivalis lectin
- MFI
- mean fluorescence intensity
- NaSCN
- sodium thiocyanate
- rhFLSC
- rhesus FLSC
- RU
- response unit
- TM
- transmembrane.
Disclosures
G. Franchini is an author on patent US 5766598 A, Recombinant attenuated ALVAC canarypoxvirus expression vectors containing heterologous DNA segments encoding lentiviral gene products (issued June 16, 1998), which is jointly held by Sanofi Pasteur and the United States government. T.F. is an author on patent US 6908612, Virus coat protein/receptor chimeras and methods of use, issued June 21, 2005. This patent is held by the University of Maryland and is licensed to Profectus BioSciences. S.P. is an employee of Sanofi Pasteur. The other authors have no financial conflicts of interest.
References
- 1.Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., et al. MOPH-TAVEG Investigators 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361: 2209–2220. [DOI] [PubMed] [Google Scholar]
- 2.Haynes B. F., Gilbert P. B., McElrath M. J., Zolla-Pazner S., Tomaras G. D., Alam S. M., Evans D. T., Montefiori D. C., Karnasuta C., Sutthent R., et al. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366: 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonsignori M., Pollara J., Moody M. A., Alpert M. D., Chen X., Hwang K. K., Gilbert P. B., Huang Y., Gurley T. C., Kozink D. M., et al. 2012. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 86: 11521–11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola J. R., Stiegler G., VanCott T. C., Katinger H., Carpenter C. B., Hanson C. E., Beary H., Hayes D., Frankel S. S., Birx D. L., Lewis M. G. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6: 207–210. [DOI] [PubMed] [Google Scholar]
- 5.Moldt B., Rakasz E. G., Schultz N., Chan-Hui P. Y., Swiderek K., Weisgrau K. L., Piaskowski S. M., Bergman Z., Watkins D. I., Poignard P., Burton D. R. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA 109: 18921–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton D. R., Hessell A. J., Keele B. F., Klasse P. J., Ketas T. A., Moldt B., Dunlop D. C., Poignard P., Doyle L. A., Cavacini L., et al. 2011. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc. Natl. Acad. Sci. USA 108: 11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alpert M. D., Harvey J. D., Lauer W. A., Reeves R. K., Piatak M., Jr., Carville A., Mansfield K. G., Lifson J. D., Li W., Desrosiers R. C., et al. 2012. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog. 8: e1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao P., Patterson L. J., Kuate S., Brocca-Cofano E., Thomas M. A., Venzon D., Zhao J., DiPasquale J., Fenizia C., Lee E. M., et al. 2012. Replicating adenovirus-simian immunodeficiency virus (SIV) recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low-dose rectal SIV(mac251) challenge. J. Virol. 86: 4644–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baum L. L., Cassutt K. J., Knigge K., Khattri R., Margolick J., Rinaldo C., Kleeberger C. A., Nishanian P., Henrard D. R., Phair J. 1996. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 157: 2168–2173. [PubMed] [Google Scholar]
- 10.Forthal D. N., Landucci G., Haubrich R., Keenan B., Kuppermann B. D., Tilles J. G., Kaplan J. 1999. Antibody-dependent cellular cytotoxicity independently predicts survival in severely immunocompromised human immunodeficiency virus-infected patients. J. Infect. Dis. 180: 1338–1341. [DOI] [PubMed] [Google Scholar]
- 11.Vargas-Inchaustegui D. A., Robert-Guroff M. 2013. Fc receptor-mediated immune responses: new tools but increased complexity in HIV prevention. Curr. HIV Res. 11: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabuka J., Nduati R., Odem-Davis K., Peterson D., Overbaugh J. 2012. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog. 8: e1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forthal D. N., Gilbert P. B., Landucci G., Phan T. 2007. Recombinant gp120 vaccine-induced antibodies inhibit clinical strains of HIV-1 in the presence of Fc receptor-bearing effector cells and correlate inversely with HIV infection rate. J. Immunol. 178: 6596–6603. [DOI] [PubMed] [Google Scholar]
- 14.Santra S., Tomaras G. D., Warrier R., Nicely N. I., Liao H. X., Pollara J., Liu P., Alam S. M., Zhang R., Cocklin S. L., et al. 2015. Human non-neutralizing HIV-1 envelope monoclonal antibodies limit the number of founder viruses during SHIV mucosal infection in rhesus macaques. PLoS Pathog. 11: e1005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt R., Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280: 1884–1888. [DOI] [PubMed] [Google Scholar]
- 16.Rizzuto C. D., Wyatt R., Hernández-Ramos N., Sun Y., Kwong P. D., Hendrickson W. A., Sodroski J. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280: 1949–1953. [DOI] [PubMed] [Google Scholar]
- 17.Guan Y., Pazgier M., Sajadi M. M., Kamin-Lewis R., Al-Darmarki S., Flinko R., Lovo E., Wu X., Robinson J. E., Seaman M. S., et al. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. USA 110: E69–E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari G., Pollara J., Kozink D., Harms T., Drinker M., Freel S., Moody M. A., Alam S. M., Tomaras G. D., Ochsenbauer C., et al. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85: 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veillette M., Désormeaux A., Medjahed H., Gharsallah N. E., Coutu M., Baalwa J., Guan Y., Lewis G., Ferrari G., Hahn B. H., et al. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J. Virol. 88: 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang S. H., Doka N., Choudhary R. K., Sodroski J., Robinson J. E. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retroviruses 18: 1207–1217. [DOI] [PubMed] [Google Scholar]
- 21.Fouts T. R., Tuskan R., Godfrey K., Reitz M., Hone D., Lewis G. K., DeVico A. L. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J. Virol. 74: 11427–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeVico A., Fouts T., Lewis G. K., Gallo R. C., Godfrey K., Charurat M., Harris I., Galmin L., Pal R. 2007. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc. Natl. Acad. Sci. USA 104: 17477–17482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouts T. R., Bagley K., Prado I. J., Bobb K. L., Schwartz J. A., Xu R., Zagursky R. J., Egan M. A., Eldridge J. H., LaBranche C. C., et al. 2015. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. [Published erratum appears in 2015 Proc. Natl. Acad. Sci. USA 112: E2413.] Proc. Natl. Acad. Sci. USA 112: E992–E999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaccari M., Gordon S. N., Fourati S., Schifanella L., Liyanage N. P. M., Cameron M., Keele B. F., Shen X., Tomaras G. D., Billings E., et al. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat. Med. 22: 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Prete G. Q., Scarlotta M., Newman L., Reid C., Parodi L. M., Roser J. D., Oswald K., Marx P. A., Miller C. J., Desrosiers R. C., et al. 2013. Comparative characterization of transfection- and infection-derived simian immunodeficiency virus challenge stocks for in vivo nonhuman primate studies. J. Virol. 87: 4584–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keele B. F., Li H., Learn G. H., Hraber P., Giorgi E. E., Grayson T., Sun C., Chen Y., Yeh W. W., Letvin N. L., et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano J. W., Williams K. G., Shurtliff R. N., Ginocchio C., Kaplan M. 1997. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Invest. 26: 15–28. [DOI] [PubMed] [Google Scholar]
- 28.Lee E. M., Chung H. K., Livesay J., Suschak J., Finke L., Hudacik L., Galmin L., Bowen B., Markham P., Cristillo A., Pal R. 2010. Molecular methods for evaluation of virological status of nonhuman primates challenged with simian immunodeficiency or simian-human immunodeficiency viruses. J. Virol. Methods 163: 287–294. [DOI] [PubMed] [Google Scholar]
- 29.Vaccari M., Keele B. F., Bosinger S. E., Doster M. N., Ma Z. M., Pollara J., Hryniewicz A., Ferrari G., Guan Y., Forthal D. N., et al. 2013. Protection afforded by an HIV vaccine candidate in macaques depends on the dose of SIVmac251 at challenge exposure. J. Virol. 87: 3538–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montefiori D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12: Unit 12.11. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Román V. R., Florese R. H., Patterson L. J., Peng B., Venzon D., Aldrich K., Robert-Guroff M. 2006. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods 308: 53–67. [DOI] [PubMed] [Google Scholar]
- 32.Kramski M., Schorcht A., Johnston A. P., Lichtfuss G. F., Jegaskanda S., De Rose R., Stratov I., Kelleher A. D., French M. A., Center R. J., et al. 2012. Role of monocytes in mediating HIV-specific antibody-dependent cellular cytotoxicity. J. Immunol. Methods 384: 51–61. [DOI] [PubMed] [Google Scholar]
- 33.Tomaras G. D., Yates N. L., Liu P., Qin L., Fouda G. G., Chavez L. L., Decamp A. C., Parks R. J., Ashley V. C., Lucas J. T., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82: 12449–12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roederer M., Keele B. F., Schmidt S. D., Mason R. D., Welles H. C., Fischer W., Labranche C., Foulds K. E., Louder M. K., Yang Z. Y., et al. 2014. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505: 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pegu P., Vaccari M., Gordon S., Keele B. F., Doster M., Guan Y., Ferrari G., Pal R., Ferrari M. G., Whitney S., et al. 2013. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J. Virol. 87: 1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartshorn K. L., Reid K. B., White M. R., Jensenius J. C., Morris S. M., Tauber A. I., Crouch E. 1996. Neutrophil deactivation by influenza A viruses: mechanisms of protection after viral opsonization with collectins and hemagglutination-inhibiting antibodies. Blood 87: 3450–3461. [PubMed] [Google Scholar]
- 37.Huber, V. C., J. M. Lynch, D. J. Bucher, J. Le, and D. W. Metzger. 2001. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 166: 7381–7388. [DOI] [PubMed]
- 38.Ackerman M. E., Moldt B., Wyatt R. T., Dugast A. S., McAndrew E., Tsoukas S., Jost S., Berger C. T., Sciaranghella G., Liu Q., et al. 2011. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottardo R., Bailer R. T., Korber B. T., Gnanakaran S., Phillips J., Shen X., Tomaras G. D., Turk E., Imholte G., Eckler L., et al. 2013. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One 8: e75665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trkola A., Dragic T., Arthos J., Binley J. M., Olson W. C., Allaway G. P., Cheng-Mayer C., Robinson J., Maddon P. J., Moore J. P. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384: 184–187. [DOI] [PubMed] [Google Scholar]
- 41.Wu L., Gerard N. P., Wyatt R., Choe H., Parolin C., Ruffing N., Borsetti A., Cardoso A. A., Desjardin E., Newman W., et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384: 179–183. [DOI] [PubMed] [Google Scholar]
- 42.Hwang S. S., Boyle T. J., Lyerly H. K., Cullen B. R. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253: 71–74. [DOI] [PubMed] [Google Scholar]
- 43.Pöhlmann S., Davis C., Meister S., Leslie G. J., Otto C., Reeves J. D., Puffer B. A., Papkalla A., Krumbiegel M., Marzi A., et al. 2004. Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors. J. Virol. 78: 3223–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C. C., Tang M., Zhang M. Y., Majeed S., Montabana E., Stanfield R. L., Dimitrov D. S., Korber B., Sodroski J., Wilson I. A., et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310: 1025–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Prete G. Q., Haggarty B., Leslie G. J., Jordan A. P., Romano J., Wang N., Wang J., Holmes M. C., Montefiori D. C., Hoxie J. A. 2009. Derivation and characterization of a simian immunodeficiency virus SIVmac239 variant with tropism for CXCR4. J. Virol. 83: 9911–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zolla-Pazner S., deCamp A., Gilbert P. B., Williams C., Yates N. L., Williams W. T., Howington R., Fong Y., Morris D. E., Soderberg K. A., et al. 2014. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS One 9: e87572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zolla-Pazner S., deCamp A. C., Cardozo T., Karasavvas N., Gottardo R., Williams C., Morris D. E., Tomaras G., Rao M., Billings E., et al. 2013. Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PLoS One 8: e53629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Rompay K. K., Abel K., Lawson J. R., Singh R. P., Schmidt K. A., Evans T., Earl P., Harvey D., Franchini G., Tartaglia J., et al. 2005. Attenuated poxvirus-based simian immunodeficiency virus (SIV) vaccines given in infancy partially protect infant and juvenile macaques against repeated oral challenge with virulent SIV. J. Acquir. Immune Defic. Syndr. 38: 124–134. [DOI] [PubMed] [Google Scholar]
- 49.Reynolds M. R., Weiler A. M., Piaskowski S. M., Piatak M., Jr., Robertson H. T., Allison D. B., Bett A. J., Casimiro D. R., Shiver J. W., Wilson N. A., et al. 2012. A trivalent recombinant Ad5 gag/pol/nef vaccine fails to protect rhesus macaques from infection or control virus replication after a limiting-dose heterologous SIV challenge. Vaccine 30: 4465–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qureshi H., Ma Z. M., Huang Y., Hodge G., Thomas M. A., DiPasquale J., DeSilva V., Fritts L., Bett A. J., Casimiro D. R., et al. 2012. Low-dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus type 5 (Ad5) and then immunized with a replication-defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the phase IIb step trial of a similar HIV-1 vaccine. J. Virol. 86: 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rolland M., Edlefsen P. T., Larsen B. B., Tovanabutra S., Sanders-Buell E., Hertz T., deCamp A. C., Carrico C., Menis S., Magaret C. A., et al. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopker M., Easlick J., Sterrett S., Decker J. M., Barbian H., Learn G., Keele B. F., Robinson J. E., Li H., Hahn B. H., et al. 2013. Heterogeneity in neutralization sensitivities of viruses comprising the simian immunodeficiency virus SIVsmE660 isolate and vaccine challenge stock. J. Virol. 87: 5477–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore P. L., Ranchobe N., Lambson B. E., Gray E. S., Cave E., Abrahams M. R., Bandawe G., Mlisana K., Abdool Karim S. S., Williamson C., Morris L., CAPRISA 002 Study. NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.