Summary
The pal/RIM ambient pH signalling pathway is crucial for the ability of pathogenic fungi to infect hosts. The Aspergillus nidulans 7‐TMD receptor PalH senses alkaline pH, subsequently facilitating ubiquitination of the arrestin PalF. Ubiquitinated PalF triggers downstream signalling events. The mechanism(s) by which PalH transduces the alkaline pH signal to PalF is poorly understood. We show that PalH is phosphorylated in a signal dependent manner, resembling mammalian GPCRs, although PalH phosphorylation, in contrast to mammalian GPCRs, is arrestin dependent. A genetic screen revealed that an ambient‐exposed region comprising the extracellular loop connecting TM4‐TM5 and ambient‐proximal residues within TM5 is required for signalling. In contrast, substitution by alanines of four aromatic residues within TM6 and TM7 results in a weak ‘constitutive’ activation of the pathway. Our data support the hypothesis that PalH mechanistically resembles mammalian GPCRs that signal via arrestins, such that the relative positions of individual helices within the heptahelical bundle determines the Pro316‐dependent transition between inactive and active PalH conformations, governed by an ambient‐exposed region including critical Tyr259 that potentially represents an agonist binding site. These findings open the possibility of screening for agonist compounds stabilizing the inactive conformation of PalH, which might act as antifungal drugs against ascomycetes.
Introduction
Regulation of gene expression by ambient pH, a transcriptional response that tailors the biosynthesis of extracellular enzymes, permeases and exported metabolites to the needs imposed by the pH of the growth medium, crucially contributes to the ability of many fungi to thrive in disparate pH environments. The molecular wiring of the pH regulatory circuit has been extensively studied in the filamentous fungus Aspergillus nidulans and in yeasts, particularly in Saccharomyces cerevisiae. It includes a dedicated signal transduction pathway, denoted the pal/RIM pathway, that triggers, with assistance of the endosomal sorting complex required for transport (ESCRT) machinery, the proteolytic activation of the executive transcription factor PacC/Rim101p in response to alkaline pH (Peñalva and Arst, 2002; 2004; Peñalva et al., 2008; 2014).
As fungal pathogens invading mammalian hosts are exposed to systemic pH values above neutrality, it is unsurprising that the pal/RIM pathway has been identified as an important factor determining pathogenicity (Davis, 2009; Cornet and Gaillardin, 2014). Since the pioneering studies revealing the involvement of this pathway in systemic candidiasis (Davis et al., 2000), studies with ascomycetes as diverse as the Saccharomycotina Candida albicans (Nobile et al., 2008) and the Pezizomycotina A. nidulans (Bignell et al., 2005) and A. fumigatus (Bertuzzi et al., 2014) have uncovered its key involvement in the ability of these fungi to infect mammalian hosts. Overwhelming evidence indicates that, in ascomycetes, the pal/RIM pathway ambient pH sensor is a transmembrane protein denoted PalH in A. nidulans and Rim21p in S. cerevisiae (Herranz et al., 2005; Hervás‐Aguilar et al., 2007; Galindo et al., 2012; Obara et al., 2012; Herrador et al., 2015). This pH sensor is the most appealing component of the pathway to serve as druggable target, although it would be unsuitable for infections caused by the basidiomycetes, for example, Cryptococcus neoformans, given that although the RIM pathway is involved in its pathogenicity (O'Meara et al., 2010; 2013; 2014), PalH/Rim21p (and their associated arrestin, see below) appear to be missing in basidiomycete lineage (Cervantes‐Chávez et al., 2010; Ost et al., 2015). However, the fact that ascomycetes including pathogenic species such as A. fumigatus, A. niger, C. albicans, C. glabrata, Penicillum marneffei and Histoplasma capsulatum account for a large share of fungal human infections makes the possibility of targeting PalH potentially very useful.
The direct or indirect activation of this PalH sensor by ambient pH is transmitted to the internal leaflet of the plasma membrane by a transducer module involving an arrestin‐like protein, denoted PalF in A. nidulans and Rim8p in S. cerevisiae, an ubiquitin ligase (Rsp5p in S. cerevisiae) and the ESCRT‐I protein Vps23. PalF becomes ubiquitinated in a PalH‐dependent manner following exposure of cells to alkaline pH (Herranz et al., 2005), and this PalF ubiquitination triggers downstream signalling events (Hervás‐Aguilar et al., 2010). This occurs because ubiquitinated PalF/Rim8 in turn recruits Vps23 to plasma membrane‐associated signalling foci, sparking ESCRT‐III polymerization in these locales, which most likely serves as an amplification step (Herrador et al., 2009; Galindo et al., 2012; Obara and Kihara, 2014) (see (Peñalva et al., 2014) for a review). ESCRT‐III recruits to these foci the downstream pal/RIM pathway components PalC/Ygr122w (Rim23) (Galindo et al., 2007; Rothfels et al., 2005), PalA/Rim20p (which serves as adaptor for PacC/Rim101p (Xu and Mitchell, 2001; Vincent et al., 2003)) and the calpain‐like protease PalB/Rim13p (Rodríguez‐Galán et al., 2009; Obara and Kihara, 2014; Lucena‐Agell et al., 2015), which results in the proteolytic processing activation of the transcription factor (Peñalva et al., 2014). It is now agreed that despite the link of pH signalling to endosomal protein complexes, the pathway functions at the plasma membrane and does not require endocytosis (Calcagno‐Pizarelli et al., 2011; Galindo et al., 2012; Obara and Kihara, 2014; Lucena‐Agell et al., 2015).
Suggestively, PalH/Rim21p shares features with members of the seven transmembrane protein family of receptors (7‐TMRs). 7‐TMRs, usually denoted G protein‐coupled receptors (GPCRs), are the ‘most druggable’ family of proteins for pharmacological intervention (30% of the top‐selling prescription drugs are directed against GPCRs (Chalmers and Behan, 2002)), in part due to the fact that small molecules are able to target them with high specificity and affinity. Therefore, should the mechanism of PalH activation resemble that of its mammalian counterparts, the possibility of targeting the fungal ambient pH receptor with molecules interfering with its normal activity appears feasible, beside being theoretically attractive.
GPCRs do not solely transduce signals by way of heterotrimeric G proteins. Arrestins are a class of proteins previously thought to act exclusively to desensitize stimulated GPCRs. However, when engaged to the cytosolic tails of stimulated receptors, ‘classical’ β‐arrestins can recruit, activate and scaffold downstream signalling complexes without involvement of heterotrimeric G‐proteins, thus playing positive roles in signalling (Lefkowitz and Shenoy, 2005; Violin and Lefkowitz, 2007). Unlike β‐arrestins, α‐arrestins, a subfamily including the fungal clade, characteristically contain docking sites for the Rsp5 ubiquitin ligase (Alvarez, 2008) and indeed many of them act as adaptors targeting integral membrane protein partners for ubiquitin‐mediated endocytic down‐regulation (Lin et al., 2008; Nikko and Pelham, 2009; Karachaliou et al., 2013). In marked contrast, the α‐arrestin PalF/Rim8p engaging the PalH/Rim21p receptor acts positively, apparently by mediating its own requisite ubiquitination (Herrador et al., 2009; Herrador et al., 2015). PalF is necessary and, if artificially ubiquitinated, sufficient for the alkaline pH‐dependent activation of the pal pathway (Hervás‐Aguilar et al., 2010; Galindo et al., 2012). However, how under normal circumstances PalH/Rim21 stimulation results in PalF/Rim8p ubiquitination is currently unknown.
GPCR receptors consist of a bundle of helices embedded in the membrane, connected by cytosolic and external loops. Agonist binding to an inactive receptor R promotes a conformational transition R > R* resulting in subtle movements of the TMs that expose the otherwise inaccessible G protein‐binding sites (Sprang, 2011; Katritch et al., 2013). In contrast, the so denoted inverse agonists displace the equilibrium towards the inactive R state, thereby suppressing basal (i.e. in the absence of agonist) activity. However, this model is an oversimplification as receptors can adopt more than two conformations in response to different ligands, a fact that gave rise to the concept of ‘biased ligands’, i.e. ligands that specifically stabilize one particular receptor conformation. This concept is ultimately translated in that, for example, a ‘biased agonist’ elicits only a subset of the full repertoire of activities of any given receptor (Violin and Lefkowitz, 2007). Thus for receptors that can signal by either G proteins or arrestins, certain agonists are biased towards arrestin signalling, strongly supporting the view that ‘arrestin‐specific conformations’ do exist (Wisler et al., 2007).
A. nidulans PalH consisting of an N‐terminal integral membrane moiety (residues 1–352) and a long C‐terminal tail (residues 353–760) (Fig. 1) not only engages an arrestin, but it appears to conform to the topology of 7‐TMRs. According to TMpred (http://www.ch.embnet.org) the membrane moiety consists of seven TMs, with TM‐1 crossing the membrane in the outside‐in direction, such that its long C‐terminal tail is exposed to the cytosol. This tail contains two PalF binding regions (Fig. 1, pink residues) one adjacent to TM‐7 (residues 349–386) and the second bound by C‐terminal residues 657‐760 (Herranz et al., 2005).
Figure 1.
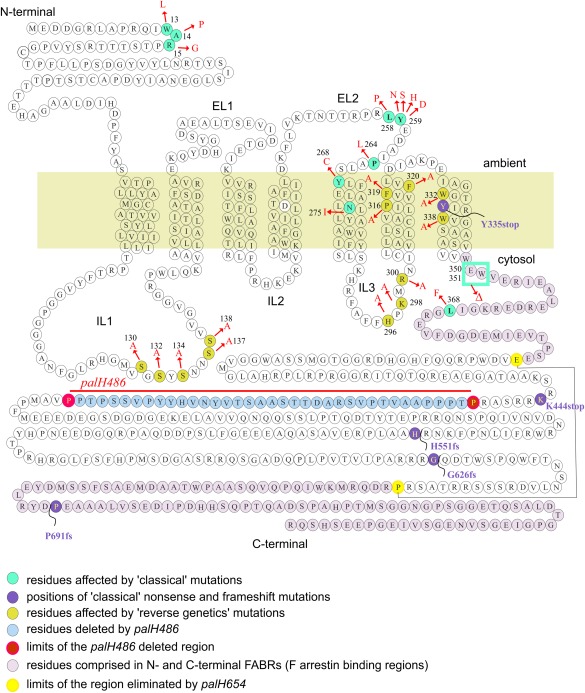
Scheme of PalH. The positions of the predicted TM domains and connecting loops relative to the lipid bilayer (olive green) are depicted. Amino acid substitutions corresponding to ‘classical’ and engineered missense mutations, the positions of truncations by ‘classical’ nonsense or frameshift alleles, the PalF arrestin binding regions (FABRs) in the cytosolic tail and the residues and borders of regions removed by deletions designed for this work are indicated.
Here, we exploit the amenability of A. nidulans to classical and reverse genetic analysis to obtain evidence that PalH is indeed a pal pathway receptor that upon exposure to alkaline pH undergoes a conformational change similar to those experienced by mammalian GPCRs, and that this change is transmitted to the cytosolic tail to transduce the alkaline pH signal by way of arrestin activation. Given that pH signalling does not involve G‐proteins, PalH/Rim21p would be a prototypical example of a class of receptors naturally biased towards arrestin signalling.
Results
Gene replacement procedure for PalH analysis
A. nidulans is haploid, facilitating the analysis of PalH function genetically, using a gene replacement procedure. We first constructed a pyrG89 palHΔ strain in which the palH coding region had been substituted by A. fumigatus pyroA (pyroAAf). This substitution/recombination event was confirmed by Southern blotting and phenotype testing. This palHΔ::pyroAAf strain was used as recipient of gene‐replacement linear DNA fragments consisting of wild‐type or mutant HA3‐tagged palH alleles and A. fumigatus pyrG as selective marker (Supporting Information Fig. 1). Transformants in which palH had been reconstituted with mutant versions were identified by their pyridoxine auxotrophy resulting from the exchange of pyroAAf by palH‐HA3::pyrGAf cassettes. Diagnostic plate tests for pH regulation demonstrated that strains carrying a wild‐type palH‐HA3 gene‐replaced allele were indistinguishable from the authentic wild‐type, and western blot (WB) analyses following a shift from acidic to alkaline conditions revealed that the proteolytic activation processing patterns of PacC in palH‐HA3 and palH+ strains were indistinguishable (Supporting Information Fig. 1), thereby showing that the genetic manipulations do not by themselves impair PalH function.
PalH is phosphorylated in an ambient pH‐ and PalF‐dependent manner
The pH signalling pathway responds to alkaline pH. Under acidic conditions the transcription factor PacC is not proteolytically activated, being present as the primary translation product, PacC72. Alkalinization of the medium activates the pal pathway, triggering the proteolytic activation of PacC72 to PacC27 via the committed intermediate PacC53 (Díez et al., 2002) (anti‐PacC western blots, Fig. 2A). In anti‐HA3 WBs of these cells cultured under acidic conditions PalH appears as a single band. However, PalH is modified upon exposing cells to alkalinity, such that an additional band(s) of reduced mobility become(s) the predominating species (Fig. 2A). This reduction in the mobility of PalH was reversed by treatment with lambda phosphatase (Fig. 2B), indicating that the mobility shift(s) result(s) from phosphorylation.
Figure 2.
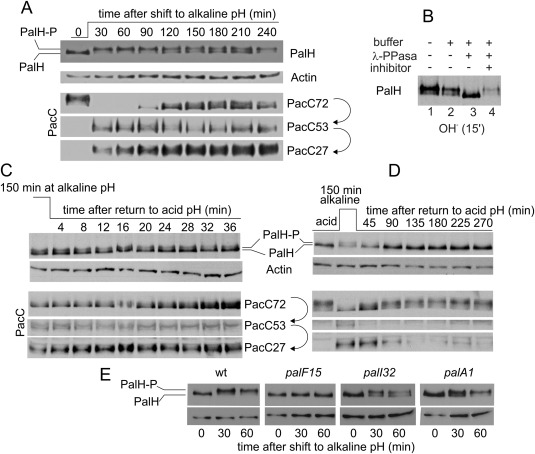
PalH phosphorylation in alkaline pH conditions.
A. Top, time course of PalH phosphorylation upon shifting cells from acidic to alkaline pH conditions for the indicated times. PalH‐P is phosphorylated PalH. Bottom, time‐course of PacC processing in the same cells. In all PacC western blots the ‘empty’ gel regions located between PacC72 and PacC53 and between the latter and PacC27 are not shown. Actin serves as loading control.
B. The two bands of PalH with different mobility coalesce into a single faster moving band after lambda bacteriophage phosphatase treatment (samples from cells shifted to alkaline pH for 15 min).
C. Time course of PalH dephosphorylation in cells that had been shifted from acidic to alkaline pH for 150 min and then back to acidic conditions for the indicated times. The bottom panel shows the corresponding anti‐PacC western blot. The relative levels of PacC53 and PacC27 change little over the time scale of this experiment, whereas PacC72 accumulates gradually, reflecting the fact that the pathway is inactive.
D. A similar time course experiment in which cells were sampled between 45 min and 4.5 h after shifting them down to acidic pH. Note how PacC27 gradually disappears due to degradation and the absence of signalling proteolysis, combined with transcriptional repression of the pacC promoter mediated by PacC72 (Bussink et al., 2015).
E. PalH phosphorylation is PalI‐ and PalA‐independent and PalF‐dependent. palF15, palI32 and palA1 are phenotypically null mutations in the corresponding genes.
PalH remained phosphorylated in cells kept at alkaline pH for as long as 4 h (Fig. 2A). When alkaline‐exposed cells were shifted back to acidic pH, the relative level of phosphorylated species declined slowly, correlating with the ‘recovery’ of PacC72 caused by the absence of signalling (i.e. the lack of proteolytic processing of freshly synthesized PacC) (Fig. 2C). Complete dephosphorylation took as long as 45–90 min after returning cells to acidic pH, a time at which the pool of PacC27, which turns over also slowly (Mingot et al., 1999), becomes barely detectable (note that PacC72 does not increase further because transcriptional autorepression counteracts the absence of its proteolytic processing (Bussink et al., 2015)) (Fig. 2D). All the above data are consistent with PalH becoming accessible to phosphorylation as a consequence of alkaline pH sensing. The correlation between pathway activation and PalH phosphorylation was suggestive as upon their agonist‐mediated activation mammalian GPCRs are rapidly phosphorylated by GRKs (G protein receptor kinases), hampering signalling (Shukla et al., 2011; Lefkowitz, 2013). Notably, although PalH phosphorylation is independent of PalI (a PalH helper) and of PalA (acting downstream of ESCRT recruitment), it is strictly dependent on PalF (Fig. 2E), indicating that PalH phosphorylation requires the functional engagement of the arrestin to the receptor.
Mutant PalH with a cytosolic tail restricted to the two PalF arrestin binding sites retains some function
Equipped with the gene replacement procedure, we first constructed an allele encoding a PalH protein in which cytosolic residues 385 through 653 separating the two PalF binding sites had been substituted by a synthetic linker consisting of a Gly‐Ala pentamer (palH654, Fig. 3A). WB showed that the protein is stably expressed (Fig. 3B). Null palH alleles such as palH72 truncating PalH after residue 12, or alleles truncating PalH within the TM domain do not grow at alkaline pH, display exacerbated sensitivity to Li+ and ion toxicity and enhanced resistance to neomycin (Negrete‐Urtasun et al., 1999; Herranz et al., 2005) (Fig. 3C). Remarkably, palH654 strains are, like palH72 strains, highly sensitive to lithium. Yet, unlike palH72, palH654 strains grow at alkaline pH and are more resistant than palH72 strains to molybdate, showing that the mutant palH654 product is, to a significant extent, functional. WB analysis of PacC processing buttressed this conclusion. In pH shift experiments the null palH72 mutation blocked the proteolytic processing activation of PacC (Hervás‐Aguilar et al., 2010). In contrast, palH654 impaired it relatively weakly (Fig. 3B, bottom), although this allele led to a reduction in the overall PacC levels. We conclude that although the 269 cytosolic residues of PalH located between the previously delimited PalF binding regions play some role, this region is not critical for pH signalling.
Figure 3.
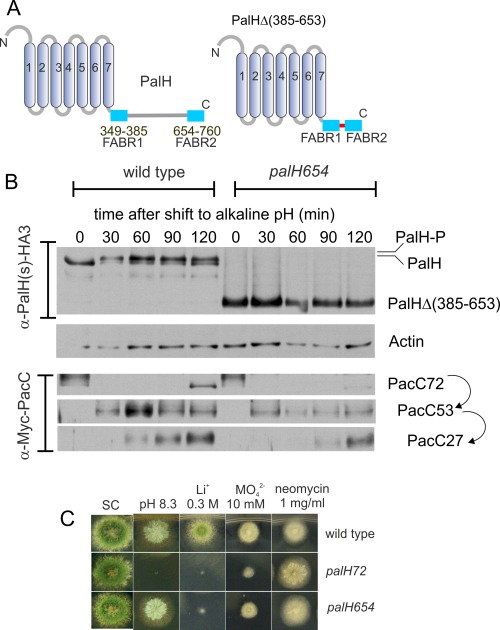
mutant PalH with a cytosolic tail containing just the arrestin binding regions (FABRs) retains substantial function.
A. Scheme of the mutant palH654 product compared to the wild‐type.
B. Time course of PalH phosphorylation and PacC processing in pH shift experiments with palH654 and wild‐type cells.
C. Diagnostic growth tests of pH signalling in the indicated strains. Note that although almost unable to grow on plates containing Li+, palH654 grows like the wild‐type at pH 8.3, a pH at which the null palH72 strain is unable to grow. palH654 also displays a noticeably stronger resistance to molybdate than palH72, although lesser than the wild‐type.
Phosphorylation is largely dispensable for PalH function
Anti‐HA3 WBs of palH654 cells strongly suggested that the phosphorylation‐mediated shift in PalH mobility requires amino acids located within the large 269 residue region deleted in the mutant (Fig. 3B). This large segment of the protein might contain phosphoacceptor residues or it might be required indirectly, for example if it contributes a docking site for the phosphorylating kinase(s). To distinguish between these possibilities, we set out to identify phosphorylatable residues throughout the protein. We first ruled out that a region containing five Ser residues between positions 130 and 138, within the first intracellular loop (IL1) connecting TM1 and TM2 (Fig. 1) serves as phosphoacceptor. We generated palH138 encoding a mutant PalH with Ala substitutions for all five of these serines. WBs revealed normal PalH phosphorylation, eliminating these serine residues as phosphoacceptors (data not shown). In passing, palH138 behaved as wt in diagnostic tests of pH regulation and showed a normal pattern of PacC72 processing in response to alkaline pH, indicating that these serines are not functionally/structurally important (data not shown).
In view of this, we focused on a highly conserved region located between Pro450 and Pro486, rich in Pro, Thr and Ser residues (Fig. 4A). We generated palH486 encoding a mutant PalH deleted for this 37‐residue motif. palH486 had four phenotypic effects. First, it resulted in an increase, albeit very modest, in the steady state levels of PalH (Fig. 4B). Second, the mutant receptor did not undergo any changes in mobility upon shifting cells to alkaline pH, showing that the deleted region is required for phosphorylation (Fig. 4C). Third, although palH486 strains behave essentially as the wild‐type in alkaline pH, molybdate and neomycin sensitivity growth tests (Fig. 4D), they display weak hypersensitivity to lithium chloride (Fig. 4E). Amongst 48 progeny obtained from a backcross between a palH486 strain and the wild‐type 20 were phenotypically wild‐type and 28 were lithium hypersensitive. According to a psi‐square test these figures are not significantly different from those expected for a single Mendelian character distribution. Six wild‐type and six lithium hypersensitive clones showed the expected PCR amplification bands for wild‐type palH and palH486, respectively, indicating that lithium hypersensitivity co‐segregates with the palH486 deletion, and thus that the phenotype is unlikely to be caused by a secondary mutation. Lastly, analysis of PacC processing showed that palH486 causes a slight delay in the activation of the pathway within 2 h after shifting cells to alkaline conditions (Fig. 4F), a defect which appears insufficient to cause detectable effects in diagnostic tests of pH regulation (with the exception of the highly sensitive Li+ toxicity test), perhaps because these growth tests are scored after a 48 h incubation. Thus phosphorylation (i.e.: those phosphorylation events detectable by mobility shift in SDS‐PAGE) is largely dispensable for PalH function.
Figure 4.
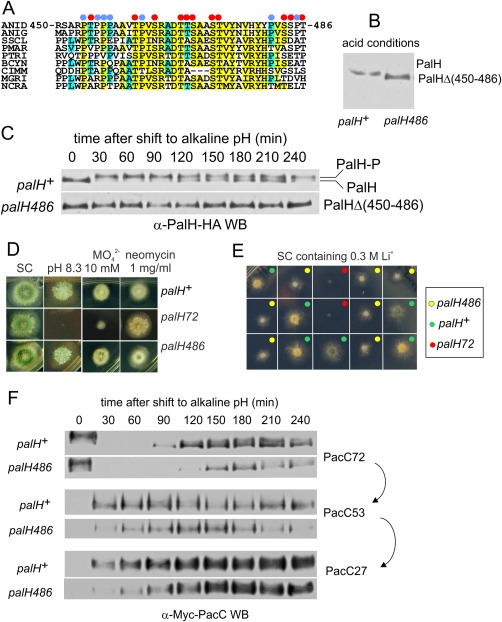
A conserved region rich in Pro, Ser and Thr residues is required for PalH phosphorylation.
A. Multiple sequence alignment showing conservation of the PalH cytosolic tail region located between resides 450 and 486. Blue and red dots indicate Pro and Ser/Thr residues, respectively, in A. nidulans PalH. Amino acid highlighting in yellow and blue indicates strong and weak conservation, respectively. ANID, A. nidulans; ANIG, A. niger; SSCL, Sclerotinia sclerotium; PMAR, Penicillium marneffei; PTRI, Pyrenophora tritici‐repentis; BCYN, Botrytis cinerea; CIMM, Coccidioides immitis; MGRI, Magnaporthe grisea; NCRA, Neurospora crassa.
B. Western blot analysis of equally loaded samples showing slightly elevated levels of the palH486 mutant product.
C. pH shift experiment showing the apparent absence of alkaline pH‐induced phosphorylation (as determined by changes in SDS‐PAGE mobility) of the palH486 mutant product.
D. palH486 is indistinguishable from the wild‐type on pH 8.3, molybdate and neomycin media.
E. palH486 is hypersensitive to Li+. Different clones of a cross between palH486 and palH+ parental strains were point inoculated onto medium containing 0.3 M Li+. Yellow and green dots indicate strains that had been genotyped as palH486 and palH +, respectively. Red dots indicate a palH72 strain included as control. Yellow and green surface palH+ colonies reflect the fact that the yellow spore marker yA2 is segregating in the cross (wild‐type spores are green; spores are not visible in palH486 clones due their impaired growth on lithium medium).
F. Comparison of the PacC proteolytic processing activation pattern of palH486 with the wild‐type during a pH shift experiment. Note the slightly delayed processing and the slower increase in PacC72 in the mutant after long times at alkaline pH.
We thus considered the possibility that PalH phosphorylation is required for the turnover of the receptor by endocytosis. To test this, we used fimAΔ ablating the endocytic patch component fimbrin, which results in a major defect in endocytosis (Upadhyay and Shaw, 2008). Figure 5 shows that in sharp contrast with the wild‐type, a proportion of phosphorylated PalH is detectable in fimAΔ cells under acidic conditions, strongly indicating that fimAΔ stabilizes this form. As fimAΔ prevents the endocytic internalization of PalH‐GFP (Lucena‐Agell et al., 2015), this build‐up of the phosphorylated form suggests that phosphorylation tags the protein for endocytic turnover, a possibility that is consistent with the increase in the steady state levels of PalH detected in fimAΔ (and in palH654) cells.
Figure 5.

iImpairment of endocytosis results in phosphorylated PalH under acidic conditions and elevates the levels of the receptor.
Western blot analysis of PalH in a null fimbrin (fimAΔ) mutant compared to the wild‐type. Two different exposures of same the blot are shown. Hexokinase was used as loading control.
Investigating PalH function by site directed mutagenesis
To gain further insight into the mechanisms of alkaline ambient pH sensing, we performed a genetic screen using the gamma amino‐butyrate (GABA) technique. Briefly, areAr mutants that cannot utilize nitrogen sources other than ammonium are unable to grow on GABA as nitrogen source unless the pH signalling pathway is switched off, because the GABA permease gene is repressible by PacC27, the end product of the PacC proteolytic activation cascade (Caddick et al., 1986; Hutchings et al., 1999; Espeso and Arst, 2000). To select pH signal‐impairing mutations specifically mapping to palH we exploited the procedure of Tilburn et al. (2005), screening for GABA utilizers among mutagenized spores of a diploid strain homozygous for areAr and heterozygous for a null mutation in palH. Besides less informative early truncating mutations, which are not discussed for simplicity, eleven novel palH missense mutations resulting in partial loss‐of‐function were identified by DNA sequencing (Figs. 1 and 6). A twelfth mutation, palH402, is a two‐codon deletion.
Figure 6.
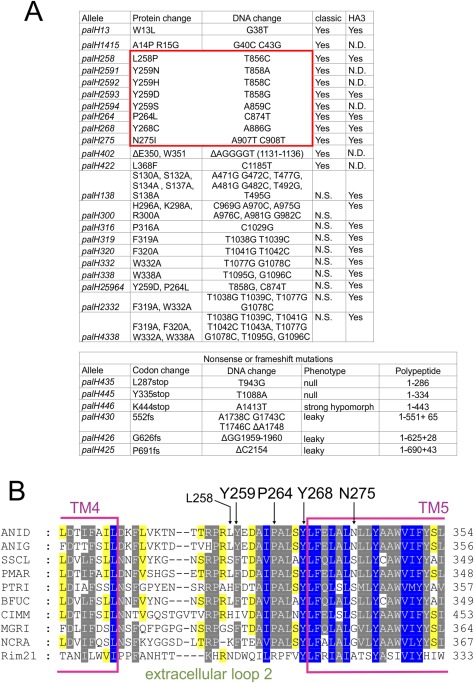
Mutational analysis of PalH: a potential agonist binding site.
A. Complete catalogue of mutations reported here (with the exception of internal deletion alleles described in the text and Fig. 1). For missense mutations (top), ‘classic’ and HA3 indicate whether the allele was obtained by UV or site‐directed mutagenesis, respectively (the latter were all HA3‐tagged). N.D. is ‘not done’ whereas N.S. is ‘not selected’. The red line boxes those missense mutations that we hypothesize comprise an agonist binding site. For nonsense/frameshifting mutations (bottom) their phenotypes are indicated. Note that mutant PalH proteins encoded by leaky truncating alleles contain the TM domain‐proximal PalF binding region (Herranz et al., 2005), which explains their partial loss‐of‐function phenotype. palH45 and palH47, described by Negrete‐Urtasun et al. (1999) and Herranz et al. (2005) are also in this category.
B. Multiple sequence alignment in the PalH region containing missense mutations hypothetically representing an agonist binding site. Species are as in Fig. 4 with the exception of S. cerevisiae, whose Rim21p is included to underline the remarkable sequence conservation existing between filamentous fungi and budding yeast homologues in TM5.
palH402 removing E350 and W351 results in complete loss‐of‐function. These residues belong to Trp349‐Glu350‐Trp351 motif located in the interface between the cytosol‐proximal C‐terminus of TM7 and the cytosolic tail (Fig. 1). The functional importance of this motif is supported by its strong conservation in ascomycetes, including PalH/Rim21 receptors from organisms as distant from filamentous ascomycetes as S. cerevisiae and Yarrowia lipolytica (Tréton et al., 2000). The Trp‐Glu‐Trp motif is contained within a 36 residue sequence (residues 349–385 of the PalH cytosolic tail) which is sufficient to interact with PalF in two‐hybrid assays (Herranz et al., 2005), strongly indicating that this motif plays a key role in receptor‐arrestin binding/coupling. Another mutation almost certainly impairing PalF binding to PalH is palH422, which affects the conserved Leu368 also located within the 36‐residue N‐terminal (i.e. membrane‐proximal) PalF binding site in the cytosolic tail. This position cannot tolerate Phe. Substitution by Ala of adjacent Gly369 debilitates the interaction with PalF (Herranz et al., 2005). The isolation of these mutations validated the efficacy of the genetic screen.
Among the remaining 10 missense mutations, two, palH13 and palH1415 affect residues located in the N‐terminal extracellular tail, indicating that this region of the protein has a functional role. Remarkably, eight of these mapped to a region encompassing the 23 residue‐long extracellular loop connecting TM4 with TM5 and the N‐terminal half (the ‘outside’ half) of TM5. Notable among these are those affecting Leu258 and Tyr259, located within a conserved motif of this loop. Tyr259 (Phe tolerated in other fungi) is highly conserved (Fig. 6B). Four different substitutions (to Asn, His, Ser, Asp) involving this residue resulted in partial loss‐of‐function, indicating that Tyr259 plays an important physiological role. Pro264Leu also affects this TM4‐TM5 loop, whereas Tyr268Cys affects the loop‐TM5 interface. Both Pro264 and Tyr268 are conserved. Lastly Asn275Ile affects a residue located in the extracellular space‐proximal half of TM5. In other PalH orthologues, a polar residue generally occupies the position of PalH Asn275. These polar residues must be protected from the lipid environment within the 3D structure, arguably by a compensating charge that would become ‘unpaired’ if position 275 were occupied by a purely aliphatic side chain residue. In any case this genetic analysis strongly indicates that residues within both TM5 and the loop connecting it with TM4 are important for PalH activity.
PalH phosphorylation can be uncoupled from PalH activation
We next examined the effects that hypomorphic palH alleles isolated in the GABA screen have on the proteolytic activation of PacC. We generated palH13 (W13L), palH258 (L258P), palH2593 (Y259D), palH264 (P264L), palH268 (Y268C) and palH275 (N275I) HA3‐tagged alleles by gene replacement. All six prevented growth on 0.3 M Li+ plates and resulted in hypersensitivity to molybdate albeit to a lesser extent than a palH72 null (Supporting Information Fig. 2), consistent with their causing partial loss‐of‐function.
Y259D, P264L, Y268C and, more conspicuously, N275I affected the persistence of signalling, as noted by the abnormal accumulation of unprocessed PacC72 at the 120 min time points (and at the 90 min time point in the case of N275I) (Fig. 7). All of these led to a reduction in PacC levels, in some cases very noticeably, possibly reflecting that the steady state levels of PacC are reduced when PacC proteolytic activation is impaired, as a consequence of the negative auto‐regulatory loop that determines the steady‐state situation (Bussink et al., 2015). Notably, none of these mutations decreased PalH levels (Fig. 7, anti‐HA blots), and thus their effects can only be attributable to PalH dysfunction. Thus, these data confirm the inferences derived from growth tests, establishing that the above missense mutations impair pH signalling.
Figure 7.
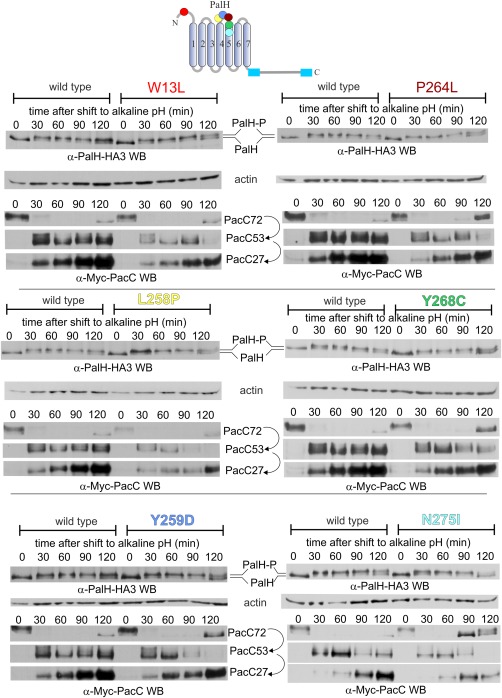
Molecular characterization of single missense mutants affecting extracellular residues.
Time courses of PalH phosphorylation and PacC processing in pH shift experiments with cells carrying single residue substitutions in residues exposed to the ambient. The colours of the dots indicating mutated residues in the PalH scheme correspond with those of the different panel labels. Mutations are denoted by the resulting substitutions, in single letter amino acid code (see Fig. 6 for standard allele nomenclature and corresponding DNA changes).
We next considered the possibility that, as in mammalian GPCRs, PalH phosphorylation occurred upon receptor stimulation. However, all of the mutant PalH proteins showed normal in vivo phosphorylation upon a shift to alkaline pH (Fig. 7), indicating that PalH phosphorylation does not correlate with receptor function. However, virtually all the above mutations led to lower levels of the PalH phosphorylated form at the 120 min time point, hinting at some degree of connection between reduced PalH function and the inability of these mutant PalH proteins to maintain the phosphorylated state.
In view of the above, we considered the possibility that the extent to which the above missense mutations impair signalling is insufficient to be reflected in the steady‐state levels of phosphorylation. Thus, we constructed the palH2264 allele encoding a double mutant protein combining Y259D and P264L. Unlike either parental, the double mutant was as sensitive to molybdate as the null and showed markedly decreased growth at alkaline pH, indicating additivity (Fig. 8A). The double substitution did not affect the steady state PalH levels nor the alkaline pH‐induced phosphorylation of PalH beyond the reduction in relative levels of phosphorylated form observed in the respective single mutants at the 120 min time point (Fig. 8B; for comparison see Fig. 7). By anti‐PacC WB, palH2264 led to an abnormal accumulation of PacC72 already at 60 min after an alkaline pH shift, which was even more striking at the 120 min time point (Fig. 8C). Thus the synthetic negative effect in pH signalling displayed by these two mutations does not correlate with decreased PalH phosphorylation, indicating that PalH phosphorylation cannot solely reflect the activation of the pathway.
Figure 8.
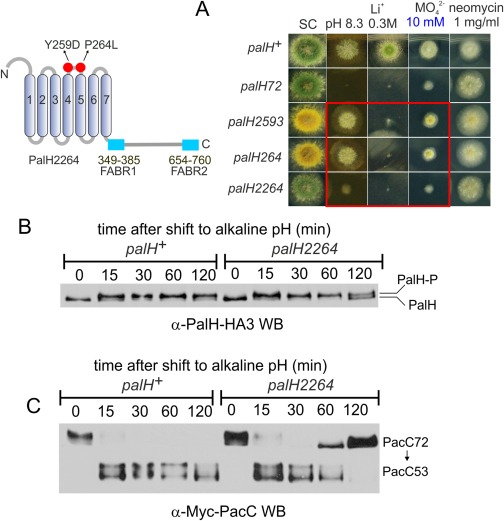
Phenotypic additivity of Y259D and P264L: molecular characterization.
A. Diagnostic growth tests showing phenotypic additivity between palH2593 (Y259D) and palH264 (P264L). palH2264 encodes a doubly‐substituted PalH. palH72 is a null palH mutation.
B. The time courses of alkaline pH‐induced wild‐type and Y259D, P264L double mutant PalH phosphorylation are very similar.
C. Effects of the double mutant substitution on the signalling proteolysis of PacC72; (Note that for emphasis only the region of the gel containing PacC72 and PacC53 is included).
Gain‐of‐function mutations in PalH obtained by site‐directed mutagenesis
Studies with GPCRs have uncovered the ‘molecular switches’, which are interactions between residues located within the heptahelical bundle that stabilize the inactive conformation of the receptor and that are disrupted after ligand binding (Xie and Chowdhury, 2013) (Schwartz et al., 2006). At the cytoplasmic side of GPCRs, activation results in conformational changes of the helical bundle generally involving shifts in the positions of helices V, VI and VII (Katritch et al., 2013). In the β2‐adrenergic receptor (β2‐AR) both TM5 and TM6 contain one Pro residue each that introduce kinks in the helices. Agonist binding modifies these kinks' angles, causing a swing in the cytosolic part of TM6 that exposes receptor epitopes to downstream transducers (Rasmussen et al., 2011; Rosenbaum et al., 2011; Warne et al., 2011). In certain types of GPCRs aromatic residues located within TM6 and TM7 modulate the basal activity of the receptor through the bend angle kink (Shi et al., 2002; Xie and Chowdhury, 2013).
Inspired by these studies we addressed, by site directed mutagenesis of TM6 and TM7 residues, the possibility that PalH undergoes conformational changes resulting in shifts in the positions of the TMs decodable by the arrestin module. TM6 contains a proline residue, Pro316, located within a conserved 316‐PVVFF‐320 motif (Fig. 9A), suggesting that, by analogy to GPCRs, this Pro may kink the helix, and that the nearby Phe residues might regulate the bending angle. Thus in a first approach we substituted Pro316 by Ala. This allele (palH316, Fig. 6) resulted in weak hypersensitivity to Li+ and ions and increased tolerance to neomycin, and impaired PacC processing markedly, indicating loss‐of‐function (Supporting Information Fig. 3). However, these phenotypic characteristics are attributable to the rapid and marked decrease in this mutant PalH levels at alkaline pH (Supporting Information Fig. 3). Encouraged by such pH‐induced instability, arguably consistent with a requirement for this Pro in pH‐mediated conformational rearrangements involving TM6, we undertook further mutagenesis studies aimed at ‘releasing’ a hypothetical molecular switch maintaining PalH in an inactive conformation. We focused on a potential aromatic toggle involving TM6 and TM7 residues, substituting by Ala four conserved aromatic amino acids including Phe319 and Phe320 in the proximity of Pro316 in TM6 and Trp332 and Trp338 in TM7 (Fig. 9B). The quadruple substitution allele was denoted palH4338. Remarkably palH4338 strains showed hyper‐resistance to both Li+ and and hypersensitivity to neomycin (Fig. 9B), strongly indicating that the quadruple substitution activates the pathway. We next established rigorously that these gain‐of‐function phenotypes result from palH4338 after outcrossing the mutant and confirming that they segregated like a single Mendelian character linked to the mutant allele, genotyped by PCR (data not shown). Furthermore, as palH is tightly linked to the cloramphenicol resistance gene camC, we demonstrated that the molybdate resistance phenotype associated with palH4338 is closely linked to camC (no recombinants obtained in n = 49 progeny of a camC108 x palH4338 heterozygous cross), largely ruling out the possibility that it is caused by an unrelated mutation.
Figure 9.
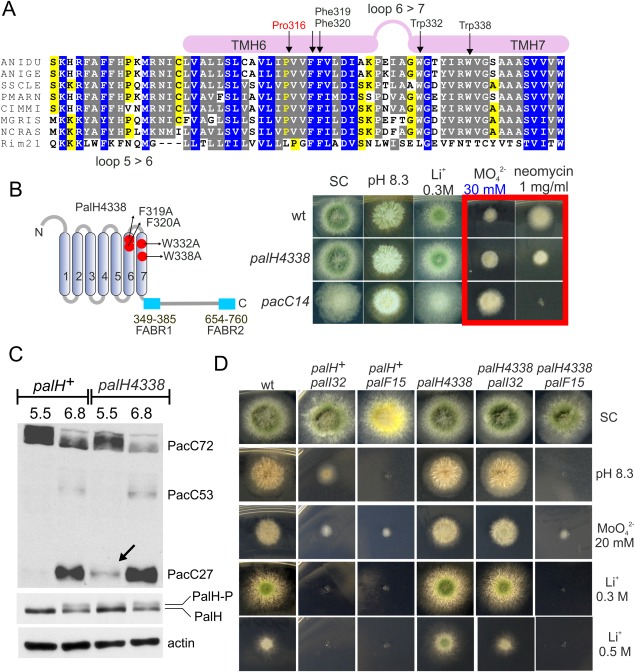
Characterization of palH4338 resulting in pH signalling pathway activation at acidic pH.
A. Multiple sequence alignment showing the region corresponding to TM6 and TM7, as in Fig. 6 (Rim21 is from S. cerevisiae). The positions of residues that were mutated to investigate the hypothetical involvement in alkaline pH signalling of conformational rearrangements of the helical bundle of transmembrane helices are indicated.
B. Diagnostic growth tests of pH signalling in which palH4338 is compared to the wild‐type and to a strong alkalinity‐mimicking pacCc14 allele. Note the intermediate response of palH4338 on molybdate and neomycin plates (boxed in red).
C. palH4338 results in weak processing of PacC72 at acidic pH (pH 5.5) (the arrow indicates PacC27, the end product of the proteolytic cascade). Note the faint degree of PalH phosphorylation at pH 5.5 seen with the mutant but not with the wild‐type. Cells were cultured overnight at ‘constant’ pH before proceeding to total protein extraction followed by anti‐PacC (Myc3) and anti‐PalH (HA3) western blotting. Actin was used as loading control.
D. Diagnostic tests of pH regulation showing that palF15 is epistatic to palH4338 whereas palI32 is hypostatic.
Thus, we studied the effect of palH4338 in PacC processing. Instead of monitoring 2 h time courses after shifting cells to alkaline pH we used overnight cultures adjusted to acidic or neutral pH to approximate the steady‐state conditions of (neutral pH) plate tests. In the wild‐type cultured under neutral conditions PacC appeared as a mixture of PacC72, PacC53 and PacC27 (Fig. 9C). In contrast, under acidic conditions wild type cells contained only PacC72, in sheer contrast with palH4338 cells, which contained detectable levels of PacC27, indicative of pathway activation (Fig. 9C). Thus, PacC processing data agree with the palH4338 gain‐of‐function phenotype. During the course of these experiments we also noticed that at pH 5.5 palH4338 cells but not palH+ cells display a faint band of phosphorylated PalH (Fig. 9C). This observation indicates that the quadruple substitution indeed affects the basal conformation of the receptor, and might suggest that such conformational change facilitates, under acidic pH conditions, the engagement of the PalF arrestin in a manner conductive to PalH phosphorylation [which is alkaline pH‐ and PalF‐dependent (Fig. 2E)]
Next, we performed epistasis analysis. palF15 and palI32 are null mutations in the genes encoding the PalF arrestin‐like and the PalI ‘helper’, respectively (PalI is a protein acting upstream of PalH assisting its plasma membrane localization (Calcagno‐Pizarelli et al., 2007); the localization of PalI to the plasma membrane is PalH‐independent, see Supporting Information Fig. 4). palF15 prevents growth at pH 8.3, whereas palI32 allows some growth at this pH because the absence of PalI does not impede pal signalling completely. We crossed palH4338 into null palI32 and palF15 backgrounds. Notably a double palH4338 palI32 mutant grew like the wild‐type, i.e. much better than the single palI32 parental mutant, at alkaline pH (Fig. 9D), confirming that palH4338 has a signal‐promoting effect even without PalI. In contrast palH4338 did not rescue to any extent the inability of palF15 strains to grow at alkaline pH, demonstrating that the pH signalling activity resulting from palH4338 requires PalF. Similar results were obtained with molybdate or lithium toxicity tests, where palH4338 suppressed the hypersensitivity phenotype resulting from palI32 but not that resulting from palF15 (Fig. 9D). Thus, all the above data strongly support the contention that palH4338 abnormally stimulates pH signalling under acidic conditions by promoting an arrestin‐activating conformation of PalH.
Contribution of individual substitutions in palH4338 to gain‐of‐function
In an attempt to identify the individual contributions of the residues mediating the effects of palH4338 we constructed four alleles each containing one of its four missense mutations. In plate tests Phe319Ala and Trp332Ala led to weak gain of function (Fig. 10A), whereas Phe320Ala led to weak loss‐of‐function and Trp338Ala behaved as wild‐type (data not shown). Thus, we combined Phe319Ala with Trp332Ala in palH2332 (Fig. 10A and B). PacC processing assays showed that palH2332 extracts displayed detectable bands of PacC27 in acidic conditions that were not seen in wild‐type controls (Fig. 10B). The double substitution resulted in slightly higher neomycin sensitivity than its constituents, suggesting gain of function, but did not increase tolerance any further, showing that its activating effect is weak (Fig. 10A). Thus, Phe319 and Trp332 in TM6 and TM7, respectively, may contribute to maintain PalH in the basal conformation, and these combined with the above data suggest that the relative position of the helices within the TM bundle participates in pH signal reception, with possible involvement of not only Phe319 in TM6 but also of Trp332 in TM7. This is very suggestive given that in the β2‐AR arrestin‐biased ligands preferentially affect the conformational states of TM7 (Liu et al., 2012).
Figure 10.
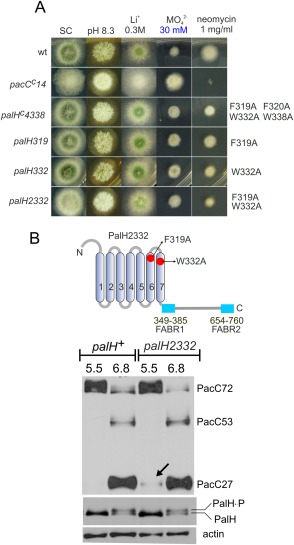
Phe319 and Trp332 seem important to maintain the basal activity of PalH.
A. Diagnostic growth tests of pH regulation in the indicated strains.
B. Scheme showing the positions of Phe319 and Trp332 in PalH.
C. Effects of the double mutant substitution on the proteolytic processing of PacC72 in the steady state situation at the indicated pH; Note the weak activation of the pathway as indicated by detection of a faint band of PacC27.
Discussion
PalH is the ascomycete alkaline pH signal sensor and PalF is the key component of a signal‐transducing module also involving ESCRT‐I Vps23, which is recruited to pH signalling complexes by ubiquitinated PalF/Rim8 (Herrador et al., 2009; Hervás‐Aguilar et al., 2010; Galindo et al., 2012; Obara and Kihara, 2014). In all likelihood Vps23 triggers an amplification step that involves ESCRT‐III polymerization, thus creating multiple docking sites for the downstream' pal/RIM signalling components PalC/Ygr122w/Rim23, PalA/Rim20 and PalB/Rim13 and, by way of its interaction with PalA/Rim20, for PacC72/Rim101 (Fig. 11) (Vincent et al., 2003; Xu et al., 2004; Galindo et al., 2007; Herrador et al., 2009; Rodríguez‐Galán et al., 2009; Calcagno‐Pizarelli et al., 2011; Galindo et al., 2012; Obara and Kihara, 2014; Peñalva et al., 2014; Herrador et al., 2015; Lucena‐Agell et al., 2015). Vps4‐mediated ESCRT‐III de‐polymerization terminates signalling (Galindo et al., 2012). However, despite this detailed understanding of the pathway, the mechanistic bases of pH signal reception (i.e. the modification(s) that PalH undergoes in response to alkaline pH) and transduction (i.e., how are these conformational changes in the PalH TM receptor result in PalF ubiquitination) are insufficiently understood.
Figure 11.
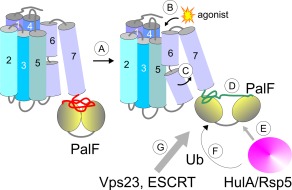
Hypothetical model for the transduction of the alkaline ambient pH signal in A. nidulans
PalH alternates between basal and active conformations. Unlike β−arrestins PalF is bound to the receptor even under acidic (i.e. non‐signalling) conditions which drive the receptor towards the basal conformation (A). (B) Alkalization of the medium generates an ‘agonist’, perhaps a lipid of the external leaflet of the plasma membrane, that upon change of its ionization status binds a region located at the ambient‐exposed interface between TM4 and TM5. Agonist binding drives the equilibrium towards the active conformation. We speculate that the transition from the basal to the active conformation results from an alkaline pH‐ and TM6 Pro316‐dependent shift in the orientation of TM6 and TM7, a shift that is also promoted by palHc4338 independently of ambient pH (C). This shift would be then transmitted to arrestin‐binding regions in the cytosolic PalH tail (D), leading to a conformational change that facilitates the recruitment of the HulA/Rsp5 ubiquitin ligase (E) and the subsequent ubiquitination of PalF (F). Next ubiquitinated PalF recruits ESCRT‐I Vps23 (G), which primes ESCRT‐III polymerization that, we hypothesize, serves as a signal amplification step because the remaining Pal proteins acting downstream of PalF (PalA, PalC and PalB) are all ESCRT‐III interactors that are transiently localized to punctate structures at the plasma membrane when ambient pH is alkalinized.
This work provides two important novel pieces of information when considered in the light of extensive knowledge available on the mechanisms of 7‐TMR activation. One is that an extracellular region comprising the TM4‐TM5 extracellular loop and residues in the ambient‐proximal half of TM5 are required for signalling. By analogy to mammalian GPCRs (Katritch et al., 2013), this region might contribute to an agonist binding site. The second is that aromatic residues embedded within TM6 and TM7 are necessary to maintain PalH in an inactive state, as their substitution by alanines weakly activates the pathway. An attractive interpretation of this observation is that TM6/TM7 substitutions result in rearrangements of the heptahelical bundle that are transmitted to PalF, promoting its transition from the basal to the active conformation. PalF activation results in ubiquitination of the arrestin (Herranz et al., 2005). Upon binding to cognate active receptors mammalian arrestins undergo conformational changes (Hirsch et al., 1999; Shukla et al., 2013), which expose otherwise masked epitopes to effectors. One such PalF effector must be the NEDD4 ubiquitin ligase (Rsp5 in S. cerevisiae) (Herrador et al., 2009), denoted HulA in A. nidulans (Karachaliou et al., 2013)). As overwhelming evidence indicates that Vps23 binds only to ubiquitinated PalF (Galindo et al., 2012), the hypothetical conformational change that PalF undergoes would be the actual transducer of the alkaline pH signal.
In both two‐hybrid assays and pull‐downs using a bacterially expressed GST‐PalH tail bait, PalF and the cytosolic tail of PalH are strong interactors (Herranz et al., 2005; Galindo et al., 2012), indicating that binding of the arrestin to the receptor does not require phosphorylation of the latter, contrary to the situation reported for mammalian GPCR/arrestin complexes. In agreement, a PalH mutant deficient in phosphorylation is largely functional (Fig. 4). Thus, even though our results cannot rule out a contribution of PalH phosphorylation to arrestin binding, this contribution would be largely inconsequential. Co‐expression experiments in vivo indicated that PalF binds to PalH even under acidic conditions (Hervás‐Aguilar et al., 2010), and Rim21p has been shown to interact with Rim8p in a ubiquitin split assay on standard yeast media (Herrador et al., 2015), supporting a scenario in which the PalH/PalF complex is preformed in vivo, irrespective of the pH conditions. Strikingly, the cytosolic tail (without the TMs) of yeast Rim21p (Rim21CT) localizes in part to the PM (Herrador et al., 2015), which might help to position Rim8 (yeast PalF) in this locale. The intrinsically unstable active conformation of GPCRs is stabilized by G protein binding at the cytosolic side of the membrane (Sprang, 2011). PalF/Rim8 binding to PalH/Rim21 might reduce the energy required by the receptor to acquire the signalling conformation, thereby increasing its basal activity (i.e. the probability that any one PalH receptor at any one moment is signalling under acidic conditions). This would ensure that there is some ‘housekeeping’ activity of the pathway under acidic circumstances. Such interpretation is consistent with the observation that PalF overexpression results in weak activation of the pathway (Hervás‐Aguilar et al., 2010).
Transduction of the alkaline pH signal essentially requires, besides the TM region of PalH, the two PalF binding sites present in its cytosolic tail (Fig. 1). The one adjacent to TM7 contains 12 acidic (Asp/Glu) residues out of a total 36. Thus a highly speculative possibility is that this region acts as a phosphomimetic epitope, playing the role of the GRK‐phosphorylated residues in mammalian GPCRs. Direct signal‐independent engagement of PalF to this region might facilitate the transmission of structural rearrangements in PalH to the PalF fold. According to our mutagenesis analysis aromatic residues in the highly conserved PalH TM7 might be implicated in structural rearrangements, which is a very suggestive finding in view of the fact that conformational changes in TM7 have been implicated in arrestin‐biased signalling by GPCRs (Wacker et al., 2013)
A major unresolved question is how is alkaline pH sensed by PalH/Rim21, including whether it is directly activated or not by alkaline pH. A potentially important observation was that the RIM pathway is activated by altering lipid asymmetry (Ikeda et al., 2008), raising the intriguing possibility that the receptor actually senses a lipid whose ionization status or whose localization changes rapidly in response to alkalinity. It has been proposed that the ability of the C‐terminal cytosolic moiety of Rim21p (Rim21CT) to bind the PM is itself modulated by pH‐dependent changes in lipid asymmetry, implying that the cytosolic region may contribute to signal reception (Nishino et al., 2015).
A GPCR receptor responding to the lipid sphingosine‐1‐phosphate has been characterized structurally (Hanson et al., 2012). Here the lipid ligand gains access to its pocket laterally, within the transmembrane moiety of the receptor, because access to the extracellular milieu from the top is blocked by the amino terminus and extracellular loops. This situation would be consistent with our genetic data, such that Trp13 and Ala14/Arg15 in the amino terminal region together with Leu258, Tyr259 and Pro264 in EL2 could form the ‘roof’ of a lipid pocket embedded in the outer leaflet of the plasma membrane and contribute to stabilizing agonist binding, thereby explaining why these residues are required for signalling. A model involving cooperation between the N‐terminal external domain and a ligand binding site contributed by the transmembrane moiety has been proposed to explain the regulation of the 7‐TM receptor Smoothened, the key transducer of the metazoan Hedgehog pathway (Rana et al., 2013). We have previously noted the similarities between the pal/RIM and Hedgehog pathways (Arst and Peñalva, 2003), similarities further highlighted by the subsequent seminal finding that β‐arrestin 2 mediates Smoothened signalling (Chen et al., 2004). It is worth emphasizing that Tyr259 within the EL2 in PalH appears to be critical. Theoretically, there are six possible missense mutations resulting from single nucleotide substitution within the TAT Tyr259 codon. Four, involving substitutions of Tyr259 by Asn, His, Asp and Ser were isolated as partial loss‐of‐function mutations, suggesting that rather than any ionization status of Tyr259 it is the aromatic moiety which is physiologically important for receptor activation.
Herrador et al. have recently reported that the activity of yeast Rim8 (PalF) is antagonized by CK1‐mediated, Rim21‐dependent phosphorylation (Herrador et al., 2015). We have shown that PalH is phosphorylated in alkaline pH‐ and PalF‐dependent manner, raising the possibility that the same kinase that is recruited by the activated arrestin phosphorylates the receptor. However, PalH phosphorylation does not seem to have any important role in promoting signalling. An instructive finding was that despite the fact that phosphorylation is normally seen only when the pathway is activated, phosphorylated PalH is detectable at acidic pH in a mutant deficient in endocytosis, indicating that the cells preferentially turnover phosphorylated PalH as compared to the non‐phosphorylated pool. Thus, we speculate that phosphorylation contributes to maintaining the steady‐state levels of PalH at the plasma membrane.
In summary, our results are consistent with a mechanism of PalH activation bearing resemblance to that of mammalian 7‐TM receptors: a ‘pocket’ accommodates an unidentified ‘ligand’ that triggers a shift in the relative positions of individual TM helices within the helical bundle. Therefore our data strongly support the possibility that PalH is a druggable target, potentially alleviating the devastating impacts of ascomycete fungi not only in animal and plant pathogenesis, but also in toxin production, food spoilage and building material damage, all of which are subordinated to the jurisdiction of the pH signalling pathway (Peñalva and Arst, 2002, 2004; Peñalva et al., 2008).
Experimental procedures
Media and growth tests
Aspergillus complete (MCA) and synthetic complete medium (SC) (Cove, 1966), containing 1% glucose and, unless otherwise indicated, 5 mM ammonium tartrate as carbon and nitrogen sources, respectively, were used for growth tests and strain maintenance. Strains, which carried markers in standard use, are listed in Table 1. pH shift experiments involving transfer of cells from acidic (pH 4) to alkaline conditions (pH ∼8) have been described (Hervás‐Aguilar et al., 2010, 2007; Lucena‐Agell et al., 2015). Overnight cultures under acidic (final pH ∼5) or neutral (final pH ∼6.5) pH conditions (Hervás‐Aguilar et al., 2010) were used for some experiments involving palH4338 (see text for details).
Table 1.
Strains used in this work.
| Strain | Genotype |
|---|---|
| MAD976 | yA2 pabaA1 argB2 palI32 |
| MAD1293 | fwA1 inoB2 palH72 argB2::[alcAp::palI::gfp::argB*] |
| MAD1349 | inoB2 palH72 pacC900 |
| MAD1445 | yA2 pabaA1 pacC14900 |
| MAD1730 | pyroA4 inoB2 nkuAΔ::bar pacC900 |
| MAD1829 | inoB2 palI32 pacC900 |
| MAD1977 | wA4 pabaA1 pacC900 palF15 |
| MAD2336 | pantoB100 palH::ha3::pyrGfum pyrG89? |
| MAD2341 | palI32 palH::ha3::pyrGfum pyrG89? |
| MAD2344 | palA1 palH::ha3::pyrGfum pyrG89? pyroA4 |
| MAD2337 | palF15 palH::ha3::pyrGfum pyrG89? pantoB100 |
| MAD2668 | palHΔ::pyroAfum pacC900 pyroA4 pyrG89? nkuAΔ::bar |
| MAD2743 | yA2 pyroA4 pantoB100 |
| MAD2885 | palH::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD2886 | palH13::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD2887 | palH258::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD2888 | palH2593::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD2889 | palH264::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD2890 | palH268::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD2891 | palH275::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD3002 | palH486::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD3319 | yA2 pabaA6 |
| MAD3877 | pyroA4 pacC900 |
| MAD3880 | palH486::ha3::pyrGfum pyrG89? pyroA4 pabaA1 pacC900 |
| MAD4256 | palH::ha3::pyrGfum pacC900 |
| MAD4258 | palH654::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4261 | palH4338::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4270 | fimA::Tn431::pyr‐4 pacC900 pyroA4 pyrG89? nkuAΔ? |
| MAD4336 | palH4338::ha3::pyrGfum pyrG89? pyroA4 |
| MAD4413 | palH316::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4416 | palH138::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4418 | palH300::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4421 | palH4338::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4422 | palH654::ha3::pyrGfum pacC900 pyrG89? pyroA4 |
| MAD4552 | yA2 palH13::ha3::pyrGfum pabaA6 pacC900 pyrG89? |
| MAD4554 | yA2 palH258::ha3::pyrGfum pabaA6 pacC900 pyrG89? |
| MAD4556 | yA2 palH2593::ha3::pyrGfum pabaA6 pacC900 pyrG89? |
| MAD4557 | yA2 palH264::ha3::pyrGfum pabaA6 pacC900 pyrG89? |
| MAD4559 | palH268:: ha3::pyrGfum pabaA6 pacC900 pyrG89? |
| MAD4576 | yA2 pantoB100 pyroA4 palF15 |
| MAD4685 | palH275::ha3::pyrGfum pabaA6 pacC900 pyrG89? |
| MAD4874 | palH319::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4876 | palH320::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4878 | palH332::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4880 | palH338::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD4882 | pyroA4 pacC900 palH72 |
| MAD5140 | palH2332::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD5142 | palH25964::ha3::pyrGfum pyroA4 pacC900 pyrG89 nkuAΔ::bar |
| MAD5205 | palI32 palH4338::ha3::pyrGfum pyroA4 pacC900 pyrG89? nkuAΔ? |
| MAD5207 | wA2 palH::ha3::pyrGfum fimA::Tn431::pyr‐4 pacC900 pyroA4 pyrG89? nkuAΔ? |
| MAD5216 | yA2 pabaA6 palH316::ha3::pyrGfum pyrG89? pacC900 |
| MAD5218 | pabaA6 palH138::ha3::pyrGfum pyrG89? pacC900 |
| MAD5220 | yA2 palH300::ha3::pyrGfum pyroA4 pabaA6 pyrG89? pacC900 |
| MAD5224 | palH654::ha3::pyrGfum pyroA4 pacC900 pyrG89? nkuAΔ::bar |
| MAD5225 | palF15 palH4338::ha3::pyrGfum pyroA4 pacC900 nkuAΔ? |
Selection and characterization of previously undescribed palH mutations by random mutagenesis
Essentially the diploid γ‐aminobutyrate (GABA) selection method of Tilburn et al. (2005) was used to select new palH mutations. A diploid of genotype pabaA1 yA2 aroC660 palH72 camC108; areAr−5; pantoB100/areAr−5; inoB2; glrA1; fwA1 was constructed by several meiotic crosses. palH (AN6886) is closely linked and centromere‐distal to aroC (AN6866) (http://www.aspgd.org/) and camC is even more tightly linked to palH (Arst et al., 1994). Inclusion of aroC660 and camC108 in coupling to palH72 minimizes the selection of mitotic recombinant palH72 homozygous diploids: The absence of aromatic amino acids in the selection medium virtually rules out obtaining single crossover events centromere‐proximal to aroC660 and screening for reduction of chloramphenicol resistance in diploids eliminates most single crossover events centromere‐distal to aroC660. (camC108 is partially dominant (Gunatilleke et al., 1975)). After UV mutagenesis, conidiospores of the above diploid were top‐layered into glucose‐minimal medium containing 5 mM GABA as sole nitrogen source and 0.054% (w/v) sodium deoxycholate and incubated for 4–5 days at 37°C. Diploids able to use GABA as nitrogen source were picked off and tested phenotypically at 37°C for sensitivity to 3.5 mg/ml chloramphenicol and GABA utilization, and at both 37°C and 20°C for growth at pH 8 and for sensitivity to 20 mM on glucose minimal medium with 5 mM ammonium tartrate as nitrogen source. Resulting diploids impaired for growth at pH 8 and hyper‐sensitive to toxicity were haploidized on complete medium containing benlate (Hastie, 1970) but without additional supplementation of pabaA1 or aroC660 and yA+ colonies were purified on otherwise supplemented glucose minimal medium lacking p‐aminobenzoate and aromatic amino acids. This ensured that virtually all haploids recovered carried the palH allele in repulsion to palH72. The haploids were tested phenotypically for other markers present in the diploids along with GABA utilization, chloramphenicol and sensitivity and growth at pH 8. As experiments to obtain palH mutations having a null phenotype yielded mainly frame‐shift and chain termination alleles lacking major regions of the protein (with the exception of palH402), we mostly focused on obtaining partial loss‐of‐function mutations in order to maximize chances of getting single residue substitutions and other phenotypically less drastic sequence changes. Phenotypically promising palH alleles were then sequenced.
Site‐directed mutagenesis and construction of palH alleles by gene replacement
This procedure is graphically summarized in Supporting Information Fig. 1. A strain (MAD1737 in our collection, MAD indicating Madrid) with genotype pyrG89; pyroA4 nkuA::bar; pacC900 [the latter encoding wild‐type Myc‐tagged PacC (Hervás‐Aguilar et al., 2007)] (Table 1), requiring both pyrimidines and pyridoxine for growth, was used to substitute the complete coding region of palH by A. fumigatus pyroA. The resulting strain, denoted MAD2668 (pyrG89 palHΔ::pyroAAf; pyroA4 nkuAΔ::bar; pacC900) still requires pyrimidines but it is prototrophic for pyridoxine. This strain was used as recipient for transformation with mutated palH alleles, all constructed after mutagenesis of plasmid p1949. This plasmid contains a DNA fragment including ∼1500 bps of palH 5′‐flanking region followed by the complete palH coding region, tagged with HA3 in the C‐terminus, followed by A. fumigatus pyrG (pyrGAf) as selective marker and then by ∼1500 bps of 3′‐flanking region facilitating recombination. The wild‐type and mutated versions of this fragment can be excised as a linear DNA molecules by appropriate restriction enzyme digestion and used to replace palHΔ::pyroAAf in MAD2668 by engineered palH alleles. As control we reconstructed a wild‐type palH::HA3 allele denoted palH806. Strains containing this allele were indistinguishable from a ‘true’ wild‐type in diagnostic tests of pH regulation (Supporting Information Fig. 1). Mutagenesis was carried out using the Stratagene QuickChange site‐directed mutagenesis kit and mutagenic primers. All mutant plasmids were confirmed by DNA sequencing and, when appropriate (internal palH deletions), by restriction enzyme digestion.
Gene replaced transformants selected by their pyrimidine‐independent growth were tested on plates lacking pyridoxine, as the gene replacement event made these transformants auxotrophic for pyridoxine. Clones thus selected were confirmed to carry gene‐replaced palH alleles by Southern blotting. A preliminary assessment of their phenotypes was carried out and, if necessary, a selected primary transformant for any given mutation was outcrossed to confirm that the phenotype segregated as a single Mendelian character, linked to the palH::HA3 allele, whose presence was diagnosed by PCR.
Protein extraction and western blots
With the single exception described below, Western blot analyses were made using ‘total protein samples’ obtained by a direct alkaline solubilization of lyophilized mycelium (Hervás‐Aguilar and Peñalva, 2010), which minimizes protein degradation and dephosphorylation. The sources of antibodies and the dilutions used for detection of Myc‐PacC, PalF‐HA3 (or PalH‐HA3), hexokinase and actin have been detailed previously (Lucena‐Agell et al., 2015). Diagnostic tests of pH regulation based on alkaline pH (pH 8.3), lithium (0.3 or 0.5 M), sodium molybdate (10, 20 or 30 mM, as required) or neomycin sulfate (1 mg/ml) sensitivity have all been described (Tilburn et al., 1995; Peñas et al., 2007; Lucena‐Agell et al., 2015).
To isolate membrane fractions used for lambda phosphatase treatment, lyophilized mycelia (collected from cultures shifted for 15 min at pH 8.3 to trigger phosphorylation of PalH) were ground to a fine powder with a FP120 FastPrep and a ceramic sphere. 80 mg of powdered mycelia were hydrated with 1 ml of buffer MPE (100 mM Tris HCl, 150 mM NaCl, 5 mM EDTA and 25 mM N‐ethylmaleimide, pH 7.5, containing Roche's EDTA‐free protease inhibitor cocktail) and mixed with 0.45 µm glass beads. The resulting slurry was FastPrep‐homogenized again. Large debris were pelleted by centrifugation at 3000 rpm for 3 min at 4°C in a microcentrifuge. The supernatant was spun for 45 min at 14,000 g and the resulting pellet was resuspended in 1 ml MPE, washed once in the same buffer and finally resuspended in 180 µl. Aliquots were mixed with lambda phosphatase buffer (New England Biolabs) and, where indicated, with lambda phosphatase. 10 mM sodium orthovanadate was used for the ‘plus inhibitor’ control reaction. Reactions were incubated for 20 min at 30°C before precipitating proteins with 10% TCA and proceeding with anti‐HA western blotting of the immunoprecipitates.
Author contributions
DL‐A, AH‐A, TM‐H, OP, JR, and HNA carried out the experimental work. DL‐A, AH‐A, AG, JT, HNA and MAP designed and interpreted experiments. MAP and HNA wrote the paper.
Supporting information
Supporting Information
Acknowledgements
We thank Elena Reoyo and Lily Stanton for technical assistance and the Wellcome Trust (grants 067878/Z/02/Z), the Spanish Ministerio de Economía y Competitividad (MINECO; grants BIO2009‐7281 and BIO2012‐30965) and the Comunidad de Madrid (grant S2010/BMD‐2414) for support. DL‐A was a predoctoral fellow of the MINECO ‘Formación de Personal Investigador’ Program. The authors declare that they do not have any conflict of interest.
References
- Alvarez, C.E. (2008) On the origins of arrestin and rhodopsin. BMC Evol Biol 8: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst, H.N., Jr , and Peñalva, M.A. (2003) pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet 19: 224–231. [DOI] [PubMed] [Google Scholar]
- Arst, H.N., Jr , Bignell, E. , and Tilburn, J. (1994) Two new genes involved in signalling ambient pH in Aspergillus nidulans . Mol Gen Genet 245: 787–790. [DOI] [PubMed] [Google Scholar]
- Bertuzzi, M. , Schrettl, M. , Alcazar‐Fuoli, L. , Cairns, T.C. , Munoz, A. , Walker, L.A. , et al (2014) The pH‐responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog 10: e1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell, E. , Negrete‐Urtasun, S. , Calcagno, A.M. , Haynes, K. , Arst, H.N., Jr , and Rogers, T. (2005) The Aspergillus pH‐responsive transcription factor PacC regulates virulence. Mol Microbiol 55: 1072–1084. [DOI] [PubMed] [Google Scholar]
- Bussink, H.J. , Bignell, E.M. , Munera‐Huertas, T. , Lucena‐Agell, D. , Scazzocchio, C. , Espeso, E.A. , et al (2015) Refining the pH response in Aspergillus nidulans: a modulatory triad involving PacX, a novel zinc binuclear cluster protein. Mol Microbiol 98: 1051–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddick, M.X. , Brownlee, A.G. , and Arst, H.N., Jr. (1986) Regulation of gene expression by pH of the growth medium in Aspergillus nidulans . Mol Gen Genet 203: 346–353. [DOI] [PubMed] [Google Scholar]
- Calcagno‐Pizarelli, A.M. , Negrete‐Urtasun, S. , Denison, S.H. , Rudnicka, J.D. , Bussink, H.J. , Munera‐Huertas, T. , et al (2007) Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell 6: 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno‐Pizarelli, A.M. , Hervás‐Aguilar, A. , Galindo, A. , Abenza, J.F. , Peñalva, M.A. , and Arst, H.N., Jr. (2011) Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT‐0, I, II and III genes by inactivation of a salt‐tolerance pathway allows examination of ESCRT gene roles in pH signalling. J Cell Sci 124: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes‐Chávez, J.A. , Ortiz‐Castellanos, L. , Tejeda‐Sartorius, M. , Gold, S. , and Ruiz‐Herrera, J. (2010) Functional analysis of the pH responsive pathway Pal/Rim in the phytopathogenic basidiomycete Ustilago maydis . Fungal Genet Biol 47: 446–457. [DOI] [PubMed] [Google Scholar]
- Chalmers, D.T. , and Behan, D.P. (2002) The use of constitutively active GPCRs in drug discovery and functional genomics. Nat Rev Drug Discov 1: 599–608. [DOI] [PubMed] [Google Scholar]
- Chen, W. , Ren, X.R. , Nelson, C.D. , Barak, L.S. , Chen, J.K. , Beachy, P.A. , de Sauvage, F. , and Lefkowitz, R.J. (2004) Activity‐dependent internalization of smoothened mediated by ß‐arrestin 2 and GRK2. Science 306: 2257–2260. [DOI] [PubMed] [Google Scholar]
- Cornet, M. , and Gaillardin, C. (2014) pH signaling in human fungal pathogens: a new target for antifungal strategies. Eukaryot Cell 13: 342–352. Correction. Eukaryot Cell 13: 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove, D.J. (1966) The induction and repression of nitrate reductase in the fungus Aspergillus nidulans . Biochim Biophys Acta 113: 51–56. [DOI] [PubMed] [Google Scholar]
- Davis, D. , Edwards, J.E., Jr , Mitchell, A.P. , and Ibrahim, A.S. (2000) Candida albicans RIM101 pH response pathway is required for host‐pathogen interactions. Infect Immun 68: 5953–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D.A. (2009) How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol 12: 365–370. [DOI] [PubMed] [Google Scholar]
- Díez, E. , Álvaro, J. , Espeso, E.A. , Rainbow, L. , Suárez, T. , Tilburn, J. , Arst, H.N., Jr , and Peñalva, M.A. (2002) Activation of the Aspergillus PacC zinc‐finger transcription factor requires two proteolytic steps. EMBO J 21: 1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeso, E.A. , and Arst, H.N., Jr. (2000) On the mechanism by which alkaline pH prevents expression of an acid‐expressed gene. Mol Cell Biol 20: 3355–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo, A. , Hervás‐Aguilar, A. , Rodríguez‐Galán, O. , Vincent, O. , Arst, H.N., Jr , Tilburn, J. , and Peñalva, M.A. (2007) PalC, one of two Bro1 domain proteins in the fungal pH signaling pathway, localizes to cortical structures and binds Vps32. Traffic 8: 1346–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo, A. , Calcagno‐Pizarelli, A.M. , Arst, H.N., Jr , and Peñalva, M.A. (2012) An ordered pathway for the assembly of ESCRT‐containing fungal ambient pH signalling complexes at the plasma membrane. J Cell Sci 125: 1784–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunatilleke, I.A.U.N. , Scazzocchio, C. , and Arst, H.N., Jr. (1975) Cytoplasmic and nuclear mutations to chloramphenicol resistance in Aspergillus nidulans . Mol Gen Genet 137: 269–276. [DOI] [PubMed] [Google Scholar]
- Hanson, M.A. , Roth, C.B. , Jo, E. , Griffith, M.T. , Scott, F.L. , Reinhart, G. , et al (2012) Crystal structure of a lipid G protein–coupled receptor. Science 335: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, A.C. (1970) Benlate‐induced instability of Aspergillus diploids. Nature 226: 771–771. [DOI] [PubMed] [Google Scholar]
- Herrador, A. , Herranz, S. , Lara, D. , and Vincent, O. (2009) Recruitment of the ESCRT machinery to a putative seven‐transmembrane‐domain receptor is mediated by an arrestin‐related protein. Mol Cell Biol 30: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador, A. , Livas, D. , Soletto, L. , Becuwe, M. , Leon, S. , and Vincent, O. (2015) Casein kinase 1 controls the activation threshold of an alpha‐arrestin by multisite phosphorylation of the interdomain hinge. Mol Biol Cell 26: 2128–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz, S. , Rodríguez, J.M. , Bussink, H.J. , Sánchez‐Ferrero, J.C. , Arst, H.N., Jr , Peñalva, M.A. , and Vincent, O. (2005) Arrestin‐related proteins mediate pH signaling in fungi. Proc Natl Acad Sci USA 102: 12141–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás‐Aguilar, A. , and Peñalva, M.A. (2010) Endocytic machinery protein SlaB is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans . Eukaryot Cell 9: 1504–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás‐Aguilar, A. , Galindo, A. , and Peñalva, M.A. (2010) Receptor‐independent ambient pH signaling by ubiquitin attachment to fungal arrestin‐like PalF. J Biol Chem 285: 18095–18102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás‐Aguilar, A. , Rodríguez, J.M. , Tilburn, J. , Arst, H.N., Jr , and Peñalva, M.A. (2007) Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J Biol Chem 282: 34735–34747. [DOI] [PubMed] [Google Scholar]
- Hirsch, J.A. , Schubert, C. , Gurevich, V.V. , and Sigler, P.B. (1999) The 2.8 a crystal structure of visual arrestin: a model for arrestin's regulation. Cell 97: 257–269. [DOI] [PubMed] [Google Scholar]
- Hutchings, H. , Stahmann, K. , Roels, S. , Espeso, E.A. , Timberlake, W.E. , Arst, H.N., Jr , and Tilburn, J. (1999) The multiply‐regulated gabA gene encoding the GABA permease of Aspergillus nidulans: a score of exons. Mol Microbiol 32: 557–568. [DOI] [PubMed] [Google Scholar]
- Ikeda, M. , Kihara, A. , Denpoh, A. , and Igarashi, Y. (2008) The Rim101 pathway is involved in Rsb1 expression induced by altered lipid asymmetry. Mol Biol Cell 19: 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachaliou, M. , Amillis, S. , Evangelinos, M. , Kokotos, A.C. , Yalelis, V. , and Diallinas, G. (2013) The arrestin‐like protein ArtA is essential for ubiquitination and endocytosis of the UapA transporter in response to both broad‐range and specific signals. Mol Microbiol 88: 301–317. [DOI] [PubMed] [Google Scholar]
- Katritch, V. , Cherezov, V. , and Stevens, R.C. (2013) Structure‐function of the G protein‐coupled receptor superfamily. Annu Rev Pharmacol Toxicol 53: 531–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz, R.J. (2013) Arrestins come of age: a personal historical perspective Prog Mol Biol Transl 118: 3–18. [DOI] [PubMed] [Google Scholar]
- Lefkowitz, R.J. , and Shenoy, S.K. (2005) Transduction of receptor signals by beta‐arrestins. Science 308: 512–517. [DOI] [PubMed] [Google Scholar]
- Lin, C.H. , MacGurn, J.A. , Chu, T. , Stefan, C.J. , and Emr, S.D. (2008) Arrestin‐related ubiquitin‐ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725. [DOI] [PubMed] [Google Scholar]
- Liu, J.J. , Horst, R. , Katritch, V. , Stevens, R.C. , and Wüthrich, K. (2012) Biased signaling pathways in β2‐ddrenergic receptor characterized by 19F‐NMR. Science 335: 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena‐Agell, D. , Galindo, A. , Arst, H.N., Jr , and Peñalva, M.A. (2015) Aspergillus nidulans ambient pH signaling does not require endocytosis. Eukaryot Cell 14: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot, J.M. , Tilburn, J. , Díez, E. , Bignell, E. , Orejas, M. , Widdick, D.A. , et al (1999) Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol Cell Biol 19: 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete‐Urtasun, S. , Reiter, W. , Díez, E. , Denison, S.H. , Tilburn, J. , Espeso, E.A. , et al (1999) Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol Microbiol 33: 994–1003. [DOI] [PubMed] [Google Scholar]
- Nikko, E. , and Pelham, H.R. (2009) Arrestin‐mediated endocytosis of yeast plasma membrane transporters. Traffic 10: 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, K. , Obara, K. , and Kihara, A. (2015) The C‐terminal cytosolic region of Rim21 senses alterations in plasma membrane lipid composition: insights into sensing mechanisms for plasma membrane lipid asymmetry. J Biol Chem 290: 30797–30805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile, C.J. , Solis, N. , Myers, C.L. , Fay, A.J. , Deneault, J.S. , Nantel, A. , et al (2008) Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol 10: 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara, T.R. , Norton, D. , Price, M.S. , Hay, C. , Clements, M.F. , Nichols, C.B. , and Alspaugh, J.A. (2010) Interaction of Cryptococcus neoformans Rim101 and protein kinase a regulates capsule. PLoS Pathog 6: e1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara, T.R. , Holmer, S.M. , Selvig, K. , Dietrich, F. , and Alspaugh, J.A. (2013) Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio 4: e00522–e00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara, T.R. , Xu, W. , Selvig, K.M. , O'meara, M.J. , Mitchell, A.P. , and Alspaugh, J.A. (2014) The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol 34: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, K. , and Kihara, A. (2014) Signaling events of the Rim101 pathway occur at the plasma membrane in a ubiquitination‐dependent manner. Mol Cell Biol 34: 3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, K. , Yamamoto, H. , and Kihara, A. (2012) Membrane protein Rim21 plays a central role in sensing ambient pH in Saccharomyces cerevisiae . J Biol Chem 287: 38473–38481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost, K.S. , O'meara, T.R. , Huda, N. , Esher, S.K. , and Alspaugh, J.A. (2015) The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator. PLoS Genet 11: e1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva, M.A. , and Arst, H.N., Jr. (2002) Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol Mol Biol Rev 66: 426–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva, M.A. , and Arst, H.N., Jr. (2004) Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu Rev Microbiol 58: 425–451. [DOI] [PubMed] [Google Scholar]
- Peñalva, M.A. , Tilburn, J. , Bignell, E. , and Arst, H.N., Jr. (2008) Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16: 291–300. [DOI] [PubMed] [Google Scholar]
- Peñalva, M.A. , Lucena‐Agell, D. , and Arst, H.N., Jr. (2014) Liaison alcaline: pals entice non‐endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol 22: 49–59. [DOI] [PubMed] [Google Scholar]
- Peñas, M.M. , Hervás‐Aguilar, A. , Múnera‐Huertas, T. , Reoyo, E. , Peñalva, M.A. , Arst, H.N., Jr. , and Tilburn, J. (2007) Further characterization of the signaling proteolysis step in the aspergillus nidulans pH signal transduction pathway. Eukaryot Cell 6: 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, R. , Carroll, C.E. , Lee, H.J. , Bao, J. , Marada, S. , Grace, C.R.R. , et al (2013) Structural insights into the role of the smoothened cysteine‐rich domain in hedgehog signalling. Nat Commun 4: 2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, S.G. , Choi, H.J. , Fung, J.J. , Pardon, E. , Casarosa, P. , Chae, P.S. , et al (2011) Structure of a nanobody‐stabilized active state of the beta(2) adrenoceptor. Nature 469: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Galán, O. , Galindo, A. , Hervás‐Aguilar, A. , Arst, H.N., Jr , and Peñalva, M.A. (2009) Physiological involvement in pH signalling of Vps24‐mediated recruitment of aspergillus PalB cysteine protease to ESCRT‐III. J Biol Chem 284: 4404–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, D.M. , Zhang, C. , Lyons, J.A. , Holl, R. , Aragao, D. , Arlow, D.H. , et al (2011) Structure and function of an irreversible agonist‐ß2 adrenoceptor complex. Nature 469: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfels, K. , Tanny, J.C. , Molnar, E. , Friesen, H. , Commisso, C. , and Segall, J. (2005) Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae . Mol Cell Biol 25: 6772–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, T.W. , Frimurer, T.M. , Holst, B. , Rosenkilde, M.M. , and Elling, C.E. (2006) Molecular mechanism of 7TM receptor activation ‐ a global toggle switch model. Annu Rev Pharmacol Toxicol 46: 481–519. [DOI] [PubMed] [Google Scholar]
- Shi, L. , Liapakis, G. , Xu, R. , Guarnieri, F. , Ballesteros, J.A. , and Javitch, J.A. (2002) ß2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem 277: 40989–40996. [DOI] [PubMed] [Google Scholar]
- Shukla, A.K. , Manglik, A. , Kruse, A.C. , Xiao, K. , Reis, R.I. , Tseng, W.C. , et al (2013) Structure of active ß‐arrestin‐1 bound to a G‐protein‐coupled receptor phosphopeptide. Nature 497: 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, A.K. , Xiao, K. , and Lefkowitz, R.J. (2011) Emerging paradigms of β‐arrestin‐dependent seven transmembrane receptor signaling. Trends Biochem Sci 36: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang, S.R. (2011) Cell signalling: binding the receptor at both ends. Nature 469: 172–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburn, J. , Sarkar, S. , Widdick, D.A. , Espeso, E.A. , Orejas, M. , Mungroo, J. , et al (1995) The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid‐ and alkaline‐expressed genes by ambient pH. EMBO J 14: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburn, J. , Sánchez‐Ferrero, J.C. , Reoyo, E. , Arst, H.N., Jr , and Peñalva, M.A. (2005) Mutational analysis of the pH signal transduction component PalC of Aspergillus nidulans supports distant similarity to BRO1 domain family members. Genetics 171: 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréton, B. , Blanchin‐Roland, S. , Lambert, M. , Lepingle, A. , and Gaillardin, C. (2000) Ambient pH signalling in ascomycetous yeasts involves homologues of the Aspergillus nidulans genes palF and paIH . Mol Gen Genet 263: 505–513. [DOI] [PubMed] [Google Scholar]
- Upadhyay, S. , and Shaw, B.D. (2008) The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans . Mol Microbiol 68: 690–705. [DOI] [PubMed] [Google Scholar]
- Vincent, O. , Rainbow, L. , Tilburn, J. , Arst, H.N., Jr , and Peñalva, M.A. (2003) YPXL/I is a protein interaction motif recognised by Aspergillus PalA and its human homologue AIP1/Alix. Mol Cell Biol 23: 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin, J.D. , and Lefkowitz, R.J. (2007) arrestin‐biased ligands at seven‐transmembrane receptors. Trends Pharmacol Sci 28: 416–422. ß‐ [DOI] [PubMed] [Google Scholar]
- Wacker, D. , Wang, C. , Katritch, V. , Han, G.W. , Huang, X.P. , Vardy, E. , et al (2013) Structural features for functional selectivity at serotonin receptors. Science 340: 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne, T. , Moukhametzianov, R. , Baker, J.G. , Nehme, R. , Edwards, P.C. , Leslie, A.G. , Schertler, G.F. , and Tate, C.G. (2011) The structural basis for agonist and partial agonist action on a ß1‐adrenergic receptor. Nature 469: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisler, J.W. , DeWire, S.M. , Whalen, E.J. , Violin, J.D. , Drake, M.T. , Ahn, S. , et al (2007) A unique mechanism of ß‐blocker action: carvedilol stimulates beta‐arrestin signaling. Proc Natl Acad Sci USA 104: 16657–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X.Q. , and Chowdhury, A. (2013) Advances in methods to characterize ligand‐induced ionic lock and rotamer toggle molecular switch in G protein‐coupled receptors In Method Enzymol 520: 153–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W. , and Mitchell, A.P. (2001) Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST‐like region and is required for its proteolytic cleavage. J Bacteriol 183: 6917–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W. , Smith, F.J., Jr , Subaran, R. , and Mitchell, A.P. (2004) Multivesicular body‐ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans . Mol Biol Cell 15: 5528–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


